Abstract
As early detection of symptoms in the subclinical to clinical psychosis spectrum may improve health outcomes, knowing the probabilistic susceptibility of developing a disorder could guide mitigation measures and clinical intervention. In this context, polygenic risk scores (PRSs) quantifying the additive effects of multiple common genetic variants hold the potential to predict complex diseases and index severity gradients. PRSs for schizophrenia (SZ) and bipolar disorder (BD) were computed using Bayesian regression and continuous shrinkage priors based on the latest SZ and BD genome-wide association studies (Psychiatric Genomics Consortium, third release). Eight well-phenotyped groups (n = 1580; 56% males) were assessed: control (n = 305), lower (n = 117) and higher (n = 113) schizotypy (both groups of healthy individuals), at-risk for psychosis (n = 120), BD type-I (n = 359), BD type-II (n = 96), schizoaffective disorder (n = 86), and SZ groups (n = 384). PRS differences were investigated for binary traits and the quantitative Positive and Negative Syndrome Scale. Both BD-PRS and SZ-PRS significantly differentiated controls from at-risk and clinical groups (Nagelkerke’s pseudo-R2: 1.3–7.7%), except for BD type-II for SZ-PRS. Out of 28 pairwise comparisons for SZ-PRS and BD-PRS, 9 and 12, respectively, reached the Bonferroni-corrected significance. BD-PRS differed between control and at-risk groups, but not between at-risk and BD type-I groups. There was no difference between controls and schizotypy. SZ-PRSs, but not BD-PRSs, were positively associated with transdiagnostic symptomology. Overall, PRSs support the continuum model across the psychosis spectrum at the genomic level with possible irregularities for schizotypy. The at-risk state demands heightened clinical attention and research addressing symptom course specifiers. Continued efforts are needed to refine the diagnostic and prognostic accuracy of PRSs in mental healthcare.
Subject terms: Clinical genetics, Schizophrenia, Bipolar disorder, Prognostic markers
Introduction
Psychosis is a mental condition characterized by disturbed contact with reality, affecting cognition, feelings, and behavior. It manifests primarily as sensory experiences in the absence of physical stimuli (hallucinations) and holding bizarre, irrational, or false beliefs (delusions) [1]. While psychotic symptoms are associated with different medical conditions, they typically occur in schizophrenia (SZ), a prototypical psychotic disorder, and as a frequent characteristic in bipolar disorder (BD). Notably, SZ and BD are highly heritable, with heritability estimates of 60–80% [2] and substantially overlapping genetic architectures [3–5]. Psychotic symptoms in SZ and BD also show clinical responses to antipsychotic medication [6]. Despite extensive research, the etiology of psychotic disorders remains unclear, with multiple putative determinants and risk-conferring factors [7]. Within this complex scenario of poly-causation, genetic burdens may predispose certain individuals to developing clinical syndromes in critical life periods [8]. Addressing the heterogeneity of SZ and BD genetics results, a recent investigation assessed the contribution of common variants to disease susceptibility [9]. In addition, the availability of large datasets enabled by high-throughput screening and genomic advances have facilitated the development of polygenic risk scores (PRSs). PRSs cumulatively estimate genome-wide effects of common variants en masse instead of the effects of individual single-nucleotide polymorphisms (SNPs) [10]. In the context of psychosis research, the value of PRSs has been substantiated across numerous applications, including their capacity to differentiate between cases and controls [11, 12] and to predict longitudinal illness courses, diagnostic subtype shifts [13], and responses to antipsychotic medication [14, 15].
Overt psychosis in SZ is often preceded by early signs and mild symptoms developing 2–5 years before a formal diagnosis [16]. Newer diagnostic manuals, such as Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition [17] and International Classification of Diseases, Eleventh Revision [18] incorporate dimensional aspects. A growing body of evidence also supports a continuum-like or extended psychosis spectrum model (cf. categorical approaches) of increasing symptom severity and persistence [19, 20]. These findings are corroborated by the modulating effect of PRS for SZ (SZ-PRS) on well-established neurocognitive intermediate phenotypes, such as prefrontal dysregulation in connection to working memory load, which has been observed in both SZ patients and healthy persons [21]. The polygenic burden indexed by SZ-PRS was also shown to be associated with cognitive–emotional, behavioral, and social impairments, known antecedents of SZ, across the developmental trajectory in large population-based cohorts [22]. Additionally, separate and joint PRSs for SZ and BD (BD-PRS) have proven useful in dissecting BD subtypes [23] and predicting progression to BD and psychotic disorders from a diagnosis of unipolar disorder [24]. As early recognition of psychosis may mitigate its negative individual and societal impacts, recent research has emphasized the importance of early detection and intervention [25]. The concepts of both schizotypy (trait-like psychosis proneness) and at-risk mental state (imminent, basic symptom criteria) have substantially informed preventive research [26, 27]. Nevertheless, the role of schizotypy in the developmental psychopathology of psychosis remains debated [26, 28], and most individuals at high risk do not transition to a diagnostic entity even years after an initial clinical presentation [29]. The molecular and genetic foundation of a broad psychosis spectrum is unclear, yet its exploration may reveal biological common denominators. In this context, the ultimate objectives of PRS analyses include the accurate prediction of disease onset and determination of tailored treatment strategies [10]. However, studies aimed at identifying associations between PRS and quantitative psychotic symptom manifestations have yielded both significant [30, 31] and inconsistent findings [32]. Notably, one study identified a linear relationship only when both cases and controls were pooled [33], underscoring the value of the continuum model. Inconsistencies may also have arisen from possible temporal symptom fluctuations and/or differences in operationalization among the clinical instruments employed. Furthermore, recent genome-wide association studies (GWASs) based on increasing discovery sample sizes and refined algorithms have contributed to the improved PRS power [34], which itself is sufficient motivation for new predictive studies.
This work investigates the associations and between-group differences of PRS computed for SZ and BD across the extended psychosis spectrum. In an early study utilizing PRS [35], SZ-PRS and BD-PRS were significantly associated with SZ and BD diagnostic spectrums. When split into separate diagnoses, SZ-PRS was associated with SZ, schizoaffective disorder (SZA), psychosis not otherwise specified, and BD type I, while BD-PRS was associated with BD type I and BD type II. However, no significant differences between any portion of the spectrum or specific groups were found after controlling for multiple comparisons (Tukey method). In contrast, the present investigation expands the phenotypic poles of normatively diagnosed disorder groups (SZ, BD type I, BD type II, SZA) and controls into the subclinical categories of high risk for developing psychosis, as well as schizotypy. If individuals across these groups differentially share genetic liability, a smooth gradient and severity-related differences in PRS should be observed. Moreover, in a transdiagnostic manner, this work examines the relationship between PRS and a well-validated continuous symptom measure, the Positive and Negative Syndrome Scale (PANSS) [36], using quantile regression modeling [37].
Methods and materials
Phenotype definition
Data included in this work come from three main projects, two completed in Switzerland [38, 39], the Zurich Program for Sustainable Development of Mental Health Services (ZinEP) project on at-risk mental state for psychosis and the Exceptional Experiences (EE) project on psychotic-like experiences, as well as the multi-center PsyCourse Study [40] on the affective-psychotic spectrum conducted in Germany and Austria. Two other smaller Swiss projects, the Zurich Family-Trio Study and the Zurich OCD Study, served as additional sources of healthy controls [41, 42]. Data from eight groups were analyzed in this study according to the pre-established criteria: SZ, SZA, BD II, BD I, at-risk state for psychosis (RISK), higher schizotypy (SCHIZ H), lower schizotypy (SCHIZ L), and non-psychiatric controls (CTRL). The at-risk categories included basic symptoms, by some conventions known as “high risk” (HR) and “ultra-high risk” (UHR) for psychosis [29]. The specific inclusion criteria were as follows: at least one cognitive–perceptive basic symptom and/or at least two cognitive disturbances based on the Schizophrenia Proneness Interview [43, 44] for HR; at least one attenuated psychotic symptom, and/or at least one brief limited intermittent psychotic symptom based on the Structured Interview for Prodromal Syndromes [45] for UHR. Additionally, the hypomanic symptoms in this group were measured using the Hypomania Checklist (HCL-32) [46]. The median-split of the Schizotypal Personality Questionnaire (SPQ) [47] score was used, as in earlier studies on higher and lower schizotypy [48, 49], with similar cut-offs and mean values. Psychiatric axis I diagnosis in these two groups was excluded using the Mini-International Neuropsychiatric Interview [50]. The diagnoses and assessments were made by clinicians and trained raters. Patients were recruited based on referrals from clinics or patient register queries. Proficiency in German was a criterion for participation. Written informed consent was obtained from all participants and, for participants under 18 years of age, their parents or legal guardians. All the participating projects were approved by the ethics committees of each respective institution (Zurich: KEK-Nr. E63/2009, KEK-Nr. 2011-0423, KEK-Nr. 2010-0340/3, KEK-Nr. 2010-0340; see detailed information on the PsyCourse Study [40] [www.psycourse.de] provided in the Supplementary Methods and Materials) and conducted in compliance with the Declaration of Helsinki. Table S1 (Supplementary Information) lists the primary inclusion criteria for the eight investigated groups. All data were anonymized for analysis.
