Abstract
Objective.
We sought a comprehensive understanding of myeloid cell types driving fibrosis in diffuse cutaneous systemic sclerosis (dcSSc) skin.
Methods.
We analyzed the transcriptomes of 2,465 myeloid cells skin biopsies from 10 healthy controls (HCs) and 12 dcSSc patients by single-cell RNA-sequencing (scRNA-seq). Monocyte-derived dendritic cells were stained by immunohistochemistry and immunofluorescence targeting FCN1.
Results.
T-stochastic neighbor embedding analysis of single cell transcriptome data revealed 12 myeloid cell clusters, nine of which paralleled previously described HC Mφ/DC clusters and three of which were dcSSc-specific myeloid cell clusters. One SSc-associated macrophage cluster, highly expressing FCGR3A, on pseudotime analysis was suggested to derive from normal CCR1+ and MARCO+ macrophages. A second SSc-associated myeloid population, highly expressed monocyte markers: FCN1, EREG, S100A8 and S100A9, but was closely related to cDC2 on pseudotime analysis and identified as Mo-DC. Mo-DC were associated with more severe skin disease. Proliferating macrophages and plasmacytoid dendritic cells were detected almost exclusively in dcSSc skin, the latter clustering with B cells and apparently derived from lymphoid progenitors.
Conclusions.
Transcriptional signatures in these and other myeloid populations indicate innate immune activation, possibly through TLRs, and highly upregulated chemokines. However, the appearance and activation of myeloid cells is variable between patients, indicating differing underlying pathogenesis and/or temporal disease activity.
Introduction
Systemic sclerosis (SSc, also known as scleroderma), is a heterogeneous autoimmune disease with unknown etiology characterized by fibrosis, vasculopathy and immune dysfunction. Current treatment remains ineffective for many complications, resulting in high morbidity and mortality (1). Diffuse cutaneous systemic sclerosis (dcSSc) is a subtype of SSc characterized by widespread skin fibrosis. Although myeloid cells, including macrophages and dendritic cells (DCs), heterogeneous immune cells in human skin, have been implicated in initiating and perpetuating SSc by releasing or activating profibrotic and proinflammatory factors, the precise mechanisms that contribute to SSc pathogenesis remain poorly understood (2, 3).
Infiltration of macrophages in SSc skin was first described in the 1990s (4, 5). More recent work has shown increased expression of specific macrophage markers in dcSSc skin, including Siglec-1 (CD169), a type I interferon-inducible gene, and CD163 and CD204, markers for activated M2 macrophages (6, 7). Other work has implicated several chemokines in SSc pathogenesis: CCL2, CCL5, CCL18, CCL19, CXCL9 and CXCL13 (8), CCL19, in particular, a strong macrophage chemoattractant (8, 9). Further supporting an important role macrophages in SSc pathogenesis, expression of macrophage markers, CD14, MS4A4A, CD163 and CCL2, correlate with clinical disease severity as assessed by the modified Rodnan skin score (MRSS) and are prognostic biomarkers for progressive skin disease in dcSSc (10, 11). Macrophage and DC genes are also seen in an inflammatory subset of skin biopsies on bulk gene expression analysis (12, 13).
Recent studies indicate that DCs may also play important roles in the pathophysiology of SSc skin disease (3, 14). The role of myeloid or conventional DCs (cDC) in SSc skin is uncertain, as markers used in previously studies are not specific for these cells (3). On the other hand several studies have shown increased plasmacytoid dendritic cells (pDCs) in the skin of SSc patients (14-16), and circulating SSc pDCs have been described to secrete CXC motif ligand (CXCL)-4, as well as IFNα (14, 16).
Despite these multiple studies, altered myeloid cell numbers and functions in SSc remain poorly understood, in part due to the complexity of macrophage and DC subsets. Here, we compare recently detailed transcriptomes of macrophage and DC subsets in normal skin (17) to discern alterations in myeloid cell subsets in dcSSc skin.
MATERIALS AND METHODS
Methods are detailed in supplementary methods.
ScRNA-seq.
For scRNA-seq 3mm dorsal mid-forearm skin punch biopsies were obtained after informed consent under protocols approved by the University of Pittsburgh or University of Michigan Institutional Review Boards. Skin was digested enzymatically and cells loaded into the Chromium instrument (10X Genomics), as described (18). Data analysis was performed with the R package: Seurat V2.3.4. Differential gene expression analysis for dcSSc versus healthy control cells for each cluster was performed using the Wilcoxon rank-sum test, using a cut-off of P < 0.05, fold change (FC) > 1.5 and further requiring the expression of genes from > 25% denoted cells. All scRNA-seq data including Gene cell UMI matrix and a BAM file containing aligned reads are available at the Gene Expression Omnibus: GSE138669
Staining of Paraffin-Embedded Skin Biopsies.
Immunohistochemistry and immunofluorescent staining using tyramide signal amplification was performed with monoclonal mouse anti-FCN1 on formalin-fixed paraffin-embedded human skin tissues from 38 SSc patients and 16 HCs.
Microarray analysis.
Microarray RNA gene expression data from 64 SSc and 15 HC skin biopsies were analyzed using Cluster 3.0 software and visualized using TreeView 1.1.6
RESULTS
Single-cell transcriptome profiles myeloid cells from dcSSc and healthy control skin.
Using scRNA-seq, we examined gene expression of all enzymatically-digested skin cells obtained from 12 patients with dcSSc and 10 healthy control subjects (HCs). All subjects have been included in another analysis for fibroblasts in fibrotic skin (Nature Comm. Manuscript under review). DcSSc patients and control subjects were balanced across sex and age (Supplemental Table S1). Transcriptomes of 28,216 cells from HC subjects and 36,983 cells from dcSSc patients were grouped and analyzed together. Similar number of cells were included from HC (mean = 2,822 cells/biopsy) and dcSSc skin (mean = 3,082 cells/biopsy; Table 1).
