Abstract
Background
Little is known about chronic cannabis smoking-associated oral microbiome and its effects on central nervous system (CNS) functions.
Methods
In the current study, we have analyzed the saliva microbiome in individuals who chronically smoked cannabis with cannabis use disorder (n = 16) and in non-smoking controls (n = 27). The saliva microbiome was analyzed using microbial 16S rRNA sequencing. To investigate the function of cannabis use-associated oral microbiome, mice were orally inoculated with live Actinomyces meyeri, Actinomyces odontolyticus, or Neisseria elongata twice per week for six months, which mimicked human conditions.
Findings
We found that cannabis smoking in humans was associated with oral microbial dysbiosis. The most increased oral bacteria were Streptococcus and Actinomyces genus and the most decreased bacteria were Neisseria genus in chronic cannabis smokers compared to those in non-smokers. Among the distinct species bacteria in cannabis smokers, the enrichment of Actinomyces meyeri was inversely associated with the age of first cannabis smoking. Strikingly, oral exposure of Actinomyces meyeri, an oral pathobiont, but not the other two control bacteria, decreased global activity, increased macrophage infiltration, and increased β-amyloid 42 protein production in the mouse brains.
Interpretation
This is the first study to reveal that long-term oral cannabis exposure is associated oral enrichment of Actinomyces meyeri and its contributions to CNS abnormalities.
Keywords: Cannabis smoking, Oral microbiome, Actinomyces meyeri, β-amyloid
Research in context.
Evidence before this study
Previous studies show oral administration of Campylobacter jejuni and Porphyromonas gingivalis resulted in central nervous system (CNS) abnormalities, suggesting a link between oral microbiome and CNS function.
Added value of this study
In the current study, we found that chronic cannabis use was associated with oral microbial dysbiosis. A. meyeri was highly enriched in the saliva from chronic cannabis smokers compared to those of non-smokers, and its oral enrichment was associated with the age of first cannabis use. We further investigated if direct administration of this bacterium, in the absence of cannabis or its components, could elicit an alteration in the oral-microbiome-brain axis in a mouse model. Long-term (∼6 months) oral inoculation of A. meyeri bacterium to mice resulted in behavioral changes, macrophage infiltration, and increased Aβ 42 protein production in the brain.
Implications of all the available evidence
Our findings reveal the importance of oral health in relation to CNS functions. This study may help develop novel treatments targeting the microbiota or its active molecules for prevention or treatment of some neurological abnormalities.
Alt-text: Unlabelled box
1. Introduction
The effects of cannabis on human health have been extensively studied, and public acceptance of cannabis for medicinal and recreational use grows worldwide. Cannabis use can result in a temporary relaxation of dysphoria and anxiety [1,2]. However, numerous studies in both humans and animals have demonstrated that cannabis use impairs neural functioning in a variety of cognitive and performance tasks, such as memory, learning, attention, perception, and motor coordination [3], [4], [5], [6]. Frequent cannabis smoking is associated with lower white-matter integrity [7], blunted psychotomimetic effects, perceptual alteration, cognitive impairment, and increasing cortisol levels [8].
Some psychoactive components of cannabis were found to affect both neural and immune systems. Δ9-Tetrahydrocannabinol (THC), a primary psychoactive component of cannabis, can directly bind to cannabinoid receptors, which are distributed extensively in the brain [9]. THC also exhibits a wide range of immunosuppressive effects, which may decrease host defense to infections [10,11]. The connection and interaction between the gut microbiome and CNS function via the microbiota-gut-brain axis have been studied [12], but the oral cavity also hosts a large community of different bacteria that interact with each other and with the host. Healthy oral microbiomes resist pathogen colonization in the mouth [13], and when the oral microbiome is compromised, opportunistic pathogenic bacteria may colonize and lead to new translocation events to the circulation that contribute to systemic disease pathogenesis [14,15]. Oral bacteria can enter into the systemic circulation through inflamed gingiva and thereby affect peripheral organs and the CNS [16,17]. In mice, oral administration of Campylobacter jejuni activated visceral sensory nuclei in the brainstem that processed gastro-intestinal sensory information [18]; Porphyromonas gingivalis, an oral pathogen contributing to the development of chronic periodontitis, may be a risk factor for developing amyloid-beta (Aβ) plaques, cognitive impairment, and dementia [19,20]. Further, exposure to viral or bacterial pathogens upregulates neuronal Aβ expression in nontransformed cell culture models and wild-type rat brains, which may represent a naive antimicrobial defense response [21].
Cannabis smoking alters the oral environment and produces numerous chemicals that directly interact with oral bacteria. Some of the chemicals are toxicants and may perturb the oral microbial ecology. Whether chronic cannabis use affects CNS function through dysbiosis of oral microbiome remains unknown. In this study, we found that saliva Actinomyces, Veillonella, Megasphaera, and Streptococcus bacteria were increased, and Neisseria bacteria were decreased in cannabis smokers compared to non-smokers. Two Actinomyces species bacteria (A. meyeri and A. odontolyticus) and one control Neisseria species bacterium (N. elongata) were inoculated to the B6 mice via oral inoculation. A. meyeri administration resulted in reduced global mouse activity, macrophage infiltration, and increased Aβ 42 protein production in the brain.
2. Materials and methods
2.1. Subjects
Non-smoking controls were recruited from the Medical University of South Carolina (MUSC) and University of Connecticut by advertisement on the campus and chosen by self-report of non-drug use. Cannabis-smoking individuals with cannabis use disorder were recruited from the Addiction Center at MUSC. This study was approved by MUSC institutional review boards. All participants provided written informed consent. The cannabis smoking cohort included 16 cannabis smokers and 27 non-smoking controls. The cannabis smokers were on current non-injection cannabis use but not on prescription drug and other illicit drug uses identified by chart reviews and urine tests. The clinical characteristics of cannabis smokers were shown in Supplementary Table 1. We conducted a Timeline Follow-back (TLFB) method [22], a web-based self-administered, to assess frequency and quantity of past 90-day cannabis uses prior to the study visit. Whether the participants used cannabis (yes/no) and the number of joints, blunts, pipes, bowls, vaporizers, spliffs, edibles, or other methods used. If participants shared a joint/blunt/etc. or otherwise did not use a full joint/blunt/etc., partial numbers were reported. Daily gram calculations of THC uses were calculated (Supplementary Table 2).
2.2. Cannabis use and psychiatric disorder diagnostic/descriptive assessment
For individuals with cannabis use disorder, we assessed exclusionary psychiatric diagnoses using appropriate modules of the Mini-International Neuropsychiatric Interview, as described in our previous study [23]. Briefly, The M.I.N.I., a brief structured interview, was to assess current Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-V) diagnoses. Because the M.I.N.I. only assesses current diagnoses and a more thorough history of substance use is needed, the substance use module of the Structured Clinical Interview for DSM-V was used for substance use disorder diagnosis. Drug screens were performed using the onTrak test cup, an in vitro diagnostic test for the qualitative detection of drug or drug metabolite in the urine. Results of urine screenings were used to substantiate self-reports of cannabis use.
2.3. Mice
C57BL/6 mice were purchased from the Jackson Laboratories and housed at the MUSC. All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the MUSC. Mice were anesthetized with isoflurane for 5 min, and the teeth were brushed to help oral bacteria colonization. The teeth brushing was performed carefully to avoid damage to the alveolar ridge. Next, 12–14-week-old female mice (10 mice for each group) were orally inoculated with live A. meyeri (ATCC, strain: VPI 10648), A. odontolyticus (BEI Resources, strain: F0309), or N. elongata (ATCC, strain: NCTC 10660), as well as PBS, twice per week for 24 weeks. The live bacteria were given 2 × 107 colony forming units (CFU) in 5 µl PBS per mice at each inoculation.
2.4. Saliva microbiome
-
•
Sample collection and DNA extraction. The volunteers were asked to rinse their mouths with water to remove food debris and to spit out the water. The subjects were asked to collect their saliva drool into a 50 mL sterile Falcon tube. This process was performed several times to collect about 2 ml. Collected saliva was centrifuged at 6000 × g at 4°C for 30 min. Total microbial DNA was extracted from saliva samples using a QIAamp DNA Microbiome Kit (QIAGEN) according to the manufacturer's standard protocol. DNA extracts were quantified using the Quant-iT PicoGreen kit (Invitrogen, ThermoFisher Scientific).
