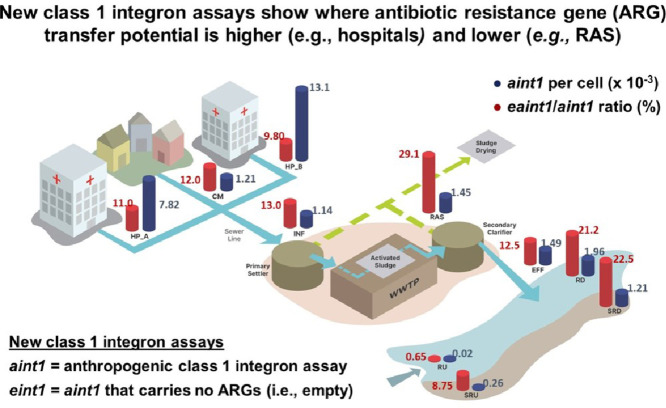
Highlights
-
•
Prevalence and types of integrons vary widely across a wastewater network.
-
•
Human-impacted class 1 integrons carrying ARGs dominate, but are highest in hospital wastewater.
-
•
New qPCR assays are reported that segregate "anthropogenic" class 1 integrons that carry ARGs (aint1) vs empty structures (eaint1).
-
•
Recycled activated sludge has triple the “empty structures” per integron relative to WWTP liquid effluents.
-
•
Integron dynamics help identify wastewater compartments with elevated ARG transfer.
Keywords: Integron dynamics, Wastewater networks, Class 1 integrons, Empty integron structures, Antibiotic resistance genes, qPCR probes
Abstract
Class 1 and other integrons are common in wastewater networks, often being associated with antibiotic resistance genes (ARGs). However, the importance of different integron structures in ARG transfer within wastewater systems has only been implied, especially between community and hospital sources, among wastewater treatment plant compartments, and in receiving waters. This uncertainty is partly because current clinical class 1 integron qPCR assays (i.e., that target human-impacted structures, i.e., clintI1) poorly delineate clintI1 from non-impacted class 1 integron structures. They also say nothing about their ARG content. To fill these technical gaps, new real-time qPCR assays were developed for “impacted” class 1 structures (called aint1; i.e., anthropogenic class 1 integrons) and empty aint1 structures (i.e., carry no ARGs; called eaint1). The new assays and other integron assays then were used to examine integron dynamics across a wastewater network. 16S metagenomic sequencing also was performed to characterise associated microbiomes. aint1 abundances per bacterial cell were about 10 times greater in hospital wastewaters compared with other compartments, suggesting aint1 enrichment with ARGs in hospital sources. Conversely, the relative abundance of eaint1 structures were over double in recycled activated sludge compared with other compartments, except receiving waters (RAS; ∼30% of RAS class 1 structures did not carry ARGs). Microbiome analysis showed that human-associated bacterial taxa with mobile integrons also differed in RAS and river sediments. Further, class 1 integrons in RAS bacteria appear to have released ARGs, whereas hospital bacteria have accumulated ARGs. Results show that quantifying integron dynamics can help explain where ARG transfer occurs in wastewater networks, and should be considered in future studies on antibiotic resistance in the environment.
Graphical abstract
1. Introduction
Increasing antibiotic resistance is an evolutionary response to the overuse of antibiotics and environmental pollution, and is a global health concern (Chen and Zhang, 2013; Devarajan et al., 2016; Quintela-Baluja et al., 2015; Xu et al., 2016). Antibiotic use selects for resistance strains in human and animal wastes, which can be released to the environment via wastewater, spreading antibiotic resistance genes (ARGs) and bacteria across nature. Selection is often through versatile gene transfer mechanisms, which allow relatively unconstrained ARG migration across phylogenetic, ecological, and geographic borders. As such, the use of antibiotics in one habitat (e.g., hospitals) can impact resistomes in other locations across the environment. Therefore, understanding the vectors and transfer mechanisms by which bacteria share and acquire ARGs is important, maybe more important than studying the levels of ARGs themselves.
A key group of ARG transmission vectors in bacteria are integrons that are carried on mobile genetic elements (MGEs), including plasmids and transposons (Cambray et al., 2010). Integrons are genetically mobile bacterial recombination systems (i.e., mobile integrons) that allow the acquisition and expression of promoterless protein-coding sequences (gene cassettes; GC). They can confer rapid adaptation and selective advantage to host bacteria (often under specific environmental pressures). A large array of GCs that carry ARGs have been found in integrons associated with MGEs (Cambray et al., 2010; Partridge et al. 2009), which are very diverse and associated with ARG transmission (Cambray et al., 2011).
Three classes of mobile integrons, i.e., class 1, 2, and 3, appear to be most important for acquiring and transmitting ARGs (Cambray et al., 2010). Class 1 integrons (intI1) are the most prevalent class among clinical and environmental isolates (Cambray et al., 2010; Cambray et al., 2011; Deng et al., 2015; Partridge et al. 2009). However, all three integron classes (intI1, intI2, and intI3) are found in human, animal, and environmental metagenomes (Cambray et al., 2010; Deng et al., 2015; Simo Tchuinte et al., 2016; Stokes and Gillings, 2011). These integrons are often associated with Proteobacteria species, although intI1 has also been found in Gram-positive strains (Cambray et al., 2010; Deng et al., 2015; Partridge et al. 2009). Over 130 GCs with ARGs have been found on mobile integrons (Stalder et al., 2012).
Class 1 integrons that carry ARGs are found in both pathogenic and non-pathogenic strains, although not all class 1 integrons are the same. A sub-group of class 1 integrons have been related with “clinical” settings (hereafter called clinical class 1 integrons, “clintI1”) (Gillings et al., 2008). clintI1 differs from other class 1 integrons in that its integron-integrase gene is conserved (5’CS end), but it carries an additional segment at its 3’CS end, which is composed of qacEΔ1 (low-level resistance to quaternary ammonium compounds), sul1 (sulfonamide resistance), and orf5 (unknown function) (Gillings et al., 2008). Like other class 1 integrons, clintI1 is co-regulated by SOS response systems, suggesting reordering and acquiring existing/exogenous cassettes may be activated under stressed conditions when genetic adaptation is needed (Barraud and Ploy, 2015; Cambray et al., 2011). However, integron regulation is not fully understood - in particular how it influences the dynamics of ARG transmission and antibiotic resistance spread. Understanding the role of integrons within a system is further confused by the fact that many integrons, including clintI1, can be found “empty”; e.g., carrying no ARGs (Chakraborty et al. 2013; Chang et al., 2007; Dillon et al., 2005; Moura et al., 2012; Sá et al., 2010; Toval et al., 2015). The incorporation of ARGs into clintI1 also occurs in the absence of antibiotics (Pérez-Valdespino et al., 2016), suggesting integron and ARG ecology are not solely driven by antibiotic use.
Here we examine integron dynamics by quantifying different integrons and microbiota across a wastewater network that includes community and hospital sources, sewer lines, a wastewater treatment plant (WWTP), and upstream and downstream receiving waters (Fig. 1). Integron types, states, and possible bacterial hosts (called here “potential bacteria carrying mobile integrons” or PBCMI) were compared across compartments. The goal was to delineate the spatial dynamics of integrons, especially human-impacted class 1 integrons, to help explain where ARG transmission really occurs within wastewater systems. However, to do this, we needed to design new probe-primer sets that better segregate clintI1 from intI1, and probes for quantifying class 1 structures that do not carry ARGs.
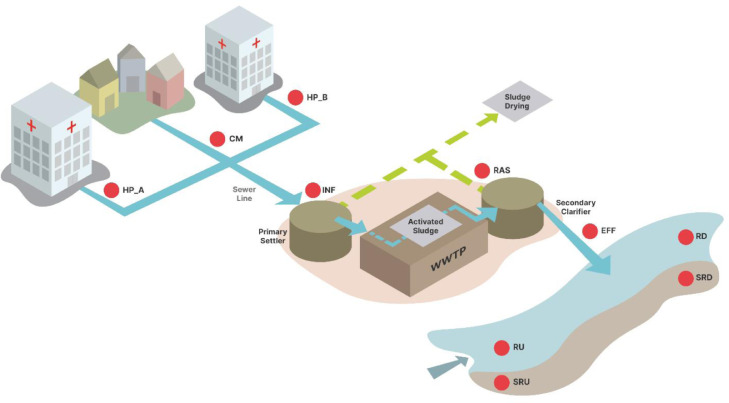
Fig. 1.
