Abstract
Epigenetic mRNA modification is an evolving field. N6-methyladenosine (m6A) is the most frequent internal transcriptional modification in eukaryotic messenger RNAs (mRNAs). This review will discuss the functions of the m6A mRNA machinery, including its “writers” which are components of the methyltransferase complex, its “readers” and its “erasers” (specifically FTO and ALKBH5) in cancer. The writers deposit the m6A and include METTL3, METTL14, WTAP, VIRMA, RBM15. This is removed by the m6A demethylases (FTO and ALKBH5). Lastly the most diverse members are the readers that can contribute to mRNA splicing, stability, translation and nuclear export. Many of these functions continue to be elucidated. The dysregulation of this machinery in various malignancies and the associated impact on tumorigenesis and drug response will be discussed herein with a focus on solid tumors. It is clear that by contributing to either mRNA stability or translation, there are down-stream targets that are impacted, contributing to cancer progression and the self-renewal ability of cancer stem cells.
Introduction:
In the past decade there have been progressive studies demonstrating that mRNA modification occurs to impact RNA stability and translation, thus impacting the control of gene expression. The most common form of over 170 RNA nucleotide modifications is N6-methyladenosine (m6A).1, 2 This modification is reversible 3, 4 and has been found to impact more than 7,000 mRNAs in mammalian cell individual transcriptomes.5, 6 There is additional data demonstrating that m6A modification in mRNAs or non-coding RNAs impact RNA translation and transcript fate/functions. These are critical for many physiologic processes included the DNA damage response, tissue development (hematopoiesis and neurogenesis) circadian rhythm regulation, sex differentiation, microRNA processes, RNA-protein exchanges and carcinogenesis.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17
Aberrant cell growth in tumorigenesis has historically been defined by abnormalities in cell division and gene expression as dictated by abnormalities in genetic and epigenetic changes. These abnormalities can be a function of genetic changes (e.g. gene mutations, deletions, amplification or chromosomal translocations) and/or epigenetic changes such as DNA or histone modification. In the past 10 years, RNA epitranscriptomics or gene regulation at the RNA level has gained more interest as an additional layer of influence in the development of malignancies. Of the various RNA modifications, m6A has been identified as a reversible RNA modification similar to the well-described histone and DNA modifications that are also reversible. With the development of high-thoroughput m6A sequencing techniques, there have been identified thousands of mRNA and non-coding RNA transcripts that are associated with m6A modifications with an additional enrichment in the 3’ untranslated regions (UTRs) in close proximity to the stop codons of mRNAs.5, 6
In the framework of mRNA m6A modification, there are methyltransferases and demethylases and in between there are proteins identified as “readers.” The readers can promote decay or enhance RNA stability, promote translation and impact splicing and nuclear export of various target mRNAs.10, 11, 12, 18, 19, 20, 21, 22 Therefore, the type of reader protein that recognizes the m6A modification of a given target mRNA can impact the stability of the target mRNA and can affect RNA translation, splicing, or nuclear transport. (Figure 1) This level of regulation – with the concept of mRNA “writers” (methyltransferases), “readers” and “erasers” (demethylases) is still a field in its infancy as it pertains to dysregulation in solid tumors.
Figure 1.
