Abstract
STAT proteins are a family of latent transcription factors that mediate the response to various cytokines and growth factors. Upon stimulation by cytokines, STAT proteins are recruited to the receptors via their SH2 domains, phosphorylated on a specific tyrosine, dimerized, and translocated into the nucleus, where they bind specific DNA sequences and activate the target gene transcription. STATs share highly conserved structures, including an N-domain, a coiled-coil domain, a DNA-binding domain, a linker domain, and an SH2 domain. To investigate the role of the coiled-coil domain, we performed a systematic deletion analysis of the N-domain and each of the α-helices and mutagenesis of conserved residues in the coiled-coil region of Stat3. Our results indicate that the coiled-coil domain is essential for Stat3 recruitment to the receptor and the subsequent tyrosine phosphorylation and tyrosine phosphorylation-dependent activities, such as dimer formation, nuclear translocation, and DNA binding, stimulated by epidermal growth factor (EGF) or interleukin-6 (IL-6). Single mutation of Asp170 or, to a lesser extent, Lys177 in α-helix 1 diminishes both receptor binding and tyrosine phosphorylation. Furthermore, the Asp170 mutant retains its ability to bind to DNA when phosphorylated on Tyr705 by Src kinase in vitro, implying a functional SH2 domain. Finally, we demonstrate a direct binding of Stat3 to the receptor. Taken together, our data reveal a novel role for the coiled-coil domain that regulates the early events in Stat3 activation and function.
Growth factors and cytokines regulate cell growth and differentiation by triggering various intracellular signaling pathways that lead to gene expression. JAK-STAT is a key pathway that mediates cellular responses to a variety of cytokines (reviewed in references 11, 20, and 30). As the name suggests, STATs (signal transducers and activators of transcription) represent a family of proteins with dual functions that transduce the signal in the cytoplasm and activate gene expression in the nucleus. Seven STATs with different functions have been identified so far in mammalian cells, and over 40 different polypeptides are known to activate one or more STATs (reviewed in reference 10). Among them, Stat3 was identified both as an acute-phase response factor activated by interleukin-6 (IL-6) in mouse liver and by homology to Stat1 (2, 46). Stat3 is also activated by other members of the IL-6 family, such as IL-11, ciliary neurotrophic factor, oncostatin M, and leukemia-inhibitory factor, which share the common transducing gp130 receptor subunit (2, 25, 33). IL-6, as a pleiotropic cytokine, exhibits various functions in immune response, hematopoiesis, and neuronal differentiation (37).
The model of activation of Stat3 by IL-6 has been established. Binding of IL-6 to its receptor gp80 (subunit α) induces homodimerization of gp130 (subunit β) and phosphorylation of the gp130-associated Janus kinases (JAKs). JAKs phosphorylate the tyrosine residues on gp130 that serve as docking sites for Stat3. Stat3 binds to the respective tyrosine residues on gp130 through its Src homology 2 (SH2) domain and is subsequently phosphorylated on a single tyrosine residue at the carboxyl terminus by the JAKs (25, 33, 34). Stat3 forms homo- or heterodimers with Stat1 via reciprocal interactions between the SH2 domain and the phosphorylated tyrosine (31) and translocates into the nucleus, where it binds to IL-6 response elements and regulates many acute-phase protein genes (2). Growth factors such as epidermal growth factor (EGF), platelet-derived growth factor, and colony-stimulating factor (CSF-1) can also activate Stat3 (29, 46). It has been reported that Stat1 interacts with the EGF receptor, and the intrinsic tyrosine kinase activity of the receptors is required for activation of Stat1 and Stat3. The EGF receptor kinase was demonstrated to phosphorylate STATs in vitro (12, 28, 32). Furthermore, the non-receptor tyrosine kinase, Src, is involved in the activation of Stat3 by CSF-1 (7). Compared to cytokine signaling, the mechanisms of Stat3 activation by growth factors are less defined.
STATs share a highly conserved structure with a number of functional domains. The three-dimensional structures of the NH2-terminal domain (N-domain) of Stat4 and the homodimers of both Stat1 and Stat3β bound to DNA have been resolved recently (3, 8, 39). The N-domain contains the NH2-terminal 130 amino acids and is composed of eight helices assembled into a hook-like structure. This domain is relatively separated from the other domains in STATs and is not essential for dimerization or binding to a single-target DNA site, but it mediates the dimer-dimer interaction and the cooperativity in binding to multiple DNA sites (38, 42). The crystal structures of Stat1 and Stat3β lacking the N-domain show several conserved domains: an NH2-terminal coiled-coil domain, a DNA-binding domain, a linker domain, and an SH2 domain. The phosphotyrosine occurs on residue 705 (3, 8). Forty to fifty amino acids at the carboxyl termini of STATs that are absent in the crystal structures comprise the transcriptional activation domain (4) and were recently reported to be involved in the proteasome-dependent turnover of Stat5 (40). The coiled-coil domain at the amino terminus contains four antiparallel α-helices and has been inferred to be involved in protein-protein interaction. For example, residues 150 to 250 of Stat1 (in particular, Lys161, located in helix α1) has been shown to interact with p48, a protein from an interferon response factor family, to form interferon (IFN)-stimulated gene factor 3 complex that regulates IFN-α-responsive genes (19). Recently, a short region in the first α-helix of the coiled-coil domain and a portion of the DNA-binding domain of Stat3 have been reported to interact with another transcription factor, c-Jun, and cooperatively activate transcription of the IL-6-inducible α2-macroglobulin gene (45). However, whether this domain plays any role in the activation and function of STATs is largely unknown.
Here we report that the coiled-coil domain of Stat3 is required for its tyrosine phosphorylation and phosphotyrosine-dependent activities, including dimerization, nuclear translocation, and DNA binding stimulated by EGF or IL-6. We further investigated the mechanism for this regulation and showed that the coiled-coil region is necessary for the recruitment of Stat3 to the cytokine receptor. Furthermore, Asp170, located in the first α-helix, is identified as one of the crucial residues for this regulation. We propose a novel role for the coiled-coil domain in regulating the receptor binding and subsequent activation of Stat3.
