Abstract
Chronic liver disease (CLD) is currently a major health problem worldwide, which is accompanied by chronic liver injury and lack of clinically effective treatment; however, systematic characterization of chronic liver injury procedures at single-cell resolution is lacking. In the present study, we established chronic liver injury mouse models and conducted single-cell RNA sequencing (scRNA-seq), including choline-deficient, ethionine-supplemented (CDE) and 3,5-diethoxycarbonyl 1,4-dihydrocollidinen (DDC) mouse models. We captured in total 16,389 high-quality cells and identified 12 main cell types in scRNA-seq data. Macrophages and endothelial cells are the largest cell populations in our dataset. Transcriptional trajectory analysis revealed different expression patterns of cells between CDE and DDC models and identified potential liver injury markers, such as Ets1, Gda, Itgam, and Sparc. Differential analysis identified 25 and 152 differentially expressed genes in CDE and DDC macrophages, respectively. In addition, 413 genes were detected to exclusively express in specific pseudotime states of macrophages. These genes were found to participate in immune-related biological processes. Further cell-cell communication analysis found extensive receding of cell-cell interactions between different cell types in the liver injury process, especially in the DDC model. Our study characterized the single-cell transcriptional landscape in the process of chronic liver injury, promoting the understanding of the underlying molecular mechanisms and providing candidate clinical strategy for effective intervention of chronic liver diseases.
Keywords: chronic liver injury, single-cell transcriptome, macrophage, cell-cell communications
Graphical abstract
We systematically characterized single-cell transcriptome in two different liver injury mouse models by single-cell RNA sequencing (scRNA-seq).
Introduction
Although the liver has a strong regenerative ability, chronic liver disease (CLD) and liver cirrhosis are growing health problems around the world. In 2018, according to data from the Centers for Disease Control and Prevention, in the United States, the population of adults diagnosed with liver diseases was approximately 4.5 million, and the number of deaths due to these complications was 44,358. The most common etiologies of CLD and cirrhosis are chronic hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol-related liver disease (ALD), and non-alcoholic fatty liver disease (NAFLD). Globally, 1.5 billion people were troubled by CLD in 2017, most of them were caused by NAFLD (60%), HBV (29%), HCV (9%), and ALD (2%).1 According to data from the Global Burden of Disease study, in 2015, the age-standardized incidence rate of cirrhosis and CLD was 20.7 per 100,000, which increased 13% from 2000. It is estimated that the incidence of liver cirrhosis in Europe is 26.0 per 100,000, 16.5 per 100,000 in East Asia, and 23.6 per 100,000 in Southeast Asia.2 Most chronic liver diseases are characterized by hepatocyte necrosis, inflammation, liver fibrosis, ductular reaction (DR), or proliferation of hepatic progenitor cells (HPCs). Current treatments for CLD are limited, and liver transplantation is the only treatment available to patients diagnosed with liver failure. Although chronic liver injury is relatively common, the cellular, molecular, and biological mechanisms that promote the progression of chronic liver injury have not been fully understood. Identifying common molecular pathways that promote liver injury in different experimental models of chronic liver injury will help in-depth understanding of the underlying molecular mechanisms and the development of better therapeutic strategy to intervene in the progression of CLDs.
NAFLD is currently the most popular liver disease in the world. Non-alcoholic steatohepatitis is a serious clinical feature of NAFLD, which is characterized by the accumulation of lipid droplets in liver cells, and undergoes liver cell death, inflammatory cell infiltration, and liver fibrosis, and can progressively develop to liver cirrhosis and liver cancer in severe cases.3,4 Therefore, it is very important to figure out the relevant molecular mechanisms of the pathological process from simple steatosis to steatohepatitis. The choline-deficient, ethionine-supplemented (CDE) diet is widely used as a chronic liver injury model in mice, which can induce steatohepatitis and HPC expansions.5 The pathogenic mechanism of the CDE diet mouse model is that the lack of choline in the diet leads to impaired assembly and secretion of very low-density lipoprotein. Combined with the hepatocarcinogen ethionine, the CDE diet will lead to hepatic fat overload, persistent inflammation, portal fibrosis, HPC response, and development of hepatocellular carcinoma.6, 7, 8, 9 Long-term feeding of the 3,5-diethoxycarbonyl 1,4-dihydrocollidinen (DDC) diet will cause intraductal porphyrin plug, duct obstruction, and cholestasis, which will lead to activation of cholangiocytes, increased DR, increased pro-inflammatory and pro-fibrotic factors, and subsequent hepatic fibrosis and cirrhosis. The DDC model is useful in studies aiming at investigating DR cell contribution in liver regeneration under injury conditions.10,11 Therefore, we chose the CDE and DDC diet models to investigate the molecular alterations in different liver injury conditions.
Herein, we performed single-cell RNA sequencing (scRNA-seq) of liver tissues derived from wild-type (WT) normal, CDE, and DDC mice. We presented a systematic characterization of the transcriptional landscape of liver injury process at single-cell resolution. CDE and DDC models showed different expression patterns along pseudotime in the process of chronic liver injury. Macrophages were found to exhibit variations of response to pathological changes. Cell-cell interactions were found to sharply decrease in both CDE and DDC liver injury models. This study shed lights on the single-cell transcriptional alterations in CDE and DDC liver injury models and suggested potential strategy for clinical intervention of CLDs.