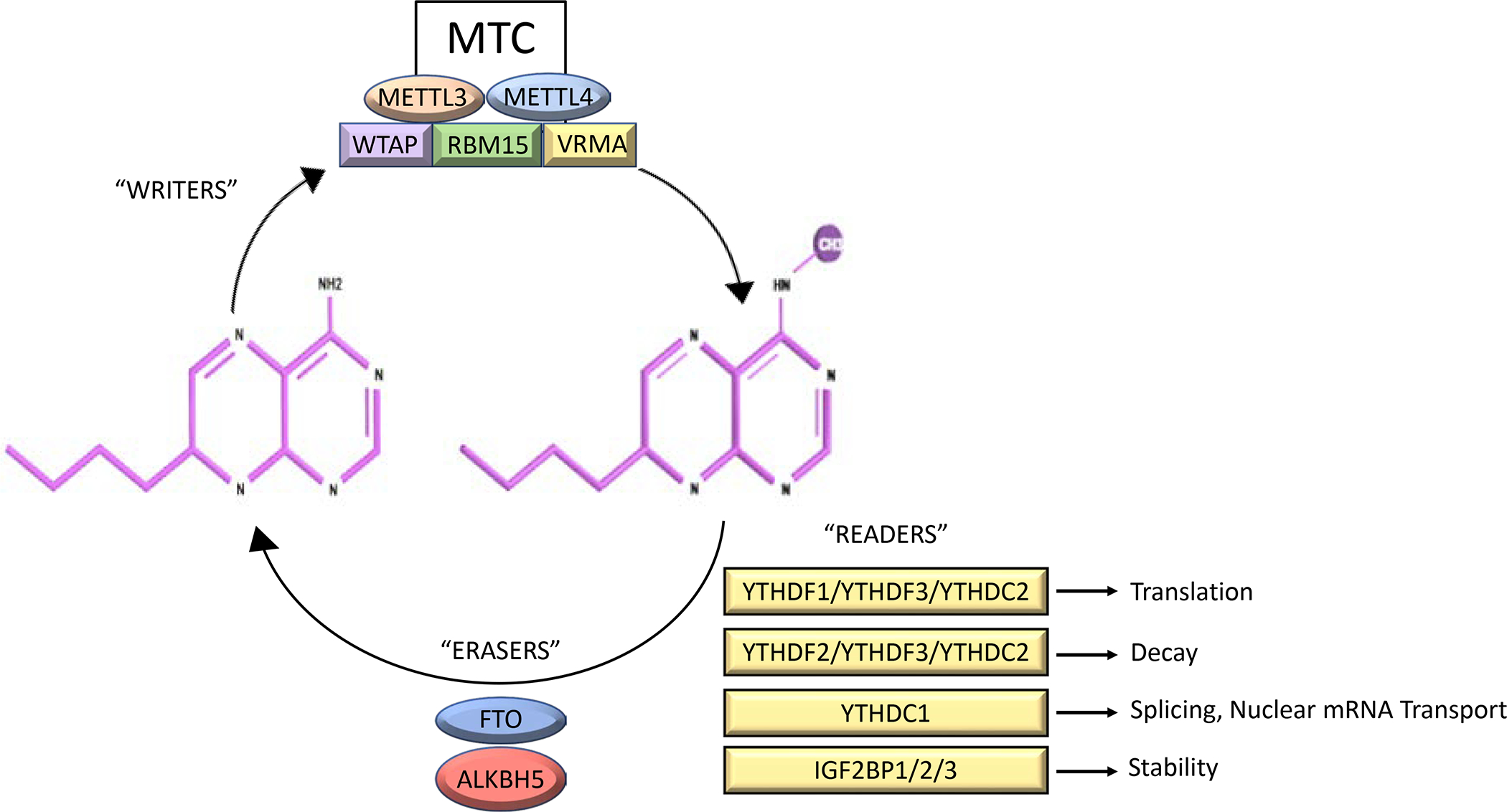
m6A modification machinery summary. The MTC is the m6A methyltransferase complex. The MTC is composed of WTAP, VIRMA, METL3, METTL14 and RM15. The MTC serves as methylase or “writer’.” FTO and ALKBH5 are demethylases or “erasers.” The “readers” have a variety of functions including translation (YTHDF1, YTHDF3, YTHDC2), decay (YTHDF2, YTHDF3, YTHDC2) splicing, nuclear export (YTHDC1), and stability (1GF2BP1/2/3).
m6A Modification Machinery
“Writers” such as Methyltransferase-like 3 and 14 (METTL3 and METTL14) and their respective cofactors, RBM15, Wilms tumor1-associated protein (WTAP), RBM15, VIRMA (KIAA1429) make up the m6A methyltransferase complex (MTC). This grouping of proteins functions as the m6A writer and catalyzes the m6A modification. 8, 23, 24, 25, 26, 27 Our recent studies have further characterized how m6A is specifically deposited in the transcriptome. Huang et al. demonstrated that histone H3 trimethylation at Lys36 (H3K36me3), a marker for transcription elongation guides m6A deposition co-transcriptionally.28 The mechanism is that H3K36me3 is recognized and bound directly by METTL14 which as noted above is a crucial part of the MTC and thus facilitates binding of the m6A MTC to RNA polymerase II, thus delivering the m6A MTC to actively transcribing RNAs to deposit m6A co-transcriptionally. This work uncovers another layer of gene expression regulation involving the communication between histone modification and RNA methylation.28 Weng et al from our group demonstrated that METTL14 is highly expressed in normal hematopoietic stem/progenitor cells (HSPCs) and acute myeloid leukemia (AML) cells carrying t(11q23), t(15;17), or t(8,21), and is down-regulated during normal myeloid differentiation.29 Inhibiting METTL14 induces terminal myeloid differentiation in AML cells and inhibits AML cell survival and growth. The pro-oncogenic role of METTL14 in AML is by regulating its mRNA targets (e.g., MYB and MYC) via m6A modification.29
Recognized demethylases or “erasers” are FTO (fat mass-and obesity associated gene) and ALKBH5.4, 9, 30 These proteins function to removed the m6A modification from mRNA and create a counter balance to the “writers.”
