Abstract
Over the past decade, artificial intelligence (AI) has contributed substantially to the resolution of various medical problems, including cancer. Deep learning (DL), a subfield of AI, is characterized by its ability to perform automated feature extraction and has great power in the assimilation and evaluation of large amounts of complicated data. On the basis of a large quantity of medical data and novel computational technologies, AI, especially DL, has been applied in various aspects of oncology research and has the potential to enhance cancer diagnosis and treatment. These applications range from early cancer detection, diagnosis, classification and grading, molecular characterization of tumors, prediction of patient outcomes and treatment responses, personalized treatment, automatic radiotherapy workflows, novel anti‐cancer drug discovery, and clinical trials. In this review, we introduced the general principle of AI, summarized major areas of its application for cancer diagnosis and treatment, and discussed its future directions and remaining challenges. As the adoption of AI in clinical use is increasing, we anticipate the arrival of AI‐powered cancer care.
Keywords: artificial intelligence, cancer diagnosis, cancer research, cancer treatment, convolutional neural network, deep learning, deep neural network, oncology
This review introduced the general principle of AI, summarize major areas of its application for cancer diagnosis, treatment, and precision medicine and discuss its future directions and remaining challenges.
Abbreviations
- AI
artificial intelligence
- ML
machine learning
- DL
deep learning
- DNN
deep neural network
- CNN
convolutional neural network
- CIN
intra‐epithelial neoplasia
- DS
dual‐stained
- AUC
area under the curve
- ctDNA
circulating tumor DNA
- cfDNA
cell‐free DNA
- HE
hematoxylin‐eosin
- PET‐CT
positron emission tomography‐CT
- WSI
whole slide imaging
- CT
computed tomography
- MRI
magnetic resonance imaging
- 3D
three dimensional
- NPC
nasopharyngeal carcinoma
- GRAIDS
gastrointestinal AI diagnostic system
- MSI
microsatellite instability
- TMB
tumor mutational burden
- NSCLC
non‐small cell lung cancer
- PD‐L1
programmed death‐ligand 1
- pCR
complete pathologic response
- OAR
organs at risk
- GTV
gross tumor volume
- CTV
clinical target volume
- RCT
randomized controlled trial
- Fbw7
F‐box/WD repeat‐containing protein 7
- DDR1
discoidin domain receptor 1
- DDL
distributed deep learning
- FDA
Food and Drug Administration
- NAC
neoadjuvant chemotherapy
- DSC
Dice similarity coefficient
1. BACKGROUND
At a workshop in Dartmouth in the summer of 1956, McCarthy et al. [1] coined the term “artificial intelligence (AI)”, also known as “machine intelligence”. To put it simply, AI is defined as a programmed machine that can learn and recognize patterns and relationships between inputs and outputs and use this knowledge effectively for decision‐making on brand‐new input data [1, 2]. Machine learning (ML) and deep learning (DL) are the predominant methods used to actualize AI and are sometimes used synonymously. In the field of computer science, ML is a subfield of AI, and DL is a specific subset of ML that focuses on deep artificial neural networks (Figure 1). Over the past decade, following advances in big data, algorithms, computer power, and internet technology, DL has achieved unprecedented success in various tasks in various fields, including facial recognition, image classification, voice recognition, automatic translation, and healthcare [3]. Given the great number of patients diagnosed with cancers each year worldwide [4], there is an acute interest in the application of AI in oncology, and such interests include making accurate diagnosis of cancers using pathological slides and radiological images, predicting patient outcomes, and optimizing treatment decisions. AI therefore has the potential to solve the problem of unbalanced distribution of medical resources and improve cancer care.
FIGURE 1.
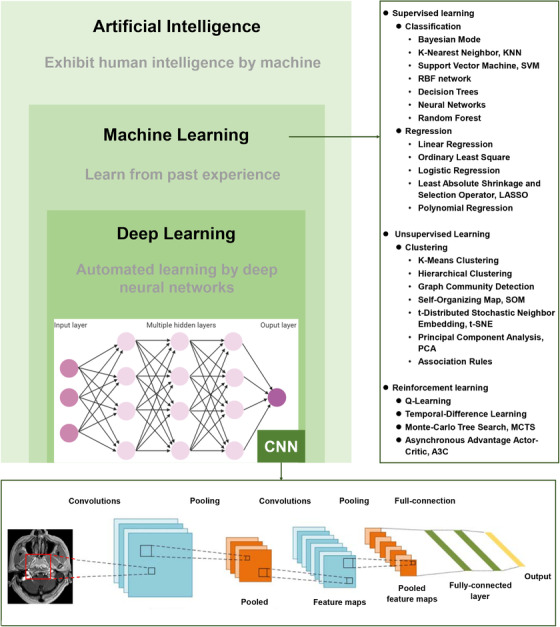
The relationship between artificial intelligence, machine learning, and deep learning and commonly used algorithms as examples. CNN, convolutional neural network
Inspired by brain neural architecture, DL uses deep neural networks (DNNs) to develop sophisticated models with multiple hidden layers to analyze various types of data and develop prediction outputs (Figure 1) [5]. Unlike conventional ML techniques, which require careful engineering to design a feature extractor that transforms raw data (such as the pixel values of an image) into relevant discriminatory features before data input, DL algorithms feed the machine with raw data with which it can automatically learn the optimal deep features that best fit the task through a training process [6, 7]. This ability likely explains the fact that DL algorithms have been consistently improved in many common AI tasks, such as image recognition, pattern recognition, speech recognition, and natural language processing. Consequently, a majority of AI research within the oncology field involves the utilization of DL.
Among DNN models, convolutional neural networks (CNNs) are the most popular DL architectures. They have been used for cancer lesion detection, recognition, segmentation and the classification of medical images [8, 9, 10]. The architecture of a typical CNN (Figure 1) is structured by stacking three main layers: convolutional layers, pooling layers, and fully‐connected layers. In doing this, CNNs transform the original images layer by layer from pixel values to the final prediction scores. The convolutional layers involve combining input data (feature map) with convolutional kernels (filters) to form a transformed feature map. The filters in the convolutional layers are automatically adjusted based on learned parameters to extract the most useful features for a specific task. Yet, there is a drawback; it is difficult to tell what features are learned by the CNNs, which is known as the “black box”.
Over the past five years, large amounts of researches have applied DL to cancer diagnosis, precision medicine, radiotherapy, and cancer research (Figure 2). Moreover, the American Food and Drug Administration (FDA) have approved a number of AI algorithms related to oncology (Table 1) and published a fast‐track approval plan for AI medical algorithms in 2018. Here, we provided an overview of the recent and enormous progresses in the application of AI in oncology in this review (Figure 3). We also highlight the limitations, challenges, and future implications of AI‐powered cancer care.
FIGURE 2.
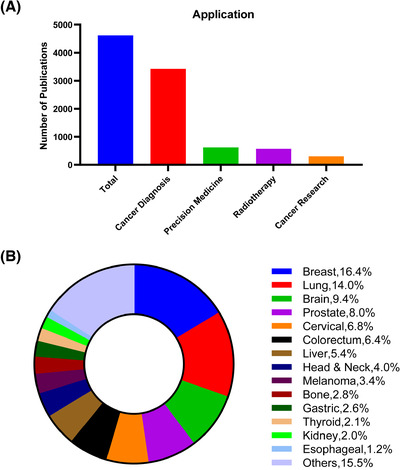
Publication statistics of deep learning by cancer area over the past five years, searched on PubMed. A. Publication statistics of deep learning by cancer diagnosis, precision medicine, radiotherapy, and cancer research. B. Publication statistics of deep learning for different cancer sites
TABLE 1.
Summary of FDA‐approved artificial intelligence devices in the field of oncology
| AI algorithm | Company | FDA approval date | Indication |
|---|---|---|---|
| ClearRead CT | Riverain Technologies | 09/09/2016 | Detection of pulmonary nodules |
| QuantX | Quantitative Insights | 07/19/2017 | Diagnosing breast cancer |
| Arterys Oncology DL | Arterys | 01/25/2018 | Liver and lung cancer diagnosis |
| cmTriage | CureMetrix | 03/08/2019 | Detection of suspicious breast lesions |
| Koios DS Breast | Koios Medical | 07/03/2019 | Breast lesion malignancy evaluation |
| ProFound AI Software V2.1 | iCAD | 10/04/2019 | Breast lesion malignancy evaluation |
| Transpara | ScreenPoint Medical BV | 03/05/2020 | Breast lesion malignancy evaluation |
| syngo.CT Lung CAD | Siemens Healthcare GmbH | 03/09/2020 | Detection of pulmonary nodules |
| MammoScreen | Therapixel | 03/25/2020 | Breast lesion malignancy evaluation |
| Rapid ASPECTS | iSchema View | 06/26/2020 | Detection of suspicious brain lesions |
| InferRead Lung CT.AI | InferRead Lung CT.AI | 07/02/2020 | Detection of pulmonary nodules |
| HealthMammo | Zebra Medical Vision | 07/16/2020 | Detection of suspicious breast lesions |
Abbreviations: FDA, Food and Drug Administration; AI, artificial intelligence; CT, computed tomography; DL, deep learning; CAD, computer‐aided diagnosis.
