Abstract
Hierarchal clustering of amino acid metabolites may identify a metabolic signature in children with pediatric acute hypoxemic respiratory failure. Seventy-four immunocompetent children, 41 (55.4%) with pediatric acute respiratory distress syndrome (PARDS), who were between 2 days to 18 years of age and within 72 h of intubation for acute hypoxemic respiratory failure, were enrolled. We used hierarchal clustering and partial least squares-discriminant analysis to profile the tracheal aspirate airway fluid using quantitative LC–MS/MS to explore clusters of metabolites that correlated with acute hypoxemia severity and ventilator-free days. Three clusters of children that differed by severity of hypoxemia and ventilator-free days were identified. Quantitative pathway enrichment analysis showed that cysteine and methionine metabolism, selenocompound metabolism, glycine, serine and threonine metabolism, arginine biosynthesis, and valine, leucine, and isoleucine biosynthesis were the top five enriched, impactful pathways. We identified three clusters of amino acid metabolites found in the airway fluid of intubated children important to acute hypoxemia severity that correlated with ventilator-free days < 21 days. Further studies are needed to validate our findings and to test our models.
Subject terms: Paediatric research, Molecular medicine, Respiratory distress syndrome
Introduction
Pediatric acute respiratory distress syndrome (PARDS) occurs in six percent of mechanically ventilated children1. Although the overall mortality for children with PARDS is 17%, children with the most severe hypoxemia have a mortality of close to 33%1. There are no targeted therapies for children with PARDS due to a poor understanding of the underlying immunologic derangements and pathobiology. Two clinical phenotypes, a hyperinflammatory (reactive) and a hypoinflammatory (uninflamed), have been identified in adults with ARDS using latent class analysis and unsupervised hierarchal clustering2–4. Although ARDS phenotypes have been described using plasma cytokine targets combined with clinically available data, several adult studies have used metabolomic approaches to understand underlying ARDS heterogeneity, identify ARDS biomarkers, and discover metabolic subgroups of patients with ARDS with different mortality rates5,6. In children, endotype identification has focused on sepsis-triggered ARDS cytokine responses and whole blood differential gene expression using microarray technology7–9.
Metabolic subtyping of children with and without PARDS is a strategy to understand underlying metabolic dysregulation of PARDS and to determine whether these responses are associated with PARDS severity and fewer ventilator-free days. Stratification on metabolic and biologic responses defining a PARDS phenotype may aid in predictive and prognostic enrichment of clinical trials of targeted interventions for PARDS. The primary objective of this study is to determine whether unsupervised hierarchal cluster analysis would identify groups of children distinguished by differences in concentrations of amino acid metabolites that would be associated with the degree of hypoxemia and a primary outcome of VFD < 21 days. We hypothesized that an unsupervised approach would identify clusters of children predicted by distinguishing patterns of airway fluid amino acid metabolite concentrations that would correlate with a primary outcome of VFD < 21 days.
Methods
Study design and clinical characterization
Children who were endotracheally intubated within the prior 72 h for acute respiratory failure were enrolled between 2018 and 2020 at the Emory University/Children’s Healthcare of Atlanta Egleston Hospital with informed consent obtained from a parent or legal guardian prior to any procedures. The Emory institutional board approved the study protocol (IRB 00034236 and IRB 00113035). All methods were carried out in accordance with relevant guidelines and regulations (Declaration of Helsinki). Children were excluded if they were immunocompromised as previously described10,11. Hypoxemia thresholds were defined using the Pediatric Acute Lung Injury and Consensus Conference (PALICC) criteria and included a group of intubated children with a level of hypoxemia that did not meet the mild PARDS threshold12. Study participants have been described11. Twenty-eight-day VFD was the primary outcome and was dichotomized as VFD < 21 days and VFD ≥ 21 days7,11,13. Overall severity of illness was quantified using the Pediatric Risk of Mortality-III (PRISM-III) score within four hours of PICU admission14. Organ failures were quantified using the Pediatric Logistic Organ Dysfunction-2 (PELOD-2) score15.
Sample collection and preparation
Tracheal aspirates were obtained within 72 h of intubation using inline suctioning with up to 10 ml of sterile saline lavage and sample collection in a sterile Lukens trap as previously described10,11. Sample were immediately placed in ice and immediately brought to lab for processing. Airway fluid was transferred to a sterile conical tube, mucus plugs and debris were dispersed by passage of the airway sample through an 18-gauge needle, followed by centrifugation in a swinging bucket rotor centrifuge at 800× G for 15 min at 4 C. Cell free supernatant was aliquoted into Eppendorf tubes in 250–500 µl aliquots and stored at −80 C until analysis.
Amino acid metabolite analysis of airway fluid
Amino acids were measured by solid phase extraction followed by derivatization and liquid/liquid extraction (EZ:faast Kit, Phenomenex, Torrance, CA). The extraction and derivatization procedures were performed according to the manufacturer’s instructions. Samples were mixed with internal standards (homoarginine, methionine-d3 and homophenylalanine), extracted, and derivatized with propyl chloroformate. The organic phase was evaporated at room temperature under a stream of nitrogen and re-dissolved in mobile phase. Samples were analyzed using a Thermo Vanquish UHPLC coupled to a Thermo TSQ Quantis triple quadrupole mass spectrometer (Thermo Scientific, Waltham, MA). Using an autosampler at 4 °C, a volume of 1 µL was injected onto a 250 × 2.0 mm × 4 µ AAA-MS column (Phenomenex) at a flow rate of 0.25 mL/min. The column was held at 35 °C. Mobile phase A was 10 mM ammonium formate in water, and mobile phase B was 10 mM ammonium formate in methanol. Samples were separated using an 18-min gradient, from 68 to 83% of mobile phase B, with a 7-min re-equilibration between samples. The ion transfer tube and vaporizer were maintained at 275 °C and 225 °C respectively. Positive electrospray ionization mode at 5000 V was used to monitor selected reaction transitions as outlined in the EZ:faast manual. Transitions were optimized for the mass spectrometer using derivatized standards, and quantitation of amino acids was performed using TraceFinder software (Thermo Scientific).
