Abstract
Non-typhoidal Salmonella enterica is an important gastrointestinal pathogen causing a considerable burden of disease. Resistance to third generation cephalosporins poses a serious threat for treatment of severe infections. In this study occurrence, phylogenetic relationship, and mechanisms of third generation cephalosporin resistance were investigated for clinical non-typhoidal S. enterica isolates in Germany. From 2017 to 2019, we detected 168 unique clinical S. enterica isolates with phenotypic resistance to third generation cephalosporins in a nation-wide surveillance. Compared to previous years, we observed a significant (P=0.0002) and consistent increase in resistant isolates from 0.41 % in 2005 to 1.71 % in 2019. In total, 34 different serovars were identified, most often S. Infantis (n=41; 24.4 %), S. Typhimurium (n=27; 16.1 %), S. Kentucky (n=21; 12.5 %), and S. Derby (n=17; 10.1 %). Whole genome analyses revealed extended-spectrum β-lactamase (ESBL) genes as main cause for third generation cephalosporin resistance, and most prevalent were bla CTX-M-1 (n=55), bla CTX-M-14 (n=25), and bla CTX-M-65 (n=23). There was no strict correlation between serovar, phylogenetic lineage, and ESBL type but some serovar/ESBL gene combinations were detected frequently, such as bla CTX-M-1 and bla CTX-M-65 in S. Infantis or bla CTX-M-14b in S. Kentucky. The ESBL genes were mainly located on plasmids, including IncI, IncA/C variants, emerging pESI variants, and a novel bla CTX-M-1harbouring plasmid. We conclude that third generation cephalosporin resistance is on the rise among clinical S. enterica isolates in Germany, and occurrence in various S. enterica serovars is most probably due to multiple acquisition events of plasmids.
Keywords: Salmonella enterica, antibiotic resistance, extended-spectrum β-lactamase (ESBL), plasmid, pESI
Data Summary
The authors confirm that all supporting data, code and protocols have been provided within the article. Illumina raw sequence reads were submitted to the European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/) and can be accessed under the bioproject PRJEB41165.
Impact Statement.
Non-typhoidal Salmonella enterica (NTS) is one of the most important agents of foodborne infections worldwide. Although in most cases salmonellosis is self-limiting, antibiotic treatment may be required. However, antibiotic resistance in NTS has been a globally emerging problem, limiting treatment options of NTS infections and further presenting a considerable threat to the public health and food safety. Knowledge about the distribution of antibiotic resistance patterns and the genetic factors in S. enterica is therefore essential for appropriate treatment of these infections and for early intervention to prevent the spread of antibiotic resistance, starting at the level of food production. In a nation-wide surveillance of third generation cephalosporin resistant S. enterica in Germany, we found that third generation cephalosporin resistance is on the rise among clinical NTS isolates. Its occurrence in various S. enterica serovars is likely due to multiple acquisition events of plasmids highlighting its facilitated spread.
Introduction
Non-typhoidal Salmonella (S.) enterica (NTS) is an important agent of foodborne infections [1]. In Europe and Germany, salmonellosis, after campylobacteriosis, represents the second most reported gastrointestinal infection caused by bacteria, encompassing 87 923 cases and 13 693 cases, respectively, in 2019 [2, 3]. S. Typhimurium and S. Enteritidis are the predominant serovars causing human infections in Germany and account for ~60 % of all reported cases (https://survstat.rki.de/). Further prevalent serovars like S. Infantis and S. Derby represented <5 % of the reported cases [3]. Although in most cases salmonellosis is self-limiting, antibiotic treatment may be required for risk group patients and more severe disease, like bloodstream infections [4]. Antibiotics for treatment of salmonellosis comprise β-lactams, fluoroquinolones, macrolides, and trimethroprim-sulfamethoxazole [4]. Challengingly, rising numbers of multidrug-resistant (MDR - resistant to three or more antimicrobials) S. enterica have been observed in recent years [5]. Including a slight increase in resistance to third generation cephalosporins, which has been reported in antimicrobial resistance surveillance studies for clinical isolates, and isolates from food (including retail meat) and animals [5–9].
Resistance to third generation cephalosporins in S. enterica is mainly due to the production of extended-spectrum β-lactamases (ESBLs) or AmpC β-lactamases [9, 10], with CTX-M-1 as the most prevalent ESBL type identified in previous studies [5, 11]. ESBL and AmpC genes are often located on plasmids, which facilitate their spread via horizontal gene transfer (HGT) between enterobacterial species and genera [12, 13]. Interestingly, ESBL production correlates with certain serovars in S. enterica . Among serovars found in human infections, ESBL genes have been more frequently detected in S. Typhimurium, S. Infantis and S. Kentucky than in S. Enteritidis [5, 14]. Furthermore, recent analyses on ESBL-producing S. Infantis from broiler chickens, broiler meat, and humans revealed a clonal lineage harbouring an especially successful conjugative pESI-like plasmid [15]. This raises the concern for transmission of ESBL genes in S. enterica along the food chain from the primary production systems to clinical cases.
In the present study, we assessed the resistance of clinical NTS in Germany and further analysed whole genomes of third generation cephalosporin resistant S. enterica for determination of phylogeny, resistance determinants, and genetic backbones to gain knowledge about the spread of this resistance mechanism in Germany.
Methods
Bacterial strains, serotyping, and antibiotic susceptibility testing
In the years 2017 – 2019, the National Reference Centre (NRC) for Salmonella and other Enteric bacterial Pathogens in Germany analysed 11 730 S . enterica isolates from human infections for pathogen surveillance. In the same time period 48 083 salmonellosis cases were notified to the German surveillance system. All S. enterica isolates were serotyped according to the White-Kauffmann-Le Minor scheme [16]. Further, antibiotic susceptibilities to 17 antibiotics (ampicillin, cefotaxime, ceftazidime, cefoxitin, meropenem, nalidixic acid, ciprofloxacin, gentamicin, amikacin, streptomycin, chloramphenicol, tetracycline and trimethoprim/sulfamethoxazole) were investigated by broth microdilution according to EUCAST criteria (v10.0, https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf). NTS isolates resistant to cefotaxime and/or ceftazidime according to EUCAST criteria (v10.0) were chosen for further analysis, however only one unique isolate per patient or infection cluster was considered. Among all NTS isolates from 2017 - 2019, 264 isolates showed phenotypic resistance to cefotaxime and/or ceftazidime and 166 unique isolates were selected for further analyses.