Table 1:
The Proportion of Mφ/DC Subpopulation in Human Skin
| subject | number of cells | number of Myeloid Cells |
percentage of Macrophages and DCs in all cells |
number of MS4A4A+ Macrophages |
percentage of MS4A4A+ Macrophages in all cells |
number of FCGR3A+ Macrophages |
percentage of FCGR3A+ Macrophages in myeloid cells |
number of FCN1+ cells |
percentage of FCN1+ Cells in myeloid cells |
number of proliferaftiing macrophage |
percentage of proliferating macropahges |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SC1nor | 1488 | 51 | 3.43% | 23 | 1.55% | 0 | 0.00% | 1 | 1.96% | 0 | 0.00% |
| SC4nor | 3887 | 56 | 1.44% | 21 | 0.54% | 4 | 7.14% | 4 | 7.14% | 0 | 0.00% |
| SC18nor | 3183 | 154 | 4.84% | 68 | 2.14% | 1 | 0.65% | 8 | 5.19% | 1 | 0.65% |
| SC32nor | 1956 | 100 | 5.11% | 35 | 1.79% | 0 | 0.00% | 2 | 2.00% | 0 | 0.00% |
| SC33nor | 2156 | 173 | 8.02% | 92 | 4.27% | 0 | 0.00% | 4 | 2.31% | 0 | 0.00% |
| SC34nor | 2118 | 24 | 1.13% | 6 | 0.28% | 2 | 8.33% | 1 | 4.17% | 0 | 0.00% |
| SC50nor | 3670 | 54 | 1.47% | 19 | 0.52% | 0 | 0.00% | 1 | 1.85% | 0 | 0.00% |
| SC68nor | 2116 | 64 | 3.02% | 33 | 1.56% | 4 | 6.25% | 1 | 1.56% | 0 | 0.00% |
| SC124nor | 4552 | 99 | 2.17% | 36 | 0.79% | 3 | 3.03% | 0 | 0.00% | 0 | 0.00% |
| SC125nor | 3090 | 62 | 2.01% | 21 | 0.68% | 0 | 0.00% | 0 | 0.00% | 0 | 0.00% |
| SC2ssc | 1608 | 10 | 0.62% | 4 | 0.25% | 0 | 0.00% | 1 | 10.00% | 0 | 0.00% |
| SC5ssc | 4120 | 158 | 3.83% | 100 | 2.43% | 2 | 1.27% | 11 | 6.96% | 1 | 0.63% |
| SC19ssc | 1920 | 127 | 6.61% | 51 | 2.66% | 4 | 3.15% | 12 | 9.45% | 0 | 0.00% |
| SC49ssc | 3091 | 15 | 0.49% | 1 | 0.03% | 0 | 0.00% | 2 | 13.33% | 0 | 0.00% |
| SC60ssc | 1785 | 38 | 2.13% | 10 | 0.56% | 0 | 0.00% | 10 | 26.32% | 0 | 0.00% |
| SC69ssc | 2407 | 169 | 7.02% | 79 | 3.28% | 44 | 26.04% | 11 | 6.51% | 1 | 0.59% |
| SC70ssc | 2714 | 25 | 0.92% | 9 | 0.33% | 0 | 0.00% | 4 | 16.00% | 0 | 0.00% |
| SC86ssc | 3663 | 88 | 2.40% | 31 | 0.85% | 1 | 1.14% | 2 | 2.27% | 0 | 0.00% |
| SC119ssc | 4696 | 35 | 0.75% | 12 | 0.26% | 0 | 0.00% | 1 | 2.86% | 1 | 2.86% |
| SC185ssc | 3443 | 262 | 7.61% | 124 | 3.60% | 2 | 0.76% | 5 | 1.91% | 0 | 0.00% |
| SC188ssc | 3993 | 219 | 5.48% | 88 | 2.20% | 2 | 0.91% | 59 | 26.94% | 13 | 5.94% |
| SC189ssc | 3543 | 482 | 13.60% | 267 | 7.54% | 155 | 32.16% | 68 | 14.11% | 10 | 2.07% |
| Mean of HC | 2822 | 84 | 2.98% | 35 | 1.24% | 1 | 1.19% | 2 | 2.38% | 0 | 0.00% |
| Mean of SSc | 3082 | 136 | 4.41% | 65 | 2.11% | 18 | 13.24% | 16 | 11.76% | 2 | 1.47% |
| p-value (Wilcoxon test)A | 0.3139 | 0.3857 | 0.4614 | 0.404 | 0.4106 | 0.3342 | 0.416 | 0.0048 | 0.001 | 0.0831 | 0.0654 |
The P-values were calculated based on the number or percentage of cells in different subpopulation from HC/SSc samples.
Combined cell-gene count matrices were analyzed by t-distributed stochastic neighbor embedding (t-SNE). 28 distinct clusters were aligned to expected cell types according to the top highly expressed genes in each cluster as described previously (18) (Figure S1a, Table S2 and S3). All clusters included cells from multiple subjects and were unbiased from V1 and V2 chemistries (Figure S1b, S2a-b). Cluster #9 was identified as myeloid cells, showing specific LIN(CD3D/CD79A/NKG7)−HLA-DQ+ and expression of myeloid-specific genes (ITGAM, ITGAX, CD14, CSF1R, CD68 and CD209); and verified by expression of MS4A4A, CD1C and CLEC9A, markers for macrophage, cDC2 and cDC1, respectively (Figure S3a-c) (17).
The percentage of myeloid cells of total cells analyzed for each subject was strikingly variable, ranging from 0.49% (15 myeloid cells of 3,091 cells for SC49ssc) to 13.60% (482 myeloid cells of 3,543 cells for SC189ssc), and the percentage of myeloid cells showed a trend towards increased percentage in SSc, from 2.98% in HC skin to 4.41% in dcSSc skin (Table 1). The percentage of myeloid cells in SSc patients correlated weakly, negatively, with disease duration but not reaching statistical significance, (R= −0.26, Figure S3B). Notably, the shared correlation strength calculated based on gene expression of all cells from each subject with Canonical Correlation Analysis (CCA) showed SC188, SC189 and SC69 patient samples deviated from other samples (Figure S4).
All myeloid cell clusters identified in healthy skin were preserved in dcSSc skin.
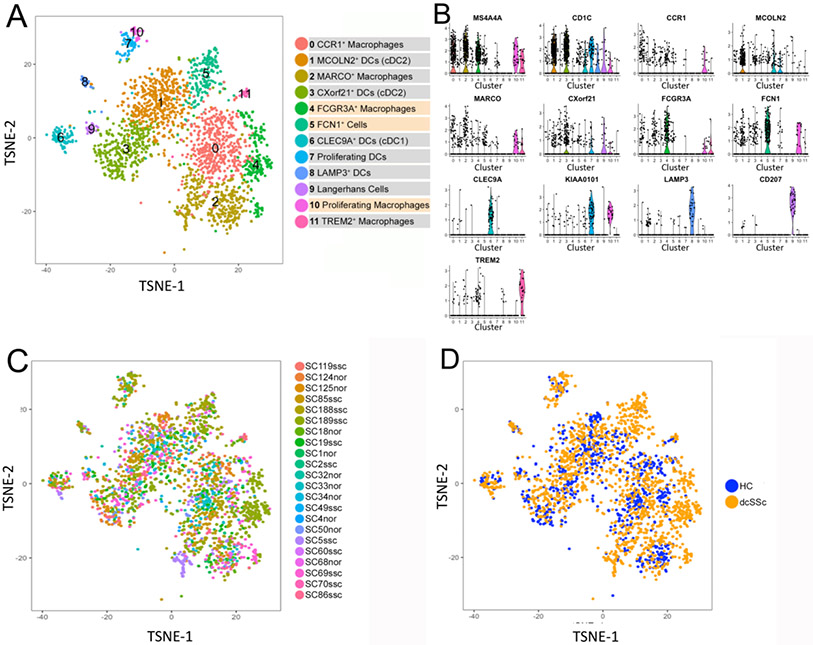
Re-analysis by t-SNE of 2,465 myeloid cells from dcSSc and HC skin showed 12 clusters of macrophage/dendritic cell (Mφ/DC, Figure 1a). Each cluster was composed of cells from six or more subjects (Figure 1c and Table S4). Nine of these 12 clusters paralleled previously described Mφ/DC clusters in healthy human skin (17).