-
•
Sequencing. 16S rRNA gene amplicons covering variable regions V3 to V4 were generated using primers (515F and 806R with Illumina adapters and 8 base pair dual indices). The PCR reaction was incubated at 94°C for 3.5 minutes, followed by 30 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 90 seconds, and a final elongation step at 72°C for 10 minutes.. PCR products were pooled for quantification and purified using the Qiaquick PCR purification kit (QIAGEN Inc.) following the manufacturer's specifications. 16S libraries were sequenced on the MiSeq using a v2 2 × 250 base-pair kit (Illumina, Inc) at the Microbial Analysis, Resources, and Services (University of Connecticut, Storrs, CT).
-
•
Data analysis. Sequences were processed using the Quantitative Insights Into Microbial Ecology (QIIME) 1 pipeline and demultiplexed [24] at the Microbial Analysis, Resources, and Services, University of Connecticut. Poor-quality sequences were removed using the default settings of the QIIME 1 script split_libraries.py (minimum average quality score = 25, minimum/maximum sequence length = 200/1000 base pairs; no ambiguous base calls and no mismatches allowed in the primer sequence). After demultiplexing and quality filtering, sequences were clustered into de novo operational taxonomic units (OTUs) based on 97% sequence similarity, and representative sequences were assigned taxonomy based on fully sequenced microbial genomes (IMG/GG GreenGenes) [24]. Chimeric sequences were identified using ChimeraSlayer [25]; sequences failing the alignment and singleton OTUs were removed.
2.5. Mouse behavioral tests
The open field spontaneous behavior and locomotor activity of mice were measured in a circular arena (45 × 60 cm in diameter and height, respectively). Dim light intensity was 60 lux throughout the arena. Animals were placed in the center of the arena for 10 min. The spontaneous behavior and activity of mice were tracked and recorded using a camera and SMART video tracking software v 3.0 (Panlab, Whitehall, PA).
2.6. Aβ measurement in brain
The Aβ measurement was performed according to methods previously described [26]. Forebrains without cerebellum were placed in 0.5 ml homogenization buffer (50mM Tris pH 8.0, 150mM NaCl, 5mM EDTA) with protease/phosphatase inhibitor cocktail (Cell Signaling, Beverly, MA) on wet ice. The brain was homogenized using a tissue homogenizer at 5,500 rpm for 30 seconds. The homogenized brain tissue was aliquoted and stored at −80°C until use. For Aβ measurements, the aliquoted homogenates (200 µl) were thawed on ice, 640 µl pre-cold formic acid (minimum 96% purity, Sigma-Aldrich, St Louis, MO) was added to each sample. The ratio of homogenates with formic acid was 1:3.2. The mixtures were sonicated for 35 s on ice, then spun at 23,000 × g at 4°C for 1 h. The supernatant was collected, and neutralization buffer (1 M Tris base, 0.5 M Na2HPO4, 0.05% NaN3 (wt/vol)) was then added at a 1:20 ratio. An observer measured aβ 40 and Aβ 42 concentrations blinded to the treatment groups with an electrochemiluminescence-linked immunoassay V-PLEX Aβ Peptide Panel 1 (4G8) Kit (Meso Scale Discovery, Gaithersburg, MD). The brain extracts were diluted using 1:2 in the buffer (Diluent 35; Meso Scale Discovery) before measurement and then performed according to the manufacturer's instructions. Data analysis was used MSD Discovery Workbench software 2.0 (Meso Scale Discovery).
2.7. Immunohistochemistry
Mice were perfused with cold PBS and 4% paraformaldehyde (PFA, Sigma-Aldrich, Saint Louis, MO). Mouse brains were post-fixed in 4% PFA for 24 h, tissue samples were processed and embedded with paraffin wax. Serial sections of 5 μm nominal thickness were used for subsequent assays. Following clearing in xylene for 10 min, sections were rehydrated in 100% ethanol, followed by 95% and 70% ethanol in water. Next, sections were incubated with 3% H2O2 for inactivation of endogenous peroxidase. Sections were washed in PBS and immune-stained with anti-mouse CD45 (RRID:AB_2799780, Cell Signaling Technology, Danvers, MA) overnight at 4 °C. Then, sections were washed and treated with peroxidase-labeled secondary antibodies for 1 h at room temperature. Visualization was accomplished with the development of 3,3 diaminobenzidine tetrahydrochloride (DAB, Vector laboratories, CA, USA). Identical conditions and reaction times were used for slides from different animals. Reactions were stopped by immersion of slides in distilled water. Slides were imaged using an AX10 Vert A1 Microscope with ZEN2 software (ZEISS, Oberkochen, Germany). Identical conditions were set in the histogram display (Black: 2800, Gama: 0.45, white: 16,000) to allow comparison between immunoreactivity optical densities. The infiltration of macrophages in the sagittal brain sections of mice was identified by CD45 high expression and macrophage morphology using IHC staining. The CD45 expression was calculated by converting each pixel to grayscale after creating a reversed image using Image J (https://imagej.nih.gov/ij/index.html).
2.8. Quantification of bacteria by Quantitative real-time PCR (qPCR)
After 1 week of last time orally inoculated with live A. meyeri, A. odontolyticus, or N. elongata, the mice were sacrificed. Oral swab samples were collected using cotton swab on the surface of gum and tongue. The stool samples were collected from colon. Total microbial DNA was extracted from oral swab or stool samples using a QIAamp DNA Microbiome Kit (Qiagen) according to the manufacturer's standard protocol. Microbial DNAs were quantified using the Quant-iT PicoGreen kit (ThermoFisher Scientific). qPCR was performed using a CFX Connect Real-time PCR Detection System (Bio-Rad). Target and reference sequences were amplified from 10 ng of DNA template in a 20 μl amplification reaction consisted of 10 μl of 2x PerfeCTa SYBR Green SuperMix (Quanta, Gaithersburg, MD) and 0.3 μM forward and reverse primers (Supplementary Table 3). The reaction conditions for DNA amplification were 95°C for 5 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. All reactions were performed in triplicate, and mean values were calculated. Cycle threshold (Ct) values of target bacteria amplification were normalized by the Ct values of total 16S rDNA using the delta Ct (ΔCt, target bacteria Ct - 16S rDNA Ct) method. The bacteria amplification results were shown using 2−ΔCt.
2.9. Plasma levels of cytokines
The human plasma levels of the following 33 cytokines were measured: Interleukin (IL)-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12/IL-23p40, IL-12p70, IL-15, IL-16, IL-17A, IL-21, IL-22, IL-27, IL-31, Eotaxin, Eotaxin-3, Granulocyte-macrophage colony-stimulating factor (GM-CSF), Interferon-γ (IFN-γ), Interferon gamma-induced protein 10 (IP-10), Monocyte chemoattractant protein (MCP)-1, MCP-4, Macrophage-derived chemokine (MDC), Macrophage inflammatory protein (MIP)- 1α, MIP-1β, MIP-3α, Thymus and activation-regulated chemokine (TARC), Tumor necrosis factor (TNF)-α, TNF-β, Vascular endothelial growth factor (VEGF). All levels were measured simultaneously using Human Th17/Cytokine/Chemokine/ProInflam kit (Meso Scale Diagnostics, Rockville, Maryland) according to the manufacturer's instructions.
The mouse plasma levels of the following 18 cytokines were measured: IL-1β, IL-2, IL-4, IL-5, IL-6, IL-9, IL-12p70, IL-15, IL-17A/F, IL-27p28/IL-30, IL-33, IFN-γ, IP-10, Keratinocyte chemoattractant/human growth-regulated oncogene (KC/GRO), MCP-1, MIP-1α, MIP-2, TNF-α. All levels were measured simultaneously using V-PLEX Mouse Cytokine 19-Plex Kit (Meso Scale Diagnostics) according to the manufacturer's instructions.
2.10. Proinflammatory cytokines detection
Peripheral blood mononuclear cells (PBMCs) from healthy individuals were isolated over a Ficoll-Hypaque cushion (GE, Pittsburgh, PA). After the bacteria were heat-inactivated at 60°C for 30 min, heat-inactivated A. meyeri, A. odontolyticus, and N. elongata were added to cell culture at a final concentration of 1 × 107 CFU/ml; LPS from E.coli 055:B5 (LPS-B5, invovoGen) was used as a positive control with a final concentration of 2 ng/ml. PBMCs were cultured with brefeldin A (5 μg/ml, BD) and incubated at 37°C for 5 h. Cells were then collected and washed with PBS, followed by a 20 min incubation with 50 μl aqua blue (Life Technologies, Carlsbad, CA) at 4°C to exclude dead cells. Next, 50 μl of an antibody cocktail containing anti-human CD3 (RRID:AB_2869824) and anti-human CD14 (RRID:AB_396848) were used for surface staining. After washing and permeabilization, cells were intracellularly stained with anti-human TNF-α (RRID:AB_2204110), anti-human IL-1β (RRID:AB_400438), and anti-human IL-6 (RRID:AB_397228). Fluorescence-labeled antibodies were purchased from BD or Biolegend (San Diego, CA). After washing, cells were collected and analyzed using a BD FACSVerse flow cytometer (BD). Data were analyzed using the FlowJo software (Version 10.0.8).
