Abstract
The introduction of the proteasome inhibitor bortezomib into treatment regimens for myeloma has led to substantial improvement in patient survival. However, whilst bortezomib elicits initial responses in many myeloma patients, this haematological malignancy remains incurable due to the development of acquired bortezomib resistance. With other patients presenting with disease that is intrinsically bortezomib resistant, it is clear that new therapeutic approaches are desperately required to target bortezomib-resistant myeloma. We have previously shown that targeting sphingolipid metabolism with the sphingosine kinase 2 (SK2) inhibitor K145 in combination with bortezomib induces synergistic death of bortezomib-naïve myeloma. In the current study, we have demonstrated that targeting sphingolipid metabolism with K145 synergises with bortezomib and effectively resensitises bortezomib-resistant myeloma to this proteasome inhibitor. Notably, these effects were dependent on enhanced activation of the unfolded protein response, and were observed in numerous separate myeloma models that appear to have different mechanisms of bortezomib resistance, including a new bortezomib-resistant myeloma model we describe which possesses a clinically relevant proteasome mutation. Furthermore, K145 also displayed synergy with the next-generation proteasome inhibitor carfilzomib in bortezomib-resistant and carfilzomib-resistant myeloma cells. Together, these findings indicate that targeting sphingolipid metabolism via SK2 inhibition may be effective in combination with a broad spectrum of proteasome inhibitors in the proteasome inhibitor resistant setting, and is an approach worth clinical exploration.
Keywords: Myeloma, Bortezomib, Resistance, Sphingolipid, Unfolded protein response
Abbreviations: ATF4, activating transcription factor 4; ATF6, activating transcription factor 6; BR, bortezomib resistant; CR, carfilzomib resistant; ER, endoplasmic reticulum; GSEA, gene set enrichment analysis; IRE1, inositol-requiring enzyme 1; PERK, protein kinase R-like ER kinase; SK2, sphingosine kinase 2; S1P, sphingosine 1-phosphate; UPR, unfolded protein response; wt, wild-type; XBP1s, X-box binding protein 1s
Introduction
Myeloma is a haematological malignancy arising from plasma cells [1]. It is currently considered incurable, although median survival rates have improved significantly over the last two decades due largely to the introduction of bortezomib into treatment regimens [2]. Bortezomib is an inhibitor of the 26S proteasome, with most potent inhibitory action against the β5 (chymotrypsin-like) subunit, encoded by the PSMB5 gene, although the other proteasome subunits (β1 and β2) are also inhibited to a lesser extent [3]. While bortezomib causes myeloma cell death through a number of mechanisms, including changes in the bone marrow microenvironment and NF-κB modulation, its main mechanism of action is thought to be through induction of endoplasmic reticulum (ER) stress and the unfolded protein response (UPR)[4,5].
The ER is responsible for folding and processing over a third of all proteins produced in the cell, including secreted proteins such as immunoglobulin, which are produced in large quantities by most myeloma cells [5,6]. Key to maintaining homeostasis within the ER is a process called ER-associated degradation, which allows for the degradation of unfolded proteins by the proteasome [7]. Inhibition of the proteasome by bortezomib leads to excessive accumulation of unfolded proteins, causing ER stress, which in turn induces the UPR [6]. The UPR is a complex signalling cascade which is activated by three ER stress sensing transmembrane proteins located in the ER membrane; inositol-requiring enzyme 1 (IRE1), protein kinase R-like ER kinase (PERK) and activating transcription factor 6 (ATF6)[8]. Their activation induces a raft of cellular changes, including decreased protein translation, expansion of the ER, and upregulation of proteins such as chaperones which modulate protein folding, in an effort to restore ER homeostasis [6]. If, however, homeostasis cannot be restored, the UPR switches to pro-apoptotic signalling, inducing cell death [6,9]. Due to their high protein load, proteasome inhibition in myeloma cells causes ER stress which cannot be resolved by the homeostatic signalling of the UPR, and thus bortezomib induces myeloma cell death [4].
Unfortunately, not all patients respond to bortezomib, and those that do inevitably develop acquired bortezomib resistance [10]. The mechanisms behind bortezomib resistance are diverse and still not fully understood, with proteasome mutations and upregulation, heat shock protein induction, autophagy activation, plasma cell de-differentiation, and changes to the bone marrow microenvironment all potentially playing a role [10]. In order to combat resistance, second generation proteasome inhibitors, such as carfilzomib, have been developed [10]. However, whilst carfilzomib has been shown to be effective in the bortezomib resistant setting, cross-resistance between proteasome inhibitors has already been observed, suggesting new therapeutic options are still required [11], [12], [13].
Sphingolipids are a family of bioactive lipids which act as both key membrane components and important signalling molecules, with modulation of sphingolipid metabolism of increasing interest as a potential avenue for cancer therapy [14]. In particular, sphingosine kinases are key targets, as they catalyse the formation of pro-survival sphingosine 1-phosphate (S1P) and decrease levels of pro-apoptotic ceramide and sphingosine [15]. We have recently shown that inhibition of sphingosine kinase 2 (SK2) induces myeloma cell death through activation of the UPR, and that combining SK2 inhibition with bortezomib induces synergistic cell death, at least in bortezomib-naïve myeloma cells [16]. Here, we examined the potential for SK2 inhibition to be employed in the bortezomib-resistant setting. We found that mouse and human bortezomib-resistant myeloma cells are also resistant to carfilzomib, and that SK2 inhibition synergises with both bortezomib and carfilzomib to induce cell death via UPR activation, effectively resensitising these cells to proteasome inhibition. This combinatorial approach reduced disease burden in a bortezomib-resistant myeloma mouse model, supporting its further exploration as a treatment option for bortezomib-resistant myeloma.
Methods
Cell culture
Drug-naïve 5TGM1 (5TGM1.wt)[17] and RPMI-8226 (8226.wt) cells were cultured as previously described [16]. Bortezomib-resistant 5TGM1 (5TGM1.BR) cells were generated by exposing 5TGM1.wt cells to increasing doses of bortezomib (Selleck Chemicals, Houston, TX), until they could be cultured in 20nM bortezomib. Bortezomib-resistant RPMI-8226 (8226.BR), Kas6 (Kas6.BR) and ANBL6 (ANBL6.BR) cells, and carfilzomib-resistant RMPI-8226 (8226.CR) cells were generated previously in a similar manner [11,18]. 5TGM1.BR cells were routinely cultured in media containing 20nM bortezomib, 8226.BR, Kas6.BR and ANBL6.BR in media containing 10nM bortezomib, and 8226.CR cells in media containing 10nM carfilzomib (Selleck Chemicals). Cells were removed from proteasome inhibitor for at least 72h before use in experiments. All human cell lines were authenticated by short tandem repeat DNA fingerprinting, tested for mycoplasma by PCR analysis [19], and used within 3 months of resuscitation.
CRISPR knockout
A single-guide RNA targeting ERN1, sequence 5′-GCAAAGGAAGTGTGCTGCCG-3′, was cloned into pLenti-U6-sgRNA-SFFV-Cas9-2A-Puro (Applied Biological Materials, Richmond, BC, Canada). Lentiviral particles were generated by transfecting HEK293T cells with pLenti plasmid and packaging plasmids pLP1 (gag/pol), pLP2 (rev) and pVSVG (envelope) using Lipofectamine 2000 (ThermoFisher Scientific, Waltham, MA). Virus was harvested 48h post transfection and used to transduce LP1 human myeloma cells that had been treated with 4µg/mL polybrene. Transduced LP1 cells were puromycin selected, and then single-cell sorted on a MoFlo Astrios Cell Sorter (Beckman Coulter Life Sciences, Indianapolis, IN). LP1.IRE1KO cells were obtained from a single clone, and IRE1 knockout validated by Western blotting.