Schematic of sampling points in study area. CM = community wastewater; HP_A, HP_B = wastewater from two different hospitals; INF = wastewater treatment plant (WWTP) influent; EFF = WWTP effluent; RAS = recycled activated sludge; RU= upstream river water column; SRU = upstream river sediment; RD = downstream river water column; and SRD = downstream river sediment. Adapted from Quintela-Baluja et al. (2019).
2. Materials and methods
2.1. Study area and sampling procedure
Sampling was performed during the summer in a city in northern Spain, which has limited heavy industry or agriculture. A schematic of the network is shown in Fig. 1. Samples were collected over three weeks, including effluents from two hospitals (HP_A and HP_B); the parallel community sewer line (CM); the WWTP influent (INF) and effluent (EFF); recycled activated sludge (RAS); and the river water column and sediments 100 m upstream (RU and SRU) and downstream (RD and SRD) of the WWTP discharge. Further details of the wastewater network, sampling regime, and sample handling and processing methods have been reported previously (Quintela-Baluja et al., 2019). Temperature, dissolved oxygen (DO), pH, and specific conductivity were measured in situ using hand-held probes (Mettler Toledo™, FG3 FiveGo™, and Jenway Model 350 pH Meter).
2.2. DNA extraction
Biomass was harvested for DNA extraction via vacuum filtration using sterile 0.22-μm membrane disc filters (Millipore, Billerica, MA, USA) or by centrifugation at 12,000 rpm for 30 min. Extraction was performed using the Fast DNA Spin Kit for Soils (MP Biomedicals, USA), according to the manufacturer's instructions. DNA was stored at −20°C prior to subsequent analysis.
2.3. Design of new primers and probes to quantify alternate clinical class 1 integron structures
The terminology of Gillings et al. (2008) has been used for many years to define class 1 integron recombination platforms. Pre-clinical class 1 integrons are elements that contain a class 1 integrase, which includes a complete tni transposition module (pclintI1). Conversely, "clinical" class 1 integrons (clintI1) contain qacEΔ, sul1, and orf5 in their structure. A random subset of recently published plasmid sequences was examined, and it was determined the old primer set for clintI1 poorly discriminated from pclintI1 structures (Figure S1, Tables S1-S3). Therefore, we designed a new set of primers that are more specific to the qacEΔ, sul1, and orf5 structure, which we call the aint1 assay.
Our new name represents “anthropogenically impacted class 1 integrons” and is an appropriate name for class 1 integrons that have been impacted by human activity and not just clinical factors. In parallel we also designed a new assay for anthropogenic class 1 integrons that do not carry ARGs, which we call empty structures (eaint1). The design process of the new assays is provided under Results.
2.4. Quantitative PCR Assays and related data analysis
IntI1, intI2, and intI3 as well as aint1 and eaint1 were quantified using the qPCR TaqMan method (Table 1). TaqMan qPCR reactions were conducted using SsoAdvanced™ Universal Probes Supermix (BioRad), employing the following thermocycler program: 3 min of initial denaturation at 95°C, and 40 cycles of 5 sec denaturation at 95°C and 30 sec annealing/extension (see temperatures in Table 1). In addition, total bacteria and total coliforms were quantified using a SYBR Green based qPCR assay. SYBR Green reactions were conducted using SsoAdvanced™ Universal SYBR® Green Supermix (BioRad), employing the following thermocycler program: 2 min of initial denaturation at 98°C, and 40 cycles of 5 sec denaturation at 98°C and 5 sec annealing/extension at 60°C (total bacteria) or 55°C (total coliforms).
Table 1.
Primers used in this study to quantify integrons (intI1, intI2, intI3), anthropogenic class 1 integrons (aint1), aint1 that contain no ARGs (eaint1), coliforms, and total bacteria (16S rRNA).
| Target | Primer | Sequence (5′–3′) | Amplicon size (bp) | Annealing (°C) | Reference |
|---|---|---|---|---|---|
| intI1 | F | GCCTTGATGTTACCCGAGAG | 196 | 60 | Barraud et al., 2010 |
| R | GATCGGTCGAATGCGTGT | ||||
| Probe | (6FAM)ATTCCTGGCCGTGGTTCTGGGTTTT(BHQ1) | ||||
| intI2 | F | TGCTTTTCCCACCCTTACC | 195 | 60 | Barraud et al., 2010 |
| R | GACGGCTACCCTCTGTTATCTC | ||||
| Probe | (TxRd)TGGATACTCGCAACCAAGTTATTTTTACGCTG(BHQ2) | ||||
| intI3 | F | GCCACCACTTGTTTGAGGA | 138 | 60 | Barraud et al., 2010 |
| R | GGATGTCTGTGCCTGCTTG | ||||
| Probe | (6FAM)CGCCACTCATTCGCCACCCA(BHQ1) | ||||
| eaint1 | F | GAGCAGCAACGATGTTAC | 131 | 60 | This study |
| R | GGCTTATTATGCACGCTTA | ||||
| Probe | (6FAM)CGCCCTAAAACAAAGTTAGATGCACT(BHQ1) | ||||
| aint1 | F | GCACTAAGCACATAATTG | 140 | 60 | This study |
| R | CCAACTATTGCGATAACA | ||||
| Probe | (6FAM)GAGATATATCATGAAAGGCTGGCTT(BHQ1) | ||||
| Coliforms | Eco1457F | CATTGACGTTACCCGCAGAAGAAGC | 190 | 60 | Bartosch et al., 2004 |
| Eco1652R | CTCTACGAGACTCAAGCTTGC | ||||
| 16S rRNA | 1055f | ATGGCTGTCGTCAGCT | 337 | 58 | Harms et al., 2003 |
| 1392r | ACGGGCGGTGTGTAC |
The limit of detection (LOD) and limit of quantification (LOQ) were calculated using a 2-fold dilution series covering the range of 1 to 500 gene copies (gc) from each standard and analysed eight times (R script available at https://github.com/cmerkes/qPCR_LOD_Calc). The LOD was defined as the lowest concentration that can be reliably detected, i.e., at which at least 95% of standard reactions are expected to amplify. The LOQ was defined as the lowest standard that can be quantified with a coefficient of variation less than 0.35 (Klymus et al., 2020) (Table S4).
All assays were carried out in triplicate using the BioRad CFX C1000 System (BioRad, Hercules, CA USA). To minimise inhibition, extracted DNA was diluted to a working concentration of 5 ng/ul and a DNA internal positive control was used in SYBR Green reactions. Briefly, an additional reaction was performed for each sample, containing 104 genes copies of the green fluorescent protein gene (gfp). Inhibition was considered to be minimal when the difference in Ct values (ΔCt) of gfp between the spiked sample and the positive control was below 1 Ct. Standard curves for each primer set were constructed using plasmid clones of the target sequences, these were loaded in triplicate in each qPCR run, in parallel with the amplification of test samples. All samples were run in triplicate, using the optimum qPCR conditions determined for each primer set.
Related statistical analyses and data manipulation were performed using the R environment (Team, 2006) with a significant cutoff of α = 0.05. Normality and the variance homoscedasticity were tested by the Shapiro-Wilk test and Levene's test, respectively. Previous conditions were not met for all integron datasets. Therefore, the Krustall-Wallis test was performed to assess statistically significant differences. Conover's-test was performed for pairwise comparison between sampling sites, and adjusted p-values were calculated with the Holm-Bonferroni method.
2.5. 16S rRNA gene amplification, sequencing, and data processing
To assess microbial communities, PCR amplification of the V4−V5 region of bacterial 16S rRNA genes in DNA extracts was conducted using fusion primers containing a PGM sequencing adaptor, a “GT” spacer, and a unique 12 base pair Golay barcode to allow multiplex analyses (primers 515F: 5′- GTGNCAGCMGCCGCGGTAA-3′, and 926R: 5′-CCGYCAATTYMTTTRAGTTT-3′). Details of PCR reactions and amplicon quantification are reported elsewhere (Quintela-Baluja et al., 2019).