MATERIALS AND METHODS
Construction of expression plasmids.
The expression plasmid of murine Stat3 was obtained from J. E. Darnell, Jr. A series of NH2-terminal deletion mutants of Stat3 were generated by PCR using primers containing a BamHI site 5′ and XhoI site 3′. The PCR products were cloned into plasmid pXJ40-FLAG, a FLAG-tagged expression vector kindly provided by Z. Zhao (26). Point mutations were prepared by using the Quikchange site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions, and mutagenesis was confirmed by standard dideoxy termination sequencing.
DNA transfection.
COS-1 and HepG2 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum (Gibco-BRL Life Technologies). Transient transfections of the expression plasmids into COS-1 and HepG2 cells were performed with Lipofectamine (Gibco-BRL Life Technologies) and FuGENE 6 transfection reagents (Boehringer Mannheim), respectively, following the manufacturer's instructions. Half the amount of ST3-ΔN was used for transfection because of its extremely high expression. The cells were lysed and extracted as crude nuclear extracts for electrophoretic mobility shift assays (EMSA) as described previously (21).
Antibodies, immunoprecipitation, and immunoblotting.
Antibodies against phospho-Tyr705 and phospho-Ser727 of Stat3 were purchased from New England Biolabs. Anti-Stat3 (C-20) and anti-FLAG M2 were purchased from Santa Cruz Biotechnology and Sigma, respectively. Immunoprecipitation and immunoblotting were performed as described previously (21). In brief, transfected cells were washed with cold phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.2], 1% deoxycholic acid, 1% Triton X-100, 0.25 mM EDTA [pH 8.0] plus 5 μg leupeptin, 5 μg of aprotinin, and 1 μg of pepstatin A per ml, with 1 mM phenylmethylsulfonyl fluoride, 5 mM NaF, and 100 μM sodium orthovanadate). The cell lysates, containing 1 mg of proteins, were incubated with the appropriate antibody overnight at 4°C, followed by incubation with protein G PLUS/protein A-agarose (Oncogene Science) for 1 h. The immunoprecipitates were washed with RIPA buffer and PBS, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with PBS containing 0.1% Tween 20 and 1% bovine serum albumin before it was incubated with the appropriate primary and secondary antibodies. The bound antibodies were visualized by using BM Chemiluminescence (Boehringer Mannheim). For a second blotting, the membrane was incubated in stripping buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 100 mM β-mercaptoethanol) for 30 min at 60°C before reblotting.
Peptide-binding assay.
Transfected cells (107) were lysed in lysis buffer (1% NP-40, 20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM sodium orthovanadate, 5 μg of aprotinin per ml, and 5 μg of leupeptin per ml). Biotinylated peptides of IL-6 receptor subunit gp130 (pY2, SSTVQ-YPO4−-STVVHS; pY3, VVHSG-YPO4−-RHQVPS; and Y3, VVHSGYRHQVPS) were purchased from Sigma-Genosys. The peptides (5 μg) were incubated with 40 μl of streptavidin-Sepharose (Pierce) at 4°C for 2 h. The beads were washed three times with 20 mM Tris-HCl (pH 7.4) and then incubated with aliquots of lysates (corresponding to ∼3 × 106 cells) or 0.5 μg of the baculovirus-produced Stat3 proteins for 1 h. The complexes were washed, boiled, fractionated on SDS-PAGE, and blotted with anti-Stat3 antibody (13, 15).
Expression, purification, and in vitro phosphorylation of baculovirus-produced Stat3.
BamHI-XhoI cDNA fragments containing full-length Stat3 and the deletion mutants that lack the N-domain and α-helices 1 to 4 were cloned into the baculovirus expression vector pFastBacHT (Gibco-BRL) so that the recombinant clones were incorporated with a six-histidine residue tag at the NH2 termini. Sf9 insect cells were transfected with recombinant bacmid DNAs for the generation of recombinant baculoviruses. The recombinant baculoviruses were then used for subsequent infection of the insect cells. For protein expression, Sf9 cells (containing 2 × 106 cells per ml) were infected with recombinant virus at a multiplicity of infection of 10. Infected cells were harvested 48 to 72 h later. They were lysed in 10 ml of lysis buffer (20 mM HEPES [pH 7.9], 100 mM NaCl, 0.5% Nonidet P-40, 15% glycerol, 2 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 1 mM Na3VO4, 1 mM Na2MoO4, and 15 mM imidazole) for 1 h. Lysates were then centrifuged for 10 min at 20,000 × g to remove the insoluble components, and the cleared supernatant was allowed to incubate with Ni2+ ProBond resin (Invitrogen) for 1 h. The mixture was packed in a column and extensively washed with lysis buffer containing 30 mM imidazole. Ni2+-bound proteins were serially eluted with imidazole at concentrations of 50, 200, 350, and 500 mM. Stat3 proteins were eluted at 200 mM imidazole. The purified protein was dialyzed against a buffer containing 10 mM HEPES (pH 7.4), 20 mM EDTA, 100 mM NaCl, and 0.5 mM dithiothreitol and stored at −70°C. Purified recombinant Stat3 proteins (2 μg) were phosphorylated by Src (Upstate Biotechnology) in a buffer containing 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7.0], 5 mM MnCl2, 1 mM NaCl, 0.1 mM dithiothreitol, and 20 μM ATP in a volume of 40 μl at 30°C for 20 min. One quarter of the reaction mixture containing 0.5 μg of Stat3 proteins was used for the Western blot analysis or EMSA using [32P]hSIE, the high-affinity binding site of Stat3, as a probe.
RESULTS
Coiled-coil domain of Stat3 is required for its tyrosine phosphorylation and subsequent activation stimulated by EGF or IL-6.
