Abstract
Background
Exercise can protect myocardial infarction (MI) and downregulate cardiac Homeodomain-Interacting Protein Kinase 2 (HIPK2). However, the role of HIPK2 in MI is unclear.
Methods
HIPK2–/– mice and miR-222–/– rats, HIPK2 inhibitor (PKI1H) and adeno-associated virus serotype 9 (AAV9) carrying miR-222 were applied in the study. Animals were subjected to running, swimming, acute MI or post-MI remodeling. HIPK2 inhibition and P53 activator were used in neonatal rat cardiomyocytes (NRCMs) and human embryonic stem cell-derived cardiomyocytes (hESC-CMs) subjected to oxygen glucose deprivation/reperfusion (OGD/R). Serum miR-222 levels were analyzed in healthy people and MI patients that were survival or readmitted to the hospital and/or died.
Findings
Cardiac HIPK2 protein levels were reduced by exercise while increased in MI. In vitro, HIPK2 suppression by lentiviral vectors or inhibitor prevented apoptosis induced by OGD/R in NRCMs and hESC-CMs. HIPK2 inhibitor-treated mice and HIPK2–/– mice reduced infarct size after acute MI, and preserved cardiac function in MI remodeling. Mechanistically, protective effect against apoptosis by HIPK2 suppression was reversed by P53 activators. Furthermore, increasing levels of miR-222, targeting HIPK2, protected post-MI cardiac dysfunction, whereas cardiac dysfunction post-MI was aggravated in miR-222–/– rats. Moreover, serum miR-222 levels were significantly reduced in MI patients, as well as in MI patients that were readmitted to the hospital and/or died compared to those not.
Interpretation
Exercise-induced HIPK2 suppression attenuates cardiomyocytes apoptosis and protects MI by decreasing P-P53. Inhibition of HIPK2 represents a potential novel therapeutic intervention for MI.
Funding
This work was supported by the grants from National Key Research and Development Project (2018YFE0113500 to JJ Xiao), National Natural Science Foundation of China (82020108002, 81722008, and 81911540486 to JJ Xiao, 81400647 to MJ Xu, 81800265 to YJ Liang), Innovation Program of Shanghai Municipal Education Commission (2017-01-07-00-09-E00042 to JJ Xiao), the grant from Science and Technology Commission of Shanghai Municipality (18410722200 and 17010500100 to JJ Xiao), the “Dawn” Program of Shanghai Education Commission (19SG34 to JJ Xiao), Shanghai Sailing Program (21YF1413200 to QL Zhou). JS is supported by Horizon2020 ERC-2016-COG EVICARE (725229).
Keywords: Exercise, Myocardial infarction, HIPK2, miR-222
Research in context.
Evidence before this study
Exercise-induced specific molecular mechanisms play vital roles in the protection of heart diseases, such as the C/EBPβ-CITED4 and IGF1-PI3K-AKT signaling pathways. The authors previous reported that miR-222 was necessary for exercise-induced cardiac growth and protected against ischemic injury. Additionally, the authors observed that homeodomain interacting protein kinase 2 (HIPK2) was a target gene of miR-222. However, the functional role of HIPK2 during post-MI recovery remains unknown.
Added value of this study
-
1.
Exercise-induced HIPK2 inhibition contributes to the protection of MI.
-
2.
HIPK2 binds to P53 and HIPK2 suppression protects OGD/R-induced NRCMs and hESC-CMs apoptosis by inhibiting P-P53.
-
3.
HIPK2 suppression represents a potential novel therapeutic intervention for MI.
-
4.
HIPK2 upstream, miR-222, protects against MI mice.
-
5.
Serum miR-222 could be a biomarker for the prognosis of MI patients.
Implications of all the available evidence
Our findings indicate that inhibition of HIPK2 represents a potential novel therapeutic intervention for MI.
Alt-text: Unlabelled box
1. Introduction
Cardiovascular disease is a major killer of human health [1]. Myocardial infarction (MI) represents the major cardiovascular disease [2]. With the acceleration of population aging, prevalence of MI patients will continue to increase [3]. Although the increasingly developed medical technology and reperfusion therapy post-MI can save patients' lives and improve clinical symptoms [4], it is still unable to fundamentally reverse or terminate the further development of heart failure post-MI. Therefore, it is urgent to develop novel strategies to attenuate post-MI remodeling and heart failure.
Exercise-induced specific molecular mechanisms play vital roles in the protection of heart diseases, such as the C/EBPβ-CITED4 and IGF1-PI3K-AKT signaling pathways [5], [6], [7], [8], [9], [10]. We previous reported that miR-222 was necessary for exercise-induced cardiac growth and protected against ischemic injury. Additionally, we observed that homeodomain interacting protein kinase 2 (HIPK2) was a target gene of miR-222 [11,12]. However, the functional role of HIPK2 during post-MI recovery remains unknown.
HIPK2 contains a conserved protein kinase domain and belongs to the serine/threonine kinase family (HIPK1-HIPK4) [13]. As a protein kinase, transcription cofactor and signal transduction element [14], HIPK2 can regulate various biological responses of cells, including cell proliferation, apoptosis, and DNA damage response. Its dysregulation contributes to diabetes, nephropathy, gastric cancer and cervical cancer [15], [16], [17]. Recent studies have shown that HIPK2 maintains basic cardiac function through mitogen activated protein kinase (ERK1/2) [18]. Inhibition of HIPK2 suppresses kidney fibrosis [19] and, although we previously reported that HIPK2 was a target gene of miR-222 [11], the role of HIPK2 in MI and its underlying mechanism are unclear.
In the present study, we identified that cardiac HIPK2 expression levels were decreased by exercise while increased upon MI. In vitro experiments showed that inhibition of HIPK2 prevented apoptosis in neonatal rat cardiomyocytes (NRCMs) and human embryonic stem cell-derived cardiomyocytes (hESC-CMs). Furthermore, HIPK2 inhibition protected acute MI or post-MI remodeling. Moreover, we identified phosphorylation of P53 mediating HIPK2’s control of cardiomyocytes’ apoptosis. Additionally, miR-222, which targets HIPK2, was involved in cardiac dysfunction after MI in mice and human. Our study indicates that HIPK2 suppression protects against apoptosis and MI through phosphorylation of P53, suggesting that inhibition of HIPK2 could be a potential therapeutic strategy for heart failure.
2. Methods
2.1. MI Patients
All human investigations conformed to the principles outlined in the Declaration of Helsinki and were approved by the institutional review committees of Tongji Hospital (2014-002). The 74 male patients diagnosed of acute MI (AMI) at the first visit were recruited with a written informed consent at Tongji Hospital (Shanghai, China) from November 2015 to December 2016. The clinical characteristics of these patients with one-year follow up were listed in Table S1. 20 male MI patients (aged 51.35±7.33) and 20 healthy people (aged 50.25±9.45) were recruited. Venous blood was collected at enrollment and was stored at -80°C until further use.
2.2. Animals and treatments
Male C57BL/6N wild type (WT) mice, aged 8 to 10-week-old, were obtained from Cavens Laboratory Animal Ltd. (Changzhou, China). Founders of C57BL/6N HIPK2–/– mice were generated in Beijing Viewsolid Bioteh Co. Ltd (Beijing, China) by embryo injection of CRISPRs targeting the second exon (guide RNA sequence: AAGTTCCAACTGGGACATGACTGGGT) of the mouse HIPK2 gene. HIPK2–/– mice with frameshift mutations (56bp deletion) were identified and crossed with C57BL/6N WT mice for colony expansion and subsequent experiments. Tail biopsies of HIPK2–/– mice were analyzed by genomic PCR (forward primer: 5’-TTCAGAGTGGAAGAACAATC-3’ and reverse primer: 5’-TTGGTAGGTGTCA AGGAG-3’).
MiR-222–/– rats were generated in Beijing Viewsolid Bioteh Co. Ltd (Beijing, China) by embryo injection of CRISPRs to cut off the target gene rno-miR-222 (guide RNA sequences: CCAGAGTCTCTCCACAGAAGG and AGAAGGAAAACCAAT CAATGG). MiR-222–/– rats were identified and crossed with Sprague Dawley WT rats for colony expansion and subsequent experiments. Tail biopsies of miR-222–/– rats were analyzed by genomic PCR (forward primer: 5’-ACTTGATCAAGGGAGCTTACCTC-3’ and reverse primer: 5’-TTGTGGCGACTTACCAATAGGTG-3’).
For HIPK2 inhibitor treatment, immediately after MI, mice were gavaged with 200 μg/kg/day PKI1H (MedChemExpress, Monmouth Junction, NJ, U.S.A) or phosphate-buffered saline (PBS) once daily for 3 weeks.
All animals were maintained on a 12 h light/dark cycle at 25°C and provided free access to commercial rodent chow and tap water. All animals were sacrificed by CO2 inhalation. Tissues were isolated and snap-frozen for future analysis or put into 4% paraformaldehyde buffer (PFA) immediately for histological study. These experiments were conducted in accordance with guidelines of laboratory animals for biomedical research published by National Institutes of Health (No. 85-23, revised 1996) and approved by the ethical committees of Shanghai University (2018011).
2.3. Exercise Training
For swimming training, mice received swimming twice per day for 1, 2, 3 or 4 weeks, and the interval between two times is more than 4 hours, starting from 10 minutes and increased by 10 minutes each day until 90 minutes. After 4-week swimming, mice were sedentary for 1, 2 or 3 weeks.
For running training, mice were placed in a treadmill, starting from 10 minutes with 5 meters per minute and we increased 10 minutes and 2 meters per minute each day until 60 minutes at 15 meters per minute. Mice were used after training with 8 weeks of running.
