Abstract
The global COVID-19 pandemic has caused substantial morbidity and mortality to humanity. Remarkable progress has been made in understanding both the innate and adaptive mechanisms involved in the host response to the causative SARS-CoV-2 virus, but much remains to be discovered. Robust upper airway defenses are critical in restricting SARS-CoV-2 replication and propagation. Further, the nasal abundance of viral uptake receptor, ACE2, and the host epithelial transcriptional landscape, are associated with differential disease outcomes across different patient cohorts. The adaptive host response to systemic COVID-19 is heterogeneous and complex. Blunted responses to interferon and robust cytokine generation are hallmarks of the disease, particularly at the advanced stages. Excessive immune cell influx into tissues can lead to substantial collateral damage to the host akin to sepsis.
This review offers a contemporary summary of these mechanisms of disease and highlights potential avenues for diagnostic and therapeutic development. These include improved disease stratification, targeting effectors of immune-mediated tissue damage, and blunting of immune cell-mediated tissue damage.
Keywords: COVID-19, Immunity, Immune system diseases, COVID-19 vaccines, Host microbial interactions
1. Introduction
The global COVID-19 pandemic has resulted in widespread socioeconomic hardship and a devastating loss of human life. At the time of writing there have been over 250 million confirmed positive cases and over 5 million associated deaths [1]. Substantial efforts have been directed towards understanding the immunological response to SARS-CoV-2 and how this influences the clinical course of COVID-19 infection. Such an understanding is essential for the development of much needed novel therapies.
SARS-CoV-2 is part of the wider family of coronaviruses, which are enveloped positive sense single stranded RNA (ssRNA) viruses [2]. Initial infection is established when viral surface spike proteins bind to the ACE2 receptor on host cells and are cleaved by the serine protease TMPRSS2. This causes fusion of the virus with the host cell and subsequent translation of the viral replication and transcription complex. Ultimately, new virions are released through exocytosis, which enables propagation of infection.
Herein we provide a contemporary summary of the complexity and diversity of the host immune responses in COVID-19 from initial viral uptake through to uncontrolled systemic infection. We summarize recent advances from clinical studies and postulate how mechanistic discoveries could continue to supply the translational pipeline for the development of more advanced targeted treatments.
2. Viral uptake and local upper airway mucosal response
The upper airway is the primary site of contact between inhaled pathogens and the host immune system. Early in the course of the pandemic, it was clear that viral loads were higher in nasal swabs than those from the throat, suggesting the nose was an important site of viral entry [3].
2.1. Mucociliary barrier function
The nasal mucosa has highly adapted physical and immunological defenses against infection. On entry to the nasal cavity, pathogens must first cross a viscous mucus layer produced by secretory goblet cells, which is usually expelled by mucociliary clearance [4]. Augmenting this physical barrier with a topical spray has been suggested as one approach to reduce viral uptake. This concept is supported by in vitro data showing a reduction in SARS-CoV-2 binding to human airway epithelial cells after pre-treatment with a protease-containing glycerin barrier spray [5]; however, data confirming its efficacy in clinical trials is lacking.
2.2. Viral uptake
The SARS-CoV-2 entry receptor, ACE2, is highly expressed in secretory nasal epithelial cells [6]. Co-expression of ACE2 with genes involved in host innate immunity suggests that nasal epithelial responses could be important in limiting viral uptake and propagation. Interestingly, ACE2 itself can be upregulated by the presence of interferon (IFN) [7]. This suggests that SARS-CoV-2 exploits the normal host response: the antiviral response of releasing IFN leads to upregulation ACE2 expression, and the subsequently greater abundance of ACE2 protein on nasal epithelial surfaces creates greater opportunities for further enhanced viral uptake.
Large population studies have identified that polymorphisms of ACE2 can contribute to disease susceptibility, presumably by altering the properties of SARS-CoV-2 viral uptake [8]. Interestingly, ACE2 abundance has also been implicated in the relatively lower risk of severe COVID-19 in children compared to adults due to lower ACE2 expression in early compared to mid- and later life [8,9]. Similarly, ACE2 is encoded on the X chromosome in humans and it has been observed clinically that male patients have worse outcomes than females - suggesting ACE2 may also have a role in differential disease outcomes by sex [10].
2.3. Nasal epithelial response
Following infection, nasal epithelial cells upregulate production of secreted immunoglobulins. It has been observed that even in healthcare workers who have negligible SARS-CoV-2 specific serum antibody titres, some do still show specific IgA in mucosal fluids, highlighting the biological importance of robust mucosal defense [11]. In this study it was noted that specific IgA titres in nasal fluid were inversely correlated with patient age, again suggesting nasal IgA may play a key role in limiting disease severity.
Other secreted proteins with antiviral effects include mucins, which are large glycoproteins that trap and expel viral particles. In severe COVID-19 infection, this physiological mucous innate defense can be hijacked to compromise the host. COVID-19 infection leads to upregulation of pro-inflammatory cytokines in a so-called ‘cytokine storm’ [11], which is associated with mucin hypersecretion, including excessive quantities of MUC1 and MUC5AC [12], causing mucous plugging in the airways. So whilst airway mucins should ordinarily contribute to host defense, hypersecretion in response to a virally-induced cytokine storm is associated with airway obstruction and consequently impaired ventilation. Excess mucin can also impair host immunity, and this has been specifically demonstrated in ventilated patients [13].
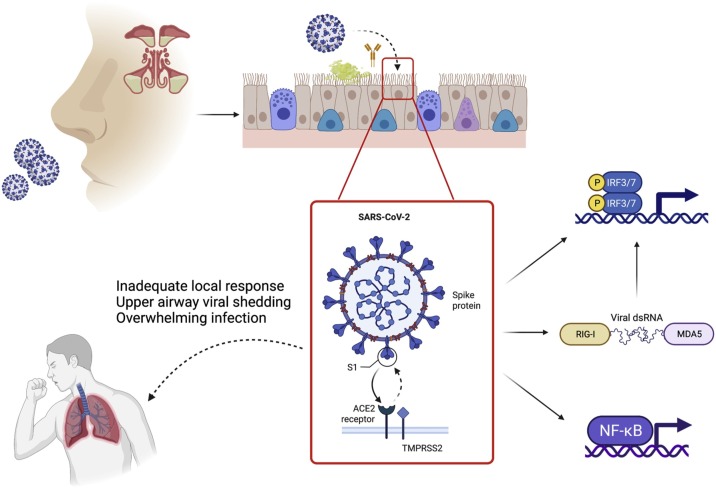
It is already known that viral infections such as SARS-CoV-2 lead to upregulation of IFN regulatory genes (IRF3 and IRF7), which subsequently upregulate production of type I IFN [15]. This can be initiated directly from ssRNA once the virus has been taken up in the endosome, or it can be triggered by intermediate double stranded RNA (dsRNA) in association with RIG-1 and MDA5 in the cytoplasm (Fig. 1 ). Indeed, when directly challenged with SARS-CoV-2, nasal epithelial cells show strong up-regulation of interferon I and III as their primary antiviral response [16]. The production of these proteins is slower than the rate of viral replication and therefore, at least in culture, seems to not substantially impact on the rate of viral replication. However, when recombinant INFβ or IFNɑ1 are given before exposure to SARS-CoV-2, then viral replication is limited. Therefore delivery of recombinant IFN, or upregulation of endogenous production in the nasal mucosa, may be an appealing strategy for interrupting progression to systemic disease. However, the timing of intranasal IFN administration around or following viral exposure will be challenging to implement in practice.
Fig. 1.
Local nasal response. Viral uptake in the nose is resisted by innate immune defenses, such as the presence of viscous mucous overlying the respiratory mucosa and secreted antiviral immunoglobulins. However, once SARS-COV-2 establishes contact with the respiratory epithelium, uptake is mediated via interaction of the viral spike protein with the host cell ACE2 receptor and cleavage by TMPRSS2. Cellular infection subsequently leads to upregulation of antiviral effectors, such as IFN, which contribute to the antiviral host response. Adapted from [14].
Transcriptional hallmarks of the IFN response are also found in nasopharyngeal swabs from patients with COVID-19 compared to healthy controls [17]. These responses are subdued compared to equivalent samples from patients with other (non-SARS-CoV-2) viral infections, and distinct from the biosignatures derived from blood samples in the same patients, highlighting the unique nature of the local versus the systemic immune response.
2.4. Nasal mucosal priming for systemic response
In addition to establishing an innate response to viral infection, the nose also has a sophisticated role in adaptive immunity. The nasopharyngeal lymphoid (adenoid) tissue is an organised mass of lymphoid tissue, which is responsible for the generation of mature T and B cells to orchestrate longer-term immune defense [18]. There are a range of specialised nasal microfold (M) cells, dendritic cells, and macrophages to capture and present relevant antigen to stimulate lymphocyte maturation. Further, migration of activated monocytes from the nasal mucosa to pulmonary lymph nodes may prepare the immune system of the lower airways prior to viral exposure [19]. However, the efficacy of the immune system is known to decline with advancing age and affects the mucosal immune system prior to the systemic compartment [20], suggesting impaired innate and adaptive immune signaling from the nasal mucosa may contribute to the greater risk of death in older patients with COVID-19 [21].