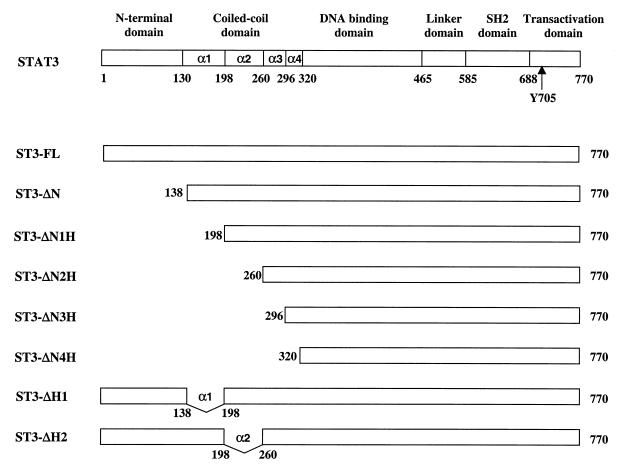
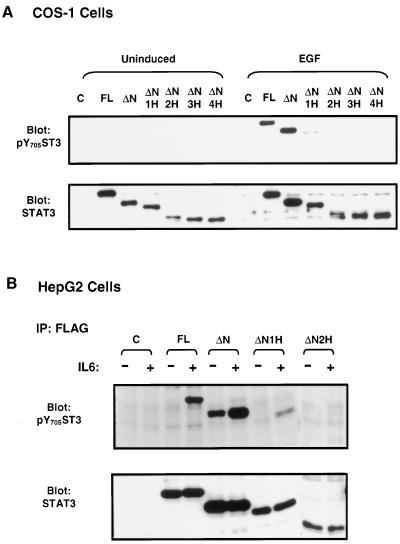
The coiled-coil domain adjacent to the N-domain of the STAT proteins consists of four antiparallel helices. To study the structure and function relationship of this domain, we generated a series of Stat3 mutants that sequentially deleted the N-domain and each of the four α helices. These constructs were tagged with the FLAG epitope at their amino termini, and the resultant mutants were named ST3-ΔN, ST3-ΔN1H, ST3-ΔN2H, ST3-ΔN3H, and ST3-ΔN4H (Fig. 1). In our initial analysis, we examined the tyrosine phosphorylation of the Stat3 mutants in response to growth factors and cytokines. COS-1 cells, which express a low level of endogenous Stat3 and respond to EGF strongly, were transfected with the expression plasmids (41). The cells were either left untreated or treated with EGF, and tyrosine phosphorylation was examined by Western blot analysis using an antibody that specifically recognizes Stat3 proteins phosphorylated on Tyr705. No tyrosine phosphorylation was detected in the untreated cells, but strong tyrosine phosphorylation was observed in cells transfected with the full-length Stat3 (ST3-FL) and the N-domain deletion mutant (ST3-ΔN) upon EGF stimulation. On the other hand, tyrosine phosphorylation was impaired in ST3-ΔN1H, in which helix α1 was deleted, and was totally abolished in mutants ST3-ΔN2H, ST3-ΔN3H, and ST3-ΔN4H, in which two, three, and four α-helices were deleted, respectively (Fig. 2A, upper panel). The expression of the mutants was comparable (lower panel).
FIG. 1.
Schematic diagrams of the structural domains and deletion mutants of Stat3. α1, α2, α3, and α4 represent the α-helices in the coiled-coil region.
FIG. 2.
Tyr phosphorylation of Stat3 deletion mutants stimulated with EGF or IL-6. (A) COS-1 cells were transfected with control (C) or Stat3 expression plasmids as labeled and either left untreated or treated with EGF for 15 min. The cell lysates were prepared and subjected to Western blot analysis using antibody against an anti-phospho-Tyr-705-Stat3 (pY705ST3), as indicated in the upper panel. The membrane was stripped and reprobed with an anti-Stat3 antibody (lower panel). (B) Transfected HepG2 cells were either left untreated (−) or treated with IL-6 (+) for 15 min. Cell lysates were immunoprecipitated (IP) with FLAG antibody, and the immunoprecipitates were subjected to Western blot analysis using the anti-pY705ST3 antibody (upper panel). The blots were stripped and reprobed with Stat3 antibody (lower panel).
We also examined the tyrosine phosphorylation of the Stat3 mutants in the human liver hepatoma cell line HepG2, in which the endogenous Stat3 is strongly tyrosine phosphorylated upon stimulation with IL-6 (2). The full-length Stat3 and the deletion mutants were transfected into HepG2 cells, and their Tyr705 phosphorylation was analyzed by immunoprecipitation and Western blotting. The results obtained were very similar to those observed in the EGF-stimulated COS-1 cells except for ST3-ΔN, which exhibited a high level of tyrosine phosphorylation in uninduced cells that was further enhanced in IL-6-treated cells (Fig. 2B, upper panel). The tyrosine phosphorylation of the deletion mutants of the coiled-coil domain was either inhibited (ST3-ΔN1H) or destroyed (ST3-ΔN2H, ST3-ΔN3H, and ST3-ΔN4H) in HepG2 cells stimulated by IL-6 (Fig. 2B and data not shown). These results indicate that the N-domain is not required for the tyrosine phosphorylation of Stat3. In contrast, the helix α1 region is essential for Stat3 tyrosine phosphorylation stimulated by EGF or IL-6.
Phosphorylation on Tyr705 is a prerequisite for STAT activities, such as dimer formation, nuclear translocation, DNA binding, and transactivation. To confirm the above results, we tested the nuclear translocation and the DNA-binding activity of these deletion mutants. In agreement with the impaired tyrosine phosphorylation, the α-helix deletion mutants ST3-ΔN1H, ST3-ΔN2H, ST3-ΔN3H, and ST3-ΔN4H failed to enter the nucleus in either EGF-treated COS-1 or IL-6-treated rat pheochromocytoma PC12 cells. These mutants were also unable to bind hSIE, the high-affinity binding site of Stat3, as examined by EMSA (data not shown). Together, these results demonstrate that the coiled-coil domain of Stat3 is required for its tyrosine phosphorylation and tyrosine phosphorylation-dependent activities, including nuclear translocation and DNA binding.
