Abstract
Advances in high resolution, nontargeted mass spectrometry allow for the simultaneous measure of thousands of metabolites in a single biosample. Application of these analytical approaches to population-scale human studies has been limited by the need for resource-intensive blood sample collection, preparation, and storage. Dried blood spotting, a technique developed decades ago for newborn screening, may offer a simple approach to overcome barriers in human blood acquisition and storage. In this study, we find that over 4,400 spectral features across diverse chemical classes may be efficiently and reproducibly extracted and relatively quantified from human dried blood spots using nontargeted metabolomic analysis employing HILIC and reversed-phase liquid chromatography coupled to Orbitrap mass spectrometry. Moreover, over 80% of metabolites were found to be chemically stable in dried blood spots stored at room temperature for up to a week. In direct relation to plasma samples, dried blood spots exhibited comparable representation of the human circulating metabolome, capturing both known and previously uncharacterized metabolites. Dried blood spot approaches provide an opportunity for rapid and facile human biosampling and storage, and will enable widespread metabolomics study of populations, particularly in resource-limited areas.
INTRODUCTION
The measurement and characterization of circulating metabolites holds enormous potential to advance and transform our understanding of human biology, providing deep insight into disease pathogenesis, drug responsiveness, and dynamic environmental exposures such as diet.1,2 Advances in high-throughput chromatography and high-resolution mass spectrometry have facilitated the measurement of thousands of metabolites within a single biosample,3,4 the majority of which remain structurally and functionally uncharacterized.5 Coupled software and computational advances have made it possible to improve the scale of experiments,6–10 a shift that will undoubtedly lead to better characterization of human chemical diversity, interactions among environmental influences and genetic factors, and ultimately personalization of dietary and pharmaceutical therapies.
These analytical and technical advances have prompted a renewed focus on optimization and streamlining of patient sample acquisition, storage, and processing, particularly for population-scale studies on the order of 10,000 to 1,000,000 biosamples. Traditional approaches are both labor and cost intensive, requiring trained health professionals, access to specialized equipment such as centrifuges, as well as immediate cold storage and refrigerated transport in specialized containers.11,12 These barriers to sample acquisition are particularly constraining in studies involving children and individuals residing in resource-poor areas.13,14 Dried blood spotting, a method developed nearly 60 years ago for blood biosampling, may represent a scalable and cost-effective solution for human blood acquisition and storage, particularly for metabolomics studies.
Dried blood spots (DBS) are currently widely employed for sampling of blood from newborns for neonatal screening. Typically, several drops of blood (~50 μL per spot) are collected from a peripheral prick from newborns and spotted on a Guthrie card. Using these samples, multiple targeted metabolites are routinely assayed to identify inborn errors of metabolism or drug levels in select cases.15 Recent studies suggest that a broader range of metabolites may be potentially assayed from DBS, beyond those traditionally examined for neonatal screening.16–20 For instance, prior studies have detailed the measurement of heavy metals and pesticides from DBS, which reflect potential in utero exposures,21,22 as well as a broad range of metabolites associated with birth weight.23 To date, however, the application of dried blood systems for nontargeted metabolomics analysis across thousands of metabolites is limited. Prior investigations of nontargeted liquid chromatography mass spectrometry (LCMS) of DBS demonstrate reliable measure of a few hundred up to one thousand unique spectral features.23–25 However, the full chemical diversity of the human metabolites that may be extracted and assayed from small volumes of blood stored as DBS, the relation of DBS to traditional venipuncture samples, the impact of blood spot location, and the stability of metabolites in stored DBS, remain incompletely characterized.
In this study, we perform a comprehensive characterization of DBS as biosample storage devices for nontargeted mass spectrometry studies employing reversed-phase and HILIC liquid chromatography coupled to an Orbitrap mass spectrometer. We present optimized sample preparation approaches for extraction of different classes of metabolites from DBS, as well as determine intra-spot variation of detectable analytes and stability of metabolites in DBS stored at room temperature over time. To determine the impact of potential sample-matrix interactions on the fidelity of metabolite measures from DBS, we perform nontargeted LCMS and of matched DBS and plasma samples.
MATERIALS AND METHODS
Chemicals and Consumables.
Whatman 903 Protein Saver Cards (GE healthcare) were used for all DBS experiments. LC-MS grade reagents were purchased from Honeywell (Methanol, Isopropyl Alcohol, Ammonium Bicarbonate and Acetic Acid), Fisher (water and Ammonium Hydroxide), and Sigma Aldrich (Acetonitrile and Ethanol). Well plates were purchased from Greiner Bio-One (96 well 1 mL Master Block plates) and Axygen (96-well V-bottom 600 μL).
Blood Samples.
Pooled whole blood collected via venipuncture were obtained from BioIVT. All BioVT whole blood samples (which were collected in EDTA tubes) were stored at 4 °C prior to use and were spotted prior to stated expiration date. DBS cards spotted with blood collected via venipuncture containing known concentrations for 29 metabolites from diverse chemical classes were obtained from the CDC (Newborn Screening Quality Assurance Program) and used for dynamic range of concentration and linearity of dilution experiments. CDC dried blood spot samples were stored at −20 °C in accordance with CDC recommendations and were used prior to stated expiration date.
Optimization of extraction of metabolites from DBS.
DBS samples were prepared by spotting 50 μL of whole blood in the center of the demarcated area of a Whatman 903 card via single channel pipette, which allowed for five 3 mm punches to be taken per spot with one central and four peripheral locations (Figure 1C). Spotted cards were then placed horizontally on a Whatman 903 card rack. The rack was placed in a Styrofoam container with Drierite desiccant chips and allowed to dry over 3.5 hours. Once dehydrated, cards were folded and placed in a freezer grade plastic bag with a silica gel desiccator pouch and a O2 absorbent packet. Finally, bags were placed in a covered box to avoid light exposure and stored at −80 °C. All spots were analyzed within a month of spotting. Plasma was prepared by spinning whole blood at 5300 rpm (~2,000 g) for 15 minutes at 4 °C and then transferring 500 μL of plasma supernatant to a fresh microfuge tube.