2.4. MI model
MI was constructed by ligating the left anterior descending artery (LAD) using a 7/0 silk thread while sham-animals were created by the same process but without LAD ligation [20].
Upon acute MI, mice were sacrificed 24 hours after operation. For pathological cardiac remodeling, mice were sacrificed 3 weeks post-MI. For exercise's protection against MI, mice were firstly subjected to MI. After 1 week, mice were trained with running for 8 weeks.
2.5. Echocardiography
Left ventricular ejection fraction (EF) and fractional shortening (FS) were evaluated by Vevo 2100 echocardiography (VisualSonics Inc, Toronto, Ontario, Canada) with a 30 MHz central frequency scan-head in mice anesthetized with 1.5% isoflurane. Measures were made from M-mode images taken from the parasternal view at the papillary muscle level.
2.6. Tissues morphological analysis
Sections of heart samples of 5 μm-thick, fixed in 4% PFA and embedded in paraffin, were stained with Masson's trichrome (MT, Servicebio, Wuhan, China) to detect the degree of collagen deposition, or used for hematoxylin and eosin staining (H&E, Servicebio, Wuhan, China) to measure cardiomyocytes cross-sectional areas. Images of the left ventricular area of each section were obtained by microscope (Nikon, Japan). Frozen sections of hearts in O.C.T Compound (optimal cutting temperature compound, Sakura) were cut at 10μm per section and stained with wheat germ agglutinin (WGA, Sigma, U.S.A) to measure cardiomyocytes cross-sectional areas. Fluorescence images were obtained using a Zeiss confocal microscope (Carl Zeiss, Oberkochen, Germany). Image J Software (National Institutes of Health) was used to quantify fibrotic region and measure cardiomyocytes cross-sectional areas in each section. The percentage of fibrosis by MT was measured as fibrosis areas/total left ventricular areas x 100%.
2.7. TTC staining
Twenty-four hours after MI, 1 mL of 1% Evans blue was slowly injected into the left ventricle and the hearts were stained with 2,3,5-triphenyltetrazolium chloride (TTC) as reported previously [21]. The area at risk/left ventricle weight (AAR/LV) ratio and the infarct size/area at risk (IS/AAR) ratio were determined to evaluate the homogeneity of surgery and the severity of MI.
2.8. Lentiviral vectors construction and administration
The DNA fragments encoding rat HIPK2 was amplified from rat heart genomic complementary DNA (cDNA) and cloned into the FUGW cloning vector. The shRNA sequence for HIPK2 is 5’- GTATGATCAGATTCGGTATAT-3’ and cloned into the pLKO.1-TRC cloning vector. Lentiviral particles were generated and packaged using psPAX2 and PMD2.G. Lentiviral vectors were diluted in PBS and administered at a dose of 107 TU/well in 12-well plates for 48 hours.
2.9. AAV9 construction and administration
The mmu-miR-222 was inserted between BamHI and EcoRI restriction enzyme sites of the pAAV vector (Hanbio Biotechnology). Adeno-associated viruses were generated using packaging plasmids AAV2/9 and Helper (Hanbio Biotechnology) together with AAV-CMV-mmu-miR-222 constructs. AAV9 virus was delivered by tail vein injection at a dose of 3*1011 TU/mice, and all mice were sacrificed after 4 weeks.
2.10. Cell isolation, culture and treatments
Neonatal rat cardiomyocytes (NRCMs) were isolated and cultured as previously reported [21,22]. To induce OGD/R, NRCMs were firstly cultured for 8 h with serum-free no glucose DMEM (Gibco, U.S.A) in an air-tight chamber with a humidified hypoxic atmosphere containing 5% CO2 and 95% N2 at 37°C. After exposure to oxygen glucose deprivation for 8 h, the culture medium was replaced with serum and glucose-containing DMEM and transferred to a normal incubator for recovery for 12 h. For HIPK2 suppression, PKI1H were incubated for 48 hours (74 nM). For P53 activation, DPBQ or CTX1 were incubated for 48 hours (1μM) or 24 hours (2μM). For miR-222 overexpression, NRCMs were exposed to miR-222 agomir or negative control (100 nM) (RiboBio, Guangzhou, China) for 48 h.
Human cardiomyocyte cell line AC16 cells were maintained in DMEM with 25 mM glucose (Gibco, U.S.A), 10% fetal bovine serum, and penicillin and streptomycin (50 mg/ml) at 37°C and 5% CO2, and incubated with DPBQ or CTX1 for indicated concentration and duration in Figures.
Human embryonic stem cells (hESCs) cell line H9 were cultured in feeder-free culture conditions on Matrigel (Corning) coated plates and mTeSR medium (Stemcell) supplemented with 10μM Y-27632 (Selleck). Differentiation was induced using CHIR99021(Selleck) at 6μM for 48 hours. On day 2, the medium was refreshed with addition of Wnt-pathway inhibitor IWP2 (Selleck) at 5 μM for 48 hours. The basal medium is RPMI 1640 medium (Life Technologies) supplemented with Albumin (0.5mg/ml) and ascorbic acid (0.2mg/ml) from day 0 to day 8. Spontaneous beating cells start to appear at day 8, and the beating cells were cultured in RPMI 1640 supplemented with B27 for further maturation. Culture medium was changed every other day. Human embryonic stem cell-derived cardiomyocytes (hESC-CMs) were used after differentiation for 30 days [23]. For OGD/R model, hESCs-CMs were exposure to oxygen glucose deprivation for 16 h, the culture medium was replaced with serum and glucose-containing DMEM and transferred to a normal incubator for recovery for 12 h. For HIPK2 suppression, PKI1H were incubated for 48 hours (74 nM). For P53 activation, DPBQ were incubated for 48 hours (1μM).
2.11. Extracellular vesicles preparation and analysis
Extracellular vesicles were collected from adult mouse cardiac fibroblasts (MCFs) which were isolated by collagenase type II (Gibco) and cultured for 48 h, and centrifuged at 3000g for 15 min at 4°C. Next, the supernatant was incubated with ExoQuickTM Exosome Precipitation Solution (System Biosciences), for 30 min and centrifuged at 1500g for 30 min at 4°C according to the manufacturer's instructions. Supernatant was then removed by aspiration and pelleted fraction was further centrifuged at 1500g for 5 min at 4°C. Extracellular vesicle pellets were resuspended with PBS and conserved at -80°C for further use. To get proteins for immunoblotting, the resuspended extracellular vesicles were heated for 7 min at 100°C with loading buffer. The diameters of extracellular vesicles were measured by Nanoparticle tracking analysis (PARTICLE METRIX). For RNA isolation and relative quantitative RT-PCR, the resuspended extracellular vesicles were extracted by miRNeasy® Mini Kit (QIAGEN) and reverse transcribed into cDNA by RevertAi First Strand cDNA Synthesis Kit (Thermo Scientific). For quantitative miRNA analysis, the Bulge-LoopTM miRNA qPCR Primer Set (RiboBio, Guangzhou, China) and iTaq Universal SYBR® Green Supermix (Bio-Rad) were used in a Real-Time PCR Detection System (Roche, Switzerland) with U6 as control. 2–ΔΔCt method was used to analyze the data.
2.12. Immunofluorescent staining
Terminal deoxynucleotidyl transferase-mediated dUTP in situ nick-end-labeling (TUNEL) staining was conducted to detect apoptotic nuclei by confocal microscopy in α-actinin-labeled cardiomyocytes, as described before [24]. NRCMs were fixed with 4% PFA for 20 min, permeabilized with 0.5% Triton X-100 for 20 min and blocked with 5% BSA in PBS-Tween for 1 h. Subsequently, NRCMs were incubated with α-actinin antibody (1:200, Sigma A7811) diluted in 5% BSA overnight at 4°C. To detect proliferation, EdU assays were performed using Click-iT Plus EdU Alexa Fluor 488 Imaging Kit (KeyGEN, Nanjing, China), according to manufacturer's instructions. Cell nuclei were counterstained with DAPI and the number of EdU-positive nuclei was calculated. Fifteen fields/sample (200 x magnification) were viewed under a confocal microscope (Leica, Germany).
NRCMs were fixed with 4% PFA for 20 min, permeabilized with 0.5% Triton X-100 for 20 min and blocked with 5% BSA in PBS-Tween for 1 h. Subsequently, NRCMs were incubated with HIPK2 (1:200, Abcam) and P53 (1:200, Cell Signaling Technology) antibody (1:200, Abcam) diluted in 5% BSA overnight at 4°C. Cell nuclei were counterstained with HOECHST. About fifty fields/sample were viewed under a confocal microscope (Leica, Germany).
2.13. RNA isolation and relative quantitative RT-PCR
Serum was collected by centrifugation of blood at 3000rpm for 10 min. The serum level of miR-222 was measured by relative quantification RT-PCR. RNA isolation and relative quantification RT-PCR were performed as described previously [22]. Briefly, exogenous cel-miR-39-3p was added as control with 50 pM final concentration in 400 μL serum, total RNA was extracted by mirVanaTM PARIS Kit (Ambion, U.S.A) and reverse transcribed into cDNA by iScript cDNA synthesis kit (Bio-Rad Laboratories, CA, U.S.A). For quantitative miRNA analysis, the Bulge-LoopTM miRNA qPCR Primer Set (RiboBio, Guangzhou, China) and Takara SYBR Premix Ex TaqTM (TliRNaseH Plus) were used in a Real-Time PCR Detection System (Roche, Switzerland) with cel-miR-39-3p as control. 2–ΔΔCt method was used to analyze the data.