Sequencing was performed using an Ion Torrent Personal Genome System (Life Technology) with processing performed using a UPARSE-QIIME pipeline (Pylro et al. 2014; Pylro et al., 2016). The Operational Taxonomic Unit (OTU) table was built using the UPARSE pipeline (Edgar, 2004). Sequences were clustered into OTUs at a 97% similarity cut off, checked for chimaeras, and reflective sequences were obtained for each phylotype. Taxonomic classification used QIIME (Caporaso et al., 2010) based on the uCLUST method (Prasad et al., 2015). See Quintela-Baluja et al. (2019) for details.
All data analysis and visualisations were conducted using R (Team 2006) through the Rstudio IDE (http://www.rstudio.com/). OTU counts and associated taxonomic assignments were imported and merged into a phyloseq object (McMurdie and Holmes, 2013). All samples were rarefied to ensure the same number of reads per sample (8,577), which corresponded to the sample with the fewest number of sequences, resulting in 5,698 unique OTUs.
Assessment of sample completeness (sample coverage) and alpha diversity were calculated with the R package iNEXT (Hsieh et al., 2016). Alpha-diversity was calculated using Hill numbers (D) with q from 0 to 3 and were expressed in units of “effective” number of species. As q increases, low-abundance OTUs are assigned less weight, while high-abundance OTUs are assigned more weight; this enables the investigation of diversity at different scales. At q=0, D is simply the number of observed OTUs (richness). At q=1, Dexp (Shannon), and at q=2, D is equal to inverse Simpson's index. A cluster dendrogram of community composition dissimilarity (Bray-Curtis, average neighbour clustering) was calculated with the R package vegan (Oksanen et al., 2019).
To determine the potential bacteria carrying mobile integrons (PBCMIs) in each compartment, consensus sequences from class 1 (anthropogenic and non-anthropogenic), 2, and 3 integrons were aligned against the NCBInr database (database accessed on June 2019) using BLAST® (McGinnis and Madden, 2004). Accession numbers from sequences with 100% identity were downloaded and taxonomy assigned with the R package taxonomizr. Only families with more than one entry in the NCBInr database were considered as potential integron hosts and used to filter the OTU table from the samples. The PBCMIs (OTU level) distribution across the network was visualized using a two-dimensional hierarchical clustering in conjunction with a heatmap of relative abundances.
2.6. Biomarker signature analysis
Linear discriminant analysis effect sizes (LEfSe) was employed to identify PBCMI that are statistically significant across sampling locations (Segata et al., 2011). With the normalised relative abundance matrix, LEfSe uses the Kruskal-Wallis rank-sum test to detect features with significantly different abundances between the assigned taxa and performs Linear Discriminant Analysis to estimate the effect size of each feature. Only taxa with average abundances >1% were considered significant. A significance level alpha of 0.05 and an effect size threshold of 2x were used for all the biomarkers evaluated in this study.
2.7. Co-occurrence between integrons and microbial taxa
A correlation matrix was developed by calculating all possible pairwise Spearman's rank correlations among 267 bacterial families, the three integrases (intI1, intI2, and intI3), and the 3’CS conserved region of aint1 present in samples from the study (n = 27). A correlation between two items was considered if the Spearman's correlation coefficient (ρ) was ≥ 0.5 and the p value was ≤ 0.05. To reduce the chances of obtaining false-positive results, the p values were adjusted with a multiple testing correction using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). The pairwise correlations of the bacterial families and integrases formed their co-occurrence networks. Network analyses were performed in R environment and was further visualised and explored to identify its topological properties (i.e., clustering coefficient, shortest average path length and modularity) in Gephi (Bastian et al., 2009).
3. Results
3.1. New qPCR probe-primer sets for "human-impacted" and "empty" class 1 integrons
Although multiple probe-primers for class 1 integrons and associated gene cassettes already exist (Gillings et al., 2009; Gillings et al., 2009; Holmes et al., 2003; Stokes et al., 2001), one set has dominated use, i.e., clintI1 (hereafter called the “old set”) (Gillings et al., 2015). This old set targets the integron-integrase gene (5’CS end). However, when one compares that target sequence with more recent sequencing data, it becomes clear the region does not clearly discriminate between “clinical” and “pre-clinical” class 1 integrons (Figure S1) (pre-clinical here meaning the evolutionary precursor of clintI1). However, we identified a conserved 3’CS region (i.e., attc/qacΔE1/sul1) that is unique to clinical (i.e., human-impacted or anthropogenic) class 1 integrons (see Figs. 2 and S2, and Tables S1-S3 for details). Using this region, we designed and validated a new set of probe-primers that better segregate human-impacted class 1 from non-impacted class 1 structures (Table 1).
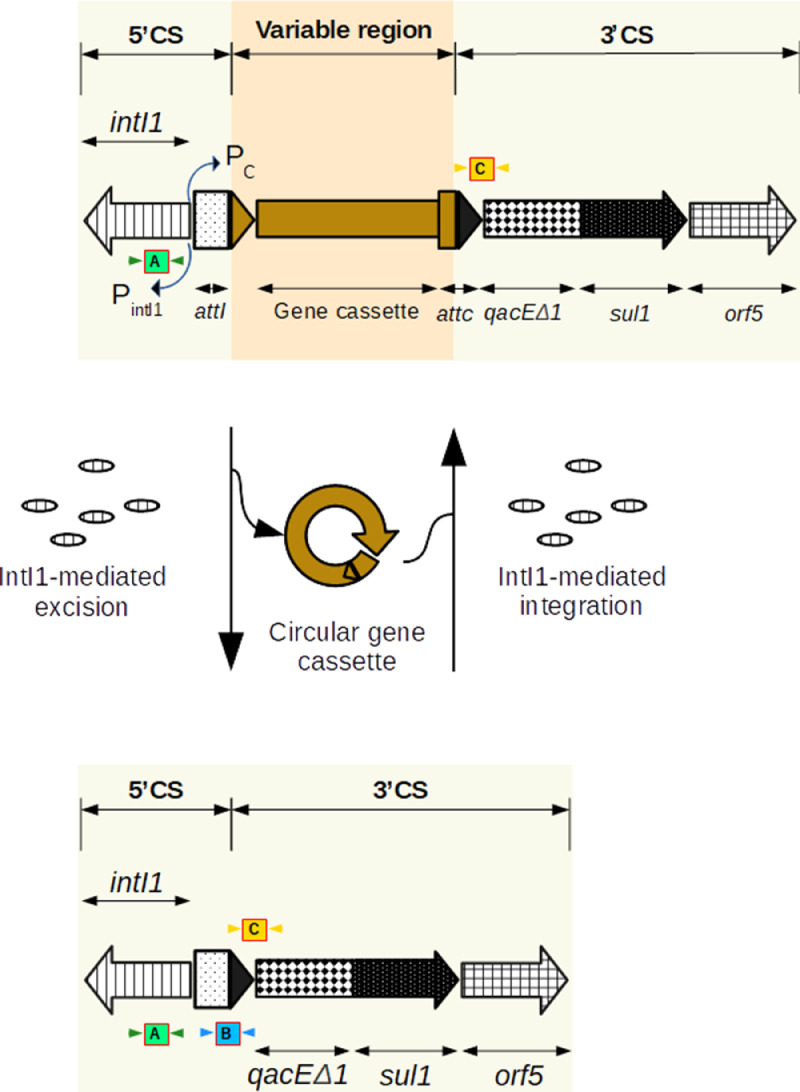
Fig. 2.
Anthropogenic class 1 integron structure with and without the variable gene cassette insertion region (top and bottom, respectively). The green, blue, and yellow boxes show the target sites of the respective primers:  - target for the class 1 integron (clintI1, Barraud et al., 2010):
- target for the class 1 integron (clintI1, Barraud et al., 2010):  - new target for the empty-class 1 integron structure (eaint1; this study); and
- new target for the empty-class 1 integron structure (eaint1; this study); and  - new target location for the clinical class 1 integron at the attc/qacΔE1 interface (aint1; this study).
- new target location for the clinical class 1 integron at the attc/qacΔE1 interface (aint1; this study).
