Summary
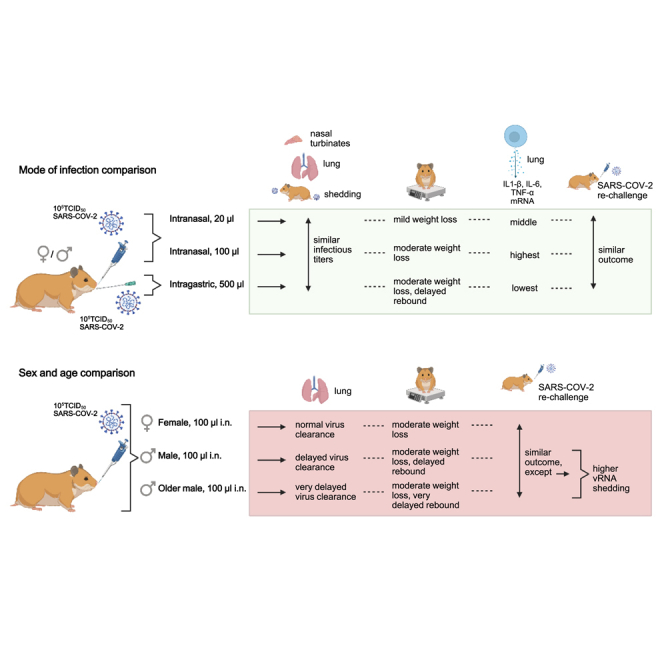
The golden hamster model of SARS-CoV-2 infection recapitulates key characteristics of COVID-19. In this work we examined the influence of the route of exposure, sex, and age on SARS-CoV-2 pathogenesis in hamsters. We report that delivery of SARS-CoV-2 by a low- versus high-volume intranasal or intragastric route results in comparable viral titers in the lung and viral shedding. However, low-volume intranasal exposure results in milder weight loss, whereas intragastric exposure leads to a diminished capacity to regain body weight. Male hamsters, and particularly older male hamsters, display an impaired capacity to recover from illness and delayed viral clearance. These factors were found to influence the nature of the host inflammatory cytokine response but had a minimal effect on the quality and durability of the humoral immune response and susceptibility to re-infection. These data further elucidate key factors that impact pre-clinical challenge studies carried out in the hamster model of COVID-19.
Subject areas: Cellular physiology, Microbiology, Virology
Graphical abstract
Highlights
-
•
Low-volume intranasal delivery of SARS-CoV-2 leads to milder weight loss in hamsters
-
•
Intragastric exposure of hamsters to SARS-CoV-2 results in delayed weight recovery
-
•
Male and older male hamsters exhibit delayed SARS-CoV-2 clearance from the lungs
-
•
Route, sex, and age only mildly influence susceptibility of hamsters to re-infection
Cellular physiology; Microbiology; Virology
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense RNA virus that first emerged in Wuhan, China in December 2019 (Zhu et al., 2020; Wu and McGoogan, 2020). It is a member of the Betacoronavirus genus and is related to the high-consequence pathogens severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome (MERS)-CoV. SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19), rapidly spread across the globe, infecting over 200 million people and killing over 5 million individuals. The COVID-19 pandemic is an unprecedented public health emergency with profound ongoing economic and societal consequences.
For the majority of people, SARS-CoV-2 infection results in asymptomatic (Poletti et al., 2021; Byambasuren et al., 2020; WHO WG 2020) or mild infection not requiring hospitalization (Chanana et al., 2020). Severe disease is often defined by hospitalization and need for high-flow oxygen or non-invasive or invasive ventilation and may result in death (Li et al., 2020). Symptoms include fever, fatigue, muscle aches, anosmia, dysgeusia, gastrointestinal (GI) dysfunction, and respiratory symptoms such as cough and difficulty breathing (Guan et al., 2020; Huang et al., 2020). Complications include pneumonia, acute respiratory distress syndrome, multi-organ dysfunction, and/or thrombotic events. Severe COVID-19 is further associated with thrombocytopenia, lymphopenia, and an aberrant pro-inflammatory cytokine response (Xu et al., 2020; Li et al., 2020). Although patients in all age groups can develop critical illness, the severity of COVID-19 is associated with advanced age and comorbidities including cardiovascular disease, diabetes, and obesity (Guan et al., 2020; Huang et al., 2020; Wu et al., 2020). Of note, males appear to be at greater risk of progressing to severe disease, which has been proposed to be a result of sex-specific differences in the immune response to infection, including auto-antibodies to type I interferons (Takahashi et al., 2020; Scully et al., 2020; Bastard et al., 2020).
SARS-CoV and SARS-CoV-2 transmission is believed to occur primarily through the deposition of short-range respiratory droplets and aerosols or from fomites/contaminated surfaces onto mucous membranes of the eyes, nose, and mouth or by direct inhalation into the lungs (Peiris et al., 2003; Prather et al., 2020; Lu et al., 2020a). Angiotensin-converting enzyme 2 (ACE2), a SARS-CoV-2 host cell receptor, and transmembrane protease serine 2 (TMPRSS2), a serine protease that facilitates viral entry, are expressed in multiple cell types, including ciliated airway epithelial cells (i.e., nasal, bronchial, and bronchiolar cells), goblet cells, alveolar type II pneumocytes, and enterocytes (Sungnak et al., 2020; Zou et al., 2020). These cell types have been shown to be among those that are susceptible to SARS-CoV-2 infection (Hou et al., 2020; Zang et al., 2020; Lamers et al., 2020). Although SARS-CoV-2 is a respiratory virus, gastrointestinal symptoms are routinely observed in patients with COVID-19 (Pan et al., 2020; Jin et al., 2020a; Cheung et al., 2020). Prolonged viral RNA (vRNA) shedding in the feces (Wang et al., 2020) and the detection of SARS-CoV-2 nucleocapsid within enterocytes in both animal models (Chan et al., 2020; Shi et al., 2020) and humans (Xiao et al., 2020) suggest that like MERS-CoV (Zhou et al., 2017) the GI tract may serve as an alternate site of SARS-CoV-2 infection.
A concerted worldwide research effort has identified a number of animal models of SARS-CoV-2 infection that are essential tools for improving our understanding of disease and for evaluating candidate vaccines and therapeutics. Several nonhuman primate (NHP) models have been described, including rhesus macaques (Macaca mulatta) (Munster et al., 2020; Lu et al., 2020b), cynomolgus macaques (Macaca fascicularis) (Rockx et al., 2020), and African green monkeys (Chlorocebus sp) (Woolsey et al., 2021). Multiple small animal models of SARS-CoV-2 infection have also been described, including transgenic mice that express human ACE2 (hACE2) (Bao et al., 2020; Hassan et al., 2020), conventional laboratory mice strains that are infected with a mouse-adapted SARS-CoV-2 (Gu et al., 2020; Dinnon et al., 2020), and several additional species that are naturally susceptible to infection with wild-type SARS-CoV-2, including tree shrews (Zhao et al., 2020), deer mice (Griffin et al., 2021; Fagre et al., 2021), hamsters (Chan et al., 2020; Sia et al., 2020), ferrets (Shi et al., 2020; Kim et al., 2020), and domesticated cats (Shi et al., 2020; Halfmann et al., 2020). Hamsters are a desirable model since upon intranasal exposure to SARS-CoV-2 they develop mild disease characterized by rapid breathing, weight loss, high viral loads in respiratory tissues, and extensive lung pathology (Chan et al., 2020; Sia et al., 2020).
Given the value of the hamster model as a tool for better understanding SARS-CoV-2 pathogenesis and for evaluating prospective anti-viral countermeasures, we have sought to extend its utility by examining the impact of route of inoculation, sex, and age on disease. Here we report that low-volume intranasal, high-volume intranasal, and intragastric routes of SARS-CoV-2 exposure result in a similar viral burden in the respiratory tract tissues and comparable levels of viral shedding; however, compared with high-volume intranasal exposure, low-volume intranasal exposure alone resulted in milder weight loss, whereas intragastric exposure led to an impaired ability to regain body weight following the acute illness period. We further demonstrate a significant male bias in disease severity, with male hamsters and to a greater extent older male hamsters displaying an impaired capacity to recover from illness. Remarkably, these differences in disease severity were not accompanied by differences in viral load in the examined tissues but did result in differential expression of inflammatory cytokines, perhaps suggesting a role for route of exposure, sex, or age-mediated immune factors in SARS-CoV-2 pathogenesis. This study provides a comprehensive characterization of the Syrian golden hamster as a model for SARS-CoV-2 infection and demonstrates that route of virus administration, sex, and age are key variables influencing the course of disease. Appreciation of these variables is critical as efforts are undertaken to further our understanding of COVID-19 pathogenesis and to develop effective vaccines and therapeutics.
Results
Influence of route of exposure on SARS-CoV-2 pathogenesis in Syrian golden hamsters
To examine the influence of different routes of exposure on SARS-CoV-2 pathogenesis, groups of five 6-week-old mixed male and female Syrian golden hamsters (Mesocricetus auratus) were exposed to 105 TCID50 of SARS-CoV-2 by one of three routes of administration: intranasal low volume (20 μL, i.n.L), intranasal high volume (100 μL, i.n.H), and intragastric oral gavage (500 μL, i.g.). These routes of administration were selected to model upper airway (i.n.L), upper/lower airway (i.n.H), and oral/gastrointestinal (i.g.) routes of initial SARS-CoV-2 exposure (Figure 1A).
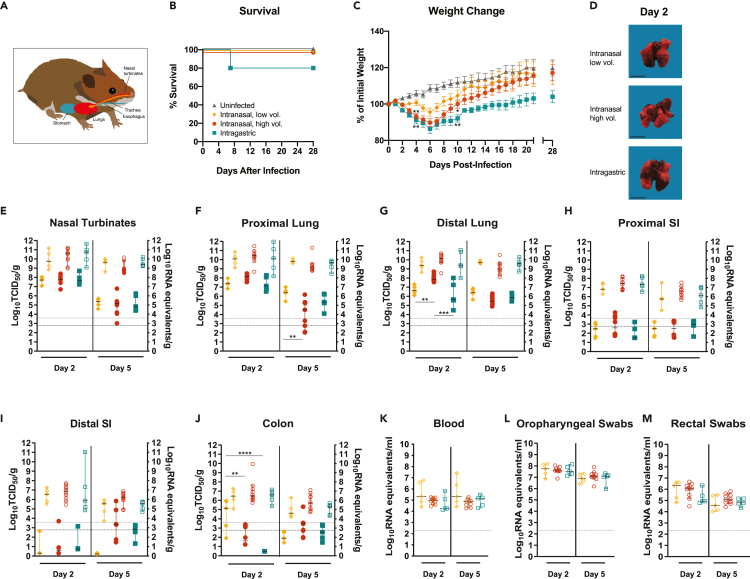
Figure 1.
