Abstract
Background
The hyperarid core of the Atacama Desert is an extremely harsh environment thought to be colonized by only a few heterotrophic bacterial species. Current concepts for understanding this extreme ecosystem are mainly based on the diversity of these few species, yet a substantial area of the Atacama Desert hyperarid topsoil is covered by expansive boulder accumulations, whose underlying microbiomes have not been investigated so far. With the hypothesis that these sheltered soils harbor uniquely adapted microbiomes, we compared metagenomes and geochemistry between soils below and beside boulders across three distantly located boulder accumulations in the Atacama Desert hyperarid core.
Results
Genome-resolved metagenomics of eleven samples revealed substantially different microbial communities in soils below and beside boulders, despite the presence of shared species. Archaea were found in significantly higher relative abundance below the boulders across all samples within distances of up to 205 km. These key taxa belong to a novel genus of ammonia-oxidizing Thaumarchaeota, Candidatus Nitrosodeserticola. We resolved eight mid-to-high quality genomes of this genus and used comparative genomics to analyze its pangenome and site-specific adaptations. Ca. Nitrosodeserticola genomes contain genes for ammonia oxidation, the 3-hydroxypropionate/4-hydroxybutyrate carbon fixation pathway, and acetate utilization indicating a chemolithoautotrophic and mixotrophic lifestyle. They also possess the capacity for tolerating extreme environmental conditions as highlighted by the presence of genes against oxidative stress and DNA damage. Site-specific adaptations of the genomes included the presence of additional genes for heavy metal transporters, multiple types of ATP synthases, and divergent genes for aquaporins.
Conclusion
We provide the first genomic characterization of hyperarid soil microbiomes below the boulders in the Atacama Desert, and report abundant and highly adapted Thaumarchaeaota with ammonia oxidation and carbon fixation potential. Ca. Nitrosodeserticola genomes provide the first metabolic and physiological insight into a thaumarchaeal lineage found in globally distributed terrestrial habitats characterized by various environmental stresses. We consequently expand not only the known genetic repertoire of Thaumarchaeota but also the diversity and microbiome functioning in hyperarid ecosystems.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-021-01177-9.
Keywords: Atacama, Hyperaridity, Archaea, Soil microbiome, Xerotolerance
Background
Deserts are widespread terrestrial ecosystems, where aridity limits the proliferation and diversification of life [1]. Despite the paucity of macrofauna in desert ecosystems, previous studies (e.g., on the Sahara Desert [2, 3], the Namib Desert [4], and the Antarctic Dry Valley [5–7]) detected highly adapted microbial communities, suggesting that the major bioprocesses in deserts are driven by microbial life [8]. The Atacama Desert is the oldest nonpolar desert on Earth [9] and its long history of hyperaridity has resulted in the accumulation of atmospheric salts in its soils [10]. In particular, the surface soils in the hyperarid core [11] of the Atacama Desert are characterized by extreme desiccation (water content < 1% by weight), low water activity as a consequence of high salt content, and high UV irradiation (~ 30 J·m−2) [12]. Low yields of DNA have been isolated and analyzed in previous studies [12–15] revealing sparse microbial communities with low diversity, dominated by Actinobacteria and Firmicutes [10]. While recent studies showed that some of these microbes are viable and actively replicating, as indicated by cultivation experiments [13] and in situ replication measures (iRep [16];) [12] respectively, very little is known about the carbon and nitrogen cycling in the hyperarid soils of the Atacama Desert. To date, only localized carbon fixation could be inferred from the findings of hypolithic and endolithic cyanobacteria [17, 18], but no information on possible pathways for the transformation of other nutrients has been obtained so far.
Thaumarchaeota mediate important environmental processes in both marine and terrestrial ecosystems and are particularly adapted to oligotrophic environments with their highly energy-efficient carbon fixation pathway [19, 20]. Thaumarchaeota have been detected in hot desert soils (e.g., Mojave Desert, California and Chihuahuan Desert, New Mexico) [21]. However, previous studies reported the general pattern of decreasing archaeal diversity with increasing aridity [22, 23] and most in-depth desert microbiome surveys focused on bacterial communities [24, 25]. In particular, microbiome studies of hyperarid deserts in Antarctica reported the absence [24, 26] or low abundance [27] of Archaea suggesting lower tolerance of Archaea to hyperaridity. The Atacama Desert soil microbiome has previously been thought to be dominated by Bacteria, with an exception of halophilic Archaea (Halobacteriales) in locations such as coastal soils [12] and salt crusts [28]. Thaumarchaeal 16S rRNA gene sequences have been detected in the Atacama region, such as in playas and alluvial fans after a heavy rainfall [29], and in high-elevation mineral soils of Volcán Llullaillaco [30]. However, the metabolic potential, adaptations and the abundance (relative to Bacteria) of these Thaumarchaeota could not be resolved in the aforementioned studies.
The Atacama Desert hyperarid core harbors many expansive boulder fields [31–33] where individual boulders are up to 37 millions of years old [34, 35]. Despite their ubiquity and the uniquely sheltered conditions beneath the boulders in the Atacama Desert, no study has determined the microbial and geochemical compositions of soils below the boulders. We compared the metagenomes and geochemistry of soils below and beside the boulders, with the hypothesis that extended periods of physical shelter and isolation by the boulder cover would result in distinct ecosystems that support a unique composition of organisms protected from harsh environmental stressors (i.e., radiation and resulting reactive oxygen species [ROS], temperature and humidity fluctuations, deposition of salts). Community structure and metabolic potentials inferred from genome-resolved metagenomics were interpreted in conjunction with geochemical measurements in order to characterize and compare the ecological significance of microbiomes found in the two sample types. Thaumarchaeota were one of the key taxa differentiating microbial communities inhabiting below and beside the boulders. Consequently, thaumarchaeal genomes were selected for an in-depth pangenomic analysis, revealing their potential for carbon and nitrogen cycling. We further compared them to their closest known relatives and to each other, to unveil possible adaptations to these uniquely protected, sparsely populated, and constantly selective environments.
Results and discussion
Hyperarid soils sheltered under the boulders are geochemically distinct and organic carbon deficient
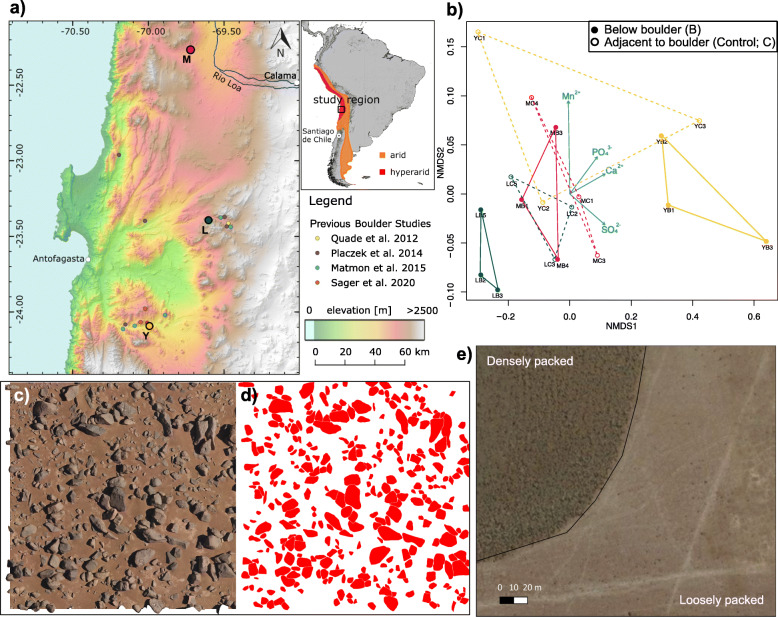
Boulder accumulations in the Atacama Desert are both frequent and expansive (see Supplementary Results R1 and Figures S1-3 for estimates of boulder coverage at different resolutions). Previous geomorphological studies [31–33] (Fig. 1a) have hypothesized a seismic origin of these boulder accumulations typically found at the base of hills and valley floors, both of which are abundant landforms in the hyperarid core (Figure S3) [36]. We used satellite images to map the presence of boulder accumulations in regions within 5 km to our sampling sites and estimated between 16 and 31% in each of the studied regions to exhibit boulder accumulations of various densities (Figure S2). For instance, in densely packed regions the boulders covered up to 21% of the topsoil (Fig. 1c–e). Soils below the boulders are subject to lower diurnal temperatures and lower fluctuations in relative humidity than those directly beside the boulders (Figure S4a-c). Based on the dew point temperature calculations, we showed that the condensation in the morning is far less likely for soil below boulders compared to soil beside boulders (Figure S4d-f), suggesting that water content below boulders may be even lower than in previously studied Atacama Desert hyperarid top soils (~ 0.2% by weight) [12].
Fig. 1.
Sampling location and soil geochemistry. a Location of three sampling sites and their abbreviations in parentheses. Locations of previously studied boulder fields are mapped and their references are shown in the legend. Top-right corner maps the arid and hyperarid climate regions across South America [11]. b Non-metric multidimensional scaling (NMDS) ordination (stress = 0.024) of anion and cation concentrations in each soil sample. Different colors represent different sampling sites (green = L, red = M, yellow = Y). Filled vs unfilled data points correspond to the sample type information (below (B) or beside boulder (C)). Blue vectors represent fitted ion species onto the ordination with adjusted p-value < 0.05. c An example drone image used to map individual boulders at high resolution; raw data collected by Sager et al. [33] and reanalyzed in this study. Image frame corresponds to 15 x 15 m. d Corresponding mapping of individual boulders imaged in c, same scale. e Satellite image distinguishing “densely” and “loosely” packed boulder accumulations
We compared soil samples of two sample types: soils taken below boulders (B) and soils taken beside the boulders (control, C) at three different sampling locations (Lomas Bayas: L, Maria Elena: M, Yungay Valley Y) (Fig. 1a, Table S1). While the collected soils were mineralogically very similar with some variation per sampling location (Figure S5), their ion concentrations showed large variance between boulder fields, individual boulders, and sample types. Interestingly, B samples clustered based on their sampling location along the NMDS1 axis, while C samples showed little pattern based on their sampling sites (Fig. 1b). In general, samples from locations L and M were enriched in F−, while Y samples were enriched in PO43−, SO42−, Mn2+ and Ca2+, suggesting boulder field specific patterns of ion concentrations. More sampling location dependent ion composition patterns among the B samples indicate that soils below the boulder are sheltered from external input of ions (i.e., atmospheric deposition of salts), thereby exhibiting a more representative ion composition patterns of the underlying soils in the sampling site. When comparing the B and C sample of each individual boulder, nitrate ion concentrations were significantly lower in B samples compared to C samples (paired Welch’s t-test; NO3−: t(8) = − 3.9, adjusted p-value = 0.027, Figure S6). Total Organic Carbon (TOC) concentrations were at or below detectable levels in both below boulder and beside boulder samples (Figure S7). Our results show that the soils below the boulders are not only hyperarid and organic carbon deficient, but also sheltered from the atmospheric input of both water (e.g., fog, dew) and salts.