Despite the nose being an immune-primed sentinel of the upper airway, and despite the negative gradient of infectivity moving distally down the airways [22], SARS-CoV-2 can spread from the upper to the lower airway to cause pneumonia and life-threatening acute respiratory distress syndrome (ARDS) [23]. Infection of the lower airways is thought to be either via direct inhalation of aerosolised viral particles or indirect via viral shedding from the upper airways [24]. Aerosols are defined as fluid particles with a diameter less than 5 μm and are small enough to be inhaled into the distal lower airways as far as the alveoli [25]. Viral particles can be aerosolised either by medical procedures, including non-invasive ventilation and cardiopulmonary resuscitation, or activities of daily living such as heavy breathing during exercise or singing; this allows them to bypass the previously discussed defense mechanisms of the upper airways and directly infect the lungs. These hypotheses of viral particle aspiration are consistent with the findings of patchy, segmental disease in lung autopsies from deceased patients with COVID-19 [22].
3. Systemic immune responses
Systemic infection leads to a wide spectrum of clinical consequences. Asymptomatic infection or mild short-lived respiratory illness in the majority, contrasts with viral pneumonitis, progressing to severe respiratory dysfunction, ARDS, thrombotic-risk, multi-organ failure and death in the most severe cases. Around 5 % of the global population are at increased risk of severe COVID-19 due to older age and comorbidity [26]. Initial observations of hyper-cytokineamia and lymphopenia in severe cases established parallels to immune dysregulation in severe sepsis [[27], [28], [29]]. Mortality reductions from broad-based immunosuppression with dexamethasone have underscored the importance of dysregulated immunity in severe disease [30,31].
Peripheral blood is the most readily accessible window into systemic immune function. Data on lung-specific immune dysfunction has been reviewed [32] and will not be discussed. Facilitated by data-rich multi-omics techniques and collaborative working, a large body of data has emerged on peripheral immune dysfunction across geographical locations and severity groups. Most data are from resource-rich countries, focusing on severity-classified hospital admissions. The next challenge is to extract from these complex datasets what immune responses are protective and what are driving pathology, to determine early biomarkers predictive of poor outcome and to identify targets for intervention to improve the clinical course of disease. Here, we will summarize the complex immune dysfunction in SARS-CoV-2 infection by dissecting the peripheral immune response into its component parts. While a siloed approach is useful for discussion, it must be borne in mind that overlap and interplay between local and systemic, innate and adaptive immune responses operate in vivo.
3.1. Blunted interferon response and broad-based inflammatory cytokine release
In a functional immune response to viral infection, pathogen-associated-molecular patterns (PAMPS), generated through viral replication, activate pattern-recognition receptor (PRR)-mediated transcription of genes for cytokines that promote local control of viral replication and mediate recruitment of immune effector cells [33]. Multiplexed protein quantification, whole blood transcriptomics, transcriptional signatures in specific immune cells and in vitro stimulation assays, while not always aligned for a given cytokine [34], together have provided insights into how this dynamic process evolves in mild and severe COVID-19 infection.
Type I IFNs are critical for controlling viral replication and supporting innate immune response in many viral infections [35]. Serum IFNɑ and IFNβ levels are elevated in individuals with SARS-CoV-2 relative to uninfected controls [[36], [37], [38], [39]]. However, the interferon-response appears to diverge between mild and severe COVID-19 infections, with lower serum IFNɑ levels [34,38] and lower interferon-stimulated gene (ISG) scores by whole blood gene expression profiling in severe/critical cases [34]. SARS-CoV-2, compared with influenza A virus, produces modest interferon responses in primary bronchial epithelial cells and ferret infection models, while inflammatory cytokine production is robust [34,40].
Several mechanisms have been outlined to account for the observed blunted interferon responses in SARS-CoV-2 infection, including inborn errors in the interferon pathways, the presence of auto-antibodies against type I IFNs, and viral interferon antagonist proteins [[41], [42], [43]]. Zhang et al. found enrichment in loss-of-function mutations at loci involved in type I IFN production in patients with life-threatening SARS-CoV-2 infection compared to healthy controls [41]; such susceptibility mutations are rare, but do contribute to the low interferon levels seen in severe disease. Neutralising autoantibodies to type I IFNs have been identified in ∼10 % of patients with severe SARS-CoV-2 infection [42,44,45]. The presence of type 1 IFN autoantibodies appear to be a poor prognostic sign [44] and have been implicated in approximately one fifth of deaths from SARS-CoV-2 [46]. Potential therapeutic strategies to manage patients with high titres of autoantibodies are in their infancy, but include plasma exchange [47] and IFN support with type 1 IFN-beta, which tends to be spared by autoantibodies [44]. Further, SARS-CoV-2 has its own escape strategies to block the interferon response with the production of accessory proteins, including Orf6, which binds to the Nup98-Rae1 complex in order to inhibit STAT1 and 2, and in turn interrupt the production of ISGs [43].
A broad-based inflammatory cytokine response is observed in patients with COVID-19 relative to healthy controls, with Th1, Th2, Th17, and anti-inflammatory cytokines involved [39]. Cytokines, chemokines and growth factors distinguishing moderate from severe or critical disease, or correlating with lung severity scores in at least one study include IL1ɑ, IL1-RA, MIP1ɑ, MCSF, GCSF, HGF, IL-6, IL10, IL7 and IP10 [[36], [37], [38],48]. IP10 (CXCL10) is similarly elevated in ARDS caused by related coronaviruses SARS and MERS [49,50]. Mouse models of viral ARDS implicate lung-infiltrating neutrophils in IP10 production and disease severity can be ameliorated through IP10 elimination [51]. Elevated IL-6 is seen in macrophage activation syndromes and immunotherapy-associated cytokine release syndromes, and IL-6 in vitro reduces HLA-DR expression on monocytes, a key feature of COVID immunoparesis discussed below [27]. IL-6 blockade with Tociluzumab may limit COVID-19 severity but does not reduce mortality [52,53].
Early separation of cytokine profiles in mild versus severe COVID-19 offers the opportunity to risk-stratify for severe disease and potentially improve outcomes by directing monitoring and supportive care to those most at risk. Considering cytokine profiles before 12 days from symptom onset, Lucas et al. found that patients who succumbed to severe COVID-19 had higher levels of IFNα, IFNλ, IL-1Ra, CCL1, CLL2, M-CSF, IL-2, IL-16 and CCL21 [39]. More tractable combinations for clinical use include the triad of elevated IP-10, IL-10 and IL-6 [48], IP-10 and MCP-2 (CCL7) [37], and IL-6 and TNFα [54]. Elevated levels of all these cytokines predicted increased disease severity and death.
3.2. Neutrophilia, immature neutrophils and myeloid-derived suppressor cells
Neutrophil accumulation in the lung is a hallmark of ARDS [55]. While neutrophils are important phagocytic cells, particularly in clearance of bacteria and fungi [56], inappropriate release of granule enzymes in the alveolar space exacerbates lung injury [57]. Neutrophilia is a recurrent observation in COVID-19 infection [48,58,59], and a positive correlation between neutrophil frequency and severe disease is seen across cohorts [28,38,39,59,60]. Immature neutrophils in moderate and severe COVID-19 can be identified as “immature granulocytes” on automated clinical haematology analysis [60,61], and as low-density neutrophils remaining in peripheral blood mononuclear cell preparations [39,62]. Characterized in detail by multi-omics [60,61], these cells span a spectrum from committed progenitors to mature neutrophils. They appear close to plasmablasts on graph based clustering, and a developmental relationship has been hypothesized [62], but trajectory inference on a large number of cells supports the conclusion that they are independent populations [60]. The emergence of immature neutrophils in COVID-19 is severity-dependent, with no such populations found in healthy controls, mild COVID-19 cases, or other flu-like illnesses, and was observed throughout the time-course of infection in severe cases [60]. The premature release of neutrophils from bone marrow suggests increased peripheral consumption of neutrophils and/or alterations in myelopoiesis in the bone marrow in severe COVID-19, as can be observed in bacterial sepsis [63].
Immature neutrophils have hallmarks of both immune activation (CD64, RANK, RANKL expression, interferon gene signature) and immune suppression (CD62 L loss, PDL1 expression, granulocytic myeloid-derived suppressor cell (gMDSC) signature) in protein and gene expression profiling [60]. Phenotypic MDSCs have also been identified by flow cytometry [64]. MDSCs arise in sepsis and cancer, when typical neutrophil maturation is subverted by inflammatory signaling, such as STAT3 activation [65]. The distinct functional properties of MDSC, including suppression of T-cell mediated immune responses, may contribute to a delay in the adaptive antiviral immune response [65,66]. Bulk neutrophils isolated from severe COVID-19 patients retained their phagocytic capacity on standardized in vitro assays, but showed impaired oxidative burst compared with neutrophils isolated from mild COVID-19 patients [60]. The consequences of neutrophil compartment abnormalities for protective immunity to SARS-CoV-2 and lung immunopathology still requires investigation.