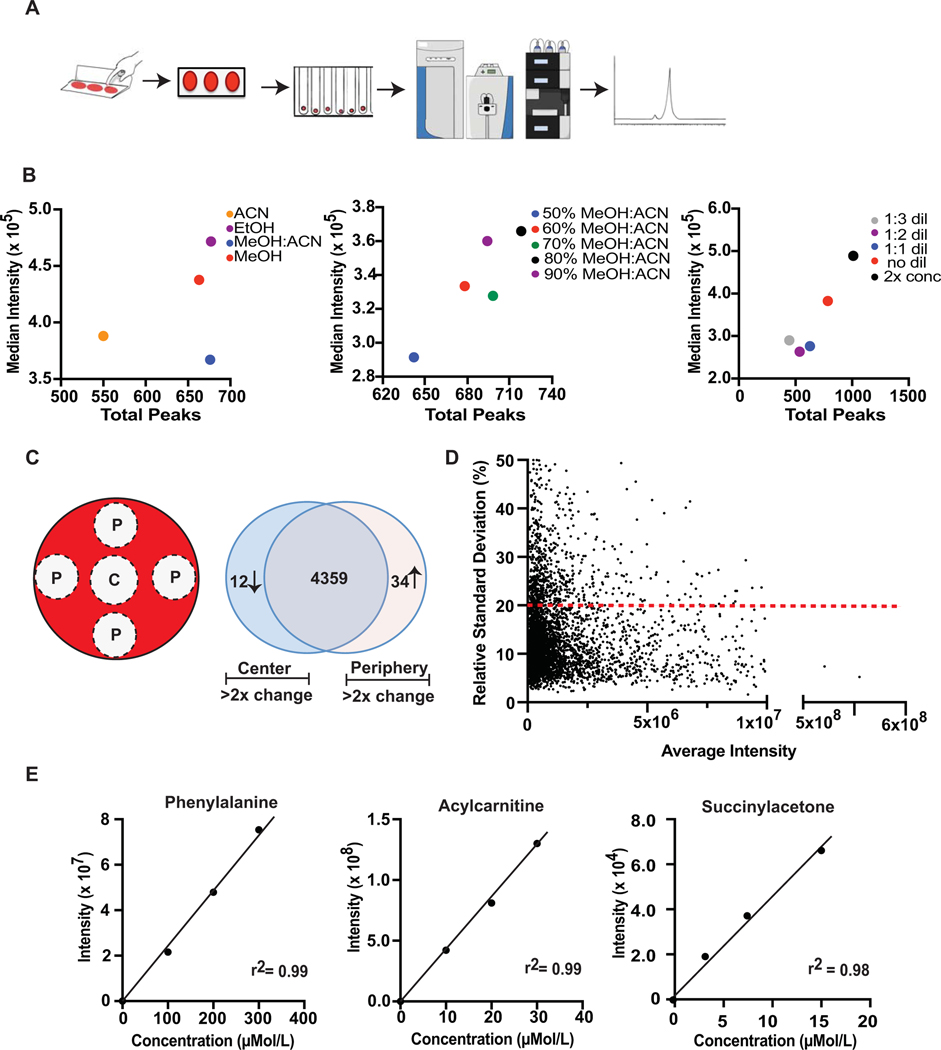
Figure 1. Non-targeted reversed-phase LC- Orbitrap MS analysis of DBS.
A. Schematic of dried blood spot collection, processing and analysis. Whole blood is spotted onto a DBS collection card and allowed to dry. A 3 mm hole is then punched from a single spot and placed in tube where metabolites are extracted and then analyzed by non-targeted mass spectrometry. Resulting data is extracted and metabolites are quantified and identified. B. Optimization of extraction procedures for Polar metabolites based on number of observed features and their corresponding signal intensities. Optimization of organic extraction solvent (center panel), organic solvent to water ratio (middle panel) and supernatant concentration (right panel). C. Schematic of DBS punch location (P, peripheral punch; C, Center punch) and variation in spectral peak intensity with punch location represented in a Venn diagram showing the number of features that vary by less than 2-fold by location (center overlap), as well the features that vary more than 2-fold in the center (center) and peripheral (right) locations. Data represents aggregated data on unique spectral peaks across all Polar, Bioactives and Lipids metabolites. D. Comparison of intensity versus relative standard deviation (RSD) for all spectral features across replicate samples (20% RSD indicated by dashed line). E. Dynamic linear range and relative quantification of metabolites extracted from control CDC DBS cards with known concentrations of target metabolites. Linearity plots for 3 representative metabolites (phenylalanine, acylcarnitine, and succinylacetone) measured from CDC DBS cards are shown.
For metabolite extraction, a 3 mm diameter punch from each blood spot (n=3 for each optimization condition) was deposited into separate wells of a Greiner Master Block 1 mL 96 well plate, which was kept on ice. Blank Whatman 903 paper was punched two times after each spot was taken to prevent carry-over between samples. To optimize the amount of water needed to initially hydrate the blood spots, various amounts of water were added to each well (100 μL, 80 μL, 60 μL, 40 μL, or 20 μL), sealed and then placed on an orbital shaker for agitation at 2000 rpm for 15 minutes at 4 °C. Various amounts of organic solvent (ethanol [EtOH], methanol [MeOH], acetonitrile [ACN], or 1:1 methanol/acetonitrile [MeOH:ACN]) were then added to each well (100 μL, 120 μL, 140 μL, 160 μL, or 180 μL, respective to the water volumes so each well contained 200 μL total volume). The plate was re-sealed and agitated again at 2000 rpm for 15 minutes at 4°C. After shaking, the plate was centrifuged at 4200 rpm for 10 minutes at 4 °C. Varying volumes of supernatant were then transferred to a fresh 1 mL 96 well plate with 200 μL glass inserts and diluted with water 1:3, 1:2, and 1:1 with 40:40:20 MeOH:ACN:H2O for polar metabolites and 35:35:30 MeOH:ACN:H2O for lipid analysis or was not diluted with water at all. For the “2x” concentration tests, 100 μL of supernatant was dried in a speed vac at 40°C for ~1 hour and then re-constituted in 50 μL of a solution of 40:40:20 MeOH:ACN:H2O for the polar metabolites and 35:35:30 MeOH:ACN:H2O for lipid analysis. The reconstitution step involved 10 minutes of orbital shaking at 650 RPM. Water hydration and organic solvent shake times were also optimized (1, 5, 10, 15, 20, and 30 minutes). For the Bioactives method, supernatant dilution was not tested but rather the amount of supernatant loaded onto the solid phase extraction (SPE) plate was evaluated (25 μL, 50 μL, 75 μL, 100 μL, 125 μL, or 150 μL).
Optimized metabolite extraction protocols.
Polar Metabolite Extraction.
After placing the 3 mm DBS punch into a 1 mL 96 well plate, 40 μL of water was transferred into the well and agitated via orbital shaking for 10 minutes at 2000 RPM. After shaking, 160 μL of ACN:MeOH (1:1) was added to the wells, and the plate was agitated again for 10 minutes at 2000 RPM. The plate was then centrifuged at 4200 RPM at 4 °C for 10 minutes. After shaking, 100 μL of supernatant was transferred into an Axygen V-bottom plate and placed in a vacuum concentrator until samples were dry. The solution was then re-constituted in 50 μL of ACN:MeOH:H2O solution (40:40:20) and shaken again in an orbital shaker at 650 RPM for 10 minutes. Then, 50 μL of supernatant was transferred to a 1 mL 96 well plate with glass inserts, and centrifuged at 4200 RPM at 4 °C for 10 minutes. Supernatant was then analyzed by LC-MS.