The assay design was as follows. Sequences of each individual region were first aligned using MUSCLE in UGENE v.33 to identify optimal target regions for more specific primers (Figure S2). This revealed selective targets within amplicon-containing sequences at intersecting loci between attc/qacΔE1 and attI/attc (Fig. 2). Using this region, primers and TaqMan probes were designed using OligoArchitect™ (http://www.oligoarchitect.com) with a melting temperature (Tm) of 60°C. The selected primers and probes were aligned against NCBI using BLAST® (McGinnis and Madden, 2004) to rule out non-specific binding to other bacterial targets. We call our new probe-primer assay aint1 to avoid confusion with the old set. The new probe-primer set was assessed in terms of historic clinical and non-clinical integron structures (Tables S2 and S3), and shows aint1 better targets the conserved 3’CS segment of the clinical structure (attc/qacΔE1/sul1) and does not overlap with pre-clinical structures. Henceforth we only refer to aint1 hereafter.
Using a similar approach, a new probe-primer assay was also designed to quantify empty anthropogenic class I integron structures (which we call eaint1) (Table 1). The need to segregate between empty structures and integrons carrying GCs was evident as all of the empty class I integron structures in our plasmid sequence dataset were associated with anthropogenic class 1 structures. The old assay did not uniquely detect anthropogenic class 1 integron structures nor indicate whether they were carrying ARGs in the GC. Combining aint1 and eaint1 allows one to quantify anthropogenic class 1 integron dynamics more exactly, segregating GCs that do not contain ARGs.
3.2. Integron pool and distribution across our urban wastewater network
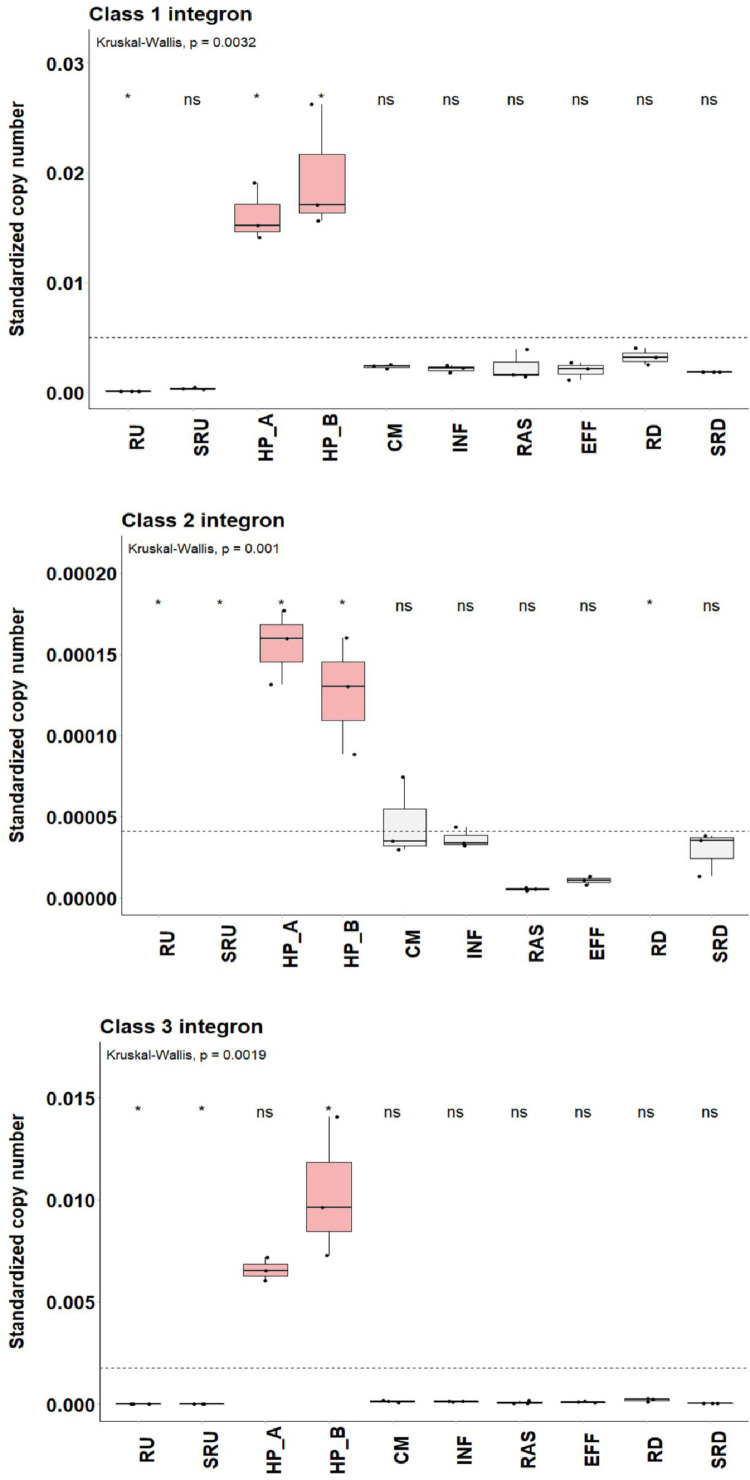
Quantification of integron structures across our wastewater network showed that class 1 integrons were present in all compartments (Table 2 and S5; Fig. 3). In contrast, class 2 integron levels were low everywhere and below detection in river samples collected upstream of the wastewater effluent discharge point or in the downstream water column. Class 3 integrons were also low in upstream samples and lower across the wastewater network, except in hospital wastewater samples. Following quantification of integron gene levels per bacterial cell across the network (i.e., as genome equivalent), hospital effluents contained significantly higher levels of integron genes per cell (p < 0.05), much greater than any other compartment (Fig. 3, Table 2). Metadata on chemical conditions in each compartment is shown in Table S6.
Table 2.
Estimated cell and coliform numbers, and relative total class 1 integron (intI1), anthropogenic class 1 integron (aint1), empty anthropogenic class 1 integron (eaint1), class 2 integron (intI2), and class 3 integron (intI3) per bacteria cell.
| Sample | 16S rRNA (copies/mL)a | Coliforms (copies/mL) | intI1 (copies/cell)b | aint1 (copies/cell) | eaint1 (copies/cell) | intI2 (copies/cell) | intI3 (copies/cell) |
|---|---|---|---|---|---|---|---|
| RU | 8.8 ± 0.11 | 0.24 ± 0.064 | 8.6E-5 ± 9.6E-6 | 1.6E-5 ± 1.2E-5 | NDc | ND | 1.4E-6 ± 2.4E-6 |
| SRU | 11.3 ± 0.052 | 0.12 ± 0.006 | 3.5E-4 ± 8.7E-5 | 2.9E-4 ± 6.8E-5 | 2.2E-5 ± 1.2E-6 | ND | 7.7E-6 ± 4.5E-6 |
| HP_A | 10.1 ± 0.053 | 3.2 ± 2.2 | 1.6E-2 ± 2.6E-3 | 7.9E-3 ± 1.3E-3 | 7.9E-4 ± 1.9E-4 | 1.6E-4 ± 2.3E-5 | 6.6E-3 ± 5.8E-4 |
| HP_B | 9.9 ± 0.038 | 5.7 ± 1.7 | 2.0E-2 ± 5.8E-3 | 1.3E-2 ± 3.4E-3 | 1.1E-3 ± 4.4E-4 | 1.3E-4 ± 3.6E-5 | 1.0E-2 ± 3.4E-3 |
| CM | 9.9 ± 0.14 | 2.3 ± 0.82 | 2.4E-3 ± 1.8E-4 | 1.2E-3 ± 1.1E-4 | 1.3E-4 ± 5.6E-5 | 4.6E-5 ± 2.4E-5 | 1.3E-4 ± 6.0E-5 |
| INF | 10.5 ± 0.37 | 0.59 ± 0.20 | 2.1E-3 ± 3.5E-4 | 1.2E-3 ± 2.7E-4 | 1.5E-4 ± 4.6E-5 | 3.6E-5 ± 6.3E-6 | 1.2E-4 ± 2.4E-5 |
| RAS | 11.7 ± 0.092 | 0.048 ± 0.013 | 2.3E-3 ± 1.4E-3 | 1.7E-3 ± 1.1E-3 | 5.4E-4 ± 4.0E-4 | 5.4E-6 ± 9.2E-7 | 7.8E-5 ± 6.8E-5 |
| EFF | 9.4 ± 0.031 | 1.3 ± 0.79 | 2.0E-3 ± 8.0E-4 | 1.4E-3 ± 6.0E-4 | 1.7E-4 ± 1.1E-4 | 1.1E-5 ± 2.7E-6 | 1.0E-4 ± 3.7E-5 |
| RD | 9.7 ± 0.056 | 0.38 ± 0.048 | 3.2E-3 ± 7.7E-4 | 1.9E-3 ± 6.5E-4 | 3.8E-4 ± 1.3E-4 | ND | 2.1E-4 ± 8.2E-5 |
| SRD | 11.6 ± 0.035 | 0.51 ± 0.17 | 1.9E-3 ± 6.3E-6 | 1.4E-3 ± 4.2E-5 | 3.1E-4 ± 1.4E-5 | 2.9E-5 ± 1.4E-5 | 2.7E-5 ± 7.9E-7 |
On the basis of the Ribosomal RNA Database, the average number of 16S rRNA-encoding genes per genome equivalent was estimated as 4.1. 16S rRNA and coliform gene copy concentrations were divided by this value to estimate bacterial cell numbers or coliforms. For sediment samples, the denominator is sediment mass in gms rather than mL.