SARS-CoV-2 infection of young adult golden Syrian hamsters by intranasal and intragastric routes of exposure
(A–M) Six-week-old mixed female and male Syrian golden hamsters (Mesocricetus auratus) were inoculated with 105 TCID50 of SARS-CoV-2 by a low- (20 μL) or high- (100 μL) volume intranasal route of administration (orange diamonds or red circles, respectively) or an intragastric route of administration (teal squares) and compared with age-matched uninfected controls (gray triangles). Bars indicate means (C) or medians (E-M). Error bars indicate SEM (C) or 95% confidence interval (CI) (E–M). Dashed lines and dotted lines indicate the limit of detection for the TCID50 assay and qRT-PCR assay, respectively. (A) Schematic depicting the routes of hamster inoculation. Kaplan-Meier curve depicting survival data (B) and weight data (C) over the course of 28 days after SARS-CoV-2 exposure. D, macroscopic images of SARS-CoV-2-infected hamster lungs. Scale bar, 1 cm. E-M, Infectious viral load (filled in shapes, left axis) and vRNA levels (empty shapes, right axis) in the (E) nasal turbinates, (F) proximal lung, (G) distal lung, (H) proximal small intestines, (I) distal small intestines, and (J) colon at the indicated days post infection. Viral RNA levels in (K) blood, (L) oropharyngeal swabs, and (M) rectal swab samples. Data were collected in a single experiment (n = 5).
Administration of SARS-CoV-2 by each of the three routes resulted in productive infection. SARS-CoV-2-exposed hamsters were monitored daily for clinical signs of illness for 28 days and showed no signs of illness other than weight loss and elevated respiratory rate and/or labored breathing in some animals. Survival data are depicted as Kaplan-Meier survival curves (Figure 1B). With the exception of a single hamster from the i.g.-exposed group (n = 15 total animals) that succumbed, all infected hamsters survived infection regardless of the route of SARS-CoV-2 exposure. Hamsters were weighed daily for 21 days post infection (dpi) and again on 28 dpi (Figure 1C). Mock-infected animals did not lose weight and gained weight throughout the course of the experiment. The majority of infected hamsters began to lose weight by 2 dpi, regardless of the route of SARS-CoV-2 exposure; however, at 4 dpi the i.n.H and i.g.-exposed hamsters showed significantly greater weight loss compared with the mock-infected control animals (one-way ANOVA, p = 0.0017 and 0.0011, respectively). Conversely, the hamsters exposed via the i.n.L route did not show significant weight loss (one-way ANOVA, p = 0.4250). At 4 dpi, the majority of the SARS-CoV-2-exposed hamsters belonging to the i.n.H and i.g. exposure groups showed weight loss ranging from 5% to 12%, and with the exception of one individual in the i.n.H group out of five hamsters, all animals had lost weight in both groups. In contrast, at the same time point three of five of the i.n.L-exposed hamsters had gained weight, and the remaining two animals showed weight loss of only 1.1% and 4.2%. At 7 dpi the mean body weight had begun to rebound in all the virus-exposure groups; however, the increase in mean body weight of the i.g.-exposed hamsters lagged behind that of either i.n.-exposed group. At 10 dpi, the mean weight of both i.n.-exposed groups had recovered to reach the initial starting weight, whereas the mean weight of the i.g.-exposed hamsters remained significantly lower at ∼90% of the initial starting weight. Compared with the uninfected hamsters, weights of the i.n.L-exposed hamsters were not significantly different (one-way ANOVA, p = 0.4243), whereas that of the i.n.H- and i.g.-exposed hamsters remained significantly lower (one-way ANOVA, p = 0.0499 and 0.038, respectively). Furthermore, at 28 dpi the i.g.-exposed hamsters had regained less weight than the hamsters belonging to the uninfected and intranasal exposure groups, although these differences did not attain statistical significance. Sex differences were noted in the i.n.H group, which are discussed in the following section.
Additional groups of 6-week-old mixed sex hamsters were similarly infected with SARS-CoV-2 by i.n.L, i.n.H, and i.g. routes and were necropsied at 2 and 5 dpi (n = 5, 10, and 5, respectively, at each time point). Macroscopic discoloration of portions of the lung tissue indicative of severe inflammation, often present in multiple lobes and sometimes encompassing more than half of the total lobe volume, was consistently observed in infected hamsters regardless of the route of exposure (Figure 1D). The viral burden in the upper and lower respiratory tract, including the nasal turbinates, proximal lung, and distal lung, was assessed (Figures 1E–1G). At 2 dpi, high levels of infectious virus (∼107–108 TCID50/g) were detected in the nasal turbinates regardless of the route of exposure (Figure 1E, solid shapes, left axis), and these values had declined similarly by several logs in all groups by 5 dpi. At 2 dpi, comparable levels of infectious virus (∼5x106–5x108 TCID50/g) were detected in the proximal lung regardless of the route of exposure (Figure 1F, solid shapes, left axis). At 5 dpi the infectious virus in the proximal lung had declined by several logs in the i.n.H- and i.g.-exposed hamsters and to a lesser extent in the i.n.L-exposed hamsters, resulting in a significantly greater infectious virus burden in the i.n.L- versus i.n.H-exposed hamsters (one-way ANOVA, p = 0.0017). At 2 dpi, high levels of infectious virus were detected in the distal lung of all hamsters, regardless of the route of exposure (Figure 1G, solid shapes, left axis); however, significantly more infectious virus was detected in the i.n.H-exposed hamsters compared with the i.n.L- and i.g.-exposed hamsters (one-way ANOVA, p = 0.0098 and 0.0002, respectively). At 5 dpi the level of infectious virus in the distal lung had significantly declined in the i.n.H.-exposed group (one-way ANOVA p < 0.0001), whereas the level of infectious virus in the i.n.L- and i.g.-exposed hamsters remained similar to the 2-dpi level. At 2 dpi, high levels of viral RNA (vRNA) were detected in the nasal turbinates, proximal lung, and distal lung (vRNA; ∼108–1012 genome equivalents/g), regardless of the route of exposure, and remained at a similar level at 5 dpi, having declined to a much lesser extent than the level of infectious virus (Figures 1E–1G, empty shapes, right axes). It is interesting to note that the i.g.-exposed hamsters displayed a notable delay in recovery to original body weights compared with the i.n.H-exposed hamsters group despite the presence of less infectious virus in the distal lung region at 2 dpi.
The infectious virus burden was also assessed in the digestive tract, including the small intestines (proximal and distal) and colon (Figures 1H–1J, solid shapes, left axes). Regardless of the route of infection, infectious virus in the GI tract tissues was below or close to the limit of detection, with the exception of some animals exposed by either the i.n. route that had moderate levels in the colon (i.n.L route group, 2 dpi) or the distal small intestine and colon (i.n.H group, 5 dpi) (Figures 1I and 1J). At 2 dpi, there was significantly more infectious virus present in the colon of the i.n.L-exposed hamsters compared with both the i.n.H- and i.g.-exposed hamsters (one-way ANOVA p = 0.0029 and <0.0001, respectively). At 2 dpi moderate levels of vRNA (ranging from ∼2x105–2x108 genome copies/g) were detected in the proximal intestine, distal intestine, and colon, regardless of the route of infection (Figures 1H–1J, empty shapes, right axes), and these values had declined by 5 dpi. Despite reports that SARS-CoV-2 can infect small intestine enterocytes in vitro (Zang et al., 2020; Lamers et al., 2020), we found that a high-titer SARS-CoV-2 challenge administered directly to the digestive tract failed to result in an increased viral burden in the small intestines or colon compared with i.n. inoculation.
Persistent SARS-CoV-2 RNAemia in humans has been linked to greater inflammation and more severe disease (Chen et al., 2020). At both 2 and 5 dpi, vRNA was detected in the blood of all hamsters, regardless of the route of exposure (Figure 1K). Low levels of infectious virus were detected in the blood at 2 dpi in a small number of samples (data not shown).
Finally, to monitor viral shedding, oropharyngeal and rectal swab samples were collected prior to euthanization at 2 and 5 dpi (Figures 1Land 1M). Moderate vRNA levels (ranging from ∼107–108 genome copies/ml in oropharyngeal swabs and 104.5–106.5 genome copies/ml in rectal swabs) were detected at 2 dpi in all groups, and vRNA levels detected in either swab sample type had declined by approximately one log at 5 dpi. The shedding of vRNA did not differ statistically between the exposure groups. Direct contact between infected and naive hamsters has been shown to result in SARS-CoV-2 transmission (Chan et al., 2020; Sia et al., 2020). The observed shedding data suggest that there may be similar shedding/transmission potential regardless of the route of exposure of an infected donor hamster. These vRNA shedding data imply that hamsters exposed by the i.n.L route that follow a milder disease course as evidenced by limited weight loss would be expected to have a similar capacity to transmit virus as the more overtly ill hamsters exposed by the i.n.H and i.g. routes.