The microbial community in the Atacama Desert hyperarid core shows evidence of in situ replication
We conducted genome-resolved metagenomics on the eleven successfully prepared metagenomes (for details, see Material and Methods and Table S2): three B and three C samples came from Lomas Bayas (LB2, LB3, LB5 and LC2, LC3, LC5), three B samples were from Maria Elena (MB1, MB3, MB4) and two B samples from Yungay Valley (YB1, YB3). Across these eleven samples, we binned 67 medium to high quality (> 75% completeness, < 10% contamination) metagenome-assembled genomes (MAGs). Eight of these genomes were thaumarchaeal with completeness ranging from 84.67 to 98.54%. The other 59 mid-to-high quality MAGs belonged to Actinobacteria (n = 28), Chloroflexi (n = 29), Firmicutes (n = 1), and Alphaproteobacteria (n = 1) (see concatenated protein trees of mid-to-high bacterial MAGs in Figure S8 and archaeal MAGs in Fig. 4). In situ replication measures [iRep [16]] and GRiD [37] were successfully calculated for 30 and 36 out of all mid-to-high-quality bacterial genomes (n = 59), respectively, indicating an active metabolism of the majority of the indexed population (filtered iRep values ranged between 1.34 and 3.47, median: 1.89; refined GRiD values ranged between 1.13 and 3.12, median: 2.07; for comparison, see Figure S9). On average, genomes recovered from below boulder metagenomes were associated with slightly higher iRep and GRiD values than the control metagenomes of the same site (p-value < 0.05, Welch’s t-test for LB and LC MAGs). A full overview of genome statistics, their taxonomic classification and corresponding iRep and GRiD values is provided in Table S3.
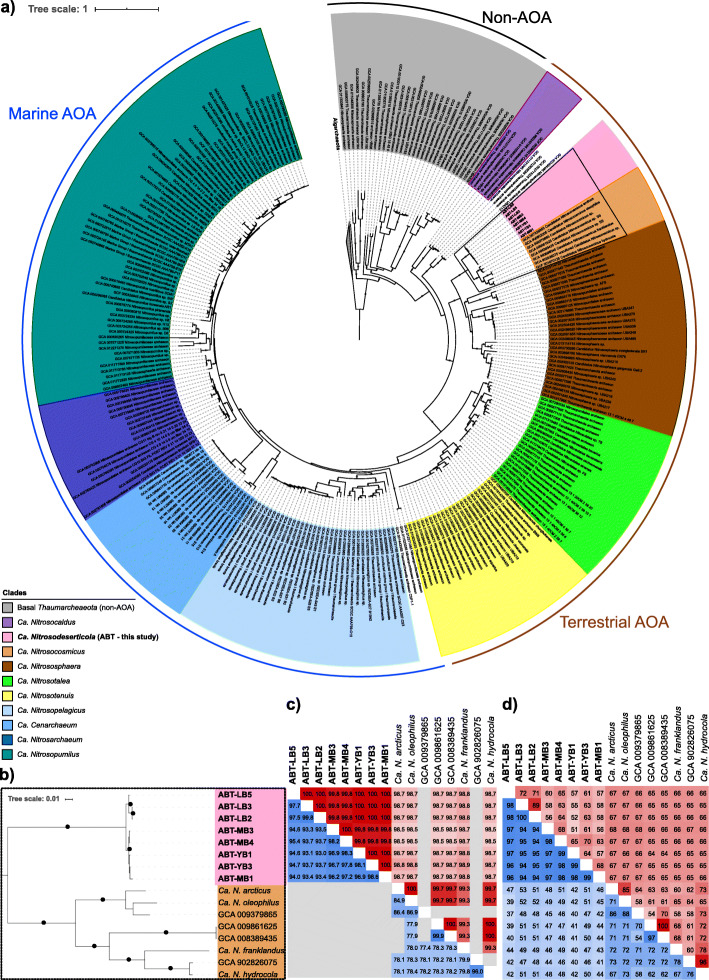
Fig. 4.
Phylogenomic placement of ABT genomes using 37 housekeeping single-copy genes. a Phylogenetic tree of 298 NCBI genomes annotated as Thaumarchaeota and eight ABT genomes. Aigarchaeota were identified and used as the outgroup. Black, brown and blue ranges distinguish whether organisms are ammonia-oxidizing Archaea (AOA) and their typical habitats (terrestrial vs marine). Strongly supported branches as described in the Materials and Methods section are indicated with black dots. b Magnified view of the branches placing the ABT (Ca. Nitrosodeserticola) genomes and its sister group Ca. Nitrosocosmicus. Strongly supported branches as described in the Materials and Methods section are indicated with black dots. c Lower-left (blue) triangle of the matrix corresponds to FastANI between genomes, where gray values indicate below calculation threshold (80% identity). Upper-right (red) triangle of the matrix corresponds to 16S rRNA gene identity values, where gray values are used for genomic bins without a 16S rRNA gene. d Lower-left (blue) triangle corresponds to the amino acid identity (AAI) and upper-right (red) triangle corresponds to the Orthologous Fraction (OF) between a pair of compared genomes
Atacama soils below boulders harbor unique microbial communities with a high relative abundance of Thaumarchaeota
We detected 147 different bacteria and Archaea based on clustering of S3 ribosomal proteins (RpS3, 99% identity, Table S4, Figure S10). Species evenness and alpha diversity were similar across samples with no statistically significant differentiation between sites or sample types (Figure S11). However, principal coordinate analysis (PCoA) of the communities (Figure S12a) demonstrated clustering of samples based on sample site (L, M, Y) as well as sample type (B, C). This was corroborated by the Multiple Response Permutation Procedure (MRPP) indicating significant influence on the community structure by both sampling location (chance corrected within group agreement A = 0.2648, significance of delta = 0.001) and sample type (A = 0.1488, significance of delta = 0.002). Using BioENV [38], we identified F− concentration to be most correlated (Spearman’s rho = 0.582) with the community composition. F− has been shown to be correlated with microbial community structures in previous Atacama soil studies [29], as well as in other environments such as groundwater [39]. Despite the low concentrations of F− (~5 mg/g soil for values above the detection limit) across soils, we identified on average 15 (± 3) putative fluoride transporters CrcB per metagenome suggesting that fluoride detoxification may play a role in shaping the Atacama Desert microbial community. Additionally, we conducted a NMDS analysis (Figure S12b), identifying additional ions (Ca2+, SO42−) that could be correlated with the community composition (adjusted p-value < 0.05).
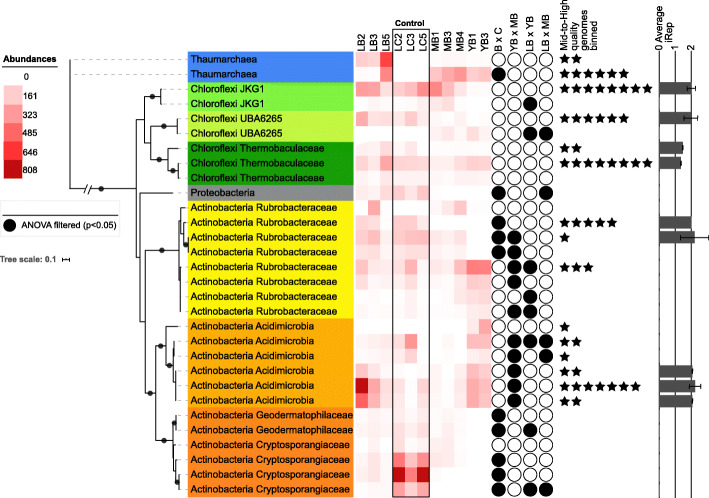
Thirty-three of the identified taxa differed significantly in their abundances (ANOVA [40], p-value < 0.05) between B and C samples. Such taxa included Actinobacteria (belonging to Cryptosporangiaceae, Streptomycetaceae, and Geodermatophilaceae), as well as one Alphaproteobacteria (Acetobacteraceae). These taxa were particularly abundant in C samples and nearly absent in B samples, suggesting specific and unknown selection processes for the two different sample types. Alternatively, some of these taxa may be deposited through aeolian transport [41]. Figure 2 shows the phylogenetic relationship among the top 30 most abundant taxa across the samples based on RpS3 proteins and links them to their respective MAGs as well as their differential coverage across the samples. We conclude that below boulder (B) and beside boulder (C) present substantially different habitats of the same ecosystem.
Fig. 2.
Phylogenetic tree of 30 most abundant taxa (rpS3 clusters) out of 147 and their normalized abundances across all samples. Filled stars represent the number of rpS3 sequences in the cluster that were successfully binned in mid-to-high quality genomes and gray bars indicate the average iRep values calculated for the mid-to-high quality genomes. Strongly supported branches as described in the “Materials and Methods” section are indicated with black dots
A thaumarchaeal OTU was the only taxon with higher abundance in below boulders samples than in control samples (ANOVA, p-value = 0.0396). All eight metagenomes constructed from samples retrieved below the boulders contained high abundances of Thaumarchaeota. Based on the ranked abundance of rpS3 gene coverages (Figure S13), Thaumarchaeota ranked among the top seven most abundant taxa across all B samples. Based on rpS3 gene coverages, the relative abundance of Thaumarchaeota varied between 4.5 and 34.6% across below boulders samples, with an average of 13.5%. In three samples (MB3, MB4, LB5), Thaumarchaeota were the most abundant organisms, e.g., in LB5, Thaumarchaeota were four-fold more abundant than the second most abundant taxon. The abundance of Thaumarchaeota under boulders and their near absence in control samples parallel the previous findings from marine environments, where thaumarchaeal abundance was observed to be anti-correlated with irradiation in the surface waters (e.g., increase in thaumarchaeal abundance in winter and decrease in summer, particularly in polar regions) [42–46]. To date, the underlying reason for the lower abundance of Thaumarchaeota in highly irradiated environments remains inconclusive. Three main hypotheses have been proposed for this phenomenon: (i) increased competition against phototrophs [44, 47], (ii) Photoinhibition of ammonia oxidation [48–50], and iii) indirect photoinhibition by reactive oxygen species (ROS) (i.e., hydrogen peroxide) [51]. When applying the first hypothesis to our study site, we posit that the near absence of Thaumarchaeota in the control samples is not due to the increased competition against phototrophs, based on the lack of phototrophs detected in the C samples. The second hypothesis is also not applicable for our study site as UV and photoradiation do not penetrate into the soil beyond the very surface of the topsoil, especially in the Atacama Desert where soils experience minimal perturbation. The third hypothesis, however, may be relevant to our study site; Although we did not directly measure ROS levels in our samples, photochemically produced ROS (H2O2 and metal superoxides and peroxides) have previously been found to accumulate in the Atacama Desert (Yungay site) top soils at levels an order of magnitude higher than in non-arid control soils [52]. Considering the limited half-life of ROS, we hypothesize that soils that have been covered under boulders for hundreds, if not thousands, of years harbor significantly lower concentrations of ROS. Further investigation correlating concentrations of specific ROS below and beside boulders with thaumarchaeal abundance (and activity) could explain the causal relationships between ROS levels [51] and thaumarchaeal abundance.