Both helices α1 and α2 are necessary for the tyrosine phosphorylation of Stat3.
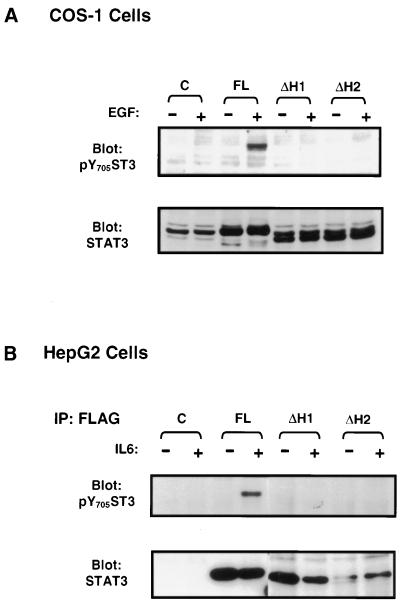
The above results showed that deletion of helix α1 abolished the tyrosine phosphorylation of Stat3. However, involvement of the other helices besides α1 in the regulation of Stat3 Tyr phosphorylation cannot be excluded. Helices α1 and α2, spanning 44 and 54 amino acid (aa) residues, respectively, are much longer than α3 (32 aa) and α4 (25 aa) and represent the major components of the coiled-coil domain (3). In order to examine the role of the other helices and to minimize possible structural disruption due to deletion of the N-domain, we produced two mutants with a deletion of either helix α1 (ST3-ΔH1) or α2 (ST3-ΔH2), with an intact N-domain (see Fig. 1), and their tyrosine phosphorylation was analyzed. Both deletions totally abrogated their tyrosine phosphorylation in EGF-induced COS-1 cells (Fig. 3A, upper panel) or in IL-6-induced HepG2 cells (Fig. 3B, upper panel). The expressions of these mutants are shown in the lower panels. These data suggest that both α1 and α2 are necessary for Stat3 tyrosine phosphorylation.
FIG. 3.
Deletion of helix α1 or α2 abolishes tyrosine phosphorylation of Stat3 in response to EGF or IL-6. COS-1 (A) or HepG2 (B) cells were transfected with the control plasmid (C), full-length Stat3 (FL), or the mutants in which either helix α1 (ΔH1) or α2 (ΔH2) was deleted and either left untreated (−) or treated with EGF or IL-6 (+). The tyrosine phosphorylation and expression of Stat3 were monitored by Western blot analysis (A) and immunoprecipitation/blotting analysis (B,) as described in the legend to Fig. 1.
Coiled-coil domain of Stat3 is required for its recruitment to the IL-6 receptor.
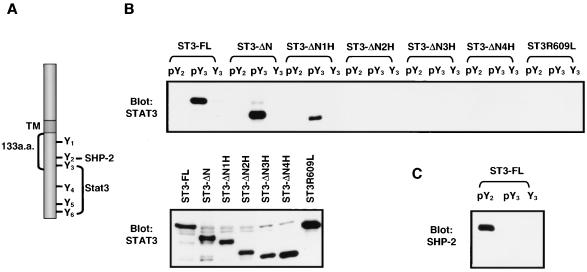
In the primary event of STAT activation, STAT proteins must be recruited to the cytokine receptors via interaction of their SH2 domains with the specific phosphorylated tyrosine residues on the cytokine receptors. The cytoplasmic domain of the IL-6 receptor subunit, gp130, contains six tyrosine residues that are phosphorylated upon stimulation by IL-6 (Fig. 4A). The second membrane-proximal tyrosine residue (Y2) is required for recruitment of the SH2-containing phosphatase-2 (SHP-2), whereas any one of the four tyrosine residues (Y3 to Y6) containing the consensus YXXQ motif in the carboxyl terminus of gp130 can mediate the tyrosine phosphorylation of Stat3 stimulated by IL-6 (1, 34, 43). The 133 amino acids, including three tyrosine residues (Y1 to Y3), from the membrane-proximal region of gp130 (shown in Fig. 4A) were reported to be sufficient to induce cell proliferation (13). One possible mechanism for the involvement of the coiled-coil domain in Stat3 phosphorylation is by affecting the protein-protein interaction between Stat3 and the receptor during the recruitment of Stat3 to the activated receptor following ligand binding. We examined the ability of the mutant Stat3 proteins to interact with the phosphorylated receptor gp130 by coprecipitation analysis using biotinylated forms of the receptor-derived peptides. The phosphorylated peptide-containing sequences surrounding the third tyrosine (pY3), coprecipitated with full-length Stat3 as well as the N-domain mutant ST3-ΔN. Surprisingly, binding was diminished in ST3-ΔN1H and was totally abolished in ST3-ΔN2H, ST3-ΔN3H, and ST3-ΔN4H. On the other hand, binding of the wild-type and mutant Stat3 proteins to the nonphosphorylated peptide Y3 and the phosphopeptide pY2 of the SHP-2 binding site was not observed (Fig. 4B, upper panel). In addition, the α1 (ST3-ΔH1) and α2 (ST3-ΔH2) deletion mutants also failed to bind the receptor peptide pY3 (data not shown). These results suggest an involvement of the coiled-coil domain in receptor binding.
FIG. 4.
Effect of the coiled-coil domain of Stat3 on its binding to the phosphorylated peptide derived from IL-6 receptor subunit gp130. (A) Diagram of the structure and Tyr phosphorylation sites of human gp130. Y1 to Y6 represent tyrosine residues 683, 759, 767, 814, 905, and 915 on the gp130 cytoplasmic tail, respectively. TM, transmembrane domain. a.a., amino acids. (B) COS-1 cells were transfected with the Stat3 plasmids as labeled on top of the upper panel and lysed after 48 h. The biotinylated peptides (pY2, SSTVQ-YPO4−-STVVHS; pY3, VVHSG-YPO4−-RHQVPS; and Y3, VVHSGYRHQVPS) derived from gp130 were incubated with streptavidin-Sepharose. The beads were washed and then incubated with aliquots of lysates. The complexes were washed, fractionated on SDS-PAGE, and immunoblotted with anti-Stat3 antibody (upper panel). The cell lysates were subjected to Western blot analysis with anti-Stat3 antibody to monitor the expression of the various Stat3 proteins in transfected cells (lower panel). (C) The blot used in panel B was stripped and reprobed with anti-SHP-2 antibody. Binding of the endogenous SHP-2 from cells transfected with full-length Stat3 to its docking site pY2 is shown.