Bioactive Metabolite Extraction.
After placing the 3 mm DBS punch into a 1 mL 96 well plate, 40 μL of water was transferred into the well and agitated via orbital shaking for 10 minutes at 2000 RPM. After shaking, 160 μL of EtOH was added to each well and the plate was again shaken for 10 minutes at 2000 RPM. The plate was then centrifuged at 4200 RPM at 4 °C for 10 minutes. Then, 150 μL of supernatant was loaded onto a Phenomenex Strata-X SPE plate and metabolites were extracted according to previously reported protocols.26
Lipid Metabolite Extraction.
After placing the 3 mm DBS punch into a 1 mL 96 well plate, 60 μL of water was transferred into the well and agitated via orbital shaking for 10 minutes at 2000 RPM. After shaking, 140 μL of ACN:MeOH (1:1) was added to the wells, and the plate was again shaken for 10 minutes at 2000 RPM. The plate was then centrifuged at 4200 RPM at 4 °C for 10 minutes after which 100 μL of supernatant was transferred into an Axygen V-bottom plate and placed in a vacuum concentrator until samples were dry. The solution was then re-constituted in 50 μL of ACN:MeOH:H2O solution (35:35:30) and shaken again in an orbital shaker at 650 RPM for 10 minutes. Then, 50 μL of supernatant was transferred to a fresh 1 mL 96 well plate containing 200 μL glass inserts, centrifuged at 500 RPM for 10 minutes, and then analyzed by LC-MS.
Quality Control Protocols.
An instrument solvent blank was determined by analyzing the 2 solvents used for the extraction protocols above, MeOH:ACN and EtOH, prior to LC-MS analysis of samples. In addition, the supernatants from blank, unspotted Whatman cards subjected to extraction with MeOH:ACN and EtOH separately were analyzed by LC-MS to determine contaminant spectral peaks related to the Whatman cards. Spectral peaks related to solvents or Whatman cards were subtracted from sample LC-MS data. A method blank was also analyzed to account for any laboratory background. Samples were analyzed in a randomized fashion and were interspaced with injections of a QC standard ( 13C glutamine for Polar method; 1-cyclohexyl-dodecanoic acid urea [CUDA] for Lipids method, and a mix of the following for the Bioactives method: 9-HODE; 13-HODE; 9,10-diHOME; 12,13-diHOME; 15-deoxy-d12,14-PGJ2; 5-oxoETE; 20-HETE; 15-HETE; 12-HETE; 5S-HETE; LTB4; CUDA; 13,14-dihydro-15-keto-PGD2; PGE2; PGE2; PGD2; 13,14-dihydro-15-keto-PGF2a; PGF2a/6-keto-PGF1a/TXB2/Resolvin D1). The QC sample was measured after every 20 sample injections.
LC-MS methods
Polar metabolites:
Polar metabolites:
Once prepared, 2 μl of sample was injected onto a SeQuant Zic-pHILIC (5 μm particle size, 100 × 2.1 mm) column and separated using mobile phases A (20 mM Ammonium Bicarbonate in water with pH 9.6) and B (100% Acetonitrile) running the following gradient: 90% B from 0 to 0.25 min, 90% to 55% B from 0.25 to 4 min, and 55% B from 4 min to 6 min, followed by a 2.5-minute re-equilibration time. The flow rate was set at 0.4 mL/min and the column compartment temperature at 45 ºC. Mass detection was performed using a Thermo QExactive orbitrap mass spectrometer equipped with a heated electrospray ionization (HESI) source and collision-induced dissociation (CID) fragmentation. The source settings used for all samples were: negative and positive ion mode profile data, sheath gas flow of 40 units, aux gas flow of 15 units, sweep gas flow of 2 units, spray voltage of +/− 3.5kV, capillary temperature of 275 ⁰C, aux gas temp of 350 ⁰C, S-lens RF at 45. For MS1 scan events, scan ranges were set to m/z 65–975, mass resolution of 35k, AGC of 1e6 and injection time of 75 ms was used. For tandem MS acquisition, mass resolution of 17.5k, AGC of 1e5, normalized collision energy (NCE) of 30 eV, loop count of 6 and injection time of 50 ms was used.
Bioactive metabolites:
Once prepared, 20 μl of sample was injected onto a Phenomenex Kinetex C18 (1.7 μm particle size, 100 × 2.1 mm) column that was pre-conditioned with plasma and separated using mobile phases A (70% water, 30% acetonitrile, 0.1% acetic acid) and B (50% acetonitrile, 50% isopropyl alcohol, 0.2% acetic acid) running the following gradient: 1% B from 0 to 0.25 min, 1% to 55% B from 0.25 to 5 min, 55% to 99% B from 5.0 to 5.5 min, 99% B from 5.5 min to 7 min, followed by a 1 min re-equilibration time. The flow rate was set at 0.375 mL/min and the column compartment temperature at 50 ºC. Mass detection was performed using a Thermo QExactive orbitrap mass spectrometer equipped with a heated electrospray ionization (HESI) source and collision-induced dissociation (CID) fragmentation. The source settings used for all samples were: negative ion mode profile data, sheath gas flow of 40 units, aux gas flow of 10 units, sweep gas flow of 2 units, spray voltage of −3.55 kV, capillary temperature of 265 ºC, aux gas temp of 350 ºC, S-lens RF at 45. For MS1 scan events, scan ranges were set to 225–650 m/z, mass resolution of 35k, AGC of 1e6 and injection time of 50 ms was used. For tandem MS acquisition, mass resolution of 17.5k, AGC of 1e5, normalized collision energy (NCE) of 30 eV, loop count of 4 and injection time of 50 ms was used.
Lipid metabolites:
Once prepared, 10 μl of sample was injected onto an Agilent EclipsePlus C18 (1.8 μm particle size, 2.1 × 50 mm) column and separated using mobile phases A (0.2% acetic acid in water) and B (50% acetonitrile, 50% isopropyl alcohol, 0.2% acetic acid) running the following gradient: 5% B from 0 to 0.5 min, 5% to 40% B from 0.5 to 3 min, 40% to 80% B from 3 to 9.5 min, 80% to 100% from 9.5 to 11 min, and 100% B from 11min to 13 min, followed by a 2 minute re-equilibration time. The flow rate was set at 0.5 mL/min and the column compartment temperature at 50 ºC. Mass detection was performed using a Thermo QExactive orbitrap mass spectrometer equipped with a heated electrospray ionization (HESI) source and collision-induced dissociation (CID) fragmentation. The source settings used for all samples were: negative and positive ion mode profile data, sheath gas flow of 40 units, aux gas flow of 10 units, sweep gas flow of 2 units, spray voltage of +/− 3.55kV, capillary temperature of 265 ⁰C, aux gas temp of 350 ⁰C, S-lens RF at 45. For MS1 scan events, scan ranges were set to 120–1800 m/z, mass resolution of 35k, AGC of 1e6 and injection time of 100 ms was used. For tandem MS acquisition, mass resolution of 17.5k, AGC of 2e5, normalized collision energy (NCE) of 30 eV, loop count of 4 and injection time of 50 ms was used.