FIGURE 3.
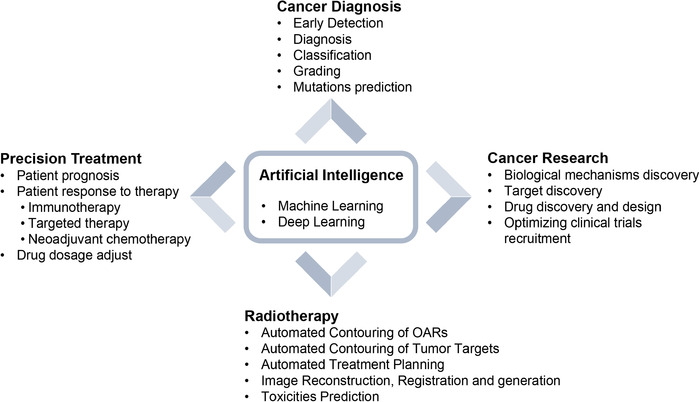
Applications of AI in cancer diagnosis, treatment and research. OARs, organs at risk
2. CANCER SCREENING, DIAGNOSIS, CLASSIFICATION, AND GRADING
Cancer screening for early detection, accurate cancer diagnosis, classification and grading are the key determinants of treatment decisions and patient outcomes. Over the past few years, there is increasing interest in the applications of AI in these critical areas (Table 2), sometimes with performance equivalent to human experts and advantages in scalability and time‐saving. More importantly, AI has shown its potential in solving challenging problems that humans simply cannot do.
TABLE 2.
Summary of key papers applying deep learning to cancer diagnosis and treatment
| Application | Reference | Task | Performance |
|---|---|---|---|
| Screening | |||
| Pathology | [14] | Automation of dual stain cytology in cervical cancer screening | Sensitivity, 87% |
| Endoscopy | [15] | Automation of polyp detection | False positive rate, 7.5% |
| Radiology | [16] | Predicting invasiveness of pulmonary adenocarcinomas | AUC, 0.788 |
| Radiology | [17] | Lung nodule classification: benign/malignant | Sensitivity, 98.45% |
| Radiology | [18] | Lung nodule classification: benign/malignant | Accuracy, 79.5% |
| Radiology | [19] | Lung nodule classification: benign/malignant | AUC, 0.944 |
| Radiology | [20] | Breast lesion classification: benign/malignant | AUC, 0.909 |
| Radiology | [21] | Breast lesion classification: benign/malignant | AUC, 0.860 |
| Radiology | [22] | Breast lesion classification: benign/malignant | AUC, 0.870 |
| Radiology | [23] | Breast lesion classification: benign/malignant | AUC, 0.860 |
| Radiology | [24] | Breast lesion classification: benign/malignant | AUC, 0.890 |
| Radiology | [25] | Breast cancer prediction | AUC, 0.8107 |
| Diagnosis | |||
| Pathology | [30] | Invasive breast cancer detection | DSC, 75.86% |
| Pathology | [31] | Breast cancer nodal metastasis detection | AUC, 0.994 |
| Pathology | [32] | Breast lesion classification: benign/malignant | Accuracy, 98.7% |
| Pathology | [33] | Detection of lymph node metastases in breast cancer | AUC, 0.994 |
| Pathology | [35] | Diagnosis of gastric cancer | AUC, 0.990‐0.996 |
| Pathology | [36] | Predicting origins for cancers of unknown primary | Accuracy, 80% |
| Pathology | [51] | Lung tumor classification: normal/ adenocarcinoma/squamous cell carcinoma | AUC, 0.97 |
| Pathology | [52] | Automated Gleason grading of prostate adenocarcinoma | Cohen's quadratic kappa statistic, 0.75 |
| Radiology | [37] | Brain tumor classification: normal/glioblastoma/sarcoma/metastatic bronchogenic carcinoma | AUC, 0.984 |
| Radiology | [38] | Liver cancer detection | Accuracy, 99.38% |
| Radiology | [39] | Prostate lesion classification: benign/malignant | AUC, 0.84 |
| Radiology | [40] | Detection of synchronous peritoneal carcinomatosis in colorectal cancer | Accuracy, 94.11% |
| Radiology | [41] | Detection of NPC using MRI | Accuracy, 97.77% |
| Radiology | [53] | Predicting grade of liver cancer | AUC, 0.83 |
| Endoscopy | [42] | Gastric lesion classification: normal/malignant | Accuracy, 96.49% |
| Endoscopy | [43] | Upper gastrointestinal cancer detection | Accuracy, 99.7% |
| Endoscopy | [44] | Polyps identification | Accuracy, 96% |
| Endoscopy | [50] | Polyps identification | AUC, 0.984 |
| Endoscopy | [45] | Invasive colorectal cancer diagnosis | Accuracy, 94.1% |
| Endoscopy | [46] | Diminutive colorectal polyps classification: hyperplastic/neoplastic | Accuracy, 90.1% |
| Endoscopy | [47] | cT1b colorectal cancer diagnosis | AUC, 0.871 |
| Endoscopy | [49] | Nasopharyngeal lesion classification: benign/malignant | Accuracy, 88% |
| Prediction of mutation | |||
| Pathology | [51] | Predicting genetic mutations of lung cancer: STK11, EGFR, FAT1, SETBP1, KRAS, and TP53 | AUC, 0.733‐0.856 |
| Pathology | [56] | Predicting genetic mutations of lung cancer: CTNNB1, FMN2, TP53, and ZFX4 | AUC>0.71 |
| Pathology | [59] | Predicting MSI status in colorectal cancer | AUC, 0.93 |
| Pathology | [60] | Predicting MSI status in colorectal cancer | AUC, 0.85 |
| Pathology | [61] | Predicting TMB status in gastric cancer | AUC, 0.75 |
| Pathology | [61] | Predicting TMB status in colon cancer | AUC, 0.82 |
| Radiology | [62] | Predicting EGFR status in NSCLC | AUC, 0.81 |
| Radiology | [63] | Predicting EGFR status in NSCLC | AUC, 0.81 |
| Radiology | [70] | Predicting TMB status in NSCLC | AUC, 0.81 |
| Predicting of prognosis | |||
| Pathology | [66] | Predicting outcome of colorectal cancer | AUC, 0.69 |
| Pathology | [67] | Predicting outcome of mesothelioma | Concordance index, 0.643 |
| Pathology | [68] | Predicting outcome of NSCLC | AUC, 0.85 |
| Immunotherapy | |||
| Radiology | [70] | Predicting response to immunotherapy in advanced NSCLC using TMB | AUC, 0.81 |
| Radiology | [74] | Predicting response to immunotherapy in NSCLC using MSI | AUC, 0.79 |
| Pathology | [72] | Predicting response to immunotherapy in advanced melanoma | AUC, 0.80 |
| Pathology | [73] | Predicting response to immunotherapy in gastrointestinal cancer using MSI | AUC > 0.99 |
| Chemotherapy | |||
| Radiology | [75] | Predicting response to NAC in breast cancer | AUC, 0.851 |
| Radiology | [76] | Predicting response to NAC in breast cancer | Accuracy, 88% |
| Radiology | [77] | Prediction response to NAC in rectal cancer | AUC, 0.83 |
| Radiology | [78] | Prediction response to NAC in NPC | Concordance index, 0.719‐0.757 |
| Radiology | [79] | Prediction response to NAC in NPC | Concordance index, 0.722 |
| Radiotherapy | |||
| Radiotherapy | [84] | Segmentation of OAR in head and neck | DSC, 37.4%‐89.5% |
| Radiotherapy | [85] | Segmentation of OAR in NPC | DSC, 86.1% |
| Radiotherapy | [86] | Segmentation of OAR in head and neck | DSC, 74% |
| Radiotherapy | [87] | Segmentation of OAR in head and neck | DSC, 60‐83% |
| Radiotherapy | [88] | Segmentation of OAR in head and neck | DSC, 53‐90% |
| Radiotherapy | [91] | 3D liver segmentation | DSC, 97.25% |
| Radiotherapy | [92] | Segmentation of CTV and OAR in rectal cancer | CTV: DSC, 87.7% |
| OAR: DSC, 61.8‐93.4% | |||
| Radiotherapy | [93] | Segmentation of OAR in esophageal cancer | DSC, 84‐97% |
| Radiotherapy | [94] | Contouring of GTV in NPC | DSC, 79% |
| Radiotherapy | [95] | Segmentation of CTV and OAR in cervical cancer | CTV: DSC, 86% |
| OAR: DSC, 82‐91% | |||
| Radiotherapy | [96] | Contouring of GTV in colorectal carcinoma | DSC, 75.5% |
| Radiotherapy | [97] | Contouring of CTV in NSCLC | DSC, 75% |
| Radiotherapy | [98] | Contouring of CTV in breast cancer | DSC, 91% |
| Radiotherapy | [99] | IMRT planning in NPC | Conformity index, 1.18‐1.42 |
| Radiotherapy | [102] | Prediction of dose distribution of IMRT in NPC | Dose difference, 4.7% |
| Radiotherapy | [103] | Prediction of three‐dimensional dose distribution of helical tomotherapy | Dose difference, 2‐4.2% |
| Radiotherapy | [104] | Prediction of dose distribution of IMRT in prostate cancer | Dose difference, 1.26‐5.07% |
| Radiotherapy | [105] | Prediction of three‐dimensional dose distribution | Dose difference < 0.5% |
Abbreviations: AUC, area under curve; NPC, nasopharyngeal carcinoma; MRI, magnetic resonance images; MSI, microsatellite instability; TMB, tumor mutation burden; NSCLC, non‐small cell lung cancer; NAC, neoadjuvant chemotherapy; DSC, Dice similarity coefficient; OAR, organs at risk; GTV, gross tumor volume; CTV, clinical target volume; IMRT, intensity‐modulated radiation therapy.