2.11. Cell migration
Cell migration was performed in 6.5 mm diameter and 5 μm pore polycarbonate filter transwell (Millipore) in 24-well plates. THP1 cells (5 × 104 cells per well) were seeded onto the upper chamber of the insert. 5 × 104 THP-1 cells (ATCC, TIB-202) in 0.5 ml RPMI-1640 medium containing 10% FBS (culture medium) were seeded onto the upper chamber, and the lower well contained 0.5 ml of culture medium. The medium in both the upper and lower chambers contained 10 ng/ml (16 nM) of 12-O-tetradecanoylphorbol-l3-acetate (PMA). The plates were incubated at 37 °C in 5% CO2. After 24 h, the THP-1 cells were washed three times with culture medium; the transwell inserts were then placed into the 24 well containing 0.5 ml culture medium. 100 µl of RPMI-1640 without FBS was added into the upper chamber. The heat-inactivated A. meyeri, A. odontolyticus, or N. elongata (1 × 107/ml), or LPS (2 ng/ml) were also added into the upper chamber in the presence or absence of 100 μM of MyD88 homodimerization inhibitory peptide (IMG2005) (IMGENEX, San Diego, CA) or control peptide. After another 24 h culture, the transwell insert was placed into the 70% ethanol solution for 10 min to allow cell fixation. The non-migrating cells were then carefully removed from the upper surface (inside) of the transwell with a wet cotton swab. 600 μl of 0.2% crystal violet was added into wells of 24-well plates to allow the membrane to stain for 5–10 min. Next, the membrane was dipped into distilled water five times to remove the excess crystal violet. After the transwell membrane dried, the cells were counted in different fields of view to obtain average cell counts migrated through the membrane.
2.12. THP-1 cell phagocytosis
The heat-inactivated A. meyeri, A. odontolyticus, or N. elongata (1 × 1010/ml) were sonicated for 60 s on ice. The bacteria lysates were treated using DNase I (20 µg/ml) in 37°C for 10 min, then treated using Proteinase K (100 µg/ml) at 55°C for 30 min and followed 70°C for 10 min. The bacteria lysates for cell phagocytosis were spun at 13,000 × g at 4°C for 20 min. The supernatant was aliquoted and stored at −80°C until use. THP-1 cells with 1 × 105/0.5ml/well were added into a 48 well-plate. Next, THP-1 cells were treated using 10 ng/ml (16 nM) PMA for 48 h and then changed with fresh media without PMA to culture for another 24 h. The bacteria lysates for cell phagocytosis (1: 1000 dilution), or LPS-B5 (2 ng/ml) were used to stimulate cells for 24 h, the THP-1 cells were washed two times with 1 x PBS. Fluorescence labeled bacteria (pHrodo Green E. coli BioParticles) were added at the final concentration of 50 ug/ml and cultured for 1 h. Cells were washed with PBS once and detached using cell scratch. Cells were collected and analyzed using a BD FACSVerse flow cytometer (BD). Data were analyzed using the FlowJo software (Version 10.0.8).
2.13. Monocyte-derived macrophage (MDM) phagocytosis
Monocytes were enriched in PBMCs using CD14 MicroBeads (positive selection, Miltenyi Biotec, Bergisch Gladbach, Germany, purity > 90%) and cultured in RPMI-1640 medium with 10% FBS and supplemented with 50 ng/ml M-CSF. After 4 days of culture, all monocytes had differentiated into macrophages (M0) with more than 90% confluency. Next, the M0 macrophages were washed twice with fresh media and cultured with bacteria lysates (1: 1000 dilution) or LPS-B5 (2 ng/ml) for 24 h. The MDM cells were washed twice with 1 x PBS and cultured with fluorescence-labeled bacteria (pHrodo Green E. coli BioParticles Conjugate for Phagocytosis) at a final concentration of 50ug/ml for 1 h. The cells were washed with PBS once and detached using a cell scraper. Cells were collected and analyzed using a BD FACSVerse flow cytometer (BD). Data were analyzed using the FlowJo software (Version 10.0.8).
2.14. Plasma levels of bacterial antigen-specific antibodies
After adding the protease/phosphatase inhibitor cocktail (Cell Signaling), the Heat-inactivated A. meyeri, A. odontolyticus, N. elongata (1 × 1010/ml) were sonicated for 60 s on ice. The bacteria lysates were spun at 13,000 × g at 4°C for 20 min. The supernatant was aliquoted and stored at −80°C until use. Microtiter plates were coated with 1:200 diluted bacteria lysates at 4°C overnight. Microwells were then washed three times with PBS wash buffer (0.1% Tween 20). The reaction was blocked using blocking buffer (KPL, Milford, MA) for 120 min at 37°C. After washing, diluted plasma was added to each well for 1 h at room temperature. After washing, horseradish peroxidase-labeled goat anti-human IgG (KPL, Gaithersburg, MD) was added at a 1:5000 dilution in PBS containing 3% BSA and incubated for 60 min at room temperature. After washing, 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) substrate solutions were used to detect binding. Absorbance was measured at 405 nm emission within 30 min.
2.15. Statistical analysis
Conventional measurements of central location and dispersion were used to describe the data, and the Multivariate Welch t-test, non-parametric Mann-Whitney's U tests, or analysis of variance (ANOVA) compared differences in continuous measurements between two or more than two groups. ANOVAs were followed by Tukey's post hoc to correct for multiple comparisons when appropriate. P values of two-group difference were adjusted for age, sex, and alcohol use by fitting log-normal linear regression models. Comparison analysis was performed using R (version 3.6.3) or GraphPad Prism 8. All tests were two-sided, and p ≤ 0.05 was considered to denote statistical significance.
2.16. Ethics approval and consent to participate
This study was approved by Medical University of South Carolina institutional review boards (IBC-2020-00948). All participants provided written informed consent. All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Medical University of South Carolina (IACUC-2019-00858).
2.17. Role of the funding source
The funders did not have any role in the design of the study; the collection, analysis, or interpretation of the data; writing of the report; or in the decision to submit the paper for publication.
3. Results
3.1. Cannabis smoking associates with oral microbial dysbiosis
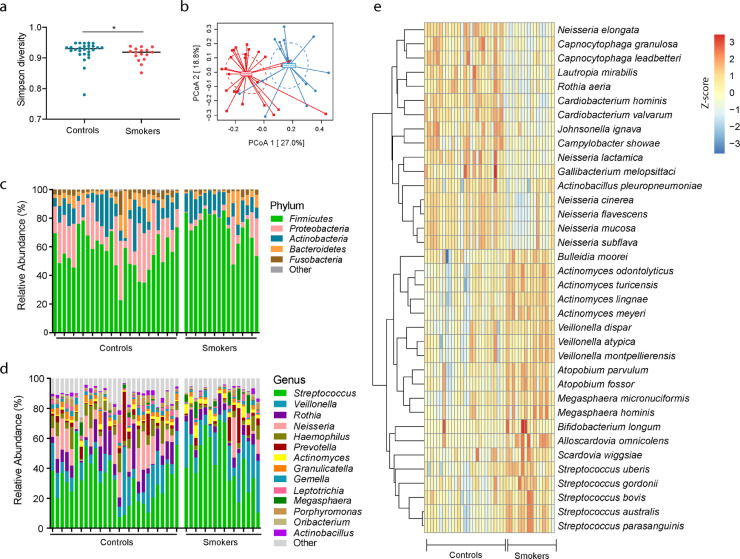
The saliva microbiome was analyzed and compared between cannabis smoking individuals and non-smoking control individuals. We found that cannabis smoking was associated with decreased oral microbial diversity compared to those in the non-smoking control group (Shannon diversity) (Fig. 1a). The oral microbial communities differed in the two study groups reflected by the β-diversity (Bray-Curtis algorithm) (Fig. 1b).
Fig. 1.
Oral microbiome in cannabis smokers (smokers) and non-smoking controls (controls). (a) The Gini Simpson diversity index (α-diversity) was used to compare the diversity of the oral microbial community between cannabis smokers and controls. (b) PCoA was conducted based on the Bray-Curtis dissimilarity distance to determine the beta diversity in cannabis smokers and controls. The statistical significance of the beta diversity was tested using the Multivariate Welch t-test. (c) The average relative abundance of each bacteria at the phylum level. (d) The average relative abundance of each bacteria at the genus level. (e) The species-level of the significantly different oral microbiome in cannabis smokers compared to controls after adjusting for FDR (p < 0.05), which included 16 species decreased and 20 species increased in cannabis smokers compared to controls.