The normalized copy numbers of integrons per bacterial cell were calculated by dividing the gene copy number of each integron by the estimated number of bacterial genome equivalents.
ND refers to not detected genes in 5 ng/ul DNA.
Fig. 3.
Comparison of the relative abundance of A) class 1; B) class 2; and C) class 3 integron genes per bacterial cell across the network. Krustall-Wallis rank sum test and the Wilcoxon-Mann-Whitney test was used to determine if each integron gene was enriched or reduced at each sample site compared with the population mean (dashed line). Significance: *0.05 and 'ns’ not significant. Sample sites: RU, upstream water column; SRU, upstream sediments; HP_A and HP_B, hospital wastewater; CM, community wastewater; INF, WWTP influent; RAS, recycled activated sludge; EFF, WWTP effluent; RD, downstream river water column; and SRD, downstream sediment.
Within the WWTP itself, integron abundances decreased between 93 to 98% from influent to effluent (log removals of 1.18 ± 0.28 [mean ± SD] for intI1, log 1.66 ± 0.26 for intI2, and log 1.21 ± 0.16 for intI3). These removals directly paralleled bacterial removals (Table 2 and S5), implying the removal of bacterial hosts and MGEs occur in tandem across the treatment steps. This is confirmed by intI1, aint1, and intI3 copies per bacterial cell being roughly constant across the system (Table S7). Despite large reductions in integrons across the WWTP, absolute class 1 and 3 integron gene abundances increased in the river downstream of the WWTP, suggesting integrons were still reaching the environment.
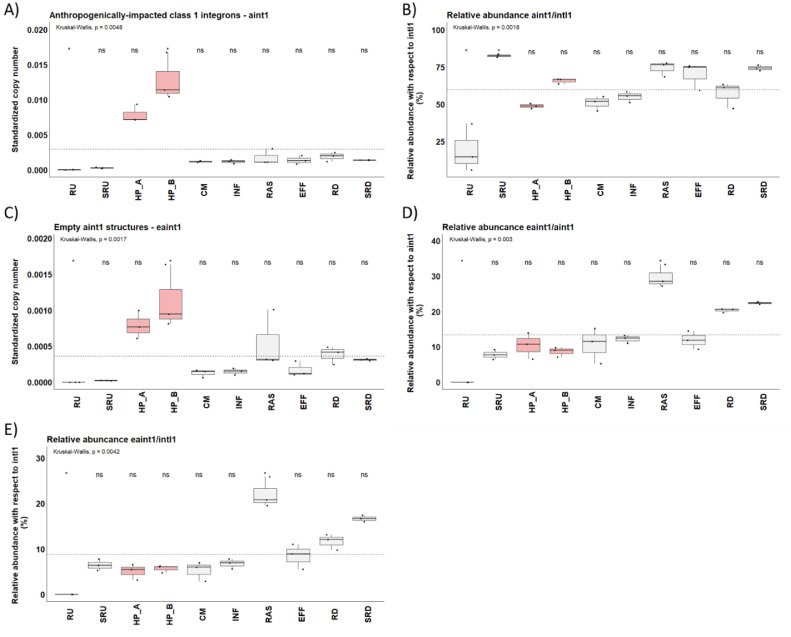
The new probe-primer design showed that the ratio of aint1 to intI1 did vary across "raw" wastewater compartments (Fig. 4A). aint1 was the dominant integron structure downstream of the biological wastewater treatment unit (p <0.05) (Fig. 4B), with relatively low levels of eaint1. In contrast, the percentage of eaint1 in RAS was consistently higher, double that in WWTP influents and hospital samples, and much higher than any other compartment (Fig. 4C-D), except downstream river locations.
Fig. 4.
Relative abundances of class 1 integron genes of different types. A) Relative number of aint1 per bacteria cell; B) Relative abundance of aint1 structure per intl1 gene; C) Relative number of eaint1 per bacteria cell; D) Relative abundance of eaint1 per aint1; and E) Relative abundance of eaint1 per intl1. Krustall-Wallis rank sum test and the Wilcoxon-Mann-Whitney test was used to determine if each integron structure was enriched or reduced at each sample site compared with the population mean (dashed line). Significance: *0.05 and 'ns’ not significant. Sample sites: RU, upstream water column; SRU, upstream sediments; HP_A and HP_B, hospital wastewater; CM, community wastewater; INF, WWTP influent; RAS, recycled activated sludge; EFF, WWTP effluent; RD, downstream river water column; and SRD, downstream sediment.
3.3. Identifying potential bacteria carrying mobile integrons (PBCMI)
Integrase gene abundance across this wastewater network correlates with the type and abundance of bacterial populations, which was previously observed for ARGs within this network (Quintela-Baluja et al., 2019). Therefore, data here lends itself well to statistical analysis on most probable populations carrying key integrons, especially intI1 vs aint1 vs eaint1.
Screening of the NCBInr database found 28 potential bacterial families known to host intI1: 14 for aint1, 6 for intI2, and 4 for intI3 (Table S8). Only intI1 has been previously found in Gram-positive bacteria, whereas the 3’CS conserved region of aint1 has only been found in Gram-negative bacteria. Of the identified families with mobile integrons (from the database), 21 were found in one or more samples in this study (Table S8).
Beta-diversity analysis was used to compare microbial community diversities among wastewater network locations. For this analysis, the dataset was re-sampled to ensure the same number of reads per sample (8,577), which corresponded to the sample with the fewest number of sequences, resulting in 5,698 OTUs. The OTUs from PBCMI families were filtered and re-sampled (from the raw reads) to the sample with the fewest number of sequences (1,266), resulting in 392 unique OTUs. The percentage of total estimated species richness in each sample resulted in 95.4 - 99.0% for the total microbial community, and 94.4 - 98.1% for the PBCMI, which suggests the overall microbial communities were adequately sampled in both cases (Table S9).
Hill number diversity indices were calculated with the parameter q from 0 to 2. Note that as q increases, rare species are given less weight and contribute less toward the “effective number of OTUs” in a sample. Hill numbers with q equal to 0 and 1 showed that raw wastewater samples had lower diversity compared with upstream river samples (both water column and sediment), and from the downstream river sediments (Fig. S3) Specifically, Hill numbers (q equal to 0 and 1) indicated that bacterial diversity was greater in samples upstream of the WWTP (RU, SRU) because of more rare taxa, which was not apparent when only PBCMI taxa were considered (Fig. S4).
Conversely, the Bray-Curtis dissimilarity dendrogram showed that total microbial community compositions differed among network compartments, which included four main clusters with distinct ecosystems (cut off = 0.72) (Fig. S5). The first cluster contained samples associated with raw wastewater (community, hospital, and WWTP influent). The second cluster contained RAS, WWTP effluent, and the downstream river water and sediment samples. The third and fourth clusters included the river water column and sediment samples upstream of the WWTP, respectively.