The “readers” are a functionally more heterogeneous family of proteins with more diverse functionality. The YT521-B homology (YTH) domain family including YTHDC1, YTHDC2, YTHDF1, YTHDF2 and YTHDF3 are considered direct readers.10, 11, 12, 18, 19, 20 YTHDF2, YTHDF3 and YTHDC2 promote degradation of their target mRNAs. YTHDF1, YTHDF3 and YTHDC2 promote translation, while YTHDC1 influences splicing and targets mRNA exportation. Our group has reported that insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs; including IGF2BP 1/2/3) as a distinct family of m6A readers.22 These target many mRNA transcripts via identifying the consensus GG(m6A)C sequence.22 In contrast to the mRNA-decay-promoting function of YTHDF2, IGF2BPs promote the stability and storage of their target mRNA (e.g., MYC) and also promote their translation in an m6A-dependent manner and therefore impact gene expression.22 Additional readers include eukaryotic initiation factor 3 (eIF3) and cytoplasmic METTL3.13, 31
Recent work has intimated that anti-tumor immunity may be in part controlled through mRNA m6A methylation and the “reader,” YTHDF1 in dendritic cells.32 In the context of anti-tumor immunity, tumor regression often correlates to the neoantigen burden. Han et al show that durable neoantigen-specific immunity is regulated by m6A methylation via the reader YTHDF1. They demonstrate that Ythdf1-deficient mice showed an amplified antigen-specific CD8+ T cell anti-tumor response. More specifically the loss of YTHDF1 in classical dendritic cells amplified the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells in vivo. Transcripts encoding lysosomal proteases are marked by m6A and identified by YTHDF1. The binding of YTHDF1 to these specific transcripts amplifies the translation of lysosomal cathepsins in dendritic cells and inhibition of the cathepsin increases cross-presentation of wild type dendritic cells. Additionally the therapeutic efficacy of PD-L1 checkpoint blockade is much more effective in Ythdf1−/− mice, implying that YTHDF1 is a potential target that can amplify anticancer immunotherapy.32
Aberrations in the m6A mRNA modification machinery have recently been associated with several malignancies (Table 1) including leukemia, glioblastoma, breast cancer, hepatocellular cancer, cervical cancer, lung cancer and gastric cancer.29, 33, 34, 35,36 There is also emerging data that targeting various aspects of this system will lead to novel therapies. The aim of this review is to highlight recent developments on the role of the m6A mRNA modification machinery as it pertains to the development, propagation and treatment of solid tumors.
Table 1.