Genotyping and genotype imputation
Genomic DNA was isolated from venous whole blood or saliva (for a small subsample of controls) and genotyped using Illumina Infinium Psych-Array BeadChips (Illumina, San Diego, CA, USA). SNP-level quality control (QC) included the removal of variants with a call rate <98%, significant deviations from Hardy–Weinberg equilibrium (HWE) (p < 0.001), or minor allele frequency (MAF) < 0.1%. In the individual-level QC, data were removed to eliminate duplicates, sex mismatches, cryptic relatives (PI-HAT > 0.125), samples with an individual genotyping rate >98%, or heterozygosity rate exceeding 3 SD from the mean. Population stratification was assessed using a principal component analysis on the pairwise genomic relationship matrix. Individuals with genotypes not clustered with 1000 Genomes Project EUR super-populations were removed from the analyses (n = 47). After QC, pre-phasing was performed using SHAPEIT [51] and imputation was conducted using IMPUTE2 [52]. The phase 3 integrated variant dataset from the 1000 Genomes Project [53] was used as the reference panel. After imputation, variants with a low information score (INFO < 0.9) or frequency (MAF < 1%) were removed. All data were processed with the same analysis pipeline. Figure S1 (Supplementary Information) visualizes the European origin of participants along the first two ancestry PCs with labeled superpopulations.
PRS computation
The PRS calculations for each individual were based on the risk alleles and the corresponding odds ratios (ORs) from the most recent GWAS of the Psychiatric Genomics Consortium (PGC3) on SZ [54] and BD [55]. The SZ GWAS included 69,369 cases and 236,642 controls, while the BD GWAS included 41,917 cases and 371,549 controls. PRSs were computed using a recently developed Bayesian regression and continuous shrinkage priors method (PRS-CS) [34, 56]. The PRS-CS method exhibited superior prediction accuracy across multiple complex diseases and traits, compared to several other methods, particularly with large training datasets [56]. In this approach, the posterior effect sizes of SNPs are inferred using information from the GWAS summary statistics and an external linkage disequilibrium reference (1000 Genomes Project phase 3, EUR super-populations), through principles of joint multivariate modeling. The global shrinkage parameter phi (φ) [57] was automatically learned from the data (PRS-CS-auto; φ1/2 ~ C+(0,1), where C+(0,1) denotes the half-Cauchy distribution). In our study, the automatically estimated global shrinkage parameter was φ = 1.8E−04 for SZ and φ = 1.3E−04 for BD. PRS-CS allowed us to use a single polygenic score for each discovery trait (SZ and BD, respectively) that considers the polygenicity of the disorder and appropriately models the genetic architecture of SZ and BD. The PRSs were standardized using z-score transformation to facilitate interpretation.
Statistical analysis: PRSs across groups
An analysis of variance (ANOVA) was conducted to assess between-group differences in SZ-PRS and BD-PRS (with group as predictor and PRS as outcome variable), using subject age, sex, and first five genetic ancestry principal components (PCs) to account for population stratification. Pairwise comparisons were corrected per analysis using a Bonferroni correction for 28 tests performed (i.e., the number of pair combinations for each PRS). Additionally, to assess the associations with group/diagnosis and variance explained by PRS, block-wise binary logistic regressions with healthy controls were conducted. For each PRS, two statistical models with case/control status as outcome were compared, one testing the covariates alone (i.e., age, sex, and the five ancestry-specific PCs as a baseline model) and the other testing the covariates with the added PRS (full model). We report the R2 value as the difference in Nagelkerke’s pseudo-R2 [58] between these two nested models as an indicator of explained variance. As these comparisons were against the control condition, no further correction was applied in these non-independent tests.
Statistical analysis: association between PRSs and symptoms
To examine the effects of PRS predicting the symptom PANSS scores (Positive Symptoms, Negative Symptoms, General Psychopathology, and Total Score) across low, medium, and high symptom score distributions, quantile regressions using three tau (τ) values (0.25, 0.50, 0.75) and accounting for age, sex, and the five ancestry-specific PCs were conducted. The quantiles indicate that 25%, 50%, and 75% of the scores, respectively, fall below each corresponding point in the distribution of scores. Quantile regression [37] was chosen over a transformation because of the floor effect of symptom data. This statistical approach estimates the effects at different locations in the criterion distribution with or without normality assumptions being met, where relying on the mean as a measure of centrality under an ordinary least squares model might be less informative or inappropriate [59].
Statistical analyses were performed using the R statistical computing environment (version 4.0.5). Nominal significance was assessed at an α < 0.05 threshold.
Results
Study sample
After QC, 1580 samples (883 males; ages 12–78 years) from the initial pool of 1752 genotyped samples, with complete information for study group, sex, and age, were available for analysis, including 6,600,214 genetic markers. PANSS data were accessible for 1134 individuals (622 males). The characteristics and symptom scores of participants are summarized in Table 1. Within the at-risk group, the following numbers of individuals met the partial (overlapping) criteria: 120 (HR or UHR), 111 (HR), 70 (UHR), 50 (HR without UHR), 86 (risk for BD).
Table 1.
Sample characteristics.
| Phenotypic spectrum | Sample size | % male |
Mean age (SD) | Mean PANSS score (SD) | |||
|---|---|---|---|---|---|---|---|
| Positive symptoms | Negative symptoms | General psychopathology | Total score | ||||
| Schizophrenia (SZ) | 384 | 64.58 | 40.54 (12.21) | 14.60 (5.87)a | 16.30 (6.82)a | 30.61 (10.36)a | 61.51 (20.58)a |
| Schizoaffective disorder (SZA) | 86 | 38.37 | 45.63 (10.77) | 12.53 (5.74)b | 14.28 (5.76)b | 28.80 (8.83)b | 55.60 (17.11)b |
| Bipolar II disorder (BD II) | 96 | 46.88 | 46.22 (13.80) | 8.69 (2.36)c | 9.82 (3.77)c | 23.56 (6.82)c | 42.07 (11.00)c |
| Bipolar I disorder (BD I) | 359 | 54.32 | 45.16 (12.72) | 9.48 (3.41)d | 10.21 (3.94)d | 23.23 (6.48)d | 42.92 (10.94)d |
| At-risk state for psychosis (RISK) | 120 | 60.00 | 21.23 (5.93) | 12.66 (4.13)e | 14.19 (5.72)e | 31.47 (8.18)e | 58.32 (15.87)e |
| Higher schizotypy (SCHIZ H) | 113 | 62.83 | 31.68 (10.20) | — | — | — | — |
| Lower schizotypy (SCHIZ L) | 117 | 72.65 | 32.51 (11.71) | — | — | — | — |
| Non-psychiatric controls (CTRL) | 305 | 43.93 | 37.29 (15.22) | 7.08 (0.32)f | 7.13 (0.50)f | 16.37 (0.83)f | 30.58 (1.01)f |
| Total sample | 1580 | 55.89 | 38.89 (14.20) | 11.28 (5.16)g | 12.42 (6.11)g | 25.96 (9.40)g | 49.67 (18.67)g |
Note: The Positive and Negative Syndrome Scale (PANSS) scores were available for the following sample subsets: an = 362, bn = 83, cn = 89, dn = 328, en = 119, fn = 153, gn = 1134. The assessments were collected during interviews at clinical presentations.
Between-group differences
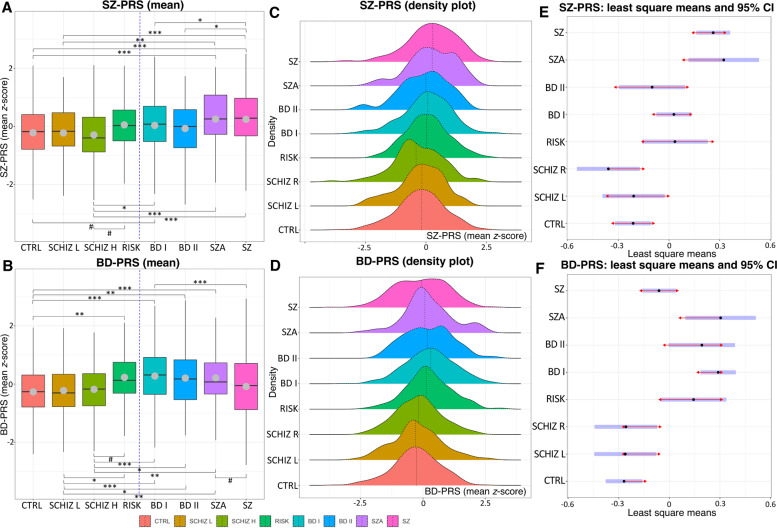
ANOVA revealed significant among-group differences for both SZ-PRS (F7,1565 = 9.30, p = 2.52 × 10−11) and BD-PRS (F7,1565 = 11.71, p = 1.37 × 10−14). Out of 28 pairwise Bonferroni-corrected tests for SZ-PRS and for BD-PRS, 9 and 12 were significant, respectively. Figure 1 depicts the mean standardized PRS, density plots, and 95% confidence intervals (CIs) with comparison arrows between groups. Notably, the controls differed from the SZ and SZA groups in SZ-PRS and from the BD I, BD II, SZA, and at-risk groups in BD-PRS. Very similar results were observed for the lower schizotypy group. Additionally, the high schizotypy group differed from most clinical groups and the at-risk group, while the latter was not distinguishable from any clinical diagnoses. Tables 2 and S2 (Supplementary Information) provide detailed statistics for this analysis.