Figure 1. Single cell RNA-sequencing analysis of human healthy control and dcSSc myeloid cell populations.
Visualization clustering by t-SNE plot of myeloid cells from all 22 combined healthy control subjects (HCs) and dcSSc skin samples, identified by cell type (panel a). Each point represents a cell. Two-dimensional t-SNE shows the dimensional reduction of cell transcriptomes. Cells were colored by K-nearest neighbors graph based on Euclidean distance in principal component analysis space using a smart local moving (SLM) algorithm to iteratively group cells. Violin plots of marker genes to distinguish each cluster of myeloid cells (panel b). Numbers in violin plots refer to clusters numbers as in Fig.1a. Cells were grouped by t-SNE as in Figure 1a, but are colored according to original subject identity in (panel c), or according to health status (panel d; HC, healthy control; dcSSc, diffuse cutaneous systemic sclerosis).
The myeloid cell clusters were highly preserved in dcSSc skin (Figure 1b), identified by the top 50 differentially expressed genes in each subcluster (Table S5). Subclusters #1 and #3 highly expressed CD1C and represented two cDC2 populations, highly expressing either markers MCOLN2 or CXorf21 (MCOLN2+ DC and CXorf21+ DC); These populations correspond closely to DC2 and DC3 circulating DCs (19) we described previously in healthy skin (17). Subcluster #6 highly expressed CLEC9A and other cDC1 markers, and was identified as cDC1 cells, corresponding to closely to circulating DC1 cells (17, 19). The cells in myeloid cluster #7, highly expressing CD1C and proliferation genes, such as MKI67, KIAA0101, TYMS, PTTG1, were identified as proliferating cDC2. Cells in subcluster #8 highly expressing LAMP3, CCL17, BIRC3 were identified as a mature subpopulation of cDC (LAMP3+ DC), and cells in subcluster #9 highly expressing CD207, FCGBP and HLA-DQB2 were identified as Langerhans cells (Figures 1b, S5 and S6). The cells in these myeloid clusters showed no noticeable shift in the clusters between HCs and dcSSc (Figure 1d).
Three previously described macrophage clusters in healthy skin, selectively expressing CCR1, MARCO or TREM2, were also detected (Figure 1a, 1b and Supplemental Table S5). The specific expression of MS4A4A, as well as other macrophage marker genes, and the absence of DC markers in subclusters #0, #2, #4, #10 and #11, indicated these as macrophage clusters (Figure 1a and S5). The percentage of all macrophage populations increased from 1.24% in HCs to 2.11% in dcSSc skin (Table 1). CCR1 and MARCO, which we described previously as good markers for two major macrophages subsets in HC skin (17), were somewhat more highly expressed by cells in clusters #0 and #2, respectively (Figures 1b and S5), but were not as discrete markers in this combined analysis of dcSSc and HC skin, apparently because the disease altered expression of these markers. However, comparing the top 15 genes distinguishing clusters in the combined database with HC clusters indicated that other markers of these two HC macrophages were preserved: HMOX1, MMP9, MMP19, CCL2, CEBPB, CTSL, CREM, THBD, EIF4E still highly expressed by cells in subcluster #0 (CCR1+ macrophages); and SEPP1, CCL13, FOLR2 and C1QA still highly expressed by cells in subcluster #2 (MARCO+ macrophages; Table S5 and Figure S5B). A distinct third macrophage population seen in both HC and SSc was distinguished by high expression of TREM2, FABP4 and FABP5, paralleled to the TREM2+ macrophages, which we previously identified in healthy skin ( (17) Figure 1b, S5A and S5B). In contrast to the nine myeloid populations common to HC and dcSSc skin samples, three other myeloid populations (cluster #4, #5 and #10) were detected almost exclusively in dcSSc skin (Figure 1a-d).
We reanalyzed the dataset using Harmony to correct for potential batch effects (20). Harmony analysis detected the same clusters shown above (Figure 1, S6A-E), except failed to identify a discrete cluster for proliferating macrophages (described further below), and indicated an additional population of macrophages (Figure S6A, cluster #4). Although this additional macrophage cluster included cells from healthy skin (Figure S6C), this population was not seen previously when analyzing only healthy skin (17).
Differential gene expression of dcSSc myeloid populations.
We compared the average differential gene expression of cells in each myeloid cluster from dcSSc skin to HC skin (Table S6). We then analyzed Gene Ontology (GO) pathways activated in each myeloid population in SSc skin after selecting differentially expressed genes showing a fold change (FC) > 1.5, detectable expression of each gene in > 25% of SSc cells in the cluster and an uncorrected p-value < 0.05 (Table S7). CCR1+ macrophages in dcSSc skin showed several statistically significant upregulated pathways, including genes associated with the interferon-gamma pathway: ICAM1, IFIT3, CCL13 and MT2A (21); and innate immune response (genes not shown) pathways. SSc MARCO+ macrophages showed upregulated genes in pathways associated with leukocyte chemotaxis: IL10, CXCL1, CCL4, GPR183, CXCL8, S100A9, IL1B and CCL3; innate immune response (genes not shown); response to IFN type 1: EGR1, ISG15 and IFITM3 and response to IFN gamma: CCL3, CCL4, HLA-DRB5, IFITM3; as well as other pathways. Both SSc cDC2 subsets showed upregulation of similar genes, but the only significant GO pathway was associated with immune response genes: C1QC, HLA-DRB5, CRIP1, IFITM3, LTB, PKM, IFITM2, S100B, CST7. SSc cDC1 showed upregulated genes in several pathways, including Fc-gamma receptor signaling pathway involved in phagocytosis: ACTR1, ACTR2, ACTR3, ACTB and CDC42; and response to wounding: ANXA1, TNFAIP3, CXCR4, GRN, ACTG1and KLF4. However, macrophage related genes previously detected highly expressed in dcSSc skin by analysis of bulk skin RNA expression (22) were not found upregulated in these clusters (Figure S6).
SIGLEC1 (CD169), a marker for IFN in SSc skin and a marker for disease severity (6, 11), was expressed only in the Macrophage/DC cluster and upregulated in this cluster. On subsetting this cluster SIGLEC1 was found mainly on subclusters #0, #1, #2, #4, #11 (all macrophage clusters except cluster #1), and upregulated in SSc mainly on cluster #0, #1 #4 and #10 (see Table S6).