Metabolite Identification and Chemical Networking.
Identification of known signals was performed using commercial standards and comparison of MS/MS spectra to reference spectra. First, accurate mass (within 10 ppm), retention time (within 0.1 minute) and MS/MS fragmentation pattern were searched against reference data from an in-house library of 852 commercial standards. Second, all experimental MS/MS spectra were searched against public repositories of MS/MS data using the GNPS Library Search function (https://gnps.ucsd.edu). Search tolerances included: Accurate Mass < 10ppm, Minimum match score of 0.80, allowed adducts of [M+H, M+Na, M+NH4, M+H-H2O, M-H, M+Acetate, M-H-H2O]. All MS/MS matches were manually inspected and confirmed. In total, 333 compounds across all 3 methods were positively identified. For generation of MS/MS networks, the GNPS Molecular Networking function was used using the following parameters: MZ tolerance of 0.01 Da, Min Matched Peaks of 6, Min Cosine Score of 0.7, TopK of 10, Max Cluster Size of 100, and MS Cluster size of 2. For library matching, the following parameters were used: Min Cosine Score of 0.8, Min Matched peaks of 6, and MS1 tolerance of <10ppm (manually trimmed post creation). Network was visualized using Cytoscape version 3.7.2 (https://cytoscape.org).
Evaluation of technical variance.
We determined relative standard deviation for measurements by analyzing 16 samples (technical replicates) and one blank. Once re-suspended, 6 samples were pooled via transfer to an Eppendorf tube. The pooled and non-pooled DBS supernatant were then run on the LC-MS protocols described above. Dynamic concentration range and linearity of dilution experiments were performed using quality control cards spotted with whole blood with known concentrations of amino acids and acylcarnitines (Lots A1715, B1715, C1715, D1715) obtained from the CDC. Samples were run in quintuplicate with one blank per concentration. Samples were then processed via the methods described above and run on the LC-MS. The studies conducted to assess the impact of location (central vs. peripheral) of blood spotting on metabolite detection and quantification were done using pooled whole blood samples from one card that was punched 5 times from each of the 5 blood spots (1 central and 4 peripheral locations) on the card. One blank was chosen for each of the 5 spot locations. Samples were then processed via the respective optimized extraction methods outlined above and analyzed using the LC-MS protocols described below.
Assessment of Metabolite Stability in DBS at Room Temperature.
Whole blood DBS samples were prepared as described above. All DBS cards were placed into separate Mylar storage bags with an oxygen absorbent packet and desiccant bags. The first sample was designated as the day 0 and immediately placed in an −80 °C freezer. The other samples were then stored at room temperature. Every 24 hours, an additional Mylar bag with samples was placed in the −80 °C freezer over a 7-day period. Metabolite stability DBS samples from day 0 to day 7 were then assessed by extracting metabolites via methods above. Samples were taken in quintuplicate. For these assays, we only selected MS spectral features that were not present in solvent and Whatman card blanks and were detectable in over 80% of quintuplicate samples on Day 0.
Comparison of Plasma Metabolite Analysis in DBS versus Plasma.
For comparison of DBS and plasma measures, plasma samples were prepared according to the following protocols. For metabolite extraction, 20 μL of plasma prepared from matched whole blood were added to 80 μL of organic solvent (ethanol for the Bioactives method and 1:1 ACN:MeOH for the Polar metabolites and Lipids protocols) and mixed in an orbital shaker at 2000 RPM for 10 minutes. Samples were then centrifuged at 4200 RPM at 4 °C for 10 minutes. For Polar metabolite and Lipids methods, 50 μL of supernatant was transferred to a 1 mL 96 well plate containing 200 μL glass inserts and analyzed by LC-MS. For the Bioactives method, samples were prepared as previously described.26
Data handling and analysis.
For data processing, in-house R-scripts were used to perform initial bulk feature alignment, MS1-MS2 data parsing, pseudo DIA-to-DDA MS2 deconvolution, and CSV-to-MGF file generation. RAW to mzXML file conversion was performed using MSconvert version 3.0.9393 (part of the ProteoWizard Software Suite). Feature extraction, secondary alignment, and compound identification were performed using both mzMine 2.21 as well as Progenesis QI software suites. Statistical analysis was performed using R (3.3.3).