Ammonia-oxidizing Thaumarchaeota occupy an important niche in the Atacama Desert carbon and nitrogen cycling
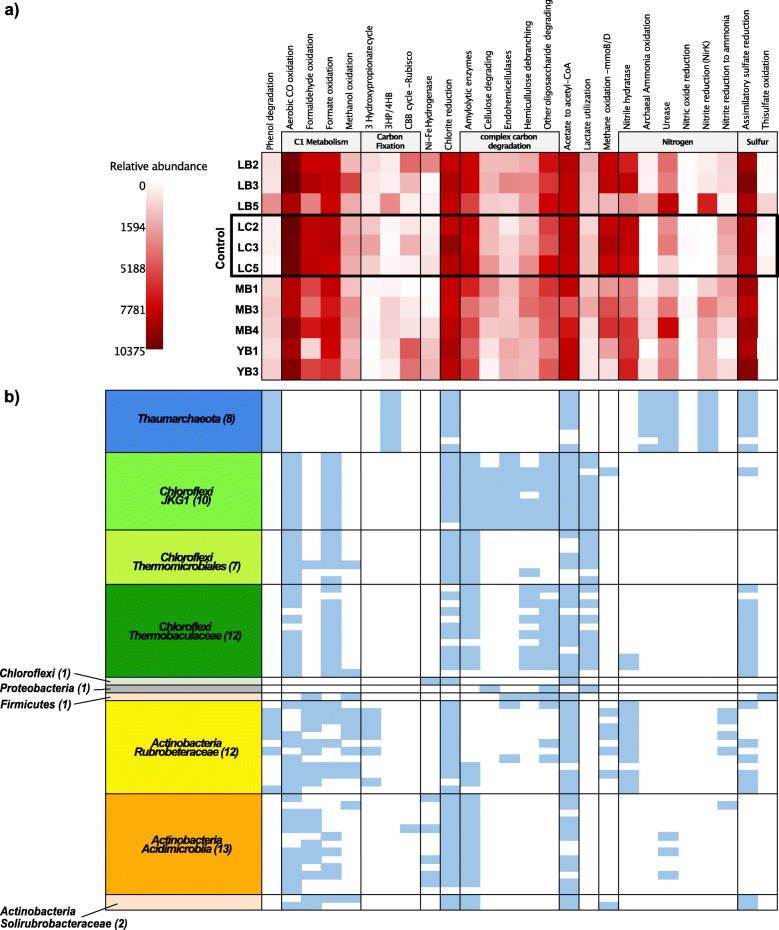
Relative abundances of key marker genes in the assembled metagenomes revealed the potential for carbon fixation, C1 metabolism, complex carbon degradation, and fermentation across all samples (Fig. 3). Notably, we detected eleven Form I RuBisCO sequences across ten out of eleven metagenomes. RuBisCO large subunits were mostly (nine out of eleven) situated next to a small subunit on very short contigs containing between one and five genes with an exception in the LC5 metagenome (Figure S14a). Regions that are difficult to assemble into longer contigs may exhibit differential kmer frequencies from the rest of the genome resulting from horizontal gene transfers, which has been observed to occur frequently for RuBisCO subunits [53, 54]. The RuBisCO large subunit sequences formed a novel subclade of type IA and were closely related to recently discovered RuBisCOs from the Negev desert [55] (Figure S14b). This phylogenetically distinct grouping of RuBisCO sequences from distantly located desert environments suggests that there exists a desert-specific ecotype of RuBisCO. We did not detect any key bacterial photosynthesis marker genes (e.g., psaA, psbA); instead, we detected 20 additional homologs of actinobacterial-type (group 1h) respiratory H2-uptake NiFe hydrogenases across all metagenomes. The presence and high abundance of RuBisCOs of subtype 1E and high-affinity Ni-Fe hydrogenase suggest that the hydrogen-driven CBB cycle may play a key role in carbon fixation in extreme conditions of the hyperarid desert soils where photosynthesis is inhibited. Our results complement recent findings by Bay et al. [55] that showed increasing H2 oxidation with increasing aridity in incubation experiments, providing evidence that hydrogenotrophic carbon fixation is more prevalent than photosynthesis in dry soils, similar to the novel chemosynthetic subclade IE RuBisCOs in antarctic soil [56] (Figure S14cd). In addition to the CBB pathway, we detected 3 hydroxypropionate cycle and 3HP/4HB pathway marker genes in our samples. The marker genes for the three carbon fixation pathways were differentially abundant between B and C samples. For instance, B samples contained higher relative abundances of 3HP/4HB pathway marker genes and lower relative abundances of 3-hydroxypropionate cycle marker genes than C samples (Kruskal-Wallis test, p-value = 0.012). Additionally, 3-hydroxypropionate cycle marker gene abundances were correlated with NO3−, Mg2+, and K+ ion concentrations (Pearson’s R = 0.75 − 0.78, adjusted p-value < 0.05).
Fig. 3.
Metabolic potential prediction across samples and mid-to-high quality genomes. a Relative abundances of chemoautolithotrophic marker genes predicted using METABOLIC for each sample. b Presence (blue) and absence (white) of chemoautolithotropic marker genes in mid-to-high quality genomes. Genomes are clustered based on taxa and the number of genomes in each cluster is shown in parentheses in the row names
Significant gaps in the potential for nitrogen cycling were observed. Archaeal ammonia oxidation potential was only found in below boulder samples (with the exception of YB1). Additionally, we found diverse sets of actinobacterial and chloroflexal ammonification proteins (Table S6). We found norB homologs in four out of eleven samples, but could not detect any norC in the same samples, possibly due to low abundance. No potential for nitrogen fixation, nitrate reduction as well as nitrite oxidation were identified in the metagenomes, indicating that these are not major processes contributing to the biological nitrogen cycling. The lack (or near absence) of nitrogen fixation and denitrification genes suggests low overall biological input and little loss of biologically available nitrogen. Although the investigated soils are known to be enriched in nitrates (particularly at ~ 1-m depth) that have accumulated over millions of years through abiotic processes (e.g., atmospheric formation through lightning followed by dry deposition and rainwater infiltration) [57], below boulder nitrate concentrations were significantly lower (Figure S6), likely due to the combined effect of microbial turnover and lack of atmospheric or hydrologic input.
Resolving the metabolic potential at the genomic level delineated the role each taxonomic group plays in these microbial communities characterized by low species diversity. The presence and absence of key metabolic genes for each mid-to-high quality genome are shown in Fig. 3b. Our analysis shows that although all samples show carbon fixation potential, taxa capable of fixing carbon are limited to Thaumarchaeota (through the 3HP/4HB pathway) and some Actinobacteria (3-hydroxypropionate pathway). Notably, one RuBisCO Form I (subtype 1A) could be binned to a high-quality Acidomicrobiia genome also containing a high-affinity H2-uptake Ni-Fe hydrogenase (group 1h). RuBisCO sequences detected in our study were also closely related to those binned in Acidimicrobiia MAGs (also encoding Ni-Fe hydrogenases) assembled from the Negev desert soil metagenomes (Figure S14b) [55], providing additional confirmation for the taxonomic assignment. Chloroflexi genomes associated with the lineage JKG1 had the broadest potential of degrading complex carbon, suggesting a fermentative lifestyle, while Actinobacteria harbored genes involved in metabolizing a wide range of C1 substrates. Nitrite reduction potential detected in below boulder sites was constrained to the nirK genes found in Thaumarchaeota. NirK in Thaumarchaeota has been hypothesized to play a key role in ammonia oxidation [58, 59] and is biochemically capable of transforming N compounds to produce nitric oxide [60]. Furthermore, thaumarchaeal genomes indicated a potential for mixotrophy, featuring ubiX genes involved in phenol metabolism, as well as acdA and acs involved in acetate metabolism. It is important to note that while the presence of the key marker genes can be used to deduce the metabolic potential of these organisms, it cannot alone determine the functional physiology without further in situ measurements (i.e., gene and protein expression, stable isotope probing). In sum, our results indicate conserved metabolic capacities across genomes that belonged to the same taxonomic family, with thaumarchaeal genomes carrying unique metabolic pathways that could contribute to poorly understood nitrogen and carbon cycling in the Atacama hyperarid core.