Results
Pathological features of chronic liver injury induced by CDE and DDC diet feeding
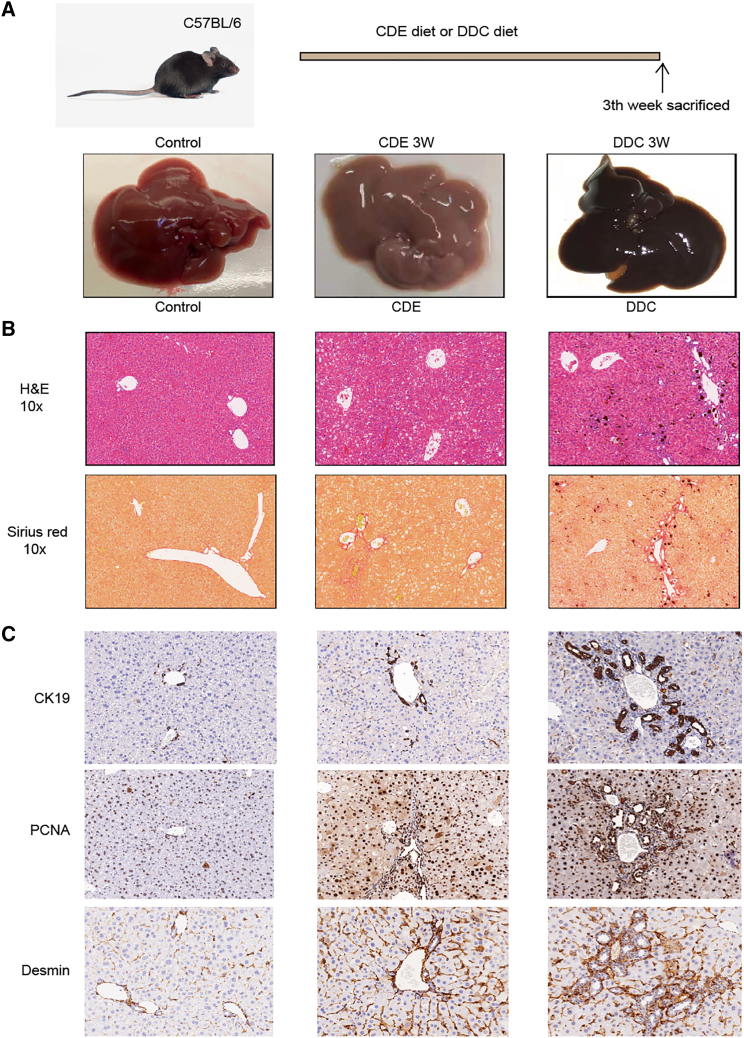
In mice, we used the CDE diet to induce steatohepatitis to create a steatohepatitis model, and fed the DDC diet to create a model of cholestatic liver disease. Fifteen mice were killed after being on the CDE and DDC diet for 3 weeks. In the gross specimen, the control group exhibited liver tissues that were normal in size, soft in texture, and ruddy in color. Liver tissues in the CDE group were enlarged, soft, and pale in color, whereas those in the DDC group were significantly enlarged, slightly harder, and brown (Figure 1A). Four micrometer-thin, paraformaldehyde-fixed, and paraffin-embedded liver tissue sections were stained with hematoxylin and eosin to assess the hepatic architecture. Healthy liver tissues displayed a normal architecture without obvious inflammatory cell infiltration and fibrous tissue deposition in the portal area. In contrast, liver tissues from CDE-treated mice showed the liver structure destruction, deposition of lipid droplets in hepatocytes, and infiltration of inflammatory cells around the portal area. However, DDC-treated liver tissues showed vastly disrupted liver architecture, plenty of inflammatory cells around the portal area, and bile thrombus formation in the bile duct. In addition, we used Sirius Red staining to assess the degree of fibrosis between the different groups. Compared with the control group, the CDE and DDC groups showed that the matrix deposition was mainly in the parenchyma around the portal vein, and the collagen deposition in the DDC group was significantly higher than that in the CDE group (Figure 1B). When C57BL/6 mice were fed the CDE and DDC diet, a strong proliferation response of liver progenitor cells (oval) was observed. In normal liver, CK19 is used as a marker of bile duct epithelium and elliptical cells, and it is stained around the portal vein. Significant expansion of CK19-positive cells was observed 21 days after receiving the CDE and DDC diet (Figure 1C). Together with the expansion of HPCs, Desmin, a marker of liver stellate cells, was also increased significantly, and the accumulation was mainly around the portal vein. Matrix deposition occurred around CK19-positive cells and surrounded the arborizing or ductlike structures, further indicating that the deposition of ECM around the portal is closely related to the increase of HSCs. In addition, PCNA, a marker of proliferation, also increased significantly after being given the CDE and DDC diet, suggesting that the proliferation ability of livers increased in the compensatory phase after injury. These observations indicate the successful establishment of chronic liver injury mouse models.
Figure 1.