Cell viability analysis
Cell viability was assessed by staining with propidium iodide and either Annexin-V-FITC for RPMI-8226 cells or Annexin-V-APC for 5TGM1 cells, with double-negative cells being counted as viable.
Western blotting
Cells were cultured in the given concentration of drug/s for 4h, and then lysed in 10mM Tris/HCl buffer (pH 7.4) containing 137mM NaCl, 10% Glycerol, 1% NP40, 10mM β-glycerophosphate, 2mM sodium vanadate, 2mM NaF, 10mM sodium pyrophosphate, and cOmpleteTM EDTA-free protease inhibitor (Sigma-Aldrich). Lysates were subjected to SDS-PAGE with pre-cast 4-20% TGX gels (ThermoFisher), proteins transferred to nitrocellulose membranes, which were blocked for at least 1h at room temperature in either 5% skim milk powder in 50mM Tris/HCl buffer (pH 7.4) containing 150mM NaCl and 1% Tween 20 if imaging on a LAS-4000 (GE Healthcare), or Li-Cor PBS Odyssey Blocking Buffer if imaging on an Odyssey (Li-Cor, Lincoln, NE). Primary antibodies used were ATF4, XBP1, PSMB5, PSMB6, PSMB7 (Cell Signaling Technology), and β-actin (Sigma-Aldrich). Secondary antibodies used were HRP-conjugated goat-anti-rabbit and HRP-conjugated goat-anti-mouse (ThermoFisher), and goat-anti-rabbit IR and goat-anti-mouse IR 680RD (Li-Cor).
Proteasome activity assay
Proteasome activity was determined in cell lysates using the Abcam proteasome activity kit according to the manufacturer's instructions, with 350/460nm fluorescence filters. MG-132 was used as a control, and all results were normalised to protein levels and maximal proteasome inhibition observed with 500nM bortezomib.
RNA sequencing and analysis
RNA was extracted from (>80% viable) cells using a Qiagen RNeasy extraction kit. Drug resistant cell lines were given a 7-day break from drug exposure prior to sequencing. PolyA+ enriched RNAseq libraries from three biological replicates for each cell line were multiplexed and sequenced on the Illumina NextSeq 500 platform. Raw data, averaging 83 and 57 million reads per sample for 5TGM1 and RPMI-8226 samples, respectively, were analysed and quality checked using the FASTQC program (http://www.bioinformatics.babraham.ac.uk/ projects/fastqc). Reads were mapped against the mouse (mm10) and human (hg19) reference genomes for 5TGM1 and RPMI-8226 cells respectively using the STAR spliced alignment algorithm [20] (version2.5.3a for 5TGM1 cells and version2.7.2c for RPMI-8226 cells with default parameters and –chimSeqmentMin 20, –quantMode GeneCounts) returning an average unique alignment rate of 78% and 89%. Differential expression analysis was evaluated from the TMM normalised gene counts using R (version 3.2.3) and edgeR (version 3.3)[21] following protocols as described [22] using a false discovery rate<0.01 and log2fold change>1. Alignments were visualised and interrogated using the Integrative Genomics Viewer v2.3.80[23]. Mutation detection was conducted by amalgamating replicate RNAseq alignment files for each group and using the resultant merged files as input for the freebayes variant detection algorithm [24] (version 1.0.2 for 5TGM1 cells and version 1.2.0 for RPMI-8226 cells). Gene-set enrichment analysis (GSEA v4.0.3) was used to look for coordinate expression of differential expression analysis results to groups of genes in the Molecular Signatures Database (MSigDB v7.1) C2 collection [25,26]. Genes were ranked for the GSEA analysis (GSEAPreranked) by calculating the “directional” negative log FDR (sign of fold change * -log10(FDR)). The RNA sequencing data has been deposited in the Gene Expression Omnibus database (accession number GSE144249).
Sphingolipidomics
5TGM1.BR cells (1 × 106/ml) were cultured in standard media with 5% FBS with K145, or vehicle, for 6h. Cells were centrifuged, and cell pellets analysed by LC-MS, as previously described [27], with sphingolipids quantified relative to internal standards.
Animal studies
Seven-week-old female NOD/SCID/IL-2Rγ−/− (NSG) mice were injected with 2 × 106 5TGM1.BR cells via the tail vein, and disease was allowed to establish for two weeks. Mice were then divided into four groups of six mice; vehicle control, bortezomib alone, K145 (3-(2-amino-ethyl)-5-[3-(4-butoxyl-phenyl)-propylidene]-thiazolidine-2,4-dione [28]; Medkoo, Morrisville, NC) alone, or dual treatment with bortezomib and K145. Mice were treated with 20mg/kg K145 or vehicle (2% DMSO, 20% PEG, 78% saline) six days a week. This was the previously determined maximum tolerated dose of K14516, but even with this the last two doses were reduced to 15mg/kg to minimise weight loss. Mice were also treated with 0.5mg/kg bortezomib or vehicle (0.5% DMSO, 99.5% saline) three times a week. Both treatments were administered i.p. over 17 days. Bioluminescence imaging was performed after injection of 100μL of 30mg/mL luciferin using an IVIS Lumina S5 on days 14 (day before first dose), 21, 28, and 32 (day after last dose). The studies were approved by the SA Pathology/Central Adelaide Local Health Network and University of South Australia Animal Ethics Committees (approval #18/17).
Statistical analysis
Analysis of in vitro EC50 was determined using sigmoidal 4PL regression in Prism 8.0.1, with other statistical differences assessed using an unpaired student t-test. Analysis of in vitro synergy was conducted using CompuSyn to generate a combination index, where synergy is indicated by a combination index of <1. Average in vivo disease burden for each treatment group and group comparisons after completion of allocated treatments were determined using a multivariable linear regression model adjusting for differences in baseline myeloma disease burden. The Kaplan-Meier method and log-rank test were used to assess differences in the survival functions between groups. A multivariate Cox proportional hazards regression model was then constructed to estimate the risk of death based on treatment received, adjusting for baseline myeloma disease burden. In vivo statistical analyses were performed using Stata version 14 (StataCorp, College Station, TX).
Results
Generation and characterisation of bortezomib-resistant myeloma cells
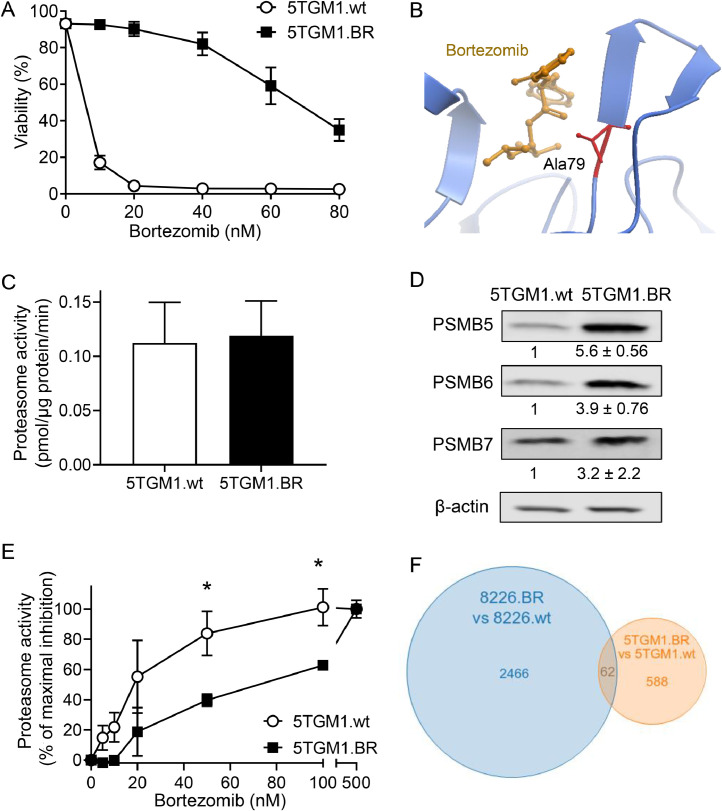
The 5TGM1 cell line is a murine myeloma cell line that can be introduced in C57BL/KaLwRij mice to generate a syngeneic mouse model of myeloma which recapitulates many facets of the human disease, making it a powerful tool for studying myeloma [17,29]. Thus, we sought to adapt this model in order to study bortezomib resistance. To do so, drug-naïve 5TGM1 (5TGM1.wt) cells were exposed to increasing doses of bortezomib until they could be cultured in 20nM bortezomib without any loss in viability. These bortezomib-resistant cells were designated 5TGM1.BR, and demonstrated an approximate 10-fold increase in resistance to bortezomib compared to 5TGM1.wt cells (Figure 1A).