3.3. Inflammatory versus anergic monocytes
Monocytes are rapidly recruited to sites of inflammation, where they differentiate into macrophages or monocyte-derived dendritic cells, contributing to inflammatory cytokine production, phagocytosis, antigen presentation, and resolution of inflammation [67]. In severe influenza virus infection, monocytes are required to maximise virus-specific CD8 T cell responses, but also contribute to immune pathology and mortality [68]. Monocytes are conventionally divided into classical (CD14++CD16-), intermediate (CD14++CD16+), and non-classical (CD14-CD16++) subsets in humans according to surface phenotype [69]. While monocytes undoubtedly accumulate in the alveoli in severe COVID-19 [70,71], peripheral blood monocyte counts are minimally deranged [28,38,60,61,72,73]. The most consistent numerical abnormalities by flow cytometry are an increased proportion of intermediate monocytes in non-severe disease [48,72], a reduced proportion of classical monocytes in severe disease [27,48,60], and a reduced proportion of non-classical monocytes in all disease severities [[60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72],72]. Marked changes in the monocyte compartment include proliferation of circulating monocytes [38,59], the presence of classical monocytes with low levels of HLA-DR [28,48,59,61,62], and widespread alteration in the transcriptional landscape [38,60].
Monocytes are differentially activated in mild and severe COVID-19, with activation in mild cases indicated by increased levels of HLA-DR and CD11c, underpinned by interferon gene signaling [38,[60], [61], [62],73]. HLA-DRlo monocytes found in severe disease express surface markers reported to promote tissue infiltration (CD69, CD226) and genes associated with alternative macrophage activation (CD163) and immaturity (MPO, PLAC8) [60]. COVID-19 HLA-DRlo monocytes show enrichment with a sepsis-associated monocyte signature, and, like sepsis-associated monocytes, have blunted responses to LPS-stimulation in vitro [60,74]. Low-level expression of HLA-DR in monocytes is an established surrogate marker of immunoparesis in sepsis, and increased proportions of HLA-DRlo monocytes correlate with secondary infection rates and mortality [75]. However, further investigation will be needed to establish whether their presence is causative in COVID-19 pathology. Comparison of alveolar space monocyte/macrophages with blood monocytes, suggests that the tissue-infiltrating cells are responsible for TNF and IL6 production [38], further questioning the functional relevance of peripheral blood monocyte changes.
A specific non-classical monocyte profile has been detected in scRNAseq data from a large cohort [38]. This small subset of cells expressing C1q complement components is seen across severity groups. Receptor-ligand interactions predict it is particularly competent to engage and activate platelets [38]. Monocyte-platelet aggregates are seen more frequently in COVID-19 and there is emerging evidence that they trigger tissue factor expression, potentially contributing to thrombosis risk [76].
3.4. Dendritic cell depletion
Dendritic cells (DCs) are innate immune effectors that initiate adaptive immune responses by presenting antigen to naive T cells and providing cytokine stimulation to tailor T cell specification [77,78]. A diverse array of DC subsets is thought to underpin flexibility of immune response to pathogens, with plasmacytoid DCs (pDCs) specialized in type I interferon production, myeloid DC1 in antigen cross-presentation to CD8 T cells, and myeloid DC2 in producing a range of CD4 T cell responses [77,78]. Both pDCs and myeloid DCs are depleted in sepsis, likely due to a combination of apoptosis and reduced production [75], but recruitment to sites of inflammation should also be considered [79]. Peripheral blood frequencies of DC1, DC2 and pDC are reduced in COVID-19 [28,34,39,62,72] and improve upon clinical recovery [28]. Depletion of pDCs is most extreme in severe disease [48,59,72]. Reductions in DC precursor frequency [72] are accompanied by increased proliferation of DC2s [48,38]. In DC2 and pDC (but not DC1), lower expression of maturation markers CD86 and HLA-DR suggests a shift towards more recently produced DCs. Reduced expression of the IL-6 receptor is seen in DC1 and inhibitory receptor CD200R in DC2 [72]. Reduced sensitivity to IL-6 potentially accelerates DC maturation and activation of T cells [80], while reduced expression of CD200R is postulated to reduce inhibitory constraints on DCs [81]. In summary, DCs are depleted in COVID-19. Whether the immature DCs that emerge are equipped for T cell activation requires further study.
3.5. Emergency myelopoiesis and megakaryopoiesis
Circulating myeloid cells are bone-marrow dependent, with half-lives of around 15 h for neutrophils and 1–7 days for monocytes [82]. The circulating half-life of DC subsets in humans is not known. When myeloid effectors are recruited to tissues or consumed in pathogen-clearance, production is expected to increase to meet this demand, a process termed demand-adapted or emergency myelopoiesis [83]. The presence of immature neutrophils in peripheral blood at a given time point is not necessarily evidence of emergency myelopoiesis. An altered cytokine environment, leading to CXCR2 upregulation on immature neutrophils can result in premature release from the bone marrow, or ‘left shift’ [82]. However, sustained neutrophilia and continuous presence of immature neutrophils seen in COVID-19 does suggest altered haematopoiesis [60,61]. The presence of immature neutrophils in COVID-19, but not flu-like illness controls, indicates either a greater magnitude of neutrophil consumption in COVID-19, or presence of specific cytokine(s) inhibiting the CXCR4:CXCL12 axis responsible for bone marrow retention of immature neutrophils.
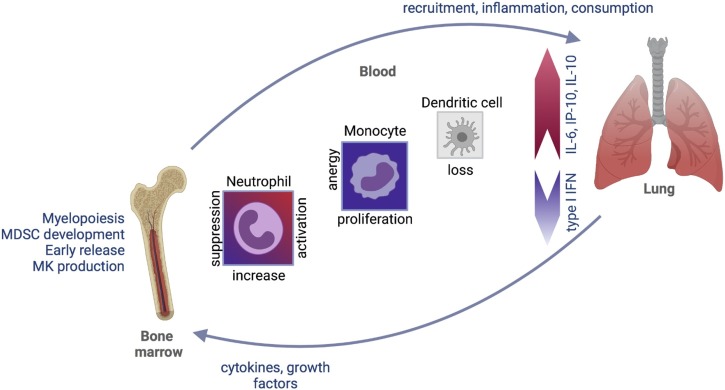
Megakaryocyte (MK) progenitors are increased in the peripheral blood in COVID-19 [38,84] and enrichment of an MK progenitor signature in the earliest hematopoietic stem/progenitor cells (HSPCs) suggests that haematopoiesis is biased towards megakaryocyte (MK) production [38]. Whether this is driven by consumption or specific signaling to the bone marrow requires further investigation (Fig. 2 ).
Fig. 2.
Innate immune response in severe COVID-19. In peripheral blood, blunted type I interferon response delays viral clearance and broad-based inflammatory cytokine response (e.g. IL-6, IP-10, IL-10) alters the functional status of circulating myeloid cells, recruitment of immune cells to the lung, and production of myeloid cells in the bone marrow.
3.6. Adaptive immune response
In addition to the initial mucosal and innate response to infection, eventual clearance of SARS coronavirus depends on a successful adaptive immune response [85]. As described above, the immunological response to infection begins with appropriate viral recognition, and secretion of interferon I/III and other inflammatory molecules by the mucosal barrier and innate immune cells [38,40,42]. This initial response ordinarily serves to limit further viral infection and replication, whilst also priming the adaptive immune system. Specific molecular signatures of immune profiling have indicated that delayed adaptive immune responses are correlated with poorer outcomes from SARS-CoV-2 infection [39,86,87]. In addition, high viral titres at a range of sampling sites (mostly naso/oropharyngeal, but also saliva, sputum, blood, plasma, urine and stool) have been associated with disease severity [88,89], although further work is required to better understand the relationship between viral titres, severity, and impaired adaptive immunity. A putative explanation for the pathophysiology of severe COVID-19 is that the immune evasion by SARS-CoV-2 prevents an adequate early adaptive immune response, allowing uncontrolled viral replication. Subsequent compensatory upregulation of the innate immune system in lieu of an adaptive response may give rise to innate immune cell proliferation, and downstream cytokine storm in severe cases, with associated lymphopenia. Lymphopenia is well established as a poor prognostic marker for outcomes in COVID-19 [62,90,91], but the mechanisms for this appear complex, including both SARS-CoV-2 and immune system specific factors [92].
In order to describe the adaptive immune response to SARS-CoV-2 infection in more detail, we have chosen four major components: CD4+ T cells, CD8+ T cells, B cells/antibodies, and other cell types such as Mucosal-associated invariant T (MAIT) cells [90,[93], [94], [95], [96], [97], [98]].
3.7. CD4+ T cells
CD4+ T cells have been shown to form a major axis of the adaptive immune response to SARS-CoV-2 [99] and are able to differentiate into several subtypes. These different physiological effectors enable CD4+ T cells to perform variable roles in the adaptive immune response to SARS-CoV-2.
Firstly, the presence of a classical anti-viral Th1 cell response in COVID-19 was first reported as early as January 2020 [90], and more recent work has further described direct anti-viral effects and protein expression profiles of IFN-γ, TNF and IL-2 [97,[100], [101], [102]]. The discovery of specific populations of spike-protein-specific Th1 CD4+ T cells has implicated this glycoprotein as a potentially critical target in a successful human response to SARS-CoV-2 [103]. The researchers in this study used a class II epitope prediction algorithm based on data from samples preceding the pandemic, to generate a ‘megapool’ of potential specific T cell epitopes, demonstrating that CD4+ T cells targeted to the spike protein form approximately 50 % of the response to SARS-CoV-2. Additional targets identified as being critical in the specific CD4+ T cell response include M, nucleocapsid and ORF3a [100,103,104]. Overall, these findings highlight the existence of immune memory to coronaviruses predating SARS-CoV-2 and the potential for cross-reactivity of spike-specific CD4+ T cells with different viral variants. An additional study by Braun et al. [97] also interrogated the role of the spike glycoprotein region and demonstrated the existence of ‘spike-reactive’ CD4+ Th1 cell populations in 83 % of individuals. SARS-CoV-2-reactive CD4+ T cells were detected in samples from 35 % of healthy donors, and the authors demonstrated increased cell populations in response to in vitro stimulation by SARS-CoV-2 as well as endemic seasonal coronaviruses (229E and OC43).