Cluster analysis, partial least squares-discriminant analysis, and pathway analysis
Forty-two metabolites for seventy-four patient samples were used in the cluster analysis. A total of 88 (2.9%) of the data were missing. By default, missing values were replaced by 1/5 of the minimum positive values of their corresponding variables. Metabolite concentrations were normalized by median, log transformed, and scaled by mean centering in MetaboAnalyst 5.016,17. Clustering was performed using the unweighted pair group method with arithmetic mean (UPGMA)18. A cluster heatmap was generated using the python package seaborn19. Patient were visually assigned to one of three cluster groups, and a partial least squares-discriminant analysis (PLS-DA) was performed using the cluster assignment to identify metabolites contributing the most variability amongst clusters using a variable importance projection. The predictive ability of the PLS-DA model was tested using leave-one-out cross-validation20.
One-way analysis of variance (ANOVA) was performed to identify significant metabolites from the three clusters using the Fisher Least Significant Difference (LSD) test and a false discovery rate (FDR) of < 0.05. Significant metabolites were used in a hypergeometric over-representation analysis test to determine metabolic pathways of importance. The importance measure for topological analysis was the relative betweenness centrality measure.
Pathway enrichment, topology analysis, and metabolic set enrichment analysis
Quantitative pathway enrichment analysis was performed using the binary classification of the primary outcome VFD < 21 days (Yes versus No) and the median normalized, log transformed, and mean center scaled metabolite concentrations using the Globaltest method. The node importance measure for topological analysis was relative betweenness centrality. The pathway impact value was calculated from pathway topology analysis. Statistical significance was determined using FDR < 0.05. Metabolic set enrichment analysis was performed using quantitative enrichment analysis using both the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Small Molecule Protein Database (SMPDB) metabolic pathway associated metabolite sets in separate queries21–23. The enrichment analysis is performed using the R package globaltest using a generalized linear model to estimate a Q-statistic for each metabolite set, which describes the correlation between the compound concentration profiles and the primary outcome, VFD < 21 days (Yes vs. No)24.
Univariate analysis of metabolites associated with VFD
Univariate analysis of metabolites for children with VFD < 21 days (Yes vs. No) was performed. Volcano plots were created and metabolites with a fold-change threshold of 2 and a FDR < 0.05 using the Wilcoxon Rank Sum Test.
Statistical analysis
Participant characteristics were described using median and 25th–75th interquartile ranges or number and percent. A Mann–Whitney-U test was used for univariate analysis for the binary outcome VFD < 21 days (Yes vs. No). One-way analysis of variance (ANOVA) was used for continuous variables and a chi-square test was used for proportions to identify significant differences amongst the three clusters. A p-value < 0.05 was statistically significant.
Results
Clinical characteristics
There were seventy-four participants in this study. Demographic and clinical features of participants in this study have been described11. Clinical features stratified by degree of hypoxemia, and low (no or mild PARDS) or high (moderate or severe PARDS) severity are shown in sTable 1.
Cluster identification
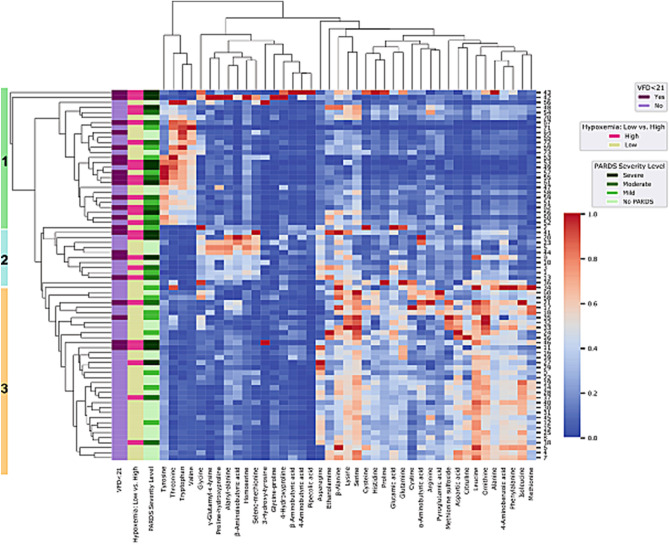
We performed an unsupervised hierarchal clustering of forty-two amino acid metabolites. Three clusters were seen by visual inspection of the heatmap (Fig. 1). Demographic and clinical characteristics of participants by cohort are show in Table 1. There were no differences in age, sex, race, ethnicity, or respiratory infection. Children in cluster 1 had more severe PARDS than children in clusters 2 and 3. Children in cluster 2 had more overall severity of illness (higher PRISM III) than children in clusters 1 and 3. Children in clusters 2 and 3 spent a longer median time of seven days versus 3.5 days on a ventilator compared with children in cluster 3 (Table 1). Increased frequency of high severity of hypoxemia (magenta bars) and a higher proportion of children with VFD < 21 days (dark purple bars) for children in clusters 1 and 2 are shown on the side of the heatmap in Fig. 1.
Figure 1.