Bacterial cultivation and whole genome sequencing
Müller Hinton broth was used for an overnight cultivation. DNA was subsequently extracted using the GenElute Bacterial Genomic DNA Kit (Sigma, St. Louis, Missouri, U.S.). DNA quantification was performed using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.) according to manufacturer’s instructions. Sequencing libraries were generated using the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, United States) and paired-end sequencing was performed using a MiSeq instrument with 2×300 MiSeq v3 reagent kit (Illumina, San Diego, CA, United States), HiSeq 2500 instrument with 2×150 with 2×300 HiSeq v2 RR reagent kit or NextSeq 550 instrument with 2×300 NextSeq v2.5 high reagent kit. Whole genome sequencing (WGS) was performed for 166 NTS isolates (Table S1, available with the online version of this article) and sequences were submitted to ENA (https://www.ebi.ac.uk/ena/) and can be accessed under the bioproject PRJEB41165.
Bioinformatics analyses
Quality of the raw sequence data was assessed using FastQC v0.11.5 (https://github.com/s-andrews/FastQC). To verify taxonomic read classification, raw data was analysed with Kraken v0.10.6 [17]. The assembly pipeline of the analysis platform SeqSphere+ v6.0.7 (Ridom, Münster, Germany) was applied for de novo assembly using SPAdes v3.12.0 with default parameter [18], including an additional FastQC and read trimming step [19].
Multilocus sequence typing (MLST) and core genome MLST (cgMLST) were performed using the de novo assembled contigs and Ridom SeqSphere+ v6.0.7 (Ridom; Münster, Germany). The EnteroBase S. enterica cgMLST v2 scheme template was applied. In silico serotyping, based on the de novo assembly genomes, was conducted, using the serotype prediction tool SISTR [20].
De novo assembled contigs were used for resistance gene and plasmid replicon type identification. For the detection of acquired antimicrobial resistance genes, the tool Abricate in combination with the NCBIres Database was used (https://github.com/tseemann/abricate, [21]). In silico replicon typing was performed to assign resistance gene contigs to plasmid replicon sequences with Abricate using the PlasmidFinder v2.0 database [22]. Identified contigs with bla gene and plasmid replicon sequence were further investigated by means of Geneious Prime v2020.0.5 (Biomatters, Ltd., Auckland, New Zealand). In case of presence of a plasmid replicon sequence marker and a bla gene on an identical contig, these contigs were additionally checked for a possible chromosomal integration via blastn (database nucleotide collection (nr/nt)) and a subsequent Mauve alignment using S. Typhimurium str. LT2 as reference (NC_003197.2). If a chromosomal integration could be ruled out, bla gene carrying contigs were assigned to the respective identified plasmid replicon and considered as the bla resistance gene carrying contig. For phylogenetically closely related isolates with identical replicon and bla resistance gene patterns but exhibiting different de novo assembled contigs carrying bla gene and replicon sequence, a reconstruction strategy as described in Weber et al. was performed [23]. If multiple replicon sequences were identified in the genome sequence of an isolate and no replicon marker sequence could be addressed as genetic backbone for the bla gene or the de novo assembled bla gene carrying contigs exhibited not sufficient surrounding genetic environment, the bla gene carrying replicon was marked as ‘not determined’.
Plasmids carrying bla CTX-M-1 (n=22) and bla CTX-M-65 (n=15) in S. Infantis isolates were compared to variants of the emerging pESI plasmid [15]. In brief, contigs of these isolates were aligned and ordered according to the pESI plasmid backbone of the S. Infantis strain 119 944 (accession no. NZ_CP047882) using the MCM algorithm of Mauve [24]. These reordered plasmid drafts were subsequently compared to the pESI backbone and to a bla CTX-M-65 carrying pESI variant (pN17S0535, accession no. CP052810). Further bla CTX-M-1 harbouring plasmids of other serovars putatively belonging to the IncI1 replicon were compared to an IncI1 reference plasmid backbone (accession no. MK181566) using Mauve. Plasmids that carried AmpC gene bla CMY-2 were compared to IncA/C and IncI1 reference plasmids (accession no. CP012929, KT186369, CP015835, CP060509) using Mauve.
Resistance transfer experiments and molecular typing
Transfer of third generation cephalosporin resistance was tested by broth mating for three selected isolates that harboured bla CTX-M-3 (18-04833), bla CTX-M-65 (19-02625) and bla CTX-M-1 (17-01817) using the sodium azide-resistant recipient strain E. coli J53 Azir. Transconjugants were cultivated on LB agar with sodium azide (200 mg l−1) and ampicillin (30 mg l−1) and screened for resistance genes as previously described [11]. The plasmid sizes of bla gene positive transconjugants were determined by S1-nuclease restriction and pulsed-field gel electrophoresis [25]. Transconjugants and the isolates 18-00018 and 18-00680 were screened for resistance genes as previously described [11].
Results
Increasing proportion of third generation cephalosporin resistant S. enterica in Germany
In the years 2017 – 2019, the German National Reference Centre for Salmonella and other Enteric Bacterial Pathogens (NRC) analysed 11 730 NTS isolates from human infections (2017=3 399, 2018=3 706, 2019=4 625 isolates). Resistance to third generation cephalosporins was detected in 264 S. enterica isolates, of which most were collected in 2019 (2017: n=36, 2018: n=56, 2019: n=172). A substantial part of the isolates (n=100) from 2019 was attributed to a single infection cluster of S. Typhimurium in a regional setting. Four unique/representative isolates of this cluster which harboured certain changes (see details below) were included into the subsequent analyses. Consequently, the number of third generation cephalosporin resistant isolates considered for further analysis in 2019 was 76, resulting in a total of 168 study isolates for the years 2017–2019 (Table S1).