Figure 1.
Characterisation of bortezomib resistant 5TGM1 cells. A) Drug-naïve 5TGM1 (5TGM1.wt) and bortezomib-resistant 5TGM1 (5TGM1.BR) cells were cultured with increasing concentrations of bortezomib for 24h and then assessed for cell viability by flow cytometry using Annexin-V and PI staining. Bortezomib has an EC50 of <10nM in 5TGM1.wt cells, and 72nM in 5TGM1.BR cells. Data shown represents mean ± SD of three independent experiments. B) Mutational analysis of RNAseq data found a highly penetrant G>C mutation in PSMB5 in 5TGM1.BR cells. This results in an Ala79→Gly substitution in the bortezomib binding pocket of PSMB5 (the proteasomal subunit β5). Effects of this mutation were analysed in silico with ICM-Pro (MolSoft, CA) using the structure of PSMB5 crystallised with bortezomib (pdb code: 5lf3_K), and shown in ribbon view. C) Chymotrypsin-like activity of the proteasome was measured by cleavage of the fluorescent substrate suc-LLVY-AMC by cell lysates from 5TGM1.wt and 5TGM1.BR cells. Activity is shown as pmol of suc-LLVY-AMC cleaved/μg protein/min. Data shown represents mean ± SD of three independent experiments. D) Levels of PSMB5, PSMB6 and PSMB7 in 5TGM1.wt and 5TGM1.BR cells was assessed by Western blot. Numbers below the blots represent quantitation of mean ±SD of three independent experiments in Image Studio normalised to β-actin and then shown as fold increase over control. E) Proteasome activity measured as in (C) with increasing concentrations of bortezomib. Activity shown as a percent of maximal inhibition normalised to control, and is the mean ± SD of three independent experiments. * p < 0.05. F) Venn diagram showing the overlap in genes up- or downregulated in the 5TGM1.BR and 8226.BR cell lines compared to the drug-naïve cell counterparts.
To examine the mechanistic basis for the acquired bortezomib resistance of the 5TGM1.BR cells, we conducted RNAseq comparing their gene expression profile to that of 5TGM1.wt cells. This analysis found 650 genes that were significantly differentially regulated between these cells (Supplementary Table 1). Gene set enrichment analysis (GSEA) found a number of altered gene sets in 5TGM1.BR cells compared to 5TGM1.wt cells (Supplementary Figure 1). Only a few of these reached a FDR<0.05, however, and apart from a decrease in ‘programmed cell death’ genes and a trend towards increased ‘repressed by BCL2’ genes, few identified pathways revealed insights into the mechanisms driving bortezomib resistance in 5TGM1.BR cells.
Along with GSEA, mutational analysis was conducted on the RNAseq data, which showed that the 5TGM1.BR cells possess acquired, homozygous mutations in seven genes (Supplementary Table 2). Notably, this included a mutation in PSMB5, the β5 proteasomal subunit, which caused an alanine to glycine substitution at residue 79. This PSMB5 mutation is particularly notable given than an Ala→Thr mutation at the same site was recently discovered clinically in a bortezomib-resistant myeloma patient, and was found to convey myeloma cell resistance to bortezomib, as well as next generation proteasome inhibitors such as carfilzomib [30]. Ala79 resides in the bortezomib-binding pocket of PSMB5 (Figure 1B), and structural modelling of the effect of the glycine mutation is predicted to reduce hydrophobic interactions between the pocket residues and bortezomib (Supplementary Figure 2). Therefore, we assessed whether this mutation altered proteasome function or sensitivity of the proteasome to inhibition by bortezomib. To do this, we measured the chymotrypsin-like activity of the proteasome in cell lysates of 5TGM1.wt and 5TGM1.BR cells. Basal proteasome activity was similar in both cell types (Figure 1C), despite PSMB5 protein levels being substantially increased in 5TGM1.BR cells, as were other proteasome subunits PSMB6 and PSMB7 (Figure 1D). This suggests the 5TGM1.BR cells have increased levels of intact 20S proteasome complex with each complex likely to have reduced chymotrypsin-like activity due to the Ala79→Gly mutation in the PSMB5 subunit. Strikingly, further analysis showed the chymotrypsin-like activity of the proteasome in the 5TGM1.BR cells was substantially less sensitive to inhibition by bortezomib (Figure 1E), providing a clear mechanism to explain the resistance these cells display to cell death induced by bortezomib.
To compare the resistance mechanisms of the 5TGM1.BR cells to other experimental models of bortezomib resistance, we analysed human RPMI-8226 myeloma cells previously made resistant to bortezomib [11], designated 8226.BR. RNAseq to compare expression profiles between 8226.BR and drug-naïve RPMI-8226 (8226.wt) cells was performed, with 2,528 genes found to be differentially regulated between these cells (Supplementary Figure 3A and Supplementary Table 3). Gene set enrichment analysis again found a number of altered pathways (Supplementary Figure 3B,C), with surprisingly little overlap between gene expression (Figure 1F) or the pathways altered in 5TGM1.BR and 8226.BR cells. Mutational analysis of the 8226.BR cells demonstrated that, unlike the 5TGM1.BR cells, the 8226.BR cells do not possess PSMB5 mutations (Supplementary Table 4), which is consistent with previous sequencing data, and findings that proteasome activity remained sensitive to bortezomib in these cells [31]. Thus, it appears that, consistent with the heterogenous nature of bortezomib resistance in the clinic [10], the 5TGM1.BR and 8226.BR cells have different mechanisms mediating bortezomib resistance, making them good complimentary models to examine potential new therapies to overcome this major clinical problem.