CDE and DDC diet-induced chronic liver injury in mice models
(A) Timeline diagram of chronic liver injury model induced by the CDE diet and DDC diet. A group of C57BL/6 male mice were given Choline-Deficiency, Ethionine supplementation diet, or 3,5-Diethoxycarbonyl-1,4-Dihydrocollidine diet, and liver samples were collected after 3 weeks. (B) Representative photomicrographs of liver tissue sections in CDE and DDC diet-induced chronic liver injury. H&E staining shows the tissue morphology, and Sirius scarlet staining shows the degree of liver fibrosis. (C) Serial liver sections from the control group and mice on the CDE diet and DDC diet for 21 days were stained with specific antibodies against CK19, PCNA, and Desmin. Original magnification, ⅹ100, as indicated.
Single-cell characterization of transcriptomic landscape of chronic liver injury in mice models
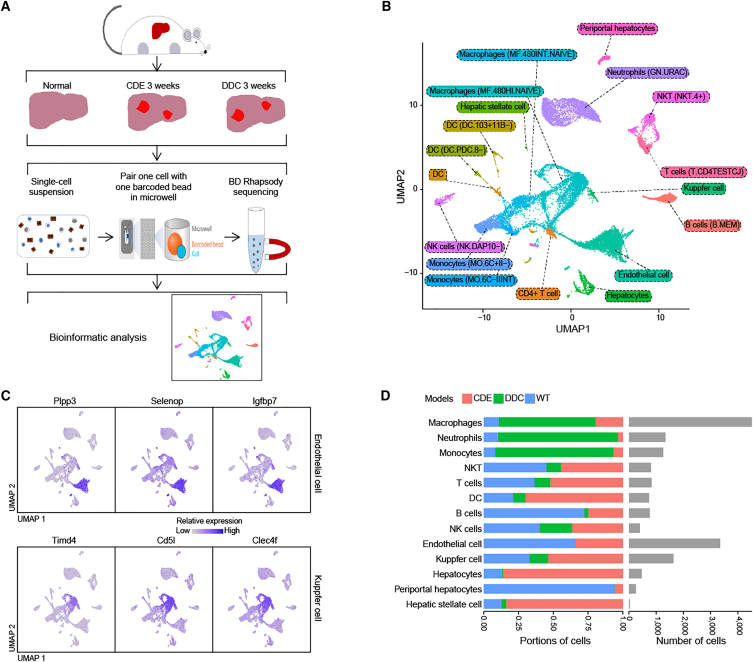
The mouse liver tissue of normal, CDE, and DDC samples were subjected to single-cell RNA library construction and sequencing (Figure 2A). In total, 16,389 cells were detected with high quality and used for further analysis. Based on the expression patterns of top variable genes, these cells were classified into 28 clusters (Figure S1 and Table S1). Most of these cell clusters showed different enriched biological functions, and some presented similar biological functions (Table S2). According to the expression profiles of mouse cell markers curated in the SingleR package and manually curated gene markers from previous studies (Table S3),12, 13, 14, 15 cell clusters were annotated as 12 main different cell types, including hepatocytes, endothelial cells, Kupffer cells, hepatic stellate cells, B cells, T cells, natural killer T cells (NKT), neutrophils, macrophages, dendritic cells (DC), natural killer cells (NK), and monocytes (Figures 2B and S2). Marker genes of different cell types showed exclusively high expression in corresponding cell types, such as Plpp3, Selenop, and Igfbp7 in endothelial cells, and Timd4, Cd5l, and Clec4f in Kupffer cells (Figure 2C). Gene markers in other immune cell types and hepatic cell types also showed exclusive expression in the corresponding cells (Figures S3 and S4). Macrophage has the largest population in captured cells, in total over 4,000 cells in all samples (Figure 2D). The second largest cell population is endothelial cells, which has about 3,500 cells in total. The most portions of macrophages, neutrophils, and monocytes were found in the DDC sample, while the majority of hepatic stellate cells, hepatocytes, DC, and Kupffer cells were in the CDE sample. These observations suggested that the damage impacted different cell types in these two liver injury models. In addition, some cell types were overrepresented in the WT normal liver sample, such as periportal hepatocytes, B cells, and endothelial cells.
Figure 2.
Characterization of single-cell transcriptomics in liver injury
(A) Experimental design for scRNA-seq experiments. (B) UMAP dimensionality reduction of all cells. (C) UMAP visualization of the expression of feature genes for identification of mouse endothelial cells (Plpp3, Selenop, and Igfbp7) and Kupffer cells (Timd4, Cd5l, and Clec4f). (D) Percentage and total numbers of different cell types in CDE, DDC, and WT samples.