Influence of sex and age on SARS-CoV-2 pathogenesis in golden Syrian hamsters
Differential disease outcomes between the sexes or different age groups in animal models of SARS-CoV-2 infection could be an important consideration for the pre-clinical evaluation of vaccines, antivirals, and candidate therapeutics (Herati and Wherry, 2018). In order to explore the contribution of sex to SARS-CoV-2 pathogenesis in hamsters we reanalyzed the i.n.H-exposed hamster data, described above, by sex, resulting in two separate groups, 6-week-old males and 6-week-old females (n = 5 for each sex). An additional group of 22-week-old older male hamsters (n = 5) were likewise infected with 105 TCID50 of SARS-CoV-2 via an i.n.H route, and similar analyses were carried out. Female, male, and older male hamsters were all productively infected, and no individuals succumbed to disease (Figure 2A). Mock-infected animals did not lose weight and gained weight throughout the course of the experiment (Figure 1B). Weight loss in all three infected hamster groups began on 2 dpi and tracked similarly until 5 dpi when the mean weights began to rebound at a faster rate in the females compared with the males and older males (Figure 2B). The SARS-CoV-2-infected female hamsters reached their pre-infection weights at 9 dpi and ultimately reached the mean weight of the uninfected hamsters at 11 dpi. Conversely, the infected males and older males had significantly lower mean weights compared with the females from 9 dpi onward (one-way ANOVA p = 0.0026 and 0.0020, respectively). Older males did not recover their initial body weights even by day 28. Uninfected young hamsters gained weight rapidly (Figure 2B), whereas aged animals, which were no longer growing, gained weight much more slowly. It is possible that this difference may contribute to the slower weight recovery in aged male animals that lag behind the young males. The macroscopic discoloration observed in large portions of the lung tissue in SARS-CoV-2-infected hamsters at 2 dpi varied greatly in individuals and did not appear to differ by sex or age (Figure 2C).
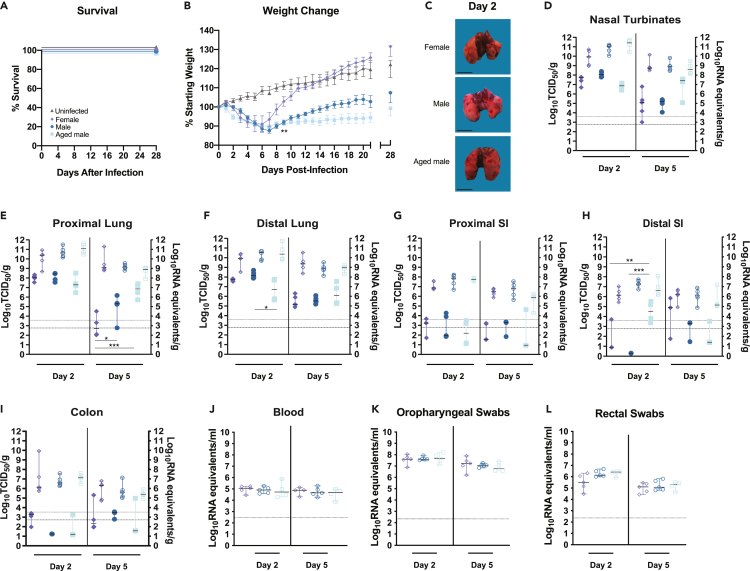
Figure 2.
SARS-CoV-2 infection of female, male, and aged golden Syrian hamsters
(A–L) Six-week-old female (lavender diamonds), six-week-old male (dark blue circles), or twenty-week-old male (light blue squares) hamsters (Mesocricetus auratus) were inoculated with 105 TCID50 of SARS-CoV-2 by a high-volume (100 μL) intranasal route (i.n.) of administration and compared with six-week-old uninfected controls (gray triangles). Bars indicate means (B) or medians (D–I). Error bars indicate SEM (b) or 95% CI (D–L). Dashed lines and dotted lines indicate the limit of detection for the TCID50 assay and qRT-PCR assay, respectively. A-B, Kaplan-Meier curve depicting survival data (A) and weight data (B) over the course of 28 days following SARS-CoV-2 exposure. C, macroscopic images of SARS-CoV-2-infected hamster lungs. Scale bars, 1 cm. D–I, Infectious viral load (filled in shapes, left axis) and vRNA levels (empty shapes, right axis) in the (D) nasal turbinates, (E) proximal lung, (F) distal lung, (G) proximal small intestines, (H) distal small intestines, and (I) colon at the indicated days post infection. Viral RNA levels in (J) blood, (K) oropharyngeal swabs, and (L) rectal swab samples. Data were collected in a single experiment (n = 5). The 6-week male and female data correspond to the data presented for the high-volume intranasal route of administration.
The infectious virus burden in the respiratory tract tissues for females, males, and older males was plotted (Figures 2D–2F, solid shapes, left axes). At 2 dpi the infectious virus burden in the respiratory tract tissues did not differ with the exception of the older males that showed a significantly lower viral burden in the distal lungs (one-way ANOVA p = 0.0364). At 5 dpi, the older males had significantly more infectious virus present in the nasal turbinates compared with females and males (one-way ANOVA p = 0.0175 and p = 0.0277, respectively). Both males and older males had significantly more infectious virus present in the proximal lungs, compared with females (one-way ANOVA p = 0.0347 and p < 0.0001, respectively). These data indicate that both sex and age differences occur in the hamster model of SARS-CoV-2 infection. These findings are consistent with the reported increased viral burden and delayed virus clearance reported in older mice compared with young mice exposed to mouse-adapted SARS-CoV-2 (Dinnon et al., 2020), but they are contrary to a prior report of SARS-CoV-2 infection in older hamsters, where no delay in viral clearance was observed (Imai et al., 2020). The level of vRNA in the respiratory tract tissues did not show statistically significant differences among the three groups (Figures 2D–2F, empty shapes, right axes). Although the levels of infectious virus generally decline in the lungs, there was persistence of high levels of vRNA in the same samples. The sources of high levels of vRNA were not examined, but the reductions in live virus titers indicate resolution of the infection that leads to the recovery in body weights that starts at around 6–7 dpi. The infectious virus burden in the digestive tract (proximal and distal small intestines and colon) was several logs lower than in the respiratory tract and was similar among the three groups (Figures 2H and 2I, solid shapes, left axis), with the exception of the older males at 2 dpi that had significantly more infectious virus in the distal small intestine compared with both the females and young males (one-way ANOVA p = 0.0096 and 0.0003, respectively). The levels of vRNA in the digestive tract did not show statistically significant differences among the three groups (Figures 2G–2I, empty shapes, right axes). At both 2 and 5 dpi, vRNA was detected in the blood of all hamsters, and the levels did not differ among the three groups (Figure 2J). The observed differences between male and female mean body weight throughout the course of illness does not appear to be due to large differences in viral burden in the tissues of the respiratory tract or GI tract and therefore may be due to the host response to infection driving immune-mediated pathology resulting in prolonged weight loss.
Moderate vRNA levels ranging from ∼107 to 108 genome copies and from 104.5 to 106.5 genome copies/ml were detected at 2 dpi in oropharyngeal and rectal swabs, respectively, and the vRNA levels did not differ among the different groups (Figure 2K and 2L). These data suggest that, regardless of the differential disease presentation in female, male, and older male hamsters, there may be similar shedding/transmission potential during acute illness irrespective of the sex or age.
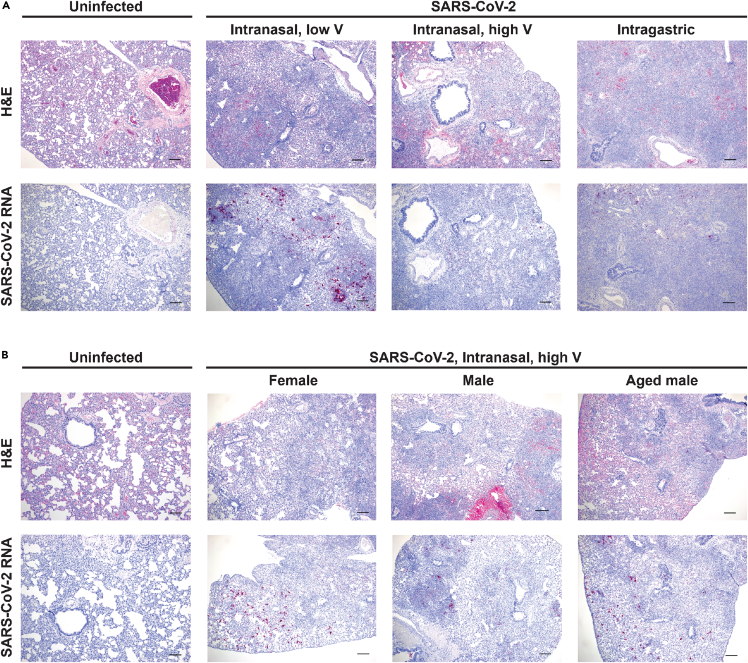
The lesions observed in the lung tissue of individual hamsters at 5 dpi were variable and ranged in severity and progression within each inoculation route group. Although the entire spectrum of lesions could be observed within each group, there were some differences between groups in the frequency in which specific lesions were observed. In some animals, lung lesions were more typical of earlier infection and characterized by patchy bronchointerstitial pneumonia with necrosis of alveolar and bronchiolar epithelial cells. Interstitial infiltrates consisted primarily of neutrophils and macrophages. There was alveolar edema/hemorrhage as well as perivascular edema. Other animals had lesions that were more typical of later infection with severe interstitial pneumonia (lymphohistiocytosis) leading to loss of alveolar spaces. There was extensive type II pneumocyte hyperplasia with characteristic tombstoning, peribronchitis, hyperplasia of bronchiolar epithelial cells, perivasculitis, presence of multinucleated syncytial cells, and occasionally vasculitis or fibrosis. Representative histology images from each group are presented (Figure 3A, upper panel). In general, more animals in the i.n.H group had lesions of patchy bronchointerstitial pneumonia compared with the i.g. group, where lesions were characterized by severe, widespread lymphohistiocytic interstitial pneumonia. In the i.n.L group, there were roughly equal numbers of each type of lesion. Owing to the limited small sample size additional experiments would be required to confirm these putative differences. In situ hybridization (ISH) staining showed the presence of vRNA, primarily detected in bronchiolar epithelial cells, which was highly variable between individual animals and/or samples (Figure 3A, lower panels). Lesions were largely absent from the nasal turbinates (data not shown), although vRNA was detected in some epithelial cells (Figure S1).
Figure 3.