Fig. 1. Mean polygenic risk score (PRS) for schizophrenia (SZ-PRS) and bipolar disorder (BD-PRS) in the eight investigated groups.
A, B Each boxplot shows the median (vertical lines) and mean (gray dots) PRS values; whiskers represent the minimum and maximum values; the vertical blue dashed lines separate the groups with subclinical versus clinical symptomatology; brackets with symbols indicate significantly different pairwise Bonferroni-corrected comparisons at p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) thresholds as well as comparisons trending toward significance (#, adjusted p value range, 0.050–0.064). The results are adjusted for age, sex, and the first five ancestry principal components. Higher scores indicate a higher polygenic burden. C, D The vertical dotted lines within the colored distributions represent the mean PRS for each group. E, F The blue bars represent confidence intervals for each group’s least squares mean. If the red arrows for two groups overlap, the difference is not significant after a Bonferroni correction. Group abbreviations: BD I bipolar I disorder, BD II bipolar II disorder, CTRL control, RISK at-risk state for psychosis, SCHIZ H higher schizotypy, SCHIZ L lower schizotypy, SZA schizoaffective disorder, SZ schizophrenia.
Table 2.
Main results of the analysis of variance using polygenic risk score (PRS) for schizophrenia (SZ-PRS) and bipolar disorder (BD-PRS) across the eight studied groups (Table S2 provides the full list of pairwise tests).
| Measure | ANOVA | Means (z-score) | Significant post hoc tests | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sum of squares | df | Mean square | F | p | Group | Mean | SD | Pairwise contrast | Beta | SE | Adjusted p | |
| SZ-PRS | 61.01 | 7 | 8.716 | 9.300 | 2.52 × 10−11 | CTRL | −0.214 | 0.988 | CTRL–SZA | −0.537 | 0.121 | 2.53 × 10−4 |
| SCHIZ L | −0.222 | 0.947 | CTRL–SZ | −0.474 | 0.076 | 1.65 × 10−8 | ||||||
| SCHIZ H | −0.323 | 1.018 | SCHIZ L–SZA | −0.534 | 0.146 | 0.007 | ||||||
| RISK | 0.055 | 0.88 | SCHIZ L–SZ | −0.471 | 0.109 | 4.50 × 10−4 | ||||||
| BD I | 0.036 | 0.986 | SCHIZ H–BD I | −0.388 | 0.112 | 0.015 | ||||||
| BD II | −0.07 | 0.977 | SCHIZ H–SZA | −0.683 | 0.146 | 9.18 × 10−5 | ||||||
| SZA | 0.253 | 0.958 | SCHIZ H–SZ | −0.621 | 0.110 | 5.73 × 10−7 | ||||||
| SZ | 0.243 | 1.012 | BD I–SZ | −0.233 | 0.072 | 0.034 | ||||||
| BD II–SZ | −0.362 | 0.112 | 0.034 | |||||||||
| CTRL–BD I | −0.242 | 0.078 | 0.054# | |||||||||
| SCHIZ H–RISK | −0.394 | 0.129 | 0.064# | |||||||||
| Residuals | 1466.67 | 1565 | 0.937 | |||||||||
| BD-PRS | 76.51 | 7 | 10.930 | 11.709 | 1.37 × 10−14 | CTRL | −0.268 | 0.943 | CTRL–RISK | −0.412 | 0.113 | 0.007 |
| SCHIZ L | −0.222 | 0.993 | CTRL–BD I | −0.558 | 0.078 | 2.86 × 10−11 | ||||||
| SCHIZ H | −0.181 | 0.905 | CTRL–BD II | −0.461 | 0.115 | 0.002 | ||||||
| RISK | 0.230 | 0.905 | CTRL–SZA | −0.572 | 0.12 | 5.99 × 10−5 | ||||||
| BD I | 0.276 | 0.984 | SCHIZ L–RISK | −0.404 | 0.128 | 0.045 | ||||||
| BD II | 0.206 | 0.96 | SCHIZ L–BD I | −0.550 | 0.111 | 2.11 × 10−5 | ||||||
| SZA | 0.213 | 0.971 | SCHIZ L–BD II | −0.453 | 0.140 | 0.033 | ||||||
| SZ | −0.095 | 1.041 | SCHIZ L–SZA | −0.565 | 0.145 | 0.003 | ||||||
| SCHIZ H–BD I | −0.547 | 0.112 | 3.10 × 10−5 | |||||||||
| SCHIZ H–BD II | −0.450 | 0.140 | 0.038 | |||||||||
| SCHIZ H–SZA | −0.561 | 0.146 | 0.004 | |||||||||
| BD I–SZ | 0.351 | 0.072 | 3.12 × 10−5 | |||||||||
| SZA–SZ | 0.365 | 0.117 | 0.050# | |||||||||
| SCHIZ H–RISK | −0.401 | 0.129 | 0.053# | |||||||||
| Residuals | 1460.92 | 1565 | 0.933 | |||||||||
Note: The analyses accounted for age, sex, and the first five genetic ancestry principal components, and statistical significance is indicated by Bonferroni-adjusted p values.
Adjusted p Bonferroni adjusted p value, beta beta coefficient, df degrees of freedom, SE standard error, BD I bipolar I disorder, BD II bipolar II disorder, CTRL controls, RISK at-risk state for psychosis, SCHIZ L lower schizotypy, SCHIZ H higher schizotypy, SZA schizoaffective disorder, SZ schizophrenia.
#Trend toward statistical significance.
Case–control status
SZ-PRS was significantly associated with the case–control status in the binary regression for the SZ (odds ratio (OR) = 1.62, 95% CI = 1.37–1.93, p = 6.88 × 10−9), SZA (OR = 1.82, 95% CI = 1.38–2.45, p = 2.04 × 10−5), BD I (OR = 1.27, 95% CI = 1.07–1.51, p = 0.006), and RISK (OR = 1.57, 95% CI = 1.14–2.20, p = 0.006) groups with the controls (change in Nagelkerke’s pseudo-R2, 1.3–6.1%). BD-PRS was also significantly associated with the case–control status for contrasts involving the BD I (OR = 1.80, 95% CI = 1.51–2.16, p = 2.53 × 10−11), BD II (OR = 1.65, 95% CI = 1.26–2.18, p = 1.98 × 10−4), SZA (OR = 1.96, 95% CI = 1.46–2.67, p = 3.41 × 10−6), SZ (OR = 1.26, 95% CI = 1.10–1.46, p = 0.011), and RISK (OR = 2.19, 95% CI = 1.53–3.20, p = 8.21 × 10−6) groups and the control group, resulting in a slightly higher Nagelkerke’s pseudo-R2 change range, 4.0–7.7% (Fig. 2 and Table S3 [Supplementary Information]).
Fig. 2. Block-wise binary logistic regressions using polygenic risk score (PRS) in the case–control comparisons.