It is well known that the SH2 domain is required for STAT protein binding to cytokine receptors. An arginine residue conserved in all known SH2 domains recognizes the phosphate group of the phosphotyrosine (23, 27). Arg602 in Stat1 and Arg609 in Stat3 are such key residues (3, 8). To confirm the essential role of the SH2 domain in receptor binding in our system, a point mutation on the SH2 domain of Stat3, ST3R609L, was produced by replacing Arg609 with Leu. This mutation totally abolished its peptide-binding ability (Fig. 4B, upper panel). The expression level of the various Stat3 proteins in the transfected COS-1 cells was comparable (lower panel of Fig. 4B). A strong binding of endogenous SHP-2 to pY2, but not to Y3 and pY3, was detected in COS-1 cells, indicating the specificity of the peptide-binding experiment (Fig. 4C). Similar binding patterns for the full-length, deletion, and SH2 mutants were also observed in transfected HepG2 cells (data not shown). These results indicate that both the SH2 domain and the coiled-coil domain are necessary for the interaction between Stat3 and the IL-6 receptor, and loss of tyrosine phosphorylation in the coiled-coil deletion mutants is due to a deficiency in receptor binding.
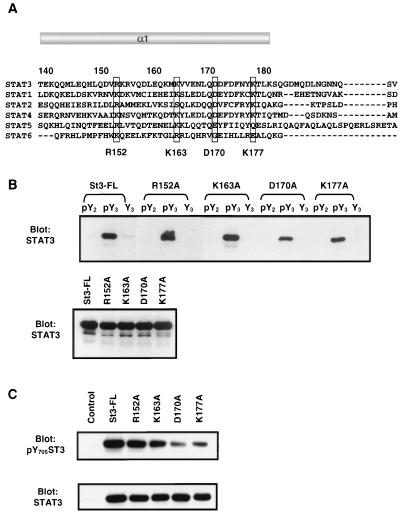
Single point mutation Asp170 in helix α1 of the coiled-coil region of Stat3 diminishes its interaction with gp130 and tyrosine phosphorylation.
How does the coiled-coil domain affect receptor binding? It is possible that the deletion mutations of the coiled-coil region used in this study may result in a change in protein conformation that alters the structure of the SH2 domain. To address this issue, four highly conserved hydrophilic amino acid residues which are distributed along the helix α1 region were chosen for site-directed mutagenesis (Fig. 5A). Each of these residues was replaced with alanine, and the receptor binding activity of the mutants was examined. Two mutants, R152A (Arg152→Ala) and K163A (Lys163→Ala), showed similar binding affinity to pY3 in comparison with wild-type Stat3. In contrast, binding of mutants D170A (Asp170→Ala) and K177A (Lys177→Ala), albeit less dramatic, was diminished. The expression levels of these mutants and the wild-type Stat3 were equivalent (Fig. 5B). In agreement with this, the tyrosine phosphorylation of D170A and K177A but not R152A and K163A was significantly reduced in response to EGF (Fig. 5C, upper panel).
FIG. 5.
Point mutant Asp170 of Stat3 diminishes its peptide-binding activity and the tyrosine phosphorylation stimulated by EGF. (A) Sequence alignment of helix α1 in the coiled-coil domain of STATs. The highly conserved hydrophilic residues are boxed and indicated at the bottom. (B) COS-1 cells were transfected with point mutants of Stat3 as labeled on top of the upper panel. The peptide-binding experiments were performed as described in the legend to Fig. 4. The expression of the mutant Stat3 proteins is examined in Western blot analysis in the lower panel. (C) COS-1 cells were transfected with control vector or Stat3 plasmids as labeled on top of the upper panel and induced by EGF for 15 min. The cell lysates were subjected to Western blot analysis with antibodies against phospho-Tyr-705-Stat3 (upper panel) or Stat3 (lower panel).
Analysis of the crystal structure of Stat3β together with molecular modeling revealed that Arg152 has a side chain pointing towards the interface between helix α1 and helix α3, whereas the other three residues have side chains pointing away from the interface and are unlikely to interact with helix α3 (not shown). The results showed that the R152A mutation has no effect on receptor binding, suggesting that its SH2 domain is intact. Therefore, the decreased receptor binding caused by mutations on surface residues Asp170 and Lys177 is unlikely due to a major conformational change of the coiled-coil domain or the SH2 domain. These data also suggest that Asp170 and, to a lesser extent, Lys177 may function as critical residues in the regulation of receptor binding.
Stat3 protein lacking helix α1 of the coiled-coil domain retains a functional SH2 domain.
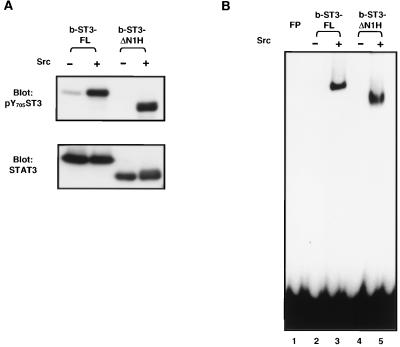
The SH2 domains of STAT proteins possess a unique dual role in their activation. They are required for recruitment to the receptors and participate in dimer formation by reciprocal interaction with the specific phosphotyrosine residue at the carboxyl termini of the STAT proteins. To further determine whether the SH2 domain is functional in the deletion mutant, mutant Stat3 which lacks the N-domain and helix α1 (b-ST3-ΔN1H) and full-length Stat3 (b-ST3-FL) were expressed and purified in the baculovirus expression system. The recombinant proteins were phosphorylated with Src kinase in vitro, and tyrosine phosphorylation and DNA-binding activity were examined. As shown in Fig. 6, strong Tyr phosphorylation by Src was detected in both the wild type and the deletion mutant. Correspondingly, upon phosphorylation, both proteins bound DNA with similar affinities, as determined by EMSA. The amounts of two proteins used in the above assays are shown in the lower panel of Fig. 6A. These results demonstrate that b-ST3-ΔN1H serves as a proper substrate for Src kinase and is able to bind DNA after phosphorylation, suggesting that its SH2 domain is functionally intact.