Histopathology and virus distribution in the lungs
Hematoxylin and eosin (H&E) staining (A and B, upper panels) and in situ hybridization (ISH) with antisense probes that detect the SARS-CoV-2 genome/mRNA (A, B, lower panels) on lung tissue of uninfected and SARS-CoV-2-infected hamsters at 5 dpi. A, Six-week-old mixed female and male golden hamsters (Mesocricetus auratus) were inoculated with 105 TCID50 of SARS-CoV-2 by a low- (20 μL) or high- (100 μL) volume intranasal route of administration or an intragastric route of administration and compared with age-matched uninfected controls. B, Six-week-old female, six-week-old male, or twenty-week-old male hamsters were inoculated with 105 TCID50 of SARS-CoV-2 by a high-volume (100 μL) intranasal route of administration and compared with six-week-old uninfected controls. Positive detection of viral genomic RNA/mRNA is indicated by magenta staining. The magnification is 5x for H&E and ISH. Scale bars, 200 μm.
Similarly, a comparison of males and females showed a wide variability in lesion severity in individual animals within each group. Overall, the spectrum of lesions observed was similar in both females and males, and no differences in lesion severity were observed (Figure 3B, upper panel). Similarly, no differences in the nature or severity of lung lesions were observed in the younger compared with the older males. Owing to the wide variability between individual animals, a larger sample size is needed to detect subtle differences between the sex groups and age groups. These observations are consistent with the variable lung pathology observed in SARS-CoV-2-infected cynomolgus macaques (Rockx et al., 2020). ISH staining showed similar levels of vRNA in females, males, and older males (Figure 3B, lower panels).
Blood biochemistry and hematological parameters were assessed for both mock- (both sexes, n = 6) and SARS-CoV-2-infected hamsters belonging to the various experimental groups described above, including the i.n.L, i.n.H, and i.g. exposure groups (both sexes, n = 5, 10, and 5 for the exposure groups, respectively); the i.n.H group broken down by sex (n = 5 of each sex); and the i.n.H-exposed older males (n = 4) (Figure S2A–S2G). At 5 dpi all the groups had similar counts of white blood cells (Figure S2A), lymphocytes (Figure S2B), and neutrophils (Figure S2C); however, the neutrophil to lymphocyte ratio (NLR), a clinical metric that has been found to be elevated in human patients during severe COVID-19 (Liu et al., 2020a), was significantly elevated in the i.n.H- (both sexes combined) and i.n.H-exposed male hamsters compared with the uninfected control hamsters (one-way ANOVA p = 0.0052, 0.0014) (Figure S2D). At 2 dpi alanine aminotransferase (ALT) values were not significantly elevated in any group, suggesting that liver damage had not occurred (Figure S2E), whereas blood albumin (ALB) was reduced in the i.n.L, i.n.H, i.g., and female i.n.H-exposed compared with uninfected hamsters (Figure S2F, one-way ANOVA p = 0.0009, 0.0048, 0.0002, 0.0005, respectively), recapitulating the ALB reductions observed in patients with severe COVID-19 (de la Rica et al., 2020). Blood urea nitrogen (BUN) was elevated in all groups, suggestive of potential kidney disfunction or pre-renal damage (dehydration), regardless of route of infection, sex, or age (Figure S2G), although the presence of vRNA was not detected in the kidneys by ISH (data not shown). Additional serum biochemical values were unremarkable (data not shown). The potential involvement of the kidneys in the disease state following infection may be an important distinguishing characteristic of the hamster model, as opposed to other reported animal models.
Influence of route of infection, sex, and age on the immune response to SARS-CoV-2 infection
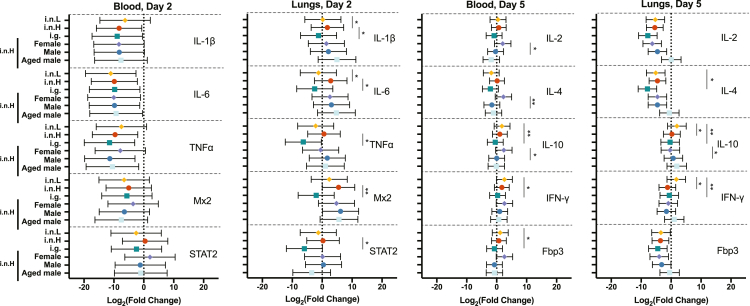
With little variation in viral burden in the tissues observed between groups of animals despite notable differences in the course of clinical disease, we sought to determine whether route, sex, and/or age differentially impacts the immune response to SARS-CoV-2 infection. We examined the relative mRNA expression of a subset of inflammatory cytokines and immune response genes. Relative gene expression in the blood and lungs of IL-1β, IL-6, TNF-α, Mx-2, and STAT-2 at 2 dpi and IL-2, IL-4, IL-10, IFN-γ, and FoxP3 at 5 dpi was quantified for all of the experimental groups described above as compared with uninfected controls for baseline levels (Figure 4). The given genes were chosen in an attempt to get a picture of potential changes in the innate immune response earlier after infection, and the adaptive immune response at, or just after, the peak of infection. The expression of inflammatory cytokines, defined here as those that promote inflammation, immune cell migration and infiltration, and/or the development and activation of specific immune cells, at 2 dpi was highly variable in the blood among individuals and did not differ significantly between any of the experimental groups (Figure 4, left panel). The route of virus administration impacted inflammatory cytokine expression in the lungs at 2 dpi (Figure 4, left middle). Overall, the mean of the examined cytokines at 2 dpi trended lower in the i.n.L group relative to the i.n.H group and was lower still in the i.g. group, although these differences did not always attain statistical significance. Expression of IL-1β and IL-6 mRNA was significantly lower in the i.n.L- versus the i.n.H-exposed hamsters (multiple t test p = 0.025 and 0.025, respectively). Furthermore, lung expression of IL-1β, IL-6, TNF-α, Mx-2, and STAT-2 mRNA was significantly less in the i.g.- versus the i.n.H-exposed hamsters (multiple t test p = 0.033, 0.014, 0.017, 0.0011, and 0.014, respectively). Sex and age were not found to have a significant effect on inflammatory cytokine expression in the lungs at this time point. In the older males, transcription of IL-1β and IL-6 trended higher and transcription of STAT-2 trended lower relative to the young males, although these differences did not attain statistical significance. Differential expression of inflammatory cytokines became more pronounced between the infection groups at 5 dpi when we examined the expression of adaptive immune response genes in the blood and lungs of infected hamsters relative to uninfected controls. Inflammatory cytokine expression in the blood at 5 dpi was similar in the two i.n.-exposed groups (i.n.L and i.n.H), with only small differences observed that did not attain statistical significance. Cytokine expression trended lower in the i.g.- relative to the i.n.-exposed groups, and the i.g. group had significantly lower IL-10, IFNγ, and Fbp3 mRNA expression relative to the i.n.L group (multiple t test p = 0.006, 0.012, 0.033, respectively). At 5 dpi, the young and older males showed similar cytokine expression in the blood, whereas cytokine expression in the females consistently trended higher, with females showing significantly elevated expression of IL-2, IL-4, and IL-10 relative to the young males (multiple t test p = 0.034, 0.002, 0.029, respectively) (Figure 4, middle right panel). Finally, consistent with overall cytokine expression observed in the blood and lung samples, IL-2 and IL-4 trended lower in the i.g. group relative to both i.n. groups, although these differences did not attain statistical significance. Inflammatory cytokine expression in the lungs at 5 dpi did not show statistically significant differences between the i.g.- and i.n.H-exposed hamsters; however, the i.n.L-exposed hamsters had elevated IL-10 and IFN-γ relative to the i.n.H-exposed hamsters (multiple t test p = 0.007 and 0.013, respectively) and elevated IL-4, IL-10, and IFN-γ relative to the i.g.-exposed hamsters (multiple t test p = 0.04, 0.01. 0.01, respectively) (Figure 4, right panel). At this time point inflammatory cytokine expression was similar in age-matched males and females, although female hamsters had significantly reduced expression of IL-10 in the lungs compared with males (multiple t test p = 0.029). In the older males, inflammatory cytokine expression in the lungs consistently trended higher relative to the young males, although these differences did not attain statistical significance. Taken together these data suggest that differences in expression of inflammatory cytokine and immune response genes may play a role in the differences seen in the SARS-CoV-2 disease course in hamsters.
Figure 4.
Hamster host response to SARS-CoV-2 infection
Six-week-old male and female hamsters (Mesocricetus auratus) were inoculated with 105 TCID50 of SARS-CoV-2 by a low-volume intranasal (orange diamonds), high-volume intranasal (red circles), or intragastric (teal squares) route of administration. The high-volume intranasal group data are further broken down by sex (female, lavender; male, dark blue) and compared with an additional group of 20-week-old males exposed to 105 TCID50 of SARS-CoV-2 by a high-volume intranasal route of exposure (light blue squares). Cytokine gene expression was measured for IL-1β, IL-6, TNFα, Mx2, and STAT2 in the blood (left panel) and lungs (middle left panel) at 2 dpi and IL-2, IL-4, IL-10, IFN ϒ, and Fbp3 in the blood (right middle panel) and lungs (right panel) at 5 dpi and displayed relative to age-matched mock-infected animals. Gene expression was normalized using RPL18 as a control. n = 5 hamsters/group. Bars indicate mean, error bars indicate 95% confidence interval (A). ∗ = p < 0.05, ∗∗p = 0.01; unpaired Student’s t test.