2.14. Antibodies, immunoblotting, and coimmunoprecipitation
Protein extraction, immunoblotting and coimmunoprecipitation assays were performed as previously described [25,26]. Briefly, equal amounts of protein were subjected to standard SDS-PAGE and transferred onto PVDF membranes by an electroblot apparatus. The primary antibodies include GAPDH (Bioworld), β-actin (Bioworld), Collagen1(Bioworld), HIPK2 (Abcam), Bax (Abclonal), Caspase3 (Abclonal), Bcl2 (Affinity), P53 (Proteintech) and phosphorylation (Ser15) of P53 (Cell Signaling Technology), phosphorylation (Ser46) of P53 (Abcam), phosphorylation (Ser33, Thr81) of P53 (ABclonal), CD63 (ABclonal). Mouse or rabbit IgG antibodies coupled to horseradish peroxidase were used as secondary antibodies.
2.15. Statistical analysis
All data were expressed as mean ± SD. Significant differences were assessed either by a two-tailed student t-test, one-way ANOVA followed by Bonferroni's post hoc test, or two-way ANOVA followed by Bonferroni's multiple comparisons test. The association of the serum level of miR-222 with clinical features was assessed by binary logistic regression analysis. The independent predictors of readmission and/or death in patients was assessed by univariate and multivariate analyses. The sensitivity and specificity of serum level of miR-222 in prediction of worse prognosis in patients was assessed by receiver-operator characteristic (ROC) curve. P value less than 0·05 was considered to be statistically different. The clinical characteristics were performed using SPSS statistics 20 and the others were analyzed using GraphPad Prism 8·0. All statistical graphs were performed by GraphPad Prism 8·0.
2.16. RRID tags
| Reagent or resource | Source | Identifier | RRID |
|---|---|---|---|
| Antibodies | |||
| HIPK2 | Abcam | ab108543 | AB_10860868 |
| GAPDH | Bioworld | AP0063 | AB_2651132 |
| β-actin | Bioworld | AP0060 | AB_2797445 |
| Collagen1 | Bioworld | BS1530 | AB_1662101 |
| p53 | Proteintech | 10442-1-AP | AB_2206609 |
| p-p53 S15 | CST | 9284S | AB_331464 |
| p-p53 S46 | Abcam | ab76242 | AB_1524137 |
| p-p53 S33 | ABclonal | AP0984 | AB_2863878 |
| p-p53 T81 | ABclonal | AP1253 | AB_2893244 |
| CD63 | ABclonal | A5271 | AB_2766092 |
| Bax | ABclonal | A12009 | AB_2861644 |
| Bcl2 | Affinity | AF6139 | AF6139 |
| Caspase3 | ABclonal | A2156 | AB_2862975 |
| Cell Lines | |||
| AC16 | Millipore | SCC109 | CVCL_4U18 |
| H9 | CVCL_9773 | ||
| Chemicals, Peptides, and Recombinant Proteins | |||
| DPBQ | MedChemExpress | HY-U00441 | |
| CTX1 | MedChemExpress | HY-U00442 | |
| PKI1H (Protein kinase inhibitors 1 hydrochloride) | MedChemExpress | HY-U00439A | |
| WGA(wheat germ agglutinin) | Sigma | L4895 | |
| Critical Commercial Assays | |||
| Western Blot lysis buffer | KeyGEN | KGP701-100 | |
| TaKaRa BCA Protein Assay Kit | Takara | T9300A | |
| Masson's trichrome staining Kit | KeyGEN BioTECH | KGMST-8003 | |
| H&E staining Kit | KeyGEN BioTECH | KGA224 | |
| TUNEL FITC Apoptosis Detection Kit | Vazyme | A111-01 | |
| Software and Algorithms | |||
| ImageJ Software | NIH | N/A | |
| SPSS20.0 | N/A | ||
| GraphPad Prism 8.0 | GraphPad | N/A | |
| Vevo2100 | VisualSonic | N/A | |
2.17. Ethics
All human investigations conformed to the principles outlined in the Declaration of Helsinki and were approved by the institutional review committees of Tongji Hospital.
All animal experiments were conducted in accordance with guidelines of laboratory animals for biomedical research published by National Institutes of Health (No. 85-23, revised 1996) and approved by the ethical committees of Shanghai University.
2.18. Role of funding source
The Funders do not play any roles in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. Cardiac HIPK2 is induced upon MI while inhibited by exercise
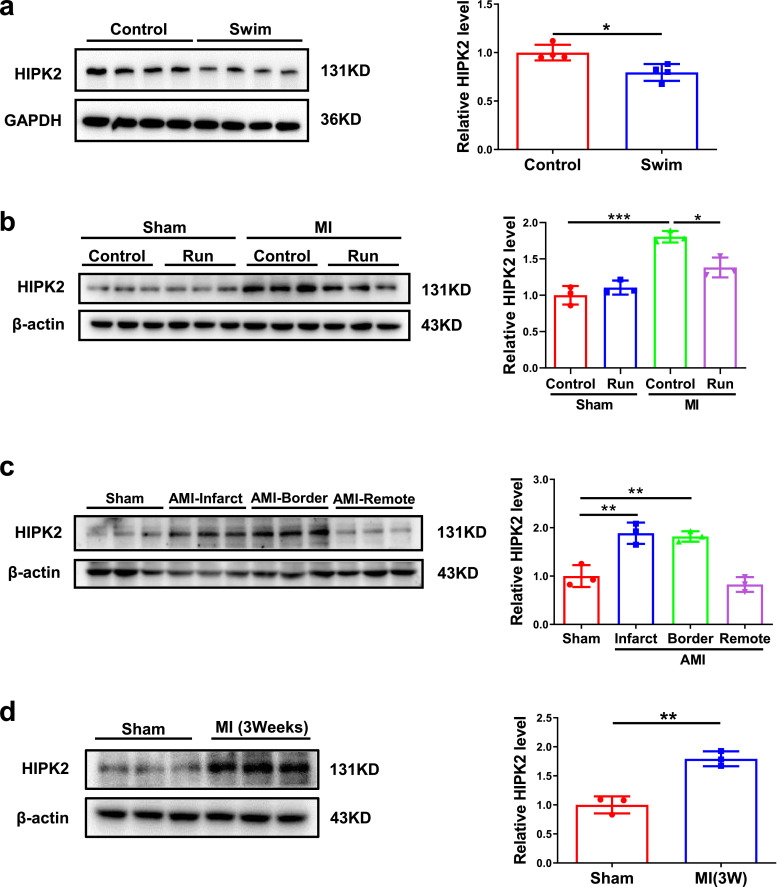
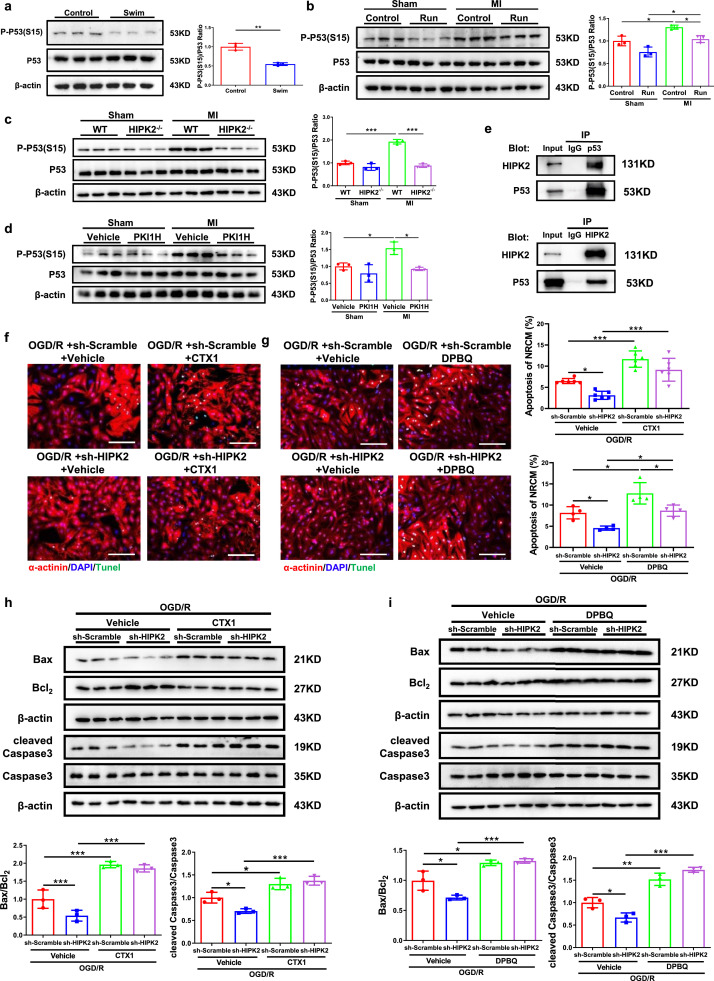
To confirm the change of HIPK2 in exercise, we subjected male C57BL/6N wild-type mice to either sedentary controls or swimming exercise and cardiac HIPK2 protein expression levels were found to be significantly decreased in exercised myocardium (Fig. 1a). Furthermore, we found that cardiac HIPK2 mRNA and protein expression levels were significantly decreased in 3-week and 4-week swimming group, while unchanged in 1-week and 2-week swimming group. The decreased HIPK2 mRNA expression levels by exercise could last for 3 weeks, while the decreased HIPK2 protein expression levels by exercise could last for 1 week (Fig. S1 and Fig. S2b, S2c, S2e and S2f). Moreover, we observed that HIPK2 was increased in myocardium 9 weeks post-MI and could also be decreased by running training (Fig. 1b), which was consistent with exercise running's protective effects for maintaining cardiac function upon MI (Fig. S3). HIPK2 protein expression levels were markedly induced in infarct and border area of acute MI (Fig. 1c), and remained elevated at 3 weeks after MI (Fig. 1d). Collectively, these data indicate that HIPK2 is induced in MI and could be inhibited by exercise, suggesting the potential functional role of HIPK2 in MI and MI-induced heart failure.