Described m6A Member Machinery in Cancer
| Component | m6A role | Role | Cancer Studied | References |
|---|---|---|---|---|
| FTO | Eraser | Oncogenic | AML, GBM | [48, 52] |
| ALKBH5 | Eraser | Oncogenic | GBM, Breast, Pancreas | [53, 66, 75] |
| METTL3 | Writer | Oncogenic, Tumor Suppressor | AML, GBM, HCC, Pancreas, Breast | [52, 67, 74, 77] |
| METTL14 | Writer | Tumor Suppressor | GBM, HCC, Endometrial | [36, 52, 73] |
| IGF3BP1/2/3 | Writer | Oncogenic | GBM | [22] |
| YTHDF1 | Reader | Oncogenic | Colorectal Cancer, Melanoma | [32] |
The oncogenic “ERASER” FTO
Approximately 10 years ago, single nucleotide polymorphisms (SNPs) in FTO were found to be strongly associated with obesity and body mass index in humans as determined by genome wide association studies (GWAS).37, 38 More recently as the m6A mRNA modification proteins have been characterized, there has been more interest in FTO as a demethylase or “eraser.” FTO catalyzes the demethylation of 3-methyl-thymine in single stranded DNA with Fe(II) and 2-oxoglutarate (2-OG) producing carbon dioxide, formaldehyde and succinate.3 Based on its jelly-roll motif protein folding structure, FTO functions with high affinity to m6A in mRNA whereby it functions as an efficient demethylase.4
The direct causality of FTO and higher BMI or obesity has not been definitively elucidated. However, it is generally believed to be associated with a greater intake of calories perhaps secondary to FTO expression in the hypothalamus.39, 40 As obesity is associated with several cancers, there are studies that correlate the connection between FTO and some obesity associated cancers.41, 42 One mechanism whereby the FTO gene is regulated is by DNA methylation. Hypomethylation of specific CpG sites in the FTO gene leads to increased FTO expression and this correlates with the presence of Type 2 Diabetes Mellitus and some cancers.43
Initial studies identified some FTO gene single-nucleotide polymorphisms (SNPs) that were associated with the risk of certain cancers including endometrial cancer and pancreatic cancer.44, 45, 46 In breast cancer, SNPs in intron 1 of FTO including rs8047395, rs9939609 and rs7206790 that have been identified as important associations with development of this disease.47 The precise mechanisms whereby these malignancy risk associated SNPs in FTO remain to be elucidated.
Our group demonstrated that FTO facilitates oncogenesis in acute myeloid leukemia (AML).48 FTO targets genes such as ASB2 and RARA as a demethylase. FTO is over-expressed in certain subtypes of AML and promotes leukemogenesis and prevents all-trans-retinoic acid-induced leukemia cell differentiation. Thus FTO functions as an oncogene in this disease by inhibiting mRNA targets such as ASB2 and RARA by reducing their m6A levels and stability.48 This work revealed a previously unidentified method of gene regulation in carcinogenesis and highlights the significance of the FTO gene and m6A mRNA demethylation in cancer.
Su et al. from our group has also demonstrated that high levels of FTO sensitize leukemic cells to the oncometabolite R-2-hydroxyglutarate (R-2HG).34 R-2HG is produced to relatively high levels by mutant isocitrate dehydrogenase1/2 (IDH1/2) which is found in 10–20% of AML patients.49 R-2HG exerts anti tumor activity via inhibition of leukemia cell proliferation/viability and induction of cell-cycle arrest and apoptosis. R-2HG inhibits FTO (demethylase) activity, and therefore increases m6A mRNA modification. This in turn decreases the stability of MYC/CEBPA transcripts and thus suppresses relevant down stream pro-tumor pathways.34 These mechanistic findings have been limited in solid tumors, however, there is emerging data of FTO and other members of the m6A mRNA modification machinery and the implications of tumorigenesis. Most recently, Huang et al. developed an effective FTO-specific inhibitor, namely FB23–2, and showed that targeting FTO by small-molecule inhibitors such as FB23–2 can significantly inhibit AML cell viability/growth, promote apoptosis, and inhibit AML progression in vivo.50 Thus, these studies provide proof-of-concept evidence suggesting that FTO is a druggable target and targeting FTO by effective inhibitors holds great therapeutic potential to treat FTO-overexpressing AML. 34, 50
Glioblastoma
Glioblastoma is the most common and aggressive primary malignant brain tumor and even with surgical resection, recurrence is common.51 Cui Q et al. found that RNA m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells (GSCs) by the regulation of mRNA m6A enrichment and expression.52 In glioblastoma, similar to in leukemia, R-2HG displays antitumor effects by inhibition of proliferation /survival in FTO-overexpressing cancer cells and targeting of the FTO/m6A/MYC/CEBPA signaling.34 These pathways are important in cell proliferation and survival. This work demonstrated why it would be reasonable to target FTO in both glioblastoma and AML as noted above.