Hierarchal cluster analysis of metabolites into three patient clusters. Children with and without pediatric acute respiratory distress syndrome (PARDS) are in blue (no PARDS) and in orange (with PARDS). Children with low hypoxemia (no or mild PARDS) are in yellow and those with high hypoxemia (moderate or severe PARDS) are in magenta. Children with no, mild, moderate, or severe PARDS are shown in green from lightest (no PARDS) to darkest (severe PARDS). Children with ventilator-free days (VFD) < 21 days (more than 7 days on a ventilator or death) are in dark purple and those with VFD ≥ 21 days are in light purple. Cluster assignments are denoted by the green (cluster 1), blue (cluster 2), and orange (cluster 3) bar to the left of the participant dendrogram.
Table 1.
Demographic and clinical characteristics of children by cluster.
| Characteristic | Cluster | |||
|---|---|---|---|---|
| 1 | 2 | 3 | p-value | |
| n = 27 (36%) | n = 11 (15%) | n = 36 (49%) | ||
| Age (years), | 1.06 | 1.67 | 0.58 | 0.0739 |
| Median (IQR) | (0.4, 2.5)) | (0.78, 2.8) | (0.12, 1.7) | |
| Sex, n (%) | ||||
| Female | 12 (44%) | 2 (18%) | 16 (44%) | 0.2619 |
| Male | 15 (56%) | 9 (82%) | 20 (56%) | |
| Race, n (%) | ||||
| Black | 15 (56%) | 8 (73%) | 15 (42%) | 0.4985 |
| White | 9 (33%) | 3 (27%) | 16 (44%) | |
| Unknown | 0 (0%) | 0 (0%) | 3 (8%) | |
| Multiple | 3 (11%) | 0 (0%) | 2 (6%) | |
| Ethnicity, n (%) | ||||
| Hispanic or Latino | 1 (4%) | 0 (0%) | 2 (6%) | 0.7111 |
| Non-Hispanic or Latino | 26 (96%) | 11 (100%) | 34 (94%) | |
| Severity of Illness Scores, median (range) | ||||
| PRISM III | 15 (9, 16) | 23 (12, 26) | 12.5 (8, 18) | 0.0125 |
| PELOD | 6 (5, 7) | 7 (4, 11) | 6 (4, 8) | 0.4333 |
| PARDS severity | ||||
| Low (No/Mild) | 13 (48%) | 7 (64%) | 30 (83%) | 0.0122 |
| High (Moderate/Severe) | 14 (52%) | 4 (36%) | 6 (17%) | |
| Ventilator Days, median (Q1-Q3) | 7 (3, 11) | 7 (6, 19) | 3.5 (2, 6) | 0.0024 |
| 28 day Ventilator-free Days, median (Q1-Q3) | 21 (16, 25) | 21 (9, 22) | 24.5 (22, 26) | 0.0026 |
| Extracorporeal Life Support, n (%) | 3 (11%) | 2 (18%) | 2 (5.6%) | 0.4462 |
| Length of Stay, median (IQR) | ||||
| PICU (days) | 8 (5, 14) | 10 (9, 21) | 6.5 (3.25, 10) | 0.0269 |
| Hospital (days) | 11 (8, 22) | 16 (10, 22) | 11 (7, 16.25) | 0.3668 |
| 28-day Mortality, n (%) | ||||
| Dead | 2 (7.4%) | 1 (9.1%) | 0 (0)% | 0.1256 |
| Respiratory Culture, n (%) | ||||
| No Growth | 2 (7.4%) | 1 (9.1%) | 9 (25%) | 0.4169 |
| Viral Only | 7 (26%) | 4 (36.4%) | 5 (14%) | |
| Bacterial Growth only | 3 (11%) | 1 (9%) | 3 (8%) | |
| Virus + Bacterial Co-detection | 15 (55%) | 5 (45%) | 19 (53%) | |
Partial least squares—discriminant analysis
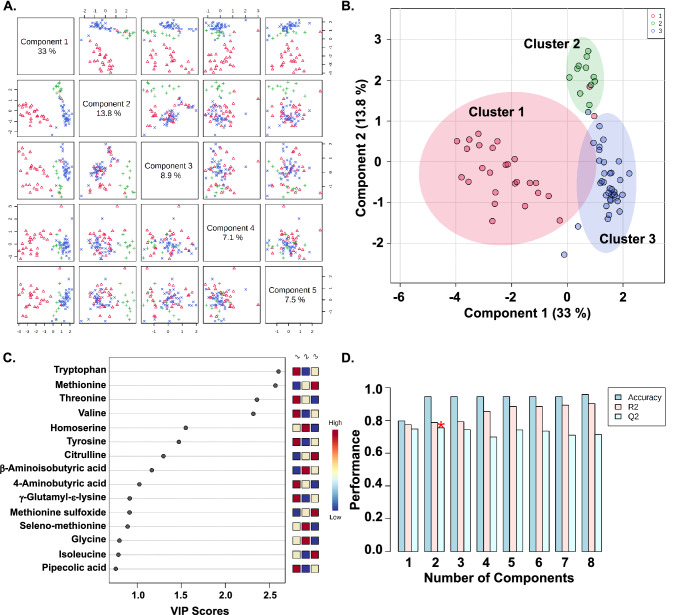
We next used the cluster assignments to perform a partial least squares-discriminant analysis (PLS-DA) to describe the amount of variability and to identify the metabolites contributing the most to cluster variability. Variability explained by the top five components are shown in Fig. 2A. Component 1 and component 2 explain 33% and 13.8% of the variability in the clusters (Fig. 2A,B). There are two outliers in cluster 1 that are misclassified in the other clusters. Both of these patients had severe PARDS and were placed on extracorporeal life support (ECLS) for an Influenza A infection. One child was coinfected with methicillin-resistant Staphylococcal aureus and died after fourteen days supported by ECLS. The child who survived was supported by ECLS for nine days and was hospitalized in the pediatric intensive care unit (PICU) for 36 days.