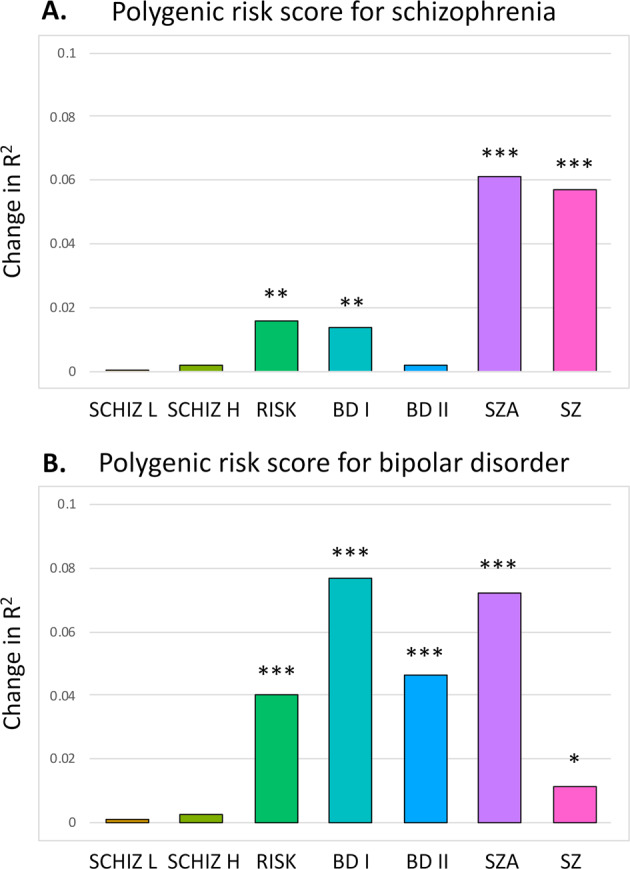
A Polygenic risk score for schizophrenia. B Polygenic risk score for bipolar disorder. Asterisks indicate statistically significant effects of the full (covariates only) versus baseline (covariates with PRS) models: *p < 0.05; **p < 0.01; ***p < 0.001. Group abbreviations: BD I bipolar I disorder, BD II bipolar II disorder, CTRL controls, RISK at-risk state for psychosis, SCHIZ H higher schizotypy, SCHIZ L lower schizotypy, SZA schizoaffective disorder, SZ schizophrenia.
PRSs and symptoms
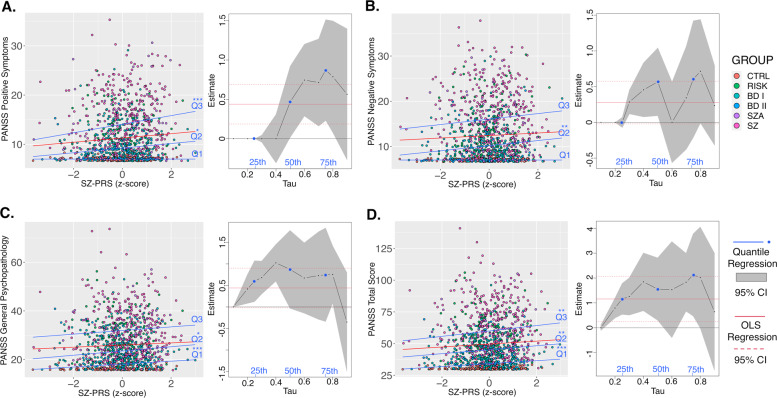
SZ-PRS was found to be a significant predictor of PANSS Total Score at the 25th (t = 3.86, p = 1.18 × 10−4), 50th (t = 3.19, p = 0.001), and 75th (t = 2.64, p = 0.008) percentiles. Significant effects were also found for Positive Symptoms at the 50th (t = 4.86, p = 1.35 × 10−6) and 75th (t = 2.50, p = 0.013) percentiles and for General Psychopathology at the 25th (t = 3.41, p = 6.70 × 10−4) and 50th (t = 2.27, p = 0.023) percentiles, while Negative Symptoms were significant only at the 50th percentile (t = 2.64, p = 0.008). In contrast, no significant effects were observed for BD-PRS across any of the PANSS measures. Figure 3 and Table S4 (Supplementary Information) report further details for this analysis.
Fig. 3. Quantile regression analysis of schizophrenia polygenic risk score (SZ-PRS) predicting symptoms.
Three quantiles of symptom score distributions (Q1 = 0.25, Q2 = 0.50, Q3 = 0.75) included three dimensions (A–C) and the total score (D) of the Positive and Negative Syndrome Scale (PANSS). The quantile regression estimates are visualized at 0.1 increments as black lines and with corresponding confidence intervals (CIs) shown as the gray shaded areas. The Q1, Q2, and Q3 quantiles are depicted in blue. For comparison, the ordinary least squares (OLS) regression estimates are shown as red solid lines with their CIs as dashed red lines. Many quantile estimates fell within the CI of the OLS regression slope, indicating overlap (though OLS does not adequately address the floor effect). *p < 0.05; **p < 0.01; ***p < 0.001. There were no significant results for BD-PRS. Group abbreviations: BD I bipolar I disorder, BD II bipolar II disorder, CTRL controls, RISK at-risk state for psychosis, SCHIZ L lower schizotypy, SCHIZ H higher schizotypy, SZA schizoaffective disorder, SZ schizophrenia.
Discussion
This work investigated the polygenic burden in four healthy/subclinical groups and four clinical groups. It aimed to quantify the associations and between-group differences of SZ-PRS and BD-PRS and their possible relationship with symptomatic presentation. Both PRSs were differentially useful in elucidating the genome-wide architecture across the extended psychosis spectrum.
Overall, the gradient of genetic risk proximity observed across groups is commensurate with the continuum model [35]. SZ-PRS and BD-PRS performed most robustly for the extremes of the psychosis spectrum, i.e., in distinguishing SZ and BD cases from controls, respectively, corroborating previous findings [11, 12]. Besides, given the significant effects for both PRSs, the existence of shared risk loci for SZA may be inferred from our results relative to this phenotype. Indeed, SZA seems genetically intermediate to BD I and SZ, with evidence from twin studies identifying the same genetic risk factors across SZA, SZ, and manic episodes [60]. Furthermore, similar results were observed for both controls and the low schizotypy group, while the at-risk group seems best positioned close to the clinical spectrum. Moreover, significant associations were seen for both BD type-I (characterized by manic episodes often alternating with depressive episodes) and BD type-II (characterized by hypomania and depression) using BD-PRS, but only for BD type-I using SZ-PRS. While we analyzed roughly four times more BD I than BD II cases, this effect is probably best explained by the putative partial genetic independence within these two BD subtypes [61]. Our findings align with prior reports confirming the ability of PRSs to accurately differentiate subtypes along diagnostic spectra, such as between SZ and other psychotic disorders [11], BD with and without psychotic features [62], and BD with manic and depressive psychosis [63].
Notably, BD-PRS values were higher, conveying a higher polygenic burden, in the at-risk group compared to controls, but not different from those of clinical groups. BD-PRS also explained a significant portion of variance in the binary logistic regression for at-risk status, an effect observed at a somewhat weaker level for SZ-PRS. These findings highlight the inherent genetic vulnerability of this subclinical group. To the best of our knowledge, there has been only one published report of a PRS analysis of a sizeable population of individuals at high risk for psychosis. Perkins et al. [64] recently demonstrated that SZ-PRSs were significantly elevated in individuals transitioning to a psychotic disorder diagnosis within a 2-year interval, compared to unaffected controls, but not in those not transitioning to a clinical diagnosis. Interestingly, our results were significant even without differentiating individuals according to their conversion status. Another key difference is the detection of effects using BD-PRS in our work. In particular, beyond the well-established high clinical risk criteria [29], we also assessed subtle subclinical hypomanic symptoms. A large majority of at-risk individuals (72%) met a cut-off value of ≥14 in the HCL-32 as a secondary measure and tentative risk criterion for BD [38]. This aligns with the common difficulties in unambiguously distinguishing SZ from BD, especially in early disorder onset [65], often continuing as diagnostic instability [66]. Given the large residual variance beyond the currently captured polygenic burden, gene × environment interactions are biologically plausible mechanisms as course specifiers for symptom development [67]. Such an interaction between BD-PRS and childhood trauma predisposing individuals to more severe disorders has indeed been shown recently [68]. As genetic risk is not modifiable, future research should thus also concentrate on psychosocial prevention of mental illness [69].
Our results inform the discussion of psychosis risk enrichment in at-risk individuals [70]. A meta-analysis has estimated a 36% transition rate within 3 years from clinical presentation in such individuals [71]. In the sample included in this study, which was a prospective investigation [38], over the same time window, the transition rate into F20 (schizophrenia), F23 (brief psychotic disorder), and BD diagnoses reached 15%. Currently, risk identification and predictors of exacerbation are primarily based on symptoms, thus lacking proper scalability [72]. In contrast to family history as a proxy for genetic risk, PRS offers a scalable quantitative metric for potential use in clinical contexts conditional on empirical validation [73]. Notably, the severity of psychopathology or clinical deterioration in the psychosis spectrum is often linked to neurocognitive deficits [74] and both SZ-PRS and BD-PRS have repeatedly been linked to cognition [75, 76]. In a differential analysis, most SZ risk alleles were associated with poorer cognition, while most BD risk alleles were linked to better cognition [77]. Accordingly, moderate neurocognitive impairments typically observed in the at-risk state [78], compared to severe impairments in SZ, align with our finding that genetic liability in this group was best characterized by risk variants covered by BD-GWAS. Downstream in the pathway towards symptom development, neurobiological mechanisms influenced by genetic variation [79] might be even more directly associated with emerging cognitive-perceptual disturbances [80]. These brain-based mechanisms may involve altered salience processing, neural coding of prediction error, and precision signaling [81]. Indeed, recent studies combining PRSs with brain imaging parameters have revealed new insights into the genetic determination of brain function and structure in psychosis spectrum [82, 83].