Fig. 1.
Cardiac HIPK2 protein expression levels are reduced by exercise while increased in MI. (a) Cardiac HIPK2 protein levels were decreased in male C57BL/6N wild-type (WT) mice received swimming training for 4 weeks compared to control (n=4 per group); (b) Cardiac HIPK2 protein levels were increased in post-MI remodeling for 9 weeks and 8-week-running training decreased HIPK2 protein expression levels during post-MI remodeling (n=3 per group); (c) Cardiac HIPK2 protein levels were increased in acute MI (n=3 per group); (d) Left ventricular HIPK2 protein levels were increased in post-MI remodeling for 3 weeks (n=3 per group). Data are represented as mean ± SD. Significant differences were assessed by two-tailed student t-test in a and d, or two-way ANOVA followed by Bonferroni's multiple comparisons test in b, or one-way ANOVA followed by Bonferroni's post hoc test in c. *: p<0.05, **: p<0.01, ***: p<0.001 versus respective control.
3.2. Inhibition of HIPK2 prevents apoptosis in NRCMs and hESC-CMs
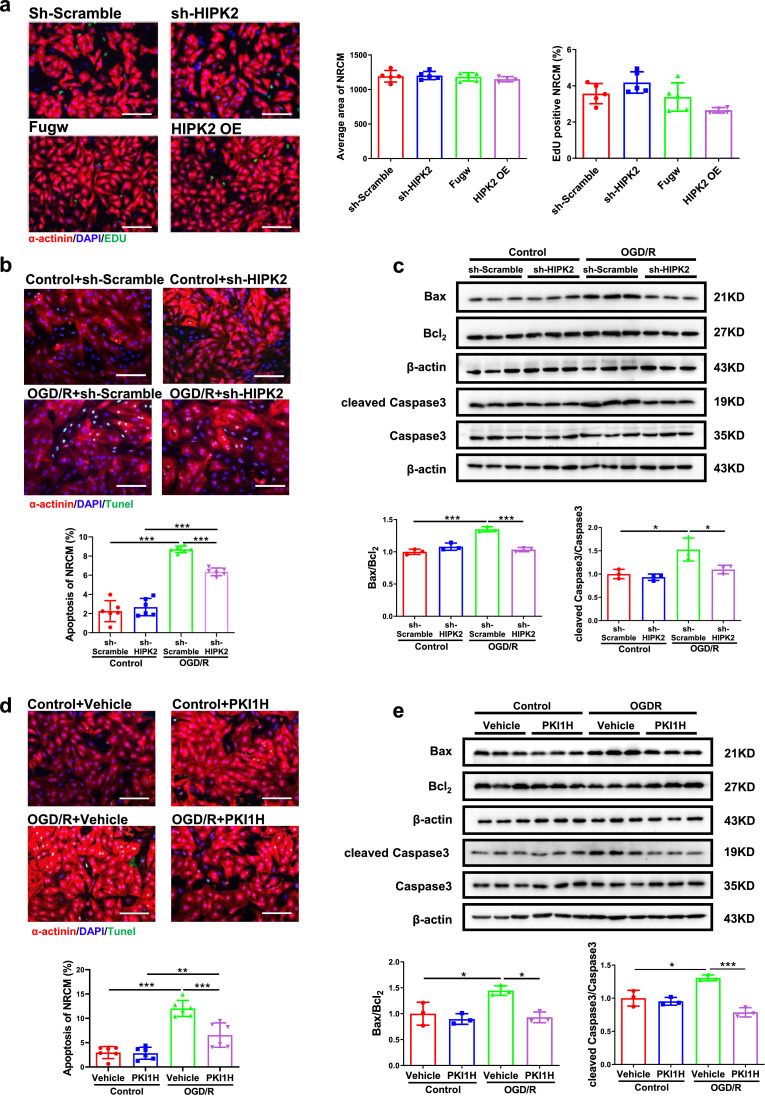
Given the response of cardiac HIPK2 to exercise and MI, we hypothesized that HIPK2 may regulate proliferation, hypertrophy or apoptosis in NRCMs. To test this possibility, we treated NRCMs with lentiviral vectors expressing shRNA against rat HIPK2 (lenti-sh-HIPK2), over-expressing HIPK2 (lenti-HIPK2), or their respective controls. HIPK2 had no effect on NRCMs proliferation and hypertrophy (Fig. 2a, S4a and S4b). Knock-down HIPK2 via lenti-sh-HIPK2 decreased NRCMs apoptosis induced by OGD/R as indicated by a reduction in TUNEL staining positive cardiomyocytes and the ratios of Bax/Bcl2 and cleaved Caspase3/Caspase3 (Fig. 2b and c). Consistently, the protective effects of HIPK2 inhibition in OGD/R-induced apoptosis was also confirmed by using HIPK2 inhibitor (PKI1H) in NRCMs and human embryonic stem cell-derived cardiomyocytes (hESC-CMs) (Fig. 2d, 2e and S5). Together, our findings illustrate that inhibition of HIPK2 prevents apoptosis in NRCMs.
Fig. 2.
Suppression of HIPK2 prevents apoptosis in NRCMs. (a) Immuno-histochemical staining for sarcomeric α-actinin and EdU followed by quantification of cardiomyocyte area in NRCMs infected with lentiviral vectors expressing scrambled (lenti-scrambled) or short-hairpin (sh) RNA against mouse HIPK2 (lenti-sh-HIPK2), or lentiviral vectors expressing Fugw (lenti-Fugw) or mouse HIPK2 (lenti-HIPK2, HIPK2 OE), and HIPK2 had no effect on NRCMs proliferation and hypertrophy (n=5 in sh-Scramble, 5 in sh-HIPK2, 5 in Fugw, and 4 in HIPK2 OE); (b and c) Lenti-sh-HIPK2 reduced the percentage of TUNEL staining positive cardiomyocytes (n=6 per group), Bax/Bcl2, cleaved Caspase3/Caspase3 (n=3 per group) in NRCMs under oxygen glucose deprivation/reperfusion (OGD/R); (d and e) HIPK2 inhibitor (PKI1H) decreased the percentage of TUNEL staining positive cardiomyocytes (n=6 per group), Bax/Bcl2, cleaved Caspase3/Caspase3 (n=3 per group) in NRCMs under OGD/R. Scale bar: 100μm. Data are represented as mean ± SD. Significant differences were assessed by two-way ANOVA followed by Bonferroni's multiple comparisons test. *: p<0.05, **: p<0.01, ***: p<0.001 versus respective control.
3.3. HIPK2 inhibition protects acute MI or MI induced heart failure
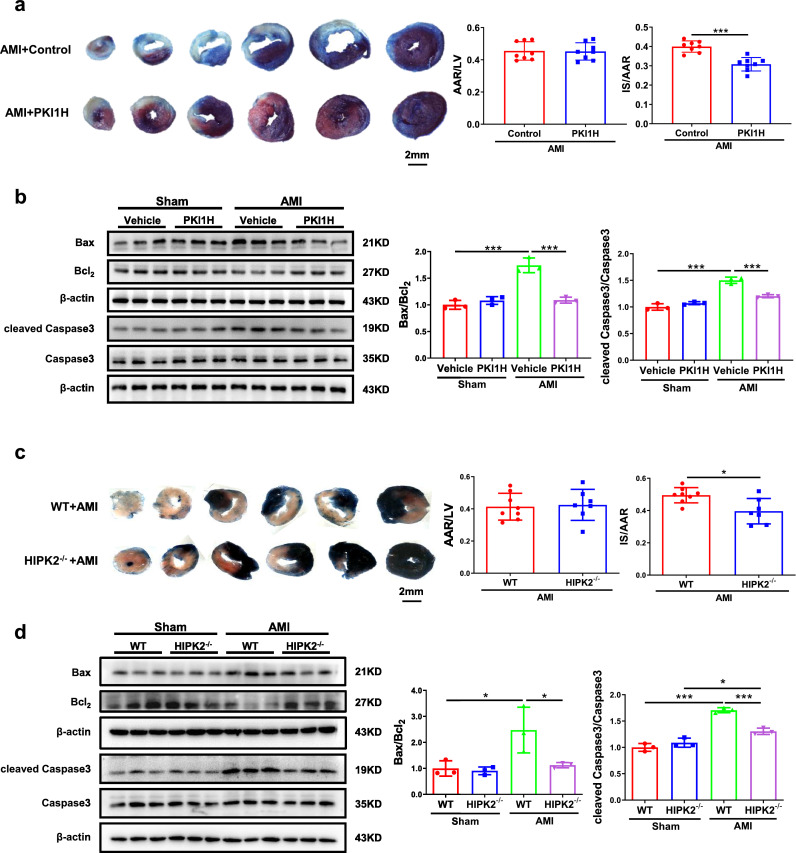
As HIPK2 was down-regulated upon exercise running in MI, which may have a protective effect for cardiac function, we explored if HIPK2 inhibition could directly protect against acute MI. We found a significant decrease in the infarct size of MI mice treated with HIPK2 inhibitor (PKI1H) (Fig. 3a). Consistently, PKI1H reduced Bax/Bcl2 and cleaved Caspase3/Caspase3 ratios in mice post-MI (Fig. 3b). Besides, we found that HIPK2–/– mice were also protected from acute MI as indicated by the decreased infarct size, Bax/Bcl2, and cleaved Caspase3/Caspase3 ratios (Fig. 3c and d).