RESULTS AND DISCUSSION
Optimization and Validation of Metabolomics Approaches for Dried Blood Spots
Currently, optimized protocols for metabolomics analysis of specific chemical classes of small molecules in DBS are not well established. We therefore developed operating workflows that allow for facile extraction and analysis of diverse metabolites from DBS, followed by nontargeted LCMS-based metabolomics using an Orbitrap mass analyzer. After spotting 50 μL of blood on a Whatman 903 card, metabolites are extracted from a 3 mm punches taken from the DBS and analyzed by reversed-phase and HILIC LC- Orbitrap MS (Figure 1A, Methods). In order to capture the widest possible range of metabolic information from DBS, three independent LC-MS methods were developed for measuring polar small molecules (Polar method), polar bioactive lipids (Bioactives method), and large nonpolar lipids (Lipids method). For each method, metabolite extraction conditions were optimized around the following parameters: choice of organic solvent (methanol, acetonitrile, ethanol, methanol:acetonitrile [MeOH:ACN]), organic solvent-dilution, and final sample/supernatant concentration. For all methods, acetonitrile showed the lowest number of total features owing to its limited solubility profile. Acetonitrile was also prone to cause clogging during liquid handling due to its adhesive precipitate. Methanol and 1:1 MeOH:ACN performed well across all methods, with MeOH:ACN showing best overall performance in the Polar and Lipids methods when reproducibility across multiple extractions, as well as the total number of peaks extracted and average peak intensity, were considered (Figure 1B, left panel-Polar method; Suppl Figure 1B, left panel – Bioactives method). Ethanol performed particularly well in the Bioactives method (Suppl Figure 1A, left panel) across all criteria, and while it did produce a high number of abundant peaks in the Polar method, it also had the highest degree of instability from replicate extractions. For organic solvent to water ratio, 70% organic solvent performed most efficiently for the Lipid method (Suppl Figure 1B, middle panel), while 80% organic solvent was optimal for the Polar (Figure 1B, middle panel) and Bioactive (Suppl Figure 1B, middle panel). Lastly, all methods favored high sample concentrations with Polar and Lipids methods performing best when the supernatant was concentrated two-fold (Figure 1B, right panel; Suppl Figure 1B, right panel) and with the Bioactives method performing best with 150 μL SPE loading (Suppl Figure 1A, right panel). Collectively, these three LC-MS approaches resulted in capture of over 4,400 spectral features from DBS samples. From these spectral features, 333 metabolites were positively identified using commercial standards and matching of experimental MS/MS spectra against reference libraries (Suppl Table 1). Identified metabolites include those related to a number of metabolic diseases, such as phenylketonuria, hyperarginemia, tyrosinemia, lysosomal storage disorders, disorders of fatty acid oxidations, carnitine transport deficiencies, and ornithine transcarbamylase deficiency, among others. In addition, we observed many endogenous metabolites involved in energetic balance including metabolites within the TCA cycle, pyruvic acid pathway, vitamin metabolism, fat oxidation, nicotinamide/NAD, and phosphorylated moieties. Metabolites related to environmental exposures, including pesticides, drugs, diet and medications, were also measured in DBS, several of which have been previously assayed using DBS samples.20
Typically, blood spotting is performed by placement of several drops of whole blood on the center of a DBS paper, with passive diffusion of the biosample through the paper matrix. As such, we next determined whether physical location in relation to the point of spotting influenced metabolite measurement. When peripheral and central blood spot locations were analyzed (Figure 1C), we found that only 36 of 4,405 spectral features (~ 1%) demonstrated greater than two-fold change according to location on the DBS (Figure 1C, Venn diagram), which was consistent across chemical classes (Suppl Figure 1C). Interestingly, among those molecules that were found to change significantly with physical location across the DBS were flavonoids, a class of plant-derived polyphenolic molecules,27 which were consistently found to be higher in the DBS periphery relative to center. While the vast majority of metabolites were highly stable across the DBS, these findings suggest a matrix-analyte interaction may promote variable physical migration for a small subset of metabolites.
To determine the robustness of metabolite measures using DBS, technical replication was evaluated. Across 48 blood replicates, over 80% of assayed metabolites, totaling over 3,500 individual features, exhibited a relative standard deviation (RSD) of <20% with an overall median RSD of 11.2% (Figure 1D), comparable to prior nontargeted LC-MS analysis of human plasma.28 RSD was related to metabolite intensity, with lower intensity features closer to the signal-to-noise threshold exhibiting greater technical variance, as expected (Figure 1D). To determine the relative contribution of metabolite extraction from individual DBS handling vs. LC-MS measure to the total technical variance, extracted metabolites from individual technical replicates were pooled together, mixed, and subjected to repeated measure. Across all metabolites, the difference in RSD between pooled and non-pooled samples was between 1–6% (data not shown), suggesting that DBS handling and metabolite extraction contribute only minimally to total technical variance.
We next evaluated the relative quantitation and dynamic linear range for metabolites measured from DBS using Center for Disease Control (CDC) DBS cards spotted with blood containing known concentrations for 28 metabolites typically assayed for newborn screening.29,30 Relative measure of these metabolites showed excellent linearity across physiologic and pathologic concentration ranges, irrespective of chemical class, with R2 values greater than 0.95 (Figure 1E, Table 1). Collectively, these results highlight optimized approaches for robust, nontargeted mass spectrometry-based metabolomic measurements of thousands of circulating human metabolites from DBS biosamples.