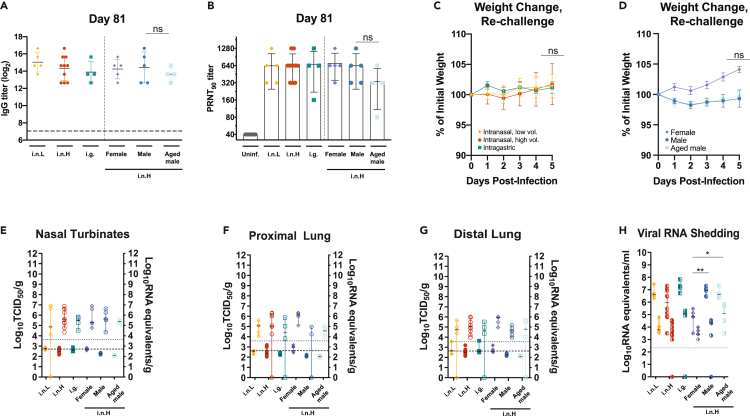
The humoral response generated following SARS-CoV-2 infection was evaluated in all the experimental groups at 21 (Figure S3) and 81 dpi (Figures 5A and 5B). All SARS-CoV-2-exposed hamsters had detectable serum IgG titers against spike antigen as assessed by ELISA at 21 dpi with endpoint titers ranging from 1,600 to 6,400, the limit of the assay, that did not differ regardless of route of exposure, sex, or age (Figure S3A). At 81 dpi IgG titers ranged from 6,400 to 102,400, and there was a high level of variation within each group (Figure 5A). Although the i.g.-exposed hamsters had lower IgG titers relative to the i.n.-exposed hamsters and the older males had lower IgG titers relative to the young males, these differences did not attain statistical significance. Similarly, neutralizing antibodies were detected by plaque reduction neutralization test (PRNT90) at 21 dpi in all infected hamsters ranging from 1:80 to 1:1,280, and no significant differences were observed regardless of route of exposure, sex, and age (Figure S3B). The neutralizing antibody levels stayed at approximately the same level when they were assessed at 81 dpi (Figure 5B). Older males generally had lower levels of neutralizing antibodies, although these differences did not attain statistical significance.
Figure 5.
Durability of humoral immune response and susceptibility to re-challenge
A–H, Six-week-old male and female hamsters (Mesocricetus auratus) were inoculated with 105 TCID50 of SARS-CoV-2 by a low-volume intranasal (orange diamonds), high-volume intranasal (red circles), or intragastric (teal squares) route of administration. The high-volume intranasal group data are further broken down by sex (female, lavender; male, dark blue) and compared with an additional group of 20-week-old males exposed to 105 TCID50 of SARS-CoV-2 by a high-volume intranasal route of exposure (light blue squares). A, IgG antibody response against SARS-CoV-2 spike antigen was assessed by ELISA using serum collected at 81 dpi. B, Neutralizing antibody against SARS-CoV-2 was measured by PRNT90 using serum collected at 81 dpi. C-H, Five hamsters belonging to each of the challenge groups described above were re-challenged at 81 dpi with 1x105 TCID50 of the same strain of SARS-CoV-2 by a high-volume intranasal route of administration. C-D, Weights were obtained daily until 5 dpi for re-challenged hamsters initially exposed by an i.n.L, i.n.H, or i.g. route (C) or female, male, and older hamsters (D). E–G, Infectious virus (left axis, shaded) and viral RNA levels (right axis, empty) were measured in the nasal turbinates (E), proximal lung (F), and distal lung (G). H, Viral RNA shedding detected in oropharyngeal (left shaded) and rectal swabs samples (right shaded). Dashed horizontal lines indicate the limit of detection. The limits of detection are indicated with dashed lines (ELISA and TCID50 assay) and dotted lines (qRT-PCR assay). Bars indicate means (A–D) or medians (E–H), error bars indicate SD (A–D) or 95% CI (E-H). ∗ = p < 0.05, ∗∗p = 0.01, ns = p > 0.05; unpaired Student’s t test.
At 81 dpi the infected hamsters belonging to each group were re-challenged by an i.n.H route with 1x105 TCID50 of the same strain of SARS-CoV-2, and swab and tissues samples were obtained at 5 days after re-infection. Upon re-challenge some hamsters were observed to lose a small amount of weight (Figure 5C). Although the route of initial exposure and age did not appear to affect post re-challenge weight loss up to 5 dpi, female hamsters gained a slight amount of weight following re-challenge, whereas the males and older males lost a small amount of weight; however, these differences did not attain statistical significance (Figures 5C and 5D). The viral burden in the respiratory tract tissues of the re-challenged hamsters were also assessed at 5 dpi (Figures 5E–5G). The amount of infectious virus and vRNA in the nasal turbinates (Figure 5E), proximal lung (Figure 5F), and distal lung (Figure 5G) were several logs lower than the levels observed during the acute period following the initial SARS-CoV-2 exposure (Figures 5E–5G versus Figures 1E–1G and 2D–2F). The level of infectious virus in the respiratory tract tissues was consistently near or below the limit of detection of the assay. An additional group of i.n.H-exposed male hamsters that were re-challenged 133 (n = 5) or 140 dpi (n = 14) showed very mild weight loss in some hamsters, although these weight changes did not attain statistical significance (Figure S4). At 5 days after re-challenge (145 dpi), only two of fourteen hamsters had infectious virus near the limit of detection in the nasal turbinates, whereas all other organs tested (proximal lung, distal lung, and small intestines) contained no detectable infectious virus (Figure S4). This apparent protection correlated with a robust antibody response that was detectable at high titers at 140 dpi (Figure S4), suggesting that protective antibody and potentially protective memory responses can persist.
Of interest, the levels of shed vRNA in oropharyngeal and rectal swab samples following the 81 dpi re-challenge were comparable with the levels observed during the acute period following the initial SARS-CoV-2 exposure (Figures 1L and 1M, 2K, and 2L versus Figure 5H). Re-challenged males and older males shed significantly more vRNA in oropharyngeal swab samples compared with females (p = 0.0053 and 0.0159, respectively) (Figure 5H). The i.g.-exposed and re-challenged hamsters shed more vRNA in rectal swabs compared with re-challenged i.n.-exposed hamsters, and re-challenged males and older males had more vRNA in rectal swabs compared with re-challenged females; however, these differences did not attain statistical significance. These data suggest that route of exposure, sex, and age do not have a meaningful impact on the protective efficacy of the host immune response resulting from a natural exposure against a homologous SARS-CoV-2 re-exposure at up to 81 days following the initial exposure; however, these data suggest that males and older males shed more vRNA in oropharyngeal swabs than females. At 5 dpi following re-challenge the IgG titer had increased in most of the hamsters (Figure S4).
Discussion
The unified global push to rapidly develop antivirals, experimental therapeutics, and vaccines that are capable of mitigating COVID-19 severity in infected individuals and/or virus transmission relies on animal models of SARS-CoV-2 infection that recapitulate with reasonable fidelity the key disease features of COVID-19. Here, we build upon previous reports that Syrian hamsters are a suitable animal model for SARS-CoV-2 infection (Chan et al., 2020; Sia et al., 2020; Imai et al., 2020) and describe the contribution of route of exposure, sex, and age to SARS-CoV-2 pathogenesis. We report that the route of inoculation influences the course of disease in golden Syrian hamsters. Compared with high-volume intranasal exposure, a low-volume intranasal exposure resulted in milder weight loss, whereas intragastric exposure resulted in slower recovery from weight loss following the period of acute illness. Furthermore, the route of SARS-CoV-2 exposure influenced the viral burden in a subset of respiratory tract and GI tract tissues, while the viral shedding and humoral immune responses were comparable. We also report that male hamsters, and to a much greater extent older male hamsters, demonstrated a reduced capacity to recover to their pre-infection body weight compared with females showing significant differences in disease course related to sex and age. Males and older males also showed delayed viral clearance from the respiratory tract tissues compared with females; however, similar humoral immune responses were identified in females, males, and older males up to 81 dpi. We further identified variable expression of inflammatory cytokines in the blood and lungs with route of exposure, sex, and age, and these differences may play a role in infection outcome, with further studies into the observed differences related to clinical presentation of disease in the various groups being warranted. Finally, we found that the route of initial exposure, sex, and age did not significantly influence the course of disease following a homologous SARS-CoV-2 re-challenge at 81 dpi or day 133/140, although we found that females shed significantly less vRNA upon re-challenge than males and older males.
Studies describing SARS-CoV-2 pathogenesis in golden hamsters have employed various routes of virus administration, including intranasal with inhalation (Chan et al., 2020; Sia et al., 2020), combined ocular and intranasal (Imai et al., 2020), and oral administration (Lee et al., 2020). Variable disease outcomes in response to different routes of inoculation, including intragastric inoculation, have been described for several highly pathogenic respiratory viruses, including H5N1 influenza A virus (Shinya et al., 2011), Nipah virus (NiV) (de Wit et al., 2014), and MERS-CoV (Zhou et al., 2017), and respiratory tract exposure has been shown to result in productive infection of the gastrointestinal tract following viremia with the enterovirus, porcine epidemic diarrhea virus (PEDV) (Li et al., 2018). The previously reported intranasal administrations of SARS-CoV-2 to hamsters employed an inoculum volume ranging from 80 to 100 μL (Chan et al., 2020; Sia et al., 2020). The volume of the inoculum delivered intranasally has been shown to dictate the efficiency of lower respiratory tract delivery, with a smaller volume more restricted to the upper respiratory tract (Southam et al., 2002). We sought to compare SARS-CoV-2 pathogenesis following a low (20 μL, i.n.L) or high (100 μL, i.n.H) inoculum volume to primarily limit the initial infectious dose to the upper respiratory tract or combined upper/lower respiratory tract, respectively. Furthermore, patients with COVID-19 often present with GI symptoms, and SARS-CoV-2 vRNA can often be detected in the feces (Pan et al., 2020; Jin et al., 2020b; Cheung et al., 2020; Wang et al., 2020), and SARS-CoV-2 nucleocapsid protein has been detected within enterocytes in both animal models (Chan et al., 2020; Shi et al., 2020) and humans (Xiao et al., 2020). We, therefore, sought to further evaluate SARS-CoV-2 pathogenesis following an intragastric route of exposure. The described data demonstrate that an i.n.L inoculation resulted in reduced weight loss compared with the standard i.n.H inoculation, whereas an i.g. inoculation also resulted in efficient infection in the respiratory tract but with much slower recovery to the original body weight following acute illness. This is in contrast to the mild disease reported following SARS-CoV-2 infection of hamsters by an oral route (Lee et al., 2020). Furthermore, the only animal to succumb to infection had been exposed by an i.g. route. The i.n.L route of exposure may serve as a useful model for the asymptomatic SARS-CoV-2 infection with viral shedding that is commonly observed in humans (Poletti et al., 2021; Byambasuren et al., 2020). Although the acidic environment of the digestive tract is expected to inactivate SARS-CoV-2 within a matter of hours (Zang et al., 2020; Lee et al., 2020), human enterocytes express both ACE2 and TMPRSS2 (Zou et al., 2020) and support virus replication in vitro (Zang et al., 2020; Lamers et al., 2020). Furthermore, we observed rare and sporadic foci of vRNA in the GI tract by ISH (data not shown) as well as infectious virus in some GI tissue samples (Figures 1 and 2). Taken together, these data suggest that a low level of viral replication may be occurring in the GI tract. As with MERS-CoV infection (Zhou et al., 2017), the GI tract may serve as a site of extrapulmonary SARS-CoV-2 replication and may contribute to viral pathogenesis or the nature of the immune response generated in response to infection. The variable levels of infectious virus detected in the digestive tract could be related in part to the feed state of the hamsters at the time of infection since simulated gastric and/or colonic fluids (particularly in the fasted state) have been shown to inactivate both MERS-CoV (Zhou et al., 2017) and SARS-CoV-2 (Zang et al., 2020). A limitation of the routes of administration described in this work is that i.n. administration results in some inoculum being swallowed and entering the gastrointestinal tract (Southam et al., 2002). Even with low-volume i.n. administration, there may be a small amount of virus that become deposited within the lower respiratory tract shortly after infection. With the i.g. instillation by oral gavage some inoculum is also expected to be introduced to the respiratory tract (de Wit et al., 2014) or oral cavity when the feeding tube is inserted or withdrawn. We questioned whether the differential pathogenesis observed with an i.g. route versus a high-volume intranasal route administration was due to the alimentary tract exposure or rather a small viral dose seeding the airway during gavage; however, a recent report describes that low-dose SARS-CoV-2 inoculation of the hamster airway results in diminished pathogenesis (Monchatre-Leroy et al., 2021). Given that the i.n.L inoculation may also be expected to result in a low-dose delivery of virus to the lower respiratory tract, the milder clinical signs of illness that we observed are consistent with the reduced pathogenesis reported by others upon low-dose exposure. Given our findings that all routes of infection resulted in similar timing and extent of virus replication in the respiratory and digestive tract tissues (Figure 1), it is possible that disease outcome is more directly related to the nature of the immune responses that are established at the initial site of greatest virus exposure and that this has a significant impact on the cytokine expression at subsequent sites of active virus replication. However, further studies looking into mechanistic explanations for this need to be performed. Furthermore, the high viral burden in the respiratory tract with viral shedding and reduced disease severity observed the i.n.L-exposed hamsters appears to recapitulate the asymptomatic disease state observed in most humans (Byambasuren et al., 2020). In contrast to a previous study (Osterrieder et al., 2020), we did not observe age-dependent differences in lung pathology (Figure 3).