Fig. 3.
HIPK2 inhibition protects acute MI. (a and b) HIPK2 inhibitor (PKI1H) reduced the infarct sizes (IS) and Bax/Bcl2, cleaved Caspase3/Caspase3 in heart of acute MI mice as determined by TTC staining (n=8 per group) and western blotting (n=3 per group); (c and d) HIPK2–/– mice reduced IS and Bax/Bcl2, cleaved Caspase3/Caspase3 in heart of acute MI mice as determined by TTC staining (n=8 in WT+AMI, 7 in HIPK2–/–+AMI) and western blotting (n=3 per group). Data are represented as mean ± SD. Significant differences were assessed by two-tailed student t-test in a and c, or two-way ANOVA followed by Bonferroni's multiple comparisons test in b and d. *: p<0.05, ***: p<0.001 versus respective control.
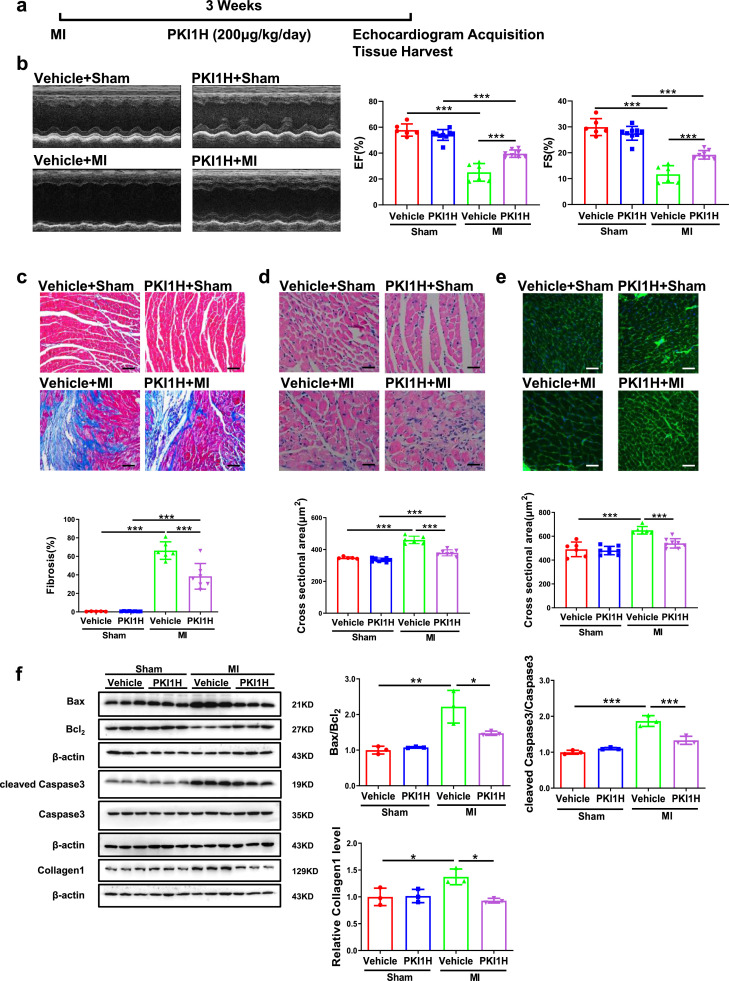
To further investigate the protective effects of HIPK2 inhibition in MI-induced heart failure, we subjected mice to either sham or MI and treated them with or without HIPK2 inhibitor (PKI1H) for 3 weeks (Fig. 4a). PKI1H preserved cardiac function post-MI of treated mice, as confirmed by increased EF and FS (Fig. 4b), reduced cardiac fibrosis and cardiomyocyte cross sectional area (Fig. 4c-e), decreased Bax/Bcl2 and cleaved Caspase3/Caspase3 ratios, as well as Collagen1 protein expression levels (Fig. 4f). Of note, PKI1H had no significant effect on cardiac function in control mice (Fig. 4). Consistently, the protective effects of HIPK2 inhibition in MI-induced heart failure was also confirmed by using HIPK2–/– mice (Fig. S4c and S6). Collectively, HIPK2 inhibition protects against cardiac dysfunction and heart failure after MI.
Fig. 4.
HIPK2 inhibitor protects against cardiac dysfunction after MI. (a) The graphical description for the experimental setup. Mice were subjected to either sham or MI and treated with or without HIPK2 inhibitor (PKI1H) for 3 weeks. Echocardiography was performed and then tissues were collected at the end of the protocol. (b) PKI1H increased ejection fractions (EF) and fractional shortening (FS) post-MI (n=6 in Vehicle+Sham, 9 in PKI1H+Sham, 6 in Vehicle+MI, and 8 in PKI1H+MI); (c and d) PKI1H reduced cardiac fibrosis and cross sectional area post-MI by H&E (n=5 in Vehicle+Sham, 8 in PKI1H+Sham, 6 in Vehicle+MI, 7 in PKI1H+MI); (e) PKI1H reduced cardiac cross sectional area post-MI by WGA (n=6 in Vehicle+Sham, 8 in PKI1H+Sham, 6 in Vehicle+MI, and 8 in PKI1H+MI); (f) PKI1H reduced Bax/Bcl2, cleaved Caspase3/Caspase3 post-MI (n=3 per group). Scale bar: 50 μm in c and 25 μm in d and e. Data are represented as mean ± SD. Significant differences were assessed by two-way ANOVA followed by Bonferroni's multiple comparisons test. *: p<0.05, **: p<0.01, ***: p<0.001 versus respective control.
3.4. Decreased phosphorylation of P53 mediates the protective effects of HIPK2 inhibition
P53 plays important roles in MI [27,28]. Phosphorylation of P53 at serine 15 (P-P53 (S15)) was identified to cause apoptosis in cardiomyocytes under ischemia [29]. However, whether phosphorylation of P53 at serine 15 could be regulated by HIPK2 and thereby mediating its’ effect in cardiomyocytes apoptosis is unclear. Phosphorylation of P53 at serine 15 was found to be significantly reduced by swimming exercise (Fig. 5a). Moreover, it was elevated by MI and subsequently decreased again by running exercise (Fig. 5b). To further investigate the possible involvement of phosphorylation of P53 in mediating HIPK2’s effect on cardiac protection, we performed western blotting to confirm that phosphorylation of P53 is regulated by HIPK2. As expected, HIPK2–/– mice showed decreased phosphorylation of P53 in hearts upon MI (Fig. 5c). Consistently, HIPK2 inhibitor (PKI1H) inhibited phosphorylation of P53 upon MI (Fig. 5d). Using a human cardiomyocyte cell line (AC16 cells) and immunoprecipitation for P53, we found that HIPK2 interacted with P53 in cardiomyocytes (Fig. 5e). Furthermore, we found that HIPK2 and P53 showed colocalization in NRCMs (Fig. S7). Besides, in hESC-CMs, we also found that phosphorylations of P53 were elevated in OGD/R and inhibited by HIPK2 suppression at Ser15, Ser46, Ser33 and Thr81 (Fig. S8)
Fig. 5.
HIPK2 inhibition mediates cardiac protection through downregulating P-P53. (a) P-P53 (S15)/P53 were reduced in the heart of swimming mice (n=3 per group); (b) Cardiac P-P53 (S15)/P53 were increased in post-MI remodeling for 9 weeks and 8-week-running training decreased cardiac P-P53 (S15)/P53 during post-MI remodeling (n=3 per group); (c) Cardiac P-P53 (S15)/P53 were increased in post-MI remodeling for 3 weeks and HIPK2–/– mice decreased cardiac P-P53 (S15)/P53 during post-MI remodeling (n=3 per group); (d) Cardiac P-P53 (S15)/P53 were increased in post-MI remodeling for 3 weeks and PKI1H decreased cardiac P-P53 (S15)/P53 during post-MI remodeling (n=3 per group); (e) Immunoprecipitation (IP) and immunoblotting were performed using the antibodies indicated and showed that HIPK2 interacted with P53; (f and g) P53 activators (CTX1 and DPBQ) increased TUNEL staining in NRCMs under OGD/R infected with lenti-sh-HIPK2 (n=6 per group in f and n=4 per group in g); (h and i) CTX1 and DPBQ increased Bax/Bcl2, cleaved Caspase3/Caspase3 in NRCMs under OGD/R infected with lenti-sh-HIPK2 (n=3 per group). Scale bar: 100μm. Data are represented as mean ± SD. Significant differences were assessed by two-tailed student t-test in a, or two-way ANOVA followed by Bonferroni's multiple comparisons test in b-i. *: p<0.05, **: p<0.01, ***: p<0.001 versus respective control.
We subsequently assessed the role for phosphorylation of P53 in HIPK2-regulated apoptosis in NRCMs. After exploring the concentration and duration of CTX1 and DPBQ (Fig.S9 and S10), we found that P53 activators, CTX1 and DPBQ could rescue the anti-apoptosis effects of HIPK2 suppression in NRCMs treated with OGD/R, as evidenced by Tunnel staining and western blotting for Bax/Bcl2 and cleaved Caspase3/Caspase3 ratios (Fig. 5f-i). Furthermore, we found that DPBQ could rescue the anti-apoptosis effects of HIPK2 suppression in human cardiomyocytes treated with OGD/R (Fig. S11). Together, we identified that decreased phosphorylation of P53 mediates the protective effects of HIPK2 inhibition.