Table 1 –
Relative Quantitation for Known Metabolites from DBS
| Metabolite | Concentration Range (μMol/L) |
R2 |
|---|---|---|
| Alanine | 200 – 600 | 0.97 |
| Arginine | 100 – 300 | 0.99 |
| Citrulline | 25 – 250 | 0.99 |
| Glycine | 300 – 900 | 0.98 |
| Leucine | 100 – 500 | 0.99 |
| Methionine | 50 – 250 | 0.99 |
| Ornithine | 100 – 300 | 0.98 |
| Phenylalanine | 100 – 300 | 0.99 |
| Succinylacetone | 2.5 – 15 | 0.98 |
| Tyrosine | 200 – 600 | 0.99 |
| Valine | 150 – 500 | 0.99 |
| C0 (Free Carnitine) | 10 – 30 | 0.99 |
| C2 (Acetylcarnitine) | 10 – 30 | 0.99 |
| C3 (Proprionylcarnitine) | 4 – 12 | 0.99 |
| C3DC (Malonylcarnitine) | 1.0 – 5.5 | 0.99 |
| C4 (Butyrylcarnitine) | 1.0 – 5.0 | 0.99 |
| C4OH (Hydroxybutyrylcarnitine) | 1.0 – 5.5 | 0.99 |
| C5 (Isovalerylcarnitine) | 0.5 – 3.0 | 0.99 |
| C5DC (Glutarylcarnitine) | 0.5 – 2.5 | 0.99 |
| C5OH (Hydroxyisovalerylcarnitine) | 0.5 – 3.0 | 0.99 |
| C6 (Hexanoylcarnitine) | 0.5 – 2.5 | 0.99 |
| C8 (Octanylcarnitine) | 0.5 – 2.5 | 0.99 |
| C10 (Decanoylcarnitine) | 0.5 – 2.5 | 0.99 |
| C12 (Dodecanoylcarnitine) | 0.5 – 3.0 | 0.99 |
| C14 (Myristoylcarnitine) | 0.5 – 3.0 | 0.99 |
| C16 (Palmitoylcarnitine) | 4.0 – 12 | 0.99 |
| C16OH (Hydroxypalmitoylcarnitine) | 0.25 – 1.5 | 0.99 |
| C18 (Stearoylcarnitine) | 1.0 – 5.0 | 0.99 |
| C18OH (Hydroxystearoylcarnitine) | 0.25 – 1.5 | 0.99 |
Stability of Metabolites in DBS at Room Temperature
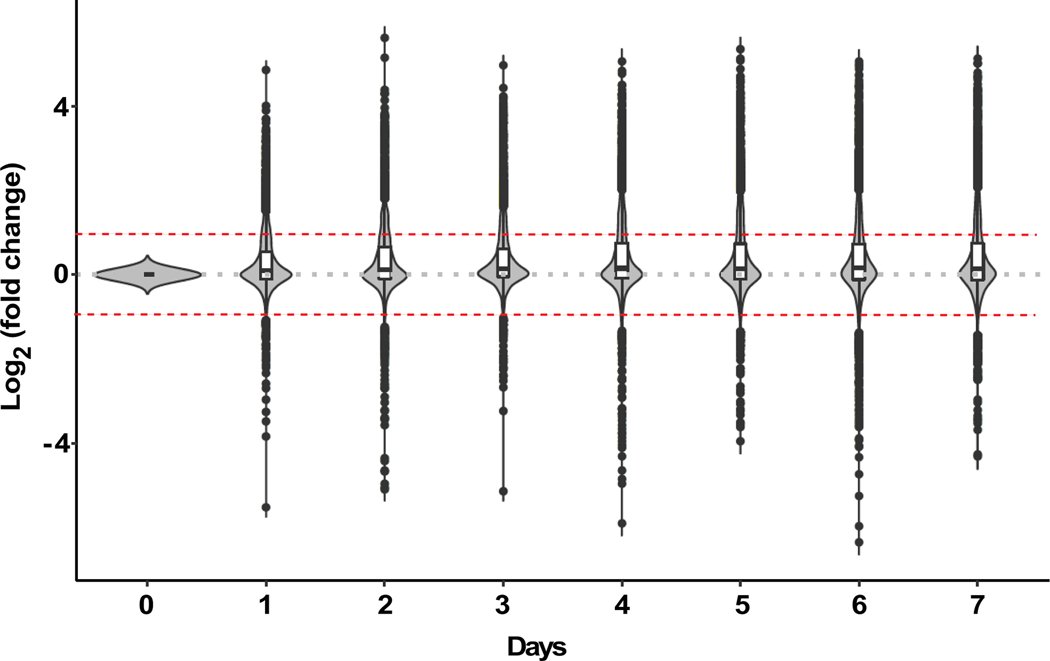
Dried blood spotting functions through rapid desiccation of samples with suspension of enzymatic reactions that may lead to the breakdown of metabolites,31 enabling potential short-term storage at room temperature (RT) prior to analysis. Prior investigation of rodent blood spots showed limited stability of metabolites with storage at RT after 1–4 weeks.32 Similar observations have been made in studies of human newborn DBS18 and adult whole blood DBS33 over periods of time ranging from 1 week up to 2 years after blood spotting; however, a detailed, high-resolution temporal profile of metabolite stability over the first week after blood spotting does not exist to date. Given that one week represents a period of time that would theoretically allow for point of care sampling and transport from most worldwide locations to central facilities for cold storage and analysis, we investigated the stability of blood metabolites in DBS at RT over the first 7 days after spotting. Metabolomic analysis revealed that across the week-long period at RT, less than 20% of the ~4,400 spectral features assayed changed more than 2-fold (Figure 2). Interestingly, among those metabolites found to be altered by RT storage over the 7-day period, significant change in relative concentration was observed primarily during the first 24 hours, suggesting inherent instability for this subset of metabolites (Figure 2). Evaluation of metabolite change across chemical classes revealed that polar metabolites, including common amino acids, creatine, hypoxanthine, uridine and carnitine among thousands of other metabolites changed minimally at RT over the 7-day period (Suppl Figure 2A). Prior work has suggested that these molecules may be susceptible to degradation over much longer periods of time at RT.34 In contrast, we find lipophilic molecules were more prone to fluctuation with RT storage over the 7-day period, with a steady increase in number of measured metabolites over time, suggestive of oxidative conversion and/or breakdown of metabolites into secondary products (Suppl Figure 2B 2C). Overall, small polar bioactive molecules revealed the greatest change at RT, consistent with their known susceptibility to non-enzymatic oxidation (Suppl Figure 2B), similar to previous reports.18 Consistent with this observation, RT storage without a desiccant pouch or an oxygen absorber exacerbated oxidation of small bioactive lipid molecules (data not shown).
Figure 2. Stability of DBS metabolites.
Violin plots showing fold change (represented as log2 fold change) in measured signal intensity for metabolites from DBS stored at room temperature over 7 days. The width of each violin plot represents the number of metabolites present at the corresponding fold-change and therefore indicate that signal intensity for most of the measured metabolites (~80%) change less than 2-fold (represented by red dashed lines) over seven days of storage relative to Day 0. The first and third quartile of the log2 fold-change is represented by the lower and upper boundaries of the white bar in the center of the violin plots, respectively. The median log2 fold-change is represented by the black line in the white bar in the center of the violin plots.
Comparison of Analytes derived from DBS versus plasma
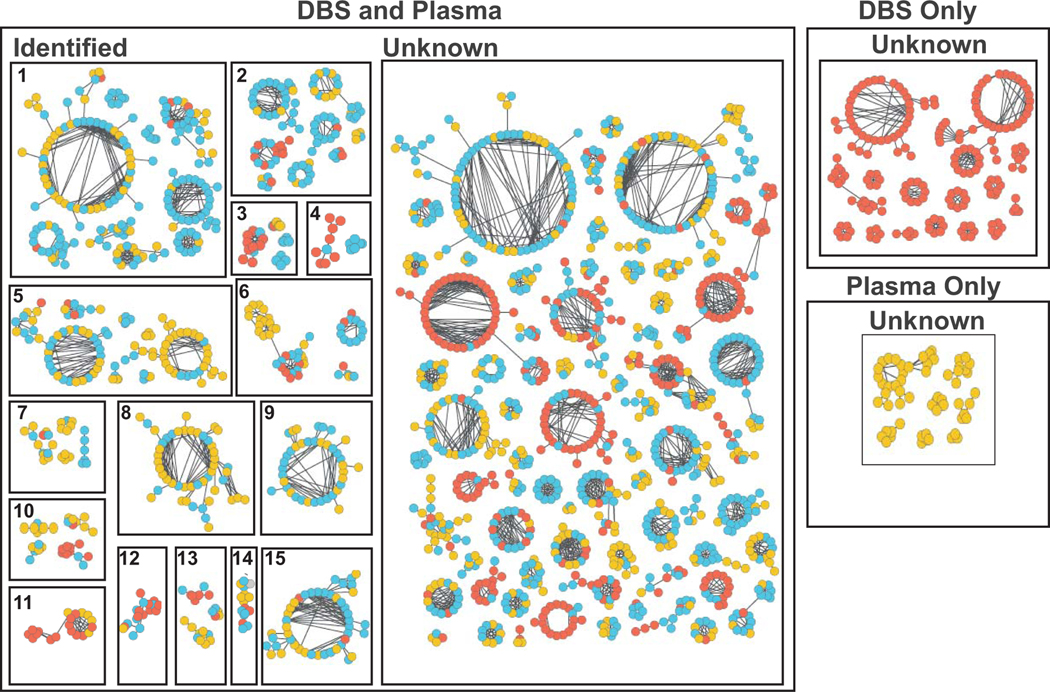
To determine whether DBS introduces sample-matrix interactions that impact metabolite measures, nontargeted LC-MS based metabolomics was performed on matched DBS and plasma samples. Chemical network analysis was then performed to compare metabolites based on MS/MS spectral similarities, with indexing of metabolites into three groups: (1) present in DBS only, (2) present in plasma samples only, and (3) present in both DBS and plasma (Figure 3).28,35,36 The vast majority of metabolite families observed in plasma samples were also observed in DBS samples, including phosphocholines, phosphoethanolamines, phosphoserines, sphingomyelins, oxylipins, phosphoinositols, polyunsaturated fatty acids, sterol and sterol-glucuronides, monosaccharides, bile acids, glutathione and glutathione-conjugates, nucleotides, amino acids, porphyrins, and carnitines, as well as hundreds to thousands of yet unidentified metabolites (Figure 3). A small subset of unknown metabolite families was uniquely present in only plasma or only DBS samples (Figure 3), likely representing metabolites resulting from clotting or enriched in red blood cells, respectively. Collectively, these results suggest that DBS sampling enables capture and measure of diverse metabolites comparable to LC-MS analysis of traditional plasma samples.