Epidemiological and clinical analyses have identified several risk factors that are associated with an increased incidence of severe and/or fatal COVID-19, including hypertension, cardiac, or pulmonary disease (Huang et al., 2020; Zhou et al., 2020); male sex (Livingston and Bucher, 2020); and advanced age (Li et al., 2020; Zhou et al., 2020; Livingston and Bucher, 2020); however, insight into how these factors contribute to SARS-CoV-2 disease progression in small animal models has only begun to be examined (Imai et al., 2020; Osterrieder et al., 2020). Several studies have reported that SARS-CoV-2 infection in male patients results in a greater risk of severe disease outcome than in females, potentially owing to differences in the host response to infection (Takahashi et al., 2020; Scully et al., 2020; Chanana et al., 2020; Jin et al., 2020a). We report that, compared with i.n.H-infected females, males regained their body weight at a slower rate following the period of acute illness. We observed a greater viral burden in the nasal turbinates and proximal lung tissues of young males and older males compared with young females. This is in agreement with sex differences observed in SARS-CoV-infected mice, where males were shown to have elevated viral titers in the respiratory tract (Channappanavar et al., 2017). In that work the sex differences were attributed to the influence of sex hormones on the innate immune response rather than the adaptive response (Channappanavar et al., 2017), and it will be interesting to determine if this is consistent in the hamster model of SARS-CoV-2 infection. It has been shown that females can often mount a stronger inflammatory response in response to viral infection and vaccination compared with males (Klein and Flanagan, 2016). Representation of both sexes will therefore be crucial for pathogenesis studies and pre-clinical evaluation of vaccines against COVID-19. A correlation between advanced age and increased risk of severe disease and mortality associated with patients with SARS-CoV-2 has been well established (Guan et al., 2020; Huang et al., 2020; Zhou et al., 2020). Greater disease severity has also been reported in both older murine and hamster models of SARS-CoV-2 infection (Gu et al., 2020; Dinnon et al., 2020; Imai et al., 2020; Sun et al., 2020). Our observation that older hamsters show a delayed recovery of their body weight following i.n. SARS-CoV-2 infection is consistent with these reported data (Imai et al., 2020; Osterrieder et al., 2020). Our findings are also consistent with the reported increased viral burden and delayed viral clearance reported in older mice compared with young mice exposed to mouse-adapted SARS-CoV-2 (Dinnon et al., 2020) but are contrary to the reported absence of delayed viral clearance in SARS-CoV-2-infected older hamsters (Imai et al., 2020).
Independent studies have identified elevated cytokine expression in the blood in severe human cases of COVID-19. Some of those that are thought to play a role in disease development include IL-1β, IL-6, TNF-α, IL-10, IL-2, and IFN-γ (Li et al., 2020; Liu et al., 2020b). Our observation that hamster lungs at 2 dpi showed that mRNA expression levels for IL-1β and IL-6 were reduced in i.n.L-exposed hamsters and IL-1β, IL-6, TNF-α, Mx-2, and STAT-2 were reduced in i.g.-exposed hamsters relative to their i.n.H-exposed counterparts indicates that initial exposure of the virus by the i.n.L or i.g. route resulted in a less robust inflammatory response in the lungs. In addition, IL4, IL10, and IFN-γ mRNA expression in the lungs was found to vary by route of infection at 5 dpi (Figure 4). The differences in adaptive immune response gene expression observed between the i.n.L, i.n.H, and i.g. groups suggest that the initial site of virus exposure has an impact on the cytokine profile and likely the nature of the immune response developed in the lungs. This observation could have important implications for understanding immune-mediated pathology, anti-viral immunity, and vaccination strategies. It is reasonable to conclude that differences in the innate immune response, including the cell types present and differential expression and activation of pattern recognition receptor (PAMP) in various tissues, as well as draining lymph nodes can dictate the nature of the adaptive immune response in response to SARS-CoV-2 infection (Netea et al., 2005; Crane and Forrester, 2005; Belyakov and Ahlers, 2009).
Of interest, although significant differences in the expression levels of cytokines between young and older hamsters were not detected, older hamsters had consistently higher expression of several examined cytokines in the lungs, consistent with a study describing a stronger inflammatory response in the lungs of aged SARS-CoV-infected NHPs (Smits et al., 2010). There remains a lack of developed, commercially available reagents for examining specific aspects of immunity in hamsters, including flow cytometric analyses and detection of cytokine production at the protein level. Further investment in the development of these types of reagents is warranted as the hamster model of SARS-CoV-2 infection has shown great utility.
Several promising discoveries have recently been achieved using the widely adopted golden hamster model of SARS-COV-2 infection to examine viral pathogenesis (Hou et al., 2020; Johnson et al., 2020), assess immunity (Boudewijns et al., 2020), and evaluate antivirals (Kaptein et al., 2020), vaccines (Brocato et al., 2021), and therapeutics (Fagre et al., 2020). Taken together, the data presented here indicate that differences in immune responses between route of exposure, sex, and age could significantly affect the outcome of SARS-CoV-2 infection in golden hamsters. It will be important to further understand the effects of each of these variables and how they may influence the outcome of pre-clinical challenge studies. Continued analysis of the immune response to SARS-CoV-2 in animal model systems could further inform future clinical interventions that could modulate the host immune response to SARS-CoV-2 infection.
Limitations of the study
In this work we studied the effect of host parameters such as age and sex as well as the mode of infection on the outcome of SARS-CoV-2 infection in Syrian hamsters. Although the work describes distinct differences in infection outcome among the groups included in the study, there are some caveats to the work described. Owing to both space and time constraints, we chose to examine five animals per group, for each of the comparisons. Larger group sizes may have allowed us to more clearly determine potential differences between groups, particularly related to efforts to determine the mechanism(s) driving the observed experimental outcomes. In addition, although Syrian golden hamsters are an established animal model for SARS-CoV-2 infection, and can recapitulate important aspects of human infection, such as respiratory distress and lung pathology, there are limitations to its use. Hamsters do not show the spectrum of disease outcome that can be seen in humans, and notably infection is not lethal in the majority of animals. Therefore, this model largely reflects mild to moderate human infection, which is the typical disease course observed in humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| SARS-CoV-2 Canada/ON/VIDO-01/2020 | VIDO-Intervac | GISAID: EPI_ISL_425177 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant SARS-CoV-2 Spike protein | Sino Biological | Cat: 40589-V08B1 |
| Experimental models: Cell lines | ||
| Vero Cells | ATCC | CCL-81; RRID:CVCL_0059 |
| Experimental models: Organisms/strains | ||
| Syrian Golden Hamster (Mesocricetus auratus) | Charles River Labs | LVG Golden Syrian Hamster 049 |
| Oligonucleotides | ||
| All oligonucleotides used are described in Table S1 | N/A | N/A |
| Software and algorithms | ||
| Prism V.9 | GraphPad | V.9.1; RRID:SCR_002798 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Dr. Darwyn Kobasa, Public Health Agency of Canada (darwyn.kobasa@phac-aspc.gc.ca).
Materials availability
This study did not generate any unique reagents or materials.
Experimental models and subject details
Ethics statement
The experiments described were carried out at the National Microbiology Laboratory (NML) of the Public Health Agency of Canada. All experiments performed under Animal User Document H-20-006, approved by the Animal Care Committee at the Canadian Science Center for Human and Animal Health in accordance with the guidelines provided by the Canadian Council on Animal Care. All procedures were performed under inhalation anesthesia using isoflurane. All efforts were made to minimize animal suffering and to reduce the number of animals used. All infectious work was performed under biosafety level 3 (BSL-3) conditions or higher.