Reconstruction of expression trajectory in chronic liver injury
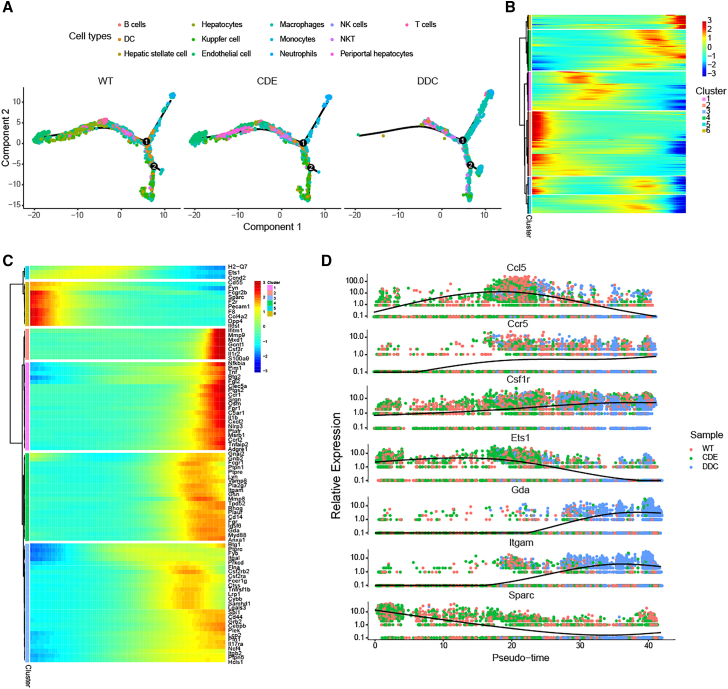
Chronic liver injury is a gradual process of liver damage. To explore distinct roles of different cell types in this progressive pathological process, expression patterns of cells were reconstructed to infer the pseudotime of different cell types across samples. The CDE model showed expression patterns more similar to the normal sample, while the DDC model showed more different transcriptional activities (Figure 3A). Different cell types showed distinct expression patterns across pseudotime tracks (Figure S5). In the early stage, T cells and NKT cells exhibited more transcriptional activities in the CDE model, whereas the normal sample shows more activity of hepatocytes. Interestingly, much fewer cells were inferred to show up in the early stage of DDC model. These observations showed that hepatocytes were damaged in the early stage of both chronic liver injury models. Genes were then classified into six different expression clusters according to the expression similarity across pseudotime (Figure 3B). Cluster 2 has the largest set of genes, wherein genes showed the highest expression level at the beginning and decreased activity in the following pseudotime. Immune-related genes also showed clustered expression patterns along the pseudotime (Figure 3C). For example, the Cd55 gene, which encodes a glycoprotein involved in the regulation of the complement cascade, showed high transcriptional activity at the early pseudotime but weak activity in the late pseudotime. Important marker genes in CLDs and liver regeneration, such as Ccl5,16 Ccr5,16 and Csf1r,17 showed high expression in middle or late pseudotime, especially in the DDC model (Figure 3D). Furthermore, we also identified several genes that exhibited remarkable changes across pseudotime. In particular, the expression levels of Ets1 and Sparc were found to decrease, whereas transcriptional activity of Gda and Itgam was found to increase in late pseudotime.
Figure 3.
Expression patterns of various cells in liver injury
(A) Pseudotime trajectory of different cells. Each point indicates one single cell. (B) DEGs along the pseudotime are hierarchically clustered into six different clusters. (C) Top DEGs in each pseudotime cluster. (D) Smoothed gene expression along pseudotime of selected markers for liver injury and immune cells.
Transcriptional disruption in macrophage in chronic liver injury models
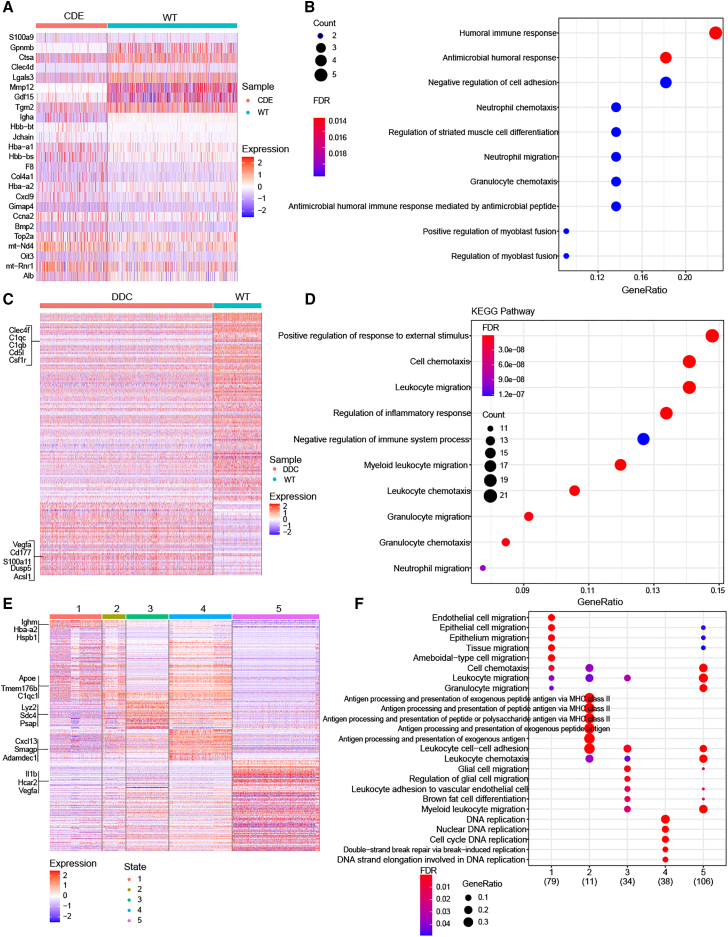
Hepatic macrophages have been shown to play key roles in the injury and repair during the progression of chronic liver diseases.18 We further compared the transcription profile of macrophages between different samples and pseudotime states. In the comparison between normal and CDE model samples, 17 up-regulated and eight down-regulated genes were identified (Figure 4A and Table S4). These dysregulated genes were found to participate in response to foreign invasions, such as "humoral immune response" and "antimicrobial humoral response," suggesting the disorder of response to exotic attack in macrophages (Figure 4B). In the DDC sample, 43 up-regulated and 109 down-regulated genes were identified when compared with the WT normal sample (Figures 4C and Table S5). These significantly altered genes were enriched in immune-related processes, such as "positive regulation of response to external stimulus," "cell chemotaxis," and "leukocyte migration" (Figure 4D). In the comparisons between different states of macrophages, 89, 22, 52, 133, and 117 genes were identified to exclusively up-regulate in state 1, 2, 3, 4, and 5, respectively (Figures 4E and Table S6). These exclusive genes among different states of macrophages were enriched in not only immune-related biological processes, such as "antigen processing" and "leukocyte migration," but also genome instability, such as "DNA replication" and "cell cycle" (Figure 4F). State 2 macrophages were found to be more dysregulated in the regulation of infectious diseases, while state 4 macrophages showed more alterations in the regulation of genome instability. Our analysis revealed that different molecular aspects of macrophages were dysregulated in CDE and DDC, and macrophages in different states showed distinct alterations.