2.1. Cancer screening and early detection
Cancer screening has contributed to decreasing the mortality of some common cancers [11, 12]. The most successful examples are the identification of precancerous lesions (e.g., cervical intra‐epithelial neoplasia [CIN] for cervical cancer screening, and adenomatous polyps for colorectal cancer screening) where the treatment leads to a decrease in the incidence of invasive cancer [13]. Given the requirement for high throughput technology and a fast turnaround, automation is being used to improve the efficiency of cancer screening.
For cervical cancer screening, Wentzensen et al. [14] developed a DL classifier for p16/Ki‐67 dual‐stained (DS) cytology slides trained on biopsy‐based gold standards. In independent testing, AI‐based DS had equal sensitivity and substantially higher specificity compared with a Pap smear and manual interpretation of DS. Most importantly, AI‐based DS reduced unnecessary colposcopies by one‐third compared with Pap smears (41.9% vs. 60.1%, P < 0.001), while it had a similar performance in identifying high‐grade CIN, which indicates immediate treatment. For colorectal cancer screening, a prospective randomized controlled trial including 1,058 patients showed that AI‐assisted colonoscopy significantly increased adenoma detection rates and the mean number of adenomas found per patient compared with conventional colonoscopy (29.1% vs. 20.3%), which was attributed to a higher number of diminutive adenomas found [15]. This is particularly important because a 1% increase in the adenoma detection rate is associated with a 3% decrease in colorectal cancer incidence [13].
Automated nodule detection and classification on low‐dose computed tomography (CT) and mammography for lung and breast cancer screening have attracted significant attention. Several successful CNN‐based models have achieved classification accuracies of 80% to 95% [16, 17, 18], which shows their transformative potential in lung cancer screening. Ardila et al. [19] proposed a DL algorithm that uses patients current and prior low‐dose CT scans to predict the risk of lung cancer with outstanding results (area under the curve [AUC] of receiver operating characteristic = 0.944). Improvement in breast cancer screening with AI mammography has also been verified in preclinical studies [20, 21, 22, 23, 24], as well as in clinical settings [25]. McKinney et al. [25] established an AI system for breast cancer screening using an ensemble of three CNN‐based models. A reduction in the numbers of false positives and false negatives was observed compared with the original decisions made in the course of clinical practice. In an independent study by six radiologists, the AUC for the AI system was 11.5% higher than the average AUC achieved by the 6 radiologists. Notably, this AI system has the ability to generalize from the training data to multicenter data.
An emerging area for the early detection of cancers is liquid biopsies for circulating tumor DNA (ctDNA) or cell‐free DNA (cfDNA) obtained via a simple blood test. These are particularly important for cancer types that currently have no effective screening method. In a promising work, Cohen et al. [26] developed CancerSEEK for the early detection and prediction of eight cancer types using ctDNA. With CancerSEEK, samples are first classified as cancer‐positive using a logistic regression model applied to 16 gene mutations and the expression levels of 8 plasma proteins. The cancer type is then predicted using a random forest classifier, with accuracies ranging from 39% to 84%. Although liquid biopsies are promising for early cancer detection, so far, they have been limited to traditional ML algorithms [27, 28]. As data acquisition from liquid biopsies increases, we anticipate that DL models will eliminate the need for manual selection and curation of discriminatory features, as well as allowing for the combination of multiple data types to enhance early cancer detection.
2.2. Cancer diagnosis, classification, and grading
CNN‐based DL models that can accurately diagnose cancers, classify cancer subtypes, and identify cancer grades using histopathology (e.g., whole slide imaging [WSI]) [29], radiology (e.g., CT and magnetic resonance imaging [MRI]), and endoscopy images (e.g., esophagogastroduodenoscopy and colonoscopy) have been extensively reported, and most of them exhibit accuracies at least equivalent to that of professionals.
For cancer diagnosis, CNN‐based DL models have exhibited exceptional accuracy in identifying malignant tumors using histopathology slides [30, 31, 32, 33, 34, 35]. In an international competition (CAMELYON16) for diagnosing breast cancer metastasis in lymph nodes using WSI with hematoxylin‐eosin (HE) staining, the best CNN algorithm (a GoogLeNet architecture‐based model) yielded an AUC of 0.994, outperforming the best pathologist with an AUC of 0.884 and in a more time‐efficient manner [33]. DL algorithms have also been adopted to predict the origin of unknown primary cancers, which is extremely challenging in cancer diagnosis [36].
The success of DL has also been consistently reported in the diagnosis of malignant diseases using CT, MRI, positron emission tomography‐CT (PET‐CT) scans [37, 38, 39, 40, 41], and endoscopy [42, 43, 44, 45, 46, 47, 48, 49, 50]. Most recently, Yuan et al. [40] used CT scans to develop a classifier using a three‐dimensional (3D) ResNet algorithm to predict occult peritoneal metastasis in colorectal cancer with an AUC of 0.922, which was substantially higher than that achieved via routine contrast‐enhanced CT diagnosis (AUC = 0.791). In another work, Ke et al. [41] used MRI images from 4,100 patients with nasopharyngeal carcinoma (NPC) to train and test a self‐constrained 3D DenseNet that could distinguish NPC from benign nasopharyngeal hyperplasia with a reported AUC of 0.95‐0.97. As for endoscopy, in a multicenter study, Luo et al. [43] developed a gastrointestinal AI diagnostic system (GRAIDS) for the diagnosis of upper gastrointestinal cancers using a CNN‐based model and tested it in a prospective study involving six different tiered hospitals. While the diagnostic accuracies varied from 0.915 to 0.977 among the six hospitals, they were similar to those of expert endoscopists and superior to those of non‐experts, thus indicating the potential benefit in improving the diagnostic effectiveness of community‐based hospitals. All in all, such models, if their performance is confirmed in multicenter prospective studies, may play an important role in making cancer diagnosis more accurate, especially in local hospitals that lack experts.
Aside from dichotomous diagnosis, DL models are used for more challenging cancer classifications and grading tasks. Coudray et al. [51] developed DeepPATH, an Inception‐v3 architecture‐based model, to classify WSI for lung tissues into three classes (normal, lung adenocarcinoma, and lung squamous cell carcinoma) with a reported AUC of 0.97. The CNN was also successfully trained to perform automated Gleason grading of prostate adenocarcinoma, with a 75% agreement between the algorithm and pathologists [52]. Cancer grading can also be done using radiology images. Zhou et al. [53] developed a DL approach (based on SENet and DenseNet) to predict liver cancer grades (low versus high) using MRI images with a reported AUC of 0.83. Overall, these studies show the promising application of AI in cancer classification and grading, with performances equal to trained experts.