3.5. MiR-222 is involved in cardiac dysfunction post-MI in mice and human
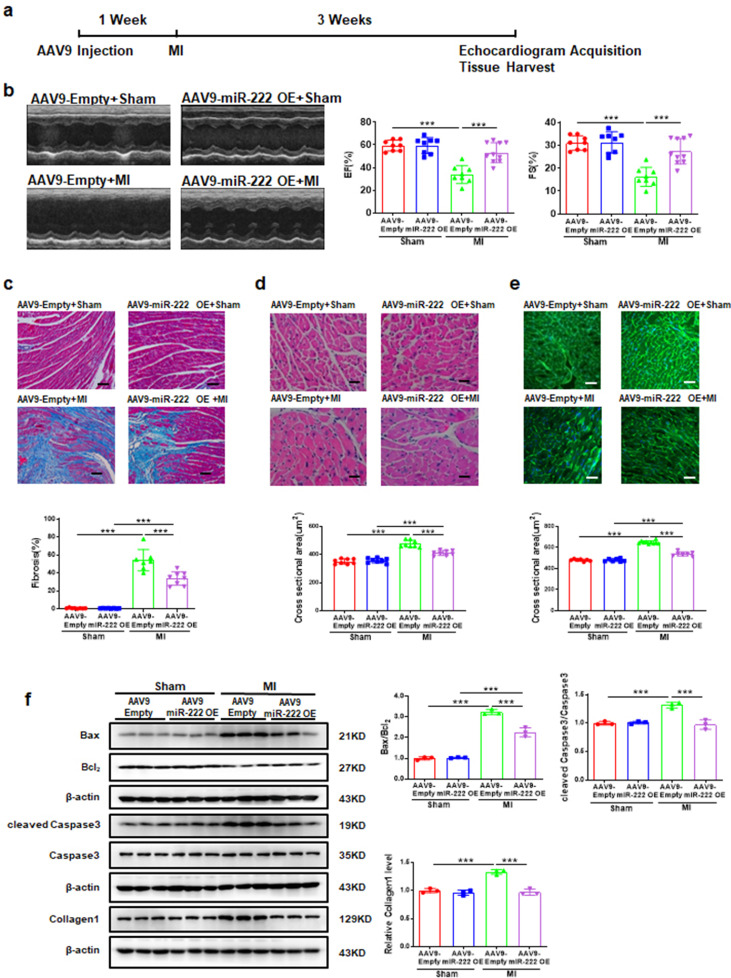
As our previous study illustrated that HIPK2 was a target of miR-222 [11] and miR-222 overexpression could down-regulated HIPK2 (Fig. S12), we explored the role of miR-222 during MI and found that mice injected with AAV9-miR-222 preserved cardiac function of MI-treated mice, as confirmed by increased EF and FS (Fig. 6a and 6b), decreased cardiac fibrosis and cardiomyocyte cross sectional area (Fig. 6c-e), decreased Bax/Bcl2 and cleaved Caspase3/Caspase3 ratios, as well as Collagen1 protein expression (Fig. 6f).
Fig. 6.
miR-222 overexpression protects against cardiac dysfunction after MI. (a) The graphical description for the experiment setup. Mice were injected with AAV9-Empty or AAV9-miR-222. After 1 week, mice were subjected to either sham or MI. After 3 weeks, echocardiography was performed and then tissues were collected at the end of the protocol; (b) AAV9-miR-222 OE increased EF and FS post-MI (n=8 in AAV9-Empty+Sham, 8 in AAV9-miR-222 OE+Sham, 8 in AAV9-Empty+MI; 10 in AAV9-miR-222 OE+MI); (c) AAV9-miR-222 OE reduced cardiac fibrosis post-MI (n=8 in AAV9-Empty+Sham, 8 in AAV9-miR-222 OE+Sham, 7 in AAV9-Empty+MI; 8 in AAV9-miR-222 OE+MI); (d) AAV9-miR-222 OE reduced cardiac cross sectional area post-MI by H&E (n=8 per group); (e) AAV9-miR-222 OE reduced cardiac cross sectional area post-MI by WGA (n=7 in AAV9-Empty+Sham, 7 in AAV9-miR-222 OE+Sham, 10 in AAV9-Empty+MI; 7 in AAV9-miR-222 OE+MI); (f) AAV9-miR-222 OE reduced cardiac Bax/Bcl2, cleaved Caspase3/Caspase3, Collagen1 protein levels post-MI (n=3 per group. Scale bar: 50 μm in c and 25 μm in d and e. Data are represented as mean ± SD. Significant differences were assessed by two-way ANOVA followed by Bonferroni's multiple comparisons test. ***: p<0.001 versus respective control.
Consistently, we observed in miR-222–/– rats that cardiac dysfunction of MI-treated rats was aggravated, as confirmed by decreased EF and FS, increased collagen deposition and cardiomyocyte cross sectional area, increased Bax/Bcl2 and cleaved Caspase3/Caspase3 ratios and Collagen1 protein expression (Fig. S13). Furthermore, we found that cardiac miR-222 expression levels were significantly increased in 3-week and 4-week swimming group, while unchanged in 1-week and 2-week swimming group. The increased miR-222 expression levels by exercise could last for 3 weeks (Fig. S2a and S2d). We also found that miR-222 expression showed no change in extracellular vesicles from culture medium of cardiac fibroblasts from swimming for 3 weeks (Fig. S14). Thus, miR-222 is probably specifically synthesized de novo from cardiomyocytes.
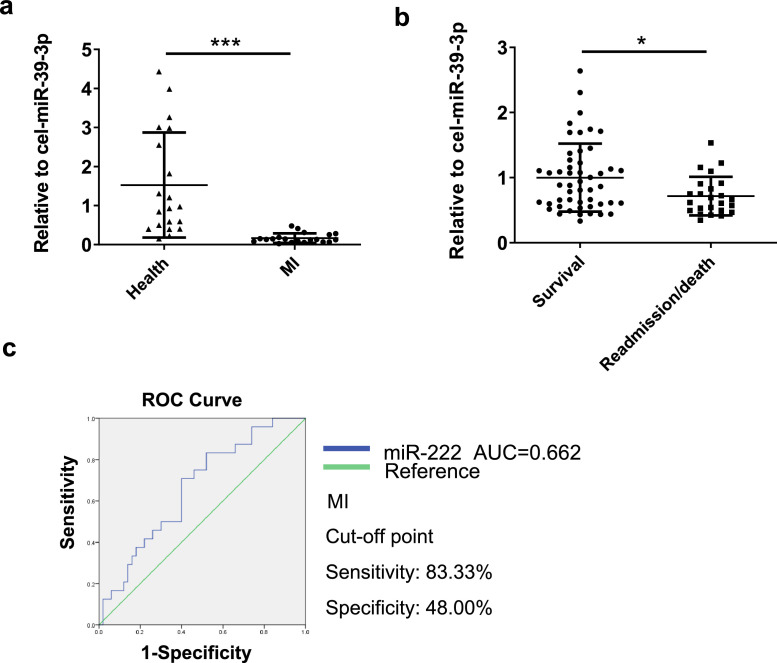
To evaluate the clinical relevance of miR-222 in human MI, serum levels of miR-222 were measured in MI patients (n=20) compared to healthy people (n=20) and we found that miR-222 serum levels were significantly reduced in MI patients (Fig. 7a). Furthermore, we analyzed serum levels of miR-222 in MI patients with readmission and/or death (n=24) compared to survival (n=50) with one-year prognosis. D-dimer serum levels were significantly higher in the miR-222 low-level group (Table S2). miR-222 serum levels were not associated with different clinical features of MI patients by binary logistic regression (Table S3). We identified serum miR-222 as independent variable related to MI readmission/death by univariate logistic regression analysis (Table S4). Serum miR-222 was further subjected to multivariate logistic regression analysis, which indicated that lower level of serum miR-222 correlated with higher risk of MI readmission/death (OR 0.109; 95% CI 0.017–0.692; P = 0.019) (Table S5). miR-222 serum levels were significantly reduced in MI patients with readmission and/or death, suggesting that lower serum miR-222 might be associated with worse prognosis in MI patients (Fig. 7b). Finally, the receiver-operator characteristic (ROC) curve analysis showed that the area under curve (AUC) for serum miR-222 was 0.662. Serum miR-222 predicted readmission and/or death with a sensitivity of 83.33% and a specificity of 48.00% in MI patients (Fig. 7c). Collectively, our results suggest that miR-222 protects against MI and lower serum miR-222 might be predictive for worse prognosis in MI patients.
Fig. 7.
The serum level of miR-222 is reduced in patients with MI
(a) Human serum miR-222 mRNA levels were decreased in MI patients (n=20) compared to healthy people (n=20); (b) Human serum miR-222 mRNA levels were decreased in patients with cardiovascular readmission and/or death (n=24) compared to those without readmission and/or death (n=50) during the 1-year follow-up; (c) The receiver-operator characteristic (ROC) curve was used to assess the sensitivity and specificity of the serum miR-222 in prediction of readmission and/or death in MI patients. Data are represented as mean ± SD. Significant differences were assessed by two-tailed student t-test in a and b. *: p<0.05, ***: p<0.001 versus respective control.