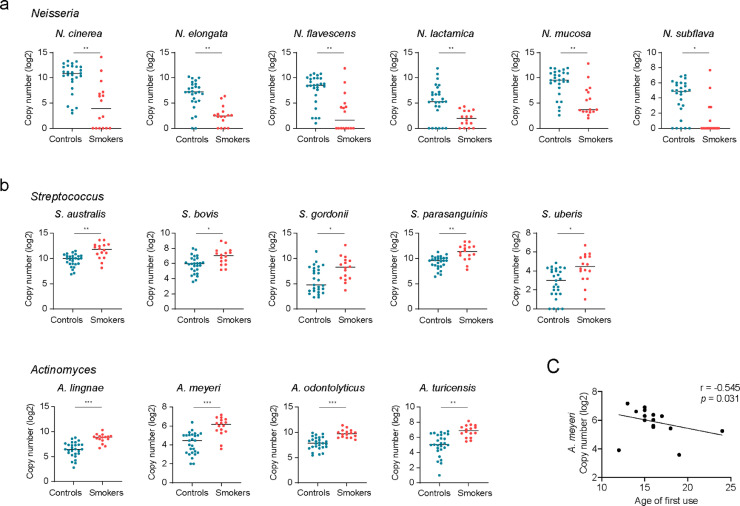
Next, we examined the relative abundance of individual bacterial taxa. The phylum Proteobacteria were decreased in cannabis smokers compared with those of non-smoking controls (Fig. 1c). At the genus level, enrichment of Neisseria was lower in cannabis smokers than in non-smokers; in contrast, Actinomyces, Veillonella, Megasphaera, and Streptococcus were found to increase among cannabis smokers (Fig. 1d). At the species level, 36 species were significantly different in cannabis smokers compared to non-smoking controls after adjusting for multiple comparisons (P < 0.05, Benjamini and Hochberg false-discovery rate, FDR), including 16 species that were decreased and 20 species that were increased in cannabis smokers compared to non-smoking controls (Fig. 1e). Among the taxa that were increased in the saliva of cannabis smokers, five belonged to the Streptococcus genus, and four belonged to the Actinomyces genus. Among the taxa that were enriched in the saliva of non-smoking controls, six belonged to the Neisseria genus (Fig. 2a, b). No differences were observed based on quantity of cannabis use or presence of neurological disease history between the heavy users (> 4 times/week) and light users (≤ 4 times/week), and between smoked cannabis containing THC and cannabis containing no THC (Supplemental Table 1). Nonetheless, the enrichment of A. meyeri was inversely correlated with the age of first cannabis use (Fig. 2c).
Fig. 2.
The top distinct oral microbiome taxa in cannabis smokers compared with non-smoking controls. (a) The most decreased oral microbiome taxa in cannabis smokers, which belonged to Neisseria genus. (b) The most increased oral microbiome taxa in cannabis smokers, which belonged to Streptococcus and Actinomyces genus. (c) A correlation between A. meyeri abundance in the oral microbiome and the age of first-time cannabis smoking. (Nonparametric Mann-Whitney's U tests and Spearman correlation test. p-values were adjusted for FDR. *p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001.)
To study which taxa may represent the cannabis smoking oral microbiome, we reanalyzed and compared the saliva microbiome from tobacco smokers and non-smoker controls from our published data [27]. Consistent with the results from previously published studies [28,29], we found increased Streptococcus and decreased Neisseria in the oral microbiome of tobacco smokers compared with those in non-smoking controls (Fig. S1a), which was similar to cannabis smokers. However, Actinomyces genus was only increased in cannabis smokers but not in tobacco smokers (Fig. S1a). To further analyze Actinomyces genus bacteria, we have shown four Actinomyces species bacteria that were significantly increased in cannabis smokers (Fig. S1b). Only Actinomyces turicensis was increased in tobacco smokers when compared with non-smoker controls (Fig. S1b). We further analyzed the difference after adjusting for age, sex, and/or alcohol use. All differences between cannabis users and controls identified in the univariate analysis shown in Fig. S1 remained significant after adjusting for sex, age, and alcohol consumption, although P values were attenuated slightly (adjusted P < 0.001 for Actinomyces and A. odontolyticus; adjusted P < 0.01 for Neisseria, A. lingnae, A. turicensis, and A meyeri; adjusted P < 0.05 for Streptococcus). When comparing tobacco users to controls, only Streptococcus (adjusted P < 0.01) and A. turicensis (adjusted P < 0.05), but not Neisseria, remained significant after adjusting for sex and age.
Cannabis smokers have shown premalignant lesions in the oral mucosa with surface decay relative to a control group [30]. The smoking and altered microbiome composition may lead to a compromised mucosal epithelial barrier, which results in the translocation of bacteria or microbial products into circulation. Thus, bacterial fragments or whole bacteria can appear in the blood from translocation and thereby influence the immune system [31,32]. To study oral microbial translocation in cannabis users, we evaluated the plasma levels of IgG antibody against antigens derived from A. meyeri, A. odontolyticus, and N. elongata. Plasma levels of IgGs against A. meyeri antigens tended to increase in the cannabis smokers compared to controls (p = 0.059, Fig. S1c), while similar levels of IgGs against antigens from the other two bacteria were observed (Fig. S1c). These results imply that A. meyeri or its antigens may preferentially translocate from the oral mucosa to the circulation in the setting of an altered oral or periodontal environment in cannabis smokers.
3.2. A. meyeri oral inoculation for six months results in CNS abnormalities in C57BL/6 mice
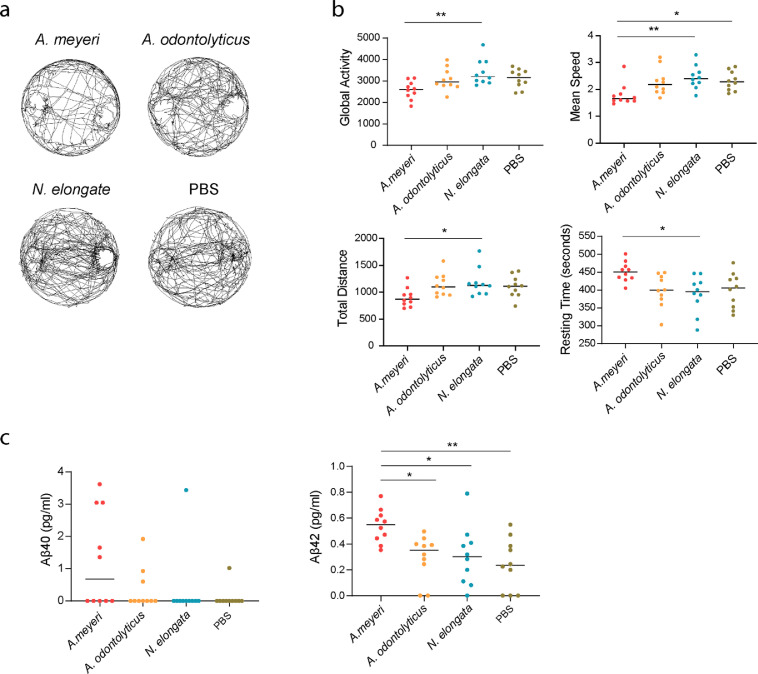
In a previous study, oral administration of Campylobacter jejuni activated state of neurons in nucleus tractus solitarius and increased c-Fos expression in the hypothalamic paraventricular nucleusas in mice [18,33]. To determine if cannabis use-associated oral microbiome affects CNS, we inoculated live A. meyeri, A. odontolyticus, and N. elongata into the oral cavity of C57BL/6 mice. A. meyeri and A. odontolyticus are oral commensal bacteria [34] and were enriched in the oral microbiome of cannabis smokers found in this study. N. elongata, which was enriched in non-smoking controls (Fig. 2a), was used as a control. We examined mouse activity through a uniformly cylindrical arena. The behaviors of mice were quantified and shown by global activity, total distance traveled (distance), average speed (speed), and resting time. The behavior of N. elongata-treated mice in the arena was comparable with the PBS-treated mice. However, compared with N. elongata-treated mice, A. meyeri-treated mice exhibited decreases in global activity, distance traveled, and mean speed, as well as increases in resting time (Fig. 3a, b).
Fig. 3.
A. meyeri oral inoculation to mice results in behavioral changes and increased β-amyloid protein in the brain. C57BL/6 mice were orally inoculated with live A. meyeri, A. odontolyticus, N. elongata, or PBS twice a week for 24 weeks (n = 10 per group). (a) Mouse activity was measured through a uniformly circular arena. (b) Mouse behavioral evaluations included global activity, total distance traveled (distance), mean speed (speed), and resting time. (c) Aβ 40 and Aβ 42 protein levels were measured in mice brain lysates by ELISA. (One-way ANOVA followed by Tukey's post hoc test, *p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001, ⁎⁎⁎⁎p < 0.0001.)