A novel genus of Thaumarchaeota with highly conserved core genome and diverse auxiliary genes
Eight mid-to-high quality Atacama Boulder Thaumarchaeal genomes (ABT) were assembled with an average GC content of 34.6% (± 0.1%) and average size of 2.5 Mbps (± 0.4 Mbps). Each genome contained on average 3,123 (± 579.7) predicted genes with a mean coding density of 71.8% (± 1.7%). The genomes were phylogenetically placed using 37 single-copy house-keeping genes, forming a monophyletic sister cluster to the recently characterized Ca. Nitrosocosmicus (Fig. 4a). The ABT clade and Ca. Nitrosocosmicus form a sister group to Ca. Nitrososphaera, a mesophilic terrestrial clade. Genomes from the same sites were more related to each other, with ABT-MB and ABT-YB genomes forming a separate branch from the ABT-LB genomes (Fig. 4b). Five ABT genomes contained a copy of the ammonia monooxygenase A (amoA) gene each (Table S6). On closer examination, two other genomes (ABT-MB3, ABT-MB4) contained conserved amoA regions, which failed in protein prediction due to ambiguous bases in scaffolding. No amoA sequences were found in ABT-YB1. Three additional unbinned amoA genes were detected across the metagenomes (MB3, MB4, YB3). Altogether, the eight amoA nucleotide sequences were 100% identical in their amino acid sequences to each other and to previously published amoA sequences from Ca. Nitrosocosmicus oleophilus and Ca. Nitrosocosmicus hydrocola, which had been phylogenetically identified to be one of the basal clades of archaeal amoA after Ca. Nitrosocaldus [61]. Figure S15a resolves the nucleotide level phylogenetic placement of binned amoA sequences as well as unbinned amoA sequences recovered from the sample metagenomes. Interestingly, one amoA recovered from a low-quality bin (68% completeness; 5.8% contamination) in the YB3 metagenome (node “ABT-YB3 (low-quality bin)” in Fig. S15a) was divergent (~ 80% ID) from the rest at the nucleotide level, while 95.8% identical to other ABT and Ca. Nitrosocosmicus amoA genes at the amino acid level. The rpS3 gene recovered from this bin was classified as thaumarchaeal, with 75% identity to other binned rpS3 in ABT, and its closest NCBI reference sequence being Ca. Nitrosocosmicus sequences at 65% identity. This divergent thaumarchaeal bin was approximately three-fold less abundant than another thaumarchaeal bin (ABT-YB3) recovered at a higher quality from the same metagenome (YB3). Due to the low quality and lower abundance of this divergent bin, our study focuses on other eight mid-to-high quality genomes that are much more closely related and found across all metagenomes under the boulders including YB3.
In order to taxonomically resolve the eight recovered Thaumarchaeota genomes, we compared them to Ca. Nitrosocosmicus genomes that had been isolated or metagenomically assembled from diverse environments from around the world (Table S7) ranging from the arctic soil [62], tar-contaminated soil [63], vegetable field [64], dinosaur fossil [65] to wastewater filters [66]. High ANI (93.0–99.8%) (Fig. 4c) between ABT genomes indicated that all ABT genomes belong to one genus. Using the ANI threshold of 95% [67, 68] for species delineation, we identified two species within the ABT clade, with genomes recovered from the LB site belonging to one species and the rest to another. The mean amino acid identity (AAI) of 53.9% between pairs of Ca. Nitrosocosmicus and ABT genomes (Fig. 4d) fell below the genus delineation threshold of 65% [69] indicating that the two clades form separate genera. Based on these findings, we propose two new species names that belong to a new genus: Ca. Nitrosodeserticola atacamae (ABT-LB2, ABT-LB3, ABT-LB) and Ca. Nitrosodeserticola subpetralis (ABT-MB1, ABT-MB3, ABT-MB4, ABT-YB1, ABT-YB3). Surveys of the recovered 16S rRNA gene in the NCBI nr database using 99% identity cut-off [70, 71] detected close relatives (no exact matches were found) of the Ca. Nitrosodeserticola genomes in diverse locations around the globe such as chromite mine in Iran, alkaline-saline soil of a former lake in Mexico [72], and high-altitude tuff in a Tibetan desert, Armenian hot spring sediment [73], high-elevation soils from a volcano in the Atacama region [74], deglaciation soils in Australia [75], and uranium-contaminated subsurface sediments in the USA [76] (Figure S15b). Our results indicate that close relatives of Ca. Nitrosodeserticola are widespread, particularly in soils that experience environmental stress. We also searched for the Ca. Nitrosodeserticola amoA genes in the NCBI nr database using 97% identity cutoff; however, only one sequence was detected from a Tibetan lake [77] at 97.5% identity.
While the eight Ca. Nitrosodeserticola genomes share a highly conserved core genome (mean AAI = 96.5 %), between 11% and 49% (mean = 37.7%) of the genes had no other orthologs in the recovered genomes despite the relatively similar and static environmental conditions that they were found in. High AAI in the orthologous fraction of the eight Ca. Nitrosodeserticola genomes and conserved amoA sequences recovered in sites more than 200 km apart from each other suggest that these organisms originated from the same strain of Thaumarchaeota.
Pangenomic comparison of ABT genomes and their sister clade reveals unique adaptations including heavy metal resistance, biofilm formation, water transport, and sodium bioenergetics
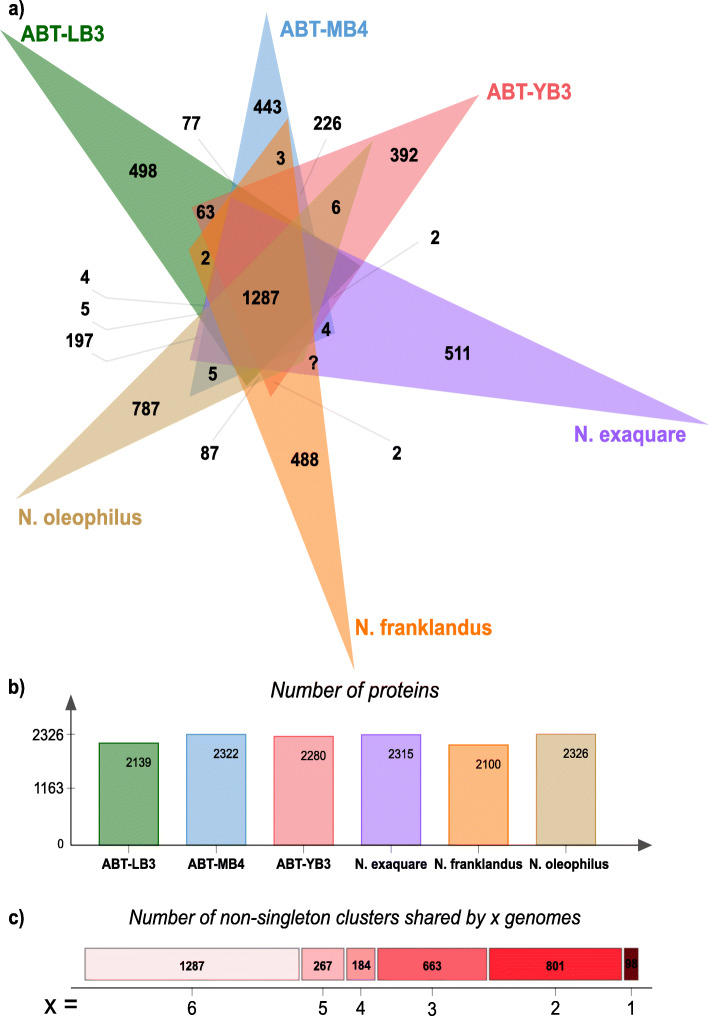
In order to understand the conserved metabolic potentials between ABT and Ca. Nitrosocosmicus, unique adaptations of the ABT in the Atacama Desert, and niche differentiations between sites, we analyzed the high quality (> 95% completeness, <5% contamination) genomes (ABT-LB3, ABT-MB4, ABT-YB3) from each of the three sites along with three (near)-complete Ca. Nitrosocosmicus reference genomes (Ca. N. franklandus, Ca. N. oleophilus, Ca. N. hydrocola). 1287 homolog clusters are shared across all six genomes (Fig. 5, Table S7). For example, all genomes contained a highly conserved AmoABX operon, although only two out of eight ABT bins contained 1–2 amoC copies. All genomes revealed the metabolic potential for mixotrophy along with important metabolic capacities for nitrogen turnover in these systems, including genes for copper-dependent nitrite reductase (nirK), urease (ureC), urea transporter, ammonium transporter, deaminases, and lyases for ammonification (Table S8). Additionally, among the shared genes, we found stress response genes such as antioxidant genes (superoxide dismutase and alkyl hydroperoxide reductase), detoxification genes (cld and arsC12 involved in chlorite and arsenate reduction, respectively), as well as bacterial-type DNA repair genes (radA, radB, xpf, herA, nreA, uvrABC) and other resistance genes (dnaJ, putative oxidoreductases) (Table S7). These stress resistance genes were found to be conserved and more prevalent in the terrestrial AOA clade than the marine counterparts [78] and a previous study [79] of the terrestrial AOA genome expansion proposed that this extensive suite of stress resistance genes acquired early in the terrestrial AOA evolutionary history provided the basis for their successful colonization of diverse niche terrestrial environments.
Fig. 5.
Shared and auxiliary protein clusters of ABT and its sister genus. a Shared orthologous protein clusters (including singletons) across six genomes (three Ca. Nitrosocosmicus, three Ca. Nitrosodeserticola [ABT]). b Number of proteins in each genome. c Number of orthologous protein clusters (excluding singletons) shared across x number of genomes
We found 296 protein clusters that were shared between ABT genomes but not present in Ca. Nitrosocosmicus genomes (Fig. 5). Interestingly, we found extracellular polymeric substances (EPS) and biofilm production genes that were specific to ABT genomes (Table S7). These EPS and biofilm production genes found in the ABT genomes appear to share little homology with genes in Ca. N. oleophilus that has previously been shown to form biofilms [63], and suggest recent gain/loss events of the EPS genes following the branching of Ca. Nitrosocosmicus and ABT lineages. EPS production and biofilm formation in general are considered major adaptation mechanisms for xerotolerant bacteria [80] and ABT genomes may also employ this mechanism to protect against desiccation. Interestingly, we did not find any genes for the production of compatible solutes (i.e., trehalose, glycine-betaine, ectoine) in any of the ABT genomes, despite it being a well-studied strategy against desiccation and high salt environments [78, 81, 82].
Of the protein-coding genes found in each of the three high-quality ABT genomes, 12.8 to 16.5% belonged to unique protein clusters or were singletons (Fig. 5, Table S9) that did not share any similarity to genes in the other ABT genomes. Among singletons of the genome ABT-LB3, many were involved in membrane transport of metals, such as magnesium, copper, and cobalt transporters as well as lead, cadmium, zinc, and mercury-transporting ATPases, potassium uptake proteins, and bacterioferritin. The presence of these genes may be an adaptation to heavy metals known to accumulate in the Atacama Desert soils [83]. Similarly, notable singletons found in genomes ABT-MB4 and ABT-YB3 included putative cobalt transporter, fluoride transporters, zinc uptake system, mercuric reductase, ferrous iron permease, and phosphite transport system (Please see Supplementary Results R3 for further findings from the pangenome analysis).