FCGR3A+ macrophages in dcSSc skin and potential progenitor populations.
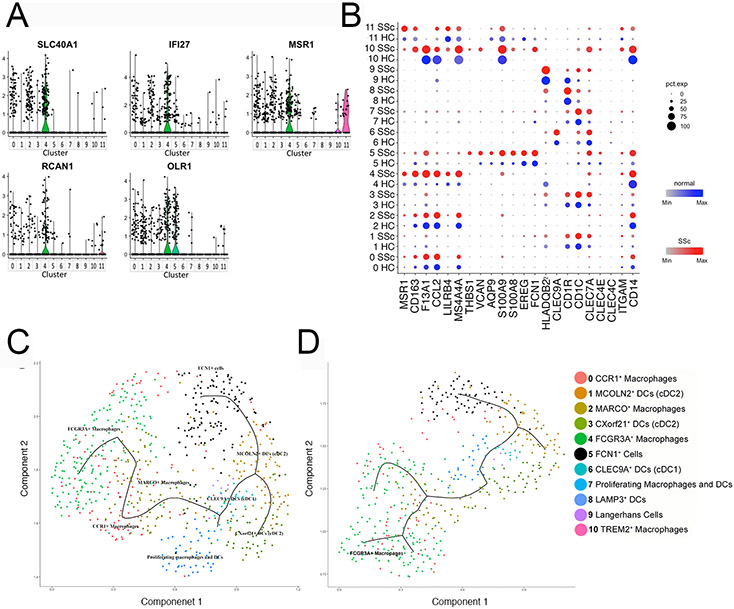
Cells in subcluster #4 (FCG3A+ macrophages) highly expressed macrophage markers, including MS4A4A and FCGR3A encoding CD16A (Figure 1a, 1b, S5 and S6). These cells also selectively expressed SLC40A1, IFI27, MSR1 (macrophage scavenger receptor 1), RCAN1 and OLR1 suggesting specialized functions of FCGR3A+ macrophages (Figure 2a). Most of the cells in this cluster were from only two of the 12 dcSSc subjects: SC69ssc (44 cells) and SC189ssc (155cells), whereas the remaining 25 cells were from five HC (14 cells) and five dcSSc (11 cells) subjects (Figure 1c, 1d and Table 1), suggesting that this subpopulation is highly expanded, but only in select dcSSc patients.
Figure 2. The signatures and putative functions of cluster #4: FCGR3A+ macrophages.
Violin plots of markers for cluster #4: FCGR3A+ macrophages (panel a). Dot plot of genes selectively expressed by FCGR3A+ macrophages (panel b). The intensity of the color indicates the level of the average scaled gene expression. The size of the dot indicates the percentage of cells expressing this gene in each cluster split by health status. Pseudo-time analysis of all myeloid cell (panel c) or only myeloid cells from dcSSc skin sample SC189 (panel d), showing FCGR3A+ macrophages trajectory from CCR1+ and MARCO+ macrophages, and FCN1+ mo-DC trajectory from MCOLN2+ cDC2.
The differential gene expression in FCGR3A+ macrophage by comparing gene expression of cells from dcSSc skin to cells from HCs showed few statistically significant genes (Table S7). We therefore compared gene expression by cells in this cluster with all the other macrophage/DC from HC clusters (Table S8), showing many significantly upregulated genes. Genes, encoding chemokines: CCL18, CXCL1, CCL8, CCL3, CCL4, CCL2, CCL13, and CXCL2 were highly expressed in this cluster. FCGR3A+ macrophages also showed upregulated expression of genes correlated with the severity of dcSSc, such as MSR1, CD163, F13A1, CCL2, LILRB4 and MS4A4A (11, 22) (Figure 2b). Based on the highly expressed genes of FCGR3A+ macrophages, Gene Ontology (GO) highly significantly enriched relatively broad terms such as “immune system process” and “immune response”, but also enriched for several more specific terms, including multiple pathways involved in chemotaxis, and TLR/innate immune activation: “response to lipopolysaccharide” and “toll-like receptor TLR6:TLR2 signaling pathway” (Table S9).
FCGR3A+ macrophages expressed the many of the most highly differentially expressed genes of CCR1+ (subcluster #0) and MARCO+ macrophages (subcluster #2) at moderate levels, suggesting a close relationship between these macrophage subpopulations (Figure 2b and S6). To further explore the relationship between FCG3A+, CCR1+ and MARCO+ macrophages, we analyzed the dataset trajectory using pseudo-time analysis, an algorithm that tracks the relationship between transcriptomes of single cells (23). Pseudo-time analysis of myeloid cells from all subjects showed that FCGR3A+ macrophages very closely related to and even admixed with CCR1+ macrophages, but also closely adjacent to MARCO+ macrophages (Figure 2c-d).
Increased FCN1+ myeloid cells in dcSSc skin.
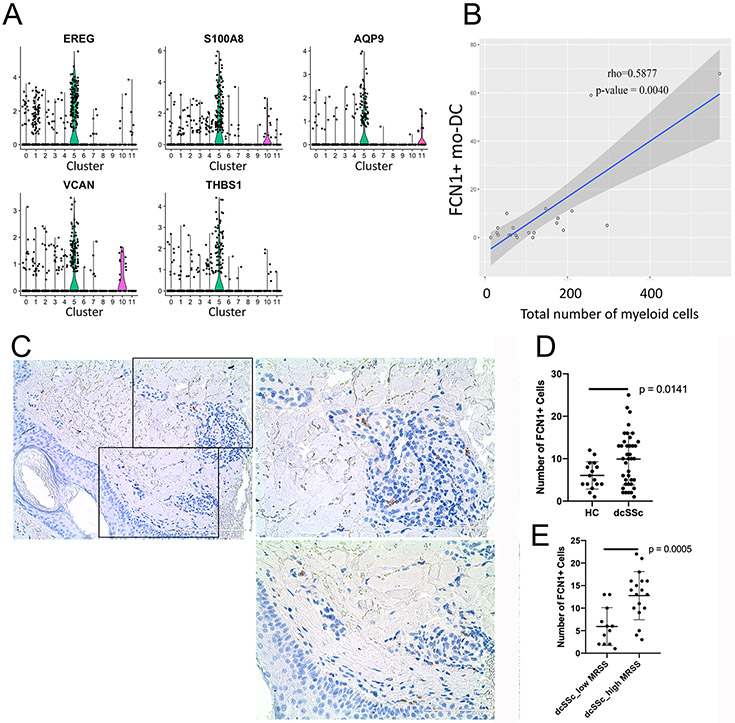
FCN1+ myeloid cells were significantly increased in dcSSc compared with HC skin (p = 0.0010), indicating this cluster as specifically expanded in dcSSc (Table 1). These cells were distributed unevenly across different dSSc samples with many of the cells coming from two patients: SC188ssc and SC189ssc (Figure 1a-d, Table 1). Cells in this cluster (cluster #5, FCN1+ mo-DC) selectively showed upregulated expression of FCN1, EREG, S100A8, AQP9, VCAN and THBS1, and expressed macrophage markers: CD163, F13A1, and MS4A4A at only moderate levels (Figure 2b, 3a and S6). As FCN1 and EREG, are monocyte markers, relative signature scores were calculated for the expression levels of CD14 monocyte and FCGR3A monocyte marker genes, previously described by scRNA-seq of human peripheral blood mononuclear cells (24). FCN1+ cells showed some similarity to CD14 monocytes, but not FCGR3A monocytes (Figure S7a-b). Among the FCN1+ myeloid cell, 50 top differentially expressed genes: FCN1, S100A8, AQP9, VCAN, CD300E, S100A9, FPR1, SOD2 and C5AR1, are markers for classical monocytes (Table S5), however other monocyte marker genes: ITGAM/CD11B, ITGB2/CD18, TLR2, and CLEC7A were not enriched in this cluster (Figure 2b).