The SK2 inhibitor K145 synergises with bortezomib to kill bortezomib-resistant myeloma
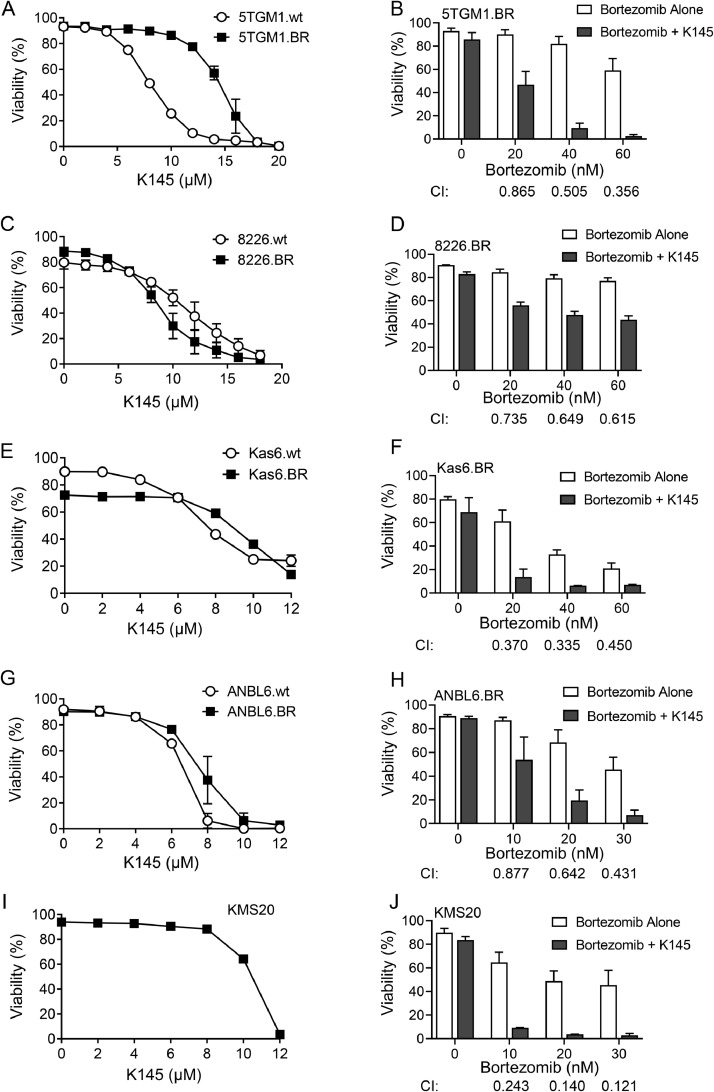
Our previous studies in bortezomib-naïve myeloma cells demonstrated that the SK2 inhibitor K145 caused apoptotic cell death and induced synergistic cell death in combination with bortezomib, both in vitro and in vivo[16]. To determine if this approach was also valid in the bortezomib-resistant setting, we examined the effect of K145 on bortezomib-resistant cells compared to their drug-naïve counterparts. Exposure to K145 resulted in death of all 5TGM1 and RPMI-8226 cells in a dose-dependent manner (Figure 2A,C). Sphingolipidomic analysis confirmed K145 decreased cellular S1P and increased sphingosine and ceramide expected from SK2 inhibition (Supplementary Figure 4). 5TGM1.BR cells showed a slightly decreased sensitivity to K145, with the EC50 for K145 approximately double that of the 5TGM1.wt cells (Figure 2A), however this was a modest change compared to the observed 10-fold change in the EC50 for bortezomib in these cells (Figure 1A). Interestingly, this modest effect was not consistent between different cell lines, with the 8226.BR cells slightly more sensitive to K145 than the 8226.wt cells (Figure 2C). A second, less potent but structurally distinct SK2 inhibitor, ABC294640, also induced death at similar doses in both drug-naïve and bortezomib resistant 5TGM1 and RPMI-8226 cells (Supplementary Figure 5A,B).
Figure 2.
K145 induces cell death and synergises with bortezomib in bortezomib-resistant myeloma cell lines. A) 5TGM1.wt and 5TGM1.BR cells were cultured with increasing concentrations of K145 for 24h and cell viability was then assessed by flow cytometry using Annexin-V and PI staining. B) 5TGM1.BR cells were cultured with increasing concentrations of bortezomib with and without 8µM K145 for 24h and cell viability was then assessed by flow cytometry. C) 8226.wt and 8226.BR cells were cultured with increasing concentrations of K145 for 24h and cell viability was then assessed by flow cytometry. D) 8226.BR cells were cultured with increasing concentrations of bortezomib with and without 4µM K145 for 24h and cell viability was then assessed by flow cytometry. E) Kas6.wt and Kas6.BR cells were cultured with increasing concentrations of K145 for 48h and cell viability was then assessed by flow cytometry. F) Kas6.BR cells were cultured with increasing concentrations of bortezomib with and without 7µM K145 for 48h and cell viability was then assessed by flow cytometry. G) ANBL6.wt and ANBL6.BR cells were cultured with increasing concentrations of K145 for 48h and cell viability was then assessed by flow cytometry. H) ANBL6.BR cells were cultured with increasing concentrations of bortezomib with and without 4µM K145 for 48h and cell viability was then assessed by flow cytometry. All data shown represents mean ± SD of three independent experiments. I) KMS20 cells were cultured with increasing concentrations of K145 for 48h and cell viability was then assessed by flow cytometry. Data shown is representative of 3 independent experiments. J) KMS20 cells were cultured with increasing concentrations of bortezomib with and without 8µM K145 for 48h and cell viability was then assessed by flow cytometry. Data shown represents mean ± SD of three independent experiments. Combination index (CI) values were calculated using CompuSyn, where a CI of <1 indicates synergy.
The combination of bortezomib with sub-cytotoxic concentrations of K145 in both bortezomib-resistant cell lines resulted in robust synergistic cell death, as indicated by the combination index (CI) scores of <1 (Figure 2B,D). This result was partially mimicked by a second, less potent SK2 inhibitor ABC294640, which synergised when combined with bortezomib in 5TGM1.BR, but not 8226.BR cells (Supplementary Figure 5C,D).
In order to confirm that this effect was broadly applicable in the bortezomib-resistant setting, K145 alone and in combination with bortezomib was examined in two additional bortezomib-resistant myeloma cell lines, ANBL6.BR and Kas6.BR, both of which are significantly more resistant to bortezomib than their bortezomib-naïve counterparts [11,32]. Both of these bortezomib-resistant cell lines had a similar sensitivity to K145 compared to their bortezomib-naïve counterparts (Figure 2E,G), and the combination of K145 and bortezomib caused synergistic cell death in both cases (Figure 2F,H). Effectively, all bortezomib-resistant cells were partially resensitised to bortezomib by the presence of K145. Furthermore, in addition to these myeloma cell models of acquired bortezomib resistance, the effects of K145 was also examined in KMS20 myeloma cells, which are known to have inherent bortezomib-resistance [33]. K145 induced cell death, and synergised with bortezomib in KMS20 cells (Figure 2I,J), suggesting that the synergy seen between K145 and bortezomib may be broadly applicable in the bortezomib-resistant setting.
Bortezomib and K145 induce synergistic UPR activation in bortezomib-resistant myeloma
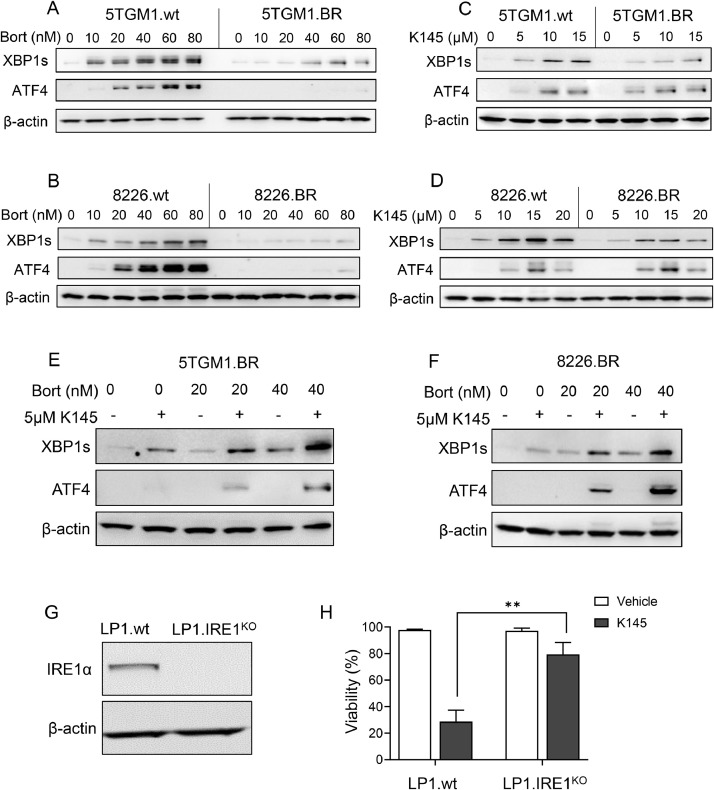
Due to their high immunoglobulin production, myeloma cells are known to have elevated basal levels of ER stress, and thus a basal activation of the UPR [5]. Bortezomib elicits its anti-myeloma effects at least partially through further induction of the UPR, resulting in myeloma cell apoptosis [5,34]. Notably, sphingolipid metabolism is intimately linked to UPR activation [35], and our previous studies have shown that K145 can also induce significant increases in UPR activation, at least in bortezomib-naïve myeloma cells [16]. Thus, we examined UPR activation in bortezomib-resistant myeloma cells in response to bortezomib, K145, or combinations of the two. To do this, we examined two markers of UPR activation, X-box binding protein 1s (XBP1s), generated in response to IRE1 activation, and activating transcription factor 4 (ATF4), produced following activation of PERK [36]. Both 5TGM1.BR and 8226.BR cells showed a drastically reduced UPR in response to treatment with bortezomib compared to their parental cell counterparts (Figure 3A,B). Notably, however, K145 caused effective upregulation of both the IRE1 and PERK arms of the UPR in both bortezomib-resistant cell lines (Figure 3C,D). K145 had no effect on proteasome activity, nor bortezomib inhibition of the proteasome in 5TGM1.BR cells (Supplementary Figure 6). Together, this suggests there are likely different modes of UPR activation induced by K145 compared to bortezomib in the bortezomib-resistant myeloma cell lines.