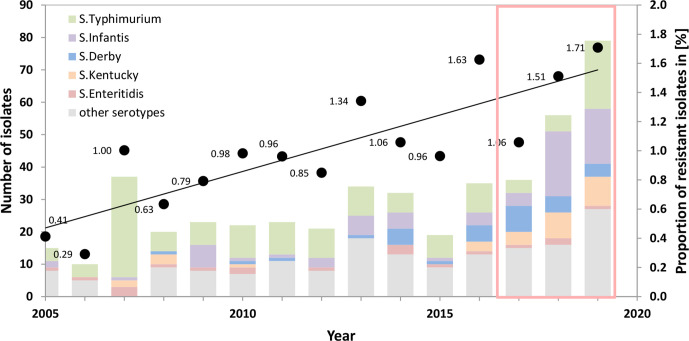
In 2019, a proportion of 1.71 % of all analysed S. enterica were resistant to third generation cephalosporins, which was an increase compared to 1.06 % in 2017. To analyse whether the increasing trend is seen in a larger time frame, NRC data from the years 2005 to 2011 [11] and 2012 to 2016 were integrated into this analysis, using identical selection criteria. Indeed, the rising trend of third generation cephalosporin resistance was corroborated for the entire time frame from 2005 to 2019. The proportion of third generation cephalosporin resistance consistently rose from 0.41 % in 2005 to 1.71 % in 2019 (Fig. 1) (P=0.0002; correlation coefficient r=0.8318; Spearman rank correlation). In addition to resistance to third generation cephalosporins, 44 of the 168 isolates (26.2 %) also showed resistance to ciprofloxacin (Table S1) and 21 isolates (12.5 %) exhibited a combined resistance to ampicillin, chloramphenicol and trimethoprim/sulfamethoxazole (Table S1). No isolate was resistant to meropenem.
Fig. 1.
Increasing proportion of German clinical S. enterica isolates showing third generation cephalosporin resistance (cefotaxime and/or ceftazidime) from 2005 to 2019. The left axis indicates the absolute numbers of third generation cephalosporin-resistant isolates (coloured bars), the right axis indicates the proportion of third generation cephalosporin-resistant isolates in percent of all analysed isolates at the NRC (black dots). Isolates from 2017 to 2019 investigated in detail in this study are highlighted by the red box. The resistant S. Typhimurium isolates from 2019 included four representative isolates of an infection cluster.
The 168 third generation cephalosporin resistant isolates were assigned to 34 different serovars, of which S. Infantis (n=41), S. Typhimurium (n=27), S. Kentucky (n=21), S. Derby (n=17) and S. Anatum (n=9) were most prevalent (Table S1 and S2).
ESBL types CTX-M-1, CTX-M-14, and CTX-M-65 are the most abundant third generation cephalosporin resistance determinants
Whole genome sequence analyses including in silico detection of the resistance determinants revealed that for most of the isolates (n=163/166) the third generation cephalosporin resistance was mediated by ESBLs (n=147) or plasmid-borne AmpC β-lactamases (n=16) (Table S1). For three isolates no β-lactamase genes were detected and analyses of porin genes revealed wild-type variants without a sign of porin loss. Therefore, the underlying mechanism of resistance of these three isolates remained unclear (Table S1). The most prevalent ESBL genes in the isolate collection were bla CTX-M-1 (n=55), followed by bla CTX-M-14 (n=25, different variants) and bla CTX-M-65 (n=23) (Table 1). The most commonly detected AmpC β-lactamase gene was bla CMY-2 (n=13), only one isolate harboured bla CMY-4, one isolate bla ACC-1, and one isolate bla DHA-1. Further identified β-lactamase genes were: bla OXA-1 (n=1), bla CARB-2 (n=2) and 39 bla TEM variants. Additionally, 38 isolates harboured plasmid-mediated quinolone resistance (PMQR) genes, mainly gene qnrS1 (n=25). Interestingly, four isolates were positive for variants of the mobile colistin resistance genes mcr-1.1 (S. Typhimurium, 17-02112), mcr-3.1 (S. Typhimurium, 19-03137) and mcr-3.11 (S. Choleraesuis and S. Bovismorbificans, 17-00186 and 17-05354) (Table 1).
Table 1.
ESBL and AmpC β-lactamase genes identified in the third generation cephalosporin resistant clinical Salmonella isolates from Germany, 2017 – 2019. Information on the co-occurrence of plasmid-mediated quinolone resistance (PMQR) genes and colistin resistance-mediating genes in the respective Salmonella isolates is given.
|
ESBL/AmpC gene |
n |
PMQR genes (n) |
Mcr genes (n) |
Majorly found in (n) |
|---|---|---|---|---|
|
bla CTX-M-1* |
55 |
qnrS1 (4) |
|
S. Infantis (15) S. Derby (17) |
|
bla CTX-M-2 |
1 |
|
|
|
|
bla CTX-M-3 |
3 |
|
|
|
|
bla CTX-M-9 |
3 |
|
|
|
|
bla CTX-M-14 |
25 |
qnrS1 (3) |
mcr-1.1 (1), mcr-3.11 (2) |
S. Kentucky (18) |
|
bla CTX-M-15 |
7 |
qnrS1 (1) |
|
|
|
bla CTX-M-32 |
1 |
|
|
|
|
bla CTX-M-55 |
14 |
qnrS1 (6) |
mcr-3.1(1) |
|
|
bla CTX-M-65 |
23 |
|
|
S. Infantis (22) |
|
bla TEM-52 |
2 |
qnrB19 (1) |
|
|
|
bla SHV-12 |
13 |
qnrS1 (10), qnrB2 (1), qnrA1 (1) |
|
S. Anatum (7) |
|
bla CMY-2 |
13 |
qnrS1 (1), qnrB19 (4), qnrB6 (1) |
|
|
|
bla CMY-4 |
1 |
qnrA1 (1) |
|
|
|
bla DHA-1 |
1 |
qnrB4 (1) |
|
|
|
bla ACC-1 |
1 |
|
|
|
*Includes four representative isolates originating from the 2019 S. Typhimurium infection cluster.