Figure 3. The signatures and putative functions of cluster #5: FCN1+ mo-DC.
Violin plots of markers for cluster #5: FCN1+ cells (panel a). The correlation between myeloid cells and FCN+ moDC in skin (panel b). Staining of FCN1+ cells showing the perivascular distribution in skin biopsies from dcSSc patients (panel a; Scale bar =100 mm). FCN1+ cell counts by immunofluorescence (IF, Figure S10), showing more FCN1+ cells in dcSSc skin biopsies compared to healthy controls (panel c). P-value was calculated by Wilcoxon test. IF staining of FCN1+ cells showing more in dcSSc skin biopsies with high mRSS (>20, panel
In SSc skin FCN1+ cells maintained expression of CD14, CD13/ANPEP, CD172a/SIRPA, as well as S100A8 and S100A9, all described as markers of mo-DC (Figure S11a,) (25), with only low expression of CD1C, CD1A and IRF4 distinguishing these cells from DCs and no expression of MS4A4A and CD163 distinguishing these cells from macrophages (Figure 2b). In pseudo-time analysis FCN1+ cells on a trajectory closest to cDC2 (Figure 2c-d), further supporting their designation as mo-DC (25).
Based on the differentially expressed genes of FCN1+ mo-DC, GO significantly enriched, relatively non-specific terms such as “inflammatory response”, “defense response”, “response to stress” and “immune response”. These and many other terms overlapped those seen in FCGR3A+ macrophages, including pathways involved in chemotaxis and response to lipopolysaccharide (Table S9).
To verify the associations of FCN1+ and FCGR3A+ myeloid cells with SSc skin, we compared 15 additional dcSSc biopsies from another cohort to the same 10 HC biopsies. FCN1+ but not FCGR3A+ cells were again identified as a discrete cluster in t-SNE plot of myeloid cells (subcluster #4, Figure S8-S9), were significantly increased in dcSSc skin (Table S10), and correlated with the total number of myeloid cells in biopsies (Figure 3b).
Perivascular localization of FCN1+ myeloid cells in dcSSc skin.
Immunohistochemistry (IHC) of dcSSc skin showed FCN1+ myeloid cells distributed primarily in perivascular regions in dcSSc skin (Figure 3c). Immunofluorescence (IF) staining of FCN1+ in 16 HC and 39 dcSSc skin biopsies showed an increased number of FCN1+ cells in dcSSc patients compared with HCs (p-value = 0.0141) (Figure 3d, S10 and Table S11). More FCN1+ myeloid cells were seen in dcSSc skin biopsies from patients with higher mRSS (> 20) compared with lower MRSS (< 20) (Figure 3e; p-value = 0.0005), FCN1+ myeloid cells also significantly correlating with the MRSS (p<0.05; Figure S10B).
FCN1+ myeloid cell markers in bulk dcSSc skin mRNA data.
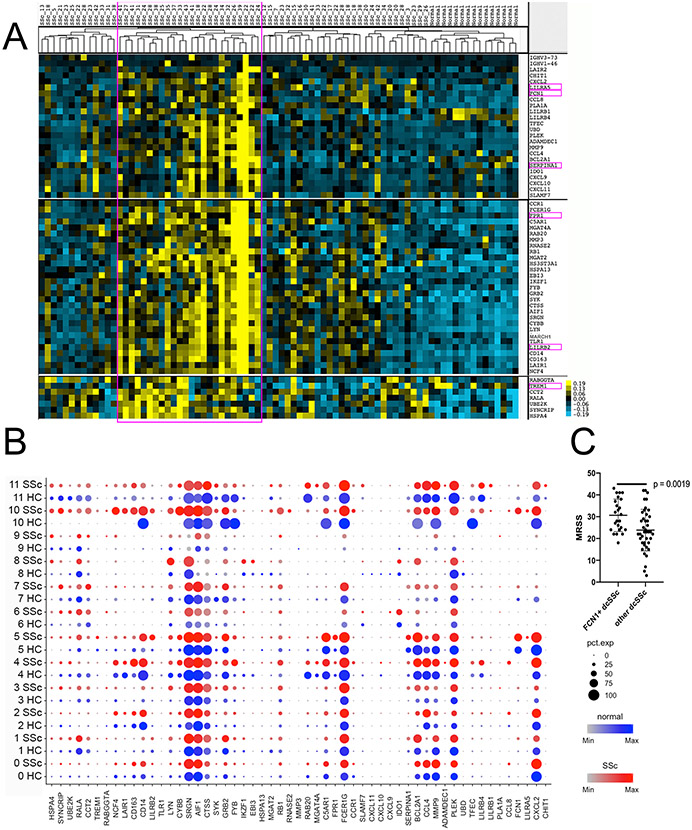
To extend our scRNA-seq observations we examined bulk microarray data from 64 SSc and 15 HC skin biopsies. Several genes: LILRA5, FCN1, SERPINA1, FPR1, LILRB2 and TREM1, were expressed by myeloid cells (cluster #9 in all cells t-SNE), most highly expressed by FCN1+ myeloid cell (myeloid subcluster #5; Figure S11a and b) and not by other skin cells or myeloid cells (Figure S12). The expression of these genes in bulk-seq data confirms the emergence of FCN1+ mo-DC in a subset of dcSSc patients (Figure 4a). Most of the genes in these clusters were expressed selectively expressed in FCN1+ mo-DC (FCN1, LILRA5, SERPINEA1, FPR1, LILRB2, TREM1), selectively in macrophages (CD14, CD163), or more broadly expressed by myeloid cells (AIF1, PLEK, Figure 4b). Moreover, the mRSS for SSc patients expressing markers of FCN1+ cells were higher than dcSSc not expressing these markers (p-value = 0.0019; Figure 4c), consistent with a general increase in macrophage/inflammatory cell infiltration in patients showing FCN1+ mo-DC, as seen histologically and by the correlation between FCN1+ mo-DC and myeloid numbers (Figure 4a).