Figure 3.
Unfolded protein response activation in response to bortezomib or K145 treatment differs between drug-naïve and bortezomib-resistant myeloma cell lines, but the combination of bortezomib and K145 still causes synergistic UPR activation in bortezomib-resistant myeloma cells. 5TGM1.wt and 5TGM1.BR cells (A), and 8226.wt and 8226.BR cells (B) were cultured with increasing concentrations of bortezomib for 4h, and then examined by Western blot for UPR proteins XBP1s and ATF4. 5TGM1.wt and 5TGM1.BR cells (C), and 8226.wt and 8226.BR cells (D) were cultured with increasing concentrations of K145 for 4h, and then examined by western blot for UPR proteins XBP1s and ATF4. 5TGM1.BR cells (E) and 8226.BR (F) were cultured with 0nM, 20nM or 40nM bortezomib, with and without 5μM K145 for 4h, and then examined by Western blot for UPR proteins XBP1s and ATF4. G) Wildtype LP1 cells (LP1.wt) or LP1 cells with IRE1 knocked out (LP1.IRE1KO) were examined by Western blot for IRE1 expression. Actin was used as a loading control. All data shown representative of three independent experiments. H) LP1.wt and LP1.IRE1KO cells were cultured with vehicle or 10μM K145 for 48h, and cell viability was then assessed by flow cytometry using Annexin-V and PI staining. Data shown is the mean ± SD of three independent experiments. ** p < 0.01.
Next, we examined the effects of combining K145 with bortezomib on UPR activation in bortezomib-resistant cells. 5TGM1.BR cells (Figure 3E) and 8226.BR cells (Figure 3F) were treated with bortezomib and/or K145 at doses that induced minimal UPR activation alone. Strikingly, in both cell lines, the combination of these low doses of bortezomib and K145 induced a strong activation of both the IRE1 and PERK arms of the UPR (Figure 3E,F). This is consistent with the notion that SK2 inhibition induces the UPR via a different mechanism to bortezomib, and suggests that the synergistic cell death seen in bortezomib-resistant cells treated with bortezomib and K145 may arise from synergistic UPR activation.
In order to examine the role UPR activation plays in K145-induced myeloma cell death, we utilised CRISPR-Cas9 to knock out IRE1 in LP1 cells, generated cells termed LP1.IRE1KO (Figure 3G). These LP1.IRE1KO cells were significantly more resistant to K145 compared to parental LP1 (LP1.wt) cells (Figure 3H), demonstrating that activation of the UPR sensor IRE1 contributes to the myeloma cell death caused by K145. This supports the hypothesis that the enhanced UPR activation seen in bortezomib-resistant cells treated with the combination of K145 and bortezomib contributes to the synergistic cell death caused by this combination.
K145 synergises with carfilzomib in both carfilzomib and bortezomib-resistant myeloma
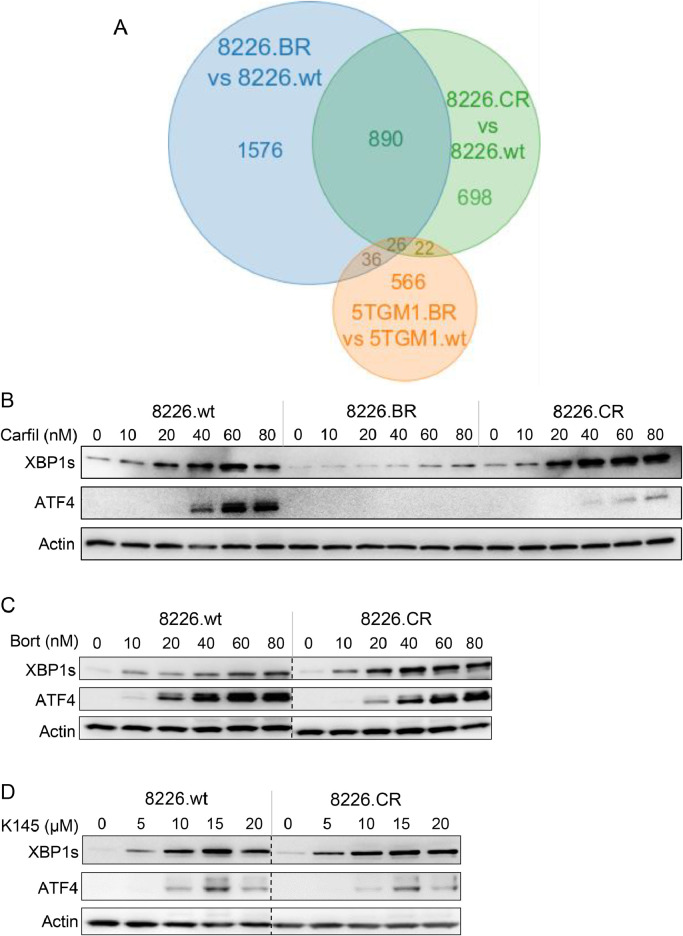
Along with bortezomib-resistant 8226.BR cells, we also examined 8226.CR cells, previously generated to be resistant to the second-generation proteasome inhibitor carfilzomib [18]. We found 1,636 genes which were differentially expressed in 8226.CR cells compared to 8226.wt cells by RNAseq (Supplementary Figure 7A and Supplementary Table 5). As with the 8226.BR cells, the 8226.CR cells do not possess mutations in PSMB5 (Supplementary Table 6). When comparing the differentially expressed genes in 8226.BR cells and 8226.CR cells, 916 genes were found to be either upregulated in both or downregulated in both compared to 8226.wt cells, with only 26 of these genes also altered in 5TGM1.BR cells (Figure 4A and Supplementary Table 7). This, along with the gene set enrichment analysis of genes differentially regulated in 8226.CR cells compared to 8226.wt cells (Supplementary Figure 7B,C), suggests that, while there is potentially a degree of overlap, resistance mechanisms between the bortezomib-resistant and carfilzomib-resistant cells are likely to be different. Indeed, this is supported by the fact that, unlike bortezomib-resistant cells, the carfilzomib-resistant cells did not show reduced UPR activation when treated with bortezomib, carfilzomib or K145 (Figure 4B-D).
Figure 4.
Unfolded protein response activation in response to bortezomib or K145 treatment is the same in wildtype and carfilzomib resistant myeloma cell lines. A) Venn diagram showing the overlap in genes up or downregulated in the 5TGM1.BR, 8226.BR, and 8226.CR cell lines compared to their drug-naïve counterparts from RNAseq analysis. B) 8226.wt, 8226.BR, and 8226.CR cells were cultured with carfilzomib for 4h, and then examined by Western blot for UPR proteins ATF4 and XBP1s. 8226.wt and 8226.CR cells were cultured with bortezomib (C) or K145 (D) for 4h, and then examined by Western blot for UPR proteins ATF4 and XBP1s. Dotted lines indicate where lanes from the same immunoblot have been spliced together to aid interpretation. Actin was used as a loading control. Data shown are representative of three independent experiments.