From a technical and practical aspect, these DL‐based diagnostic tools integrate features for fine‐tuning and enhancing performance, which simplifies the pipelines of conventional computer‐aided diagnosis and reduces false positive rates [54]. Although preclinical assessments of AI tools have paved the way for clinical trials to improve the accuracy and efficiency of cancer diagnoses, the robustness and generalizability of DL models need to be improved [55].
2.3. Predicting gene mutations in cancer
DL algorithms have also been used to characterize the underlying genetic and epigenetic heterogeneity using histopathology images. Using HE‐stained WSI of lung cancer, a CNN was trained to predict six different genetic mutations with an AUC from 0.733 to 0.856 as measured on a held‐out testing cohort [51]. Using WSI, the CNN model (Inception‐V3) also identified common mutations in liver cancer with AUCs >0.71 [56]. Using WSI, DL tools have also been developed for the prediction of whole‐genome duplications, chromosome arm gains and losses, focal amplifications and deletions, and gene variations for pan‐cancer [57, 58]. Expanded from predicting mutations in individual genes, DL models have been used to predict mutational footprints, such as microsatellite instability (MSI) status and tumor mutational burden (TMB) status, which are the most important biomarkers for responses to checkpoint immunotherapy. Most recently, Yamashita et al. [59] trained and tested MSINet, a transfer learning model based on MobileNetV2 architecture, to classify MSI status in HE‐stained WSI in a colorectal cancer cohort of 100 primary tumors and reported an AUC of 0.93. Using multiple instances of learning‐based DL, Cao et al. [60] also tried to classify MSI status using WSI in a colorectal cancer cohort and achieved an AUC of 0.85. In a work to classify TMB status using WSI, Wang et al. [61] compared eight different DL models and reported GoogLeNet as the best model for gastric tumors (AUC = 0.75) and VGG‐19 as the best for colon cancer (AUC = 0.82). The results indicate that features from histopathology images can be used to predict genetic mutations in cases in which obtaining tumor specimens for mutation analysis are not possible. Notably, it may be more cost‐effective than direct sequencing.
In addition to histopathology images, identifying cancer mutations using noninvasive radiology images such as CT or MRI scans has been explored. For example, the prediction of EGFR mutation status in non‐small cell lung cancer (NSCLC) can be achieved using CT and PET/CT scans using DL models both with AUCs >0.81 [62, 63]. In another work, Shboul et al. [64] introduced a ML approach to predict O6‐methylguanine‐DNA methyltransferase methylation, isocitrate dehydrogenase mutation, 1p/19q co‐deletion, alpha‐thalassemia/mental retardation syndrome X‐linked mutation, and telomerase reverse transcriptase mutation of low‐grade gliomas with radiomics, and achieved AUCs from 0.70 to 0.84. CT scans have also been used to predict TMB status in NSCLC (AUC = 0.81). The results were promising, but understanding what features are being learned by the CNN models to determine mutation status remains under researched.
3. PATIENT PROGNOSIS, RESPONSE TO THERAPY, AND PRECISION MEDICINE
Precision medicine refers to the tailoring of treatment to individual patients [65]. It aims to classify individuals into subgroups with differences in their disease prognosis or in their response to a specific treatment and thus make therapeutic interventions for those who will benefit and sparing expense and side effects for those who will not. DL algorithms are used to automatically extract features from medical data to build models that can accurately predict risk of tumor relapse and patients’ responses to treatments [66, 67, 68]. Based on the prediction results, physicians can provide more precise and suitable treatments.
Immunotherapy drugs have been approved for the treatment of metastatic melanoma, lung cancer, and other malignancies. However, more than 50%‐80% of cancer patients fail to respond to checkpoint inhibitor therapy. Currently, response prediction for immunotherapies is based on biomarkers of the immunogenic tumor microenvironment, such as programmed death‐ligand 1 (PD‐L1) expression, TMB, MSI, and somatic copy number alterations. However, these biomarker data were acquired via a biopsy, which is invasive, difficult to perform longitudinally, and limited to a single tumor region. Furthermore, the predictive value of biomarkers may be limited. In the KEYNOTE‐189 clinical trial, immunotherapy with pembrolizumab combined with standard chemotherapy provided survival benefits for all patients regardless of their PD‐L1 expression [69]. To achieve the goal of precision medicine, many researchers have established DL models to predict patient biomarkers related to immunotherapy using radiomics and pathomics data [70, 71, 72, 73]. Johannet el al. developed a pipeline that integrates DL on histology specimens with clinical data to predict immunotherapy response in advanced melanoma [72]. The results showed that the classifier accurately stratified patients into responders and non‐responders with an AUC of 0.80. Most excitingly, Arbour et al. [74] developed a DL model that directly predicts the best overall response and progression‐free survival using radiology text reports for patients with NSCLC treated with a programmed cell death protein‐1 blockade. These studies underscore the potential ability of AI to identify individuals who may benefit from immunotherapy without the aforementioned negatives of biopsies.
In addition to immunotherapy, other therapies (e.g., targeted therapy and neoadjuvant chemotherapy [NAC]) have achieved prominent clinical success in specific populations, driving the need for accurate predictive assays to inform patient selection. This requirement can be met by a combination of big data and AI. AI predictive models can identify imaging phenotypes that are associated with a targeted mutation. This AI‐based approach has the advantage of identifying the mutation status repeatedly and noninvasively. This approach was supported by a PET/CT‐based DL model for patients with NSCLC, which uses radiomic features to discriminate EGFR‐mutant types from wild‐type with an AUC of 0.81 [62]. Moreover, with a large amount of radiomics data, DL algorithms have shown power in estimating responses to NAC for patients with breast cancer [75, 76], rectal cancer [77], and NPC [78, 79]. After NAC, about 35% of patients with locally advanced breast cancer achieved a pathologic complete response (pCR), which was associated with improved survival [80]. Whereas, a poor response to NAC was associated with an adverse prognosis [81]. Therefore, the accurate prediction of treatment response is warranted, which can avoid unnecessary toxicity and delays to surgery. Using pretreatment MRI from patients with locally advanced breast cancer, Ha et al. [76] trained a CNN to predict pCR, and no response/progression after NAC, reaching an overall accuracy of 88%. In addition to predicting patient responses to therapies, AI now offers additional avenues to adjust drug dosage for single or combinational therapies for individual patients in a dynamic manner using patient‐specific data collected over time [82].
4. DEEP LEANING IN RADIOTHERAPY
Radiotherapy constitutes an integral modality in the treatment of cancers with half of patients receiving it. The image‐, data‐driven and quality assurance frameworks of radiotherapy provide an excellent foundation for the development of AI algorithms and their integration into radiotherapy workflows. There has been an acute interest in exploring AI to facilitate radiotherapy for target volume and organs at risk (OAR) delineation and automated treatment planning [83].
Target volume and OAR delineation is a labor‐intensive process, and its accuracy depends heavily on the experience of the radiation oncologists. CNN‐based semantic segmentation has been consistently established as a state‐of‐the‐art tool in the automated delineation of OAR in head and neck [84, 85, 86, 87, 88], thorax [89], abdomen [90, 91] and pelvic regions [92]. OAR is usually delineated on CT images, and the runtime for each patient lasts only several seconds. From these published studies, the segmentation accuracies of organs with large volumes, rigid and regular shapes were rather high, such as those of the mandible (Dice similarity coefficient [DSC] = 0.94), parotid (DSC = 0.84), kidney (DSC = 0.96), and liver (DSC = 0.97), while for organs with small volumes, movable and irregular shapes, the segmentation accuracies decreased, such as those of the optic nerve (DSC = 0.69), chiasm (DSC = 0.37), intestine (DSC = 0.65), and esophagus (DSC = 0.83). Of note, preliminary studies have shown that differences in dosimetry parameters between automatic and manual delineations were small, and automatic segmentations performed sufficiently well for treatment planning purposes [87, 93].
Given the variety of shapes, locations, and internal morphologies of tumors, automated contouring of tumor targets by DL is still a great challenge. Nonetheless, automatic contouring speeds up the process and improves consistency among radiation oncologists. Automated delineation of the gross tumor volume (GTV) and clinical target volume (CTV) have been investigated in many cancers, such as nasopharyngeal [94], cervical [95], colorectal [92, 96], lung [97] and breast cancers [98]. Lin et al. [94] first constructed an automated contouring tool for NPC by applying a 3D CNN model to MRI. In this independent test, they found acceptable concordance between the AI tool and human experts, with an overall accuracy of 79%. Moreover, in a multicenter test involving eight radiation oncologists from seven hospitals, the AI tool outperformed half of the physicians and was equal to the other four. With AI's assistance, substantial improvement in the contouring accuracy among five of the eight physicians as well as significant reductions in the interobserver variation (by 54.5%) and contouring time (by 39.4%) were observed.