Zhang et al queried the TCGA to assess which components of the m6A machinery were associated with poor patient prognosis.53 They found that the “eraser” ALKBH5 predicted poor prognosis in all data sets.53 They showed that targeting ALKBH5 impairs self renewal, decreased proliferation and tumorigenesis in glioma stem cells (GSCs). The downstream targets of ALKBH5 in GSC were evaluated and FOXM1 a key transcription factor important in GSCs was identified.53 Mechanistically, ALKBH5 found was found to demethylate FOXM1 nascent transcripts leading to increased expression and this may be an avenue to therapy for glioblastoma.53
RNA Methylation in Gastric Cancer
Gastric cancer is the third most frequent cause of cancer related mortality and is the fifth most common cancer in the world.54, 55 The management for early disease is surgery with or without systemic therapy. Advanced disease is managed with chemotherapy and several targeted therapies as a function of tumor characteristics. Xu et al. demonstrated that by immunohistochemistry tissue microarray, FTO is markedly increased in gastric cancer tissues compared to adjacent non-tumor tissues (56vs 38%).56 FTO expression was significantly associated with poor differentiation and lymph node metastases and positively correlated with worse stage. High FTO expression was also significantly associated with poor prognosis. Down-regulation of FTO expression inhibited the proliferation, migration and invasion of GC cell lines in vitro.56
Mechanistically, Zhang et al demonstrated that reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer.33 Utilizing a proteomics-based gastric cancer cohort they had previously generated and the TCGA-GC cohort, they merged expression of canonical m6A writers (METTL3/METTLE14), readers (YTHDF1/YTHDF2/YTHDF3), and erasers (ALKBH5/FTO), respectively as W,R, and E signatures to represent the m6A modification. They stratified patients according to those signatures to decipher m6A’s associations with critical mutations, prognosis and clinical indices. M6A’s biological function in gastric cancer was predicted by gene set enrichment analysis (GEA) and validated via in vitro experiments. W and R were potential tumor suppressive signatures and E was a potential oncogenic signature in gastric cancer. Based on W/R/E stratifications, patients with low m6A were associated with higher mutations of specific genes (CDH1, AR, GLI3, SETBP1, RHOA, MUC6 and TP53) and also demonstrated worse clinical outcomes.33 Via in vitro experiments they demonstrated that m6A suppression (as METTL14 knockdown) promoted cell proliferation and invasiveness via activating Wnt and PI3K-Akt signaling, while m6A elevation (ie FTO knockdown) reversed these changes.33 Additionally, the findings implied that m6A modification may be involved in interferon signaling and immune responses in gastric cancer. These data imply that targeting the “erasers” such as FTO or amplifying the “writers” such as METTL14 may be therapeutic avenues to pursue as it pertains to the m6A machinery in gastric cancer.33
Writers and Erasers in Breast Cancer
Breast cancer is the most prevalent malignancy in women. Although there has been great progress in this disease, the primary cause of mortality is secondary to distant metastases.57 The population of tumor-initiating cells or breast cancer stem cells (BCSCs) have the capability of self-renewal.58 The phenotype of these cells is characterized by the expression of several core pluripotency proteins including Kruppel-like factor 4 (KLF4), Octamer-binding transcription factor 4 (OCT4), SRY-box 2 (SOX2), and NANOG.59, 60, 61, 62, 63 In the context of metastatic disease, intratumor hypoxia leads to the expression of the transcription factor hypoxia-inducible factor-1α.64 There is recent work indicating that HIFs are necessary for the maintenance of BCSCs via transcriptional regulation of genes encoding the pluripotency associated genes NANOG, SOX2, and KLF4.65 Pluripotency factors have been associated with changes in mRNA stability as dictated by m6A mRNA methylation. Zhang et al. have reported that exposure of breast cancer cells to hypoxia induces m6A demethylation and stabilization of NANOG mRNA, thus supporting the BSCS phenotype.66 Additionally, they showed that down regulating the expression of the ALKBH5 (coding a demethylase or eraser) or HIF-1s (which activate ALKBH5 gene transcription in hypoxic breast cancer cells) led to decreased NANOG expression and growth inhibition of BCSCs in vivo.66 Further work is needed to assess if competitive agonists of ALKBH5 may be useful as therapy that targets BCSCs.
In other work, Cai et al demonstrated that METTL3 over expression in breast cancer drives the progression of breast cancer via inhibiting tumor suppressor let-7g.67 Initial observations were that the overexpression of both METTL3 and the oncoprotein Mammalian Hepatitis B X-Interacting Protein (HBXIP) were associated with breast cancer.67, 68, 69 Mechanistically they were able to demonstrate that HBXIP modulates METTL3 by inhibiting miRNA let-7g which down regulates the expression of METTL3. Interestingly they found that METTL3 promoted the expression of HBXIP via m6A modification essentially creating a feedback loop.69 These findings provided new insights into the mechanism of m6A mRNA modification in the progression of breast cancer and more work remains to be done in in this field.