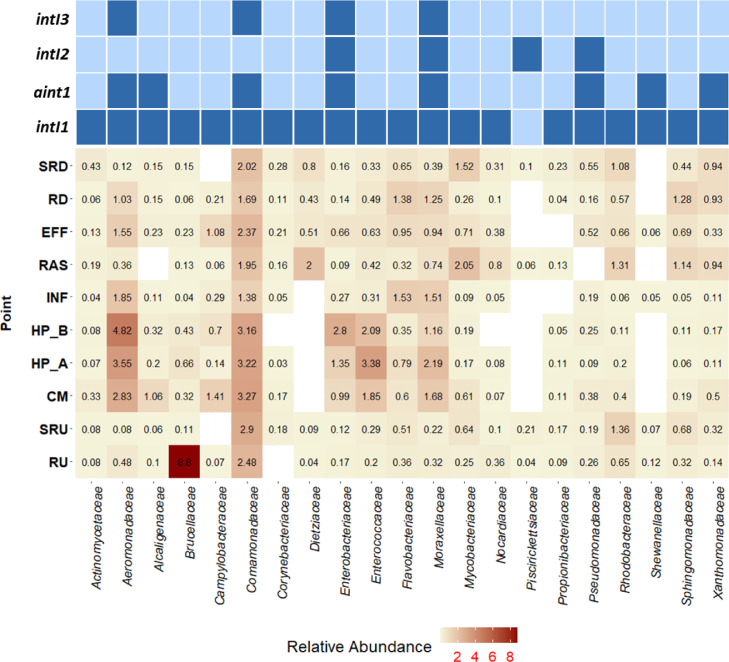
The PBCMI sub-community followed similar patterns, but the cut off value of the cluster structure was significantly lower (cut off = 0.53). This observation, in conjunction with evident two-dimensional hierarchical clustering seen in the heatmap of PBCMI relative abundances (Fig. S6), suggests that a PBCMI core sub-community exists in all network compartments. However, Fig. S6 shows that WWTP releases clearly impact river microbial communities, increasing PBCMIs in the downstream environment (Figs. 5 and S6). The family Comamonadaceae is ubiquitous, which is often associated with wastewater (e.g., Aeromonadaceae and Enterobacteriaceae), and those more associated with the environment (e.g., Rodobacteriaceae or Sphingomonadaceae).
Fig. 5.
Relative abundance of potential bacteria carrying integron genes in wastewater network compartments. The right side of the table shows the bacterial families which host integron genes (dark blue). The numbers show the relative abundance of the potential bacteria hosting integrons in each compartment. RU, upstream river water column; SRU, upstream river sediments; HP, hospital wastewater (HP_A and HP_B); CM, community wastewater; INF, WWTP influent; RAS, recycled activated sludge; EFF, WWTP effluent; RD, downstream river water column; and SRD, downstream river sediment.
3.4. Microbial communities and their connectivity across the network
Characterising microbial communities across a wastewater network (in terms of diversity, evenness, and taxonomic composition) is key to identifying linkages among compartments and determining microbial contributions from internal versus external sources. LEfSE analysis showed the relative presence of the family Erysipelotrichaceae, Lachnospiraceae, Bacteroidaceae, and Ruminococcaceae tend to best define community wastewater. In contrast, hospital wastewater was characterised by higher abundances of Prevotellaceae, Enterobacteriaceae, and Desulfovibrionaceae. Weeksellaceae and Moraxellaceae were associated with WWTP influent, whereas RAS was best defined by Rhodobacteraceae, Frankiaceae, Dietziaceae, Gordoniaceae, and Intrasporangiaceae (Fig. S7).
Each compartment of the network had characteristic, reflective signature families, suggesting local enrichment and-or selection. To examine this, we combined the biomarker discovery tool LefSE and co-occurrence network analysis to assess the relative transmission of ARGs associated with mobile integrons across compartments. The co-correlation network consists of 259 nodes (255 taxa and 4 mobile integrons) and 4.465 edges with an average degree or node connectivity of 34.479. The average network distance between all pairs of nodes (average path length) was 2.94 edges with a network diameter of 7 edges.
Bacterial families, comprising PBCMI, that most explain differences between network compartments (LefSE analysis) are highlighted in Fig. S7. Network analysis produced four different modules (Fig. S8), equivalent to the microbial composition dissimilarity clusters. The network showed possible ARG dissemination pathways via mobile integrons from human-associated taxa (Module I) to bacteria in human-impacted environments (Module II) and non-human environmental bacteria (Modules III and IV). LEfSE analysis revealed that Module I include bacterial families defining raw wastewater (CM, HP, and INF) and all mobile integrons (Fig. S9A). Conversely, Module II mostly contains RAS and river sediment taxa with limited PBCMI contributions (RAS, RD, SRD, and EFF), emphasising the connectivity of RAS bacteria with river sediments (Fig. S8B). Module III contains taxa defining the river water column (Fig. S8C). Finally, Module IV contains mostly bacterial families in upstream river sediments (SRU). However, some bacteria families from the downstream river sediments also are present in Module IV (Fig. S8D).
4. Discussion
Here we show that intI1 and aint1 dominated all compartments in our wastewater network, with intI2 and intI3 being much less abundant at all sampling points. All integron markers, except for empty structures (eaint1), were highest in hospital wastewater where antibiotic use is more intensive (Escudero-Oñate et al., 2017; Rodriguez-Mozaz et al., 2015). This is consistent with previous data that showed the relative number of ARGs per bacterial cell were at least one order of magnitude higher in hospital wastewater compared with the other compartments (Quintela-Baluja et al., 2019). Interestingly, while intI2 levels per bacterial cell declined along the wastewater process stream, the relative levels of intI1, aint1, and intI3 per cell did not change amongst compartments, except in upstream river samples. Consistently low levels of intI2 are probably due to it typically lacking a functional integrase, meaning it cannot catalyse recombination reactions alone (Deng et al., 2015).
It is noteworthy that intI3 was detected across the whole wastewater network and roughly mirrored intI1 patterns, although abundances were about 20 times lower than intI1. The ubiquitous presence of intI3 here contrasts with results of Stalder et al. (2014) who found intI3 levels decreased across their WWTP. This may be due to differences in intI3 found in the two studies or differences between the local wastewater systems. IntI3 performs a similar function to intI1, although its recombination frequencies have historically been lower (Stokes and Gillings, 2011), which makes intI3 less associated with bacterial adaptation under adverse conditions.
Empty human-impacted class 1 integron structures have been found previously (Malek et al., 2015; Mokracka et al., 2012; Moura et al., 2012; Sá et al., 2010; Toval et al., 2015), but here we more exactly quantified levels of eaint1 vs aint1 and intI1. We had expected to find higher relative abundances of eaint1 cassettes in hospital wastewater. It was hypothesized that the presence of higher concentrations of antibiotics and other bioactive agents would select for cellular defence GCs and trigger SOS responses that would activate integrase aint1 expression (Cambray 2011). But instead, the highest relative abundances of eaint1 cassettes were found in RAS samples, suggesting this is where empty cassettes accumulate or, more likely, ARG carriage is not a selection factor for bacterial hosts that reside in RAS. RAS appears to have a reduced tendency for class 1 integrons or their hosts to retain ARGs.
Although the scale of differences may be specific to our network, we suggest hospital wastewaters tend to select and retain a greater array of “defence” genes in their aint1 due to greater stressors in their environment (e.g., therapeutic drugs) and-or richer organic carbon levels (wastes dominated by faeces from patients). Similar uniqueness of hospital wastewater was also seen in Sweden (Kraupner et al., 2021). Conversely, bacteria in RAS grow under nutrient-limited conditions and under longer residence times (e.g., than a gut environment); thus, RAS bacteria are under persistent nutrient stress compared to the gut. Most activated sludge WWTPs recycle RAS for days rather than hours under comparatively dilute but fluctuating biotic and abiotic conditions. Such conditions might promote rearrangement in aint1 and loss of ARGs, but apparently not reacquisition, possibly because selective agents (e.g., antibiotics) are more dilute or not bioavailable.