4. Discussion
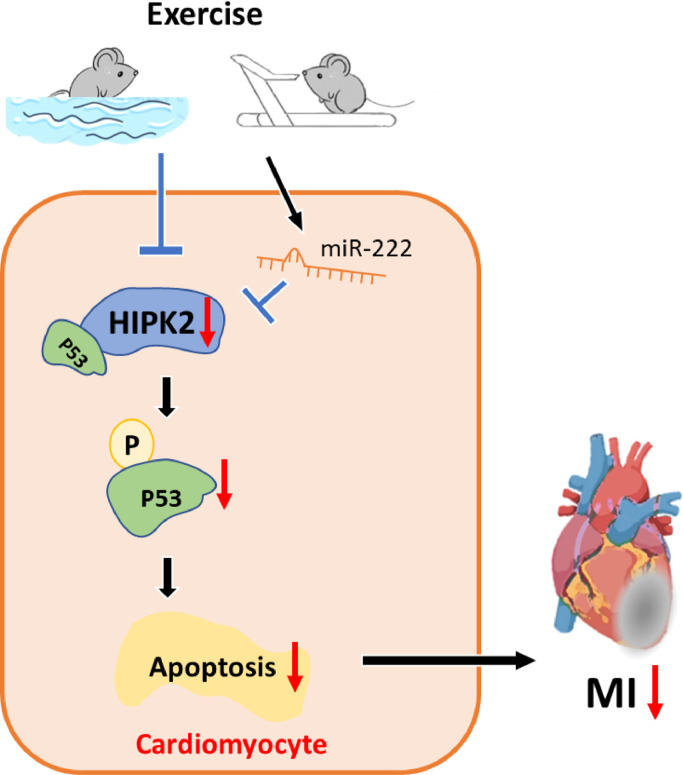
The number of MI patients increases rapidly and although the acute mortality has been significantly decreased, MI may finally lead to heart failure development, causing great burden to the society [30]. Hence, it is of great significance to explore new strategies to prevent and treat MI and post-MI heart failure. In the present study, we demonstrated that inhibition of HIPK2 could attenuate cardiomyocytes apoptosis and MI, which is supported by both genetic and pharmacological interventions. As summarized in Figure 8, cardiac HIPK2 protein expression levels were reduced by exercise while increased post-MI. Knockdown of HIPK2 prevented apoptosis of cardiomyocytes and preserved cardiac function which is impaired by MI through phosphorylation of P53. Finally, miR-222, which targets HIPK2, protected against cardiac dysfunction after MI.
Fig. 8.
Graphical abstract
Exercise induces HIPK2 suppression. Inhibition of HIPK2 decreases phosphorylation of P53 and attenuates cardiomyocytes apoptosis and myocardial infarction (MI). MiR-222, which targets HIPK2, is involved in cardiac dysfunction after MI in mice and human.
HIPK2, a protein kinase, is involved in various biological processes and plays crucial roles in regulating apoptosis in many cell types. HIPK2 downregulation significantly reduces the level of apoptosis in glioblastoma cells, chondrocytes and gastric cancer cells [31], [32], [33]. HIPK2 overexpression promotes seam cell apoptosis, as well as colorectal cancer cell apoptosis [34,35]. Consistent with the effect of HIPK2 on apoptosis in other types of cells, our study showed that inhibition of HIPK2 by lentiviral vectors prevented apoptosis in NRCMs induced by OGD/R. The anti-apoptosis effects of HIPK2 were also confirmed in MI mice models. Contrarily, HIPK2 overexpression was reported to relieve hypoxia/reoxygenation-induced apoptosis and oxidative damage of new born mouse cardiomyocytes through enhancement of the Nrf2/ARE signaling pathway [36]. It may be due to different species’ cardiomyocytes and apoptosis models. On the other hand, results from Guo et al suggested that HIPK2 maintained basal cardiac function by activating phosphorylation of ERK1/2 [18]. Our findings confirmed the previous results that HIPK2–/- mice showed no change in cardiac fibrosis and pathological hypertrophy compared with the control mice at 2-month-old. However, we found that HIPK2–/– mice prevented cardiac dysfunction after MI. We speculate it is because in wild-type mice the heart function is good and cannot be further improved, the protective effect of HIPK2–/– mice can only be obviously found in damaged conditions. Nevertheless, our study combined animal experiments with cell experiments and tried to generate a complete overview how the inhibition of HIPK2 can protect against MI by reducing apoptosis. Our current study provides new experimental models and deeper mechanistic insight. We found that the types of exercise are important. As previously reported, swimming could induce physiological hypertrophy [11] while treadmill running did not show the same phenotype [37]. Consistently, swimming reduced HIPK2 whereas running did not in healthy mice.
Phosphorylation of P53 was involved in apoptosis [38] and serine 46 participated in MI rats [39] while serine 15 was identified to cause apoptosis in cardiomyocytes under ischemia [29]. Phosphorylation of P53 at serine 46 was reported to be a target of HIPK2 and involved in apoptosis during DNA damage, and colorectal cancer [35,40]. However, whether phosphorylation of P53 is regulated by HIPK2 and involved in mediating HIPK2’s effect on cardiac protection was unknown. Here we identified phosphorylation of P53 was regulated by HIPK2, as well as mediating the protection of HIPK2 suppression against cardiomyocytes apoptosis. In summary, our findings proved that activation of P53 reversed the apoptosis of cardiomyocytes inhibited by HIPK2 suppression.
MiR-222 has been reported to mediate exercise-induced physiological cardiac hypertrophy, inhibit cardiomyocytes apoptosis and target HIPK2 [11]. Besides, miR-222 inhibited cardiac fibrosis in diabetic mice via negatively regulating Wnt/β-catenin-mediated endothelium to mesenchymal transition [41]. Here, we found that miR-222 protected against MI by AAV9 overexpression while miR-222–/– rats aggravated cardiac dysfunction post-MI. To determine if the critical importance of miR-222 was relevant to human MI, we assessed the expression levels of miR-222 in human subjects and found that miR-222 serum levels were significantly reduced in MI patients with readmission and/or death, suggesting that lower serum miR-222 might be associated with worse prognosis in MI patients. Our data suggest that serum miR-222 could be a biomarker for the prognosis of MI.
Several limitations of the present study should be highlighted. As we used a standard swimming protocol, we are not clear about how often does it need for the exercise (i.e. per day or per week) to maintain the downregulated levels of HIPK2 and the upregulated levels of miR-222. Further exploration should be tested if HIPK2 could mediate exercise-induced physiological hypertrophy and cardio-protection. We will explore the modulation of the miR-222-HIPK2 axis in exercise's protection against MI. The age of MI patients involved in this study is about 50. In order to exclude the hormone effects of female menopause, we selected male patients as the research object. In the future, we will further analyze the relationship of miR-222 and female patients. Besides, cardiac-specific knockout of HIPK2 should be added.
In conclusion, exercise-induced HIPK2 suppression attenuates cardiomyocytes apoptosis and MI by decreasing phosphorylation of P53. Inhibition of HIPK2 might be a novel therapy for MI and MI-related heart failure.
Contributors
QL Zhou, JL Deng and JH Yao designed the research studies and conducted experiments. JX Song, DN Meng, YJ Zhu, MJ Xu and YJ Liang acquired and analyzed the data. JH Xu and JS contributed to the helpful discussion. JJ Xiao directed the project, wrote and edited the manuscript. All authors read, verified the underlying data and approved the manuscript.
Declaration of Competing Interest
The authors have declared that no conflict of interest exists.
Acknowledgments
Acknowledgments
This work was supported by the grants from National Key Research and Development Project (2018YFE0113500 to JJ Xiao), National Natural Science Foundation of China (82020108002, 81722008, and 81911540486 to JJ Xiao, 81400647 to MJ Xu, 81800265 to YJ Liang), Innovation Program of Shanghai Municipal Education Commission (2017-01-07-00-09-E00042 to JJ Xiao), the grant from Science and Technology Commission of Shanghai Municipality (18410722200 and 17010500100 to JJ Xiao), the “Dawn” Program of Shanghai Education Commission (19SG34 to JJ Xiao), Shanghai Sailing Program (21YF1413200 to QL Zhou). JS is supported by Horizon2020 ERC-2016-COG EVICARE (725229).
Data Sharing Statement
The data for this study are available by contacting the corresponding author upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103713.