This work also reveals a possible irregularity within the continuum model at the genomic level, suggesting that schizotypy may share less genetic architecture with severe psychotic disorders than formerly thought. This tentative interpretation can be inferred from the lack of significant differences between the high and low schizotypy groups and between either schizotypy group or controls. Notably, in absolute terms, the high schizotypy group had the most negative mean SZ-PRS among all groups and marginally lower SZ-PRS and BD-PRS compared to the at-risk group (trend level). In the binary logistic regressions, neither high nor low schizotypy was significantly associated with either of the two PRSs. It should be emphasized that this sample in our study was composed of healthy participants by excluding individuals with clinical diagnoses. Drawing participants without restriction from a general population could lead to different results. Previously, findings indicating high overlaps between schizotypy and SZ have been made for single candidate genes [84, 85]. Our observation aligns with a recent study by Nenadić et al. reporting no association between SZ-PRS, BD-PRS, or MDD-PRS and symptom SPQ scores in non-clinical subjects [28]. Similarly, another PRS study identified a counter-intuitive negative relationship between SZ-PRS and schizotypal dimensions [86]. These studies challenge the key role of genetic predisposition and emphasize environmental stress as a moderator of schizotypy-related phenotypic expression [86], further informing the discussion on competition versus synergism of genetic–environmental influences [87]. Thus, schizotypy may be better positioned as a qualitatively distinct taxon-like cluster rather than along a unidimensional liability scale [88]. Furthermore, the possibility of “benign” schizotypy [89] supported by some brain-based endophenotype findings [90, 91] suggests that some protective mechanisms may impede a psychotic decomposition of schizotypal traits. While this claim merits further investigation, our results encourage a local reappraisal of the genetic continuum model for schizotypy.
Our findings also contribute to the ongoing discussion of the symptom- and function-level correlates of PRS in support of the continuum model. To the best of our knowledge, this study is the largest to investigate relations between PRS and the gold standard PANSS and is also unique in its transdiagnostic approach. We observed significant associations with SZ-PRS across all symptom measures. However, we were unable to detect any effect whatsoever using BD-PRS, suggesting its lack of specificity in this application and/or its limited power. Previous studies varied in their capacity to find associations with continuous symptom measures. These earlier results included no significant effects [32], links with negative symptoms [92], general psychopathology [30], and a mixture of positive, negative, and cognitive trait dimensions [93]. It has been suggested that a higher SZ-PRS typically predisposes individuals to a general vulnerability [94]. Our results from 1134 data points extend this conclusion based on the observed effect for overall symptomology (PANSS total score) across all three quantiles examined. Still, the strongest effect was identified at the high quantile for specific and more severe positive symptoms, a hallmark of psychosis. Overall, the distribution-dependent effects, especially as linked to different symptom facets and their interactions with environmental factors, may be a source of interesting findings in the field [68, 95].
This study has two particularly promising clinical applications of PRSs in the context of psychotic disorders and their antecedents: as potential support or a stratification tool in early diagnostic decision-making; in informing pre-emptive or full treatment strategies. Substantial clinical and research-based evidence demonstrates the utility of early interventions in reducing the highly debilitating character of psychotic disorders [96]. Indeed, long periods of untreated illness are among the most negative predictors of outcome, progression into chronic forms, and total time spent disabled [97]. Variation in help-seeking behavior may also delay presentation to healthcare specialists [98]. This further motivates the search for objective methods of early detection, which may be as easy as genotyping a blood sample and performing an analysis with a sufficiently large training dataset. For example, integrating PRS with other risk-conveying variables has been shown to improve the prediction of psychosis [64]. Proposals for refining this approach have been incorporated into current pilot projects aiming to create poly-risk scores that consider idiosyncratic combinations of risk and protective factors [99]. PRS-based approaches might also guide drug targeting [100] or explain resistance to pharmacotherapy [15]. For example, PRSs have been shown to be associated with responses to antipsychotic medication [14] and lithium [101]. Such PRS-informed bio-typing may identify patients who will benefit more from one pharmacological treatment among alternatives.
We acknowledge some limitations of this work. The numbers of participants varied substantially among groups. Inclusion of more polarized schizotypy groups (i.e., more extreme phenotypes) might also have been better suited to the goals of this study. The cross-sectional nature of these findings implies the possibility of a progression across the phenotypic spectrum, development of comorbidities, or dampening of genetic liability later in life. Indeed, the at-risk group had the lowest mean age. We also did not consider the role of medication. Finally, as GWASs exclude rare variants, we only investigated the effects of common variants with population frequencies of 1% or more. There are also more general limitations to polygenic risk profiling discussed in the current scientific debate on PRS, which has included articles covering its potential clinical utility [10] and bioethics [102], with some expressly critical tones [103]. Notably, provided the ancestral differences in linkage disequilibrium and polymorphisms, the predictive accuracy of PRS is currently constrained to individuals of European descent, owing to the lack of statistically powerful GWASs of psychiatric disorders and traits from ethnically diverse populations [104].
While PRS is certainly not immediately ready for routine clinical application, this study highlights the value of PRS for SZ and BD in elucidating the genetic architecture of psychosis and identifying vulnerable individuals who may benefit from early attention and monitoring. Future research should focus on possible differential developmental trajectories and factors precipitating or mitigating pre-existing genetic liability.
Supplementary information
Acknowledgements
We greatly thank the study participants, interviewers, and the technical and administrative personnel, without whom this work would not have been possible.
Author contributions
LS and SP performed data analyses. EG, TGS and PF jointly provided supervision. EG, TGS, PF, WR, SW, LS and SP contributed to the study design and analysis concept. PH and SH provided technical support for genotyping. TFMA supported data analysis and interpretation. Responsible for planning, site coordination, recruitment or data collection (in alphabetical order): KA, HA-S, I-GA, VA, BB, MB, RB, TB, ALC, DED, FD, UD, BE, AJF, AJF, CF, LF-N, AG, KG, MG, ISH, KH, MH, UH, GJ, MJ, CK, JLK, FK-S, FUL, MM, VN, ER, JR, MR, DR-E, CS, ECS, FS, MS, SKS, AT, MVH, DW, JW, MEW, SHW, JZ. Acquired funding: TGS, MR, MN, WR and SW. LS drafted the first manuscript version and all authors contributed to the critical revisions and approved the final manuscript.
Funding
This work was financially supported by the Zurich Program for Sustainable Development of Mental Health Services (ZInEP), the Donald C. Cooper-Fonds, and the German Research Foundation (DFG) (Project: Genotype-Phenotype Relationships and Neurobiology of the Longitudinal Course of Psychosis and Pathomechanisms and Signatures in the Longitudinal Course of Psychosis; SCHU 1603/4-1, 5-1, 7-1; FA241/16-1). The genotyping for the PsyCourse Study was in part funded by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Program with grants awarded to TGS (01ZX1614K), MR (01ZX1614G), and MMN (01ZX1614A). TGS received additional support from the German Federal Ministry of Education and Research (BMBF) within the framework of the BipoLife network and the Dr. Lisa Oehler Foundation, Kassel (Germany). MR received support from the German Federal Ministry of Education and Research (BMBF) within the framework of the ASD-Net (01EE1409C), AERIAL (01EE1406C), EMBED (01EW1904), Synschiz (01EW1810), TargetOxy (031L0190A), and SysmedSUD (01ZX01909A) and the German Research Foundation (DFG, grant FOR2107, RI 908/11-2, and TRR 265/1). UD was funded by the German Research Foundation (DFG, grant FOR2107 DA1151/5‐1; SFB‐TRR58, Project C09) and the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant Dan3/012/17). FD received support from the BONFOR Programme of the University of Bonn, Germany. AJF has received a grant from the Else Kröner-Fresenius-Stiftung (2019_A127). UH was supported by European Union’s Horizon 2020 Research and Innovation Programme (PSY-PGx, grant agreement No 945151). ECS is supported by the Munich Clinician Scientist Program (MCSP). SW’s research has been supported by the Swiss National Science Foundation (SNSF), several EU FP7s, HSM High Specialized Medicine of the Canton Zurich, Switzerland, Bfarm Germany, the Hartmann Müller Foundation, the Olga Mayenfisch Foundation, and the Gertrud Thalmann Foundation. PD MEW was supported by the Clinician Scientist Programme of the Medical Faculty of Ulm University. JW is supported by an Ilídio Pinho professorship and iBiMED (UID/BIM/04501/2013), at the University of Aveiro. SHW was funded by the German Research Foundation (DFG, grant FOR2107, WI 3439/3-2 and grant GRK 2350). Study design, data collection, analysis, interpretation, and preparation of the manuscript were conducted independently from the funding sources.
Data availability
PsyCourse data are available according to a mutually agreed memorandum of understanding. Other data and analysis code are available upon reasonable request.