Figure 2.
Partial least squares-discriminant analysis (PLS-DA) defined by three clusters. (A) Pairwise score plots for the first five components of the PLS-DA analysis. The first component explains 33% of the variability in the three groups. The second and third component explains 13.8% and 8.9% of the variability in the three groups. (B) The scores plot for the first two components labeled by the three clusters: cluster 1 (red), cluster 2 (green), and cluster 3 (blue). (C) Variable importance of projection (VIP) score plot of normalized metabolites by cluster. Higher concentrations are red. Intermediate concentrations are yellow. Lower concentrations are blue. (D) Values of the classification performance assessed by accuracy, goodness of fit (R2), and predictive ability (Q2) for the top six components. Two components best classify the model shown with the red asterisk using leave-one-out cross-validation.
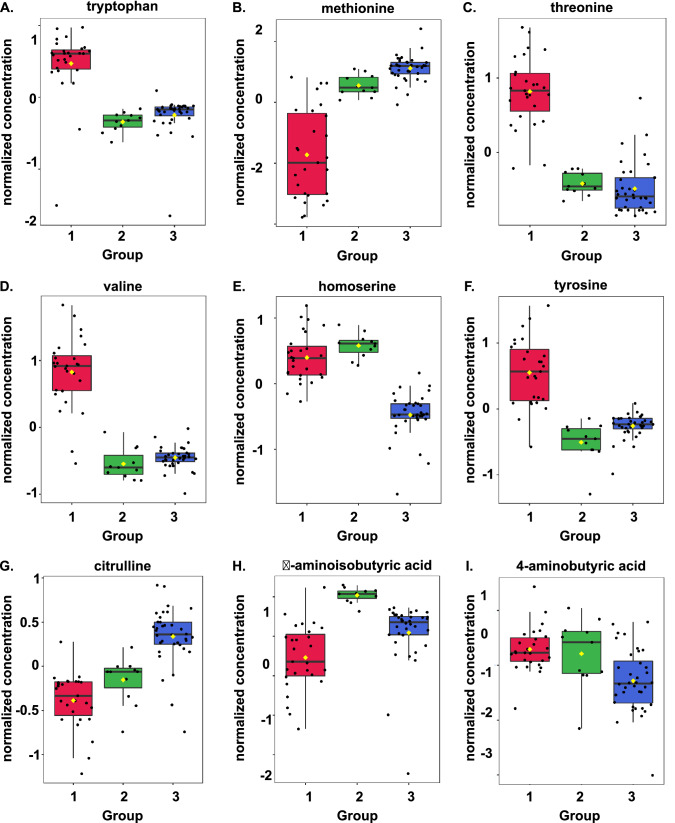
The metabolites contributing the most variability to each cluster are shown by the variable importance of projection (VIP) score plot of normalized metabolites by cluster (Fig. 2C). Classification performance of the PLS-DA model were assessed by accuracy, goodness of fit (R2), and predictive ability (Q2) for the top eight components (Fig. 2D). The second component best classifies the model shown with the red asterisk using leave-one-out cross-validation with an accuracy of 0.95, an R2 of 0.79, and a Q2 of 0.76 (Fig. 2D, sTable 2). Boxplots of the normalized concentrations of the top nine metabolites shown in the VIP score plot in Fig. 2C are shown by cluster (group) in Fig. 3A–I.
Figure 3.
Normalized concentration of the top nine metabolites explaining the variable importance of projection (VIP) in the partial least squares-discriminant analysis (PLS-DA) by cluster or group. (A) Methionine, (B) Tryptophan, (C) Threonine, (D) Valine, (E) Homoserine, (F) Tyrosine, (G) Citrulline, (H) 4-aminobutyric acid, and (I) β-aminoisobutyric acid.
Pathway analysis of clusters
Thirty out of forty-two metabolites were significantly different from each other amongst the three clusters (sTable 3). Over-representation analysis testing was used to explore the metabolic pathways important to distinguishing the three clusters. Significant metabolic pathways with an impact ≥ 0.1 and a FDR < 0.05 for children in the cohort are shown in sFig. 1. A two-cluster solution was also explored and did not reveal meaningful differences in the results (sFigs. 2 & 3).