Next, we have evaluated amyloid production in mouse brain tissues as it is a marker of neurodegenerative diseases. Although Aβ 40 tended to be increased in A. meyeri-treated mice, there was no statistical difference between any two groups (Fig. 3c). Notably, the Aβ 42 peptide in the brain from A. meyeri-treated mice was increased significantly compared to the control groups (Fig. 3c). To validate alterations in oral and gut microbiome, we collected the samples from oral swab and stool one week after the final oral administration of live bacteria. Specific bacteria were quantified using qPCR, and the abundance of each bacterium was normalized by total 16S rDNA. We confirmed oral inoculated bacteria by qPCR (Fig. S2a). Notably, both A. odontolyticus and N. elongata, but not A. meyeri, presented in the oral swab of some mice from the PBS group, suggesting A. odontolyticus and N. elongata may be oral commensal microbiome in mice (Fig. S2a). A. meyeri did not present in any group except the orally inoculated A. meyeri group. Furthermore, N. elongata presented in the stool samples from some mice after oral inoculation, but was not significantly elevated compared to the controls; elevation of A. meyeri in stool after oral inoculation was extremely limited (Fig. S2b). These results indicate that A. meyeri may not be an oral commensal bacterium in B6 mice, but it can colonize well on the surface of the oral cavity during oral inoculation.
3.3. Actinomyces affects myeloid cell function
Long-term heightened systemic inflammation affects neuroinflammation, and neuroinflammation may result in CNS damage and Aβ protein production [35]. To evaluate systemic inflammation after 6-month oral bacterial inoculation, we tested plasma levels of 33 cytokines or chemokines in mice. Among these inflammatory markers, only MIP-1α and TNF-α levels were increased in the A. meyeri group compared to the control group (Fig. S3). Plasma levels of MIP-1α were also increased in the A. odontolyticus group compared to the control N. elongata group (Fig. S3). However, blood levels of the other microbial TLR-downstream proinflammatory cytokines including IL-1β, IL-6, IFN-γ, IP-10, MCP-1, and MIP-2 were similar among the four study groups. In humans, plasma levels of IL-4, IL-7, IL-8, IL-12/IL-23p40, IL-16, IL-22, IP-10, MCP-1, MIP-1α, MIP-1β, and TNF-β were decreased in cannabis smokers, whereas levels of IL-15, IL-21, IL-31, MDC, and MIP-3α were increased in cannabis smokers compared to levels in non-smokers (Fig. S4). We analyzed the correlation of A. meyeri, A. odontolyticus, and N. elongata with cytokine levels, and found that N. elongata enrichment in the saliva was directly correlated with levels of IFN-γ (p = 0.008, r = 0.637) in the cannabis smokers. However, after FDR adjustment, the p value was not significant. No correlation was found between plasma cytokine levels and saliva enrichment of A. meyeri or A. odontolyticus.
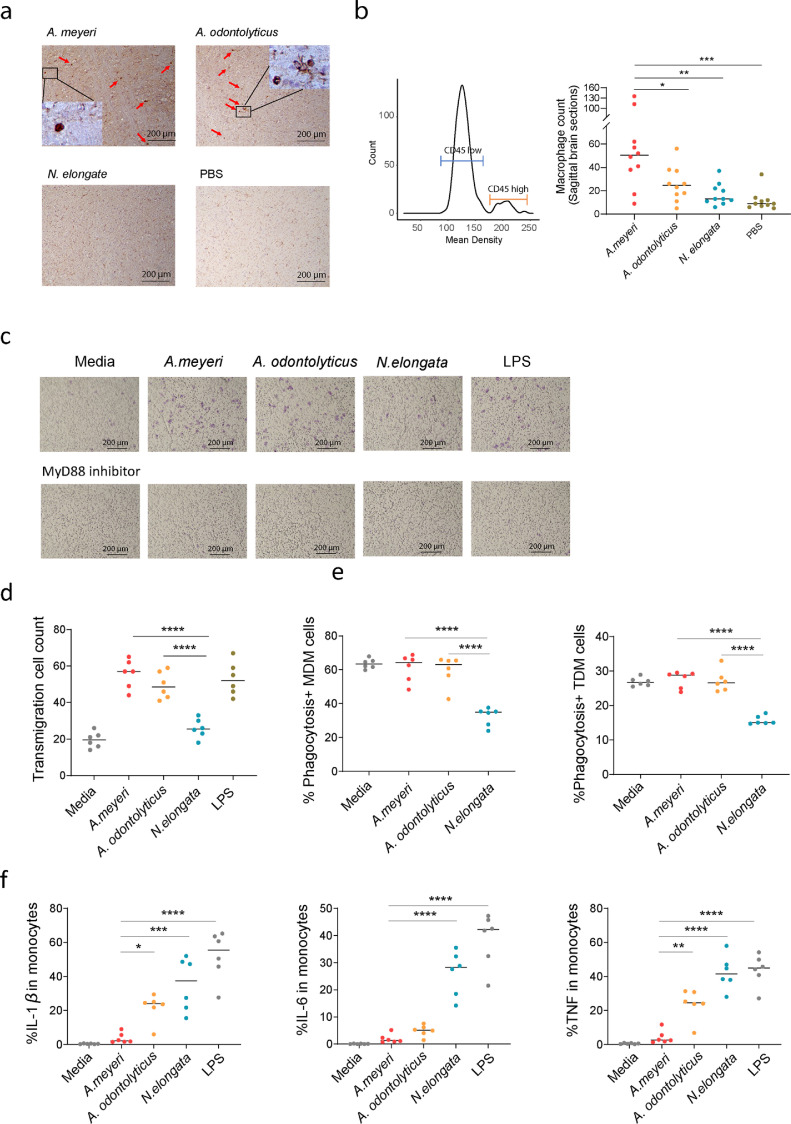
Infiltrating monocyte-derived macrophages in the CNS was related to the progression of neurodegenerative disease pathology. [36]. To investigate the potential mechanism of oral A. meyeri-mediated CNS abnormalities, we evaluated myeloid cell migration and infiltration by IHC using mouse brain tissues after oral inoculation of bacteria. Notably, macrophage infiltration (identified by high CD45 expression and enlarged morphology) was increased in the brain of A. meyeri-treated mice when compared to N. elongata or PBS-treated mice (Fig. 4a, b). In in vitro studies, A. meyeri, A. odontolyticus, and LPS (a positive control) significantly enhanced cell transmigration through microporous membranes in human macrophages (THP-1 cells) via the MyD88 cell signaling pathway (Fig. 4c, d).
Fig. 4.
A. meyeri induced macrophage migration and/or infiltration to the brain in vivo and in vitro. (a) Macrophage infiltration was assessed in sagittal brain sections of mice by CD45 and macrophage morphology using IHC staining. Macrophages were labeled using red arrows. (b) The mean density and median levels of macrophage infiltration in the sagittal section of mouse brain using CD45 staining and macrophage morphology. (c) A. meyeri and A. odontolyticus, but not N. elongata induced THP-1 cell transmigration in the transwell assay which was prevented by MyD88 inhibitor. (d) Transmigrated THP-1 cell counts after treatment with A. meyeri, A. odontolyticus, N. elongata, LPS (E. coli 055:B5), or medium control. (e) The percentages of phagocytosis in THP-1 derived macrophage (TDM) and human monocyte-derived macrophage (MDM) in each condition. (f) The percentages of IL-1β, IL-6, and TNF-α producing monocytes were evaluated using flow cytometry after stimulation with 1 × 107 units/ml of each bacteria or 2 ng/ml of LPS for 6 h. (One-way ANOVA followed by Tukey's post hoc test, *p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001, ⁎⁎⁎⁎p < 0.0001.)
Phagocytosis is a major function of antigen-presenting cells. Bacterial products activate antigen-presenting cells and decrease phagocytic capacity [37]. We found that treatment of N. elongata significantly decreased phagocytosis in both human primary monocyte-derived macrophages (MDM) and THP-1-derived macrophages (TDM) (Fig. 4e). However, treatment of Actinomyces species bacteria maintained macrophage phagocytic capacity similar to those of unstimulated cells (Fig. 4e). Moreover, as expected, A. odontolyticus or N. elongata induced TNF-α, IL-1β, and IL-6 production in human primary monocytes (Fig. 4f). Unexpectedly, A. meyeri did not activate monocytes to produce TNF-α, IL-1β, and IL-6 (Fig. 4f).