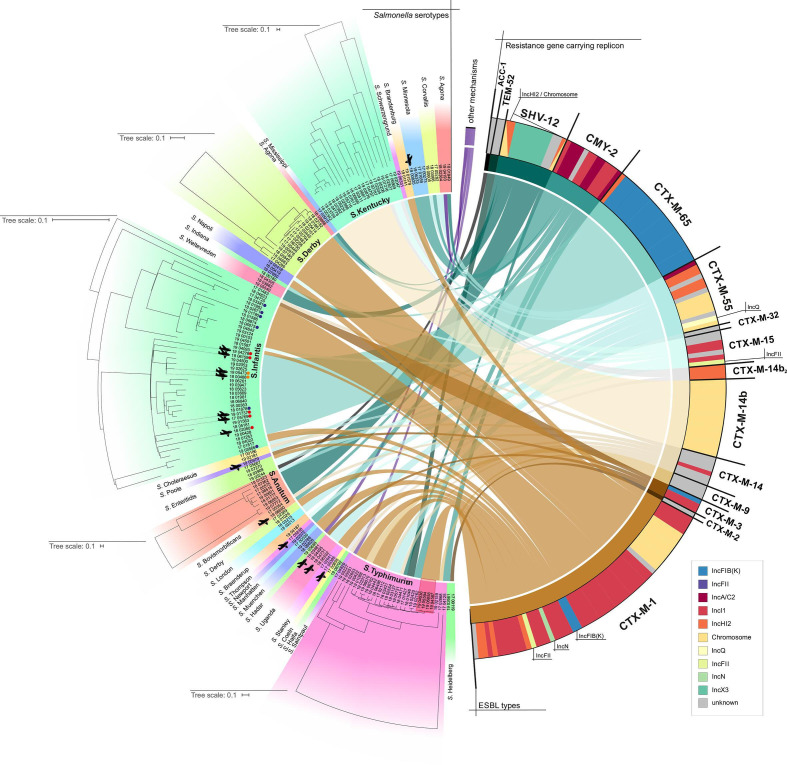
Although a strict correlation of serovar and ESBL/AmpC type was not detected, some ESBL types were observed in high proportions in specific serovars. For example, bla CTX-M-65 (n=22/23) was mainly identified in S. Infantis. Additionally, bla CTX-M-14 was differentiated into three subgroups (bla CTX-M-14, n=6, bla CTX-M-14b, n=16 and bla CTX-M-14b2, n=3): While bla CTX-M-14b was solely detected in S. Kentucky (Fig. 2, Table 1), the remaining bla CTX-M-14 variants were present in various serovars.
Fig. 2.
Attribution of the Salmonella serovar to the respective ESBL/AmpC gene of 166 third generation resistant clinical Salmonella isolates from Germany 2017 – 2019. The attributed resistance gene of each isolate is indicated via coloured links. The resistance gene carrying replicon is additionally indicated in the coloured bars next to the resistance gene (see legend). The respective phylogenetic relationships of S. Anatum, S. Derby, S. Infantis, S. Kentucky and S. Typhimurium are highlighted by independent phylogenetic trees. Four representative isolates belonging to the S. Typhimurium infection cluster are shown with a red background. Isolates with possible travel history are highlighted by airplanes and coloured dots, indicating the origin of the isolates (orange: South-East Asia, red: South America, blue: Germany). The figure was created with Circos [47].
Plasmids as the main carriers of genes encoding ESBLs and AmpC β-lactamases
For 137 of 163 of the ESBL/AmpC-positive isolates an attribution of the resistance gene to a respective genetic backbone / replicon was possible. The majority of the resistance genes was located on mobile genetic elements (n=105, 65.2 %), particularly on plasmids, irrespective of their hosting S. enterica serovar. However, a substantial number of isolates (n=32, 18.8 %) revealed chromosomal integration of the ESBL genes. The chromosomal integration of bla CTX-M-14 in all S. Kentucky isolates was identical as described by Coipan et al: bla CTX-M-14 was flanked by an upstream ISEcp1 element, downstream of the hcp1 gene in the type VI secretion system region of the genome [14]. In all S. Derby isolates the integration of bla CTX-M-1 with an upstream ISEcp1, as presumable mobilizing element, took place in a different genetic region than in S. Kentucky. Bla CTX-M-1 was integrated into the kduI gene, which was inferred from homology, causing a truncation of this pectin degradation pathway gene (accession no. A0A379QY07).
For 26 isolates it was only possible to recover the direct genetic environment but not to attribute the respective replicon type.
While for some of the ESBL genes a correlation of resistance gene and the gene carrying replicon was observed (bla CTX-M-65 and IncFIB(K) (22/23), bla CTX-M-14b and chromosome (16/19), bla CMY-2 and IncI1 (6/13), bla CMY-2 and IncA/C (6/13)), other genes showed a more diverse genetic background. For example, bla CTX-M-1 was found on IncI1, IncHI2, IncN and IncQ plasmids.