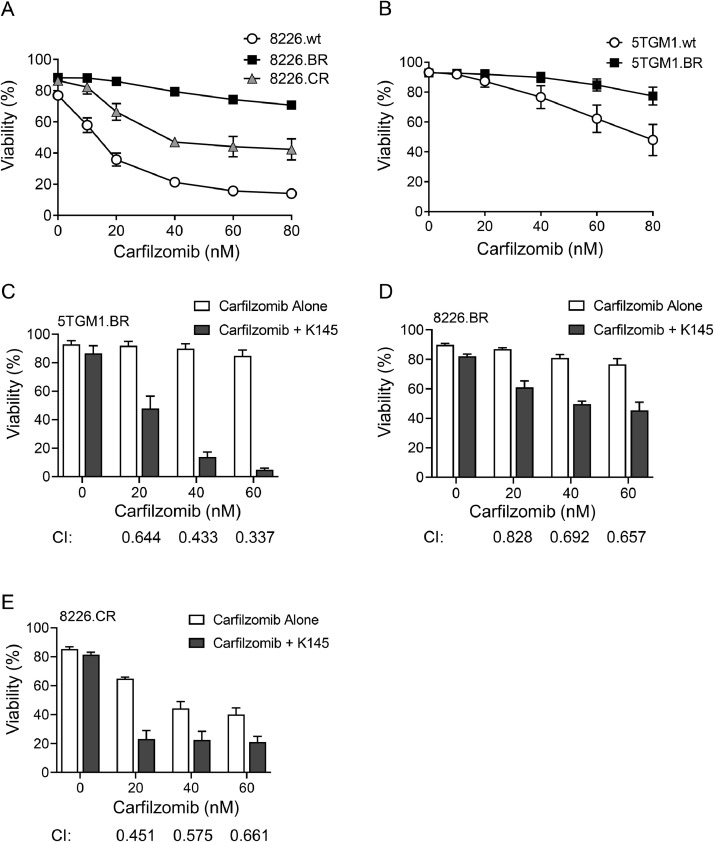
It has previously been noted that, whilst carfilzomib can be effective in the bortezomib resistant setting, there is some cross-resistance between proteasome inhibitors [11,13,37]. Cross resistance was also observed in the cell lines we examined; both the 8226.CR and 8226.BR cells were more resistant to carfilzomib than the 8226.wt cells (Figure 5A), and cross resistance was also seen in the 5TGM1.BR cells (Figure 5B). However, sensitivity to carfilzomib was significantly increased by the addition of sub-cytotoxic doses of K145, both in bortezomib-resistant (Figure 5C,D) and carfilzomib-resistant (Figure 5E) myeloma cell lines.
Figure 5.
K145 induces cell death and synergises with carfilzomib in bortezomib-resistant and carfilzomib-resistant myeloma cell lines. 8226.wt, 8226.BR and 8226.CR cells (A), and 5TGM1.wt and 5TGM1.BR (B) were cultured in increasing concentrations of carfilzomib for 24h, and then cell viability was assessed by flow cytometry using Annexin-V and PI staining. Data shown represents mean ± SD of three independent experiments. C) 5TGM1.BR cells were cultured with increasing concentrations of carfilzomib with or without 8µM K145 for 24h and cell viability was then assessed by flow cytometry. D) 8226.BR cells were cultured with increasing concentrations of carfilzomib with or without 4µM K145 for 24h and cell viability was then assessed by flow cytometry. E) 8226.CR cells were cultured with increasing concentrations of carfilzomib with or without 6µM K145 for 24h and cell viability was then assessed by flow cytometry. Data shown represents mean ± SD of three independent experiments. Combination index (CI) values were calculated using CompuSyn, where a CI of <1 indicates synergy.
Combination of bortezomib and K145 attenuates progression of bortezomib-resistant myeloma in vivo
Since the combination of bortezomib and K145 in bortezomib-resistant myeloma cells demonstrated synergism in vitro, we next examined these effects in vivo. The 5TGM1.BR cells were engrafted into the bone marrow of mice via tail vein injection, and the disease was allowed to establish for two weeks. Mice were randomised into groups with equal disease burden (Supplementary Figure 8), then treated for 17 days with bortezomib alone, K145 alone, or a combination of the two. Disease burden was followed by bioluminescence imaging, and the mice monitored until they reached their ethical end point.
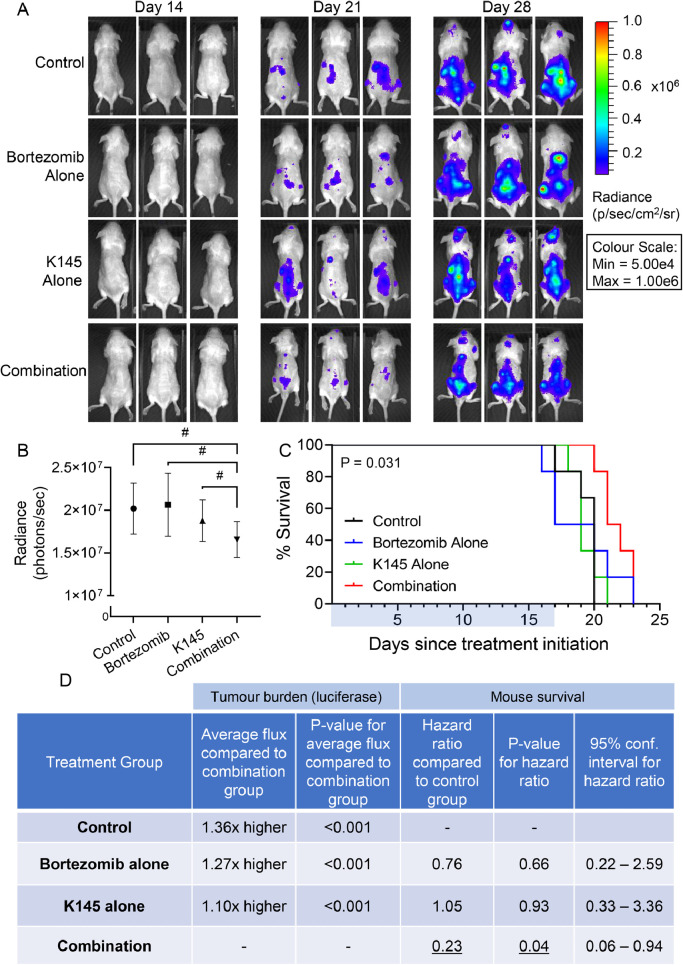
Consistent with the bortezomib resistance of this model, bortezomib alone showed little effect on the myeloma burden (Figure 6A,B), despite having anti-myeloma effects in vivo at these doses when the 5TGM1.wt cells were employed [16]. K145 alone also showed only minor effects (Figure 6). The combination of both bortezomib and K145, however, resulted in a modest but significant reduction in disease burden compared to the control, bortezomib alone, and K145 alone (Figure 6B,D). Furthermore, the combination of bortezomib and K145 also resulted in a modest but significant survival advantage for mice in this aggressive model, whilst either treatment alone had no effect (Figure 6C,D).
Figure 6.
Dual treatment with K145 and bortezomib shows efficacy in an aggressive, bortezomib-resistant murine model of myeloma. A) Representative bioluminescence images showing disease burden in each treatment group of NSG mice after luciferin injection 14 days (pre-treatment), 21 days (after one week of treatment), and 28 days (after 2 weeks of treatment) post injection of two million 5TGM1.BR cells via the tail vein. B) Quantitation of myeloma disease burden (as average bioluminescent flux, measured in photons/sec ± SEM, of each treatment group after 28 days (two weeks of treatment)). # refer to panel D C) Mouse Kaplan-Meier survival curves for each treatment group from the day of treatment starting. Treatment was administered for 17 days (as indicated by the blue bar), and then the mice were monitored until their ethical end points were reached (p = 0.031, log-rank test). D) Average disease burden was compared to the combination group using a multivariable linear regression model adjusting for baseline disease burden, with the p-value shown alongside. A multi-variable Cox regression survival analysis adjusting for baseline disease burden was used to assess the risk of death between treatment groups. Hazard rations for each treatment group compared to the control group are shown, together with the p-values and 95% confidence intervals.