Another important application of AI in radiotherapy is automated treatment planning. Radiotherapy planning is a complex process that involves “trial‐and‐error” based on physicists’ subjective priorities to achieve specific dosimetry objectives. As a result, treatment planning quality depends heavily on the experience of the clinical physicists. While automated planning using knowledge‐based techniques, such as RapidPlan in Eclipse, have improved the consistency of planning quality [99, 100], these methods are suboptimal since they cannot provide estimations of patient‐specific achievable dose distributions. Recently, DL‐based methods have become a promising approach for individualized 3D dose prediction and optimization [101, 102, 103, 104]. Fan et al. [105] first developed an automated treatment planning strategy based on ResNet to achieve an accurate 3D dose prediction and voxel‐by‐voxel dose optimization for head and neck cancers. The results showed no significant difference between the predicted and real clinical plans for most clinically relevant dosimetry indices. More importantly, with this strategy, patients with different prescription doses can be learned and predicted in a single framework.
Other applications of AI in radiotherapy include the prediction of radiation‐induced toxicities [106, 107, 108], image reconstruction [109, 110, 111], synthetic CT generation [112, 113, 114], image registration [115, 116, 117], and intra‐ and inter‐fraction motion monitoring [118, 119, 120]. In summary, AI has the potential to improve the accuracy, efficiency and quality of radiotherapy. Furthermore, MRI‐only radiotherapy [121] and real‐time adaptive radiotherapy [109] could be achieved with the implementation of effective and efficient automated segmentation, image processing, and automated treatment planning tools based on DL, which are significantly faster than standard approaches.
5. DL IN CANCER RESEARCH
DL approaches have been applied in various aspects of cancer research, including investigating biological underpinnings, developing anti‐cancer therapeutics, and implementing randomized controlled trials (RCTs). To uncover the biological mechanisms of cancer, studies have used DL to analyze the relationship between genotypes and phenotypes with a large number of achievements already reported. In a recent study leveraging DL algorithms, the role of F‐box/WD repeat‐containing protein 7 (Fbw7) in cancer cell oxidative metabolism was discovered via gene expression signatures from The Cancer Gene Atlas dataset [122]. Watson for Genomics also recognized genomic alterations with potential clinical effects that were not identified by the conventional molecular tumor boards across a spectrum of cancer types [123]. Identification of these genetic variants not only pinpoints relevant biological pathways but also suggests targets for drug discovery. ML methods have also been employed to accelerate the early discovery of potential anti‐cancer agents [124, 125, 126, 127, 128, 129]. Valeria et al. [129] reported the first perturbation model combined with ML to enable the design and prediction of dual inhibitors cyclin‐dependent kinases 4 and human epidermal growth factor receptor 2 with sensitivity and specificity higher than 80%. Another key aspect of drug discovery is the determination of compounds with good on‐target effects and minimal off‐target effects. Zhavoronkov et al. [130] developed a DL model and discovered powerful inhibitors of the discoidin domain receptor 1 (DDR1, a kinase target implicated in multiple cancers) in just 21 days versus conventional timelines of approximately one year. All in all, DL AI is accelerating drug discovery and is already successfully predicting drug behavior.
The adoption of novel cancer treatments is dependent on successful RCTs. However, successful recruitment of appropriate patients into these trials is regarded as one of the most challenging aspects. Matching complicated eligibility criteria to potential subjects is a tedious, labor‐intensive, and difficult task [131]. To automate this, Hassanzadeh et al. [132] used natural language processing and a Multi‐Layer Perceptron model to extract meaningful information from patient records to help collate evidence for better decision making on the eligibility of patients according to certain inclusion and exclusion criteria,. It achieved an overall micro‐F1 score of 84%. Selecting top‐enrolling investigators is also essential for the efficient execution of RCTs. To facilitate the automation of selection, Gligorijevic et al. [133] proposed a DL approach to learn from both investigator‐ and trial‐related heterogeneous data sources and rank investigators based on their expected enrollment performance in new RCTs. Here, DL shows the potential to optimize clinical cancer trials.
6. CHALLENGES AND FUTURE IMPLICATIONS
While AI is widely investigated in oncology, studies need to be performed to translate DL models into real‐world applications. Barriers to improving doctors’ acceptance and performance of clinically applied DL include the generalizability of its applications, the interpretability of algorithms, data access, and medical ethics.
6.1. Generalizability and real‐world application
Because of the great heterogeneity in medical data across institutions, the performance of DL models tends to decrease when applied at different hospitals, therefore, external validation sets may be required to confirm their performance [55]. Additionally, the extremely large number of parameters in DL results in a high likelihood of overfitting and limiting of the generalizability across different populations [134]. More importantly, in clinical settings, to make a precise decision, oncologists need to consider a variety of data, including clinical manifestations, laboratory examinations, imaging data, and epidemiological histories. However, most recent studies have only adopted one type of data (such as imaging) as the input model. To mimic real clinical settings, a multimodal DL model incorporating the aforementioned information plus imaging data needs to be constructed in future studies.
6.2. Interpretability: the black‐box problem
DL has been criticized for being a “black box” that does not explain how the model generates outputs from given inputs. The large number of parameters involved makes it difficult for oncologists to understand how DL models analyze data and make decisions. However, some efforts have been made to make this black box more transparent [135, 136]. For example, the heat map‐like class activation algorithm, visualizes which image regions are taken into account with DL models when making decisions and to what degree. These innovative studies render DL tools more interpretable and applicable in clinical oncology settings.
6.3. Data access and medical ethics
DL studies not only face technological challenges but also resource and ethical challenges. The power and believability of DL relies on a large amount of training data. Limited data may cause overfitting, yielding an inferior performance in an external test cohort [134]. Given the concerns of protecting patient information, medical data are often the property of individual institutions, and there is a lack of data‐sharing systems to link institutions. Fortunately, this obstacle is beginning to be overcome, with privacy‐preserving distributed DL (DDL) and multicenter data‐sharing agreements [137, 138, 139]. DDL provides a privacy‐preserving solution to enable multiple parties to jointly learn via a deep model without explicitly sharing local datasets. The Cancer Imaging Archive, which collects clinical images from different institutes and hospitals, also provides a good example of data sharing and may promote radiomic studies [140]. In the future, an authoritative framework should be developed by governments and enterprises to realize secure data sharing. In addition, several ethical issues need to be addressed prior to the clinical implementation of DL tools. First, the degree of supervision required from physicians should be determined. Second, the responsible party for incorrect decisions made by DL tools should also be determined.
7. CONCLUSIONS
DL is a newly developed AI method in oncology which is rapidly progressing. With the growth of high‐quality medical data and the development of algorithms, DL methods have great potential in improving the precision and efficiency of cancer diagnosis and treatment. Moreover, the positive attitude of the FDA towards AI medical devices further increases the prospect of DL's practical application in oncology. For the realization of clinical implementation, future researches should focus on the reproducibility and interpretability to make DL methods more applicable.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
ZHC, LL, YS, and RHX conceived this study. ZHC, LL, and CFW drafted the manuscript. YS, RHX, and CFL revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
Not applicable.
Chen Z‐H, Lin L, Wu C‐F, Li C‐F, Xu R‐H, Sun Y. Artificial intelligence for assisting cancer diagnosis and treatment in the era of precision medicine. Cancer Commun. 2021;41:1100–1115. 10.1002/cac2.12215
Contributor Information
Rui‐Hua Xu, Email: xurh@sysucc.org.cn.
Ying Sun, Email: sunying@sysucc.org.cn.
REFERENCES
- 1. McCarthy J, Minsky ML, Rochester N, Shannon CE. A proposal for the dartmouth summer research project on artificial intelligence, august 31, 1955. AI Magazine. 2006;27(4):12. [Google Scholar]
- 2. Yasser E‐M, Honavar V, Hall A. Artificial Intelligence Research Laboratory. 2005.