Pathway analysis by primary outcome
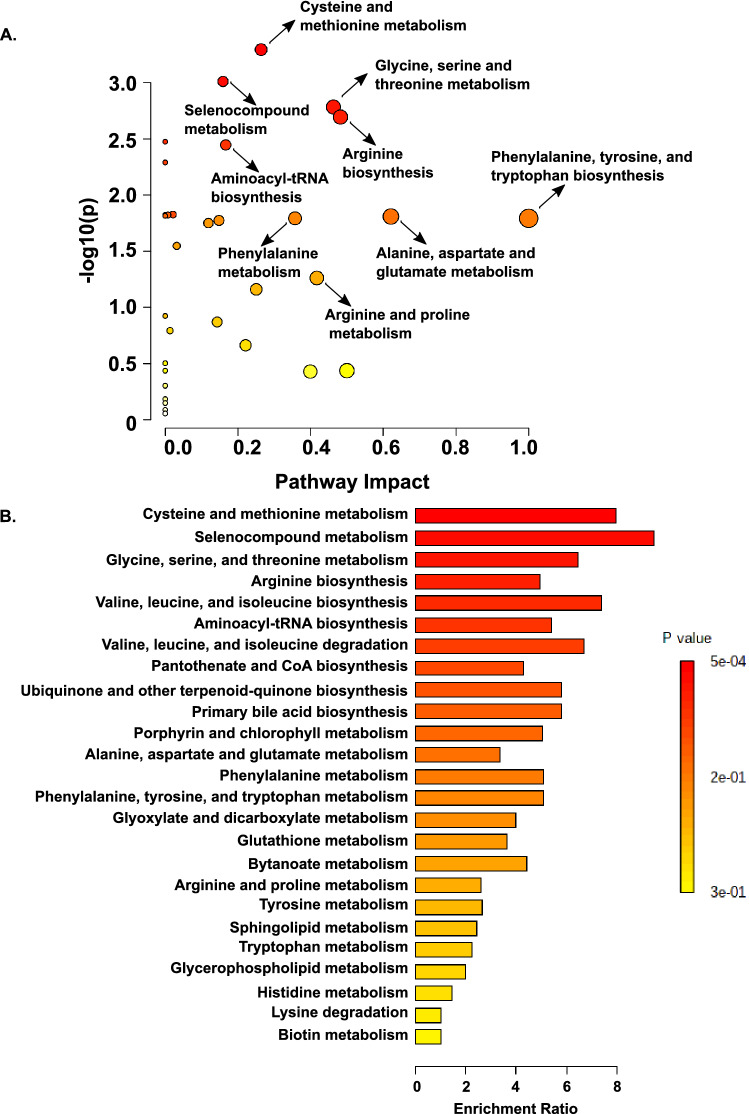
We next performed pathway analysis combining results from a pathway enrichment analysis with a topology analysis using the KEGG database to identify the most relevant metabolic pathways involved in distinguishing children with VFD < 21 days (Yes vs. No) (Fig. 4A)21-23. Pathways with the highest impact and enrichment ratios include: cysteine and methionine metabolism, selenocompound metabolism, glycine, serine and threonine metabolism, arginine biosynthesis, and valine, leucine, and isoleucine biosynthesis (Fig. 4A-B). We also performed a metabolic set enrichment analysis using an alternative database, the Small Molecule Protein Database (SMPDB), using the compound concentration values to explore the metabolic differences between children with VFD < 21 days (Yes vs. No) and found similarly enriched pathways (sFig. 4A). Network analysis of the quantitative pathway enrichment analysis is shown in sFig. 4B. We performed two sensitivity analyses using only samples collected within thirty-six hours of intubation (61/74, 82% of all samples) and forty-eight hours of intubation (70/74, 95% of all samples). There were four pathways with a FDR < 0.05 for the analysis performed using only the tracheal aspirate samples collected within forty-eight hours of intubation, and these top four pathways were the same as the whole cohort (sTable 4). There were no pathways with a FDR < 0.05 for the analysis performed using only the tracheal aspirate samples collected within thirty-six hours of intubation likely due to sample size; however, the top three pathways were the same as the whole cohort (sTable5). To identify individual metabolites that were significantly different between children with VFD < 21 days (Yes vs. No) we performed a univariate analysis. Metabolites with a fold change of 2 or more and a FDR < 0.05 are shown in the volcano plot are shown (sFig. 5A). Boxplots of the normalized concentrations of the six significant metabolites alanyl-alanine, citrulline, homoserine, methionine, selenomethionine, and threonine are shown in sFig. 5B-G.
Figure 4.
Quantitative pathway enrichment analysis using the compound concentration values to explore the metabolic differences between children with ventilator-free days (VFD) < 21 days (more than 7 days on a ventilator or death; Yes) versus those with VFD ≥ 21 days (No). (A) Significant metabolic pathways with an impact ≥ 0.1 for children with VFD < 21 days (Yes vs. No). (B) Quantitative metabolic set enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database for children with VFD < 21 days (Yes vs. No) using normalized metabolic concentrations from airway fluid21–23. Significant pathways are red and orange. The enrichment ratio is calculated as the observed hits/expected hits in the pathway.
Discussion
We used a targeted amino acid metabolite strategy along with unsupervised hierarchal clustering and PLS-DA to discover metabolic airway fluid signatures in children with acute hypoxemic respiratory failure within 72 h of endotracheal intubation. We identified three clusters in our cohort that were defined by differences in thirty metabolites with the most significant and impactful pathways including arginine biosynthesis, glycine, serine, and threonine metabolism, and cysteine and methionine metabolism using over-representation analysis. Children with no or mild acute hypoxemic respiratory failure predominated in Cluster 1; Clusters 2 and 3 were metabolically distinct endotypes made up predominantly of children with moderate or severe acute hypoxemic respiratory failure who spent a median of a week or more on invasive mechanical ventilation. A quantitative metabolic set enrichment analysis identified pathways important for distinguishing children with versus without VFD < 21 days (death or 7 or more days of invasive mechanical ventilation) identified pathway important for oxidative stress (cysteine and methionine metabolism), substrates for one-carbon metabolism (glycine, serine and threonine metabolism), branched-chain amino acids (isoleucine, leucine, and valine metabolism), and the arginine biosynthesis pathways.
There are several studies that have compared the metabolic profiles of plasma, pulmonary edema fluid, bronchoalveolar lavage fluid (BALF), and non-bronchoscopic alveolar lavage (mini BALF or mBALF) in adults with acute respiratory distress syndrome (ARDS) to healthy controls using various untargeted and targeted techniques including 1H-nuclear magnetic resonance (1-H NMR), gas chromatography-mass spectrometry (GC–MS) and liquid chromatography-mass spectrometry (LC–MS)5,6,25,26. Others have used metabolomic analyses to aid diagnosis, stratify ARDS patients by severity, and predict survival outcome5,27–29. Pathways distinguishing survivors from non-survivors with ARDS included glutamine and glutamate metabolism, phenylalanine, tyrosine, and tryptophan biosynthesis, and phenylalanine metabolism6,29. Adult endotyping studies using mBALF showed differences in lysine, arginine, tyrosine, threonine, and branched chain amino acids6. The findings and limitations of these adult studies have been reviewed30. No definitive diagnostic metabolic pattern of ARDS has emerged due to small sample sizes, variability in technique used (1H-NMR, GC–MS, LC–MS/MS) and fluid sampled (plasma, BALF, mBALF), lack of an external validation cohort, and the use of non-mechanically ventilated controls30.