Figure 4.
Transcriptional differential analysis of macrophages in CDE, DDC, and different cell states
(A) Heatmap shows the differential genes between CDE and WT cells. (B) Enriched biological processes of differential genes in CDE. (C) DEGs between DDC and WT cells. (D) Biological processes enriched in CDE cells. (E) Heatmap shows the differential genes in cells of different states. (F) Enriched biological processes in different cells of states.
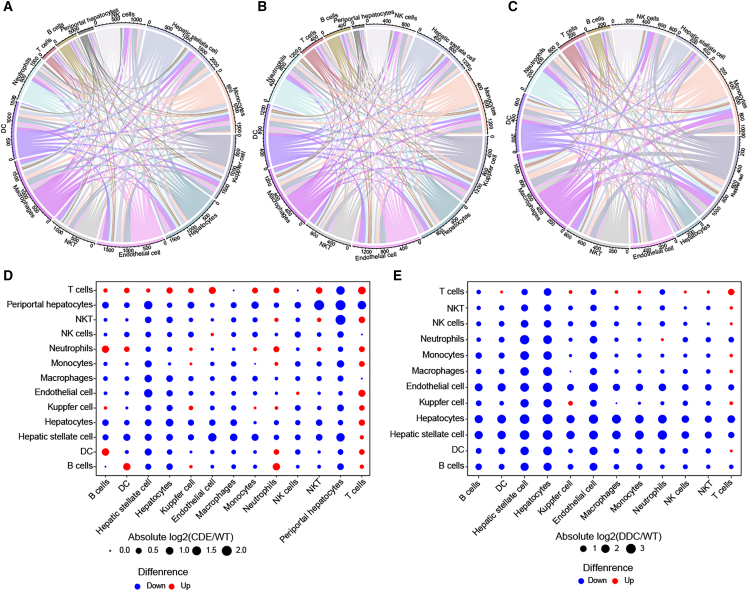
Analysis of cell-cell communications in chronic liver injury
To further investigate the changes of cell-cell interactions in chronic liver injury, we evaluated the interactions between each two different cell types. By mapping the expression of paired genes of ligands and receptors, cell-cell communications between different cell types in WT normal, CDE, and DDC samples were individually constructed (Figures 5A–5C). In the WT normal sample, hepatic stellate cells and macrophages showed the most interactions with other cells. Kupffer cells and endothelial cells exhibited the most intense interactions with other cells in the CDE sample, whereas the DDC sample showed the most cell communications in Kupffer cells and macrophages. To further investigate changes of cell-cell communications between different samples, we compared the interaction density by using the number of expressed ligand-receptor pairs. In the comparison between CDE and normal samples, hepatic stellate cells and periportal hepatocytes showed the most reducing of interactions with other cells, while interactions between T cells and other cells were extensively enhanced (Figure 5D). The DDC sample was found to show widespread decline in cell-cell interactions, especially for hepatic stellate cells, hepatocytes, and endothelial cells (Figure 5E). These analyses uncovered prevailing variations of cell-cell interactions in chronic liver injury.
Figure 5.
Communications between different cell types in WT, CDE, and DDC samples
Interactions between different types of cells in WT (A), CDE (B), and DDC samples (C). (D) The changes of cell-cell interaction intensity of different cell types between CDE and WT samples. (E) The changes of cell-cell interaction intensity of different cell types between DDC and WT samples.
Discussion
Chronic liver disease is threatening human health worldwide, of which chronic liver injury is the major pathological feature. The chronic pathological process often causes unremarkable clinical manifestations until very late stages. Therefore, in-depth analysis, especially in molecular levels, is needed to unveil subtle changes in the slow progress of chronic liver injury. In the present analysis, we introduced a systematic description of single-cell transcriptome in chronic liver injury CDE and DDC models. We performed a series of bioinformatics analysis to reveal the expression patterns, transcriptional changes of macrophages, and cell-cell communications in liver injury samples.