4. Discussion
Long-term cannabis users may suffer disturbed brain connectivity, cognitive impairment, and psychological disorders [38], but the exact mechanisms are not fully understood. In the current study, we found that chronic cannabis use correlated with alterations of several taxa of the oral microbiome. A. meyeri was highly enriched in the saliva from chronic cannabis smokers compared to those of non-smokers, and oral enrichment of A. meyeri was associated with the age of first cannabis use. We further investigated if direct administration of this bacterium, in the absence of cannabis or the psychoactive THC component, could elicit alteration in the brain-immune axis in a mouse model. Long-term (∼6 months) oral inoculation of A. meyeri bacterium to mice resulted in behavioral changes, macrophage infiltration into the brain, and increased Aβ 42 protein production in the brain.
Oral flora plays a role in maintaining oral health; however, environmental changes can result in dysbiosis [39]. Prior results have shown tobacco smoking alter the oral microbiome [29,40]. Decreased abundance of Neisseria and Capnocytophaga and increased abundance of Streptococcus were found in tobacco smokers compared with those of non-smokers [28,29]. Consistent with these findings, here we found that the abundance of Neisseria, Capnocytophaga, and Cardiobacterium genera was reduced and the abundance of Streptococcus was increased in cannabis smokers compared to non-smokers. In contrast, cannabis use was associated with increases in the genera, Actinomyces, Atopobium, Megasphaera, and Veillonella. A previous study demonstrated a strikingly similarity in the physical and chemical properties produced by cannabis and tobacco smoking, which contained large amounts of hydrocarbon and changed the acidity of saliva [41]. Thus, Streptococcus and Actinomyces, acid-tolerant and facultative anaerobes, may preferentially grow in a smoking-mediated environment. In contrast, bacteria such as Neisseria sp. and Corynebacterium sp. were decreased in cannabis smokers, suggesting that smoking renders an unfavorable environment to facultative or strict anaerobes. Although some oral microbiome is shared between tobacco smokers [27], [28], [29] and cannabis smokers, including increased Streptococcus and decreased Neisseria genus bacteria compared to non-smokers, A. meyeri was only increased in cannabis smokers. Moreover, the younger the age of first cannabis use, the more A. meyeri was orally enriched.
Although the gut microbiome has been shown to play a crucial role in the CNS activities via the bidirectional gut-brain axis [42], strong connections between the oral microbiome and the CNS have been reported as well. P. gingivalis, a key pathogen in chronic periodontitis was identified to contribute to Alzheimer's disease [43]. Besides P. gingivalis, other oral resident microbes were shown to associate with neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease [44,45]. Moreover, oral bacteria Treponema and N. meningitidis can infect the brain through the trigeminal or olfactory nerve [46,47]. The perturbations of intestinal microbiota resulted in the impairment of memory formation and cognition in the hippocampus [48]. To test the effect of cannabis use-associated oral dysbiosis on CNS functions, we performed various behavioral tests in mice following bacteria oral inoculations but did not observe significant memory changes. Actinomyces are gram-positive facultatively anaerobic bacteria. Some Actinomyces species are commensal bacteria in the skin, oral, gut, and vagina of humans [49], but can also become opportunistic pathogens leading to infections in the dental cavity and other systemic sites [50]. A. meyeri is associated with brain infection or dysfunction [51,52], but the causality and mechanisms of A. meyeri-mediated CNS dysfunction have never been reported. In this study, A. meyeri enrichment in the oral microbiome was inversely correlated with the age of first cannabis use, which indicates that the longer duration of cannabis exposure, the more enrichment of A. meyeri in the oral cavity. A previous study found younger age of first cannabis use was associated with decreased orbital prefrontal cortex volume [53]. Another study found negative correlations between the age of initiation of cannabis use and altered thickness of the right superior frontal gyrus [54]. Our study implies that the age of first cannabis use may be critical for particular oral microbiome development and its potential impact on cognitive function. Given the many toxicant components found in cannabis smokers, it is not surprising that cannabis smoking notably alters the oral microbial ecology. Importantly, long-term repeated oral inoculation of A. meyeri, which mimicked cannabis exposure-increased oral A. meyeri in humans, resulted in the development of CNS abnormalities.
Recent studies have found correlations between Actinomyces and Alzheimer's disease. For example, brains from patients with Alzheimer's disease have been reported to have strikingly large bacterial loads compared to controls [55]. Actinobacteria, a phylum of Actinomyces, were exclusively detected in the post mortem brain samples from patients with Alzheimer's disease compared with those of normal brains [55]. Actinobacteria were also found enriched in the gut microbiota of patients with Alzheimer's disease [56]. Another study using 16S rDNA sequencing in the brain cell lysates further found Actinomycetales, Prevotella, Treponema, and Veillonella were exclusively present in the brain of patients with Alzheimer's disease [57]. In a previous study, oral microbiome and resting-state functional magnetic resonance imaging (fMRI) scans were conducted in cannabis smokers; the enrichment of Actinomyces in the oral microbiome was positively correlated with brain resting-state functional networks [58] which are significantly perturbed with Alzheimer's disease. Neuropathological hallmarks of Alzheimer's disease include loss of neurons, progressive impairments in synaptic function, and deposition of amyloid plaques within the neuropil. Although mice do not readily develop amyloid plaques, our results show Aβ 42 deposition was increased in the brain from A. meyeri-treated mice compared with controls, suggesting oral microbiome-induced neuronal responses that have relevance to Alzheimer's disease neuropathology.
Previous studies have suggested that bacteria in the oral cavity were initially taken up by tissue macrophages which may facilitate CNS infection [59]. In the current study, A. meyeri treatment resulted in increased myeloid cell migration and phagocytosis in vitro and elevated macrophage infiltration into the mouse brain in vivo, compared with those of N. elongata treatment. The cytokines that differed in cannabis users and non-users and in A. meyeri-treated mice and control mice are related to monocyte/macrophage functions. The TNF superfamily cytokine promoted a compromised blood-brain barrier (BBB), and monocytes migrated across the BBB into the brain in response to MCP-1 [60,61]. Although it is not clear if macrophage infiltration results in CNS abnormalities in the setting of disease-associated immune perturbations, macrophage infiltration into the brain has been demonstrated in the pathogenesis of several diseases [62,63]. MIP-1 cytokines are induced in myeloid cells in response to bacterial endotoxins or membrane components [64]. In the current study, A. meyeri administration increased plasma levels of MIP-1α in some mice. However, cannabis smoking altered oral microbiome not limited to A. meyeri; thus, the decreased plasma levels of MIP-1α in cannabis users may stem from myeloid cell activation by other bacteria or by reduced total bacterial translocation due to cannabis-reduced barrier permeability [65]. In general, bacterial stimulation (e.g., N. elongata) reduces phagocytosis and promotes proinflammatory cytokine production by myeloid cells. Unexpectedly, A. meyeri did not affect phagocytosis and did not induce proinflammatory cytokines but did increase myeloid cell infiltration and amyloid production in the brain. It is possible that A. meyeri maybe a new exposure to mice which induces the immune responses and CNS effect. However, there is no evidence on the causal link between a new bacterial exposure in the oral cavity and neuropathology in mice. Thus, we believe that A. meyeri is a unique oral bacterium that is linked to CNS function.
We have tested novel object recognition in C57/B6 mice after 6-month exposure to A. meyeri, but did not find significant memory deficits. The reasons for the null finding are as follows: 1) more than 6-month exposure is necessary to see memory changes, 2) the nature of wildtype C57/B6 mice, and 3) the age of mice might play an important role with our mice being too young to detect any changes. To date, there were few to no published studies measuring effects of a specific oral microbial dysbiosis pathobiont on behavior in wildtype mice. In 2018, the study of P. gingivalis [66] found that this pathobiont (via oral gavage, similar to our administration procedure) induces memory impairment in 13-month-old mice and not 2-month-old mice suggesting an age-related effect, but without enough age cross sections to determine when susceptibility occurred. Thus, we have refined our future strategy to analyze other neurological defects or pathological signs (e.g., myelination, α-synuclein aggregation) and started to conduct studies that use mice at different ages and include memory-related longitudinal measures, such as the Novel Object Recognition (NOR) task that focuses on the hippocampus and prefrontal cortex memory functions, the Novel Tactile Recognition (NOT) task that focuses on the hippocampus and parietal cortex memory functions, and finally the Water radial arm maze that focuses on spatial memory and cognitive flexibility. We have identified a rotarod (general motor skill) and marble burying (anxiety, perseverance) methods and will add these to the battery of tests in future studies.