Airway fluid obtained from readily accessible tracheal aspirate sampling of intubated children represents the metabolic derangements of the injured lung compared with plasma metabolites due to the proximity of the airway fluid to the damaged alveolar epithelial lining of the lung. We focused on amino acid based metabolic pathways because many of the predominant signatures from prior adult studies found significant differences in amino acid metabolic, degradation, and biosynthetic pathways in their analyses as previously reviewed30.
Increased amino acid cellular uptake and biosynthesis drive cell proliferation and control energy-generating metabolic switches by regulating glycolysis, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation (OXPHOS)31. Branched-chain amino acids provide acetyl-CoA and succinyl-CoA substrates for energy and reducing equivalent production via the TCA cycle31. Sulfur-containing amino acids, such as cysteine and methionine, and their metabolic pathways are central regulators of cellular and extracellular redox status31. Amino acids supply methyl and acetyl groups to accomplish post-translational modification of proteins and epigenetic modification of histones to regulate gene expression, acute immune responses and immune cell memory31. For example, leucine-supplied acetyl-CoA acetylates and activates mTORC1, a key intermediate in the nutrient-sensing mTOR pathway, increasing glycolysis32.
Amino acids support protein synthesis and metabolic programming critical for immune cell activation31. T cell proliferation, activation and survival depends on rapid one-carbon metabolism, or the transfer of a methyl group to various substrates, for the biosynthesis of purine nucleotides, amino acid homeostasis of glycine, serine and methionine, epigenetic maintenance, and reduction–oxidation (redox) defense. Glycine is a precursor for many essential biosynthetic pathways including glutathione, purine, creatine, and heme synthesis. When glycine supply is unavailable, serine is metabolized to glycine to fuel purine and glutathione synthesis33. One-carbon reactions use and create redox equivalents, such as NADPH, that are important to mitochondrial redox homeostasis33,34.
Cysteine is one of the amino acid precursors of glutathione, and cysteine’s metabolism and redox state is altered during acute lung injury and inflammation35. The uptake of cystine, the oxidized form of cysteine, is needed for T cell activation, proliferation, and DNA synthesis31,36–38. The requirement for sulfur amino acids increases during trauma, sepsis, and other critical illnesses39. T cell proliferation is impaired and activation is reduced with a deficiency of cysteine and intracellular glutathione40. Selenoproteins play an important role in antioxidant defense and oxidative metabolism and modulation of reactive oxygen species and inflammatory signaling pathways affecting the ability to respond to viral infections such as influenza41,42.
Reduced levels of arginine result in a failure of T cells to proliferate43. Activated neutrophils recruited to the airways of children with PARDS deplete arginine from the airway environment due to degranulate and release arginase 144–47. Arginase 1 metabolism of arginine results in the synthesis of polyamines such as spermidine that limit inflammatory activity of M1-like macrophages and enhance to function of wound-healing M2-like macrophages31. Increased arginine metabolism by macrophages and myeloid-derived suppressor cells restricts arginine supply to T cells limiting T cell activation and interferon-γ production31,48. Similar to arginine, tryptophan also promotes T cell proliferation and catabolism of tryptophan by macrophages inhibits T cell activation, proliferation and cytokine production in response to infection49.
The pediatric acute respiratory distress incidence and epidemiology (PARDIE) study shows that many children with mild hypoxemia at onset will progress to meeting the Berlin criteria within seventy-two hours of meeting PARDS criteria1. It is not known how supportive treatment over the first seventy-two hours following the initiation of mechanical ventilation influences metabolic pathways in the lung environment. It is likely that some metabolites reflect treatment including the introduction of enteral and parenteral nutrition and the effect this likely has on amino acid, lipid and glucose-containing products. Significant resuscitation measures, including ventilator setting manipulation, fluid resuscitation, and initiation of antibiotics, analgosedation, and neuromuscular blockage, are likely to be ongoing within the first twenty-four to forty-eight hours following intubation and diagnosis of acute hypoxemic respiratory failure. We performed sensitivity analyses that included only samples collected within thirty-six hours and forty-eight hours after intubation. The top four pathways were the same and remained significant between the samples collect within forty-eight hours versus seventy-two hours. Due to a smaller sample size, the samples collected within thirty-six hours had the same top 3 pathways identified, but did not reach a FDR < 0.05. Severity stratification following twenty-four hours of resuscitation and stabilization may more accurately represent the true degree of lung injury due to improvement in ventilation/perfusion mismatch and lung recruitment with positive pressure ventilation50; however, there is little data on the temporal changes of metabolites, protein, or transcriptional biomarkers within the first days to week after onset of pediatric acute hypoxemic respiratory failure or PARDS.