Animal models are crucial for investigating the molecular pathogenesis and improving our understanding of human chronic liver diseases.19 Multiple mouse and rat models have been established for chronic liver diseases. The CDE and DDC mouse models used in our study are classic chronic liver disease models, which well recapitulate human chronic liver injury, especially in the process of NAFLD and non-alcoholic steatohepatitis. More animal models and even clinical patient samples will be needed to further validate our findings in this study, but our findings in the present study still offered useful insights for deeper understanding of transcriptional changes in chronic liver injury and suggestions for potential markers indicating the pathological process. After DDC and CDE diet-induced injury, hepatocytes around the portal vein are damaged, and the transcription profile of hepatocytes after injury is significantly changed. In addition, some hepatocytes also express bile duct cell markers, which may be HPCs, suggesting that liver injury can promote the activation of HPCs and its repair to the liver. However, the origin of HPCs is not clear at present.
Accumulated evidence has shown that the disorder of the immune system, including changes in the number or function of immune cells, is important during the development of chronic liver injury. For example, macrophages play an important role in the activation and expansion of HPCs, and liver regeneration mediated by HPCs in chronic liver injury. Our results also show that the increase in the proportion of Kupffer cells and dendritic cells may promote the activation of HPCs, while the increase in the proportion of inflammatory cells promotes the progression of liver inflammation and fibrosis in chronic liver injury. Besides, in the DDC model, the expression of vascular endothelial growth factor A in macrophages was significantly increased, which can stimulate endothelial cell capillarization and accelerate the progression of liver fibrosis.
The liver has a strong ability to regenerate and repair. Our study found that a small population of cells highly expressed genes related to cell cycle, chromosome stability, and proliferation. These genes are closely related to cell proliferation and cell cycle division, and the proportion of cells in CDE and DDC groups is higher than that in WT group, indicating that the proliferation ability of hepatocytes is enhanced after injury. This result is also consistent with our PCNA staining results, and whether this group of cells are liver stem cells that originally existed in the liver, remains to be verified. But from the perspective of grouping, it is obviously different from HPCs.
Our transcriptional trajectory analysis revealed the difference of expression patterns between CDE and DDC models and identified some gene markers of chronic liver injury. To validate these findings, not only more samples will be needed, but more time intervals during the pathological process will also be necessary. Analysis of samples in different time points along pathological process of chronic liver injury will enlarge molecular changes and largely facilitate our understanding of the occurrence and development of chronic liver diseases. Notably, a considerable portion of hepatocytes were badly damaged, which might not be captured in the scRNA-seq library. After being fed with CDE or DDC diet, the damage of liver cells in mice is aggravated, and the viability of isolated liver cells is reduced, which greatly reduces the proportion of liver cell populations. Therefore, the changing features of hepatocytes may not be well recapitulated in our dataset because of technical issues. Although the number of hepatocytes is not very large, it does not affect our major observations and analysis of the results. This could be resolved by using single-cell nucleic sequencing in the further study. In WT and CDE mice, endothelial cells and Kupffer cells are the starting points of differentiation, indicating that endothelial cells and the immune environment serve the goalkeepers of the liver's homeostasis. When faced with injury, they are the first to respond. In the CDE model, hepatocytes around the portal vein can be activated at an early stage, and at the same time T cells, NK cells, and stellate cells are also activated to respond to injury; however, it is particularly interesting that stellate cells and CDE-derived hepatocytes in the early stage of WT account for more, which is significantly different from CDE. It shows that the hepatocytes regenerated after injury in the CDE model are similar to the original hepatocytes in the WT, and can differentiate into periportal hepatocytes. This phenomenon further confirms the contribution of HPCs to liver regeneration. This is also in line with the physiological process of the liver's damage repair.
Our study offered a systematic view on single-cell transcriptional alterations in chronic liver injury models. Although our findings are based on mouse models and need validation in clinical patient samples, these analyses provided organized cognition of molecular pathological changes and potential therapeutic targets for chronic liver diseases. With more diverse sources of samples, including those from different time points of chronic liver injury and surgical specimen of clinical patients, we will be able to describe a more refined transcriptional landscape and identify markers at early stage for early clinical intervention.
Materials and methods
Establishment of in vivo mouse models
C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA). Mice used in this study were aged 6 weeks and weighed around 19 g. For the CDE diet, mice were given unlimited access to choline-deficient diet (TROPHIC, Nantong, China) and drinking water supplement with 0.15% (w/v) DL-ethionine (Sigma-Aldrich, St. Louis, MO) for 3 weeks. For the DDC diet, 6-week-old male mice were fed a diet supplemented with 0.1% DDC (TROPHIC) for 3 weeks, while control mice received normal chow and drinking water for the entire experimental period and were killed at the same time as mice fed with the CDE and DDC diet. All animal studies were approved by the Committee on the Ethics of animal experiments of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. All animal experiments in the present study were performed under protocols following the Guide for the Care and Use of Laboratory Animals.