Discussion
Bortezomib resistance continues to present a major hurdle for the treatment of myeloma [10]. Whilst the introduction of next generation proteasome inhibitors such as carfilzomib have shown efficacy in the bortezomib resistant setting, a level of cross-resistance has also been observed [11], [12], [13],37]. This was also seen in our study, with both bortezomib-resistant cell lines demonstrating not only significant resistance to bortezomib, but also carfilzomib. Furthermore, RNAseq and mutational analysis of the 5TGM1.BR cell line revealed that these cells possess a highly penetrant mutation in PSMB5 which results in an amino acid substitution in the bortezomib binding pocket of the β5 subunit of the proteasome. A mutation at the same residue has recently been discovered in a bortezomib-resistant patient, and analysis of this mutation revealed it conveyed resistance to bortezomib and carfilzomib, as well as the newest FDA-approved proteasome inhibitor ixazomib [30]. Whilst the PSMB5 mutation found in the 5TGM1.BR cells causes a smaller change in the bortezomib binding pocket than that found in the patient, it is still enough to reduce sensitivity to bortezomib inhibition, demonstrating that this 5TGM1.BR model represents disease that is relevant to the clinic. Furthermore, the cross resistance to carfilzomib seen in both bortezomib-resistant cell lines demonstrates how new, non-proteasome targeting therapies are still required for the bortezomib-resistant setting.
We have previously found that SK2 is a valid target in bortezomib-naïve myeloma, and that inhibition of SK2 by K145 is able to cause synergistic cell death when combined with bortezomib via UPR activation [16]. In this current study we demonstrated the significant finding that this combination remains valid even in the clinically-relevant bortezomib-resistant setting, and that synergistic cell death still appears to occur via the UPR despite the fact that these cells show little UPR activation when treated with bortezomib alone. This, combined with our findings that K145 had no effect on proteasome activity, nor proteasome inhibition by bortezomib suggested that SK2 inhibition activates the UPR via a different mechanism. Although the UPR is classically activated by unfolded proteins, changes in lipid composition are also known to activate the UPR. This can occur indirectly, with changes to ER lipid composition resulting in altered calcium levels or protein trafficking, which increases the unfolded protein burden in the ER [35,[38], [39], [40], [41]. However, it has also been found that changes in lipid composition, specifically increases in saturated lipids, of which sphingolipids are a major class, are able to activate the ER stress sensors IRE1 and PERK directly, independent of unfolded protein levels [35,[42], [43], [44]. More recently it was found that IRE1 possesses an amphipathic helix that mediates its activation in response to changes in lipid composition of the ER membrane [45]. Several sphingolipid enzymes, including SK2, are known to localise to the ER, the site of de novo sphingolipid synthesis, where their activity, or inhibition, can influence the composition of the ER membrane [46], [47], [48]. Furthermore, a recent study has found that the ER stress sensor ATF6 can be directly activated by increased levels of dihydroceramide and dihydrosphingosine [49], two sphingolipids upstream of SK2 that are known to be elevated in response to SK2 inhibition [50]. Indeed, our findings with IRE1 knockout myeloma cells clearly show this arm of the UPR to be critical for cell death induced by SK2 inhibition with K145. Thus, K145-induced activation of the UPR through a mechanism independent of unfolded proteins would explain why bortezomib-resistant myeloma cells remain more sensitive to UPR activation induced by K145 compared to bortezomib, where UPR activation is markedly attenuated. Furthermore, the potential difference in UPR activation mechanisms likely explains why the combination of K145 and bortezomib still results in a synergistic UPR activation in the bortezomib-resistant setting.
Our findings of synergism between SK2 inhibition and bortezomib in 5TGM1.BR cells makes mechanistic sense as these cells possess a clinically relevant proteasome mutation that we found decreases the sensitivity of the proteasome to inhibition by bortezomib. Notably, however, these findings were replicated in multiple other bortezomib-resistant cell lines, including several that do not possess proteasome mutations [31]. Indeed, the little concordance in differentially regulated genes or pathways in these cell lines indicate that they likely have different mechanisms of bortezomib resistance. Despite this, K145 synergistically enhanced sensitivity to bortezomib in all bortezomib-resistant myeloma cell lines examined. Furthermore, K145 also synergised with carfilzomib in both the bortezomib-resistant and the carfilzomib-resistant setting, despite the fact that carfilzomib has a different active moiety to bortezomib [3]. This suggests that SK2 inhibition may be effective in combination with any proteasome inhibitor in the proteasome-inhibitor resistant setting.
In summary, we have shown that the SK2 inhibitor K145 is effective in resensitising proteasome-inhibitor resistant myeloma to bortezomib or carfilzomib. This suggests that modulation of sphingolipid signalling could assist in circumventing proteasome inhibitor resistance in myeloma. While K145 suffers from dose-limiting toxicity, these findings warrant development of more potent and pharmacokinetically favourable modulators of sphingolipid metabolism that could be therapeutic options in the future.
Author contribution
M.K.B., C.T.W. and S.M.P designed experiments; M.K.B., M.L, M.N.T., D.A., D.J.C, J.A.P. and B.L.G. performed experiments; M.K.B., M.R.P., J.T., P.P.-S.W, and J.A.P analysed results; M.K.B., M.L., J.T. and C.T.W. performed statistical analysis; R.Z.O. provided vital new reagents and advice; M.K.B. and S.M.P. wrote the manuscript with input from all other authors.
Declaration of competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by an MF and MH Joyner Scholarship, the Royal Adelaide Hospital (RAH) Research Fund Dawes Scholarship, and an Australian Government Research Training Program Scholarship (to M.K.B.); a RAH Research Fund Florey Fellowship (to M.R.P); the National Cancer Institute (RO1s CA184464 and CA194264), the Leukemia & Lymphoma Society Specialized Center of Research (SCOR-12206-17), and the Dr. Miriam and Sheldon G. Adelson Research Foundation (to R.Z.O.); a National Health and Medical Research Council of Australia (NHMRC) Peter Doherty Biomedical Early Career Fellowship (1071945), a Servier Staff "Barry Young" Research Establishment Fellowship through the Royal Australasian College of Physicians, and a Viertel Foundation Clinical Investigator Award (to C.T.W.); the Fay Fuller Foundation, a NHMRC Project Grant (1162954), and Senior Research Fellowships from the NHMRC (1042589 and 1156693; to S.M.P.).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.11.009.
Contributor Information
Craig T. Wallington-Beddoe, Email: craig.wallingtonbeddoe@flinders.edu.au.
Stuart M. Pitson, Email: stuart.pitson@unisa.edu.au.