There are several limitations to our single-center study. We sampled the tracheal aspirate fluid without measuring paired plasma analytes. While the majority of our samples were collected within twenty-four hours of intubation, we extended the sampling window to seventy-two hours to increase enrollment into our study. We acknowledge that the first seventy-two hours following intubation is a dynamic period where pathophysiology is evolving due to underlying the disease process, changes in ventilator settings, fluid balance, development of other organ dysfunctions, and ongoing resuscitation and clinical interventions, including blood transfusions, pressor requirements and changes in medications could confound the interpretation of our observations. Despite these limitations, in the largest multi-institutional, international epidemiologic study of PARDS, the prediction of mortality stratified by severity of hypoxemia was relatively stable from 6 to 48 h after an intubated child met PARDS criteria1. We did not collect serial time-points to correlate metabolite levels with clinical trajectory, and this should be a focus of future studies. While an accepted and validated non-pulmonary multiple organ dysfunction score does not exist, we acknowledge that other organ dysfunctions influence global metabolism and could impact our study findings in unknown ways despite our sampling of tracheal aspirate fluid rather than serum or plasma. We used a targeted metabolomic approach that focused on amino acids and dipeptide analytes limiting the metabolites measured. Lipid metabolites are an important component of surfactant and signaling pathways in the lung that we were unable to capture using our approach. While this is the only metabolomic study to date in intubated children with and without acute hypoxemic respiratory failure, the sample size is small, although it is larger than several adult ARDS studies30. There is no external validation cohort, and replication of our findings in a larger independent cohort of children at risk for and with varying severity of PARDS is needed.
In summary, we identified three patient groups using an unsupervised clustering method and explored the amino acid metabolites and pathways important to acute hypoxemic respiratory failure. We then identified metabolites and pathways that differentiated children with and without ventilator-free days < 21 days. Further studies are needed to validate our findings and to test our models.
Supplementary Information
Acknowledgements
We acknowledge the support and contributions from the Children’s Healthcare of Atlanta and Emory University Pediatric Biomarkers Core.
Author contributions
J.G., A.F., and R.K. conceived and developed the study, supervised the acquisition of the biological data, analyzed and interpreted the data. J.G. drafted and edited the manuscript. R.K., M.R., and A.F. assisted with drafting and editing the manuscript. S.S. and A.M. helped with patient sample processing, performed experiments and edited the manuscript. C.O. assisted in identifying, consenting, acquiring patient samples and assisted in collecting clinical information about the patients. All authors edited and approved the final version of this manuscript.
Funding
Funding was provided by NIH grants K12HD072245 (Atlanta Pediatric Scholars Program), K23 HL151897-01, and an Emory University Pediatrics Research Alliance Junior Faculty Focused Pilot award to JG. Funding was provided by the NIH grant K24 NR018866 to AF. RK is funded by UL1TR002378. National Institutes of Health, K12HD072245, National Institutes of Health, NHLBI K23 HL151897-01, Emory University Pediatrics Research Alliance, Junior Faculty Focused Pilot, National Institutes of Health, NINR, K24NR018866, National Center for Advancing Translational Sciences (HHS—NIH), UL1TR002378.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-02354-4.
References
- 1.Khemani RG, et al. Paediatric acute respiratory distress syndrome incidence and epidemiology (PARDIE): An international, observational study. Lancet Respir. Med. 2019;7:115–128. doi: 10.1016/S2213-2600(18)30344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinha P, et al. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: A secondary analysis of randomised controlled trials. Lancet Respir. Med. 2020;8:247–257. doi: 10.1016/S2213-2600(19)30369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha P, et al. Latent class analysis of ARDS subphenotypes: A secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intens. Care Med. 2018;44:1859–1869. doi: 10.1007/s00134-018-5378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos LD, et al. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax. 2017;72:876–883. doi: 10.1136/thoraxjnl-2016-209719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers AJ, et al. Profiling of ARDS pulmonary edema fluid identifies a metabolically distinct subset. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;312:L703–L709. doi: 10.1152/ajplung.00438.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viswan A, Ghosh P, Gupta D, Azim A, Sinha N. Distinct metabolic endotype mirroring acute respiratory distress syndrome (ARDS) subphenotype and its heterogeneous biology. Sci. Rep. 2019;9:2108. doi: 10.1038/s41598-019-39017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yehya N, Thomas NJ, Wong HR. Evidence of endotypes in pediatric acute hypoxemic respiratory failure caused by sepsis. Pediatr. Crit. Care Med. 2019;20:110–112. doi: 10.1097/PCC.0000000000001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yehya N, et al. Peripheral blood transcriptomic sub-phenotypes of pediatric acute respiratory distress syndrome. Crit. Care. 2020;24:681. doi: 10.1186/s13054-020-03410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yehya N, Wong HR. Adaptation of a biomarker-based sepsis mortality risk stratification tool for pediatric acute respiratory distress syndrome. Crit. Care Med. 2018;46:e9–e16. doi: 10.1097/CCM.0000000000002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunwell JR, et al. Neutrophil dysfunction in the airways of children with acute respiratory failure due to lower respiratory tract viral and bacterial coinfections. Sci. Rep. 2019;9:2874. doi: 10.1038/s41598-019-39726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunwell JR, et al. Differential type I interferon response and primary airway neutrophil extracellular trap release in children with acute respiratory distress syndrome. Sci. Rep. 2020;10:19049. doi: 10.1038/s41598-020-76122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khemani, R. G., Smith, L. S., Zimmerman, J. J., Erickson, S. & Pediatric Acute Lung Injury Consensus Conference, G. Pediatric acute respiratory distress syndrome: definition, incidence, and epidemiology: proceedings from the Pediatric Acute Lung Injury Consensus Conference. Pediatr. Crit. Care Med.16, S23–40, 10.1097/PCC.0000000000000432 (2015). [DOI] [PubMed]
- 13.Sauthier MS, Jouvet PA, Newhams MM, Randolph AG. Machine learning predicts prolonged acute hypoxemic respiratory failure in pediatric severe influenza. Crit. Care Explor. 2020;2:e0175. doi: 10.1097/CCE.0000000000000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollack MM, Patel KM, Ruttimann UE. PRISM III: An updated pediatric risk of mortality score. Crit. Care Med. 1996;24:743–752. doi: 10.1097/00003246-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Leteurtre S, et al. PELOD-2: an update of the PEdiatric logistic organ dysfunction score. Crit. Care Med. 2013;41:1761–1773. doi: 10.1097/CCM.0b013e31828a2bbd. [DOI] [PubMed] [Google Scholar]
- 16.Pang, Z. et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucl. Acids Res. 10.1093/nar/gkab382 (2021). [DOI] [PMC free article] [PubMed]
- 17.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 18.Sokal & Michener A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 1958;38:1409–1438. [Google Scholar]
- 19.Waskom, M. L. Seaborn: statistical data visualization. J. Open Source Softw.6, 3021–3024. 10.21105/joss (2021).