Figure 4. Hierarchical clustering of bulk sequence data showing gene clusters of showing increased expression of myeloid, macrophage and FCN1+ mo-DC markers.
Heat map of hierarchical clustering of bulk sequence data from 64 dcSSc patients and 15 healthy controls showing clusters of SSc patients (panel a). The six highlighted markers of cluster #5: FCN1+ cells were used to identify the gene clusters. Dot plots of all the genes in the clusters showing expression in each myeloid cell types from scRNA-seq analysis (panel b). MRSS for dcSSc patients highly expressed the specific marker of are higher than others (panel c; p-value was calculated by Wilcoxon test).
Proliferating macrophages and DC in dcSSc skin
Proliferating myeloid cells (subclusters #7 and #10), predicted to be in the G2/S phase by cell phase analysis (Figure S13a), clustered separately by their uniquely high expression of genes associated with active cell proliferation, including KIAA0101, MKI67, TYMS, PTTG1, CDK1 and PCNA (Figure S6 and Table S5). Cells in the lower corner, in subcluster #7, were identified as proliferating cDC2 cells, as we have previously described in healthy skin, expressing both KIAA0101 and CD1C (Figure S13b), These were found in similar numbers in dcSSc and HC skin (Figure 1d). In addition, we identified a group of proliferating macrophages based on co-expression of cell proliferation genes and MS4A4A (subcluster #10, Figure S13c). Cells making up this cluster were mainly from SC188ssc (13/25 cells) and SC189ssc (10/25 cells), the same patient samples showing the most FCN1+ mo-DC (Table S4).
To track which macrophage subpopulation(s) were proliferating, we examined expression of single marker genes of each macrophage cluster and module scores of each cluster based on the top 10 most highly expressed genes for each macrophage cluster in the proliferating cell cluster (Figure S14). It appeared that all the macrophage subsets, as well as FCN1+ mo-DC, were represented in the proliferating cell cluster.
Plasmacytoid dendritic cells cluster with B cells and are increased in dcSSc skin.
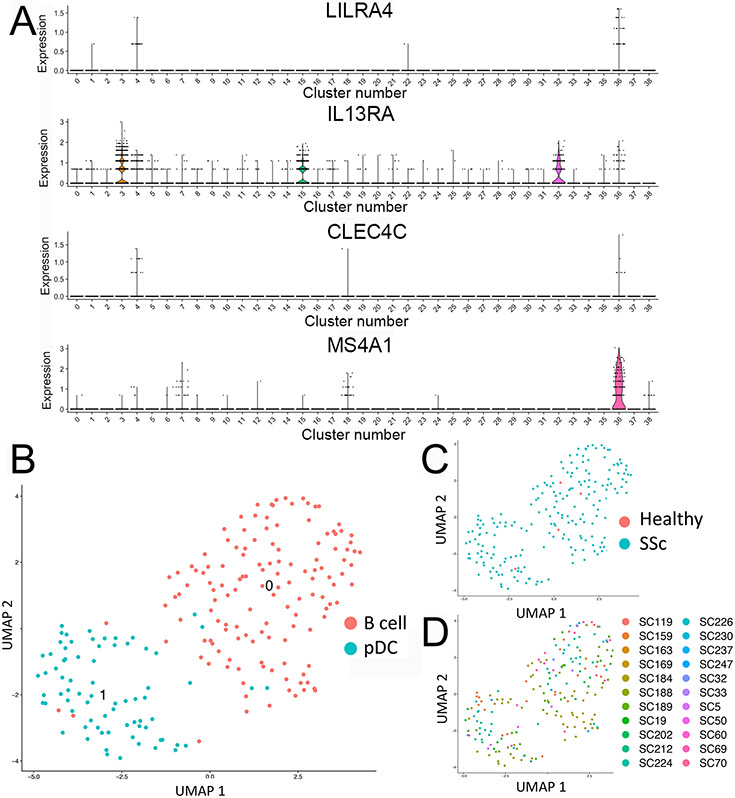
Because we did not find pDC in our initial clustering, we clustered all 27 dcSSc samples with the 10 normal skin samples (data not shown) and searched again for markers of these cells. LILRA4, IL3RA and CLEC4C were co-expressed in a cluster comprising mostly B cells (Figure 5). Re-clustering these cells detected two clusters, one prominently expressing B cell markers (MS4A1A, CD79A and immunoglobulin genes) and the other markers of pDC (LILRA4, IL3RA, CLEC4C), described previously as circulating DC6 (17, 19). The pDC came nearly exclusively from dcSSc patients (73 cells of 74 total cells) and only 1 pDC from the10 heathy controls. We detected upregulated expression of transcription factors important in pDC differentiation: IRF4, IRF7, IRF8 and ZEB2 compared to other skin cells or B cells, but not expression of IFNA1-11 or CXCL4 (Figure S15 and Table S12).
Figure 5. pDC in SSc and HC skin.
Feature plot analysis of 27 dcSSc and 10 HC samples, showing expression of pDC markers, LILRA4, IL3RA (CD123), CLEC4C (BDCA2); and B cell marker MS4A1 (CD20) by cluster #36 (panel a). Feature plot re-clustering of cluster 36, showing two cells subclusters, representing B cells and pDC (panel b). Feature plot of cluster #36 re-clustered cells colored by health status (panel c) or by sample source (panel d).
DISCUSSION
We have previously implicated markers of macrophages by bulk RNA analyses in both the severity and progressivity of SSc skin disease (11, 22, 26). ScRNA-seq studies here provide a comprehensive analysis of myeloid cells populations in dcSSc skin. While all myeloid clusters detected in healthy skin were preserved in dcSSc skin, four dcSSc-specific myeloid populations were discovered: FCGR3A+ macrophages, FCN1+ mo-DC, pDC and proliferating macrophages. The transcriptomes of each of these populations revealed gene signatures and putative altered functions of each cluster. Importantly, FCN1+ mo-DC showed a perivascular distribution along with other inflammatory cells, a higher frequency in dcSSc and an association with the severity of skin disease.
Fibrotic SSc skin preserved all of the myeloid populations, we previously reported in healthy skin: CCR1+, MARCO+ and TREM2+ macrophages; and six clusters of DCs: cDC1, two cDC2 populations, Langerhans cells, a mature subpopulation of LAMP3+ cDC, and a population of proliferating cDC2 (17). Upregulated genes in these clusters indicated pathways of innate immune and toll-like receptor activation, and response to both type I and type II interferons. consistent with several previous studies (6, 10, 27, 28) .