FIG. 6.
Phosphorylation and DNA binding of the helix α1 deletion mutant in vitro. Baculovirus-expressed full-length Stat3 (b-ST3-FL) and Stat3 with deletions of the N-domain and helix α1 (b-ST3-ΔN1H) were purified from Sf9 cells and incubated with Src kinase. The reaction mixture was divided so that each part contained 0.5 μg of Stat3 proteins and subjected to either Western blot analysis with the antibodies indicated (A) or EMSA using [32P]hSIE as a probe (B). FP, free probe.
Direct interaction of Stat3 with the gp130-derived phosphopeptide.
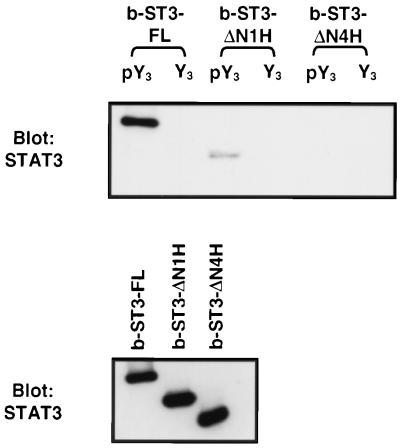
To further determine whether Stat3 interacts directly with the gp130 receptor, baculovirus-produced b-ST3-FL, b-ST3-ΔN1H, and b-ST3-ΔN4H (deleting four α-helices) were purified, and their receptor-binding abilities were analyzed by the peptide-binding assay. Similar to the results obtained in vivo (Fig. 4), we observed a strong interaction between wild-type Stat3 and the gp130-derived phosphopeptide pY3, but not Y3 (Fig. 7, lanes 1 and 2). The binding was significantly diminished for b-ST3-ΔN1H (lane 3) and totally abrogated for b-ST3-ΔN4H (lane 5). These data demonstrate that Stat3 is able to interact directly with its phosphorylated docking site on the gp130 receptor without the participation of an adaptor protein. These results also indicate that the deficiency of b-ST3-ΔN1H in receptor binding is unlikely to be due to a lack of interaction with other proteins.
FIG. 7.
Direct interaction of Stat3 and the gp130-derived phosphopeptide. Baculovirus-expressed full-length Stat3 (b-ST3-FL) and the deletion mutants (b-ST3-ΔN1H and b-ST3-ΔN4H) were purified from Sf9 cells, and 0.5 μg of each protein was subjected to peptide binding with either pY3 or Y3 as described in the legend to Fig. 4 (upper panel) or Western blot analysis (lower panel).
DISCUSSION
The crystal structures of Stat1 and Stat3β exhibit highly conserved structural features, including newly defined domains such as the linker domain and the coiled-coil domain (3, 8). These data provide insight into the function of each domain. The coiled-coil domain is commonly related to the protein-protein interaction (22). Although this domain of Stat3 consists of 182 amino acids that occupy approximately one fourth of the whole molecule, its function is largely unknown. Our systematic studies on this region for the first time reveal its pivotal role in the regulation of Stat3 tyrosine phosphorylation stimulated by EGF or IL-6. It is well established that tyrosine phosphorylation is a prerequisite for STAT activation. In our experiments, tyrosine phosphorylation-dependent activities, such as dimerization, nuclear translocation, and the DNA binding of Stat3, were all abrogated by deletion of one to four α helices (data not shown). Although such inhibitory effects could be due simply to a deficiency of tyrosine phosphorylation but not to the effect on the activities per se, the possibility that the coiled-coil region has other functions cannot be excluded and requires further investigation.
The first step for STATs becoming tyrosine phosphorylated is their recruitment to the receptors of the polypeptide ligands. The SH2 domains of the STAT proteins interact with specific tyrosine residues on the cytokine receptors, which facilitate their subsequent tyrosine phosphorylation by the receptor-associated JAKs (16, 17, 34). To understand the mechanisms of the coiled-coil region regulating the tyrosine phosphorylation of Stat3, we examined the ability of the wild-type and the mutant Stat3 proteins binding to the IL-6 receptor subunit, gp130. Our results demonstrate that deletion of helix α1 and/or α2 abolishes the binding of Stat3 to the receptor. Although the binding experiments were performed with peptides derived from gp130, activation of Stat3 by EGF could be mediated in a similar way. Two autophosphorylated tyrosine residues containing the consensus YXXQ binding site of Stat3 on the EGF receptor have been identified as critical for Stat3 activation (9). Correspondingly, we have observed that the SH2 domain is also required for Stat3 induction by EGF (data not shown), suggesting that the binding of Stat3 to the EGF receptor by its SH2 domain is necessary for its subsequent tyrosine phosphorylation. Moreover, the tyrosine phosphorylation profiles of the deletion and point mutants of Stat3 induced by EGF and IL-6 were similar. These data suggest that in addition to the SH2 domain, the coiled-coil domain is also required for the recruitment of Stat3 to both the cytokine and growth factor receptors.
We further addressed the mechanisms of how the coiled-coil domain is involved in the binding of Stat3 to the receptor. One possibility is that the binding is independently mediated either by the coiled-coil domain or by the SH2 domain. However, this is unlikely because our results indicate that either the coiled-coil mutations or the SH2 mutation eliminates binding. Another possibility is that both are required for efficient receptor binding. The coiled-coil domain may modulate SH2 domain binding activity or vice versa. Given the considerable evidence on critical interaction between the SH2 domain and phosphotyrosine residue on the receptor and the binding specificity of the Stat3 SH2 domain to the phosphopeptide pY3 shown in our experiments (Fig. 4 and 5), it is more likely that the coiled-coil domain affects the SH2 domain binding to the receptor, but not the reverse.