Appendix. Supplementary materials
References
- 1.Chou R, Dana T, Blazina I, Daeges M, Jeanne TL. Statins for Prevention of Cardiovascular Disease in Adults: Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA. 2016;316(19):2008–2024. doi: 10.1001/jama.2015.15629. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the. Circulation. 2017;135(10):e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richart AL, Reddy M, Khalaji M, Natoli AK, Heywood SE, Siebel AL, et al. ApoA-I Nanoparticles Delivered Post Myocardial Infarction Moderate Inflammation. Circ Res. 2020 doi: 10.1161/CIRCRESAHA.120.316848. [DOI] [PubMed] [Google Scholar]
- 4.Deng J, Guo M, Li G, Xiao J. Gene therapy for cardiovascular diseases in China: basic research. Gene Ther. 2020;27(7-8):360–369. doi: 10.1038/s41434-020-0148-6. [DOI] [PubMed] [Google Scholar]
- 5.Bostrom P, Mann N, Wu J, Quintero PA, Plovie ER, Panakova D, et al. C/EBPbeta controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell. 2010;143(7):1072–1083. doi: 10.1016/j.cell.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou J, Li H, Chen X, Zeng S, Ye J, Zhou C, et al. C/EBPbeta knockdown protects cardiomyocytes from hypertrophy via inhibition of p65-NFkappaB. Mol Cell Endocrinol. 2014;390(1-2):18–25. doi: 10.1016/j.mce.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Lerchenmuller C, Rabolli CP, Yeri A, Kitchen R, Salvador AM, Liu LX, et al. CITED4 Protects Against Adverse Remodeling in Response to Physiological and Pathological Stress. Circ Res. 2020;127(5):631–646. doi: 10.1161/CIRCRESAHA.119.315881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezzerides VJ, Platt C, Lerchenmuller C, Paruchuri K, Oh NL, Xiao C, et al. CITED4 induces physiologic hypertrophy and promotes functional recovery after ischemic injury. JCI Insight. 2016;1(9) doi: 10.1172/jci.insight.85904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weeks KL, Bernardo BC, Ooi JYY, Patterson NL, McMullen JR. The IGF1-PI3K-Akt Signaling Pathway in Mediating Exercise-Induced Cardiac Hypertrophy and Protection. Adv Exp Med Biol. 2017;1000:187–210. doi: 10.1007/978-981-10-4304-8_12. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Yan W, Mao Y, Ni Y, Zhou L, Song H, et al. Effect of Aerobic Exercise on Treg and Th17 of Rats with Ischemic Cardiomyopathy. J Cardiovasc Transl Res. 2018;11(3):230–235. doi: 10.1007/s12265-018-9794-0. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Xiao J, Zhu H, Wei X, Platt C, Damilano F, et al. miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 2015;21(4):584–595. doi: 10.1016/j.cmet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Lv Y, Li G, Xiao J. MicroRNAs in heart and circulation during physical exercise. J Sport Health Sci. 2018;7(4):433–441. doi: 10.1016/j.jshs.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem. 1998;273(40):25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 14.Wook Choi D, Yong Choi C. HIPK2 modification code for cell death and survival. Mol Cell Oncol. 2014;1(2) doi: 10.1080/23723548.2014.955999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Tong J, He F, Yu X, Fan L, Hu J, et al. miR-141 regulates TGF-beta1-induced epithelial-mesenchymal transition through repression of HIPK2 expression in renal tubular epithelial cells. Int J Mol Med. 2015;35(2):311–318. doi: 10.3892/ijmm.2014.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conte A, Pierantoni GM. Update on the Regulation of HIPK1, HIPK2 and HIPK3 Protein Kinases by microRNAs. Microrna. 2018;7(3):178–186. doi: 10.2174/2211536607666180525102330. [DOI] [PubMed] [Google Scholar]
- 17.Oh HJ, Kato M, Deshpande S, Zhang E, Das S, Lanting L, et al. Inhibition of the processing of miR-25 by HIPK2-Phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy. Sci Rep. 2016;6:38789. doi: 10.1038/srep38789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Sui JY, Kim K, Zhang Z, Qu XA, Nam YJ, et al. Cardiomyocyte Homeodomain-Interacting Protein Kinase 2 Maintains Basal Cardiac Function via Extracellular Signal-Regulated Kinase Signaling. Circulation. 2019;140(22):1820–1833. doi: 10.1161/CIRCULATIONAHA.119.040740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Das B, Xiao W, Li Z, Li H, Lee K, et al. A Novel Inhibitor of Homeodomain Interacting Protein Kinase 2 Mitigates Kidney Fibrosis through Inhibition of the TGF-beta1/Smad3 Pathway. J Am Soc Nephrol. 2017;28(7):2133–2143. doi: 10.1681/ASN.2016080841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen S, Jiang H, Bei Y, Zhang J, Zhang H, Zhu H, et al. Qiliqiangxin Attenuates Adverse Cardiac Remodeling after Myocardial Infarction in Ovariectomized Mice via Activation of PPARgamma. Cell Physiol Biochem. 2017;42(3):876–888. doi: 10.1159/000478641. [DOI] [PubMed] [Google Scholar]
- 21.Bei Y, Pan LL, Zhou Q, Zhao C, Xie Y, Wu C, et al. Cathelicidin-related antimicrobial peptide protects against myocardial ischemia/reperfusion injury. BMC Med. 2019;17(1):42. doi: 10.1186/s12916-019-1268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao L, Bei Y, Chen P, Lei Z, Fu S, Zhang H, et al. Crucial Role of miR-433 in Regulating Cardiac Fibrosis. Theranostics. 2016;6(12):2068–2083. doi: 10.7150/thno.15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Maimaitiaili R, Yao J, Xie Y, Qiang S, Hu F, et al. Percutaneous Intracoronary Delivery of Plasma Extracellular Vesicles Protects the Myocardium Against Ischemia-Reperfusion Injury in Canis. Hypertension. 2021 doi: 10.1161/HYPERTENSIONAHA.121.17574. HYPERTENSIONAHA12117574. [DOI] [PubMed] [Google Scholar]
- 24.Shi J, Bei Y, Kong X, Liu X, Lei Z, Xu T, et al. miR-17-3p Contributes to Exercise-Induced Cardiac Growth and Protects against Myocardial Ischemia-Reperfusion Injury. Theranostics. 2017;7(3):664–676. doi: 10.7150/thno.15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Chan MC, Yu Y, Bei Y, Chen P, Zhou Q, et al. miR-29b contributes to multiple types of muscle atrophy. Nat Commun. 2017;8:15201. doi: 10.1038/ncomms15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng J, Guo Y, Yuan F, Chen S, Yin H, Jiang X, et al. Autophagy inhibition prevents glucocorticoid-increased adiposity via suppressing BAT whitening. Autophagy. 2020;16(3):451–465. doi: 10.1080/15548627.2019.1628537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong Y, Chen H, Gao J, Liu Y, Li J, Wang J. Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. J Mol Cell Cardiol. 2019;136:27–41. doi: 10.1016/j.yjmcc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Crow MT, Mani K, Nam YJ. Kitsis RN. The mitochondrial death pathway and cardiac myocyte apoptosis. Circ Res. 2004;95(10):957–970. doi: 10.1161/01.RES.0000148632.35500.d9. [DOI] [PubMed] [Google Scholar]
- 29.Kim YA, Kim MY, Yu HY, Mishra SK, Lee JH, Choi KS, et al. Gadd45beta is transcriptionally activated by p53 via p38alpha-mediated phosphorylation during myocardial ischemic injury. J Mol Med (Berl) 2013;91(11):1303–1313. doi: 10.1007/s00109-013-1070-9. [DOI] [PubMed] [Google Scholar]
- 30.Pan W, Zhu Y, Meng X, Zhang C, Yang Y, Bei Y. Immunomodulation by Exosomes in Myocardial Infarction. J Cardiovasc Transl Res. 2019;12(1):28–36. doi: 10.1007/s12265-018-9836-7. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Roos WP, Wu Q, Hofmann TG, Kaina B. The SIAH1-HIPK2-p53ser46 Damage Response Pathway is Involved in Temozolomide-Induced Glioblastoma Cell Death. Mol Cancer Res. 2019;17(5):1129–1141. doi: 10.1158/1541-7786.MCR-18-1306. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Li S, Chen P, Yang B, Yang J, Liu R, et al. MicroRNA-27b-3p inhibits apoptosis of chondrocyte in rheumatoid arthritis by targeting HIPK2. Artif Cells Nanomed Biotechnol. 2019;47(1):1766–1771. doi: 10.1080/21691401.2019.1607362. [DOI] [PubMed] [Google Scholar]
- 33.Tan X, Tang H, Bi J, Li N, Jia Y. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J Cell Biochem. 2018;119(7):5153–5162. doi: 10.1002/jcb.26542. [DOI] [PubMed] [Google Scholar]
- 34.Ke CY, Mei HH, Wong FH, Lo LJ. IRF6 and TAK1 coordinately promote the activation of HIPK2 to stimulate apoptosis during palate fusion. Sci Signal. 2019;12(593) doi: 10.1126/scisignal.aav7666. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, Feng Y, Jin Y, Liu X, Sui H, Chai N, et al. Verbascoside promotes apoptosis by regulating HIPK2-p53 signaling in human colorectal cancer. BMC Cancer. 2014;14:747. doi: 10.1186/1471-2407-14-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang X, Zhang R, Peng Z, Qin Y, Sun J, Niu Z, et al. HIPK2 overexpression relieves hypoxia/reoxygenation-induced apoptosis and oxidative damage of cardiomyocytes through enhancement of the Nrf2/ARE signaling pathway. Chem Biol Interact. 2020;316 doi: 10.1016/j.cbi.2019.108922. [DOI] [PubMed] [Google Scholar]
- 37.Fewell JG, Osinska H, Klevitsky R, Ng W, Sfyris G, Bahrehmand F, et al. A treadmill exercise regimen for identifying cardiovascular phenotypes in transgenic mice. Am J Physiol. 1997;273(3 Pt 2):H1595–H1605. doi: 10.1152/ajpheart.1997.273.3.H1595. [DOI] [PubMed] [Google Scholar]
- 38.Wang B, Li D, Kovalchuk O. p53 Ser15 phosphorylation and histone modifications contribute to IR-induced miR-34a transcription in mammary epithelial cells. Cell Cycle. 2013;12(13):2073–2083. doi: 10.4161/cc.25135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Richards AM, Wang P. MicroRNA-221 Is Cardioprotective and Anti-fibrotic in a Rat Model of Myocardial Infarction. Mol Ther Nucleic Acids. 2019;17:185–197. doi: 10.1016/j.omtn.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancini F, Pieroni L, Monteleone V, Luca R, Fici L, Luca E, et al. MDM4/HIPK2/p53 cytoplasmic assembly uncovers coordinated repression of molecules with anti-apoptotic activity during early DNA damage response. Oncogene. 2016;35(2):228–240. doi: 10.1038/onc.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Wang Z, Gao L, Xiao L, Yao R, Du B, et al. miR-222 inhibits cardiac fibrosis in diabetic mice heart via regulating Wnt/beta-catenin-mediated endothelium to mesenchymal transition. J Cell Physiol. 2020;235(3):2149–2160. doi: 10.1002/jcp.29119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.