Further genome comparisons across all eight ABT genomes revealed additional key adaptations to desiccation and osmotic stress. We identified up to seven different copies of water channel membrane proteins (aquaporin Z2) [84] per genome. Interestingly, some of these proteins were highly divergent from each other at the AA sequence level, while others were truncated (Figure S17) despite being found mid-contig with relatively conserved surrounding genes. Multiple copies of aquaporin genes per genome as well as the divergent and truncated subset indicate possible genome-specific adaptations to desiccation and osmotic stress. We also recovered two distinct types of ATP synthases (namely A-type and V-type [85, 86]) from the eight ABT genomes. Three ABT genomes (ABT-LB2, ABT-LB3, ABT-YB1) contained only A-type ATP synthase, while the rest contained both the A-type and the V-type ATP synthases often in multiple copies. Wang et al. [85] concluded that the V-type ATP synthases were horizontally transferred from Euryarchaeota to Thaumarchaeota and conserved among the acidophilic and hadopelagic Thaumarchaeota, potentially playing a key role in their adaptations to acidic environments and elevated pressure through proton extrusion. Considering that Atacama Desert soils are slightly alkaline (average pH = 7.7 Figure S18), it is surprising that the V-type ATP synthase is found and conserved across five ABT genomes. The hypothesis by Zhong et al. [86] proposing that V-type ATP synthases may be coupled with Na+ motive force instead of proton pumping, seems to be a more probable scenario for the ABT. Indeed, Atacama Desert soils present high salt stress, and therefore the V-type ATP synthase could perform Na+ pumping and provide protection against high sodium stress (Table S10). In order to understand possible ecological implications of site-specific strains of ABT in soils of varying geochemistry, we created eight genome-based metabolic models using sample-specific geochemical information and ABT genomes binned from the respective samples (see Supplementary Methods M7 for detail) and our results suggest that the geochemical heterogeneity between samples coupled with the flexibility in ABT genomes result in different potentials of N and C flux ratios in each sample, with possible signatures of differentiation by site (Figure S17). Our modeling efforts provide directions for further in situ measurements and experimentation to characterize different levels of C and N turnover by Ca. Nitrosodeserticola in the Atacama.
Conclusions
We report here the first evidence of highly adapted ammonia-oxidizing Thaumarchaeota inhabiting the soil beneath the expansive boulder accumulations of the hyperarid Atacama Desert in high relative abundance. This is also the first systematic comparison of microbial communities found below boulders of the Atacama Desert hyperarid core with the microbial communities present in the open, unprotected desert soil. Our study revealed the remarkable adaptability and resilience of Thaumarchaeota, expanding the physical limits of thaumarchaeal habitat range to include hyperarid, high-salt, and extremely low-carbon environments. In-depth characterization of these Ca. Nitrosodeserticola genomes suggests their potential niche roles in N and C cycling in highly nutrient-deficient Atacama Desert soils, as well as resilience against oxidative stress and hyperaridity. We compared eight closely related Ca. Nitrosodeserticola genomes were retrieved from these isolated and disconnected habitats and found that they harbor highly conserved shared genes and large numbers of site-specific auxiliary genes. Our results indicate genomic plasticity of Ca. Nitrosodeserticola, whose closest relatives have been found across the globe in terrestrial environments characterized by high oxidative stress and high toxicity. Beyond the Atacama Desert, this study provides a blueprint for future studies of extreme terrestrial environments (i.e., Antarctic and extraterrestrial) where finding pockets of pristine, sheltered, and contained environments, as simple as below boulders, could lead to a discovery of uniquely adapted organisms.
Material and methods
Sampling location and procedure
Sampling was conducted in March 2019, during a dry period with the last recorded rain event occurring in June 2017 in the Yungay region. Three sampling sites, Y, M, and L, were chosen based on a previous study [12] that identified inland hyperarid sites using the threshold of water content < 1% by weight (Fig. 1a). The coordinates of the three sample sites can be found in Table S1. Sampling was conducted in previously described characteristic boulder fields [31, 32]. At each boulder field, six boulders of diameter ~ 50 cm and height ~ 20 cm were chosen within a radius of ~100 m from each other. For each boulder, two types of samples were taken: one below boulder (B) and one control sample (C) in the open soil ~ 10 cm away from the boulder (Figure S19), to compare the effect of physical shelter and isolation provided by the boulder on soil microbiomes while minimizing the effect of the spatial heterogeneity in soil geochemistry and mineralogy as observed in previous studies [15, 87]. All chosen boulders were well distanced from other boulders to make sure that the C samples were never constantly shadowed by the sampled boulder or other boulders. In total, 36 samples were collected, twelve (six pairs of B and C samples) across three sampling sites. Samples were collected aseptically from the top 1-cm layer of soil into sterile 50-ml falcon tubes ( ~ 60 g per sample), which were then flash frozen in a liquid nitrogen dry shipper within half an hour of sampling. Control soil samples were taken first and then boulders were flipped over to sample below boulder soil as soon as possible to avoid aerial contamination. Additional samples were taken for geochemical analyses with a small shovel ( ~ 100 g per sample) into a PE-sample bag (Whirl-Pak®, WI, USA) which were then stored at room temperature in the dark. Please see Supplementary Materials and Methods M1 for additional field measurements.
Geochemical and mineralogical analysis
Detailed methods for pH and electrical conductivity, anion and cation analysis, total organic carbon analysis, and bulk mineralogy can be found in the Supplementary Materials and Methods M2-5.
Estimation of boulder coverage
Detailed methods for the estimation of boulder field coverage, density, and abundance are described in the Supplementary Materials and Methods M6.
DNA extraction, Illumina library preparation, and sequencing
Metagenomic DNA was extracted from 10 g of soil as described previously [12]. Briefly, the soil was mixed for 30 minutes in 40 mL of cell extraction buffer (1% PEG 8000; 1M NaCl, pH 9,2) [88]. The supernatant was ultra-centrifuged 2 h at 44,000 × g at 4 °C and DNA was extracted from the pellet using a bead-beating and phenol/chloroform/isoamylalcohol based protocol [89]. DNA was resuspended in 30 μL of DEPC treated water. Two extractions were performed per sample and the resulting DNA was combined. DNA concentration was measured using the Qubit 1x dsDNA HS Assay Kit and Qubit 4 Fluorometer (both Thermo Fisher Scientific, USA). 10 mL of the cell extraction buffer was used as a negative control for DNA extraction. Approximately 5-15 ng of DNA was sheared with a E220 Focused-ultrasonicator (Covaris® Inc., USA), targeting 300–400-bp fragment size, and used to prepare the metagenomic libraries. The libraries were constructed using the NEBNEXT® Ultra II DNA library prep kit for Illumina and the NEBNext® primer set 1 (Dual index, New England BioLabs, UK) with three modifications: 1) the primer adapters were diluted 1:50 v/v, 2) the primers were diluted 1:2 v/v and 3) a second cleaning step was performed after PCR amplification. Purification and size selection were conducted using magnetic beads Agencourt® AMPure® XP (Beckman-Coulter, USA). Inserts between 400 and 500 bp were kept and their quality evaluated using a Fragment Analyzer™ (Advanced Analytical, USA). Library concentration was measured with the Qubit 1x dsDNA HS Assay Kit and Qubit 4 Fluorometer. The metagenomic libraries were sequenced on an Illumina HiSeq 2500 (Illumina, USA) as 2 × 250-bp reads using the HiSeq Rapid SBS Kit v2 (500 cycles, Illumina, USA) and loading 12 pM including 1% v/v PhiX.
Metagenome assembly, binning, and annotation
Due to low biomass and high levels of inhibitors in the samples, only 15 samples yielded measurable amounts of DNA (Table S2, Figure S20). Of those, eleven DNA extracts successfully yielded metagenomic libraries and subsequent metagenomic analyses were performed. HiSeq reads were quality filtered using BBduk (https://sourceforge.net/projects/bbmap/) and sickle (https://github.com/najoshi/sickle). MetaSPADES 3.13 [90] was used to assemble the reads and the resulting scaffolds were filtered for length (≥ 1000 bp) for gene prediction using Prodigal [91] in meta mode and for annotation using Diamond version 0.9.9 [92] against the UniRef100 database [93] with an e-value cut-off of 1E−5. Scaffold coverages were calculated by mapping reads using Bowtie2 in sensitive mode [94]. Genomes were binned using abawaca (github.com/CK7/abawaca), ESOM [95] and MaxBin2 [96], and the resulting bins were aggregated using DAS Tool [97]. Each genomic bin was manually curated using coverage, gene-based taxonomy, and GC content information for each scaffold. ra2 [98] was used to fix assembly errors in all binned scaffolds. CheckM [99] was used to estimate the quality of the bins and only mid-to-high quality bins [100] with completeness > 75% and contamination < 10% were considered for further analysis. For all mid-to-high quality genomes, GTDB-Tk (v1.5.0) classify_wf [101] was used for a broad taxonomic classification and bacterial concatenated protein tree generation. In situ genome replication measures (iRep) [16] and growth rate index (GRiD) [37] were calculated for all bacterial mid-to-high quality genomes. Filtered iRep values (default parameters including genome coverage > 5×) were computed using --mm 3 flag after mapping the reads to scaffolds with Bowtie2 [102]. GRiD values were refined as recommended using the suggested strain heterogeneity threshold (< 0.3). Correlation between the two measures was established using Spearman correlation in R [103] (p < 0.03, rho = 0.561, Figure S10b) and iRep was chosen for subsequent analysis and interpretation in this study based on the comparability between the two measures. Further functional and metabolic capacities of mid-to-high quality genomes and metagenomes were determined using METABOLIC [104]. METABOLIC output was further expanded upon using hidden Markov model (HMM) search results for the archaeal AmoA protein (PF12942) (HMMER v3.2.1 (http://hmmer.org/) hmmscan -E 1e−5) and other genes previously annotated using the UniREF100 database [93]. Relative abundances of key metabolic genes were calculated by identifying scaffolds carrying the gene in question, summing up their coverages and then normalizing the summed coverage with the sequencing depth of each sample. 16S rRNA sequences were detected using HMMs (https://github.com/christophertbrown/bioscripts/blob/master/ctbBio/16SfromHMM.py). Hydrogenases were verified and classified using HydDB [105]. RuBisCO sequences were classified using phylogenetic placement. RuBisCO reference sequences were extracted from a local database of dereplicated NCBI genomes (accessed 2019/06/01) with HMMER using the hmm profile (PF00016.22) with 1e−10 e-value cutoff. Form IV and most form III RuBisCO sequences were removed and fragmented contiguous sequences were fused. RuBisCO sequences from this study and the Negev desert [55] were added for alignment using MUSCLE [106] followed by BMGE v1.12 trimming [107] with default conditions, and the tree was calculated using IQ-TREE v2.1.2 [108] with flags -m MFP -alrt 1000 -bb 1000.