Single-cell collections
The mouse liver tissues were surgically removed and kept in MACS Tissue Storage Solution (Miltenyi Biotec) until processing. The tissue samples were processed as described below. Briefly, samples were first washed with PBS, minced into small pieces (approximately 1 mm3) on ice and enzymatically digested with 50 U/mL collagenase I (Worthington) and 30 U/mL DNase I (Worthington) for 45 min at 37°C, with agitation. After digestion, samples were sieved through a 70-μm cell strainer, and centrifuged at 300 × g for 5 min. After the supernatant was removed, the pelleted cells were suspended in red blood cell-lysis buffer (Miltenyi Biotec) to lyse red blood cells. After washing with PBS containing 0.04% BSA, the cell pellets were re-suspended in PBS containing 0.04% BSA and re-filtered through a 40-m cell strainer. Dissociated single cells were then stained for viability assessment using Calcein-AM (Thermo Fisher Scientific) and Draq7 (BD Biosciences). The single-cell suspension was further enriched with an MACS dead cell removal kit (Miltenyi Biotec).
Single-cell RNA-seq
The BD Rhapsody system was used to obtain transcriptomic information of single cells. Single-cell capture was achieved by random distribution of a single-cell suspension across >200,000 microwells using a limited dilution approach. Beads with oligonucleotide barcodes were added to saturation to pair the beads with the cells in microwells. Cell-lysis buffer was added to hybridize poly-adenylated RNA molecules to the beads. Beads were collected into a single tube for reverse transcription.
Single-cell RNA-seq data processing
The raw scRNA-seq data were aligned and quantified against the mouse reference genome (mm10) by applying the BD Rhapsody Analysis pipeline (Version 1.8, https://hub.docker.com/r/bdgenomics/rhapsody) with default parameters. Particularly, sequencing reads were mapped to by using STAR software (version 2.7.6a) with customized settings from BD Rhapsody standard pipeline.20 Low-quality cells were filtered out as previously described.21 In particular, quality of cells was evaluated based on the number of total UMI counts, the number of detected genes, and the proportion of mitochondrial gene counts in each cell. Cells were filtered out as low-quality cells if total UMI counts or detected gene numbers (in log10 scale) were lower than the median of all cells minus three times of median absolute deviation. In addition, cells with mitochondrial gene proportions higher than the median of all cells minus three times of median absolute deviation were also removed in the following analysis.
Cell doublet detection and removal
The Scrublet software was applied to detect cell doublets, which are formed from the situation that two or more cells entered the same microfluid droplet and were labeled with undistinguished barcodes.22 Specifically, a doublet score was calculated for each single cell by using default settings. Then the threshold was inferred from the bimodal distribution calculation. All cells with doublet scores lower than the threshold were removed as cell doublets.
Data integration of all samples
The Seurat package (version 4.0) was used to integrate all cells across samples.23 In particular, reciprocal (RPCA) was employed to determine anchors between samples, wherein each sample was projected to other principal components analysis (PCA) space and constrained by the same mutual neighborhood requirement. Anchors were identified by using the FindIntegrationAnchors function, which takes Seurat objects of all samples as input. These anchors were then used to integrate together by IntegrateData function.
Unsupervised clustering analysis
The unsupervised graph-based clustering algorithm implemented in the Seurat package was used to cluster cells by their gene expression. The top 2,000 variable genes were first generated with appropriate threshold of the mean expression and dispersion. The PCA was then performed by using these variable genes. The FindCluster function implemented in Seurat was used with default parameters to perform clustering.
Dimensionality reduction using UMAP
The dimensionality of all sample datasets was further reduced by using Uniform Manifold Approximation and Projection (UMAP) implemented in Seurat with RunUMAP function. The same number of PCs as those in clustering was used to calculate the embedding. The UMAP dimensionality reduction was used for visualization.
Differential expression analysis
Differentially expressed genes (DEGs) in specific cell types or clusters between different samples were identified by using the FindMarkers function embedded in Seurat package. In each comparison, genes with fold change >1.5 and Benjamini-Hochberg adjusted p value <0.05 were considered statistically significant. Additionally, the clusterProfiler package was utilized to detect enriched Kyoto Encyclopedia of Genes and Genome pathways or Gene Ontology biological functions from each set of DEGs.24
Trajectory inference
The pseudotime trajectory analysis was performed by using the Monocle2 package with default settings.25 Monocle2 is an unsupervised algorithm designed to recover single-cell gene expression kinetics during cellular processes. DEGs (Q value <0.001) were identified by the DifferentialGeneTest function. The trajectory in this study was visualized in 2D tSNE graphs and the expression dynamic expression heatmaps were constructed by using the plot_pseudotime_heatmap function.
Cell-cell interactions analysis
The cell-cell interaction analysis at molecular level was conducted by using CellPhoneDB with default parameters according to the official protocols.26 CellPhoneDB is a computational framework to predict cell-type specific ligand-receptor complexes based on well-curated complexes.27 Significant cell-cell interactions were identified by performing 1,000 interactions for the statistical analysis (p < 0.05). In this study, the ligand-receptor interactions between each pair of different cell types was based on expression profile.