5. Conclusions
In this study, we found that chronic cannabis smoking may associate with oral A. meyeri and A. odontolyticus enrichment specially. This is due to the evidence of oral A. species bacterial enrichment in cannabis smokers compared to tobacco smoking (similar oral environments), as well as the inverse correlation between the age of first cannabis use and A. meyeri enrichment in cannabis users. In mice, oral inoculation of A. meyeri resulted in decreased global activity, increased macrophage infiltration, and Aβ 42 protein deposition in the brain. Unlike most bacterial-induced monocyte/macrophage activation, treatment of A. meyeri maintained cell phagocytic capacity and promoted monocyte/macrophage transmigration without TNF-α, IL-1β, and IL-6 proinflammatory cytokine production in vitro. Thus, oral enrichment of A. meyeri may contribute to CNS abnormalities.
There are several limitations in this study: (1) There is no significant difference of A. meyeri enrichment in the saliva from cannabis heavy users versus light users, with or without neurological disease history, or cannabis containing THC versus cannabis containing no THC. This may be due to the small sample size in this study. (2) The clinical evaluation of oral health and oral diseases (e.g., periodontal disease) is not available for the participants in the current study. (3) There is no information on cannabidiol (CBD) or metabolites. (4) The small sample size and less accurate information on cannabis use dose and duration limited further conclusions. Nonetheless, our findings emphasize the importance of oral health and its associated brain function. This study may also help develop potentially novel treatments targeting the microbiota or its active molecules for prevention or treatment of some neurological abnormalities.
Author's contributions
Z.L., W.J., and E.D.H. conceived the study. Z.L. wrote the manuscript. Z.L., S.F., C.R., A.B., M.L., Y.W., X.F., W.N., and X.W. performed experiments. Z.L. and M.L. analyzed data. Z.Z. designed qPCR primers. D.A., N.F., X.Y., A.W., E.D.H., L.H., X.C., W.X., K.M., A.S., J.Y., A.M.C., E.D.H, and W.J. revised the manuscript. S.F., W.N., and B.J.W. have verified the underlying data. All authors reviewed and approved the final version of the manuscript.
Data sharing statement
The full oral microbiome sequencing data sets can be available at http://www.ncbi.nlm.nih.gov/bioproject/685840.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
We would like to thank all the investigators and patients who participated in the present study. This study was supported by grants from the National Institute on Aging K23 AG044434 (A.B.), R01 AG054159 (A.B.), National Institute of Drug Abuse K24DA038240 (A.M.) and R01 DA045596 (S.F.), NINDS R01NS099595 (C.R.), NIGMS P20GM109040 (C.R.), NIA/NICHD 3R01HD096501-02S1 (J.Y.), NR016928 (X.C.), AG074331 (A.S.), P30 AR072582 (B.J.W.), ONF RE01 and P20NR016605 (Starkweather)—Pilot 3 sub-award (W.X.), the National Natural Science Foundation of China (81772185, L.H.), the Deutsche Forschungsgemeinschaft (AM 488/1-1, 1-2, D.A.) and the Brain & Behavior Research Foundation (NARSAD Young Investigator Award 2018, D.A.), and the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001450 (the pilot grant, W.J.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103701.
Contributor Information
Eric D. Hamlett, Email: hamlette@musc.edu.
Wei Jiang, Email: jianw@musc.edu.
Appendix. Supplementary materials
References
- 1.D'Souza D.C., Perry E., MacDougall L., et al. The psychotomimetic effects of intravenous delta-9-tetrahydrocannabinol in healthy individuals: implications for psychosis. Neuropsychopharmacology. 2004;29(8):1558. doi: 10.1038/sj.npp.1300496. [DOI] [PubMed] [Google Scholar]
- 2.Wade D.T., Robson P., House H., Makela P., Aram J. A preliminary controlled study to determine whether whole-plant cannabis extracts can improve intractable neurogenic symptoms. Clin Rehabil. 2003;17(1):21–29. doi: 10.1191/0269215503cr581oa. [DOI] [PubMed] [Google Scholar]
- 3.Abood M.E., Martin B.R. Neurobiology of marijuana abuse. Trends Pharmacol Sci. 1992;13:201–206. doi: 10.1016/0165-6147(92)90064-d. [DOI] [PubMed] [Google Scholar]
- 4.Zalesky A., Solowij N., Yücel M., et al. Effect of long-term cannabis use on axonal fibre connectivity. Brain. 2012;135(7):2245–2255. doi: 10.1093/brain/aws136. [DOI] [PubMed] [Google Scholar]
- 5.Ferland J.N., Hurd Y.L. Deconstructing the neurobiology of cannabis use disorder. Nat Neurosci. 2020;23(5):600–610. doi: 10.1038/s41593-020-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran H.V., Freeman T.P., Mokrysz C., Lewis D.A., Morgan C.J., Parsons L.H. Keep off the grass? Cannabis, cognition and addiction. Nat Rev Neurosci. 2016;17(5):293–306. doi: 10.1038/nrn.2016.28. [DOI] [PubMed] [Google Scholar]
- 7.Jacobus J., F Tapert S. Effects of cannabis on the adolescent brain. Curr Pharm Des. 2014;20(13):2186–2193. doi: 10.2174/13816128113199990426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DC D.'., Ranganathan M., Braley G., et al. Blunted psychotomimetic and amnestic effects of Δ-9-tetrahydrocannabinol in frequent users of cannabis. Neuropsychopharmacology. 2008;33(10):2505. doi: 10.1038/sj.npp.1301643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mechoulam R., Parker L.A. The endocannabinoid system and the brain. Annu Rev Psychol. 2013;64:21–47. doi: 10.1146/annurev-psych-113011-143739. [DOI] [PubMed] [Google Scholar]
- 10.Eisenstein T.K., Meissler J.J., Wilson Q., Gaughan J.P., Adler M.W. Anandamide and Δ9-tetrahydrocannabinol directly inhibit cells of the immune system via CB2 receptors. J Neuroimmunol. 2007;189(1-2):17–22. doi: 10.1016/j.jneuroim.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croxford J.L., Yamamura T. Cannabinoids and the immune system: potential for the treatment of inflammatory diseases? J Neuroimmunol. 2005;166(1-2):3–18. doi: 10.1016/j.jneuroim.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Sharon G., Sampson T.R., Geschwind D.H., Mazmanian S.K. The central nervous system and the gut microbiome. Cell. 2016;167(4):915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker J.L., Edlund A. Exploiting the oral microbiome to prevent tooth decay: has evolution already provided the best tools? Front Microbiol. 2019;9:3323. doi: 10.3389/fmicb.2018.03323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu D., Lu Q., Deitch E.A. Elemental diet-induced bacterial translocation associated with systemic and intestinal immune suppression. J Parenter Enter Nutr. 1998;22(1):37–41. doi: 10.1177/014860719802200137. [DOI] [PubMed] [Google Scholar]
- 15.El Kholy K., Genco R.J., Van Dyke T.E. Oral infections and cardiovascular disease. Trends Endocrinol Metab. 2015;26(6):315–321. doi: 10.1016/j.tem.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Han Y., Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92(6):485–491. doi: 10.1177/0022034513487559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukasaki M., Komatsu N., Nagashima K., et al. Host defense against oral microbiota by bone-damaging T cells. Nat Commun. 2018;9(1):1–11. doi: 10.1038/s41467-018-03147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaykema R.P., Goehler L.E., Lyte M. Brain response to cecal infection with Campylobacter jejuni: analysis with Fos immunohistochemistry. Brain Behav Immun. 2004;18(3):238–245. doi: 10.1016/j.bbi.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Noble J.M., Borrell L.N., Papapanou P.N., Elkind M., Scarmeas N., Wright C.B. Periodontitis is associated with cognitive impairment among older adults: analysis of NHANES-III. J Neurol Neurosurg Psychiatry. 2009;80(11):1206–1211. doi: 10.1136/jnnp.2009.174029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominy S.S., Lynch C., Ermini F., et al. Porphyromonas gingivalis in Alzheimer's disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5(1):eaau3333. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar D.K.V., Choi S.H., Washicosky K.J., et al. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med. 2016;8(340):340ra72. doi: 10.1126/scitranslmed.aaf1059. -ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomko R.L., Baker N.L., McClure E.A., et al. Incremental validity of estimated cannabis grams as a predictor of problems and cannabinoid biomarkers: evidence from a clinical trial. Drug Alcohol Depend. 2018;182:1–7. doi: 10.1016/j.drugalcdep.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W., Luo Z., Martin L., et al. Drug use is associated with anti-CD4 IgG-mediated CD4+ T cell death and poor CD4+ T cell recovery in viral-suppressive HIV-infected individuals under antiretroviral therapy. Curr HIV Res. 2018;16(2):143–150. doi: 10.2174/1570162X16666180703151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso J.G., Kuczynski J., Stombaugh J., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju F., Zhang T. 16S rRNA gene high-throughput sequencing data mining of microbial diversity and interactions. Appl Microbiol Biotechnol. 2015;99(10):4119–4129. doi: 10.1007/s00253-015-6536-y. [DOI] [PubMed] [Google Scholar]