The highly increased frequency of FCN1+ mo-DC in dcSSc skin was verified by scRNA-seq, microarray analysis, IHC and IF staining for FCN1. The expression of monocyte marker genes, FCN1, S100A8 and S100A9 by these cells indicates that these cells are recruited from circulating monocytes. Trajectory analysis, as well as low level expression of CD1a and CD1c placed FCN+ cells most closely as mo-DCs (29). Cells with very similar gene expression profile have been identified in blister fluid of saline and house dust mite stimulated human skin (30). In humans mo-DC are one of several myeloid cell types capable of cross-presentation (25). The association of these cells with perivascular inflammation and high expression of multiple chemokines suggest they may promote the migration of other inflammatory cells.
THBS1 and FPR1, which are both pharmacodynamic biomarkers for the extent of skin disease and prognostic for progressive skin disease in dcSSc (11, 22), were predominantly expressed by FCN1+ mo-DC cells (Figure S15). FCN1+ mo-DC also showed a significant association with dcSSc severity (MRSS>20), suggesting a key role in disease pathogenesis. In addition, these cells likely play a prominent role in previous bulk RNA SSc skin expression studies, which describe an inflammatory patient subset (12, 31), as patients overexpressing FCN1+ markers also overexpressed both IFN-regulated and Immunoglobulin genes (Figure S16).
FCGR3A+ macrophages expressed most other macrophage markers, failed to express monocyte markers and trajectory analysis suggested that CCR1+ macrophages and/or MARCO+ macrophages are the progenitors of these cells. As CCR1+ and MARCO+ macrophages are present in normal skin and proliferate in SSc skin it is likely that these are resident cell populations that differentiate into FCGR3A+ macrophages in SSc skin. Enriched GO terms suggest FCGR3A+ macrophages are activated through TLR signaling. Several recent studies have placed TLR signaling upstream of inflammatory and profibrotic changes in SSc (32-34) Potentially TLR stimulates FCGR3A+ macrophages to highly express many cytokines and chemokines that attract and activate other immune cells, including CCL18 and IL6. We have previously reported that CCL18, the gene showing the highest increase in expression (4.4 fold-increase), is upregulated in SSc skin and SSc-related interstitial lung disease (8, 10). Serum levels of CCL18 are rapidly reduced to normal and CCL18 expression in the skin was blocked upon inhibition of IL-6, suggesting IL-6 may play a role in differentiation/activation of FCGR3A+ macrophages in select patients showing infiltration with this macrophage population (35).
We identified proliferating macrophages in dcSSc, but not normal skin. It appeared that all macrophages subsets and FCN1+ mo-DC cells were proliferating in some dcSSc patients. The majority of the proliferating cells were from SC188ssc and SC189ssc, the same patients contributing most of the cluster #5, FCN1+ mo-DCs. We have recently characterized proliferating macrophages in idiopathic pulmonary fibrosis and seen similar cells in SSc-ILD (36, 37), suggesting that common cytokine signals may drive macrophage proliferation in both SSc skin and lung.
Previous studies have detected perivascular pDC in the affected skin of SSc, but not control skin by co-staining of CD123 and in situ IFNA expression (15), or by expression of CLEC4 (BCDA-2) (16). Consistent with these studies, we found that pDC were very rare in normal skin (1/20,073 cells), and increased in numbers but still rare in dcSSc skin (73 of 74,607 cells). We were unable to detect expression by pDC or any other cell type in SSc skin of IFNA1-IFNA11 or CXCL4 genes, the former classically secreted by these pDC, the latter described as upregulated in SSc pDC (16) and implicated in co-activation of TLR9 (14, 38). This likely represents the relatively low level of expression of these genes though we are able to easily detect CXCL4 in platelets in a different dataset (R. Lafyatis, unpublished observation). Although rare, these pDC are likely the main source for type I, interferons in SSc skin and resulting effects on multiple other cell types. The functional importance of these cells in skin fibrosis is supported by markedly ameliorated skin bleomycin-induced fibrosis upon pDC depletion (14). Our data confirm the expression of IRF8 in dermal pDC, previously shown upregulated in pDC in SSc PBMC (14), as well as upregulated expression of IRF4 and IRF7 compared to other dermal myeloid cells or their closest transcriptome relative, B cells. The clustering of SSc pDC with B cells is intriguing in terms of recent understanding that pDC can differentiate along a myeloid or lymphoid pathway with a common B cell and pDC progenitor (39, 40). The clustering of these cells in SSc skin with B cells suggest that in SSc skin pDC differentiate exclusively along the lymphoid pathway from an IL-7R+ FLT3+ lymphoid progenitor.
Not all patients show a dramatic myeloid expansion, so it remains possible that myeloid cells drive pathogenesis in only a subset of patients, consistent with bulk microarray studies (31). As macrophage subsets are also involved in repair it is also possible that their presence reflects repair processes in the skin (41). The lack of local skin score data is a limitation to this study.
In summary, our data reveal several populations of myeloid cells in dcSSc that are likely driving SSc vascular pathology and skin fibrosis. This more profound understanding of inflammatory dcSSc patient subsets provides new insights into stratifying patients for targeted therapies and possibly for predicting responses to immunosuppressives.