- 3. Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2(10):719–31. [DOI] [PubMed] [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 5. Wainberg M, Merico D, Delong A, Frey BJ. Deep learning in biomedicine. Nat Biotechnol. 2018;36(9):829–38. [DOI] [PubMed] [Google Scholar]
- 6. Meyer P, Noblet V, Mazzara C, Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med. 2018;98:126–46. [DOI] [PubMed] [Google Scholar]
- 7. Samuel A. Some studies in machine learning using the game of checkers. IBMJ Res Dev. 1959;3:210–29. [Google Scholar]
- 8. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist‐level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. [Google Scholar]
- 10. Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69(2):127–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung‐cancer mortality with low‐dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Byers T, Wender RC, Jemal A, Baskies AM, Ward EE, Brawley OW. The American Cancer Society challenge goal to reduce US cancer mortality by 50% between 1990 and 2015: Results and reflections. CA Cancer J Clin. 2016;66(5):359–69. [DOI] [PubMed] [Google Scholar]
- 13. Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370(14):1298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wentzensen N, Lahrmann B, Clarke MA, Kinney W, Tokugawa D, Poitras N, et al. Accuracy and efficiency of deep‐learning‐based automation of dual stain cytology in cervical cancer screening. J Natl Cancer Inst. 2021;113(1):72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, et al. Real‐time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68(10):1813–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao W, Yang J, Sun Y, Li C, Wu W, Jin L, et al. 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018;78(24):6881–9. [DOI] [PubMed] [Google Scholar]
- 17. Kang G, Liu K, Hou B, Zhang N. 3D multi‐view convolutional neural networks for lung nodule classification. PLoS One. 2017;12(11):e0188290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ciompi F, Chung K, van Riel SJ, Setio AAA, Gerke PK, Jacobs C, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep. 2017;7:46479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End‐to‐end lung cancer screening with three‐dimensional deep learning on low‐dose chest computed tomography. Nat Med. 2019;25(6):954–61. [DOI] [PubMed] [Google Scholar]
- 20. Swiderski B, Kurek J, Osowski S, Kruk M, Barhoumi W, editors. Deep learning and non‐negative matrix factorization in recognition of mammograms. Eighth International Conference on Graphic and Image Processing (ICGIP 2016); 2017: International Society for Optics and Photonics. [Google Scholar]
- 21. Arevalo J, González FA, Ramos‐Pollán R, Oliveira JL, Lopez MAG, editors. Convolutional neural networks for mammography mass lesion classification. 2015 37th Annual international conference of the IEEE engineering in medicine and biology society (EMBC); 2015: IEEE. [DOI] [PubMed] [Google Scholar]
- 22. Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K. Mass detection in digital breast tomosynthesis: Deep convolutional neural network with transfer learning from mammography. Med Phys. 2016;43(12):6654–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. Journal of Medical Imaging. 2016;3(3):034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Medical Physics. 2017;44(10):5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94. [DOI] [PubMed] [Google Scholar]
- 26. Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science. 2018;359(6378):926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating genomic features for non‐invasive early lung cancer detection. Nature. 2020;580(7802):245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br J Cancer. 2021;124(4):686‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cruz‐Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih N. Accurate and reproducible invasive breast cancer detection in whole‐slide images: a deep learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. Epub 2017/04/19. 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y, Kohlberger T, Norouzi M, Dahl GE, Smith JL, Mohtashamian A, et al. Artificial intelligence–based breast cancer nodal metastasis detection: Insights into the black box for pathologists. Arch Pathol Lab Med. 2019;143(7):859‐68. [DOI] [PubMed] [Google Scholar]
- 32. Jannesari M, Habibzadeh M, Aboulkheyr H, Khosravi P, Elemento O, Totonchi M, et al., editors. Breast cancer histopathological image classification: a deep learning approach. 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2018: IEEE. [Google Scholar]
- 33. Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang Y, Yang M, Wang S, Li X, Sun Y. Emerging role of deep learning‐based artificial intelligence in tumor pathology. Cancer Commun (Lond). 2020;40(4):154–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song Z, Zou S, Zhou W, Huang Y, Shao L, Yuan J, et al. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun. 2020;11(1):4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, et al. AI‐based pathology predicts origins for cancers of unknown primary. Nature. 2021;594(7861):106–10. [DOI] [PubMed] [Google Scholar]
- 37. Mohsen H, El‐Dahshan E‐SA, El‐Horbaty E‐SM, Salem A‐BM. Classification using deep learning neural networks for brain tumors. Future Computing and Informatics Journal. 2018;3(1):68–71. [Google Scholar]
- 38. Das A, Acharya UR, Panda SS, Sabut S. Deep learning based liver cancer detection using watershed transform and Gaussian mixture model techniques. Cognitive Systems Research. 2019;54:165–75. [Google Scholar]
- 39. Liu S, Zheng H, Feng Y, Li W, editors. Prostate cancer diagnosis using deep learning with 3D multiparametric MRI. Medical imaging 2017: computer‐aided diagnosis; 2017: International Society for Optics and Photonics. [Google Scholar]
- 40. Yuan Z, Xu T, Cai J, Zhao Y, Cao W, Fichera A, et al. Development and Validation of an Image‐based Deep Learning Algorithm for Detection of Synchronous Peritoneal Carcinomatosis in Colorectal Cancer. Ann Surg. 2020. [DOI] [PubMed] [Google Scholar]
- 41. Ke L, Deng Y, Xia W, Qiang M, Chen X, Liu K, et al. Development of a self‐constrained 3D DenseNet model in automatic detection and segmentation of nasopharyngeal carcinoma using magnetic resonance images. Oral Oncol. 2020;110:104862. [DOI] [PubMed] [Google Scholar]
- 42. Lee JH, Kim YJ, Kim YW, Park S, Choi Y‐i, Kim YJ, et al. Spotting malignancies from gastric endoscopic images using deep learning. Surg Endosc. 2019;33(11):3790–7. [DOI] [PubMed] [Google Scholar]
- 43. Luo H, Xu G, Li C, He L, Luo L, Wang Z, et al. Real‐time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case‐control, diagnostic study. Lancet Oncol. 2019;20(12):1645–54. [DOI] [PubMed] [Google Scholar]
- 44. Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, et al. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology. 2018;155(4):1069–78. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, et al. Accuracy of diagnosing invasive colorectal cancer using computer‐aided endocytoscopy. Endoscopy. 2017;49(8):798–802. [DOI] [PubMed] [Google Scholar]
- 46. Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate classification of diminutive colorectal polyps using computer‐aided analysis. Gastroenterology. 2018;154(3):568–75. [DOI] [PubMed] [Google Scholar]
- 47. Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic diagnostic support system for cT1b colorectal cancer using deep learning. Oncology. 2019;96(1):44–50. [DOI] [PubMed] [Google Scholar]
- 48. Trasolini R, Byrne MF. Artificial intelligence and deep learning for small bowel capsule endoscopy. Dig Endosc. 2020;33:290–7. [DOI] [PubMed] [Google Scholar]
- 49. Li C, Jing B, Ke L, Li B, Xia W, He C, et al. Development and validation of an endoscopic images‐based deep learning model for detection with nasopharyngeal malignancies. Cancer Commun (Lond). 2018;38(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, et al. Development and validation of a deep‐learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng. 2018;2(10):741–8. [DOI] [PubMed] [Google Scholar]
- 51. Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non‐small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Q, Zhou Z, Chen C, Fan G, Chen G, Heng H, et al. Grading of hepatocellular carcinoma using 3D SE‐DenseNet in dynamic enhanced MR images. Comput Biol Med. 2019;107:47–57. [DOI] [PubMed] [Google Scholar]
- 54. Liu B, Chi W, Li X, Li P, Liang W, Liu H, et al. Evolving the pulmonary nodules diagnosis from classical approaches to deep learning‐aided decision support: three decades' development course and future prospect. J Cancer Res Clin Oncol. 2020;146(1):153–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross‐sectional study. PLoS Med. 2018;15(11):e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol. 2020;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fu Y, Jung AW, Torne RV, Gonzalez S, Vöhringer H, Shmatko A, et al. Pan‐cancer computational histopathology reveals mutations, tumor composition and prognosis. Nature Cancer. 2020;1(8):800–10. [DOI] [PubMed] [Google Scholar]