Predominance of emerging pESI plasmids in S. Infantis harbouring bla CTX-M
Detailed comparative analyses of bla CTX-M-65 carrying plasmids revealed a high sequence identity with the previously described pESI-like plasmid identified in S. Infantis. Specifically, 22 of 23 bla CTX-M-65 positive isolates showed high resemblance with the chimeric backbone of the pESI-plasmid p119944 (accession no. NZ_CP047882) and all belonged to the serovar S. Infantis. The non-matching isolate belonged to S. Indiana (18-00180) and the respective plasmid revealed no similarities to the pESI plasmid backbone. The integration of bla CTX-M-65 usually takes place upstream of a previously described drug and heavy metal resistance gene carrying region and this was also observed for the S. Infantis isolates in this study [26]. Further analyses showed that the presence of bla CTX-M-1 on a pESI-like plasmid backbone was observed only in two out of 15 S. Infantis isolates (17-01817 and 18-04022). We found that the integration of bla CTX-M-1 took place in the conserved IncI1 backbone structure of pESI. The remaining 13 S. Infantis isolates however exhibited a high sequence identity (>95 %) with the IncI1 plasmid p15078279 (accession no. MK181566), which was previously identified in E. coli [27]. Additionally, in one isolate (18-04833) bla CTX-M-3 was suspected to be located on a pESI-like plasmid. While confirmation of the insertion of bla CTX-M-3 into the pESI based on short read sequencing data was not possible, the sequence data however suggested the presence of the full pESI-like backbone. Additional mating experiments and subsequent S1-nuclease restriction and pulsed-field gel electrophoresis revealed transferability of the bla CTX-M-3 gene and its presence on an approx. 290 kb sized plasmid for the bla CTX-M-3 positive transconjugant of 18-04833. In comparison, the size of the transferred bla gene carrying plasmids of bla positive transconjugants of other pESI-like plasmid carrying strains, such as S. Infantis 17-01817 (bla CTX-M-1) and 19-02625 (bla CTX-M-65), was approx. 290 kb and approx. 315 kb, respectively.
bla CMY-2 harbouring plasmids share high sequence similarity with IncI1 and IncA/C plasmids from Europe
AmpC β-lactamase CMY-2 was the main determinant of resistance to third generation cephalosporins present in 13 isolates (mainly serovars S. Minnesota n=4 and S. Typhimurium n=3). The bla CMY-2 gene was found to be located on plasmids of the replicon type IncI1 (n=6) and IncA/C replicon type (n=6) and for one isolate (19-00825) was not assignable. The plasmids of IncI1 replicon type showed a high sequence identity to two previously described variants: IncI1 belonging to the plasmid sequence type pST12 (accession no. CP012929) (n=4) and IncI1 belonging to pST2 (accession no. KT186369) (n=2) and shared >98 %, respective >99 % sequence identity with these reference plasmids. Isolates 17-05838 and 19-02072 resembled the backbone of IncA/C2 plasmids of type 1 (accession no. CP015835) [28], while the other four isolates (17-00273, 17-01659, 19-02661, 19-06333) showed a high similarity with a IncA/C reference plasmid (accession no. CP060509), which was identified in NTS isolates from Russia.
The S. Typhimurium infection cluster isolates from 2019 harboured a novel plasmid carrying bla CTX-M-1
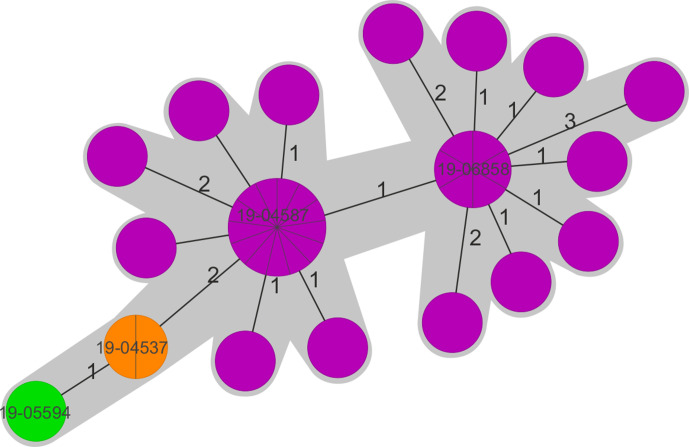
As depicted above, a substantial part of the S. Typhimurium isolates (n=100) of the year 2019 originated from a single infection cluster in a regional setting. Genome analyses of 36 of these isolates were performed and confirmed their close genetic relationship. The maximum distance in the cgMLST analysis were five alleles; the majority of the genomes depicted a distance of zero or one alleles (Fig. 3). The resistance gene content and plasmid replicon analyses showed almost identical patterns (bla CTX-M-1 : 36/36, qnrS1 : 35/36, IncHI2 replicon type: 35/36). The identified resistance gene carrying plasmid showed no sequence similarity to previously published bla gene carrying plasmids. Interestingly, one isolate (19-05594) showed absence of the qnrS1 gene and revealed a unique plasmid replicon background (IncI1), indicating a potential independent plasmid acquisition event. Two further isolates (19-04536 and 19-04537) exhibited a slight alteration of the bla CTX-M-1 carrying plasmid, varying in IS element integration sites. One representative isolate from each group (19-04587, 19-06858, 19-04536: IncHI2 plasmid alteration; 19-05594: IncI1 plasmid) was included in all here presented analyses (Figs. 2 and 3).
Fig. 3.
cgMLST-based Minimum Spanning Tree of 36 S. Typhimurium isolates of the 2019 infection cluster. Isolates with an identical IncHI2 plasmid structure are shown in purple, IncHI2 plasmid variants with the additional IS element in orange and the isolate with the unique IncI1 plasmid in green. Only the four representative isolates are highlighted with strain numbers.