Supplementary Material
ACKNOWLEDGEMENTS
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases under award number 2P50 AR060780. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. D.X. was sponsored by China Scholarship Council (File No. 201706370258) and the third Xiangya hospital, central south university and acknowledge the encouragement of Dr. Weiru Zhang and Dr. Xiaoxia Zu
Footnotes
Conflict of interest. R.L. has served as a consultant for Bristol Myers Squibb, Formation, Sanofi, Biocon, Boehringer-Mannheim, Merck and Genentech/Roche, and holds or recently had research grants from Corbus, Formation, Elpidera, Regeneron, Pfizer and Kiniksa. Dr. Robyn Domsic has worked as a consultant for Eicos Sciences Inc. and Boehringer-Ingelheim. Dr. Dinesh Khanna has served as a consultant for Actelion, Acceleron, BMS, Blade Therapeutics, Boehringer Ingelham, Bayer, ChemomAB, Cytori, Celgene, Curzion, Corbus Pharmaceuticals, CSL Behring, GSK, Genentech, Mitsubishi Tanabe Pharma Development America, Sanofi-Aventis, and UCB; he has stocks in Eicos Sciences, Inc. and has employment with CiviBio Pharma, Inc; he has received grant support from Bristol Myers Squibb, Pfizer, Bayer, and Horizon.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi:10.1002/ART.41813
REFERENCES
- 1.Denton CP, Khanna D Systemic sclerosis. Lancet. 2017;390(10103):1685–99. [DOI] [PubMed] [Google Scholar]
- 2.Toledo DM, Pioli PA Macrophages in Systemic Sclerosis: Novel Insights and Therapeutic Implications. Current rheumatology reports. 2019;21(7):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Affandi AJ, Carvalheiro T, Radstake T, Marut W Dendritic cells in systemic sclerosis: Advances from human and mice studies. Immunol Lett. 2018;195:18–29. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa O, Ishikawa H Macrophage infiltration in the skin of patients with systemic sclerosis. The Journal of rheumatology. 1992;19(8):1202–6. [PubMed] [Google Scholar]
- 5.Kraling BM, Maul GG, Jimenez SA Mononuclear cellular infiltrates in clinically involved skin from patients with systemic sclerosis of recent onset predominantly consist of monocytes/macrophages. Pathobiology. 1995;63(1):48–56. [DOI] [PubMed] [Google Scholar]
- 6.York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum. 2007;56(3):1010–20. [DOI] [PubMed] [Google Scholar]
- 7.Higashi-Kuwata N, Jinnin M, Makino T, Fukushima S, Inoue Y, Muchemwa FC, et al. Characterization of monocyte/macrophage subsets in the skin and peripheral blood derived from patients with systemic sclerosis. Arthritis research & therapy. 2010;12(4):R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathes AL, Christmann RB, Stifano G, Affandi AJ, Radstake TR, Farina GA, et al. Global chemokine expression in systemic sclerosis (SSc): CCL19 expression correlates with vascular inflammation in SSc skin. Annals of the rheumatic diseases. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xuan W, Qu Q, Zheng B, Xiong S, Fan GH The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. Journal of leukocyte biology. 2015;97(1):61–9. [DOI] [PubMed] [Google Scholar]
- 10.Christmann RB, Sampaio-Barros P, Stifano G, Borges CL, de Carvalho CR, Kairalla R, et al. Association of Interferon- and transforming growth factor beta-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis. Arthritis & rheumatology. 2014;66(3):714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice LM, Ziemek J, Stratton EA, McLaughlin SR, Padilla CM, Mathes AL, et al. A Longitudinal Biomarker for the Extent of Skin Disease in Patients With Diffuse Cutaneous Systemic Sclerosis. Arthritis & rheumatology. 2015;67(11):3004–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milano A, Pendergrass SA, Sargent JL, George LK, McCalmont TH, Connolly MK, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PloS one. 2008;3(7):e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assassi S, Swindell WR, Wu M, Tan FD, Khanna D, Furst DE, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis & rheumatology. 2015;67(11):3016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ah Kioon MD, Tripodo C, Fernandez D, Kirou KA, Spiera RF, Crow MK, et al. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Science translational medicine. 2018;10(423). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming JN, Nash RA, McLeod DO, Fiorentino DF, Shulman HM, Connolly MK, et al. Capillary regeneration in scleroderma: stem cell therapy reverses phenotype? PloS one. 2008;3(1):e1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Bon L, Affandi AJ, Broen J, Christmann RB, Marijnissen RJ, Stawski L, et al. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. The New England journal of medicine. 2014;370(5):433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue D, Tabib T, Morse C, Lafyatis R Transcriptome landscape of myeloid cells in human skin reveals diversity, rare populations and putative DC progenitors. Journal of dermatological science. 2020;97(1):41–9. [DOI] [PubMed] [Google Scholar]
- 18.Tabib T, Morse C, Wang T, Chen W, Lafyatis R SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. J Invest Dermatol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature methods. 2019;16(12):1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivashkiv LB IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018;18(9):545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stifano G, Sornasse T, Rice LM, Na L, Chen-Harris H, Khanna D, et al. Skin Gene Expression Is Prognostic for the Trajectory of Skin Disease in Patients With Diffuse Cutaneous Systemic Sclerosis. Arthritis & rheumatology. 2018;70(6):912–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology. 2014;32(4):381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology. 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collin M, Bigley V Human dendritic cell subsets: an update. Immunology. 2018;154(1):3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stifano G, Affandi AJ, Mathes AL, Rice LM, Nakerakanti S, Nazari B, et al. Chronic Toll-like receptor 4 stimulation in skin induces inflammation, macrophage activation, transforming growth factor beta signature gene expression, and fibrosis. Arthritis research & therapy. 2014;16(4):R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharyya S, Wang W, Qin W, Cheng K, Coulup S, Chavez S, et al. TLR4-dependent fibroblast activation drives persistent organ fibrosis in skin and lung. JCI Insight. 2018;3(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skaug B, Assassi S Type I interferon dysregulation in Systemic Sclerosis. Cytokine. 2019:154635. [DOI] [PubMed] [Google Scholar]
- 29.Kashem SW, Haniffa M, Kaplan DH Antigen-Presenting Cells in the Skin. Annu Rev Immunol. 2017;35:469–99. [DOI] [PubMed] [Google Scholar]
- 30.Chen YL, Gomes T, Hardman CS, Vieira Braga FA, Gutowska-Owsiak D, Salimi M, et al. Re-evaluation of human BDCA-2+ DC during acute sterile skin inflammation. The Journal of experimental medicine. 2020;217(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pendergrass SA, Lemaire R, Francis IP, Mahoney JM, Lafyatis R, Whitfield ML Intrinsic gene expression subsets of diffuse cutaneous systemic sclerosis are stable in serial skin biopsies. J Invest Dermatol. 2012;132(5):1363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya S, Midwood KS, Yin H, Varga J Toll-Like Receptor-4 Signaling Drives Persistent Fibroblast Activation and Prevents Fibrosis Resolution in Scleroderma. Adv Wound Care (New Rochelle). 2017;6(10):356–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farina G, York M, Collins C, Lafyatis R dsRNA activation of endothelin-1 and markers of vascular activation in endothelial cells and fibroblasts. Annals of the rheumatic diseases. 2011;70(3):544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farina GA, York MR, Di Marzio M, Collins CA, Meller S, Homey B, et al. Poly(I:C) drives type I IFN- and TGFbeta-mediated inflammation and dermal fibrosis simulating altered gene expression in systemic sclerosis. J Invest Dermatol. 2010;130(11):2583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khanna D, Denton CP, Jahreis A, van Laar JM, Frech TM, Anderson ME, et al. Safety and efficacy of subcutaneous tocilizumab in adults with systemic sclerosis (faSScinate): a phase 2, randomised, controlled trial. Lancet. 2016. [DOI] [PubMed] [Google Scholar]
- 36.Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J. 2019;54(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valenzi E, Bulik M, Tabib T, Morse C, Sembrat J, Trejo Bittar H, et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Annals of the rheumatic diseases. 2019;78(10):1379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lande R, Lee EY, Palazzo R, Marinari B, Pietraforte I, Santos GS, et al. CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-alpha production in systemic sclerosis. Nature communications. 2019;10(1):1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues PF, Alberti-Servera L, Eremin A, Grajales-Reyes GE, Ivanek R, Tussiwand R Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat Immunol. 2018;19(7):711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musumeci A, Lutz K, Winheim E, Krug AB What Makes a pDC: Recent Advances in Understanding Plasmacytoid DC Development and Heterogeneity. Frontiers in immunology. 2019;10:1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brancato SK, Albina JE Wound macrophages as key regulators of repair: origin, phenotype, and function. The American journal of pathology. 2011;178(1):19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.