M6A Machinery in the Liver: Non-Alcoholic Steatohepatitis (NASH) and Hepatocellular Cancer (HCC)
NASH is a rising etiology of liver failure world-wide. With the rise of obesity, the prevalence of NASH continues to increase and also correlates with the incidence of hepatocellular carcinoma in this population.70 At present there are no FDA approved medications for the treatment of NASH. NASH is histologically characterized by hepatocyte ballooning, inflammation, focal fibrosis and steatosis.71 It is further described as lipotoxicity in hepatocytes. There are limited treatment options. Lim et al demonstrate that the expression of FTO is significantly increased in the livers of NASH patients as well as in a rodent model.71 They demonstrated that genetic silencing of FTO protects against palmitate-induced oxidative stress, mitochondrial dysfunction, ER stress and apoptosis in vitro.71 These results indicate that FTO over expression may have a deleterious role in hepatocytes and perhaps this contributes to the increased liver damage in NASH. Studies need to be done further explore FTO targeting as a route to mitigate NASH and perhaps decrease the risk of HCC development in the future in these patients. Additionally, it is not clear if the lipotoxicity is inducing the FTO expression or is FTO expression facilitating lipotoxicity.
In liver cancer, it has been reported that the writer components function as a tumor-suppressors. Ma et al found that in HCC, METTL14 and m6A levels were decreased relative to normal or paratumor controls with unchanged levels of the other writers METTL3 and WTAP.36 Additionally, they found that METTL14 expression correlated with poor prognosis. METTL14 knockdown facilitated HCC metastasis and over-expression amplified tumor invasion and metastasis via m6A-dependent modulation of microRNA (ie, mir-126) processing via interaction with DGCR8. 36 In another study, Chen et al. found that METTL3 was actually overexpressed in HCC compared to normal control, with WTAP levels unchanged.72 METTL3 overexpression in this cohort correlated with worse prognosis. Furthermore, they demonstrated that overexpression of METTL3 augmented growth of HCC both in vitro and in vivo while down-regulation of METTL3 inhibited tumorigenesis and lung metastasis in vivo.72 These findings were associated with negative regulation of SOCS2 expression by an m6A- and YTHDF2-dependent mechanism.72 These data therefore imply that potential therapeutic targets in HCC development include FTO in the context of NASH and the writers METTL3 and METTL14 as they pertain to HCC growth and metastasis.
M6A mRNA Methylation in Endometrial Cancer: A METTL14 Hot Spot
Sequencing studies have identified that 70% of endometrial tumors have reduced total m6A mRNA methylation compared to adjacent normals.73 However, the functional significance was largely unknown. The specific METTL14 (R298P) mutation occurs at the RNA-binding groove and leads to inhibiting m6A mRNA methylation in tumors.73 Additionally, endometrial tumors have significantly reduced METTL3 m6A methyltransferase. It appears that these abnormalities are mutually exclusive.73 Liu et al. have found that either METTL14 mutation or reduced expression of METTL3 led to increased proliferation and tumorigenicity of endometrial cancer cells via activation of the AKT pathway.73 Similar to several other solid and liquid tumors, they conclude that changes in m6A methylation is an oncogenic mechanism in endometrial cancer. A great deal needs to be studied about these mechanisms in order to develop relevant therapeutics for the treatment of endometrial cancer.
RNA Modification in Pancreatic Cancer
Pancreatic ductal adenocarcinoma (PDAC) is a fatal malignancy with a 5 year survival of 9%. 74 He et al. found that ALKBH5 was down-regulated in pancreatic cancer cells, in which a lncRNA, KCNK15-AS1 is a direct target of ALKBH5 and thus is also down-regulated; forced expression of ALKBH5 or KCNK15-AS1 could inhibit pancreatic cancer cell migration and invasion.75 More recently, Zhang et al. reported that cigarette smoke condensate (CSC) could cause hypomethylation in the METTL3 promoter region and thereby up-regulate expression of METTL3, which in turn promotes the maturation process of primary microRNA-25 (miR-25) in pancreatic duct epithelial cells. The excessive miR-25–3p maturation results in the activation of the oncogenic AKT-p70S6K signaling, which promotes malignant phenotypes of pancreatic cancer cells.74 This study revealed a previously unappreciated link between cigarette smoke, m6A modification, microRNA maturation, and the pathogenesis of pancreatic cancer.