Competing interests
VA has received personal fees from Janssen Pharmaceuticals, AstraZeneca, Lundbeck, Sanofi, and Servier Laboratories. PF has received research support and honorarium for lectures or advisory activity from Abbott Laboratories, Boehringer Ingelheim, Janssen Pharmaceuticals, Essex Pharma, Lundbeck, Otsuka Pharmaceutical, Recordati, Richter Pharma, Servier Laboratories, and Takeda. CK has received personal fees from Aristo Pharmaceuticals, Janssen-Cilag, Eli Lilly and Company, MagVenture, Trommsdorff GmbH, Lundbeck, Neuraxpharm, and Servier Laboratories. MMN has received personal fees from the Lundbeck Foundation, the Robert-Bosch-Stiftung, HMG Systems Engineering GmbH, and Shire Deutschland GmbH. He receives salary payments from Life & Brain GmbH and holds shares in Life & Brain GmbH. JR has received personal fees from Lundbeck and Otsuka Pharmaceutical. MS has received personal fees for being a member of the speaker’s bureau and advisory board from Aristo Pharmaceuticals, Janssen-Cilag, Lundbeck, Neuraxpharm, and Recordati. SW has received royalties from Thieme Hogrefe, Kohlhammer, Springer, and Beltz and lecture honoraria from Opopharma in the past 5 years. JW has received grants from the German Research Foundation and personal fees from Abbott Laboratories, Boehringer Ingelheim, Immungenetics, Eli Lilly and Company, MSD Sharp & Dohme, Roche, Actelion, Amgen, Janssen-Cilag, Pfizer, and Med Update and has patents PCT/EP 2011001724 and PCT/EP 2015052945 issued. The other authors declare no biomedical financial interests or potential conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lukasz Smigielski, Sergi Papiol, Peter Falkai, Thomas G. Schulze, Edna Grünblatt.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01720-0.
References
- 1.Strain E, Lofwall M, Jaffe J. Kaplan and Sadock’s comprehensive textbook of psychiatry. Philadelphia, PA: Lippincott Williams and Wilkins; 2009.
- 2.Pepper E, Cardno AG. Genetics of schizophrenia and other psychotic disorders. Curr Psychiatry Rev. 2014;10:133–42. [Google Scholar]
- 3.Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J, Duncan L, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–9. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muntané G, Farré X, Bosch E, Martorell L, Navarro A, Vilella E. The shared genetic architecture of schizophrenia, bipolar disorder and lifespan. Hum Genet. 2021;140:441–55. doi: 10.1007/s00439-020-02213-8. [DOI] [PubMed] [Google Scholar]
- 6.Singh J, Chen G, Canuso CM. Antipsychotics in the treatment of bipolar disorder. Handb Exp Pharmacol. 2012;187–212. [DOI] [PubMed]
- 7.Radua J, Ramella-Cravaro V, Ioannidis JPA, Reichenberg A, Phiphopthatsanee N, Amir T, et al. What causes psychosis? An umbrella review of risk and protective factors. World Psychiatry. 2018;17:49–66. doi: 10.1002/wps.20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwicker A, Denovan-Wright EM, Uher R. Gene–environment interplay in the etiology of psychosis. Psychol Med. 2018;48:1925–36. doi: 10.1017/S003329171700383X. [DOI] [PubMed] [Google Scholar]
- 9.Kavanagh DH, Tansey KE, O’Donovan MC, Owen MJ. Schizophrenia genetics: emerging themes for a complex disorder. Mol Psychiatry. 2015;20:72–6. doi: 10.1038/mp.2014.148. [DOI] [PubMed] [Google Scholar]
- 10.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–90. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 11.Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J, et al. An examination of polygenic score risk prediction in individuals with first-episode psychosis. Biol Psychiatry. 2017;81:470–7. doi: 10.1016/j.biopsych.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 12.Calafato MS, Thygesen JH, Ranlund S, Zartaloudi E, Cahn W, Crespo-Facorro B, et al. Use of schizophrenia and bipolar disorder polygenic risk scores to identify psychotic disorders. Br J Psychiatry. 2018;213:535–41. doi: 10.1192/bjp.2018.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonas KG, Lencz T, Li K, Malhotra AK, Perlman G, Fochtmann LJ, et al. Schizophrenia polygenic risk score and 20-year course of illness in psychotic disorders. Transl Psychiatry. 2019;9:300. doi: 10.1038/s41398-019-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J-P, Robinson D, Yu J, Gallego J, Fleischhacker WW, Kahn RS, et al. Schizophrenia polygenic risk score as a predictor of antipsychotic efficacy in first-episode psychosis. Am J Psychiatry. 2018;176:21–28. doi: 10.1176/appi.ajp.2018.17121363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank J, Lang M, Witt S, Strohmaier J, Rujescu D, Cichon S, et al. Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol Psychiatry. 2015;20:150–1. doi: 10.1038/mp.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodberry KA, Shapiro DI, Bryant C, Seidman LJ. Progress and future directions in research on the psychosis prodrome: a review for clinicians. Harv Rev Psychiatry. 2016;24:87. doi: 10.1097/HRP.0000000000000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narrow WE, Kuhl EA. Dimensional approaches to psychiatric diagnosis in DSM-5. J Mental Health Pol Econ. 2011;14:197–200. [PubMed] [Google Scholar]
- 18.Gaebel W, Stricker J, Kerst A. Changes from ICD-10 to ICD-11 and future directions in psychiatric classification. Dialog Clin Neurosci. 2020;22:7. doi: 10.31887/DCNS.2020.22.1/wgaebel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Os,J, Linscott RJ, Myin-Germeys I, Delespaul P, Krabbendam L. A systematic review and meta-analysis of the psychosis continuum: evidence for a psychosis proneness–persistence–impairment model of psychotic disorder. Psychol Med. 2009;39:179–95. doi: 10.1017/S0033291708003814. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Peng X, Pelletier-Baldelli A, Orlov N, Farabaugh A, Nasr S, et al. Altered temporal, but intact spatial, features of transient network dynamics in psychosis. Mol Psychiatry. 2021;26:2493–503. doi: 10.1038/s41380-020-00983-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kauppi K, Westlye LT, Tesli M, Bettella F, Brandt CL, Mattingsdal M, et al. Polygenic risk for schizophrenia associated with working memory-related prefrontal brain activation in patients with schizophrenia and healthy controls. Schizophr Bull. 2015;41:736–43. doi: 10.1093/schbul/sbu152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riglin L, Collishaw S, Richards A, Thapar AK, Maughan B, O’Donovan MC, et al. Schizophrenia risk alleles and neurodevelopmental outcomes in childhood: a population-based cohort study. Lancet Psychiatry. 2017;4:57–62. doi: 10.1016/S2215-0366(16)30406-0. [DOI] [PubMed] [Google Scholar]
- 23.Coombes BJ, Markota M, Mann JJ, Colby C, Stahl E, Talati A, et al. Dissecting clinical heterogeneity of bipolar disorder using multiple polygenic risk scores. Transl Psychiatry. 2020;10:1–8. doi: 10.1038/s41398-020-00996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musliner KL, Krebs MD, Albiñana C, Vilhjalmsson B, Agerbo E, Zandi PP, et al. Polygenic risk and progression to bipolar or psychotic disorders among individuals diagnosed with unipolar depression in early life. Am J Psychiatry. 2020;177:936–43. doi: 10.1176/appi.ajp.2020.19111195. [DOI] [PubMed] [Google Scholar]
- 25.Schultze-Lutter F, Michel C, Schmidt SJ, Schimmelmann BG, Maric NP, Salokangas R, et al. EPA guidance on the early detection of clinical high risk states of psychoses. Eur Psychiatry. 2015;30:405–16. doi: 10.1016/j.eurpsy.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Debbané M, Eliez S, Badoud D, Conus P, Flückiger R, Schultze-Lutter F. Developing psychosis and its risk states through the lens of schizotypy. Schizophr Bull. 2015;41:S396–407. doi: 10.1093/schbul/sbu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrantes-Vidal N, Grant P, Kwapil TR. The role of schizotypy in the study of the etiology of schizophrenia spectrum disorders. Schizophr Bull. 2015;41:S408–16. doi: 10.1093/schbul/sbu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nenadić I, Meller T, Schmitt S, Stein F, Brosch K, Mosebach J, et al. Polygenic risk for schizophrenia and schizotypal traits in non-clinical subjects. Psychol Med. 2020. 1–10. [Epub ahead of print]. [DOI] [PubMed]
- 29.Fusar-Poli P, Borgwardt S, Bechdolf A, Addington J, Riecher-Rössler A, Schultze-Lutter F, et al. The psychosis high-risk state: a comprehensive state-of-the-art review. JAMA Psychiatry. 2013;70:107–20. doi: 10.1001/jamapsychiatry.2013.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta SM, MacDonald K, Fathalli F, Yim A, Lepage M, Iyer S, et al. Polygenic Risk Score associated with specific symptom dimensions in first-episode psychosis. Schizophr Res. 2017;184:116–21. doi: 10.1016/j.schres.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 31.Santoro ML, Ota V, de Jong S, Noto C, Spindola LM, Talarico F, et al. Polygenic risk score analyses of symptoms and treatment response in an antipsychotic-naive first episode of psychosis cohort. Transl psychiatry. 2018;8:1–8. doi: 10.1038/s41398-018-0230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sieradzka D, Power RA, Freeman D, Cardno AG, McGuire P, Plomin R, et al. Are genetic risk factors for psychosis also associated with dimension-specific psychotic experiences in adolescence? PLoS ONE. 2014;9:e94398. doi: 10.1371/journal.pone.0094398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derks EM, Vorstman JA, Ripke S, Kahn RS, Schizophrenia Psychiatric Genomic Consortium, Ophoff RA. Investigation of the genetic association between quantitative measures of psychosis and schizophrenia: a polygenic risk score analysis. PLoS ONE. 2012;7:e37852. [DOI] [PMC free article] [PubMed]
- 34.Ni G, Zeng J, Revez JA, Wang Y, Zheng Z, Ge T, et al. A comparison of ten polygenic score methods for psychiatric disorders applied across multiple cohorts. Biol Psychiatry. 2021;90:611–20. doi: 10.1016/j.biopsych.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tesli M, Espeseth T, Bettella F, Mattingsdal M, Aas M, Melle I, et al. Polygenic risk score and the psychosis continuum model. Acta Psychiatr Scand. 2014;130:311–7. doi: 10.1111/acps.12307. [DOI] [PubMed] [Google Scholar]
- 36.Kay SR, Fiszbein A, Opler LA. The Positive and Negative Syndrome Scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–76. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 37.Koenker R, Chernozhukov V, He X, Peng L. Handbook of quantile regression. London: Chapman and Hall/CRC; 2017.