- 58. Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan‐cancer image‐based detection of clinically actionable genetic alterations. Nat Cancer. 2020;1(8):789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. 2021;22(1):132–41. [DOI] [PubMed] [Google Scholar]
- 60. Cao R, Yang F, Ma SC, Liu L, Zhao Y, Li Y, et al. Development and interpretation of a pathomics‐based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics. 2020;10(24):11080–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang L, Yudi J, Qiao Y, Zeng N, Yu R. A novel approach combined transfer learning and deep learning to predict TMB from histology image. Pattern Recognit Lett. 2020;135:244–8. [Google Scholar]
- 62. Mu W, Jiang L, Zhang J, Shi Y, Gray JE, Tunali I, et al. Non‐invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun. 2020;11(1):5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53(3):1800986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shboul ZA, Chen J, K MI. Prediction of Molecular Mutations in Diffuse Low‐Grade Gliomas using MR Imaging Features. Sci Rep. 2020;10(1):3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Timmerman L. What's in a name? A lot, when it comes to ‘precision medicine’. Xconomy; 2013. [Google Scholar]
- 66. Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8(1):3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Courtiol P, Maussion C, Moarii M, Pronier E, Pilcer S, Sefta M, et al. Deep learning‐based classification of mesothelioma improves prediction of patient outcome. Nat Med. 2019;25(10):1519–25. [DOI] [PubMed] [Google Scholar]
- 68. Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, et al. Predicting non‐small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016;7:12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gandhi L, Rodríguez‐Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus Chemotherapy in Metastatic Non‐Small‐Cell Lung Cancer. N Engl J Med. 2018;378(22):2078–92. [DOI] [PubMed] [Google Scholar]
- 70. He B, Di Dong YS, Zhou C, Fang M, Zhu Y, Zhang H, et al. Predicting response to immunotherapy in advanced non‐small‐cell lung cancer using tumor mutational burden radiomic biomarker. J Immunother Cancer. 2020;8(2):e000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bao X, Shi R, Zhao T, Wang Y. Immune landscape and a novel immunotherapy‐related gene signature associated with clinical outcome in early‐stage lung adenocarcinoma. J Mol Med (Berl). 2020;98(6):805–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johannet P, Coudray N, Donnelly DM, Jour G, Illa‐Bochaca I, Xia Y, et al. Using machine learning algorithms to predict immunotherapy response in patients with advanced melanoma. Clin Cancer Res. 2021;27(1):131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Arbour KC, Luu AT, Luo J, Rizvi H, Plodkowski AJ, Sakhi M, et al. Deep learning to estimate RECIST in patients with NSCLC treated with PD‐1 blockade. Cancer Discov. 2021;11:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huynh BQ, Antropova N, Giger ML, editors. Comparison of breast DCE‐MRI contrast time points for predicting response to neoadjuvant chemotherapy using deep convolutional neural network features with transfer learning. Medical imaging 2017: computer‐aided diagnosis; 2017: International Society for Optics and Photonics. [Google Scholar]
- 76. Ha R, Chin C, Karcich J, Liu MZ, Chang P, Mutasa S, et al. Prior to initiation of chemotherapy, can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J Digit Imaging. 2019;32(5):693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shi L, Zhang Y, Nie K, Sun X, Niu T, Yue N, et al. Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre‐treatment and mid‐radiation multi‐parametric MRI. Magn Reson Imaging. 2019;61:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Qiang M, Li C, Sun Y, Sun Y, Ke L, Xie C, et al. A prognostic predictive system based on deep learning for locoregionally advanced nasopharyngeal carcinoma. J Natl Cancer Inst. 2021;113:606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Peng H, Dong D, Fang M‐J, Li L, Tang L‐L, Chen L, et al. Prognostic value of deep learning PET/CT‐based radiomics: potential role for future individual induction chemotherapy in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2019;25(14):4271–9. [DOI] [PubMed] [Google Scholar]
- 80. Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long‐term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–72. [DOI] [PubMed] [Google Scholar]
- 81. Cortazar P, Geyer CE, Jr . Pathological complete response in neoadjuvant treatment of breast cancer. Ann Surg Oncol. 2015;22(5):1441–6. [DOI] [PubMed] [Google Scholar]
- 82. Blasiak A, Khong J, Kee T. CURATE.AI: Optimizing personalized medicine with artificial intelligence. SLAS Technol. 2020;25(2):95–105. [DOI] [PubMed] [Google Scholar]
- 83. Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, et al. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46(1):e1–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ibragimov B, Xing L. Segmentation of organs‐at‐risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44(2):547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liang S, Tang F, Huang X, Yang K, Zhong T, Hu R, et al. Deep‐learning‐based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur Radiol. 2019;29(4):1961–7. [DOI] [PubMed] [Google Scholar]
- 86. van Dijk LV, Van den Bosch L, Aljabar P, Peressutti D, Both S, JHMS R, et al. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiother Oncol. 2020;142:115–23. [DOI] [PubMed] [Google Scholar]
- 87. van Rooij W, Dahele M, Brandao HR, Delaney AR, Slotman BJ, Verbakel WF. Deep learning‐based delineation of head and neck organs at risk: Geometric and dosimetric evaluation. Int J Radiat Oncol Biol Phys. 2019;104(3):677–84. [DOI] [PubMed] [Google Scholar]
- 88. Brunenberg EJL, Steinseifer IK, van den Bosch S, Kaanders J, Brouwer CL, Gooding MJ, et al. External validation of deep learning‐based contouring of head and neck organs at risk. Phys Imaging Radiat Oncol. 2020;15:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126(2):312–7. [DOI] [PubMed] [Google Scholar]
- 90. Kline TL, Korfiatis P, Edwards ME, Blais JD, Czerwiec FS, Harris PC, et al. Performance of an artificial multi‐observer deep neural network for fully automated segmentation of polycystic kidneys. J Digit Imaging. 2017;30(4):442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61(24):8676. [DOI] [PubMed] [Google Scholar]
- 92. Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44(12):6377–89. [DOI] [PubMed] [Google Scholar]
- 93. Zhu J, Chen X, Yang B, Bi N, Zhang T, Men K, et al. Evaluation of Automatic Segmentation Model With Dosimetric Metrics for Radiotherapy of Esophageal Cancer. Front Oncol. 2020;10:564737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lin L, Dou Q, Jin Y‐M, Zhou G‐Q, Tang Y‐Q, Chen W‐L, et al. Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal carcinoma. Radiology. 2019;291(3):677–86. [DOI] [PubMed] [Google Scholar]
- 95. Liu Z, Liu X, Guan H, Zhen H, Sun Y, Chen Q, et al. Development and validation of a deep learning algorithm for auto‐delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother Oncol. 2020;153:172–9. [DOI] [PubMed] [Google Scholar]
- 96. Huang YJ, Dou Q, Wang ZX, Liu LZ, Jin Y, Li CF, et al. 3‐D RoI‐Aware U‐Net for Accurate and Efficient Colorectal Tumor Segmentation. IEEE Trans Cybern. 2020. [DOI] [PubMed] [Google Scholar]
- 97. Bi N, Wang J, Zhang T, Chen X, Xia W, Miao J, et al. Deep Learning Improved Clinical Target Volume Contouring Quality and Efficiency for Postoperative Radiation Therapy in Non‐small Cell Lung Cancer Front Oncol. 2019;9:1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Men K, Zhang T, Chen X, Chen B, Tang Y, Wang S, et al. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med. 2018;50:13–9. [DOI] [PubMed] [Google Scholar]
- 99. Chang ATY, Hung AWM, Cheung FWK, Lee MCH, Chan OSH, Philips H, et al. Comparison of Planning Quality and Efficiency Between Conventional and Knowledge‐based Algorithms in Nasopharyngeal Cancer Patients Using Intensity Modulated Radiation Therapy. Int J Radiat Oncol Biol Phys. 2016;95(3):981–90. [DOI] [PubMed] [Google Scholar]