Community analysis based on metagenomics
Operational taxonomic units (OTUs) were determined by extracting all genes encoding for the S3 ribosomal protein (rpS3) using hmmsearch (HMMER v.3.2.1, http://hmmer.org/) as described previously [109] across all assembled metagenomes. Retrieved RpS3 amino acid sequences were clustered using USEARCH [110] at 99% identity [106] and centroid sequences were extracted. Coverages of OTUs across all samples were calculated by mapping reads from each sample to the scaffolds of the centroids using Bowtie2 in sensitive mode [94] and filtering for a maximum of 5 mismatches (2% error rate) per read for both reads in a read pair. Coverages were then normalized by the total number of reads per sample. OTUs were placed into a phylogenetic tree by aligning using MUSCLE [107], alignment trimming using BMGE [108] in default mode, and tree construction using iqtree v1.3.11.1 [111] with flags -m TEST -alrt 1000 -bb 1000. The phylogenetic tree was visualized using iToL [112]. Shannon-Wiener Indices were calculated using the Vegan package [113] in R [103]. All univariate and multivariate analyses including Bray-Curtis [114] distance matrices for principal coordinate analyses (PCoA), BioENV [38], Non-metric multidimensional scaling (NMDS), and Multiple response permutation procedures (MRPP, permutations = 999) [115] were calculated and subsequently visualized in R v.4.0.2 [103]. Percent relative abundance was calculated using coverage of individual rpS3 sequences divided by the total coverages of all rpS3 genes detected in a sample [116].
Phylogenomic analysis
Phylogenomic placements of the thaumarchaeal metagenome-assembled genomes (MAGs) were determined using a supermatrix of 37 single-copy marker genes with all NCBI genomes annotated as Thaumarchaeota as of 4/6/2020 [117]. The fact that the Thaumarchaeota classification on NCBI includes the recently reclassified phylum Aigarchaeota [118] allowed us to use the latter as an outgroup. CheckM [99] was used to quality filter genomes with thresholds <5% contamination, >50% completeness. Two local databases were created from the Atacama Desert and NCBI Thaumarchaeota MAGs (Table S11) respectively. Homologs of Phylosift marker genes [117] were searched in both databases using HMMER 3.2.1 (http://hmmer.org/) with an e-value cutoff of 1e−5. The resulting datasets were aligned with MUSCLE with default parameters [107] and curated manually to fuse contiguous fragmented sequences and remove extra gene copies. Ultimately, two genomes (GCA_011605725, GCA_011773305) were removed, since they contained multiple sequences that were too distant from both Thaumarchaeota and Aigarchaeota, respectively. The resulting datasets were realigned as above, trimmed with BMGE (BLOSUM30) [108], and concatenated into a supermatrix of 312 operational taxonomic units (OTUs) and 7426 positions. Phylogenies were reconstructed with IQ-TREE 2 [111]; first, a tree with ModelFinder [119] (-m MFP -bb 1000 -alrt 1000 -abayes) that served as a guide tree for a run with the PMSF model [120] (-m LG+C60+F+G -bb 1000 -alrt 1000 -abayes). AmoA nucleotide sequences were phylogenetically placed using reference AOA genomes with Ca. Nitrosocaldus amoA as an outgroup [61]. Nucleotide sequence phylogeny for thaumarchaeal 16S rRNA (with references from NCBI nr and SILVA 138.1 Ref NR databases) and amino acid sequence phylogenies for AmoA amino acid phylogenetic trees were created as follows: Sequences were aligned using MUSCLE with default parameters [107] and trimmed using BMGE [108] in default mode, and trees we constructed using iqtree v1.3.11.1 [111] with flags -m MFP -alrt 1000 -bb 1000. As per the suggestion of the IQ-TREE authors, we considered those branches strongly supported with at least 95 for ultrafast bootstrap [121] and 80 for the SH-aLRT test [122].
Comparative genomics
The predicted protein sequences of eight NCBI Ca. Nitrosocosmicus reference genomes (Table S3) were compared with the recovered thaumarchaeal MAGs. The CompareM package (github.com/dparks1134/CompareM) was used to identify the orthologous fraction (OF) and calculate the average amino acid identity (AAI) of orthologous genes between a pair of genomes, and fastANI [123] was used to calculate the average nucleotide identity between genomes using default parameters. OrthoVenn2 [124] was used to identify and visualize orthologous clusters across genomes.
Genome-based metabolic modeling
Detailed methods for the construction of genome-based metabolic models and simulations of carbon uptake and nitrogen output can be found in Supplementary Materials and Methods M7.
Supplementary Information
Acknowledgements
We thank Pedro Zamorano and Bernardita Valenzuea for providing support in sample collection and transportation. We thank Bärbel Fösel for insightful discussions, Manuela Alt and Kirstin Weiß for TOC measurements, Thomas Neumann for providing access to the XRD laboratory, and Iris Pieper and Claudia Kuntz for technical assistance and measurement of water-soluble ion concentrations. We thank Bernhard Schink for his advice on naming the archaeal Candidatus species.
Authors’ contributions
DSM and YH conceived the project; YH and AJP designed the study; YH, FLA, and AA planned and conducted sampling; JSS and MS prepared metagenomic libraries and performed sequencing as well as initial quality filtering of reads; YH assembled, curated, and analyzed sequence data with contribution from TLVB and AJP; AJP provided computational resources; FLA performed geochemical analyses and AA provided input in data interpretation; CS conducted boulder coverage analysis; PSA performed phylogenetic analysis; WZ and YZ constructed metabolic models and conducted simulations; YH and AJP wrote the manuscript with input from all co-authors. The authors read and approved the final manuscript.
Funding
This work was funded by ERC Advanced Grant HOME (# 339231) to DSM. AJP and TVLB were supported by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen (“Nachwuchsgruppe Dr. Alexander Probst”). Genome-scale metabolic modeling was supported by the National Science Foundation under Grant No. 1553211. PSA is supported by a postdoctoral fellowship from the Alexander von Humboldt Foundation. We acknowledge support by the German Aerospace Center (DLR) under contract DISPERS (50WB1922). Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
SRA accession numbers are available in Table S2 and all genomes investigated in this study were deposited to NCBI under BioProject PRJNA665391. Newick treefiles for the concatenated thaumarchaeal tree (Fig. 4), thaumarchaeal 16S rRNA gene tree, and RuBisCO genes are provided in Additional Files 1, 3, and 4 respectively.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dirk Schulze-Makuch, Email: schulze-makuch@tu-berlin.de.
Alexander J. Probst, Email: alexander.j.probst@gmail.com
References
- 1.Maestre FT, Delgado-Baquerizo M, Jeffries TC, Eldridge DJ, Ochoa V, Gozalo B, et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci U S A. 2015;112:15684–15689. doi: 10.1073/pnas.1516684112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prigent M, Leroy M, Confalonieri F, Dutertre M, DuBow MS. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles. 2005;9:289–296. doi: 10.1007/s00792-005-0444-5. [DOI] [PubMed] [Google Scholar]
- 3.Benslama O, Boulahrouf A, et al. Impact of glyphosate application on the microbial activity of two Algerian soils. Int J Curr Microbiol App Sci. 2013;2:628–635. [Google Scholar]
- 4.Cowan DA, Hopkins DW, Jones BE, Maggs-Kölling G, Majewska R, Ramond J-B. Microbiomics of Namib Desert habitats. Extremophiles. 2020;24:17–29. doi: 10.1007/s00792-019-01122-7. [DOI] [PubMed] [Google Scholar]
- 5.Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol. 2010;8:129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 6.Chan Y, Van Nostrand JD, Zhou J, Pointing SB, Farrell RL. Functional ecology of an Antarctic Dry Valley. Proc Natl Acad Sci U S A. 2013;110:8990–8995. doi: 10.1073/pnas.1300643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowan DA, Pointing SB, Stevens MI, Craig Cary S, Stomeo F, Tuffin IM. Distribution and abiotic influences on hypolithic microbial communities in an Antarctic Dry Valley. Polar Biol. 2011;34:307–311. [Google Scholar]
- 8.Makhalanyane TP, Valverde A, Gunnigle E, Frossard A, Ramond J-B, Cowan DA. Microbial ecology of hot desert edaphic systems. FEMS Microbiol Rev. 2015;39:203–221. doi: 10.1093/femsre/fuu011. [DOI] [PubMed] [Google Scholar]
- 9.Hartley AJ, Chong G, Houston J, Mather AE. 150 million years of climatic stability: evidence from the Atacama Desert, northern Chile. J Geol Soc Lond. 2005;162:421–424. [Google Scholar]
- 10.Ewing SA, Sutter B, Owen J, Nishiizumi K, Sharp W, Cliff SS, et al. A threshold in soil formation at Earth’s arid--hyperarid transition. Geochim Cosmochim Acta. 2006;70:5293–5322. [Google Scholar]
- 11.Houston J, Hartley AJ. The central Andean west-slope rainshadow and its potential contribution to the origin of hyper-aridity in the Atacama Desert. Int J Climatol. 2003:1453–64 Available from: 10.1002/joc.938.