One possible mechanism by which the coiled-coil domain regulates the SH2 domain binding to receptor is that the coiled-coil domain may affect the structure of the SH2 domain so that in its absence the conformation of the SH2 domain is altered. The data we have presented so far argue against this. Based on the structure of Stat3 dimer bound to DNA, the coiled-coil domain is physically separated from the SH2 domain by two other structural domains, the DNA-binding domain and the linker domain. It is unlikely that any local structural perturbation that happens in the coiled-coil region at the amino terminus directly affects the SH2 domain at the carboxyl terminus. Furthermore, to minimize possible structural disruption, we mutated four highly conserved, hydrophilic amino acid residues in the helix α1 region and analyzed the side chain interactions involving these residues. We believe that if there is any effect of these mutations on the structure of Stat3, it is likely to be restricted to the coiled-coil region and unlikely to cause a global conformational change that leads to distortion of the SH2 domain. First, these four amino acids are exposed residues and do not form part of the buried residues contributing to the interaction essential for structural integrity of the four-helix bundles of the coiled-coil domain (24). Second, three of the four residues have side chains which do not point towards the interface between either helices 1 and 3 or helices 1 and 2. Mutations in any of these residues are therefore unlikely to disrupt the interactions between helices. Arg152, the only residue among the four with a side chain that forms hydrogen bonds with Ser269 and Glu272 of helix 3 (data not shown), has no detectable effect on the SH2 function when it is mutated. In addition, we demonstrate that a baculovirus-expressed Stat3 mutant, b-ST3-ΔN1H, carrying the helix α1 deletion, is capable of binding DNA after Tyr705 phosphorylation by Src kinase in vitro (Fig. 6B). These results confirm that the deletion mutant of the α1 helix retains its capacity to form dimer, which indirectly suggests that its SH2 domain remains functional.
The latter result, on the other hand, also raises the question of why ST3-ΔN1H fails to bind to the receptor and yet can form dimers. We compared the binding affinities of the wild-type and mutant Stat3 to the gp130-derived phosphopeptide (pY3) and to the Stat3 Tyr705-derived phosphopeptide. The results showed that both transfected and baculovirus-expressed Stat3 binds to its own peptide with affinities that are 25- and 50-fold lower than those of gp130 peptide, respectively. In the case of the mutant ST3-ΔN1H, binding to the gp130 peptide was diminished and to the Stat3 Y705 peptide was almost undetectable (data not shown). These results are consistent with those of similar experiments performed in Stat1 (15). The affinity of Stat1 binding to the IFN-γ receptor phosphopeptide is 110 times higher than binding to the Stat1 phosphopeptide, indicating that the latent form of Stat1 interacts preferentially with its docking site on the receptor. However, the phosphorylated Stat1 subsequently forms a high-avidity complex through reciprocal interactions between the SH2 domains and the phosphotyrosines of two monomers, and the dimerized Stat1 is unlikely to rebind to the receptor. It has been suggested that Stat1 recruitment to the receptor, dissociation from the receptor after phosphorylation, and subsequent dimerization are an ordered process driven by affinity. The unidirectional release of activated Stat1 from the IFN-γ receptor indicates the preference of free tyrosine-phosphorylated Stat1 monomers to form high-avidity reciprocal homodimers rather than reassociating with the receptor binding site. Our experiments suggest that Stat3 activation may follow a similar process. The activation process involves two phases, before and after phosphorylation. Prior to phosphorylation, Stat3 is recruited to the receptor with high affinity. Once Tyr705 is phosphorylated, disassociation from the receptor is driven perhaps by the formation of an energetically more favorable dimer. In our experiment, after Stat3 and mutant ST3-ΔN1H are phosphorylated by Src in vitro, dimerization and subsequent DNA binding occur. This could partially explain why ST3-ΔN1H can form a dimer although it fails to bind to the receptor.
The second mechanism could be that the deletion and point mutations in the coiled-coil domain cause the protein to fold in such a manner that the binding of the SH2 domain pocket by the peptide is precluded or masked. Based on the structural studies, the coiled-coil domain of Stat1 and Stat3β presents a predominantly hydrophilic surface area for interaction with other proteins (3, 8). Although it is unlikely that the coiled-coil domain interacts directly with the receptor peptide, it is possible that this domain regulates the accessibility of the SH2 domain for peptide binding via interactions with either another protein(s) or another domain(s) within the Stat3 protein. Since we have shown that the receptor binding of Stat3 does not require other proteins in vitro and the unlikely destruction of the SH2 domain in the point mutants, our hypothesis is that regulation of SH2 domain binding to the receptor may be mediated by an intramolecular interaction that involves the coiled-coil domain. Removal of the first α-helix, or more specifically mutation of the two surface residues Asp170 and Lys177, may result in loss of the intramolecular interaction which is essential to maintain SH2 domain accessibility to receptor binding but not for dimer formation. The proposed hypothesis is currently under investigation.
Stat3 plays an essential role in embryonic development, as well as in growth and differentiation of hematopoietic cells (18, 30, 35). Increasing evidence also shows an important role for Stat3 in oncogenesis. For example, Stat3 is constitutively activated in v-Src-transformed cells (7, 44) and plays a crucial role in the transformation of cells by v-Src (5, 36). Furthermore, constitutively activated Stat3 has been found in many tumor cell lines and primary tumors (14). More strikingly, Stat3 itself has been identified as an oncogene when it is spontaneously dimerized (6). Therefore, understanding the mechanisms that control Stat3 activation may provide useful information for cancer therapies, and the coiled-coil region of Stat3 may be a useful target for drug design and screening of small molecules that inhibit Stat3 activity by disruption of this domain.