This finding of specific pre-existing memory CD4+ Th1 cells with coronavirus cross-reactivity implicates the role of prior exposure in determining successful response to SARS-CoV-2. Furthermore, recent work on comparing CD4+ (and CD8+) T cell epitopes between vaccinated (Pfizer, Moderna) and unvaccinated individuals has demonstrated effective recognition of several SARS-CoV-2 variants (B.1.1.7, B.1.351, P.1, and CAL.20C) [105].
Secondly, virus-specific populations of T follicular helper (Tfh) cells are implicated in a range of functions including aiding B cell function in SARS-CoV-2 infection [102,106]. Higher levels of circulating Tfh cells are associated with milder severity of SARS-CoV-2 disease [102], and are crucial in stimulating B cell production. The association between disease severity, Tfh cell count and neutralising antibody titres is complex and not yet fully understood [93,102].
In addition to these main CD4+ T cell functions, there is also a growing body of evidence for other subtypes of CD4+ effectors, including a CD8+ helper cell, a cytotoxic-like phenotype, and a CCR6 expressing cell postulated to have a role in tissue healing via IL-22 expression [38,101,102,104,107,108]. Single-cell RNA sequencing studies have been crucial in identifying key cell subtypes implicated in different presentations of disease severity. A key study in this field by Zheng et al. described populations of CD4+ effector-granulysin (GNLY) as well as CD8+ effector GNLY and a specific natural killer (NK) cell population. These populations were associated with a convalescent outcome in patients with mild-moderate disease. In addition, the authors describe a state of ‘T-cell exhaustion’ in cases of severe or critical COVID-19, already postulated by an earlier publication using flow cytometry [94,108].
3.8. CD8+ T cells
CD8+ T cells are crucial effectors in the response to several viral infections, and increased levels in the peripheral blood have been correlated with reduced disease severity [99,100]. Their role is to facilitate cytotoxicity through expression of molecules such as IFN-γ, granzyme B, perforin, and CD107a [96,102]. Similarly as has been shown for CD4+ T cells, the existence of SARS-CoV-2 specific circulating CD8+ effector T cells displaying immunological memory and cross-reactivity has been demonstrated. In particular, Schulien et al. describe the presence of such CD8+ T cells as early as 1 day post-symptom onset, and in 70 % of individuals in convalescence [96]. The targets for specific CD8+ T cell subpopulations have been identified as largely formed of spike, nucleocapsid, M and ORF3a, appearing to corroborate with work done for CD4+ T cells [100,104].
3.9. B cells and antibodies
As is the case for most viruses, the B cell response to SARS-CoV-2 is curated by Tfh cells and ultimately results in clonal B-cell proliferation, with production of plasmablast subsets for specific neutralising antibody production. B cell response to SARS-CoV-2 can occur particularly swiftly, in many cases without need for affinity maturation [99], and also demonstrates a diverse and complex picture in the literature, with conflicting reports of correlation with severity [38,84,109]. Initial consensus was driven towards an increased plasmablast subset response in more severe COVID-19 [93], but later studies have demonstrated more overwhelming evidence of an impaired B cell proliferation and plasmablast population in severe COVID-19 [38,109].
Neutralising antibodies to SARS-CoV-2 infection have been shown to emerge through seroconversion at day 5–15 of symptom onset, with 90 % by day 10 [99,110]. The main antigenic targets of SARS-CoV-2 infection IgG are the spike and nucleocapsid proteins, with the receptor binding domain (RBD) of spike representing the specific target for >90 % of antibodies [111]. A recent study by Planas et al. highlights a selective advantage of the ‘Delta’ variant (B1.617.1-3) to be less susceptible to antibodies targeting the spike protein, especially in unvaccinated convalescence. It also highlights that two doses of vaccination may achieve a neutralising response in 95 % of cases [112]. The diversity of immune response with neutralising antibodies to SARS-CoV-2 infection is complex; however, a general trend appears to be higher antigen load drives higher antibody titres [113]. Although the association between neutralising antibodies and COVID-19 severity is not fully understood, higher titres are not correlated with improved outcomes [114].
Longer term, B cell responses are critical to the development of durable host protection from reinfection. Longitudinal studies have revealed that memory B cells against SARS-CoV-2 spike protein increase between months 1 and 8 post infection, and the majority of these are IgG with a smaller proportion of IgA [84]. RBC-specific memory B cell responses also become more robust over time, with greater somatic hypermutation and increased potency by 6 months as compared with 1 month post infection [115], indicating the development of increasingly sophisticated immune maturation over time.
3.10. MAIT cells
Innate lymphoid cells (ILCs) such as Mucosal associated invariant T (MAIT) cells, are enriched in tissues such as the airway, and take on a variety of roles, from initial response to pathogens to tissue repair [116,117]. MAIT cells bridge the innate and adaptive immune system - they can be stimulated both through TCR-dependent and TCR independent mechanisms in conjunction with relevant cytokines, and also expand upon activation to trigger a substantial immune response [116]. Stephenson et al. highlighted MAIT cell activity as an important correlate of milder outcomes from COVID-19 using single-cell RNA sequencing analysis [38], building on earlier flow cytometry studies indicating a similar pattern, and purporting increased MAIT-cell tropism for the lung tissue [118]. Flament et al. analysed MAIT cell transcriptional profiles to further support an association of COVID-19 severity with MAIT activation and cytotoxicity, attributed to cytokines such as IFN-α–IL-18 [98].
4. Conclusions
The host response to SARS-CoV-2 is complex. Various innate immune mechanisms in the upper airways, including protective layers of viscous mucous and secreted immunoglobulins, may help to restrict initial viral uptake. Once viral particles are in contact with epithelial cells, surface protein ACE2 is critical for establishing infection and recent work suggests that various polymorphisms and differential abundances of ACE2 may help explain some of the different clinical outcomes seen in different patient groups. Further, nasal epithelial responses themselves, such as the production of IFN, are an important antiviral activity but can lead to increased ACE2 expression which may facilitate further viral uptake. The nasal mucosa also has an important role in priming the adaptive systemic response.
Once the virus escapes local control in the upper airways, a systemic response is generated. This response varies from host to host and is influenced by several well-described clinical risk factors. Typically, in COVID-19, there is a blunted response to IFN and rigorous cytokine production, increasingly so in more severe disease. The pro-inflammatory milieu leads to excessive immune cell recruitment, skewing of their functional capabilities, and often significant collateral tissue damage; all characteristics that draw parallels to immune dysfunction in sepsis. Differences in immune cell function clearly exist in different tissue compartments, such as upper airways, blood, and lung. Studies are ongoing to harness these findings to improve patient risk stratification and to identify pathways amenable to manipulation for therapeutic benefit.
Funding
MM is supported by an Action Medical Research Clinical Fellowship (GN2779). LJ is an NIHR Academic Clinical Lecturer. MH is funded by the Wellcome, the Lister Institute and the NIHR Newcastle Biomedical Research Centre.
Declaration of Competing Interest
None to declare.
Acknowledgements
Figures were created using www.biorender.com.
References
- 1.COVID-19 map - Johns Hopkins Coronavirus Resource Centre, (n.d.). https://coronavirus.jhu.edu/map.html (accessed July 7, 2021).