- 12.Schulze-Makuch D, Wagner D, Kounaves SP, Mangelsdorf K, Devine KG, de Vera J-P, et al. Transitory microbial habitat in the hyperarid Atacama Desert. Proc Natl Acad Sci U S A. 2018;115:2670–2675. doi: 10.1073/pnas.1714341115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azua-Bustos A, Caro-Lara L, Vicuña R. Discovery and microbial content of the driest site of the hyperarid Atacama Desert, Chile. Environ Microbiol Rep. 2015:388–94 Available from: 10.1111/1758-2229.12261. [DOI] [PubMed]
- 14.Navarro-González R, Rainey FA, Molina P, Bagaley DR, Hollen BJ, de la Rosa J, et al. Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science. 2003;302:1018–1021. doi: 10.1126/science.1089143. [DOI] [PubMed] [Google Scholar]
- 15.Crits-Christoph A, Robinson CK, Barnum T, Fricke WF, Davila AF, Jedynak B, et al. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome. 2013;1:28. doi: 10.1186/2049-2618-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol. 2016:1256–63 Available from: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed]
- 17.Azúa-Bustos A, González-Silva C, Mancilla RA, Salas L, Gómez-Silva B, McKay CP, et al. Hypolithic Cyanobacteria Supported Mainly by Fog in the Coastal Range of the Atacama Desert. Microb Ecol. 2011:568–81 Available from: 10.1007/s00248-010-9784-5. [DOI] [PubMed]
- 18.Moreno ML, Piubeli F, Bonfa MRL, García MT, Durrant LR, Mellado E. Analysis and characterization of cultivable extremophilic hydrolytic bacterial community in heavy-metal-contaminated soils from the A tacama D esert and their biotechnological potentials. J Appl Microbiol. 2012;113:550–559. doi: 10.1111/j.1365-2672.2012.05366.x. [DOI] [PubMed] [Google Scholar]
- 19.Könneke M, Schubert DM, Brown PC, Hügler M, Standfest S, Schwander T, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci U S A. 2014;111:8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 21.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A. 2012;109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neilson JW, Califf K, Cardona C, Copeland A, van Treuren W, Josephson KL, et al. Significant Impacts of Increasing Aridity on the Arid Soil Microbiome. mSystems. 2017;2 Available from: 10.1128/mSystems.00195-16. [DOI] [PMC free article] [PubMed]
- 23.Huang M, Chai L, Jiang D, Zhang M, Zhao Y, Huang Y. Increasing aridity affects soil archaeal communities by mediating soil niches in semi-arid regions. Sci Total Environ. 2019;647:699–707. doi: 10.1016/j.scitotenv.2018.07.305. [DOI] [PubMed] [Google Scholar]
- 24.Pointing SB, Chan Y, Lacap DC, Lau MCY, Jurgens JA, Farrell RL. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci U S A. 2009;106:19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maza F, Maldonado J, Vásquez-Dean J, Mandakovic D, Gaete A, Cambiazo V, et al. Soil Bacterial Communities From the Chilean Andean Highlands: Taxonomic Composition and Culturability. Front Bioeng Biotechnol. 2019;7:10. doi: 10.3389/fbioe.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh BK. Archaea in a hyper-arid polar desert. Proc Natl Acad Sci. 2010:E1 Available from: 10.1073/pnas.0912316107. [DOI] [PMC free article] [PubMed]
- 27.Zhang E, Thibaut LM, Terauds A, Raven M, Tanaka MM, van Dorst J, et al. Lifting the veil on arid-to-hyperarid Antarctic soil microbiomes: a tale of two oases. Microbiome. 2020;8:37. doi: 10.1186/s40168-020-00809-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finstad KM, Probst AJ, Thomas BC, Andersen GL, Demergasso C, Echeverría A, et al. Microbial Community Structure and the Persistence of Cyanobacterial Populations in Salt Crusts of the Hyperarid Atacama Desert from Genome-Resolved Metagenomics. Front Microbiol. 2017;8:1435. doi: 10.3389/fmicb.2017.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández-Martínez MÁ, Dos Santos SR, Moreno-Paz M, Gallardo-Carreño I, Blanco Y, Warren-Rhodes K, et al. Prokaryotic Community Structure and Metabolisms in Shallow Subsurface of Atacama Desert Playas and Alluvial Fans After Heavy Rains: Repairing and Preparing for Next Dry Period. Front Microbiol. 2019;10:1641. doi: 10.3389/fmicb.2019.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch RC, King AJ, Farías ME, Sowell P, Vitry C, Schmidt SK. The potential for microbial life in the highest-elevation (>6000 m.a.s.l.) mineral soils of the Atacama region. J Geophys Res Biogeosci. 2012; Available from: 10.1029/2012jg001961.
- 31.Matmon A, Quade J, Placzek C, Fink D, Copeland A, Neilson JW, et al. Seismic origin of the Atacama Desert boulder fields. Geomorphology. 2015:28–39 Available from: 10.1016/j.geomorph.2014.11.008.
- 32.Quade J, Reiners P, Placzek C, Matmon A, Pepper M, Ojha L, et al. Seismicity and the strange rubbing boulders of the Atacama desert, Northern Chile. Geology. 2012;40:851–854. [Google Scholar]
- 33.Sager C, Airo A, Arens FL, Rabethge C, Schulze-Makuch D. New types of boulder accumulations in the hyper-arid Atacama Desert. Geomorphology. 2020;350:106897. [Google Scholar]
- 34.Placzek CJ, Matmon A, Granger DE, Quade J, Niedermann S. Evidence for active landscape evolution in the hyperarid Atacama from multiple terrestrial cosmogenic nuclides. Earth Planet Sci Lett. 2010;295:12–20. [Google Scholar]
- 35.Dunai TJ, González López GA, Juez-Larré J. Oligocene–Miocene age of aridity in the Atacama Desert revealed by exposure dating of erosion-sensitive landforms. Geology. 2005:321 Available from: 10.1130/g21184.1.
- 36.Kramm, Kramm, Hoffmeister. A Relief Dependent Evaluation of Digital Elevation Models on Different Scales for Northern Chile. ISPRS Int J Geo-Inf. 2019:430 Available from: 10.3390/ijgi8100430.
- 37.Emiola A, Oh J. High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat Commun. 2018;9:4956. doi: 10.1038/s41467-018-07240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke KR, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205. [Google Scholar]
- 39.Zhang X, Gao X, Li C, Luo X, Wang Y. Fluoride contributes to the shaping of microbial community in high fluoride groundwater in Qiji County, Yuncheng City, China. Sci Rep. 2019;9:14488. doi: 10.1038/s41598-019-50914-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher RA. XV.—The Correlation between Relatives on the Supposition of Mendelian Inheritance. Earth Environ Sci Trans R Soc Edinb. 1919;52:399–433. [Google Scholar]
- 41.Azua-Bustos A, González-Silva C, Fernández-Martínez MÁ, Arenas-Fajardo C, Fonseca R, Martín-Torres FJ, et al. Aeolian transport of viable microbial life across the Atacama Desert, Chile: Implications for Mars. Sci Rep. 2019;9:11024. doi: 10.1038/s41598-019-47394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massana R, Taylor LT, Murray AE, Wu KY, Jeffrey WH, DeLong EF. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antarctica, during early spring. Limnol Oceanogr. 1998;43:607–617. [Google Scholar]
- 43.Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K, et al. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Church MJ, DeLong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. Abundance and distribution of planktonicArchaeaandBacteriain the waters west of the Antarctic Peninsula. Limnol Oceanogr. 2003:1893–902 Available from: 10.4319/lo.2003.48.5.1893.
- 45.Alonso-Sáez L, Sánchez O, Gasol JM, Balagué V, Pedrós-Alio C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ Microbiol. 2008;10:2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x. [DOI] [PubMed] [Google Scholar]
- 46.Grzymski JJ, Riesenfeld CS, Williams TJ, Dussaq AM, Ducklow H, Erickson M, et al. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012;6:1901–1915. doi: 10.1038/ismej.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith JM, Chavez FP, Francis CA. Ammonium Uptake by Phytoplankton Regulates Nitrification in the Sunlit Ocean. PLoS ONE. 2014:e108173 Available from: 10.1371/journal.pone.0108173. [DOI] [PMC free article] [PubMed]
- 48.Merbt SN, Stahl DA, Casamayor EO, Martí E, Nicol GW, Prosser JI. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett. 2012;327:41–46. doi: 10.1111/j.1574-6968.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 49.Merbt SN, Bernal S, Proia L, Martí E, Casamayor EO. Photoinhibition on natural ammonia oxidizers biofilm populations and implications for nitrogen uptake in stream biofilms. Limnol Oceanogr. 2017:364–75 Available from: 10.1002/lno.10436.
- 50.Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci U S A. 2014;111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tolar BB, Powers LC, Miller WL, Wallsgrove NJ, Popp BN, Hollibaugh JT. Ammonia Oxidation in the Ocean Can Be Inhibited by Nanomolar Concentrations of Hydrogen Peroxide. Front Mar Sci. 2016; Available from: 10.3389/fmars.2016.00237.
- 52.Georgiou CD, Sun HJ, McKay CP, Grintzalis K, Papapostolou I, Zisimopoulos D, et al. Evidence for photochemical production of reactive oxygen species in desert soils. Nat Commun. 2015;6:7100. doi: 10.1038/ncomms8100. [DOI] [PubMed] [Google Scholar]
- 53.Delwiche CF, Palmer JD. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 54.Jaffe AL, Castelle CJ, Dupont CL, Banfield JF. Lateral Gene Transfer Shapes the Distribution of RuBisCO among Candidate Phyla Radiation Bacteria and DPANN Archaea. Mol Biol Evol. 2019;36:435–446. doi: 10.1093/molbev/msy234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bay SK, Waite DW, Dong X, Gillor O, Chown SL, Hugenholtz P, et al. Chemosynthetic and photosynthetic bacteria contribute differentially to primary production across a steep desert aridity gradient. ISME J. 2021:1–18 Nature Publishing Group. [DOI] [PMC free article] [PubMed]
- 56.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA, et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature. 2017;552:400–403. doi: 10.1038/nature25014. [DOI] [PubMed] [Google Scholar]
- 57.Reich M, Bao H. Nitrate Deposits of the Atacama Desert: A Marker of Long-Term Hyperaridity. Elements. 2018:251–6 Available from: 10.2138/gselements.14.4.251.