No strict correlation of ESBL genes with certain serovars was observed, but some serovar/resistance gene combinations, such as S. Infantis with bla CTX-M-1 or bla CTX-M-65 or S. Kentucky with bla CTX-M-14b, were detected most frequently
cgMLST-based analysis revealed that the majority of the isolates showed no close phylogenetic relationship but clustered according to the serovar. However, among the serovars various (distinctly related) subclades were observed (Fig. 2, subclades shown for S. Anatum, S. Derby, S. Infantis, S. Kentucky, and S. Typhimurium). As expected, the four representative S. Typhimurium isolates from the infection cluster formed one cluster (sequence type ST19) (Figs 2 and 3), while the other S. Typhimurium isolates belonged to the ST34. Linking data on bla gene and plasmid content to phylogenetic information, a tendency regarding the presence of specific ESBL genes in certain serovars was observed. The most apparent correlations were seen for S. Infantis with bla CTX-M-1 and bla CTX-M-65, S. Typhimurium with bla CTX-M-1, S. Derby with bla CTX-M-1 and S. Kentucky with bla CTX-M-14b (Fig. 2).
In depth cgMLST analyses of the S. Infantis clade showed that these strains can be differentiated into at least two different subclades: subclade one exhibited the IncI1 bla CTX-M-1 carrying plasmid while subclade two is associated with bla CTX-M-65 and bla CTX-M-1 carrying pESI-like plasmids (Fig. S1). Additionally, one isolate (18-04833) exhibited bla CTX-M-3 presumably integrated into the pESI backbone. Little information on travel background of patients was available. However, for thirteen of the 41 S. Infantis isolates a definite exposition region was determined. Interestingly, especially isolates from patients from the subclade carrying the bla CTX-M-65 pESI-like plasmid exhibited travel history to South East Asia or South America (Figs 2 and S1).
Discussion
The analyses of clinical NTS from Germany showed a trend towards increasing resistance proportions for third generation cephalosporins (Fig. 1). This observation is in accordance with the worldwide emergence of antibiotic resistance in NTS. European data for clinical NTS isolates (period 2018 – 2019) by EFSA and ECDC [5] uncovered high resistance rates for different classes of antibiotics, especially ampicillin, sulfonamides, and tetracyclines. Increasing resistance was further reported for fluoroquinolones, while resistance to third generation cephalosporins was still on low levels but with rising proportions in certain serovars [5]. In Germany, five serovars (S. Typhimurium, S. Derby, S. Kentucky, S. Infantis and S. Anatum) accounted for over 65 % of the resistant isolates in this study [5, 29, 30]. S. Enteritidis, while being one of the most prevalent S. enterica serovars, remains mostly susceptible to third generation cephalosporins [31].
In the present study we identified ESBL and AmpC β-lactamase genes as most important determinants of third generation resistance in clinical NTS. These observations are in concordance to reports from the EU, where in 2018/19 ESBLs were reported in 16 different serovars (among those S. Infantis, S. Kentucky, S. Typhimurium) and AmpC β-lactamases in ten different serovars, mostly in S. Anatum, S. Bredeny and S. Thompson [5]. The majority of isolates in this study harboured ESBL genes encoding CTX-M enzymes (n = 132; 80.0%) with CTX-M-1, CTX-M-14 and CTX-M-65 as the most prevalent types. Similar results were found in studies on S. enterica and E. coli isolated from animals, especially poultry [11, 32, 33] and in reports from the EU, where, if tested by member states, the ESBL variants CTX-M-1, CTX-M-14/14b and CTX-M-9 were frequently reported [5]. In the present study bla CTX-M-1 was detected in various serovars, including S. Typhimurium, S. Infantis and S. Derby. However, bla CTX-M-14b and bla CTX-M-65, were almost exclusively found in S. Kentucky and S. Infantis, respectively. Detailed sequence analyses of the 23 bla CTX-M-65 carrying plasmids of the study isolates revealed a high sequence identity among them: out of the 23 isolates, 22 belonged to the serovar S. Infantis and showed an identical plasmid structure, which additionally was highly similar to the chimeric backbone of the pESI-plasmid p119944 (accession no. NZ_CP047882).
In recent studies the spread of third generation cephalosporin resistant S. Infantis in food-producing animals associated with the presence of a specific resistance plasmid type, the pESI-like megaplasmid, was reported [15, 34, 35]. This plasmid type/backbone was associated with the carriage of ESBL genes (bla CTX-M-65 and bla CTX-M-1), several other resistance genes (tet(A), sul1, dfrA1, dfrA14, aadA1) and heavy metal resistance genes [26]. The pESI-like megaplasmid was first identified by Aviv et al. in Israel[34], however, not yet harbouring third generation cephalosporin resistance genes. Subsequently findings of pESI harbouring third generation cephalosporin resistance genes were reported from Italy [15], the U.S. [28], Denmark, Netherlands and the UK [15, 34–36]. In 2018, Brown et al. observed the linkage of S. Infantis pESI-like plasmids carrying bla CTX-M-65 in the USA with travel activity to Latin America [37]. Despite too little information on the travel history of the patients in our study for statistical analyses, the present data partly indicate a correlation between travel activity (Asia and South America) and bla CTX-M-65 harbouring S. Infantis.
While the pESI plasmid with bla CTX-M-65 was highly prevalent in S. Infantis strains from Germany, only two of 15 S. Infantis strains exhibiting bla CTX-M-1 (17-01817 and 18-04022) harboured the resistance gene on the pESI plasmid. In Germany the occurrence of pESI was described in S. Infantis isolates from broiler farms since the 2010s [38]. However, these plasmids did not carry ESBL genes and were identified in S. Infantis strains of another lineage (ST2283) than the bla CTX-M-1 carrying isolates from this study (ST32). In our study one additional isolate of S. Infantis exhibited bla CTX-M-3 on a pESI-like plasmid, highlighting the potential of this particular plasmid backbone to acquire further resistance genes. Phylogenetic analysis revealed that isolates harbouring pESI with bla CTX-M-3, bla CTX-M-65 and bla CTX-M-1 are more related among each other than to the clade of the non-pESI bla CTX-M-1 carrying S. Infantis isolates. It can be speculated, that certain genetic factors enable the plasmid to establish in this specific S. Infantis bla CTX-M-1/-65 clade. The presence of multiple post-segregational killing determinants on the plasmid may be of advantage for this process [34].