Statistical analysis
Statistical analysis and data visualization in the present study was performed by using R software (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org). Unless specific statements, all statistical tests were two-tailed and p value or false discovery rate <0.05 was considered as statistically significant.
Acknowledgments
This study was supported by National Natural Science Foundation of China (No. 81970528) and Shanghai General Hospital Startup Funding (02.06.01.20.06 and 02.06.02.21.01).
Author contributions
X.C., S.L., and L.L. conceived and supervised the project. J.W. and Z.S. performed the mouse model construction. J.W. and W.H. performed the single-cell sequencing and data analysis. T.L., W.D., and B.S. performed the statistical analysis and assisted in plotting. X.L. and J.W. interpreted the results. S.L. and X.C. wrote the manuscript with comments from all authors. All authors reviewed the manuscript and consented for publication.
Declaration of interests
The authors declare no potential conflicts of interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.11.008.
Contributor Information
Lungen Lu, Email: lulungen1965@126.com.
Shengli Li, Email: shengli.li@shsmu.edu.cn.
Xiaobo Cai, Email: caixiaobo1979@hotmail.com.
Supplemental information
References
- 1.James S.L., Abate D., Abate K.H., Abay S.M., Abbafati C., Abbasi N., Abbastabar H., Abd-Allah F., Abdela J., Abdelalim A., et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and Injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong M.C.S., Huang J.L.W., George J., Huang J., Leung C., Eslam M., Chan H.L.Y., Ng S.C. The changing epidemiology of liver diseases in the Asia–Pacific region. Nat. Rev. Gastroenterol. Hepatol. 2019;16:57–73. doi: 10.1038/s41575-018-0055-0. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M. Non-alcoholic fatty liver disease – a global public health perspective. J. Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 4.Musso G., Cassader M., Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat. Rev. Drug Discov. 2016;15:249–274. doi: 10.1038/nrd.2015.3. [DOI] [PubMed] [Google Scholar]
- 5.Miyajima A., Tanaka M., Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Van Hul N.K.M., Abarca-Quinones J., Sempoux C., Horsmans Y., Leclercq I.A. Relation between liver progenitor cell expansion and extracellular matrix deposition in a CDE-induced murine model of chronic liver injury. Hepatology. 2009;49:1625–1635. doi: 10.1002/hep.22820. [DOI] [PubMed] [Google Scholar]
- 7.Knight B., Tirnitz-Parker J.E.E., Olynyk J.K. C-kit inhibition by imatinib mesylate attenuates progenitor cell expansion and inhibits liver tumor formation in mice. Gastroenterology. 2008;135:969–979. doi: 10.1053/j.gastro.2008.05.077. [DOI] [PubMed] [Google Scholar]
- 8.Akhurst B., Croager E.J., Farley-Roche C.A., Ong J.K., Dumble M.L., Knight B., Yeoh G.C. A modified choline-deficient, ethionine-supplemented diet protocol effectively induces oval cells in mouse liver. Hepatology. 2001;34:519–522. doi: 10.1053/jhep.2001.26751. [DOI] [PubMed] [Google Scholar]
- 9.Yao Z., Vance D.E. The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J. Biol. Chem. 1988;263:2998–3004. [PubMed] [Google Scholar]
- 10.Mariotti V., Strazzabosco M., Fabris L., Calvisi D.F. Animal models of biliary injury and altered bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:1254–1261. doi: 10.1016/j.bbadis.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigo-Torres D., Affò S., Coll M., Morales-Ibanez O., Millán C., Blaya D., Alvarez-Guaita A., Rentero C., Lozano J.J., Maestro M.A., et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology. 2014;60:1367–1377. doi: 10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong X., Kuang H., Ansari S., Liu T., Gong J., Wang S., Zhao X.Y., Ji Y., Li C., Guo L., et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell. 2019;75:644–660.e5. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halpern K.B., Shenhav R., Matcovitch-Natan O., Tóth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E., et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Zhang S., Liu Y., He X., Qu M., Xu G., Wang H., Huang M., Pan J., Liu Z., et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 2020;6:22. doi: 10.1038/s41421-020-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crinier A., Milpied P., Escalière B., Piperoglou C., Galluso J., Balsamo A., Spinelli L., Cervera-Marzal I., Ebbo M., Girard-Madoux M., et al. High-dimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity. 2018;49:971–986.e5. doi: 10.1016/j.immuni.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams D.H., Ju C., Ramaiah S.K., Uetrecht J., Jaeschke H. Mechanisms of immune-mediated liver injury. Toxicol. Sci. 2010;115:307–321. doi: 10.1093/toxsci/kfq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meunier L., Larrey D. Drug-induced liver injury: biomarkers, requirements, candidates, and validation. Front. Pharmacol. 2019;10:1–8. doi: 10.3389/fphar.2019.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan Z., Ju C. Hepatic macrophages in liver injury. Front. Immunol. 2020;11:1–9. doi: 10.3389/fimmu.2020.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nevzorova Y.A., Boyer-Diaz Z., Cubero F.J., Gracia-Sancho J. Animal models for liver disease – a practical approach for translational research. J. Hepatol. 2020;73:423–440. doi: 10.1016/j.jhep.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 20.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845.e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 27.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.