The high prevalence of empty cassettes in RAS and their apparent low presence in the WWTP effluents has important implications to pathways of integron-mediated ARG spread from WWTPs to the environment. Until now, only two studies have used high-throughput sequencing to study intI1 cassette diversity in environmental samples (Gatica et al., 2016; Stalder et al., 2014). While Stalder et al. (2014) sequenced clone libraries of intI1 cassettes from wastewater compartments, Gatica et al. (2016) targeted recombination sites that flank the gene-cassettes of intI1 genes in WWTP effluents. Both studies found a high diversity of gene cassettes; however, many of their detected sequences had unknown functions. Stalder et al. (2014) found their WWTP tended to reduce the diversity of gene cassette arrays relative to raw wastewater and had elevated levels of empty cassettes. Whereas Gatica et al. (2016) showed a marginal persistence of β-lactam resistance gene cassettes in soil microbiomes impacted by WWTP effluents.
Work here shows the nature and content of integron structures, and associated microbial communities differ widely across compartments, further confirming “local” evolutionary ecosystems drive selection in sub-ecosystems embedded within larger networks (Baquero et al., 2008; Quintela-Baluja et al., 2019). Local ecosystems and their communities are spatially separate, and each is driven by different habitat and other selective factors. This includes specific community selection (Munck et al., 2015) and ARG carriage (Quintela-Baluja et al., 2019). Parallel differences exist among prevalent mobile integrons, and their state relative to ARG carriage (empty or not). However, even though different wastewater compartments have different communities, continual hydraulic interconnection allows microbial dispersal and gene flow, which explains linkages among local resistomes, including local mobile integrons.
Co-occurrence analysis (Fig.s S8 and S9) highlights the association of PBCMIs within microbial communities in raw wastewater, especially compared with communities less impacted by raw wastewater. The importance of this observation is most evident in river sediments, where resident bacteria and their mobile integrons reflect unsettled floc in WWTP effluents. As such, sediments may be place of environmental ARG transfer, although there would need to be selective drivers for gene acquisition to occur, such as other pollutants in the sediments. Our previous work supported this hypothesis where downstream sediments were largely comprised of RAS bacteria (Quintela-Baluja et al., 2019). A recent study by Di Cesare et al. (2020) supported this hypothesis by showing that the vertical distribution of intI1 in lake sediments mirrored the deposition of bacteria from the water column and to sediments below.
Consistent with previous work, sediments downstream of our WWTP were enriched in ARGs, probably due to outputs from the WWTP (Di Cesare et al., 2020; Quintela-Baluja et al., 2019), which is consistent with greater abundances of aint1 compared with upstream sediments. Although it is speculation, we suspect the co-release of potentially selective agents in WWTP effluents might further promote such persistence. Specifically, the dynamic process of gene cassette acquisition in aint1 and the presence of a qac gene in its structure (resistance to biocides) potentially confers advantageous phenotypes that may be behind the fixation and spread of this integron structure (Gillings et al., 2009).
A noteworthy new observation here, which supports the potential for ARG transfer in sediments downstream of WWTPs, is elevated levels of eaint1. The two compartments with the highest relative levels of empty clinical class 1 integron cassettes are the RAS and downstream sediments, which implies they are hydraulically connected and may be locations of reacquisition of ARGs due to higher levels of empty mobile integrons. A comparison of microbial communities between RAS and downstream sediments (Quintela-Baluja et al., 2019) furthers shows this connection and suggests downstream sediments could be locations of elevated horizontal gene transfer.
5. Conclusions
Here we show that integrons have their own ecology within a wastewater network and can be used to speculate on the dynamics of ARGs within such systems. However, integron dynamics appear more complex than previously reported, especially the spatial distribution of ARGs, and relationships with human-impacted and empty class 1 integron cassettes. This recognition was only possible by developing a more exact probe-primers for anthropogenic class 1 integrons that better segregate class 1 integron types and determine whether they carry ARGs or not. We are clearly getting closer to understanding what really happens in wastewater networks relative to AMR spread, but new evidence here suggests ARG transmission in wastewater networks may differ than previously beleived. Regardless, we hope this work will guide new and improved WWTPs and their operations in the future.
Data availability
The 16S rRNA sequencing data were submitted to EBI with accession number PRJEB46784 http://www.ebi.ac.uk/ena/data/view/PRJEB3216.
Declaration of Competing Interest
NA.
Acknowledgments
The work was funded by MERMAID; An Initial Training Network in the People Programme (Marie Skłodowska-Curie Actions) of the EU's Seventh Framework Programme FP7/2007-2013/under REA grant agreement n°607492 and by the UK Medical Research Council (MR/P028195/1). We thank Dr. Myra J Giesen and Mr Zach Hammerlsey for performing structural revisions and developing visualizations, respectively.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2021.117720.
Contributor Information
Marcos Quintela-Baluja, Email: marcosquintelab@gmail.com.
David W. Graham, Email: david.graham@newcastle.ac.uk.
Appendix. Supplementary materials
References
- Baquero F., Martínez J-L., Cantón R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008;19(3):260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Barraud O., Ploy M-C. Diversity of class 1 integron gene cassette rear-rangements selected under antibiotic pressure. J. Bacteriol. 2015;197(13):2171–2178. doi: 10.1128/JB.02455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud O., Baclet M.C., Denis F., Ploy M.C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010;65(8):1642–1645. doi: 10.1093/jac/dkq167. [DOI] [PubMed] [Google Scholar]
- Bartosch S., Fite A., Macfarlane G.T., McMurdo M.E.T. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004;70(6):3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M., Heymann S., Jacomy M. International AAAI Conference on Weblogs and Social Media. 2009. Gephi: An open source software for exploring and manipulating networks. [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate : a practical and powerful approach to multiple testing. J R Stat. Soc. Series B Stat. Methodol. 1995;57(1):289–300. [Google Scholar]
- Cambray G., Guerout A-M., Mazel D. Integrons. Annu. Rev. Genet. 2010;44(1):141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- Cambray G., Sanchez-Alberola N., Campoy S., Guerin E., Da Re S., González-Zorn B., Ploy M-C., Barbé J., Mazel D., Erill I. Prevalence of SOS-mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mob. DNA. 2011;2(1):6. doi: 10.1186/1759-8753-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M.…Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Kumar A, Bhowal SS, Mandal AK, Tiwary BK, Mukherjee S. Diverse gene cassettes in class 1 integrons of facultative oligotrophic bacteria of River Mahananda, West Bengal, India. PLoS One. 2013;8(8):e71753. doi: 10.1371/journal.pone.0071753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L.L., Chang T.M., Chang C.Y. Variable gene cassette patterns of class 1 integron-associated drug-resistant Escherichia coli in Taiwan. Kaohsiung J. Med. Sci. 2007;23(6):273–280. doi: 10.1016/S1607-551X(09)70409-7. [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang M. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ. Int. 2013;55:9–14. doi: 10.1016/j.envint.2013.01.019. [DOI] [PubMed] [Google Scholar]
- Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., Chen D., Bian H., Li Y., Yu G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015;14(1):45. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan N., Laffite A., Mulaji C.K., Otamonga J-P., Mpiana P.T., Mubedi J.I., Prabakar K., Ibelings B.W., Poté J. Occurrence of antibiotic resistance genes and bacterial markers in a tropical river receiving hospital and urban wastewaters. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0149211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cesare A, Eckert EM, Cottin C, Bouchez A, Callieri C, Cortesini M, Lami A, Corno G. The vertical distribution of tetA and intI1 in a deep lake is rather due to sedimentation than to resuspension. FEMS Microbiol. Ecol. 2020;96(2) doi: 10.1093/femsec/fiaa002. fiaa002. [DOI] [PubMed] [Google Scholar]