The majority of S. Infantis isolates with bla CTX-M-1 (13/15) exhibited a highly similar plasmid background to an IncI1 plasmid backbone. Identical or highly similar bla CTX-M-1 carrying plasmids were identified in the present study also in S. Derby, S. Enteritidis, S. Typhimurium, S. Stanley and S. Brandenburg. In contrast to the pESI plasmid, which appears to be more adapted to the S. Infantis host - but still can be transferred into other S. enterica serovars and E. coli [15, 39] - these non-pESI IncI1 plasmids exhibited a wider host range and were also reported in previous studies in E. coli [27].
Further resistance genes detected in this study were bla CTX-M-15 and bla CMY-2. ESBL type CTX-M-15, which is highly prevalent in E. coli from outpatient and hospitalized patients in Germany [40], was identified only in seven clinical S. enterica isolates of different serovars. AmpC β-lactamase CMY-2, which was detected in <1 % of clinical E. coli isolates with third generation cephalosporin resistance in Germany [41] were detected in 12 S. enterica isolates in the present study (7.45 %). The observed higher proportions of CTX-M-1 and CMY-2 genes with comparable lower proportions of CTX-M-15 in our clinical S. enterica isolates may on the one hand indicate a possible transmission route between livestock and humans via the food chain. The active acquisition of plasmids via horizontal gene transfer along the food production chain seems rational, as identical resistance gene carrying plasmids were observed in S. enterica of different serovars. This is strengthened by the observation of identical resistance gene carrying plasmids in E. coli, Klebsiella pneumoniae and other Enterobacterales species in Germany, Europe, and worldwide [42–44]. Furthermore, it has been reported that approx. 6 % of the healthy population of Germany carries ESBL positive E. coli [45]. Ingestion of other ESBL-positive Enterobacterales and transfer of resistance genes from commensal bacteria therefore might have an impact on the treatment options of severe salmonellosis, as the resistance genes may be acquired during treatment [46]. However, since CTX-M-15 is the most common ESBL type in E. coli/ K. pneumoniae in the human population, these events might be limited in occurrence rather than being the main route for ESBL gene transmission.
We conclude that third generation cephalosporin resistance, is on the rise among clinical S. enterica isolates in Germany and that its occurrence in various S. enterica serovars is likely due to multiple acquisition events of plasmids. However, specific adaption processes, as observed for S. Infantis with pESI plasmids harbouring different bla CTX-M genes, could facilitate the spread of resistant S. enterica isolates.
Supplementary Data
Funding information
The study was supported by a grant from the German Ministry of Health for the project ‘Integrated Genome-Based Surveillance of Salmonella’ (GenoSalmSurv, decision ZMVI1-2518FSB709 of 26 November 2018).
Acknowledgements
We thank Monique Duwe, Susanne Kulbe, Marita Wahnfried, and Sibylle Müller-Bertling for excellent technical assistance. We also thank Andrea Thürmer and Aleksander Radonic on behalf of the core facility for genome sequencing MF2 at the Robert Koch Institute. We are also thankful to all laboratories that sent strains to the NRC for further analysis.
Author contributions
We describe author contributions to the paper using the CRedit taxonomy. Conceptualization: Conceptualization: M.P. and A.F. Data curation: M.P., S.S. and A.M. Formal Analysis: M.P. Funding acquisition: S.S. and A.F. Investigation: M.P., Y.P. and S.S. Methodology: M.P. Project administration: A.F. Resources: S.S. E.T. and A.F. Software: M.P. and E.T. Supervision: A.F. Validation: M.P., S.S. and A.F. Visualization: M.P. Writing – original draft: M.P. and A.F. Writing – review and editing: all authors.
Conflicts of interest
The authors have no conflicts of interest.
Ethical statement
The study does not include human subjects, human material, or human data. Solely, bacterial isolates were studied and were obtained as part of routine submissions to the National Reference Centre for Salmonella and other Enteric Bacterial Pathogens.
Footnotes
Abbreviations: cgMLST, Core genome multilocus sequence typing; ENA, European Nucleotide Archive; ESBL, extended-spectrum β-lactamase; HGT, horizontal gene transfer; MDR, multidrug-resistant; MLST, Multilocus sequence typing; NRC, National Reference Centre (for Salmonella); NTS, Non-typhoidal Salmonella enterica; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary tables and one supplementary figure are available with the online version of this article.