- 20.Szymanska E, Saccenti E, Smilde AK, Westerhuis JA. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics. 2012;8:3–16. doi: 10.1007/s11306-011-0330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Prot. Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucl. Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: Testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 25.Lin S, et al. Explore potential plasma biomarkers of acute respiratory distress syndrome (ARDS) using GC-MS metabolomics analysis. Clin. Biochem. 2019;66:49–56. doi: 10.1016/j.clinbiochem.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Evans CR, et al. Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J. Proteome Res. 2014;13:640–649. doi: 10.1021/pr4007624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswan A, Sharma RK, Azim A, Sinha N. NMR-based metabolic snapshot from minibronchoalveolar lavage fluid: An approach to unfold human respiratory metabolomics. J. Proteome Res. 2016;15:302–310. doi: 10.1021/acs.jproteome.5b00919. [DOI] [PubMed] [Google Scholar]
- 28.Viswan A, Singh C, Kayastha AM, Azim A, Sinha N. An NMR based panorama of the heterogeneous biology of acute respiratory distress syndrome (ARDS) from the standpoint of metabolic biomarkers. NMR Biomed. 2020;33:e4192. doi: 10.1002/nbm.4192. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, et al. Increased mortality of acute respiratory distress syndrome was associated with high levels of plasma phenylalanine. Respir. Res. 2020;21:99. doi: 10.1186/s12931-020-01364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metwaly, S. M. & Winston, B. W. Systems Biology ARDS Research with a Focus on Metabolomics. Metabolites10. 10.3390/metabo10050207 (2020). [DOI] [PMC free article] [PubMed]
- 31.Kelly B, Pearce EL. Amino assets: How amino acids support immunity. Cell Metab. 2020;32:154–175. doi: 10.1016/j.cmet.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Son, S. M. et al. Leucine Signals to mTORC1 via Its Metabolite Acetyl-Coenzyme A. Cell Metab.29, 192–201 e197. 10.1016/j.cmet.2018.08.013 (2019). [DOI] [PMC free article] [PubMed]
- 33.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan J, et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyer SS, Jones DP, Brigham KL, Rojas M. Oxidation of plasma cysteine/cystine redox state in endotoxin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 2009;40:90–98. doi: 10.1165/rcmb.2007-0447OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010;70:68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levring TB, et al. Activated human CD4+ T cells express transporters for both cysteine and cystine. Sci. Rep. 2012;2:266. doi: 10.1038/srep00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levring TB, et al. Human CD4+ T cells require exogenous cystine for glutathione and DNA synthesis. Oncotarget. 2015;6:21853–21864. doi: 10.18632/oncotarget.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grimble RF. The effects of sulfur amino acid intake on immune function in humans. J. Nutr. 2006;136:1660S–1665S. doi: 10.1093/jn/136.6.1660S. [DOI] [PubMed] [Google Scholar]
- 40.Li P, Yin YL, Li D, Kim SW, Wu G. Amino acids and immune function. Br. J. Nutr. 2007;98:237–252. doi: 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- 41.Beck MA, Levander OA, Handy J. Selenium deficiency and viral infection. J. Nutr. 2003;133:1463S–1467S. doi: 10.1093/jn/133.5.1463S. [DOI] [PubMed] [Google Scholar]
- 42.Beck MA, et al. Selenium deficiency increases the pathology of an influenza virus infection. FASEB J. 2001;15:1481–1483. doi: 10.1096/fj.00-0721fje. [DOI] [PubMed] [Google Scholar]
- 43.Vonwirth V, et al. Inhibition of arginase 1 liberates potent T cell immunostimulatory activity of human neutrophil granulocytes. Front. Immunol. 2020;11:617699. doi: 10.3389/fimmu.2020.617699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munder M, et al. Suppression of T-cell functions by human granulocyte arginase. Blood. 2006;108:1627–1634. doi: 10.1182/blood-2006-11-010389. [DOI] [PubMed] [Google Scholar]
- 45.Rotondo R, et al. Exocytosis of azurophil and arginase 1-containing granules by activated polymorphonuclear neutrophils is required to inhibit T lymphocyte proliferation. J. Leukoc. Biol. 2011;89:721–727. doi: 10.1189/jlb.1109737. [DOI] [PubMed] [Google Scholar]
- 46.Zea AH, et al. L-Arginine modulates CD3zeta expression and T cell function in activated human T lymphocytes. Cell Immunol. 2004;232:21–31. doi: 10.1016/j.cellimm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Caldwell RW, Rodriguez PC, Toque HA, Narayanan SP, Caldwell RB. Arginase: A multifaceted enzyme important in health and disease. Physiol. Rev. 2018;98:641–665. doi: 10.1152/physrev.00037.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fletcher M, et al. l-Arginine depletion blunts antitumor T-cell responses by inducing myeloid-derived suppressor cells. Cancer Res. 2015;75:275–283. doi: 10.1158/0008-5472.CAN-14-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yehya N, Servaes S, Thomas NJ. Characterizing degree of lung injury in pediatric acute respiratory distress syndrome. Crit. Care Med. 2015;43:937–946. doi: 10.1097/CCM.0000000000000867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.