- 58.Stahl DA, de la Torre JR. Physiology and Diversity of Ammonia-Oxidizing Archaea. Annu Rev Microbiol. 2012:83–101 Available from: 10.1146/annurev-micro-092611-150128. [DOI] [PubMed]
- 59.Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–1845. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kobayashi S, Hira D, Yoshida K, Toyofuku M, Shida Y, Ogasawara W, et al. Nitric Oxide Production from Nitrite Reduction and Hydroxylamine Oxidation by Copper-containing Dissimilatory Nitrite Reductase (NirK) from the Aerobic Ammonia-oxidizing Archaeon, Nitrososphaera viennensis. Microbes Environ. 2018;33:428–434. doi: 10.1264/jsme2.ME18058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alves RJE, Minh BQ, Urich T, von Haeseler A, Schleper C. Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat Commun. 2018;9:1517. doi: 10.1038/s41467-018-03861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alves RJE, Kerou M, Zappe A, Bittner R, Abby SS, Schmidt HA, et al. Ammonia Oxidation by the Arctic Terrestrial Thaumarchaeote Nitrosocosmicus arcticus Is Stimulated by Increasing Temperatures. Front Microbiol. 2019;10:1571. doi: 10.3389/fmicb.2019.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung M-Y, Kim J-G, Sinninghe Damsté JS, Rijpstra WIC, Madsen EL, Kim S-J, et al. A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environ Microbiol Rep. 2016;8:983–992. doi: 10.1111/1758-2229.12477. [DOI] [PubMed] [Google Scholar]
- 64.Liu L, Li S, Han J, Lin W, Luo J. A Two-Step Strategy for the Rapid Enrichment of Nitrosocosmicus-Like Ammonia-Oxidizing Thaumarchaea. Front Microbiol. 2019; Available from: 10.3389/fmicb.2019.00875. [DOI] [PMC free article] [PubMed]
- 65.Liang R, Lau MCY, Saitta ET, Garvin ZK, Onstott TC. Genome-centric resolution of novel microbial lineages in an excavated Centrosaurus dinosaur fossil bone from the Late Cretaceous of North America. Environ Microbiome. 2020;15:8. doi: 10.1186/s40793-020-00355-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sauder LA, Albertsen M, Engel K, Schwarz J, Nielsen PH, Wagner M, et al. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J. 2017;11:1142–1157. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA--DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 69.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC. Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics. 2018;34:2371–2375. doi: 10.1093/bioinformatics/bty113. [DOI] [PubMed] [Google Scholar]
- 72.Valenzuela-Encinas C, Neria-González I, Alcántara-Hernández RJ, Arturo Enríquez-Aragón J, Estrada-Alvarado I, Hernández-Rodríguez C, et al. Phylogenetic analysis of the archaeal community in an alkaline-saline soil of the former lake Texcoco (Mexico). Extremophiles. 2008:247–54 Available from: 10.1007/s00792-007-0121-y. [DOI] [PubMed]
- 73.Hedlund BP, Dodsworth JA, Cole JK, Panosyan HH. An integrated study reveals diverse methanogens, Thaumarchaeota, and yet-uncultivated archaeal lineages in Armenian hot springs. Antonie Van Leeuwenhoek. 2013;104:71–82. doi: 10.1007/s10482-013-9927-z. [DOI] [PubMed] [Google Scholar]
- 74.Lynch RC, King AJ, Farías ME, Sowell P, Vitry C, Schmidt SK. The potential for microbial life in the highest-elevation (> 6000 masl) mineral soils of the Atacama region. J Geophys Res Biogeosci. 2012;117 Available from: https://agupubs.onlinelibrary.wiley.com/doi/abs/10.1029/2012JG001961. Wiley Online Library.
- 75.Nicol GW, Tscherko D, Chang L, Hammesfahr U, Prosser JI. Crenarchaeal community assembly and microdiversity in developing soils at two sites associated with deglaciation. Environ Microbiol. 2006;8:1382–1393. doi: 10.1111/j.1462-2920.2006.01031.x. [DOI] [PubMed] [Google Scholar]
- 76.Lin X, Kennedy D, Fredrickson J, Bjornstad B, Konopka A. Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ Microbiol. 2012;14:414–425. doi: 10.1111/j.1462-2920.2011.02659.x. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, Jiang H, Dong H, Wang H, Wu G, Hou W, et al. amoA-encoding archaea and thaumarchaeol in the lakes on the northeastern Qinghai-Tibetan Plateau, China. Front Microbiol. 2013;4:329. doi: 10.3389/fmicb.2013.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kerou M, Offre P, Valledor L, Abby SS, Melcher M, Nagler M, et al. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc Natl Acad Sci U S A. 2016;113:E7937–E7946. doi: 10.1073/pnas.1601212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheridan PO, Raguideau S, Quince C, Holden J, Zhang L, Thames Consortium et al. Gene duplication drives genome expansion in a major lineage of Thaumarchaeota. Nat Commun. 2020;11:5494. doi: 10.1038/s41467-020-19132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lebre PH, De Maayer P, Cowan DA. Xerotolerant bacteria: surviving through a dry spell. Nat Rev Microbiol. 2017;15:285–296. doi: 10.1038/nrmicro.2017.16. [DOI] [PubMed] [Google Scholar]
- 81.Czech L, Hermann L, Stöveken N, Richter A, Höppner A, Smits S, et al. Role of the Extremolytes Ectoine and Hydroxyectoine as Stress Protectants and Nutrients: Genetics, Phylogenomics, Biochemistry, and Structural Analysis. Genes. 2018:177 Available from: 10.3390/genes9040177. [DOI] [PMC free article] [PubMed]
- 82.Burg MB, Ferraris JD. Intracellular Organic Osmolytes: Function and Regulation. J Biol Chem. 2008:7309–13 Available from: 10.1074/jbc.r700042200. [DOI] [PMC free article] [PubMed]
- 83.Moreno ML, Piubeli F, Bonfá MRL, García MT, Durrant LR, Mellado E. Analysis and characterization of cultivable extremophilic hydrolytic bacterial community in heavy-metal-contaminated soils from the Atacama Desert and their biotechnological potentials. J Appl Microbiol. 2012;113:550–559. doi: 10.1111/j.1365-2672.2012.05366.x. [DOI] [PubMed] [Google Scholar]
- 84.Calamita G. The Escherichia coli aquaporin-Z water channel: MicroReview. Mol Microbiol. 2000;37:254–262. doi: 10.1046/j.1365-2958.2000.02016.x. [DOI] [PubMed] [Google Scholar]
- 85.Wang B, Qin W, Ren Y, Zhou X, Jung M-Y, Han P, et al. Expansion of Thaumarchaeota habitat range is correlated with horizontal transfer of ATPase operons. ISME J. 2019;13:3067–3079. doi: 10.1038/s41396-019-0493-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhong H, Lehtovirta-Morley L, Liu J, Zheng Y, Lin H, Song D, et al. Novel insights into the Thaumarchaeota in the deepest oceans: their metabolism and potential adaptation mechanisms. Microbiome. 2020;8:78. doi: 10.1186/s40168-020-00849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mörchen R, Lehndorff E, Diaz FA, Moradi G, Bol R, Fuentes B, et al. Carbon accrual in the Atacama Desert. Glob Planet Chang. 2019;181:102993. [Google Scholar]
- 88.Narayan A, Jain K, Shah AR, Madamwar D. An efficient and cost-effective method for DNA extraction from athalassohaline soil using a newly formulated cell extraction buffer. 3 Biotech. 2016;6:62. doi: 10.1007/s13205-016-0383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Töwe S, Wallisch S, Bannert A, Fischer D, Hai B, Haesler F, Kleineidam K, Schloter M. Improved protocol for the simultaneous extraction and column-based separation of DNA and RNA from different soils. J Microbiol Methods. 2011;84:406–12. 10.1016/j.mimet.2010.12.028. [DOI] [PubMed]
- 90.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015:59–60 Available from: 10.1038/nmeth.3176. [DOI] [PubMed]
- 93.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 94.Langdon WB. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015;8:1. doi: 10.1186/s13040-014-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 97.CMK S, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018:836–43 Available from: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed]
- 98.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015:208–11 Available from: 10.1038/nature14486. [DOI] [PubMed]
- 99.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bowers RM, The Genome Standards Consortium, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017:725–31 Available from: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed]
- 101.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019; Available from: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed]
- 102.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Core Team R, et al. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 104.Zhou Z, Tran PQ, Breister AM, Liu Y, Kieft K, Elise S. Cowley, Karaoz U, Anantharaman K. METABOLIC: High-throughput profiling of microbial genomes for functional traits, biogeochemistry, and community-scale metabolic networks. BioRxiv. 761643. Available from: 10.1101/761643. [DOI] [PMC free article] [PubMed]
- 105.Søndergaard D, Pedersen CNS, Greening C. HydDB: A web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23:111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010:210 Available from: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed]
- 109.Probst AJ, Elling FJ, Castelle CJ, Zhu Q, Elvert M, Birarda G, et al. Lipid analysis of CO2-rich subsurface aquifers suggests an autotrophy-based deep biosphere with lysolipids enriched in CPR bacteria. ISME J. 2020;14:1547–1560. doi: 10.1038/s41396-020-0624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 111.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P. The vegan package: Community Ecology Package. R package version 2.0--2. 2011. [Google Scholar]
- 114.Bray JR, Curtis JT. An ordination of upland forest communities of southern Wisconsin. Ecological Monographs (27). Change Mar Communities. 1957;27(4):325–49 PRIMER-E Plymouth.
- 115.Mielke PW, Berry KJ, Johnson ES. Multi-response permutation procedures for a priori classifications. Commun Stat Theory Methods. 1976:1409–24 Available from: 10.1080/03610927608827451.
- 116.Bouma-Gregson K, Olm MR, Probst AJ, Anantharaman K, Power ME, Banfield JF. Impacts of microbial assemblage and environmental conditions on the distribution of anatoxin-a producing cyanobacteria within a river network. ISME J. 2019;13:1618–1634. doi: 10.1038/s41396-019-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Darling AE, Jospin G, Lowe E, Matsen FA, 4th, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hua Z-S, Qu Y-N, Zhu Q, Zhou E-M, Qi Y-L, Yin Y-R, et al. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nat Commun. 2018; Available from: 10.1038/s41467-018-05284-4. [DOI] [PMC free article] [PubMed]
- 119.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang H-C, Minh BQ, Susko E, Roger AJ. Modeling Site Heterogeneity with Posterior Mean Site Frequency Profiles Accelerates Accurate Phylogenomic Estimation. Syst Biol. 2018;67:216–235. doi: 10.1093/sysbio/syx068. [DOI] [PubMed] [Google Scholar]
- 121.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. [DOI] [PubMed]
- 123.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu L, Dong Z, Fang L, Luo Y, Wei Z, Guo H, Zhang G, Gu YQ, Coleman-Derr D, Xia Q, Wang Y. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47(W1):W52–W58. 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SRA accession numbers are available in Table S2 and all genomes investigated in this study were deposited to NCBI under BioProject PRJNA665391. Newick treefiles for the concatenated thaumarchaeal tree (Fig. 4), thaumarchaeal 16S rRNA gene tree, and RuBisCO genes are provided in Additional Files 1, 3, and 4 respectively.