References
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. The global burden of nontyphoidal Salmonella gastroenteritis . Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 2.European food safety A, European Centre for Disease P, control The European Union one Health 2019 zoonoses report. EFSA J. 2021;19:e06406. doi: 10.2903/j.efsa.2021.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch-Institut R Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2019. Robert Koch-Institut; 2020. [Google Scholar]
- 4.Hagel S, Epple HJ, Feurle GE, Kern WV, Lynen Jansen P, et al. S2k-guideline gastrointestinal infectious diseases and Whipple’s disease. Z Gastroenterol. 2015;53:418–459. doi: 10.1055/s-0034-1399337. [DOI] [PubMed] [Google Scholar]
- 5.European Food Safety Authority, and European Centre for Disease Prevention and Control The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021;19:e06490. doi: 10.2903/j.efsa.2021.6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afema JA, Mather AE, Sischo WM. Antimicrobial resistance profiles and diversity in salmonella from humans and cattle, 2004-2011. Zoonoses Public Health. 2015;62:506–517. doi: 10.1111/zph.12172. [DOI] [PubMed] [Google Scholar]
- 7.Pardos de la Gandara M, Seral C, Castillo Garcia J, Rubio Calvo C, Weill FX. Prevalence and characterization of extended-spectrum beta-lactamases-producing Salmonella enterica isolates in Saragossa, Spain (2001-2008) Microb Drug Resist. 2011;17:207–213. doi: 10.1089/mdr.2010.0105. [DOI] [PubMed] [Google Scholar]
- 8.EFSA Panel on Biological Hazards (BIOHAZ) Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA Journal. 2011;9:2322. doi: 10.2903/j.efsa.2011.2322. [DOI] [Google Scholar]
- 9.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. beta-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother. 2005;56:115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 10.Livermore DM. Beta-lactamase-mediated resistance and opportunities for its control. J Antimicrob Chemother. 1998;41:25–41. doi: 10.1093/jac/41.suppl_4.25. [DOI] [PubMed] [Google Scholar]
- 11.Eller C, Simon S, Miller T, Frick JS, Prager R, et al. Presence of beta-lactamases in extended-spectrum-cephalosporin-resistant Salmonella enterica of 30 different serovars in Germany 2005-11. J Antimicrob Chemother. 2013;68:1978–1981. doi: 10.1093/jac/dkt163. [DOI] [PubMed] [Google Scholar]
- 12.Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae . J Antimicrob Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 14.Coipan CE, Westrell T, van Hoek A, Alm E, Kotila S, et al. Genomic epidemiology of emerging ESBL-producing Salmonella Kentucky bla CTX-M-14b in Europe. Emerg Microbes Infect. 2020;9:2124–2135. doi: 10.1080/22221751.2020.1821582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, et al. Supplement 2003-2007 (no.47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010;161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, et al. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLOS ONE. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. 2019;63:11. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber RE, Pietsch M, Fruhauf A, Pfeifer Y, Martin M, et al. IS26-Mediated Transfer of bla NDM-1 as the main route of resistance transmission during a polyclonal, multispecies outbreak in a German Hospital. Front Microbiol. 2019;10:2817. doi: 10.3389/fmicb.2019.02817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton BM, Harding GP, Zuccarelli AJ. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 26.Cohen E, Rahav G, Gal-Mor O. Genome sequence of an emerging Salmonella enterica serovar infantis and genomic comparison with other S. Genome Biol Evol. 2020;12:151–159. doi: 10.1093/gbe/evaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valcek A, Roer L, Overballe-Petersen S, Hansen F, Bortolaia V, et al. IncI1 ST3 and IncI1 ST7 plasmids from CTX-M-1-producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids from E. coli of animal origin. J Antimicrob Chemother. 2019;74:2171–2175. doi: 10.1093/jac/dkz199. [DOI] [PubMed] [Google Scholar]
- 28.Hancock SJ, Phan MD, Peters KM, Forde BM, Chong TM, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother. 2017;61:e01740-16. doi: 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Food Safety Authority, and European Centre for Disease Prevention and Control The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020;18:e06007. doi: 10.2903/j.efsa.2020.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.European Food Safety Authority, and European Centre for Disease Prevention and Control The European Union One Health 2018 Zoonoses Report. EFSA J. 2019;17:12. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su LH, Chiu CH, Chu C, Ou JT. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis. 2004;39:546–551. doi: 10.1086/422726. [DOI] [PubMed] [Google Scholar]
- 32.Kola A, Kohler C, Pfeifer Y, Schwab F, Kuhn K, et al. High prevalence of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat. J Antimicrob Chemother. 2012;67:2631–2634. doi: 10.1093/jac/dks295. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, et al. Extended-spectrum {beta}-lactamases and AmpC {beta}-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J Antimicrob Chemother. 2009;64:301–309. doi: 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- 34.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, et al. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol. 2014;16:977–994. doi: 10.1111/1462-2920.12351. [DOI] [PubMed] [Google Scholar]
- 35.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, et al. Comparative analysis of extended-spectrum-beta-lactamase CTX-M-65-producing Salmonella enterica serovar infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother. 2017;61:e00488-17. doi: 10.1128/AAC.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gymoese P, Kiil K, Torpdahl M, Osterlund MT, Sorensen G, et al. WGS based study of the population structure of Salmonella enterica serovar Infantis. BMC Genomics. 2019;20:870. doi: 10.1186/s12864-019-6260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown AC, Chen JC, Watkins LKF, Campbell D, Folster JP, et al. CTX-M-65 extended-spectrum beta-lactamase-producing Salmonella enterica serotype infantis, United States(1) Emerg Infect Dis. 2018;24:2284–2291. doi: 10.3201/eid2412.180500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Soto S, Abdel-Glil MY, Tomaso H, Linde J, Methner U. Emergence of multidrug-resistant Salmonella enterica subspecies enterica serovar infantis of multilocus sequence type 2283 in German broiler farms. Front Microbiol. 2020;11:1741. doi: 10.3389/fmicb.2020.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aviv G, Rahav G, Gal-Mor O. Horizontal transfer of the Salmonella enterica serovar infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio. 2016;7:e01395–16. doi: 10.1128/mBio.01395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietsch M, Eller C, Wendt C, Holfelder M, Falgenhauer L, et al. Molecular characterisation of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet Microbiol. 2017;200:130–137. doi: 10.1016/j.vetmic.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 41.Pietsch M, Irrgang A, Roschanski N, Brenner Michael G, Hamprecht A, et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics. 2018;19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen F, Olsen SS, Heltberg O, Justesen US, Fuglsang-Damgaard D, et al. Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb Drug Resist. 2014;20:316–324. doi: 10.1089/mdr.2013.0157. [DOI] [PubMed] [Google Scholar]
- 43.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, et al. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 2014;10:12. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae . J Antimicrob Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 45.Valenza G, Nickel S, Pfeifer Y, Eller C, Krupa E, et al. Extended-spectrum-beta-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob Agents Chemother. 2014;58:1228–1230. doi: 10.1128/AAC.01993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakkeren E, Huisman JS, Fattinger SA, Hausmann A, Furter M, et al. Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature. 2019;573:276–280. doi: 10.1038/s41586-019-1521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.