- 38.Theodoridou A, Heekeren K, Dvorsky D, Metzler S, Franscini M, Haker H, et al. Early recognition of high risk of bipolar disorder and psychosis: an overview of the ZInEP “early recognition” study. Front Public Health. 2014;2:166. doi: 10.3389/fpubh.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unterrassner L, Wyss TA, Wotruba D, Ajdacic-Gross V, Haker H, Rössler W. Psychotic-like experiences at the healthy end of the psychosis continuum. Front Psychol. 2017;8:775. doi: 10.3389/fpsyg.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budde M, Anderson-Schmidt H, Gade K, Reich-Erkelenz D, Adorjan K, Kalman JL, et al. A longitudinal approach to biological psychiatric research: the PsyCourse study. Am J Med Genet B Neuropsychiatr Genet. 2019;180:89–102. doi: 10.1002/ajmg.b.32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grünblatt E, Marinova Z, Roth A, Gardini E, Ball J, Geissler J, et al. Combining genetic and epigenetic parameters of the serotonin transporter gene in obsessive-compulsive disorder. J Psychiatr Res. 2018;96:209–17. doi: 10.1016/j.jpsychires.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Hauser TU, Iannaccone R, Dolan RJ, Ball J, Hattenschwiler J, Drechsler R, et al. Increased fronto-striatal reward prediction errors moderate decision making in obsessive-compulsive disorder. Psychol Med. 2017;47:1246–58. doi: 10.1017/S0033291716003305. [DOI] [PubMed] [Google Scholar]
- 43.Schultze-Lutter F, Addington J, Ruhrmann S, Klosterkötter J. Schizophrenia proneness instrument, adult version (SPI-A). Rome: Giovanni Fioriti; 2007.
- 44.Schultze-Lutter, F & Koch, E Schizophrenia Proneness Instrument: child and youth version (SPI-CY). Rome: Fioriti; 2010. [DOI] [PubMed]
- 45.McGlashan T, Miller T, Woods S, Rosen J, Hoffman R, Davidson L. Structured interview for prodromal syndromes. New Haven, CT: PRIME Research Clinic, Yale School of Medicine; 2001.
- 46.Angst J, Adolfsson R, Benazzi F, Gamma A, Hantouche E, Meyer TD, et al. The HCL-32: towards a self-assessment tool for hypomanic symptoms in outpatients. J Affect Disord. 2005;88:217–33. doi: 10.1016/j.jad.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 47.Raine A. The SPQ: a scale for the assessment of schizotypal personality based on DSM-III-R criteria. Schizophr Bull. 1991;17:555–64. doi: 10.1093/schbul/17.4.555. [DOI] [PubMed] [Google Scholar]
- 48.Oestreich LK, Mifsud NG, Ford JM, Roach BJ, Mathalon DH, Whitford TJ. Subnormal sensory attenuation to self-generated speech in schizotypy: electrophysiological evidence for a ‘continuum of psychosis’. Int J Psychophysiol. 2015;97:131–8. doi: 10.1016/j.ijpsycho.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Cruz AL, Ali OM, Asare G, Whyte MS, Walpola I, Segal J, et al. Embrained drives to perform extraordinary roles predict schizotypal traits in the general population. NPJ Schizophr. 2016;2:1–7. doi: 10.1038/npjschz.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59:22–33. [PubMed] [Google Scholar]
- 51.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 52.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Consortium GP. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S, Walters JT, O’Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv:10.1101/2020.09.12.20192922 [Preprint]. 2020.
- 55.Mullins N, Forstner AJ, O'Connell KS, Coombes B, Coleman JR, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29. doi: 10.1038/s41588-021-00857-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polson NG, Scott JG. Shrink globally, act locally: sparse Bayesian regularization and prediction. Bayesian Stat. 2010;9:105. [Google Scholar]
- 58.Smith TJ, McKenna CM. A comparison of logistic regression pseudo R2 indices. Mult Linear Regres Viewp. 2013;39:17–26. [Google Scholar]
- 59.Le Cook B, Manning WG. Thinking beyond the mean: a practical guide for using quantile regression methods for health services research. Shanghai Arch Psychiatry. 2013;25:55. doi: 10.3969/j.issn.1002-0829.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cardno AG, Owen MJ. Genetic relationships between schizophrenia, bipolar disorder, and schizoaffective disorder. Schizophr Bull. 2014;40:504–15. doi: 10.1093/schbul/sbu016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Charney A, Ruderfer D, Stahl E, Moran J, Chambert K, Belliveau R, et al. Evidence for genetic heterogeneity between clinical subtypes of bipolar disorder. Transl Psychiatry. 2017;7:e993. doi: 10.1038/tp.2016.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aminoff SR, Tesli M, Bettella F, Aas M, Lagerberg TV, Djurovic S, et al. Polygenic risk scores in bipolar disorder subgroups. J Affect Disord. 2015;183:310–4. doi: 10.1016/j.jad.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 63.Markota M, Coombes BJ, Larrabee BR, McElroy SL, Bond DJ, Veldic M, et al. Association of schizophrenia polygenic risk score with manic and depressive psychosis in bipolar disorder. Transl Psychiatry. 2018;8:1–7. doi: 10.1038/s41398-018-0242-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perkins DO, Olde Loohuis L, Barbee J, Ford J, Jeffries CD, Addington J, et al. Polygenic risk score contribution to psychosis prediction in a target population of persons at clinical high risk. Am J Psychiatry. 2020;177:155–63. doi: 10.1176/appi.ajp.2019.18060721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angst J. Historical aspects of the dichotomy between manic–depressive disorders and schizophrenia. Schizophr Res. 2002;57:5–13. doi: 10.1016/s0920-9964(02)00328-6. [DOI] [PubMed] [Google Scholar]
- 66.Fusar-Poli P, Cappucciati M, Rutigliano G, Heslin M, Stahl D, Brittenden Z, et al. Diagnostic stability of ICD/DSM first episode psychosis diagnoses: meta-analysis. Schizophr Bull. 2016;42:1395–406. doi: 10.1093/schbul/sbw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geoffroy PA, Etain B, Houenou J. Gene x environment interactions in schizophrenia and bipolar disorder: evidence from neuroimaging. Front Psychiatry. 2013;4:136. doi: 10.3389/fpsyt.2013.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aas M, Bellivier F, Bettella F, Henry C, Gard S, Kahn JP, et al. Childhood maltreatment and polygenic risk in bipolar disorders. Bipolar Disord. 2020;22:174–81. doi: 10.1111/bdi.12851. [DOI] [PubMed] [Google Scholar]
- 69.Collins PY, Patel V, Joestl SS, March D, Insel TR, Daar AS, et al. Grand challenges in global mental health. Nature. 2011;475:27. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fusar-Poli P, de Pablo GS, Correll CU, Meyer-Lindenberg A, Millan MJ, Borgwardt S, et al. Prevention of psychosis: advances in detection, prognosis, and intervention. JAMA Psychiatry. 2020;77:755–65. doi: 10.1001/jamapsychiatry.2019.4779. [DOI] [PubMed] [Google Scholar]
- 71.Fusar-Poli P, Bonoldi I, Yung AR, Borgwardt S, Kempton MJ, Valmaggia L, et al. Predicting psychosis: meta-analysis of transition outcomes in individuals at high clinical risk. Arch Gen Psychiatry. 2012;69:220–9. doi: 10.1001/archgenpsychiatry.2011.1472. [DOI] [PubMed] [Google Scholar]
- 72.Oliver D, Reilly TJ, Baccaredda Boy O, Petros N, Davies C, Borgwardt S, et al. What causes the onset of psychosis in individuals at clinical high risk? A meta-analysis of risk and protective factors. Schizophr Bull. 2020;46:110–20. doi: 10.1093/schbul/sbz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikeda M, Saito T, Kanazawa T, Iwata N. Polygenic risk score as clinical utility in psychiatry: a clinical viewpoint. J Hum Genet. 2021;66:53–60. doi: 10.1038/s10038-020-0814-y. [DOI] [PubMed] [Google Scholar]