Cells
Vero cells (ATCC) were cultured in Minimal Essential Medium (MEM) (Hyclone) supplemented with 5% Bovine Growth Serum Supplemented Calf (Hyclone) and L-glutamine. Cells were cultured at 37°C with 5% CO2. Vero cells were not authenticated prior to use and the sex of the cells is unknown.
Viruses
The SARS-CoV-2 (Canada/ON-VIDO-01/2020) was isolated from a positive patient sample from early in the pandemic and stocks of virus were grown in VeroE6 cells. This stock was fully sequenced by Next Generation sequencing and the sequence has been submitted to the GISAID database (GISAID: EPI_ISL_425177). Virus stocks were titered by TCID50 assay before being used for subsequent in vivo experiments. All virus used for in vivo experiments was from passage 2.
Hamster challenge and re-challenge experiments
All hamsters used for experiments were purchased from Charles River. Four-six week old hamster, classified as young adults were used for most experiments. “Aged” hamsters included in some studies were at least 22 weeks of age at the start of the experiment. All animals were in healthy condition upon arrival and acclimated for at least one week prior to the experiments. Groups of male or female golden Syrian hamsters (n=5) were anaesthetized and exposed to 1x105 TCID50 SARS-CoV-2 by an intranasal route of inoculation with the inoculum distributed evenly into both nares in a volume of 20 μl (intranasal low volume) or in a volume of 100 μl to allow for inhalation of the inoculum (intranasal high volume), or by an intragastric route of infection via oral gavage. Mock infected animals were inoculated by an intranasal route with 100 μL of neat DMEM. Animals were monitored daily for clinical signs of disease including lethargy, hunched posture, inactivity, and labored breathing. Temperatures were measured daily via implanted transponders, and all animals were weighed once daily until 21 dpi and again on 28 dpi. Animals meeting the pre-determined endpoint criteria were anaesthetized and humanely euthanized. Previously exposed hamsters were re-infected with the same strain of SARS-CoV-2 at 81, 133, or 140 dpi and were necropsied at 5 dpi to assess the viral burden in the tissues, viral shedding, and the humoral immune response.
Method details
Hematology and blood biochemistry
Complete blood counts (CBC) and hematological analysis was performed using a VetScan HM5 (Abaxis Veterinary Diagnostics), as per manufacturer's instructions. Serum biochemistry values were determined with a VetScan VS2 (Abaxis Veterinary Diagnostics) using complete diagnostic profile disks according to manufacturer’s instructions. Blood and serum obtained from uninfected hamsters were used to establish baseline values. For both hematological and serum biochemistry analysis, fresh whole blood or serum from animals was used, immediately after collection.
Measurement of viral burden in the tissues
For measurement of viral titers in the blood and tissues of infected animals, TCID50 assays were performed. Following necropsy, blood and tissue samples were frozen at −80°C for storage. For infectious assays, tissue samples were thawed and placed in MEM, supplemented with 1x L-glutamine and 1% FBS, and homogenized with 5 mm stainless steel beads in a Bead Ruptor Elite Tissue Homogenizer (Omni). Homogenates and whole blood samples were clarified by centrifugation at 1500 x g for 10 minutes and ten-fold serial dilutions of tissue homogenates were made in MEM. GI tract tissues were additionally processed to minimize the presence of fecal bacteria: feces were carefully removed from the GI tract lumen and dilutions and infectious assays were carried out in MEM supplemented with 1x L-glutamine, 1% FBS, and a 2x dose of penicillin/streptomycin. Dilutions were added to 90-100% confluent Vero cells in triplicate wells and cytopathic effect was read at 5 dpi. TCID50 values per mL or gram of tissue were calculated using a modified Reed and Muench method (Reed and Muench, 1938).
For determination of viral RNA, collected tissues were stored in RNAlater. RNA was extracted using an RNeasy mini plus kit (Qiagen), according to manufacturer’s instructions. For viral RNA present in blood, RNA was extracted from whole blood using a viral RNA mini kit (Qiagen). RT-qPCR detection of SARS-CoV-2 was performed on a QuantStudio 5 instrument (Applied Biosystems) using a TaqPath 1-step RT-qPCR Master Mix (Applied Biosystems) and primers specific for the E gene of SARS-CoV-2 as per the diagnostic protocol recommended by the World Health Organization (Forward – ACAGGTACGTTAATAGTTAATAGCGT; Reverse – ATATTGCAGCAGTACGCACACA; Probe – FAM-ACACTAGCCATCCTTACTGCGCTTCGBBQ) (Table S1). For viral RT-qPCR, 4μL of extracted RNA was used in duplicate wells. Oligonucleotide concentrations were 400nM for the primers and 200nM for the probe. RT-qPCR stages were as follows: UNG incubation (25°C for 2 minutes), reverse transcription (53°C for 10 minutes), polymerase activation (95°C for 2 minutes), followed by amplification (40 cycles of 95°C for 3 seconds and 60°C for 30 seconds).
Detection of virus in mucosal swab samples
Oropharyngeal and rectal swabs as well as nasal washes were obtained from animals during necropsy. Swabs were stored in MEM + 2% penicillin-streptomycin. Prior to titration procedures, tubes containing swabs were vortexed and centrifuged briefly. For viral RNA detection, 140μL of the medium containing the swab was used for viral lysis and extraction using a viral RNA mini kit as above.
Transcriptional profiling of host responses
Tissue RNA was extracted as described above using an RNeasy mini plus kit, which includes a genomic DNA eliminator step. RNA previously extracted from whole blood was used for analysis in blood samples. Host mRNA expression of various genes including IL1β, IL6, TNFα, IL2, IFNγ, IL4, IL10, FoxP3, STAT2, and Mx2 was quantified using a previously designed primer set with RPL18 as an internal reference gene (Table S1, Safronetz et al., 2011). Briefly, RT-qPCR was performed using a TaqPath 1-step RT-qPCR kit similar to above using 1μL of extracted RNA for RT-qPCR, using the primers and probes listed in Table S1. Following RT-qPCR, fold change in gene expression compared to RPL18 was calculated using the delta-delta Ct method. The mean and 95% confidence intervals of the Log2 fold change in gene expression for each gene, in each group of animals, is depicted.
SARS-CoV-2-S-specific enzyme-linked immunosorbent assay (ELISA)
Ninety-six-well flat-bottom high-binding microplates (Corning, New York, USA) were coated with recombinant SARS-CoV-2 spike protein in PBS at concentration of 25 ng/well overnight at 4°C. The next day, plates were washed four times with PBS-T (PBS + 0.1% Tween 20) and then blocked with blocking buffer (PBS-T + 5% skim milk powder) for 1 hour at 37°C. Following blocking, plates were washed four times with PBS-T and serum samples from infected or mock infected hamsters were serially diluted and added to the plates in triplicate for 1 hours at 37°C. Plates were then washed with PBS-T four times and secondary peroxidase AffiniPure Goat Anti-Syrian Hamster IgG (H+L) (Jackson IR, #107-035-142) was added to the plates at a dilution of 1:1000 for 1 hour at 37°C. Plates were once again washed with PBS-T and ABTS substrate was added to the plates for 30 minutes at room temperature and then OD405nm readings were taken.
Plaque reduction virus neutralization test (PRNT-90)
Hamster serum samples were collected and stored at −80°C. SARS-CoV-2 stocks were titrated and used in the PRNT-90 (Mendoza et al., 2020). In short, serum was heat-inactivated at 56°C for 30 min and diluted 2-fold (dilution range 1:40 to 1:1280) in DMEM supplemented with 2% FBS. Diluted sera was incubated with 50 plaque forming units of SARS-CoV-2 at 37°C with 5% CO2 for 1 hour. The sera-virus mixtures were added to VeroE6 cells at 100% confluence in 24-well plate format, followed by incubation at 37°C and 5% CO2 for 1 hour. After adsorption, 1.5% carboxymethylcellulose diluted in MEM supplemented with 4% FBS, L-glutamine, non-essential amino acids, and sodium bicarbonate was added to each well. Plates were then incubated at 37°C and 5% CO2 for 72 hours. The liquid overlay was removed and the cells were fixed with 10% neutral-buffered formalin for 1 hour at room temperature. The monolayers were then stained with 0.5% crystal violet for 10 minutes and washed with 20% ethanol. Plaques were counted and compared to a 90% neutralization control. The PRNT-90 endpoint titre was defined as the highest dilution of serum resulting in a 90% reduction of plaques. PRNT-90 titers ≥1:40 were considered positive for neutralizing antibodies.
Histopathology and vRNA in situ hybridization
Tissues were fixed in 10% neutral phosphate buffered formalin for a minimum of 7 days. Routine processing was carried out and tissue samples were sectioned at 5 μm. A set of slides was stained with hematoxylin and eosin for histopathologic examination. RNA in situ hybridization (ISH) was carried out using RNAscope 2.5 HD Detection Reagent-Red (Advanced Cell Diagnostics), according to the manufacturer’s instructions. Briefly, formalin-fixed, paraffin-embedded tissue samples were mounted on slides, baked in a dry oven for 1 hour at 60°C, and deparaffinized. Tissue sections were then pre-treated with RNAscope H2O2 for 10 minutes at room temperature, and target retrieval was carried out using the RNAscope Target Retrieval Reagent for 15 minutes. RNAscope Protease Plus Reagent was then applied for 15 minutes at 40°C. The probes targeting SARS-CoV-2 RNA (Table S1; V-nCoV2019-S probe, ref#848561) or anti-genomic RNA (Table S1; V-nCoV2019-S-sense ref#845701) were designed and manufactured by Advanced Cell Diagnostics, and the negative probe was also obtained from Advanced Cell Diagnostics (Reference # 310034). The stained tissues were counterstained with Gills I Hematoxylin. The final images were captured using a light microscope equipped with a digital camera.
Quantification and statistical analysis
Data analysis
Results were analyzed and graphed using Prism 8 software (GraphPad Software). As appropriate, statistical analyses were performed using ANOVA with multiple comparison correction, the multiple t test (two-tailed), or the unpaired t test (two-tailed) with Welch’s correction or Mann-Whitney test.