Colon Cancer: METTLE3 Associated with Tumor Progression.
The role of m6A methylation in colorectal cancer (CRC) remains largely unexplored. Li et al. have found via the TCGA that METTL3 expression correlated with poor prognosis in CRC.76 METTL3 knockdown led to decreased CRC cell self-renewal, stem cell frequency and migration in vitro and inhibited growth and metastases in vivo.76 METTLE3 was also found to target SRY (sec determining region Y)-box 2 (SOX2). Mechanistically they found that when SOX2 transcripts were methylated, they were then recognized by a specific m6A reader, insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2) to prevent SOX2 mRNA degradations.76 Additionally, they found that the combination of “writer” METTL3, “reader” IGF2BP2 and “target” SOX2 correlated with better prognostic accuracy for CRC patient than the individual components.76 More work is needed to assess the efficacy of targeting components of this combination as a therapeutic strategy.
Conclusion
In summary, it is clear that the m6A mRNA machinery is an important mechanism in gene regulation and expression. In nearly all malignancies studied there appears to be a role in contributing to cancer stem cell self-renewal. Targeting the various functions of “writers,” “readers,” and “erasers” is a field of great interest and the oncogenic roles of the m6A RNA methylation machinery needs to be further elucidated. There is a great deal to be learned by this novel epigenetic regulation at the RNA level. Development of effective and selective small-molecule compounds or other agents/tools targeting the dysregulated m6A machinery components is urgently needed as these may provide more effective novel therapies for cancer treatment.
Acknowledgements
We apologize to colleagues whose work could not be included due to space limitations.
Funding
This work was supported in part by the National Institute of Health (NIH) Grants R01 CA214965 (J.C.), R01 CA236399 (J.C.), R01 CA211614 (J.C.)., R56 DK120282 (J.C.) as well as the Norman and Sadie Lee Grant City of Hope (L.M.) PanCan Translational Grant 2019 (L.M.). J.C. is a Leukemia & Lymphoma Society (LLS) Scholar.
Footnotes
Conflict of Interest
J.C has a patent filed based on his R-2HG/FTO work. J.C. is a scientific founder and the chief scientific officer of Genovel Biotech Corp. and also holds equity with this company.
REFERENCES
- 1.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic acids research 2018, 46(D1): D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science 2018, 361(6409): 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends in genetics : TIG 2013, 29(2): 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology 2011, 7(12): 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485(7397): 201–206. [DOI] [PubMed] [Google Scholar]
- 6.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149(7): 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng X, Su R, Feng X, Wei M, Chen J. Role of N(6)-methyladenosine modification in cancer. Current opinion in genetics & development 2018, 48: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature cell biology 2014, 16(2): 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell 2013, 49(1): 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505(7481): 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161(6): 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell research 2017, 27(9): 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163(4): 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519(7544): 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526(7574): 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543(7646): 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549(7671): 273–276. [DOI] [PubMed] [Google Scholar]
- 18.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Molecular cell 2016, 61(4): 507–519. [DOI] [PubMed] [Google Scholar]
- 19.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell research 2017, 27(3): 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell research 2017, 27(3): 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m(6)A Transcripts by the 3’-->5’ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Molecular cell 2017, 68(2): 374–387 e312. [DOI] [PubMed] [Google Scholar]
- 22.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature cell biology 2018, 20(3): 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna 1997, 3(11): 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology 2014, 10(2): 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research 2014, 24(2): 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell reports 2014, 8(1): 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537(7620): 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019, 567(7748): 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell stem cell 2018, 22(2): 191–205 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nature reviews Molecular cell biology 2017, 18(1): 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Molecular cell 2016, 62(3): 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 2019, 566(7743): 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer medicine 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 2018, 172(1–2): 90–105 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell research 2018, 28(5): 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017, 65(2): 529–543. [DOI] [PubMed] [Google Scholar]
- 37.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature genetics 2007, 39(6): 724–726. [DOI] [PubMed] [Google Scholar]