- 2.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellings P.W., Steelant B. Epithelial barriers in allergy and asthma. J. Allergy Clin. Immunol. 2020;145:1499–1509. doi: 10.1016/j.jaci.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Posch W., Vosper J., Zaderer V., Noureen A., Constant S., Bellmann-Weiler R., Lass-Flörl C., Wilflingseder D. ColdZyme maintains integrity in SARS-CoV-2-Infected airway epithelia. MBio. 2021;12 doi: 10.1128/mBio.00904-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sungnak W., HCA Lung Biological Network, Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F., Pippucci T., Torella A., Trezza A., Valentino F., Baldassarri M., Brusco A., Asselta R., Bruttini M., Furini S., Seri M., Nigro V., Matullo G., Tartaglia M., Mari F., GEN-COVID Multicenter Study, Renieri A., Pinto A.M. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunyavanich S., Do A., Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagliardi M.C., Tieri P., Ortona E., Ruggieri A. ACE2 expression and sex disparity in COVID-19. Cell Death Discov. 2020;6:37. doi: 10.1038/s41420-020-0276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervia C., Nilsson J., Zurbuchen Y., Valaperti A., Schreiner J., Wolfensberger A., Raeber M.E., Adamo S., Weigang S., Emmenegger M., Hasler S., Bosshard P.P., De Cecco E., Bächli E., Rudiger A., Stüssi-Helbling M., Huber L.C., Zinkernagel A.S., Schaer D.J., Aguzzi A., Kochs G., Held U., Probst-Müller E., Rampini S.K., Boyman O. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J. Allergy Clin. Immunol. 2021;147:545–557. doi: 10.1016/j.jaci.2020.10.040. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W., Liu X., Wang T., Liu F., Zhu A., Lin Y., Luo J., Ye F., He J., Zhao J., Li Y., Zhong N. Elevated MUC1 and MUC5AC mucin protein levels in airway mucus of critical ill COVID-19 patients. J. Med. Virol. 2021;93:582–584. doi: 10.1002/jmv.26406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell J., Garnett J.P., Mather M.W., Cooles F.A.H., Nelson A., Verdon B., Scott J., Jiwa K., Ruchaud-Sparagano M.-H., Cummings S.P., Perry J.D., Wright S.E., Wilson J.A., Pearson J., Ward C., Simpson A.J. Excess mucin impairs subglottic epithelial host defense in mechanically ventilated patients. Am. J. Respir. Crit. Care Med. 2018;198:340–349. doi: 10.1164/rccm.201709-1819OC. [DOI] [PubMed] [Google Scholar]
- 14.Gallo O., Locatello L.G., Mazzoni A., Novelli L., Annunziato F. The central role of the nasal microenvironment in the transmission, modulation, and clinical progression of SARS-CoV-2 infection. Mucosal Immunol. 2021;14:305–316. doi: 10.1038/s41385-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowie A.G., Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatton C.F., Botting R.A., Duenas M.E., Haq I.J., Verdon B. Delayed induction of type I and III interferons and nasal epithelial cell permissiveness to SARS-CoV-2. bioRxiv. 2021 doi: 10.1038/s41467-021-27318-0. https://www.biorxiv.org/content/10.1101/2021.02.17.431591v1.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng D.L., Granados A.C., Santos Y.A., Servellita V., Goldgof G.M., Meydan C., Sotomayor-Gonzalez A., Levine A.G., Balcerek J., Han L.M., Akagi N., Truong K., Neumann N.M., Nguyen D.N., Bapat S.P., Cheng J., Martin C.S.-S., Federman S., Foox J., Gopez A., Li T., Chan R., Chu C.S., Wabl C.A., Gliwa A.S., Reyes K., Pan C.-Y., Guevara H., Wadford D., Miller S., Mason C.E., Chiu C.Y. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onal M., Onal O., Turan A. Can secondary lymphoid organs exert a favorable effect on the mild course of COVID-19 in children? Acta Otolaryngol. 2021;141:83–84. doi: 10.1080/00016489.2020.1814965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua X., Vijay R., Channappanavar R., Athmer J., Meyerholz D.K., Pagedar N., Tilley S., Perlman S. Nasal priming by a murine coronavirus provides protective immunity against lethal heterologous virus pneumonia. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujihashi K., Kiyono H. Mucosal immunosenescence: new developments and vaccines to control infectious diseases. Trends Immunol. 2009;30:334–343. doi: 10.1016/j.it.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schäfer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O’Neal W.K., Randell S.H., Boucher R.C., Baric R.S. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matricardi P.M., Negro R.W.D., Nisini R. The first, holistic immunological model of COVID‐19: implications for prevention, diagnosis, and public health measures. Pediatr. Allergy Immunol. 2020;31:454–470. doi: 10.1111/pai.13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richard M., van den Brand J.M.A., Bestebroer T.M., Lexmond P., de Meulder D., Fouchier R.A.M., Lowen A.C., Herfst S. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat. Commun. 2020;11:766. doi: 10.1038/s41467-020-14626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernstrom A., Goldblatt M. Aerobiology and its role in the transmission of infectious diseases. J. Pathog. 2013;2013 doi: 10.1155/2013/493960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark A., Jit M., Warren-Gash C., Guthrie B., Wang H.H.X., Mercer S.W., Sanderson C., McKee M., Troeger C., Ong K.L., Checchi F., Perel P., Joseph S., Gibbs H.P., Banerjee A., Eggo R.M. Centre for the Mathematical Modelling of Infectious Diseases COVID-19 working group, Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob. Health. 2020;8:e1003–e1017. doi: 10.1016/S2214-109X(20)30264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000. doi: 10.1016/j.chom.2020.04.009. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A., Kuthuru O., Apostolidis S.A., Bershaw L., Dougherty J., Greenplate A.R., Pattekar A., Kim J., Han N., Gouma S., Weirick M.E., Arevalo C.P., Bolton M.J., Goodwin E.C., Anderson E.M., Hensley S.E., Jones T.K., Mangalmurti N.S., Luning Prak E.T., Wherry E.J., Meyer N.J., Betts M.R. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., Gong X., Wang Z., Chen R., Li T., Zeng D., Li M. Clinical features of familial clustering in patients infected with 2019 novel coronavirus in Wuhan, China. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kewan T., Covut F., Al–Jaghbeer M.J., Rose L., Gopalakrishna K.V., Akbik B. Tocilizumab for treatment of patients with severe COVID–19: a retrospective cohort study. EClinicalMedicine. 2020;24 doi: 10.1016/j.eclinm.2020.100418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T.R.C. Group, The RECOVERY Collaborative Group Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/nejmoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelaia C., Tinello C., Vatrella A., De Sarro G., Pelaia G. Lung under attack by COVID-19-induced cytokine storm: pathogenic mechanisms and therapeutic implications. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620933508. 1753466620933508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 34.J. Hadjadj, N. Yatim, L. Barnabei, A. Corneau, J. Boussier, H. Péré, B. Charbit, V. Bondet, C. Chenevier-Gobeaux, P. Breillat, N. Carlier, R. Gauzit, C. Morbieu, F. Pène, N. Marin, N. Roche, T.-A. Szwebel, N. Smith, S.H. Merkling, J.-M. Treluyer, D. Verer, L. Mouthon, C. Blanc, P.-L. Tharaux, F. Rozenberg, A. Fischer, D. Duffy, F. Rieux-Laucat, S. Kernéis, B. Terrier, Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients, (n.d.). 10.1101/2020.04.19.20068015. [DOI]
- 35.Müller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., Zhang Z., Qin Y., Li X., Zhao D., Li S., Tan S., Wang Z., Li J., Shen C., Li J., Peng L., Wu W., Cao M., Xing L., Xu Z., Chen L., Zhou C., Liu W.J., Liu L., Jiang C. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Sci. Rev. 2020;7:1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., Peng L., Yang M., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Gao G.F., Jiang C., Liu L., Liu Y. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127. doi: 10.1016/j.jaci.2020.04.027. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephenson E., Reynolds G., Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., Bach K., Sungnak W., Worlock K.B., Yoshida M., Kumasaka N., Kania K., Engelbert J., Olabi B., Spegarova J.S., Wilson N.K., Mende N., Jardine L., Gardner L.C.S., Goh I., Horsfall D., McGrath J., Webb S., Mather M.W., Lindeboom R.G.H., Dann E., Huang N., Polanski K., Prigmore E., Gothe F., Scott J., Payne R.P., Baker K.F., Hanrath A.T., Schim van der Loeff I.C.D., Barr A.S., Sanchez-Gonzalez A., Bergamaschi L., Mescia F., Barnes J.L., Kilich E., de Wilton A., Saigal A., Saleh A., Janes S.M., Smith C.M., Gopee N., Wilson C., Coupland P., Coxhead J.M., Kiselev V.Y., van Dongen S., Bacardit J., King H.W., Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research (CITIID-NIHR) COVID-19 BioResource Collaboration, Rostron A.J., Simpson A.J., Hambleton S., Laurenti E., Lyons P.A., Meyer K.B., Nikolić M.Z., Duncan C.J.A., Smith K.G.C., Teichmann S.A., Clatworthy M.R., Marioni J.C., Göttgens B., Haniffa M. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., Takahashi T., Tokuyama M., Lu P., Venkataraman A., Park A., Mohanty S., Wang H., Wyllie A.L., Vogels C.B.F., Earnest R., Lapidus S., Ott I.M., Moore A.J., Muenker M.C., Fournier J.B., Campbell M., Odio C.D., Casanovas-Massana A., Yale IMPACT Team, Herbst R., Shaw A.C., Medzhitov R., Schulz W.L., Grubaugh N.D., Dela Cruz C., Farhadian S., Ko A.I., Omer S.B., Iwasaki A. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A. B.R. tenOever, imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.-H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.-L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L.R., D’Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.-C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.-E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., COVID-STORM Clinicians, COVID Clinicians, Imagine COVID Group, French COVID Cohort Study Group, CoV-Contact Cohort, Amsterdam UMC Covid-19 Biobank, COVID Human Genetic Effort, NIAID-USUHS/TAGC COVID Immunity Group, Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.-Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.-L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Lab H., N.-U.I.R. to C. Group, C. Clinicians, C.-S. Clinicians, I.C. Group, F.C.C.S. Group, M.I. Consortium, C.-C. Cohort, A.U.C.-19 Biobank, C.H.G. Effort, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., Danziger O., White K.M., Rathnasinghe R., Uccellini M., Gao S., Aydillo T., Mena I., Yin X., Martin-Sancho L., Krogan N.J., Chanda S.K., Schotsaert M., Wozniak R.W., Ren Y., Rosenberg B.R., Fontoura B.M.A., García-Sastre A. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Troya J., Bastard P., Planas-Serra L., Ryan P., Ruiz M., de Carranza M., Torres J., Martínez A., Abel L., Casanova J.-L., Pujol A. Neutralizing autoantibodies to type I IFNs in &10% of patients with severe COVID-19 pneumonia hospitalized in Madrid, Spain. J. Clin. Immunol. 2021;41:914–922. doi: 10.1007/s10875-021-01036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solanich X., Rigo-Bonnin R., Gumucio V.-D., Bastard P., Rosain J., Philippot Q., Perez-Fernandez X.-L., Fuset-Cabanes M.-P., Gordillo-Benitez M.-Á., Suarez-Cuartin G., Boza-Hernandez E., Riera-Mestre A., Parra-Martínez A., Colobran R., Antolí A., Navarro S., Rocamora-Blanch G., Framil M., Calatayud L., Corbella X., Casanova J.-L., Morandeira F., Sabater-Riera J. Pre-existing autoantibodies neutralizing high concentrations of type I interferons in almost 10% of COVID-19 patients admitted to intensive care in Barcelona. J. Clin. Immunol. 2021 doi: 10.1007/s10875-021-01136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.-H., Eto S., Garcia-Prat M., Bizien L., Parra-Martínez A., Yang R., Haljasmägi L., Migaud M., Särekannu K., Maslovskaja J., de Prost N., Tandjaoui-Lambiotte Y., Luyt C.-E., Amador-Borrero B., Gaudet A., Poissy J., Morel P., Richard P., Cognasse F., Troya J., Trouillet-Assant S., Belot A., Saker K., Garçon P., Rivière J.G., Lagier J.-C., Gentile S., Rosen L.B., Shaw E., Morio T., Tanaka J., Dalmau D., Tharaux P.-L., Sene D., Stepanian A., Megarbane B., Triantafyllia V., Fekkar A., Heath J.R., Franco J.L., Anaya J.-M., Solé-Violán J., Imberti L., Biondi A., Bonfanti P., Castagnoli R., Delmonte O.M., Zhang Y., Snow A.L., Holland S.M., Biggs C., Moncada-Vélez M., Arias A.A., Lorenzo L., Boucherit S., Coulibaly B., Anglicheau D., Planas A.M., Haerynck F., Duvlis S., Nussbaum R.L., Ozcelik T., Keles S., Bousfiha A.A., El Bakkouri J., Ramirez-Santana C., Paul S., Pan-Hammarström Q., Hammarström L., Dupont A., Kurolap A., Metz C.N., Aiuti A., Casari G., Lampasona V., Ciceri F., Barreiros L.A., Dominguez-Garrido E., Vidigal M., Zatz M., van de Beek D., Sahanic S., Tancevski I., Stepanovskyy Y., Boyarchuk O., Nukui Y., Tsumura M., Vidaur L., Tangye S.G., Burrel S., Duffy D., Quintana-Murci L., Klocperk A., Kann N.Y., Shcherbina A., Lau Y.-L., Leung D., Coulongeat M., Marlet J., Koning R., Reyes L.F., Chauvineau-Grenier A., Venet F., Monneret G., Nussenzweig M.C., Arrestier R., Boudhabhay I., Baris-Feldman H., Hagin D., Wauters J., Meyts I., Dyer A.H., Kennelly S.P., Bourke N.M., Halwani R., Sharif-Askari N.S., Dorgham K., Sallette J., Sedkaoui S.M., AlKhater S., Rigo-Bonnin R., Morandeira F., Roussel L., Vinh D.C., Ostrowski S.R., Condino-Neto A., Prando C., Bonradenko A., Spaan A.N., Gilardin L., Fellay J., Lyonnet S., Bilguvar K., Lifton R.P., Mane S., HGID Lab, COVID Clinicians, COVID-STORM Clinicians, NIAID Immune Response to COVID Group, NH-COVAIR Study Group, Danish CHGE, Danish Blood Donor Study, St. James’s Hospital, SARS CoV2 Interest group, French COVID Cohort Study Group, Imagine COVID-Group, Milieu Intérieur Consortium, CoV-Contact Cohort, Amsterdam UMC Covid-19, Biobank Investigators, COVID Human Genetic Effort, CONSTANCES cohort, 3C-Dijon Study, Cerba Health-Care, Etablissement du Sang study group, Anderson M.S., Boisson B., Béziat V., Zhang S.-Y., Vandreakos E., Hermine O., Pujol A., Peterson P., Mogensen T.H., Rowen L., Mond J., Debette S., de Lamballerie X., Duval X., Mentré F., Zins M., Soler-Palacin P., Colobran R., Gorochov G., Solanich X., Susen S., Martinez-Picado J., Raoult D., Vasse M., Gregersen P.K., Piemonti L., Rodríguez-Gallego C., Notarangelo L.D., Su H.C., Kisand K., Okada S., Puel A., Jouanguy E., Rice C.M., Tiberghien P., Zhang Q., Cobat A., Abel L., Casanova J.-L. Autoantibodies neutralizing type I IFNs are present in 4% of uninfected individuals over 70 years old and account for 20% of COVID-19 deaths. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Prost N., Bastard P., Arrestier R., Fourati S., Mahévas M., Burrel S., Dorgham K., Gorochov G., Tandjaoui-Lambiotte Y., Azzaoui I., Fernandes I., Combes A., Casanova J.-L., Mekontso-Dessap A., Luyt C.-E. Plasma exchange to rescue patients with autoantibodies against type I interferons and life-threatening COVID-19 pneumonia. J. Clin. Immunol. 2021;41:536–544. doi: 10.1007/s10875-021-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J.D., Gee S., Chan J.N.E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmägi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M.A.A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K.J., Di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A.C. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 49.Huang K.-J., Su I.-J., Theron M., Wu Y.-C., Lai S.-K., Liu C.-C., Lei H.-Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin H.-S., Kim Y., Kim G., Lee J.Y., Jeong I., Joh J.-S., Kim H., Chang E., Sim S.Y., Park J.-S., Lim D.-G. Immune responses to middle east respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin. Infect. Dis. 2019;68:984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichikawa A., Kuba K., Morita M., Chida S., Tezuka H., Hara H., Sasaki T., Ohteki T., Ranieri V.M., dos Santos C.C., Kawaoka Y., Akira S., Luster A.D., Lu B., Penninger J.M., Uhlig S., Slutsky A.S., Imai Y. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am. J. Respir. Crit. Care Med. 2013;187:65–77. doi: 10.1164/rccm.201203-0508OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamed D.M., Belhoul K.M., Al Maazmi N.A., Ghayoor F., Moin M., Al Suwaidi M., Narainen M., Makki M., AbdulRahman M. Intravenous methylprednisolone with or without tocilizumab in patients with severe COVID-19 pneumonia requiring oxygen support: a prospective comparison. J. Infect. Public Health. 2021;14:985–989. doi: 10.1016/j.jiph.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salama C., Han J., Yau L., Reiss W.G., Kramer B., Neidhart J.D., Criner G.J., Kaplan-Lewis E., Baden R., Pandit L., Cameron M.L., Garcia-Diaz J., Chávez V., Mekebeb-Reuter M., Lima de Menezes F., Shah R., González-Lara M.F., Assman B., Freedman J., Mohan S.V. Tocilizumab in patients hospitalized with Covid-19 pneumonia. N. Engl. J. Med. 2021;384:20–30. doi: 10.1056/NEJMoa2030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Juss J.K., House D., Amour A., Begg M., Herre J., Storisteanu D.M.L., Hoenderdos K., Bradley G., Lennon M., Summers C., Hessel E.M., Condliffe A., Chilvers E.R. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-Kinase inhibition. Am. J. Respir. Crit. Care Med. 2016;194:961–973. doi: 10.1164/rccm.201509-1818OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segal A.W. How neutrophils kill microbes. Annu. Rev. Immunol. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paunel-Görgülü A., Kirichevska T., Lögters T., Windolf J., Flohé S. Molecular mechanisms underlying delayed apoptosis in neutrophils from multiple trauma patients with and without sepsis. Mol. Med. 2012;18:325–335. doi: 10.2119/molmed.2011.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., ’an Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mann E.R., Menon M., Knight S.B., Konkel J.E., Jagger C., Shaw T.N., Krishnan S., Rattray M., Ustianowski A., Bakerly N.D., Dark P., Lord G., Simpson A., Felton T., Ho L.