ACKNOWLEDGMENTS
We thank J. E. Darnell, Jr., for plasmid pRC/CMV-Stat3 and E. Manser and L. Lim for the pXJ40-FLAG vector, Cheh Peng Lim for reading the manuscript, Robert Qi for helpful suggestions for the peptide-binding experiments, and R. Tham for photography.
K. T. Seow is a Structural Bioinformatics Scientist. This work was supported by the National Science and Technology Board of Singapore.
REFERENCES
- 1.Adachi M, Fischer E H, Ihle J, Imai K, Jirik F, Neel B, Pawson T, Shen S, Thomas M, Ullrich A, Zhao Z. Mammalian SH2-containing protein tyrosine phosphatases. Cell. 1996;85:15. doi: 10.1016/s0092-8674(00)81077-6. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Nishio Y, Inoue M, Wang X J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 3.Becker S, Groner B, Müller C M. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 5.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bromberg J F, Wrzeszczynska M H, Devgan G, Zhao Y, Pestell R G, Albanese C, Darnell J E., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Tay A, Guy G R, Tan Y H. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 9.Coffer P J, Kruijer W. EGF receptor deletions define a region specifically mediating STAT transcription factor activation. Biochem Biophys Res Commun. 1995;210:74–81. doi: 10.1006/bbrc.1995.1629. [DOI] [PubMed] [Google Scholar]
- 10.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 11.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 12.Fu X-Y, Zhang J-J. Transcription factor p91 interacts with the epidermal growth factor receptor and mediates activation of the c-fos gene promoter. Cell. 1993;74:1135–1145. doi: 10.1016/0092-8674(93)90734-8. [DOI] [PubMed] [Google Scholar]
- 13.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 14.Garcia R, Jove R. Activation of STAT transcription factors in oncogenic tyrosine kinase signaling. J Biomed Sci. 1998;5:79–85. doi: 10.1007/BF02258360. [DOI] [PubMed] [Google Scholar]
- 15.Greenlund A C, Morales M O, Viviano B L, Yan H, Krolewski J, Schreiber R D. Stat recruitment by tyrosine-phosphorylated cytokine receptors: an ordered reversible affinity-driven process. Immunity. 1995;2:677–687. doi: 10.1016/1074-7613(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 16.Heim M H, Kerr I M, Stark G R, Darnell J E., Jr Contribution of STAT SH2 groups to specific interferon signaling by the Jak-STAT pathway. Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 17.Hemmann U, Gerhartz C, Heesel B, Sasse J, Kurapkat G, Grötzinger J, Wollmer A, Zhong Z, Darnell J E, Jr, Graeve L, Heinrich P C, Horn F. Differential activation of acute phase response factor/Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. II. Src homology SH2 domains define the specificity of Stat factor activation. J Biol Chem. 1996;271:12999–13007. doi: 10.1074/jbc.271.22.12999. [DOI] [PubMed] [Google Scholar]
- 18.Hirano T, Nakajima K, Hibi M. Signaling mechanisms through gp130: a model of the cytokine system. Cytokine Growth Factor Rev. 1997;8:241–252. doi: 10.1016/s1359-6101(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 19.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 21.Jain N, Zhang T, Kee W H, Li W, Cao X. Protein kinase C δ associates with and phosphorylates Stat3 in an interleukin-6-dependent manner. J Biol Chem. 1999;274:24392–24400. doi: 10.1074/jbc.274.34.24392. [DOI] [PubMed] [Google Scholar]
- 22.Kammerer R A. Alpha-helical coiled-coil oligomerization domains in extracellular proteins. Matrix Biol. 1997;15:555–565. doi: 10.1016/s0945-053x(97)90031-7. [DOI] [PubMed] [Google Scholar]
- 23.Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 24.Lumb K J, Kim P S. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 25.Lütticken C, Wegenka U M, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur A G, Wilks A F, Yasukawa K, Taga T, Kishimoto T, Barbieri G, Pellegrini S, Sendtner M, Heinrich P C, Horn F. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 26.Manser E, Loo T H, Koh C G, Zhao Z S, Chen X Q, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;2:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 27.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 28.Quelle F W, Thierfelder W, Witthuhn B A, Tang B, Cohen S, Ihle J N. Phosphorylation and activation of the DNA binding activity of purified Stat1 by the Janus protein-tyrosine kinases and the epidermal growth factor receptor. J Biol Chem. 1995;270:20775–20780. doi: 10.1074/jbc.270.35.20775. [DOI] [PubMed] [Google Scholar]
- 29.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 30.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 31.Shuai K, Horvath C M, Huang L H, Qureshi S A, Cowburn D, Darnell J E., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 32.Silvennoinen O, Schindler C, Schlessinger J, Levy D E. Ras-independent growth factor signaling by transcription factor tyrosine phosphorylation. Science. 1993;261:1736–1739. doi: 10.1126/science.8378775. [DOI] [PubMed] [Google Scholar]
- 33.Stahl N, Boulton T G, Farruggella T, Ip N Y, Davis S, Witthuhn B A, Quelle F W, Silvennoinen O, Barbieri G, Pellegrini S, Ihle J N, Yancopoulos G D. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science. 1994;263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 34.Stahl N, Farruggella T J, Boulton T G, Zhong Z, Darnell J E, Jr, Yancopoulos G D. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 35.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot R P, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Snick J. Interleukin-6: an overview. Annu Rev Immunol. 1990;8:253–278. doi: 10.1146/annurev.iy.08.040190.001345. [DOI] [PubMed] [Google Scholar]
- 38.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 39.Vinkemeier U, Moarefi I, Darnell J E, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 40.Wang D M, Moriggl R, Stravopodis D, Carpino N, Marine J-C, Teglund S, Feng J, Ihle J N. A small amphipathic α-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 2000;19:392–399. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Sun Y L, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 43.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. Differentiation and growth arrest signals are generated through the cytoplasmic region of gp130 that is essential for Stat3 activation. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 44.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Wrzeszczynska M H, Horvath C M, Darnell J E., Jr Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol Cell Biol. 1999;19:7138–7146. doi: 10.1128/mcb.19.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z, Wen Z, Darnell J E., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]