- Dillon B., Thomas L., Mohmand G., Zelynski A., Iredell J. Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods. 2005;62(2):221–232. doi: 10.1016/j.mimet.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids. Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero-Oñate C., Ferrando-Climent L., Rodríguez-Mozaz S., Santos L.H.M.LM. In: Hospital Wastewaters. The Handbook of Environmental Chemistry. Verlicchi P., editor. Springer; Cham: 2017. Occurrence and risks of contrast agents, cytostatics, and antibiotics in hospital effluent; p. 60. [Google Scholar]
- Gatica J., Tripathi V., Green S., Manaia C.M., Berendonk T., Cacace D., Merlin C., Kreuzinger N., Schwartz T., Fatta-Kassinos D., Rizzo L., Schwermer C.U., Garelick H., Jurkevitch E., Cytryn E. High throughput analysis of integron gene cassettes in wastewater environments. Environ. Sci. Technol. 2016;50(21):11825–11836. doi: 10.1021/acs.est.6b03188. [DOI] [PubMed] [Google Scholar]
- Gillings M., Boucher Y., Labbate M., Holmes A., Krishnan S., Holley M., Stokes H.W. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008;190(14):5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings M.R., Holley M.P., Stokes H.W. Evidence for dynamic exchange of qac gene cassettes between class 1 integrons and other integrons in freshwater biofilms. FEMS Microbiol. Lett. 2009;296(2):282–288. doi: 10.1111/j.1574-6968.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- Gillings M.R., Xuejun D., Hardwick S.A., Holley M.P., Stokes H.W. Gene cassettes encoding resistance to quaternary ammonium compounds: A role in the origin of clinical class 1 integrons? ISME J. 2009;3(2):209–215. doi: 10.1038/ismej.2008.98. [DOI] [PubMed] [Google Scholar]
- Gillings M.R., Gaze W.H., Pruden A., Smalla K., Tiedje J.M., Zhu Y-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015;9(6):1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms G., Layton A.C., Dionisi H.M., Gregory I.R., Garrett V.M., Hawkins S.A., Robinson K.G., Sayler G.S. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 2003;37(2):343–351. doi: 10.1021/es0257164. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Gillings MR, Nield BS, Mabbutt BC, Nevalainen KMH, Stokes HW. The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 2003;5(5):383–394. doi: 10.1046/j.1462-2920.2003.00429.x. [DOI] [PubMed] [Google Scholar]
- Hsieh T.C., Ma K.H., Chao A. iNEXT: An R package for rarefaction and extrapolation of species diversity (H ill numbers) Methods Ecol. Evol. 2016;7(12):1451–1456. [Google Scholar]
- Klymus K.E., Merkes C.M., Allison M.J., Goldberg C.S., Helbing C.C., Hunter M.E., Richter C.A. Reporting the limits of detection and quantification for environmental DNA assays. Environmental DNA. 2020;2(3):271–282. ... [Google Scholar]
- Kraupner N., Hutinel M., Schumacher K., Gray D.A., Genheden M., Fick J., Flach C-F., Larsson D.G.J. Evidence for selection of multi-resistant E. coli by hospital effluent. Environ Int. Online early. 2021 doi: 10.1016/j.envint.2021.106436. [DOI] [PubMed] [Google Scholar]
- Malek M.M., Amer F.A., Allam A.A., El-Sokkary R.H., Gheith T., Arafa M.A. Occurrence of classes I and II integrons in Enterobacteriaceae collected from Zagazig University Hospitals, Egypt. Front. Microbiol. 2015;6:601. doi: 10.3389/fmicb.2015.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic. Acids. Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One; 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokracka J., Koczura R., Kaznowski A. Multiresistant Enterobacteriaceae with class 1 and class 2 integrons in a municipal wastewater treatment plant. Water Res. 2012;46(10):3353–3363. doi: 10.1016/j.watres.2012.03.037. [DOI] [PubMed] [Google Scholar]
- Moura A., Jové T., Ploy M.C., Henriques I., Correia A. Diversity of gene cassette promoters in class 1 integrons from wastewater environments. Appl. Environ. Microbiol. 2012;78(15):5413–5416. doi: 10.1128/AEM.00042-12. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck C., Albertsen M., Telke A., Ellabaan M., Nielsen P.H., Sommer M.O. Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 2015;6:8452. doi: 10.1038/ncomms9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., Minchin, P. R., O'Hara, R. B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., and Wagner, H. (2019). Vegan: Community ecology package. http://CRAN.R-project.org/package=vegan.
- Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 2009;33(4):757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- Pérez-Valdespino A., Lazarini-Martínez A., Rivera-González A.X., García-Hernández N., Curiel-Quesada E. Dynamics of a class 1 integron located on plasmid or chromosome in two Aeromonas spp. strains. Front. Microbiol. 2016;7:1556. doi: 10.3389/fmicb.2016.01556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad D.V.V., Madhusudanan S., Jaganathan S. uCLUST - a new algorithm for clustering unstructured data. ARPN J. Eng. Appl. Sci. 2015;10(5):2108–2217. [Google Scholar]
- Pylro V.S., Morais D.K., de Oliveira F.S., Dos Santos F.G., Lemos L.N., Oliveira G., Roesch L.F. BMPOS: a flexible and user-friendly tool sets for microbiome studies. Microb. Ecol. 2016;72(2):443–447. doi: 10.1007/s00248-016-0785-x. [DOI] [PubMed] [Google Scholar]
- Pylro VS, Roesch LF, Morais DK, Clark IM, Hirsch PR, Tótola MR. Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J. Microbiol. Methods. 2014;107:30–37. doi: 10.1016/j.mimet.2014.08.018. [DOI] [PubMed] [Google Scholar]
- Quintela-Baluja M., Chan C.W., Alnakip M.E., Abouelnaga M., Graham D.W. In: The Battle Against Microbial Pathogens: Basic Science, Technological Advances And Educational Programs. Méndez-Vilas A., editor. Formatex Research Center; Spain: 2015. Sanitation, water quality and antibiotic resistance dissemination; pp. 965–975. Microbiology Series N° 5. [Google Scholar]
- Quintela-Baluja M., Abouelnaga M., Romalde J., Su J-Q., Yu Y., Gomez-Lopez M., Smets B., Zhu Y.G., Graham D.W. Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters. Water Res. 2019;162:347–357. doi: 10.1016/j.watres.2019.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S., Chamorro S., Marti E., Huerta B., Gros M., Sànchez-Melsió A., Borrego C.M., Barceló D., Balcázar J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015;69:234–342. doi: 10.1016/j.watres.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Sá L.L., Fonseca E.L., Pellegrini M., Freitas F., Loureiro E.C., Vicente A.C. Occurrence and composition of class 1 and class 2 integrons in clinical and environmental O1 and non-O1/non-O139 Vibrio cholerae strains from the Brazilian Amazon. Mem. Inst. Oswaldo Cruz. 2010;105(2):229–232. doi: 10.1590/s0074-02762010000200021. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo Tchuinte P.L., Stalder T., Venditti S., Ngandjio A., Dagot C., Ploy M.C., Barraud O. Characterisation of class 3 integrons with oxacillinase gene cassettes in hospital sewage and sludge samples from France and Luxembourg. Int. J. Antimicrob. Agents. 2016;48(4):431–434. doi: 10.1016/j.ijantimicag.2016.06.018. [DOI] [PubMed] [Google Scholar]
- Stalder T., Barraud O., Casellas M., Dagot C., Ploy M.C. Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 2012;3:119. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder T., Barraud O., Jové T., Casellas M., Gaschet M., Dagot C., Ploy M.C. Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J. 2014;8(4):768–777. doi: 10.1038/ismej.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes H.W., Gillings M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011;35(5):790–819. doi: 10.1111/j.1574-6976.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- Stokes H.W., Holmes A.J., Nield B.S., Holley M.P., Nevalainen K.M., Mabbutt B.C., Gillings M.R. Gene cassette PCR: Sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 2001;67(11):5240–5246. doi: 10.1128/AEM.67.11.5240-5246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC. Available at The R Project for Statistical Computing. 2006. A language and environment for statistical computing.http://www.R-project.org [Google Scholar]
- Toval F., Guzmán-Marte A., Madriz V., Somogyi T., Rodríguez C., García F. Predominance of carbapenem-resistant Pseudomonas aeruginosa isolates carrying blaIMP and blaVIM metallo-β-lactamases in a major hospital in Costa Rica. J. Med. Microbiol. 2015;64(1):37–43. doi: 10.1099/jmm.0.081802-0. [DOI] [PubMed] [Google Scholar]
- Xu L., Ouyang W., Qian Y., Su C., Su J., Chen H. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ. Pollut. 2016;213:119–126. doi: 10.1016/j.envpol.2016.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequencing data were submitted to EBI with accession number PRJEB46784 http://www.ebi.ac.uk/ena/data/view/PRJEB3216.