-P., NIHR Respiratory TRC, Feldmann M., CIRCO, Grainger J.R., Hussell T. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., De Domenico E., Wendisch D., Grasshoff M., Kapellos T.S., Beckstette M., Pecht T., Saglam A., Dietrich O., Mei H.E., Schulz A.R., Conrad C., Kunkel D., Vafadarnejad E., Xu C.-J., Horne A., Herbert M., Drews A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., Müller-Redetzky H., Heim K.-M., Machleidt F., Uhrig A., Bosquillon de Jarcy L., Jürgens L., Stegemann M., Glösenkamp C.R., Volk H.-D., Goffinet C., Landthaler M., Wyler E., Georg P., Schneider M., Dang-Heine C., Neuwinger N., Kappert K., Tauber R., Corman V., Raabe J., Kaiser K.M., Vinh M.T., Rieke G., Meisel C., Ulas T., Becker M., Geffers R., Witzenrath M., Drosten C., Suttorp N., von Kalle C., Kurth F., Händler K., Schultze J.L., Aschenbrenner A.C., Li Y., Nattermann J., Sawitzki B., Saliba A.-E., Sander L.E. Deutsche COVID-19 OMICS Initiative (DeCOI), severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440. doi: 10.1016/j.cell.2020.08.001. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., Rameau P., Catelain C., Alfaro A., Dussiau C., Friedrich C., Sourdeau E., Marin N., Szwebel T.-A., Cantin D., Mouthon L., Borderie D., Deloger M., Bredel D., Mouraud S., Drubay D., Andrieu M., Lhonneur A.-S., Saada V., Stoclin A., Willekens C., Pommeret F., Griscelli F., Ng L.G., Zhang Z., Bost P., Amit I., Barlesi F., Marabelle A., Pène F., Gachot B., André F., Zitvogel L., Ginhoux F., Fontenay M., Solary E. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418. doi: 10.1016/j.cell.2020.08.002. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honda T., Uehara T., Matsumoto G., Arai S., Sugano M. Neutrophil left shift and white blood cell count as markers of bacterial infection. Clin. Chim. Acta. 2016;457:46–53. doi: 10.1016/j.cca.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 64.Rendeiro A.F., Casano J., Vorkas C.K., Singh H., Morales A., DeSimone R.A., Ellsworth G.B., Soave R., Kapadia S.N., Saito K., Brown C.D., Hsu J., Kyriakides C., Chiu S., Cappelli L.V., Cacciapuoti M.T., Tam W., Galluzzi L., Simonson P.D., Elemento O., Salvatore M., Inghirami G. Profiling of immune dysfunction in COVID-19 patients allows early prediction of disease progression. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J., Nefedova Y., Lei A., Gabrilovich D. Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin. Immunol. 2018;35:19–28. doi: 10.1016/j.smim.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinha P., Clements V.K., Bunt S.K., Albelda S.M., Ostrand-Rosenberg S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J. Immunol. 2007;179:977–983. doi: 10.4049/jimmunol.179.2.977. [DOI] [PubMed] [Google Scholar]
- 67.Shi C., Pamer E.G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aldridge J.R., Moseley C.E., Boltz D.A., Negovetich N.J., Reynolds C., Franks J., Brown S.A., Doherty P.C., Webster R.G., Thomas P.G. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziegler-Heitbrock L., Ancuta P., Crowe S., Dalod M., Grau V., Hart D.N., Leenen P.J.M., Liu Y.-J., MacPherson G., Randolph G.J., Scherberich J., Schmitz J., Shortman K., Sozzani S., Strobl H., Zembala M., Austyn J.M., Lutz M.B. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 70.Carsana L., Sonzogni A., Nasr A., Rossi R.S., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M., Galli M., Catena E., Tosoni A., Gianatti A., Nebuloni M. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 2020;20:1135–1140. doi: 10.1016/s1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 72.Kvedaraite E., Hertwig L., Sinha I., Ponzetta A., Hed Myrberg I., Lourda M., Dzidic M., Akber M., Klingström J., Folkesson E., Muvva J.R., Chen P., Gredmark-Russ S., Brighenti S., Norrby-Teglund A., Eriksson L.I., Rooyackers O., Aleman S., Strålin K., Ljunggren H.-G., Ginhoux F., Björkström N.K., Henter J.-I., Svensson M., Karolinska KI/K COVID-19 Study Group Major alterations in the mononuclear phagocyte landscape associated with COVID-19 severity. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2018587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao C., Bora S.A., Parimon T., Zaman T., Friedman O.A., Palatinus J.A., Surapaneni N.S., Matusov Y.P., Chiang G.C., Kassar A.G., Patel N., Green C.E.R., Aziz A.W., Suri H., Suda J., Lopez A.A., Martins G.A., Stripp B.R., Gharib S.A., Goodridge H.S., Chen P. Cell-type-specific immune dysregulation in severely ill COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reyes M., Filbin M.R., Bhattacharyya R.P., Billman K., Eisenhaure T., Hung D.T., Levy B.D., Baron R.M., Blainey P.C., Goldberg M.B., Hacohen N. An immune-cell signature of bacterial sepsis. Nat. Med. 2020;26:333–340. doi: 10.1038/s41591-020-0752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venet F., Demaret J., Gossez M., Monneret G. Myeloid cells in sepsis-acquired immunodeficiency. Ann. N. Y. Acad. Sci. 2020 doi: 10.1111/nyas.14333. [DOI] [PubMed] [Google Scholar]
- 76.Hottz E.D., Azevedo-Quintanilha I.G., Palhinha L., Teixeira L., Barreto E.A., Pão C.R.R., Righy C., Franco S., Souza T.M.L., Kurtz P., Bozza F.A., Bozza P.T. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reynolds G., Haniffa M. Human and mouse mononuclear phagocyte networks: a tale of two species? Front. Immunol. 2015;6:330. doi: 10.3389/fimmu.2015.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jardine L., Wiscombe S., Reynolds G., McDonald D., Fuller A., Green K., Filby A., Forrest I., Ruchaud-Sparagano M.-H., Scott J., Collin M., Haniffa M., Simpson A.J. Lipopolysaccharide inhalation recruits monocytes and dendritic cell subsets to the alveolar airspace. Nat. Commun. 2019;10:1999. doi: 10.1038/s41467-019-09913-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park S.-J., Nakagawa T., Kitamura H., Atsumi T., Kamon H., Sawa S.-I., Kamimura D., Ueda N., Iwakura Y., Ishihara K., Murakami M., Hirano T. IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J. Immunol. 2004;173:3844–3854. doi: 10.4049/jimmunol.173.6.3844. [DOI] [PubMed] [Google Scholar]
- 81.Minas K., Liversidge J. Is the CD200/CD200 receptor interaction more than just a myeloid cell inhibitory signal? Crit. Rev. Immunol. 2006;26:213–230. doi: 10.1615/critrevimmunol.v26.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patel A.A., Ginhoux F., Yona S. Monocytes, macrophages, dendritic cells and neutrophils: an update on lifespan kinetics in health and disease. Immunology. 2021;163:250–261. doi: 10.1111/imm.13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takizawa H., Boettcher S., Manz M.G. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 84.Bernardes J.P., Mishra N., Tran F., Bahmer T., Best L., Blase J.I., Bordoni D., Franzenburg J., Geisen U., Josephs-Spaulding J., Köhler P., Künstner A., Rosati E., Aschenbrenner A.C., Bacher P., Baran N., Boysen T., Brandt B., Bruse N., Dörr J., Dräger A., Elke G., Ellinghaus D., Fischer J., Forster M., Franke A., Franzenburg S., Frey N., Friedrichs A., Fuß J., Glück A., Hamm J., Hinrichsen F., Hoeppner M.P., Imm S., Junker R., Kaiser S., Kan Y.H., Knoll R., Lange C., Laue G., Lier C., Lindner M., Marinos G., Markewitz R., Nattermann J., Noth R., Pickkers P., Rabe K.F., Renz A., Röcken C., Rupp J., Schaffarzyk A., Scheffold A., Schulte-Schrepping J., Schunk D., Skowasch D., Ulas T., Wandinger K.-P., Wittig M., Zimmermann J., Busch H., Hoyer B.F., Kaleta C., Heyckendorf J., Kox M., Rybniker J., Schreiber S., Schultze J.L., Rosenstiel P. HCA lung biological network, deutsche COVID-19 omics initiative (DeCOI), longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53:1296–1314. doi: 10.1016/j.immuni.2020.11.017. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li C.K.-F., Wu H., Yan H., Ma S., Wang L., Zhang M., Tang X., Temperton N.J., Weiss R.A., Brenchley J.M., Douek D.C., Mongkolsapaya J., Tran B.-H., Lin C.-L.S., Screaton G.R., Hou J.-L., McMichael A.J., Xu X.-N. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F., Gan R., Zhen Z., Hu X., Li X., Zhou F., Liu Y., Chen C., Xie S., Zhang B., Wu X., Huang Z. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther. 2020;5:156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Butler D., Mozsary C., Meydan C., Foox J., Rosiene J., Shaiber A., Danko D., Afshinnekoo E., MacKay M., Sedlazeck F.J., Ivanov N.A., Sierra M., Pohle D., Zietz M., Gisladottir U., Ramlall V., Sholle E.T., Schenck E.J., Westover C.D., Hassan C., Ryon K., Young B., Bhattacharya C., Ng D.L., Granados A.C., Santos Y.A., Servellita V., Federman S., Ruggiero P., Fungtammasan A., Chin C.