Acknowledgments
We thank Darryl Falzarano of Vaccine and Infectious Disease Organization - International Vaccine Centre (VIDO-InterVac) for isolation of the SARS-CoV-2 strain used in these studies. The authors also thank Michelle French, Kimberly Azaransky, Stephanie Kucas, Amanda Stevenson, and Christine DeGraff of the Veterinary Technical Services at the National Microbiology Laboratory (NML) for their technical assistance during the course of this work. The work described in this paper was financially supported by the Public Health Agency of Canada and the Canadian Food Inspection Agency.
Author contributions
Conceptualization, B.D.G., B.M.W., A.S.B., D.K.; methodology, B.D.G., B.M.W., E.V., D.K.; formal analysis, B.D.G., B.M.W., J.A., A.S.B., C.E.-H.; investigation, B.D.G., B.M.W., M.C., E.V., N.T., A.L., S.H., W.C., E.M., L.G., K.T., K.N.T., A.A., K.M., G.S., A.S.B., R.V., J.L., Y.D., W.Z., D.K.; resources, L.B., S.M., D.S., H.W., J.E.S., C.E.-H., D.K.; writing – original draft, B.D.G., B.M.W.; writing – review and editing, B.D.G., B.M.W., A.S.B., D.K.; visualization, B.D.G., B.M.W.; supervision, L.B., S.M., D.S., H.W., S.M., J.E.S., C.E.-H., D.K.; funding acquisition, L.B., D.S., H.W., S.M., J.E.S., C.E.-H., D.K.
Declaration of interests
The authors declare no competing interests.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103530.
Supplemental information
Data and code availability
-
•
All relevant data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyakov I.M., Ahlers J.D. What role does the route of immunization play in the generation of protective immunity against mucosal pathogens? J. Immunol. 2009;183:6883–6892. doi: 10.4049/jimmunol.0901466. [DOI] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocato R.L., Kwilas S.A., Kim R.K., Zeng X., Principe L.M., Smith J.M., Hooper J.W. Protective efficacy of a SARS-CoV-2 DNA vaccine in wild-type and immunosuppressed Syrian hamsters. NPJ Vaccin. 2021;6:16. doi: 10.1038/s41541-020-00279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byambasuren O., Cardona M., Bell K., Clark J., McLaws M.-L., Glasziou P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. JAMMI. 2020;5:223–234. doi: 10.3138/jammi-2020-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020:ciaa325. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanana N., Palmo T., Sharma K., Kumar R., Graham B.B., Pasha Q. Sex-derived attributes contributing to SARS-CoV-2 mortality. Am. J. Physiol. Endocrinol. 2020;319:E562–E567. doi: 10.1152/ajpendo.00295.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fett C., Mack M., Ten Eyck P.P., Meyerholz D.K., Perlman S. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (Rnaemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane I.J., Forrester J.V. Th1 and th2 lymphocytes in autoimmune disease. Crit. Rev. Immunol. 2005;25:75–102. doi: 10.1615/CritRevImmunol.v25.i2.10. [DOI] [PubMed] [Google Scholar]
- Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagre A.C., Manhard J., Adams R., Eckley M., Zhan S., Lewis J., Rocha S.M., Woods C., Kuo K., Liao W., et al. A potent SARS-CoV-2 neutralizing human monoclonal antibody that reduces viral burden and disease severity in Syrian hamsters. Front. Immunol. 2020;11:614256. doi: 10.3389/fimmu.2020.614256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagre A., Lewis J., Eckley M., Zhan S., Rocha S.M., Sexton N.R., Burke B., Geiss B., Peersen O., Bass T., et al. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: implications for spillback to New World rodents. PLoS Pathog. 2021;17:e1009585. doi: 10.1371/journal.ppat.1009585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B.D., Chan M., Tailor N., Mendoza E.J., Leung A., Warner B.M., Duggan A.T., Moffat E., He S., Garnett L., et al. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat. Commun. 2021;12:3612. doi: 10.1038/s41467-021-23848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., Kinoshita N., Hattori S., Sakai-Tagawa Y., Iwatsuki-Horimoto K., et al. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020;383:592–594. doi: 10.1056/NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herati R.S., Wherry E.J. What is the predictive value of animal models for vaccine efficacy in humans?: consideration of strategies to improve the value of animal models. CSH Perspect. Biol. 2018;10:a031583. doi: 10.1101/cshperspect.a031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020 doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M., Liu S., Yang J.-K. Gender differences in patients with covid-19: focus on severity and mortality. Front. Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Lian J.-S., Hu J.-H., Gao J., Zheng L., Zhang Y.-M., Hao S.-R., Jia H.-Y., Cai H., Zhang X.-L., et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., Zhang L., et al. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.08.26.268854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S.J.F., Jacobs S., Langendries L., Seldeslachts L., ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Barthelemy K., et al. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2−infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. U S A. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I., Kim S.-G., Kim S.-M., Kim E.-H., Park S.-J., Yu K.-M., Chang J.-H., Kim E.J., Lee S., Casel M.A.B., et al. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.C.-Y., Zhang A.J., Chan J.F.-W., Li C., Fan Z., Liu F., Chen Y., Liang R., Sridhar S., Cai J.-P., et al. Oral SARS-CoV-2 inoculation establishes subclinical respiratory infection with virus shedding in golden syrian hamsters. Cell Rep. Med. 2020;1:100121. doi: 10.1016/j.xcrm.2020.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu Q., Huang L., Yuan C., Wang J., Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018;9:3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xu S., Yu M., Wang K., Tao Y., Zhou Y., Shi J., Zhou M., Wu B., Yang Z., et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020;146:110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston E., Bucher K. Coronavirus disease 2019 (COVID-19) in Italy. JAMA. 2020;323:1335. doi: 10.1001/jama.2020.4344. [DOI] [PubMed] [Google Scholar]
- Lu C., Liu X., Jia Z. 2019-nCoV transmission through the ocular surface must not be ignored. Lancet. 2020;395:e39. doi: 10.1016/S0140-6736(20)30313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C., et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Sig. Transduct. Target Ther. 2020;5:157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza E.J., Manguiat K., Wood H., Drebot M. Two detailed plaque assay protocols for the quantification of infectious sars-cov-2. Curr. Prot. Microbiol. 2020;57 doi: 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchatre-Leroy E., Lesellier S., Wasniewski M., Picard-Meyer E., Richomme C., Boué F., Lacôte S., Murri S., Pulido C., Vulin J., et al. Hamster and ferret experimental infection with intranasal low dose of a single strain of SARS-CoV-2. J. Gen. Virol. 2021;102 doi: 10.1099/jgv.0.001567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Van der Meer J.W.M., Sutmuller R.P., Adema G.J., Kullberg B.-J. From the th1/th2 paradigm towards a toll-like receptor/t-helper bias. Antimicrob. Agents Chemother. 2005;49:3991–3996. doi: 10.1128/AAC.49.10.3991-3996.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., Li P., Hu B., Wang J., Hu C., et al. Clinical characteristics of covid-19 patients with digestive symptoms in hubei, China: a descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Poletti P., Tirani M., Cereda D., Trentini F., Guzzetta G., Sabatino G., Marziano V., Castrofino A., Grosso F., Del Castillo G., et al. Association of age with likelihood of developing symptoms and critical disease among close contacts exposed to patients with confirmed SARS-CoV-2 infection in Italy. JAMA Netw. Open. 2021;4:e211085. doi: 10.1001/jamanetworkopen.2021.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-CoV-2. Science. 2020;368:1422–1424. doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints12. Am. J. Trop. Med. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- de la Rica R., Borges M., Aranda M., del Castillo A., Socias A., Payeras A., Rialp G., Socias L., Masmiquel L., Gonzalez-Freire M. Low albumin levels are associated with poorer outcomes in a case series of covid-19 patients in Spain: a retrospective cohort study. Microorganisms. 2020;8:1106. doi: 10.3390/microorganisms8081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronetz D., Zivcec M., LaCasse R., Feldmann F., Rosenke R., Long D., Haddock E., Brining D., Gardner D., Feldmann H., et al. Pathogenesis and host response in Syrian hamsters following intranasal infection with andes virus. PLoS Pathog. 2011;7:e1002426. doi: 10.1371/journal.ppat.1002426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K., Makino A., Tanaka H., Hatta M., Watanabe T., Le M.Q., Imai H., Kawaoka Y. Systemic dissemination of h5n1 influenza a viruses in ferrets and hamsters after direct intragastric inoculation. Virol. J. 2011;85:4673–4678. doi: 10.1128/JVI.00148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., de Lang A., van den Brand J.M.A., Leijten L.M., van IJcken W.F., Eijkemans M.J.C., van Amerongen G., Kuiken T., Andeweg A.C., Osterhaus A.D.M.E., et al. Exacerbated innate host response to sars-cov in aged non-human primates. PLoS Pathog. 2010;6:e1000756. doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam D.S., Dolovich M., O’Byrne P.M., Inman M.D. Distribution of intranasal instillations in mice: effects of volume, time, body position, and anesthesia. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L83C3–L839. doi: 10.1152/ajplung.00173.2001. [DOI] [PubMed] [Google Scholar]
- Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., HCA Lung Biological Network, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Prescott J., Falzarano D., Bushmaker T., Scott D., Feldmann H., Munster V.J. Foodborne transmission of nipah virus in syrian hamsters. PLoS Pathog. 2014;10:e1004001. doi: 10.1371/journal.ppat.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey C., Borisevich V., Prasad A.N., Agans K.N., Deer D.J., Dobias N.S., Heymann J.C., Foster S.L., Levine C.B., Medina L., et al. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat. Immunol. 2021;22:86–98. doi: 10.1038/s41590-020-00835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Zhou Q., Xu J. Mechanism of thrombocytopenia in COVID-19 patients. Ann. Hematol. 2020;99:1205–1208. doi: 10.1007/s00277-020-04019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang J., Kuang D., Xu J., Yang M., Ma C., Zhao S., Li J., Long H., Ding K., et al. Susceptibility of tree shrew to SARS-CoV-2 infection. Sci. Rep. 2020;10:16007. doi: 10.1038/s41598-020-72563-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Zhao G., Chu H., Wang D., Yan H.H.-N., Poon V.K.-M., Wen L., Wong B.H.-Y., Zhao X., et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci. Adv. 2017;3:eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All relevant data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.