Appendix. Supplementary materials
References
- 1.Rajkumar S.V., Kumar S. Multiple myeloma: diagnosis and treatment. Mayo Clinic proceedings. 2016;91:101–119. doi: 10.1016/j.mayocp.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merin N.M., Kelly K.R. Clinical use of proteasome inhibitors in the treatment of multiple myeloma. Pharmaceuticals. 2014;8:1–20. doi: 10.3390/ph8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandolfi S., et al. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. 2017;36:561–584. doi: 10.1007/s10555-017-9707-8. [DOI] [PubMed] [Google Scholar]
- 4.Kubiczkova L., Pour L., Sedlarikova L., Hajek R., Sevcikova S. Proteasome inhibitors - molecular basis and current perspectives in multiple myeloma. Journal of cellular and molecular medicine. 2014;18:947–961. doi: 10.1111/jcmm.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vincenz L., Jager R., O'Dwyer M., Samali A. Endoplasmic reticulum stress and the unfolded protein response: targeting the Achilles heel of multiple myeloma. Molecular cancer therapeutics. 2013;12:831–843. doi: 10.1158/1535-7163.MCT-12-0782. [DOI] [PubMed] [Google Scholar]
- 6.Hetz C., Papa F.R. The unfolded protein response and cell fate control. Mol Cell. 2018;69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Smith M.H., Ploegh H.L., Weissman J.S. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroder M., Kaufman R.J. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 9.Corazzari M., Gagliardi M., Fimia G.M., Piacentini M. Endoplasmic reticulum stress, unfolded protein response, and cancer cell fate. Front Oncol. 2017;7:78. doi: 10.3389/fonc.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallington-Beddoe C.T., Sobieraj-Teague M., Kuss B.J., Pitson S.M. Resistance to proteasome inhibitors and other targeted therapies in myeloma. Br J Haematol. 2018;182:11–28. doi: 10.1111/bjh.15210. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn D.J., et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimopoulos M.A., et al. Carfilzomib and dexamethasone versus bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma (ENDEAVOR): a randomised, phase 3, open-label, multicentre study. Lancet Oncol. 2016;17:27–38. doi: 10.1016/S1470-2045(15)00464-7. [DOI] [PubMed] [Google Scholar]
- 13.Khan M.L., Stewart A.K. Carfilzomib: a novel second-generation proteasome inhibitor. Future Oncol. 2011;7:607–612. doi: 10.2217/fon.11.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truman J.P., Garcia-Barros M., Obeid L.M., Hannun Y.A. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim Biophys Acta. 2014;1841:1174–1188. doi: 10.1016/j.bbalip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitson S.M. Regulation of sphingosine kinase and sphingolipid signaling. Trends in biochemical sciences. 2011;36:97–107. doi: 10.1016/j.tibs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Wallington-Beddoe C.T., et al. Sphingosine kinase 2 inhibition synergises with bortezomib to target multiple myeloma by enhancing endoplasmic reticulum stress. Oncotarget. 2017;8:43602–43616. doi: 10.18632/oncotarget.17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dallas S.L., et al. Ibandronate reduces osteolytic lesions but not tumor burden in a murine model of myeloma bone disease. Blood. 1999;93:1697–1706. [PubMed] [Google Scholar]
- 18.Lee H.C., et al. RNA polymerase I inhibition with CX-5461 as a novel therapeutic strategy to target MYC in multiple myeloma. Br J Haematol. 2017;177:80–94. doi: 10.1111/bjh.14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopert A., Uphoff C.C., Wirth M., Hauser H., Drexler H.G. Mycoplasma detection by PCR analysis. In Vitro Cell Dev Biol Anim. 1993;29:819–821. doi: 10.1007/BF02634350. [DOI] [PubMed] [Google Scholar]
- 20.Dobin A., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lun A.T.L., Chen Y., Smyth G.K. In: Statistical Genomics: Methods in Molecular Biology. Mathé E., Davis S., editors. Springer; New York, New York, NY: 2016. It's DE-licious: A recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR; pp. 391–416. [DOI] [PubMed] [Google Scholar]
- 23.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 1207.3907 [q-bio.GN] [Google Scholar]
- 25.Mootha V.K., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aurelio L., et al. From sphingosine kinase to dihydroceramide desaturase: A structure-activity relationship (SAR) study of the enzyme inhibitory and anticancer activity of 4-((4-(4-Chlorophenyl)thiazol-2-yl)amino)phenol (SKI-II) J Med Chem. 2016;59:965–984. doi: 10.1021/acs.jmedchem.5b01439. [DOI] [PubMed] [Google Scholar]
- 28.Liu K., et al. Biological characterization of 3-(2-amino-ethyl)-5-[3-(4-butoxyl-phenyl)-propylidene]-thiazolidine-2,4-dione (K145) as a selective sphingosine kinase-2 inhibitor and anticancer agent. PLoS One. 2013;8:e56471. doi: 10.1371/journal.pone.0056471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asosingh K., Radl J., Van Riet I., Van Camp B., Vanderkerken K. The 5TMM series: a useful in vivo mouse model of human multiple myeloma. Hematol J. 2000;1:351–356. doi: 10.1038/sj.thj.6200052. [DOI] [PubMed] [Google Scholar]
- 30.Barrio S., et al. Spectrum and functional validation of PSMB5 mutations in multiple myeloma. Leukemia. 2018 doi: 10.1038/s41375-018-0216-8. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn D.J., et al. Targeting the insulin-like growth factor-1 receptor to overcome bortezomib resistance in preclinical models of multiple myeloma. Blood. 2012;120:3260–3270. doi: 10.1182/blood-2011-10-386789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B., et al. The nuclear factor (erythroid-derived 2)-like 2 and proteasome maturation protein axis mediate bortezomib resistance in multiple myeloma. The Journal of biological chemistry. 2015;290:29854–29868. doi: 10.1074/jbc.M115.664953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsubaki M., et al. Activation of serum/glucocorticoid regulated kinase 1/nuclear factor-kappaB pathway are correlated with low sensitivity to bortezomib and ixazomib in resistant multiple myeloma cells. Biomedicines. 2021;9 doi: 10.3390/biomedicines9010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obeng E., et al. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennett M.K., Wallington-Beddoe C.T., Pitson S.M. Sphingolipids and the unfolded protein response. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:1483–1494. doi: 10.1016/j.bbalip.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 37.Moreau P., et al. Impact of prior treatment on patients with relapsed multiple myeloma treated with carfilzomib and dexamethasone vs bortezomib and dexamethasone in the phase 3 ENDEAVOR study. Leukemia. 2017;31:115–122. doi: 10.1038/leu.2016.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunha D.A., et al. Initiation and execution of lipotoxic ER stress in pancreatic beta-cells. J Cell Sci. 2008;121:2308–2318. doi: 10.1242/jcs.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu S., et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boslem E., et al. A lipidomic screen of palmitate-treated MIN6 beta-cells links sphingolipid metabolites with endoplasmic reticulum (ER) stress and impaired protein trafficking. The Biochemical journal. 2011;435:267–276. doi: 10.1042/BJ20101867. [DOI] [PubMed] [Google Scholar]
- 41.Boslem E., et al. Alteration of endoplasmic reticulum lipid rafts contributes to lipotoxicity in pancreatic beta-cells. The Journal of biological chemistry. 2013;288:26569–26582. doi: 10.1074/jbc.M113.489310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Promlek T., et al. Membrane aberracy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532. doi: 10.1091/mbc.E11-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou N.S., et al. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci U S A. 2014;111:E2271–E2280. doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volmer R., van der Ploeg K., Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halbleib K., et al. Activation of the unfolded protein response by lipid bilayer stress. Mol Cell. 2017;67:673–684. doi: 10.1016/j.molcel.2017.06.012. e678. [DOI] [PubMed] [Google Scholar]
- 46.Maceyka M., et al. SphK1 and SphK2, sphingosine kinase isoenzymes with opposing functions in sphingolipid metabolism. The Journal of biological chemistry. 2005;280:37118–37129. doi: 10.1074/jbc.M502207200. [DOI] [PubMed] [Google Scholar]
- 47.Breslow D.K. Sphingolipid homeostasis in the endoplasmic reticulum and beyond. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gault C.R., Obeid L.M., Hannun Y.A. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol. 2010;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam A.B., et al. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev Cell. 2018;46:327–343. doi: 10.1016/j.devcel.2018.04.023. e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitman M.R., et al. A selective ATP-competitive sphingosine kinase inhibitor demonstrates anti-cancer properties. Oncotarget. 2015;6:7065–7083. doi: 10.18632/oncotarget.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.