- 26.Schelle J., Wegenast-Braun B.M., Fritschi S.K., et al. Early Aβ reduction prevents progression of cerebral amyloid angiopathy. Ann Neurol. 2019;86(4):561–571. doi: 10.1002/ana.25562. [DOI] [PubMed] [Google Scholar]
- 27.Luo Z., Alekseyenko A., Ogunrinde E., et al. Rigorous plasma microbiome analysis method enables disease association discovery in clinic. Front Microbiol. 2020;11:3350. doi: 10.3389/fmicb.2020.613268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris A., Beck J.M., Schloss P.D., et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187(10):1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J., Peters B.A., Dominianni C., et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10(10):2435–2446. doi: 10.1038/ismej.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz-Katterbach M., Imfeld T., Imfeld C. Cannabis and caries–does regular cannabis use increase the risk of caries in cigarette smokers? Schweiz Monatsschrift Zahnmed. 2009;119(6):576–583. = Revue mensuelle suisse d'odonto-stomatologie= Rivista mensile svizzera di odontologia e stomatologia. [PubMed] [Google Scholar]
- 31.Brenchley J.M., Price D.A., Schacker T.W., et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 32.Karin M., Lawrence T., Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124(4):823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Tengeler A.C., Dam S.A., Wiesmann M., et al. Gut microbiota from persons with attention-deficit/hyperactivity disorder affects the brain in mice. Microbiome. 2020;8:1–14. doi: 10.1186/s40168-020-00816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma N., Bhatia S., Sodhi A.S., Batra N. Oral microbiome and health. AIMS Microbiol. 2018;4(1):42–66. doi: 10.3934/microbiol.2018.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinney J.W., Bemiller S.M., Murtishaw A.S., Leisgang A.M., Salazar A.M., Lamb B.T. Inflammation as a central mechanism in Alzheimer's disease. Alzheimer's Dement Transl Res Clin Interv. 2018;4:575–590. doi: 10.1016/j.trci.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q., Barres B.A. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol. 2018;18(4):225. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 37.Jiang W., Lederman M.M., Harding C.V., Sieg S.F. Presentation of soluble antigens to CD8+ T cells by CpG oligodeoxynucleotide-primed human naive B cells. J Immunol. 2011;186(4):2080–2086. doi: 10.4049/jimmunol.1001869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall W., Degenhardt L., Teesson M. Cannabis use and psychotic disorders: an update. Drug Alcohol Rev. 2004;23(4):433–443. doi: 10.1080/09595230412331324554. [DOI] [PubMed] [Google Scholar]
- 39.Wade W.G. The oral microbiome in health and disease. Pharmacol Res. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Kumar P.S., Matthews C.R., Joshi V., de Jager M., Aspiras M. Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect Immun. 2011;79(11):4730–4738. doi: 10.1128/IAI.05371-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graves B.M., Johnson T.J., Nishida R.T., et al. Comprehensive characterization of mainstream marijuana and tobacco smoke. Sci Rep. 2020;10(1):7160. doi: 10.1038/s41598-020-63120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Kasper L.H. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominy S.S., Lynch C., Ermini F., et al. Porphyromonas gingivalis in Alzheimer's disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5(1):eaau3333. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira P.A., Aho V.T., Paulin L., Pekkonen E., Auvinen P., Scheperjans F. Oral and nasal microbiota in Parkinson's disease. Parkinsonism Relat Disord. 2017;38:61–67. doi: 10.1016/j.parkreldis.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 45.Shoemark D.K., Allen S.J. The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer's disease. J Alzheimers Dis. 2015;43(3):725–738. doi: 10.3233/JAD-141170. [DOI] [PubMed] [Google Scholar]
- 46.Riviere G.R., Riviere K., Smith K. Molecular and immunological evidence of oral Treponema in the human brain and their association with Alzheimer's disease. Mol Oral Microbiol. 2002;17(2):113–118. doi: 10.1046/j.0902-0055.2001.00100.x. [DOI] [PubMed] [Google Scholar]
- 47.Sjölinder H., Jonsson A.B. Olfactory nerve-a novel invasion route of Neisseria meningitidis to reach the meninges. PLoS One. 2010;5(11):e14034. doi: 10.1371/journal.pone.0014034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gareau M.G., Wine E., Rodrigues D.M., et al. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60(3):307–317. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 49.Dewhirst F.E., Chen T., Izard J., et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clarridge J.E., Zhang Q. Genotypic diversity of clinical Actinomyces species: phenotype, source, and disease correlation among genospecies. J Clin Microbiol. 2002;40(9):3442–3448. doi: 10.1128/JCM.40.9.3442-3448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smego R.A. Actinomycosis of the central nervous system. Rev Infect Dis. 1987;9(5):855–865. doi: 10.1093/clinids/9.5.855. [DOI] [PubMed] [Google Scholar]
- 52.Könönen E., Wade W.G. Actinomyces and related organisms in human infections. Clin Microbiol Rev. 2015;28(2):419–442. doi: 10.1128/CMR.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Churchwell J.C., Lopez-Larson M., Yurgelun-Todd D.A. Altered frontal cortical volume and decision making in adolescent cannabis users. Front Psychol. 2010;1:225. doi: 10.3389/fpsyg.2010.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez-Larson M.P., Bogorodzki P., Rogowska J., et al. Altered prefrontal and insular cortical thickness in adolescent marijuana users. Behav Brain Res. 2011;220(1):164–172. doi: 10.1016/j.bbr.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emery D.C., Shoemark D.K., Batstone T.E., et al. 16S rRNA next generation sequencing analysis shows bacteria in Alzheimer's post-mortem brain. Front Aging Neurosci. 2017;9:195. doi: 10.3389/fnagi.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhuang Z.Q., Shen L.L., Li W.W., et al. Gut microbiota is altered in patients with Alzheimer's disease. J Alzheimers Dis. 2018;63(4):1337–1346. doi: 10.3233/JAD-180176. [DOI] [PubMed] [Google Scholar]
- 57.Siddiqui H., Eribe E.R., Singhrao S.K., Olsen I. High throughput sequencing detect gingivitis and periodontal oral bacteria in Alzheimer's disease autopsy brains. J Neurosci Res. 2019;1(3) [Google Scholar]
- 58.Lin D., Hutchison K.E., Portillo S., et al. Association between the oral microbiome and brain resting state connectivity in smokers. Neuroimage. 2019;200:121–131. doi: 10.1016/j.neuroimage.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drevets D.A., Leenen P.J., Greenfield R.A. Invasion of the central nervous system by intracellular bacteria. Clin Microbiol Rev. 2004;17(2):323–347. doi: 10.1128/CMR.17.2.323-347.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatovic S.M., Shakui P., Keep R.F., et al. Monocyte chemoattractant protein-1 regulation of blood-brain barrier permeability. J Cereb Blood Flow Metab. 2005;25(5):593–606. doi: 10.1038/sj.jcbfm.9600055. [DOI] [PubMed] [Google Scholar]
- 61.Wen J., Doerner J., Weidenheim K., et al. TNF-like weak inducer of apoptosis promotes blood brain barrier disruption and increases neuronal cell death in MRL/lpr mice. J Autoimmun. 2015;60:40–50. doi: 10.1016/j.jaut.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamasaki R., Lu H., Butovsky O., et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med. 2014;211(8):1533–1549. doi: 10.1084/jem.20132477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varvel N.H., Neher J.J., Bosch A., et al. Infiltrating monocytes promote brain inflammation and exacerbate neuronal damage after status epilepticus. Proc Natl Acad Sci. 2016;113(38):E5665. doi: 10.1073/pnas.1604263113. -E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prinz M., Kann O., Draheim H.J., et al. Microglial activation by components of gram-positive and-negative bacteria: distinct and common routes to the induction of ion channels and cytokines. J Neuropathol Exp Neurol. 1999;58(10):1078–1089. doi: 10.1097/00005072-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 65.Perisetti A., Rimu A.H., Khan S.A., Bansal P., Goyal H. Role of cannabis in inflammatory bowel diseases. Ann Gastroenterol. 2020;33(2):134–144. doi: 10.20524/aog.2020.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding Y., Ren J., Yu H., Yu W., Zhou Y. Porphyromonas gingivalis, a periodontitis causing bacterium, induces memory impairment and age-dependent neuroinflammation in mice. Immun Ageing. 2018;15:6. doi: 10.1186/s12979-017-0110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.