-S., Pearson N.M., Langhorst B.W., Tanner N.A., Kim Y., Reeves J.W., Hether T.D., Warren S.E., Bailey M., Gawrys J., Meleshko D., Xu D., Couto-Rodriguez M., Nagy-Szakal D., Barrows J., Wells H., O’Hara N.B., Rosenfeld J.A., Chen Y., Steel P.A.D., Shemesh A.J., Xiang J., Thierry-Mieg J., Thierry-Mieg D., Iftner A., Bezdan D., Sanchez E., Campion T.R., Jr., Sipley J., Cong L., Craney A., Velu P., Melnick A.M., Shapira S., Hajirasouliha I., Borczuk A., Iftner T., Salvatore M., Loda M., Westblade L.F., Cushing M., Wu S., Levy S., Chiu C., Schwartz R.E., Tatonetti N., Rennert H., Imielinski M., Mason C.E. Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Nat. Commun. 2021;12:1660. doi: 10.1038/s41467-021-21361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shenoy S. SARS-CoV-2 (COVID-19), viral load and clinical outcomes; lessons learned one year into the pandemic: a systematic review. Pediatr. Crit. Care Med. 2021;10:132–150. doi: 10.5492/wjccm.v10.i4.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsukagoshi H., Shinoda D., Saito M., Okayama K., Sada M., Kimura H., Saruki N. Relationships between viral load and the clinical course of COVID-19. Viruses. 2021;13 doi: 10.3390/v13020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.J. Naf, G. I, de N. Ah, B. Ck, K. Vacm, M. V, K. V, H. X, K. M, K. Hjpm, S. Rl, J. I, B. Rjm, K. Ije, van der H. Hg, S. Ja, F. T, R. Mhe, H. W, D. Asm, van A. Mj, B. Mjt, V. K, M. C, S. Ah, H. Ie, D. Lpg, van D. M, van der M. Jwm, van C. R, G.-B. Ej, J. Lab, van den H. Mm, H. J, de M. Q, P. P, N. Mg, van de V. Fl Dysregulated Innate and Adaptive Immune Responses Discriminate Disease Severity in COVID-19. J. Infect. Dis. 2021;223:1322–1333. doi: 10.1093/infdis/jiab065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jafarzadeh A., Jafarzadeh S., Nozari P., Mokhtari P., Nemati M. Lymphopenia an important immunological abnormality in patients with COVID-19: possible mechanisms. Scand. J. Immunol. 2021;93:e12967. doi: 10.1111/sji.12967. [DOI] [PubMed] [Google Scholar]
- 93.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S.A., Peters B., Krammer F., Simon V., Saphire E.O., Smith D.M., Weiskopf D., Sette A., Crotty S. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/SCIENCE.ABF4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. 17 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li S., Jiang L., Li X., Lin F., Wang Y., Li B., Jiang T., An W., Liu S., Liu H. E. al., Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Lago M.S., Decker A., Luxenburger H., Binder B., Bettinger D., Sogukpinar O., Rieg S., Panning M., Huzly D., Schwemmle M., Kochs G., Waller C.F., Nieters A., Duerschmied D., Emmerich F., Mei H.E., Schulz A.R., Llewellyn-Lacey S., Price D.A., Boettler T., Bengsch B., Thimme R., Hofmann M., Neumann-Haefelin C. Characterization of pre-existing and induced SARS-CoV-2-specific CD8 + T cells. Nat. Med. 2020;27(1):78–85. doi: 10.1038/s41591-020-01143-2. 27 (2020) [DOI] [PubMed] [Google Scholar]
- 97.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Röhmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Müller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 98.Flament H., Rouland M., Beaudoin L., Toubal A., Bertrand L., Lebourgeois S., Rousseau C., Soulard P., Gouda Z., Cagninacci L., Monteiro A.C., Hurtado-Nedelec M., Luce S., Bailly K., Andrieu M., Saintpierre B., Letourneur F., Jouan Y., Si-Tahar M., Baranek T., Paget C., Boitard C., Vallet-Pichard A., Gautier J.-F., Ajzenberg N., Terrier B., Pène F., Ghosn J., Lescure X., Yazdanpanah Y., Visseaux B., Descamps D., Timsit J.-F., Monteiro R.C., Lehuen A. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021;22(3):322–335. doi: 10.1038/s41590-021-00870-z. 22 (2021) [DOI] [PubMed] [Google Scholar]
- 99.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., López-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C., Grimes J.M., Antson A.A., Bayfield O.W., Hawkins D.E.D.P., Ker D.S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B.M., Fry J.W., Schwabe N.F., Semple M.G., Baillie J.K., Moore S.C., Openshaw P.J.M., Ansari M.A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.P., Dong T., Klenerman P., McMichael A., Ogg G., Kenneth Baillie J., Cornall R.J., Conlon C.P., Screaton G.R., Mongkolsapaya J., Knight J.C. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Akber M., Berglin L., Bergsten H., Brighenti S., Brownlie D., Butrym M., Chambers B., Chen P., Jeannin M.C., Grip J., Gomez A.C., Dillner L., Lozano I.D., Dzidic M., Tullberg M.F., Färnert A., Glans H., Haroun-Izquierdo A., Henriksson E., Hertwig L., Kalsum S., Kokkinou E., Kvedaraite E., Loreti M., Lourda M., Maleki K., Malmberg K.J., Marquardt N., Maucourant C., Michaelsson J., Mjösberg J., Moll K., Muva J., Mårtensson J., Nauclér P., Norrby-Teglund A., Medina L.P., Persson B., Radler L., Ringqvist E., Sandberg J.T., Sohlberg E., Soini T., Svensson M., Tynell J., Varnaite R., Kries A.V., Unge C., Rooyackers O., Eriksson L.I., Henter J.I., Sönnerborg A., Allander T., Albert J., Nielsen M., Klingström J., Gredmark-Russ S., Björkström N.K., Sandberg J.K., Price D.A., Ljunggren H.G., Aleman S., Buggert M. Robust t cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., Kato Y., Crotty E.G., Kim C., Rawlings S.A., Mateus J., Tse L.P.V., Frazier A., Baric R., Peters B., Greenbaum J., Ollmann Saphire E., Smith D.M., Sette A., Crotty S. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., Mallal S., Lammers M., Rubiro P., Quiambao L., Sutherland A., Yu E.D., Da Silva Antunes R., Greenbaum J., Frazier A., Markmann A.J., Premkumar L., De Silva A., Peters B., Crotty S., Sette A., Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370 doi: 10.1126/SCIENCE.ABD3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.G. A, W. D, R. Si, M. J, D. Jm, M. Cr, R. Sa, S. A, P. L, J. Rs, M. D, de S. Am, F. A, C. Af, G. Ja, P. B, K. F, S. Dm, C. S, S. A Targets of t cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., Frazier A., Ramirez S.I., Rawlings S.A., Smith D.M., da Silva Antunes R., Peters B., Scheuermann R.H., Weiskopf D., Crotty S., Grifoni A., Sette A. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/SCIIMMUNOL.ABD2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang J.-Y., Wang X.-M., Xing X., Xu Z., Zhang C., Song J.-W., Fan X., Xia P., Fu J.-L., Wang S.-Y., Xu R.-N., Dai X.-P., Shi L., Huang L., Jiang T.-J., Shi M., Zhang Y., Zumla A., Maeurer M., Bai F., Wang F.-S. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 109.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., Manne S., Chen Z., Huang Y.J., Reilly J.P., Weisman A.R., Ittner C.A.G., Kuthuru O., Dougherty J., Nzingha K., Han N., Kim J., Pattekar A., Goodwin E.C., Anderson E.M., Weirick M.E., Gouma S., Arevalo C.P., Bolton M.J., Chen F., Lacey S.F., Ramage H., Cherry S., Hensley S.E., Apostolidis S.A., Huang A.C., Vella L.A., Betts M.R., Meyer N.J., Wherry E.J., Alam Z., Addison M.M., Byrne K.T., Chandra A., Descamps H.C., Kaminskiy Y., Hamilton J.T., Noll J.H., Omran D.K., Perkey E., Prager E.M., Pueschl D., Shah J.B., Shilan J.S., Vanderbeck A.N. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369 doi: 10.1126/SCIENCE.ABC8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 111.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Joost Wiersinga W., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 113.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., Acton O.J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J.C., Mele F., Jovic S., Rodriguez B.F., Gupta S.V., Jin F., Piumatti G., Lo Presti G., Pellanda A.F., Biggiogero M., Tarkowski M., Pizzuto M.S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H.W., Lanzavecchia A., Corti D., Veesler D. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baumgarth N., Nikolich-Žugich J., Lee F.E.-H., Bhattacharya D. Antibody responses to SARS-CoV-2: let’s stick to known knowns. J. Immunol. 2020;205:2342–2350. doi: 10.4049/jimmunol.2000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., Cipolla M., Viant C., Barnes C.O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K.G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Robbiani D.F., Zhao Z., Gazumyan A., Schwartz R.E., Hatziioannou T., Bjorkman P.J., Mehandru S., Bieniasz P.D., Caskey M., Nussenzweig M.C. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hinks T.S.C., Zhang X.-W. MAIT cell activation and functions. Front. Immunol. 2020;11:1014. doi: 10.3389/fimmu.2020.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Godfrey D.I., Koay H.-F., McCluskey J., Gherardin N.A. The biology and functional importance of MAIT cells. Nat. Immunol. 2019;20:1110–1128. doi: 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- 118.Parrot T., Gorin J.-B., Ponzetta A., Maleki K.T., Kammann T., Emgård J., Perez-Potti A., Sekine T., Rivera-Ballesteros O., Karolinska COVID-19 Study Group, Gredmark-Russ S., Rooyackers O., Folkesson E., Eriksson L.I., Norrby-Teglund A., Ljunggren H.-G., Björkström N.K., Aleman S., Buggert M., Klingström J., Strålin K., Sandberg J.K. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]