Figure 3. Chemical networking of metabolites from DBS and plasma.
LC-MS/MS data collected from DBS and matched plasma samples were subjected to spectral chemical networking and clustered according to MS/MS spectral similarity. Metabolites present in DBS only, plasma only, and both plasma and DBS were denoted in red, yellow, and blue, respectively. Known metabolites include 1) Phosphocholines, 2) Phosphoethanolamines, 3) Phosphoserines, 4) Sphingomyelins, 5) Oxylipins, 6) Phosphoinositols, 7) Polyunsaturated fatty acids, 8) Sterol / Sterol-glucuronides, 9) Monosaccharides, 10) Bile acids, 11) Glutathione / Glutathione-conjugates, 12) Nucleotide phosphates, 13) Amino acids, 14) Porphyrins, and 15) Carnitines.
CONCLUSION
Dried blood spot sampling offers unique opportunities for facile and efficient collection and storage of blood biosamples with subsequent measure of hundreds to thousands of metabolites across diverse chemical classes. Importantly, DBS-based metabolomics was found to be highly robust with technical variance comparable to that of traditional plasma-based LC-MS,10,28 despite additional technical considerations related to puncture of blood spots and extraction of metabolites from the paper matrix. In addition, DBS sampling enables measure of diverse metabolites, from polar small molecules to nonpolar lipids, with comparability to metabolites assayed from traditional plasma samples using nontargeted LC-MS approaches. Finally, metabolites in DBS appear relatively stable at RT, enabling sampling and facile handling in resource-poor areas or remote point-of-care sites, prior to cold storage and analysis in centralized facilities. DBS sampling has established utility in specific clinical contexts such as newborn screening and given the observed applications for broad interrogation of the circulating metabolome. Our data support the need for future investigation of DBS generated from finger stick as a blood collection and storage strategy which may be of high utility in resource-limited areas as well as in population- scale efforts aimed at relating metabolomics measures to indices of health and disease.
Supplementary Material
Supplemental Figure 1. A. Effect of solvent choice (center panel), solvent dilution (middle panel), and supernatant volume (right panel) on number of peaks observed and overall median intensity for small polar lipids (Bioactives). B. Effect of solvent choice (center panel), solvent dilution (middle panel) and supernatant dilution (right panel) on number of peaks observed and overall median intensity for large hydrophilic lipid (Lipids). C. Composition of log2 (fold intensity) changes between center and periphery spots across all 3 classes of metabolites.
Supplemental Figure 2. Fold Change (represented as log2 fold change) in measured signal intensity for different chemical classes of metabolites extracted from DBS stored at room temperature over 7 days. The first and third quartile of the log2 fold-change is represented by the lower and upper boundaries of the white bar in the center of the violin plots, respectively. The median log2 fold-change is represented by the black line in the white bar in the center of the violin plots. Wider sections of the violin plot represent higher frequency of measures in the corresponding range of fold-change. Dashed red lines represent a 2 fold increase or decrease compared to day 0. Although a few analytes show a large fold-change, more change is noted in lipophilic analytes as opposed to hydrophilic analytes (Polar and Bioactives).
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (NIH), including NIH K01DK116917 and P30DK063491 pilot award to J.D.W.; F31CA236405 to I.M., R01CA199376 to S.S., R01HL134168 and R01HL143227 to S.C., and S10OD020025, R01ES027595, and P42ES010337 to M.J.
Footnotes
Notes: The authors declare no competing financial interest.
REFERENCES
- 1.Nicholson JK & Lindon JC Systems biology: Metabonomics. Nature 455, 1054–1056 (2008). [DOI] [PubMed] [Google Scholar]
- 2.van der Greef J. Systems biology, connectivity and the future of medicine. Syst Biol (Stevenage) 152, 174–178 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Yates JR A Century of Mass Spectrometry: from Atoms to Proteomes. Nature Methods 8, 633–637 (2011). [Google Scholar]
- 4.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD & McLean JA Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom 27, 1897–1905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Silva RR, Dorrestein PC & Quinn RA Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A 112, 12549–12550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CA, Want EJ, O’Maille G, Abagyan R & Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Brunius C, Shi L. & Landberg R. Large-scale untargeted LC-MS metabolomics data correction using between-batch feature alignment and cluster-based within-batch signal intensity drift correction. Metabolomics 12, 173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thonusin C, IglayReger HB, Soni T, Rothberg AE, Burant CF & Evans CR Evaluation of intensity drift correction strategies using MetaboDrift, a normalization tool for multi-batch metabolomics data. J Chromatogr A 1523, 265–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizuno H, Ueda K, Kobayashi Y, Tsuyama N, Todoroki K, Min JZ & Toyo’oka T. The great importance of normalization of LC-MS data for highly-accurate non-targeted metabolomics. Biomed Chromatogr 31(2017). [DOI] [PubMed] [Google Scholar]
- 10.Watrous JD, Henglin M, Claggett B, Lehmann KA, Larson MG, Cheng S. & Jain M. Visualization, Quantification, and Alignment of Spectral Drift in Population Scale Untargeted Metabolomics Data. Anal Chem 89, 1399–1404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dried Blood Spots: Applications and Techniques, (John Wiley and Sons, Inc., Hoboken, NJ, 2014). [Google Scholar]
- 12.Hernandes VV, Barbas C. & Dudzik D. A review of blood sample handling and pre-processing for metabolomics studies. Electrophoresis 38, 2232–2241 (2017). [DOI] [PubMed] [Google Scholar]
- 13.McDade TW Development and validation of assay protocols for use with dried blood spot samples. Am J Hum Biol 26, 1–9 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Bell L, Calder B, Hiller R, Klein A, Soares NC, Stoychev SH, Vorster BC & Tabb DL Challenges and Opportunities for Biological Mass Spectrometry Core Facilities in the Developing World. J Biomol Tech 29, 4–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDade TW, Williams S. & Snodgrass JJ What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography 44, 899–925 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Zakaria R, Allen KJ, Koplin JJ, Roche P. & Greaves RF Advantages and Challenges of Dried Blood Spot Analysis by Mass Spectrometry Across the Total Testing Process. EJIFCC 27, 288–317 (2016). [PMC free article] [PubMed] [Google Scholar]