- 38.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS genetics 2007, 3(7): e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 2007, 316(5826): 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McTaggart JS, Lee S, Iberl M, Church C, Cox RD, Ashcroft FM. FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PloS one 2011, 6(11): e27968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang Y, Liu F, Liu Y. Is FTO gene variant related to cancer risk independently of adiposity? An updated meta-analysis of 129,467 cases and 290,633 controls. Oncotarget 2017, 8(31): 50987–50996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng X, Su R, Stanford S, Chen J. Critical Enzymatic Functions of FTO in Obesity and Cancer. Frontiers in endocrinology 2018, 9: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnik BC. Milk: an epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. Journal of translational medicine 2015, 13: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delahanty RJ, Beeghly-Fadiel A, Xiang YB, Long J, Cai Q, Wen W, et al. Association of obesity-related genetic variants with endometrial cancer risk: a report from the Shanghai Endometrial Cancer Genetics Study. American journal of epidemiology 2011, 174(10): 1115–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Zhao J, Yang M, Li M, Zheng J. Association between FTO gene polymorphism (rs9939609 T/A) and cancer risk: a meta-analysis. European journal of cancer care 2017, 26(5). [DOI] [PubMed] [Google Scholar]
- 46.Lurie G, Gaudet MM, Spurdle AB, Carney ME, Wilkens LR, Yang HP, et al. The obesity-associated polymorphisms FTO rs9939609 and MC4R rs17782313 and endometrial cancer risk in non-Hispanic white women. PloS one 2011, 6(2): e16756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaklamani V, Yi N, Sadim M, Siziopikou K, Zhang K, Xu Y, et al. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC medical genetics 2011, 12: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer cell 2017, 31(1): 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell 2010, 18(6): 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer cell 2019, 35(4): 677–691 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia CR, Slone SA, Dolecek TA, Huang B, Neltner JH, Villano JL. Primary central nervous system tumor treatment and survival in the United States, 2004–2015. Journal of neuro-oncology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell reports 2017, 18(11): 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer cell 2017, 31(4): 591–606 e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGuire S World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Advances in nutrition 2016, 7(2): 418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians 2015, 65(2): 87–108. [DOI] [PubMed] [Google Scholar]
- 56.Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncology reports 2017, 38(4): 2285–2292. [DOI] [PubMed] [Google Scholar]
- 57.O’Shaughnessy J Extending survival with chemotherapy in metastatic breast cancer. The oncologist 2005, 10 Suppl 3: 20–29. [DOI] [PubMed] [Google Scholar]
- 58.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer research 2009, 69(4): 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics 2008, 40(5): 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer research 2008, 68(16): 6533–6540. [DOI] [PubMed] [Google Scholar]
- 61.Yu F, Li J, Chen H, Fu J, Ray S, Huang S, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 2011, 30(18): 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31(11): 1354–1365. [DOI] [PubMed] [Google Scholar]
- 63.Iv Santaliz-Ruiz LE, Xie X, Old M, Teknos TN, Pan Q. Emerging role of nanog in tumorigenesis and cancer stem cells. International journal of cancer 2014, 135(12): 2741–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148(3): 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer research 2011, 71(13): 4640–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proceedings of the National Academy of Sciences of the United States of America 2016, 113(14): E2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer letters 2018, 415: 11–19. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Cui M, Cai X, Sun B, Liu F, Zhang X, et al. The oncoprotein HBXIP up-regulates SCG3 through modulating E2F1 and miR-509–3p in hepatoma cells. Cancer letters 2014, 352(2): 169–178. [DOI] [PubMed] [Google Scholar]
- 69.Yue L, Li L, Liu F, Hu N, Zhang W, Bai X, et al. The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis 2013, 34(4): 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cusi K Role of obesity and lipotoxicity in the development of nonalcoholic steatohepatitis: pathophysiology and clinical implications. Gastroenterology 2012, 142(4): 711–725 e716. [DOI] [PubMed] [Google Scholar]
- 71.Lim A, Zhou J, Sinha RA, Singh BK, Ghosh S, Lim KH, et al. Hepatic FTO expression is increased in NASH and its silencing attenuates palmitic acid-induced lipotoxicity. Biochemical and biophysical research communications 2016, 479(3): 476–481. [DOI] [PubMed] [Google Scholar]
- 72.Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell research 2017, 27(9): 1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nature cell biology 2018, 20(9): 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25–3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nature communications 2019, 10(1): 1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 2018, 48(2): 838–846. [DOI] [PubMed] [Google Scholar]
- 76.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Molecular cancer 2019, 18(1): 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018, 67(6): 2254–2270. [DOI] [PubMed] [Google Scholar]