- 74.Seidman LJ, Shapiro DI, Stone WS, Woodberry KA, Ronzio A, Cornblatt BA, et al. Association of neurocognition with transition to psychosis: baseline functioning in the second phase of the North American Prodrome Longitudinal Study. JAMA Psychiatry. 2016;73:1239–48. doi: 10.1001/jamapsychiatry.2016.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shafee R, Nanda P, Padmanabhan JL, Tandon N, Alliey-Rodriguez N, Kalapurakkel S, et al. Polygenic risk for schizophrenia and measured domains of cognition in individuals with psychosis and controls. Transl Psychiatry. 2018;8:1–9. doi: 10.1038/s41398-018-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richards AL, Pardiñas AF, Frizzati A, Tansey KE, Lynham AJ, Holmans P, et al. The relationship between polygenic risk scores and cognition in schizophrenia. Schizophr Bull. 2020;46:336–44. doi: 10.1093/schbul/sbz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smeland OB, Bahrami S, Frei O, Shadrin A, O’Connell K, Savage J, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2019;25:844–53. doi: 10.1038/s41380-018-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodberry KA, Seidman LJ, Giuliano AJ, Verdi MB, Cook WL, McFarlane WR. Neuropsychological profiles in individuals at clinical high risk for psychosis: relationship to psychosis and intelligence. Schizophr Res. 2010;123:188–98. doi: 10.1016/j.schres.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH. The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci. 2016;19:1397. doi: 10.1038/nn.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wotruba D, Michels L, Buechler R, Metzler S, Theodoridou A, Gerstenberg M, et al. Aberrant coupling within and across the default mode, task-positive, and salience network in subjects at risk for psychosis. Schizophr Bull. 2014;40:1095–104. doi: 10.1093/schbul/sbt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Howes OD, Hird EJ, Adams RA, Corlett PR, McGuire P. Aberrant salience, information processing and dopaminergic signalling in people at clinical high risk for psychosis. Biol Psychiatry. 2020;88:304–14. doi: 10.1016/j.biopsych.2020.03.012. [DOI] [PubMed] [Google Scholar]
- 82.Alnæs D, Kaufmann T, van der Meer D, Córdova-Palomera A, Rokicki J, Moberget T, et al. Brain heterogeneity in schizophrenia and its association with polygenic risk. JAMA Psychiatry. 2019;76:739–48. doi: 10.1001/jamapsychiatry.2019.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao H, Zhou H, Cannon TD. Functional connectome-wide associations of schizophrenia polygenic risk. Mol Psychiatry. 2020;26:2553–61. doi: 10.1038/s41380-020-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meller T, Schmitt S, Stein F, Brosch K, Mosebach J, Yueksel D, et al. Associations of schizophrenia risk genes ZNF804A and CACNA1C with schizotypy and modulation of attention in healthy subjects. Schizophr Res. 2019;208:67–75. doi: 10.1016/j.schres.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 85.Stefanis NC, Trikalinos TA, Avramopoulos D, Smyrnis N, Evdokimidis I, Ntzani EE, et al. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62:784–92. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 86.Hatzimanolis A, Avramopoulos D, Arking DE, Moes A, Bhatnagar P, Lencz T, et al. Stress-dependent association between polygenic risk for schizophrenia and schizotypal traits in young army recruits. Schizophr Bull. 2018;44:338–47. doi: 10.1093/schbul/sbx074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wigman JT, van Winkel R, Ormel J, Verhulst FC, van Os J, Vollebergh WA. Early trauma and familial risk in the development of the extended psychosis phenotype in adolescence. Acta Psychiatr Scand. 2012;126:266–73. doi: 10.1111/j.1600-0447.2012.01857.x. [DOI] [PubMed] [Google Scholar]
- 88.Schultze-Lutter F, Nenadic I, Grant P. Psychosis and schizophrenia-spectrum personality disorders require early detection on different symptom dimensions. Front Psychiatry. 2019;10:476. doi: 10.3389/fpsyt.2019.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mohr C, Claridge G. Schizotypy—do not worry, it is not all worrisome. Schizophr Bull. 2015;41:S436–S443. doi: 10.1093/schbul/sbu185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kühn S, Schubert F, Gallinat J. Higher prefrontal cortical thickness in high schizotypal personality trait. J Psychiatr Res. 2012;46:960–5. doi: 10.1016/j.jpsychires.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 91.Smigielski L, Wotruba D, Treyer V, Rössler J, Papiol S, Falkai P, et al. The interplay between postsynaptic striatal D2/3 receptor availability, adversity exposure and odd beliefs: a [11C]-raclopride PET study. Schizophr Bull. 2021;47:1495–508. doi: 10.1093/schbul/sbab034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones HJ, Stergiakouli E, Tansey KE, Hubbard L, Heron J, Cannon M, et al. Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry. 2016;73:221–8. doi: 10.1001/jamapsychiatry.2015.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pain O, Dudbridge F, Cardno AG, Freeman D, Lu Y, Lundstrom S, et al. Genome‐wide analysis of adolescent psychotic‐like experiences shows genetic overlap with psychiatric disorders. Am J Med Genet Part B Neuropsychiatr Genet. 2018;177:416–25. doi: 10.1002/ajmg.b.32630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kendler KS. The schizophrenia polygenic risk score: to what does it predispose in adolescence? JAMA Psychiatry. 2016;73:193–4. doi: 10.1001/jamapsychiatry.2015.2964. [DOI] [PubMed] [Google Scholar]
- 95.Nelemans SA, Boks M, Lin B, Oldehinkel T, van Lier P, Branje S, et al. Polygenic risk for major depression interacts with parental criticism in predicting adolescent depressive symptom development. J Youth Adolescence. 2021;50:159–76. doi: 10.1007/s10964-020-01353-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaur T, Cadenhead KS. Treatment implications of the schizophrenia prodrome. Curr Top Behav Neurosci. 2010;4:97–121. doi: 10.1007/7854_2010_56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Penttilä M, Jääskeläinen E, Hirvonen N, Isohanni M, Miettunen J. Duration of untreated psychosis as predictor of long-term outcome in schizophrenia: systematic review and meta-analysis. Br J Psychiatry. 2014;205:88–94. doi: 10.1192/bjp.bp.113.127753. [DOI] [PubMed] [Google Scholar]
- 98.Murphy J, Shevlin M, Houston J, Adamson G. A population based analysis of subclinical psychosis and help-seeking behavior. Schizophr Bull. 2012;38:360–7. doi: 10.1093/schbul/sbq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oliver D, Uher R, Fusar-Poli P. Psychosis polyrisk score (PPS) for the detection of individuals at-risk and the prediction of their outcomes. Front Psychiatry. 2019;10:174. doi: 10.3389/fpsyt.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gibson G. On the utilization of polygenic risk scores for therapeutic targeting. PLoS Genet. 2019;15:e1008060.. doi: 10.1371/journal.pgen.1008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, Cearns M, et al. Association of polygenic score for major depression with response to lithium in patients with bipolar disorder. Mol Psychiatry. 2020;26:2457–70. doi: 10.1038/s41380-020-0689-5. [DOI] [PubMed] [Google Scholar]
- 102.Palk AC, Dalvie S, De Vries J, Martin AR, Stein DJ. Potential use of clinical polygenic risk scores in psychiatry–ethical implications and communicating high polygenic risk. Philos Ethics Humanit Med. 2019;14:4. doi: 10.1186/s13010-019-0073-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wald NJ, Old R. The illusion of polygenic disease risk prediction. Genet Med. 2019;21:1705–7. doi: 10.1038/s41436-018-0418-5. [DOI] [PubMed] [Google Scholar]
- 104.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PsyCourse data are available according to a mutually agreed memorandum of understanding. Other data and analysis code are available upon reasonable request.