- 17.Drolet J, Tolstikov V, Williams BA, Greenwood BP, Hill C, Vishnudas VK, Sarangarajan R, Narain NR & Kiebish MA Integrated Metabolomics Assessment of Human Dried Blood Spots and Urine Strips. Metabolites 7(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koulman A, Prentice P, Wong MCY, Matthews L, Bond NJ, Eiden M, Griffin JL & Dunger DB The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics 10, 1018–1025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prentice P, Koulman A, Matthews L, Acerini CL, Ong KK & Dunger DB Lipidomic analyses, breast- and formula-feeding, and growth in infants. J Pediatr 166, 276–281.e276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman JD, Rosman LM, Ratcliff JD, Strickland PT, Graham DR & Silbergeld EK State of the Science in Dried Blood Spots. Clin Chem 64, 656–679 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Funk WE, McGee JK, Olshan AF & Ghio AJ Quantification of arsenic, lead, mercury and cadmium in newborn dried blood spots. Biomarkers 18, 174–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma WL, Gao C, Bell EM, Druschel CM, Caggana M, Aldous KM, Louis GM & Kannan K. Analysis of polychlorinated biphenyls and organochlorine pesticides in archived dried blood spots and its application to track temporal trends of environmental chemicals in newborns. Environ Res 133, 204–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrick L, Edmands W, Schiffman C, Grigoryan H, Perttula K, Yano Y, Dudoit S, Whitehead T, Metayer C. & Rappaport S. An untargeted metabolomics method for archived newborn dried blood spots in epidemiologic studies. Metabolomics 13(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denes J, Szabo E, Robinette SL, Szatmari I, Szonyi L, Kreuder JG, Rauterberg EW & Takats Z. Metabonomics of newborn screening dried blood spot samples: a novel approach in the screening and diagnostics of inborn errors of metabolism. Anal Chem 84, 10113–10120 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Chepyala D, Kuo HC, Su KY, Liao HW, Wang SY, Chepyala SR, Chang LC & Kuo CH Improved Dried Blood Spot-Based Metabolomics Analysis by a Postcolumn Infused-Internal Standard Assisted Liquid Chromatography-Electrospray Ionization Mass Spectrometry Method. Anal Chem 91, 10702–10712 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Lagerborg KA, Watrous JD, Cheng S. & Jain M. High-Throughput Measure of Bioactive Lipids Using Non-targeted Mass Spectrometry. Methods Mol Biol 1862, 17–35 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Panche AN, Diwan AD & Chandra SR Flavonoids: an overview. J Nutr Sci 5, e47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watrous JD, Niiranen TJ, Lagerborg KA, Henglin M, Xu YJ, Rong J, Sharma S, Vasan RS, Larson MG, Armando A, Mora S, Quehenberger O, Dennis EA, Cheng S. & Jain M. Directed Non-targeted Mass Spectrometry and Chemical Networking for Discovery of Eicosanoids and Related Oxylipins. Cell Chem Biol 26, 433–442.e434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C.f.D.C.a. Prevention. Laboratory Quality Assurance and Standardization Programs. (2018). [Google Scholar]
- 30.C.f.D.C.a. Prevention. Laboratory Quality Assurance and Standardization Programs. (2019). [Google Scholar]
- 31.Elbin CS, Olivova P, Marashio CA, Cooper SK, Cullen E, Keutzer JM & Zhang XK The effect of preparation, storage and shipping of dried blood spots on the activity of five lysosomal enzymes. Clin Chim Acta 412, 1207–1212 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Michopoulos F, Theodoridis G, Smith CJ & Wilson ID Metabolite profiles from dried blood spots for metabonomic studies using UPLC combined with orthogonal acceleration ToF-MS: effects of different papers and sample storage stability. Bioanalysis 3, 2757–2767 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Prentice P, Turner C, Wong MC & Dalton RN Stability of metabolites in dried blood spots stored at different temperatures over a 2-year period. Bioanalysis 5, 1507–1514 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Kyle JE, Casey CP, Stratton KG, Zink EM, Kim Y-M, Zheng X, Monroe ME, Weitz KK, Bloodsworth KJ, Orton DJ, Ibrahim YM, Moore RJ, Lee CG, Pedersen C, Orwoll E, Smith RD, Burnum-Johnson KE & Baker ES Comparing Identified and Statistically Significant Lipids and Polar Metabolites in 15-Year old Serum and Dried Blood Samples for Longitudinal Studies. Rapid communications in mass spectrometry 5, 447–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N. & Dorrestein PC Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci U S A 109, E1743–1752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, Jarmusch AK, Vargas F, Aksenov AA, Gauglitz JM, Weldon K, Petras D, da Silva R, Quinn R, Melnik AV, van der Hooft JJJ, Caraballo-Rodríguez AM, Nothias LF, Aceves CM, Panitchpakdi M, Brown E, Di Ottavio F, Sikora N, Elijah EO, Labarta-Bajo L, Gentry EC, Shalapour S, Kyle KE, Puckett SP, Watrous JD, Carpenter CS, Bouslimani A, Ernst M, Swafford AD, Zúñiga EI, Balunas MJ, Klassen JL, Loomba R, Knight R, Bandeira N. & Dorrestein PC Mass spectrometry searches using MASST. Nat Biotechnol 38, 23–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. A. Effect of solvent choice (center panel), solvent dilution (middle panel), and supernatant volume (right panel) on number of peaks observed and overall median intensity for small polar lipids (Bioactives). B. Effect of solvent choice (center panel), solvent dilution (middle panel) and supernatant dilution (right panel) on number of peaks observed and overall median intensity for large hydrophilic lipid (Lipids). C. Composition of log2 (fold intensity) changes between center and periphery spots across all 3 classes of metabolites.
Supplemental Figure 2. Fold Change (represented as log2 fold change) in measured signal intensity for different chemical classes of metabolites extracted from DBS stored at room temperature over 7 days. The first and third quartile of the log2 fold-change is represented by the lower and upper boundaries of the white bar in the center of the violin plots, respectively. The median log2 fold-change is represented by the black line in the white bar in the center of the violin plots. Wider sections of the violin plot represent higher frequency of measures in the corresponding range of fold-change. Dashed red lines represent a 2 fold increase or decrease compared to day 0. Although a few analytes show a large fold-change, more change is noted in lipophilic analytes as opposed to hydrophilic analytes (Polar and Bioactives).