- 100. Tseng M, Ho F, Leong YH, Wong LC, Tham IW, Cheo T, et al. Emerging radiotherapy technologies and trends in nasopharyngeal cancer. Cancer Commun (Lond). 2020;40(9):395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang C, Zhu X, Hong JC, Zheng D. Artificial Intelligence in Radiotherapy Treatment Planning: Present and Future. Technol Cancer Res Treat. 2019;18:1533033819873922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen X, Men K, Li Y, Yi J, Dai J. A feasibility study on an automated method to generate patient‐specific dose distributions for radiotherapy using deep learning. Med Phys. 2019;46(1):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liu Z, Fan J, Li M, Yan H, Hu Z, Huang P, et al. A deep learning method for prediction of three‐dimensional dose distribution of helical tomotherapy. Med Phys. 2019;46(5):1972–83. [DOI] [PubMed] [Google Scholar]
- 104. Nguyen D, Long T, Jia X, Lu W, Gu X, Iqbal Z, et al. A feasibility study for predicting optimal radiation therapy dose distributions of prostate cancer patients from patient anatomy using deep learning. Sci Rep. 2019;9(1):1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fan J, Wang J, Chen Z, Hu C, Zhang Z, Hu W. Automatic treatment planning based on three‐dimensional dose distribution predicted from deep learning technique. Med Phys. 2019;46(1):370–81. [DOI] [PubMed] [Google Scholar]
- 106. Valdes G, Solberg TD, Heskel M, Ungar L, Simone CB, 2nd . Using machine learning to predict radiation pneumonitis in patients with stage I non‐small cell lung cancer treated with stereotactic body radiation therapy. Phys Med Biol. 2016;61(16):6105–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhen X, Chen J, Zhong Z, Hrycushko B, Zhou L, Jiang S, et al. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: a feasibility study. Phys Med Biol. 2017;62(21):8246–63. [DOI] [PubMed] [Google Scholar]
- 108. Ibragimov B, Toesca D, Chang D, Yuan Y, Koong A, Xing L. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Med Phys. 2018;45(10):4763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Terpstra ML, Maspero M, d'Agata F, Stemkens B, Intven MPW, Lagendijk JJW, et al. Deep learning‐based image reconstruction and motion estimation from undersampled radial k‐space for real‐time MRI‐guided radiotherapy. Phys Med Biol. 2020;65(15):155015. [DOI] [PubMed] [Google Scholar]
- 110. Jiang Z, Chen Y, Zhang Y, Ge Y, Yin FF, Ren L. Augmentation of CBCT Reconstructed From Under‐Sampled Projections Using Deep Learning. IEEE Trans Med Imaging. 2019;38(11):2705‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Madesta F, Sentker T, Gauer T, Werner R. Self‐contained deep learning‐based boosting of 4D cone‐beam CT reconstruction. Med Phys. 2020;47(11):5619–31. [DOI] [PubMed] [Google Scholar]
- 112. Maspero M, Savenije MHF, Dinkla AM, Seevinck PR, Intven MPW, Jurgenliemk‐Schulz IM, et al. Dose evaluation of fast synthetic‐CT generation using a generative adversarial network for general pelvis MR‐only radiotherapy. Phys Med Biol. 2018;63(18):185001. [DOI] [PubMed] [Google Scholar]
- 113. Liu Y, Lei Y, Wang Y, Wang T, Ren L, Lin L, et al. MRI‐based treatment planning for proton radiotherapy: dosimetric validation of a deep learning‐based liver synthetic CT generation method. Phys Med Biol. 2019;64(14):145015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bird D, Nix MG, McCallum H, Teo M, Gilbert A, Casanova N, et al. Multicentre, deep learning, synthetic‐CT generation for ano‐rectal MR‐only radiotherapy treatment planning. Radiother Oncol. 2020;156:23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lei Y, Fu Y, Wang T, Liu Y, Patel P, Curran WJ, et al. 4D‐CT deformable image registration using multiscale unsupervised deep learning. Phys Med Biol. 2020;65(8):085003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fu Y, Wang T, Lei Y, Patel P, Jani AB, Curran WJ, et al. Deformable MR‐CBCT prostate registration using biomechanically constrained deep learning networks. Med Phys. 2020;48:253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Duan L, Ni X, Liu Q, Gong L, Yuan G, Li M, et al. Unsupervised learning for deformable registration of thoracic CT and cone‐beam CT based on multiscale features matching with spatially adaptive weighting. Med Phys. 2020;47(11):5632–47. [DOI] [PubMed] [Google Scholar]
- 118. Mylonas A, Keall PJ, Booth JT, Shieh CC, Eade T, Poulsen PR, et al. A deep learning framework for automatic detection of arbitrarily shaped fiducial markers in intrafraction fluoroscopic images. Med Phys. 2019;46(5):2286–97. [DOI] [PubMed] [Google Scholar]
- 119. Kim KH, Park K, Kim H, Jo B, Ahn SH, Kim C, et al. Facial expression monitoring system for predicting patient's sudden movement during radiotherapy using deep learning. J Appl Clin Med Phys. 2020;21(8):191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Roggen T, Bobic M, Givehchi N, Scheib SG. Deep Learning model for markerless tracking in spinal SBRT. Phys Med. 2020;74:66–73. [DOI] [PubMed] [Google Scholar]
- 121. Florkow MC, Guerreiro F, Zijlstra F, Seravalli E, Janssens GO, Maduro JH, et al. Deep learning‐enabled MRI‐only photon and proton therapy treatment planning for paediatric abdominal tumours. Radiother Oncol. 2020;153:220–7. [DOI] [PubMed] [Google Scholar]
- 122. Davis RJ, Gönen M, Margineantu DH, Handeli S, Swanger J, Hoellerbauer P, et al. Pan‐cancer transcriptional signatures predictive of oncogenic mutations reveal that Fbw7 regulates cancer cell oxidative metabolism. Proc Natl Acad Sci USA. 2018;115(21):5462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Patel NM, Michelini VV, Snell JM, Balu S, Hoyle AP, Parker JS, et al. Enhancing Next‐Generation Sequencing‐Guided Cancer Care Through Cognitive Computing. Oncologist. 2018;23(2):179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Speck‐Planche A, Scotti MT. BET bromodomain inhibitors: fragment‐based in silico design using multi‐target QSAR models. Mol Divers. 2019;23(3):555–72. [DOI] [PubMed] [Google Scholar]
- 125. Kleandrova VV, Scotti MT, Scotti L, Nayarisseri A, Speck‐Planche A. Cell‐based multi‐target QSAR model for design of virtual versatile inhibitors of liver cancer cell lines. SAR QSAR Environ Res. 2020;31(11):815–36. [DOI] [PubMed] [Google Scholar]
- 126. Speck‐Planche A. Multicellular Target QSAR Model for Simultaneous Prediction and Design of Anti‐Pancreatic Cancer Agents. ACS Omega. 2019;4(2):3122–32. [Google Scholar]
- 127. Speck‐Planche A, Cordeiro MNDS. Fragment‐based in silico modeling of multi‐target inhibitors against breast cancer‐related proteins. Mol Divers. 2017;21(3):511–23. [DOI] [PubMed] [Google Scholar]
- 128. Speck‐Planche A. Combining Ensemble Learning with a Fragment‐Based Topological Approach To Generate New Molecular Diversity in Drug Discovery: In Silico Design of Hsp90 Inhibitors. ACS Omega. 2018;3(11):14704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kleandrova VV, Scotti MT, Scotti L, Speck‐Planche A. Multi‐target Drug Discovery via PTML Modeling: Applications to the Design of Virtual Dual Inhibitors of CDK4 and HER2. Curr Top Med Chem. 2021;21(7):661–75. [DOI] [PubMed] [Google Scholar]
- 130. Zhavoronkov A, Ivanenkov YA, Aliper A, Veselov MS, Aladinskiy VA, Aladinskaya AV, et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol. 2019;37(9):1038–40. [DOI] [PubMed] [Google Scholar]
- 131. Kadam RA, Borde SU, Madas SA, Salvi SS, Limaye SS. Challenges in recruitment and retention of clinical trial subjects. Perspect Clin Res. 2016;7(3):137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hassanzadeh H, Karimi S, Nguyen A. Matching patients to clinical trials using semantically enriched document representation. J Biomed Inform. 2020;105:103406. [DOI] [PubMed] [Google Scholar]
- 133. Gligorijevic J, Gligorijevic D, Pavlovski M, Milkovits E, Glass L, Grier K, et al. Optimizing clinical trials recruitment via deep learning. J Am Med Inform Assoc. 2019;26(11):1195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mummadi SR, Al‐Zubaidi A, Hahn PY. Overfitting and Use of Mismatched Cohorts in Deep Learning Models: Preventable Design Limitations. Am J Respir Crit Care Med. 2018;198(4):544‐5. [DOI] [PubMed] [Google Scholar]
- 135. Yang JH, Wright SN, Hamblin M, McCloskey D, Alcantar MA, Schrübbers L, et al. A white‐box machine learning approach for revealing antibiotic mechanisms of action. Cell. 2019;177(6):1649–61. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Samek W, Wiegand T, Müller K‐R. Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv preprint arXiv:170808296. 2017.
- 137. London JW. Cancer Research Data‐Sharing Networks. JCO Clin Cancer Inform. 2018;2:1–3. [DOI] [PubMed] [Google Scholar]
- 138. Ross JS, Waldstreicher J, Bamford S, Berlin JA, Childers K, Desai NR, et al. Overview and experience of the YODA Project with clinical trial data sharing after 5 years. Sci Data. 2018;5:180268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Duan J, Zhou J, Li Y. Privacy‐Preserving distributed deep learning based on secret sharing. Information Sciences. 2020;527:108–27. [Google Scholar]
- 140. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


