Abstract
In 2010, Brazil introduced the 10-valent pneumococcal conjugate vaccine (PCV10) into the national children’s immunization programme. This study describes the genetic characteristics of invasive Streptococcus pneumoniae isolates before and after PCV10 introduction. A subset of 466 [pre-PCV10 (2008–2009): n=232, post-PCV10 (2012–2013): n=234;<5 years old: n=310, ≥5 years old: n=156] pneumococcal isolates, collected through national laboratory surveillance, were whole-genome sequenced (WGS) to determine serotype, pilus locus, antimicrobial resistance and genetic lineages. Following PCV10 introduction, in the <5 years age group, non-vaccine serotypes (NVT) serotype 3 and serotype 19A were the most frequent, and serotypes 12F, 8 and 9 N in the ≥5 years old group. The study identified 65 Global Pneumococcal Sequence Clusters (GPSCs): 49 (88 %) were GPSCs previously described and 16 (12 %) were Brazilian clusters. In total, 36 GPSCs (55 %) were NVT lineages, 18 (28 %) vaccine serotypes (VT) and 11 (17 %) were both VT and NVT lineages. In both sampling periods, the most frequent lineage was GPSC6 (CC156, serotypes 14/9V). In the <5 years old group, a decrease in penicillin (P=0.0123) and cotrimoxazole (P<0.0001) resistance and an increase in tetracycline (P=0.019) were observed. Penicillin nonsusceptibility was predicted in 40 % of the isolates; 127 PBP combinations were identified (51 predicted MIC≥0.125 mg l−1); cotrimoxazole (folA and/or folP alterations), macrolide (mef and/or ermB) and tetracycline (tetM, tetO or tetS/M) resistance were predicted in 63, 13 and 21.6 % of pneumococci studied, respectively. The main lineages associated with multidrug resistance in the post-PCV10 period were composed of NVT, GPSC1 (CC320, serotype 19A), and GPSC47 (ST386, serotype 6C). The study provides a baseline for future comparisons and identified important NVT lineages in the post-PCV10 period in Brazil.
Keywords: Brazil, genomic surveillance, global pneumococcal sequence cluster, Multi-locus sequencing typing, PCV10, Streptococcus pneumoniae
Data Summary
Genome sequences are deposited in the European Nucleotide Archive (ENA), the accession number and the sample data is available in the supplementary material (Data_Summary_GPS_Brazil.xlsx). The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
This study, based on WGS analysis, makes several noteworthy contributions to understanding the genetic structure of the S. pneumoniae population in Brazil. We analysed genomic data from invasive pneumococcal isolates collected in Brazil between 2008 and 2013 to provide a detailed description of the population structure during that sampling period. We identified globally spreading lineages that also included non-vaccine serotype components, indicating that they potentially might contribute to vaccine evasion. The data generated by this study can be used as a baseline to determine vaccine impact during the following years.
Introduction
Streptococcus pneumoniae is the main cause of otitis media and community-acquired pneumonia as well as invasive pneumococcal disease (IPD), including meningitis, sepsis and bacteremia [1]. Antimicrobials and vaccines are tools currently available to treat and prevent pneumococcal diseases, respectively.
The indiscriminate use of antimicrobial agents in community settings results in the selection of resistant pneumococcal strains and impacts IPD by resulting in antibiotic treatment failure [2]. In addition, pressure due to vaccination can change resistance patterns temporally and geographically [1].
Pneumococcal conjugate vaccines (PCV, 7-valent PCV, 10-valent PCV, and 13-valent PCV) are highly effective in preventing IPD caused by serotypes present in its composition [3, 4]. Brazil was the first country to introduce the 10-valent pneumococcal conjugate vaccine (PCV10, target serotypes 1, 4, 5, 6B, 7F, 9V, 14, 18C, 19F and 23F) into their national childhood immunization programme in March 2010 [5]. The vaccine schedule was three primary doses at ages 2, 4 and 6 months and a booster dose for children aged 12–15 months. During the first year of PCV10 introduction, a catch-up campaign with two primary doses for children at 7 to 11 months of age plus a booster dose at 12–15 months, and a single dose for children aged 12 to <24 months was adopted [5]. In 2016, the primary schedule was changed to two primary doses at ages 4 and 6 months and a booster dose at 12 months [6].
It is well documented that PCV introduction was followed by changes in S. pneumoniae epidemiology due to (i) vaccine pressure leading to evidence of serotype replacement by the capsular switch, (ii) through the expansion of common strains and (iii) by increases in newly emerging non-vaccine type strains [3, 4]; as well as reductions in the transmission of vaccine types (VT) resulting in the indirect effect of herd immunity in the unvaccinated population. Therefore, surveillance is essential to detect potential temporal changes in the epidemiological and genetic characteristics of pneumococcal isolates [1, 7, 8]. Genomic methods have proved to be excellent tools for understanding the biology and epidemiology of important bacterial pathogens, including S. pneumoniae . Multi-locus sequence typing (MLST) is a molecular method historically important, and still widely used in pneumococcal epidemiology that consists of sequencing seven housekeeping genes as a sample of genomic variation and is used to define the sequence type (ST) and clonal complexes (CCs) [1]. This present study aimed to describe the genetic characteristics of invasive pneumococcal disease (IPD) isolates sampled from ongoing routine laboratory surveillance in Brazil during the pre- (2008–2009) and post- (2012–2013) PCV10 introduction periods. The whole-genome sequence data was used for in silico analysis of the serotype, antimicrobial resistance predictions, to identify possible changes in genetic lineages, and circulation of multi-drug-resistant clones following PCV10 introduction. The data generated will provide a baseline for continued vaccine impact monitoring and support future vaccination strategies for pneumococcal disease control.
Methods
Bacterial strain collection
The study collection consisted of a random subset of 466 IPD isolates recovered through a national laboratory surveillance network led by Institute Adolfo Lutz (IAL), the Brazilian National Reference Laboratory for Meningitis and Pneumococcal Infections. This study included pneumococcal isolates from pre- (n=232, 2008–2009) and post- (n=234, 2012–2013) PCV10 introduction periods collected in 20 of 26 Brazilian States (Table S1, available in the online version of this article) and corresponding to 14 % (n=466/3342) of the total S. pneumoniae isolates received by IAL in the period analysed. Isolates from the years 2010 and 2011 corresponding to the first years of PCV10 introduction were excluded from the study. Table S2 shows the pneumococcal study collection isolates stratified by age groups, clinical diagnosis and vaccine periods. Brandileone and collaborators [9] included in their publication the detailed phenotypical analyses of the pneumococcal serotypes that caused IPD before and after the introduction of PCV10 using data from the laboratory surveillance system in Brazil from a larger period (2005 to 2015) of time. From this dataset, data of 3342 isolates corresponding to the periods from 2008 to 2009 and 2012 to 2013 were used in our study as a basis for selection and comparison of serotype distribution of the 466 isolates whole-genome sequencing (WGS) subset (Figs S1 and S2). We presented serotype data for the entire collection of 3342 isolates and restrict other analyses to the 466 randomly sampled WGS.
The IAL receives strains previously identified as S. pneumoniae by the laboratory of origin and confirms this identification using classical methodologies described by WHO [10]. For routine surveillance, serotyping was performed by Quellung reaction and antimicrobial susceptibility profiles were determined by the disc diffusion and/or broth microdilution to determine minimal inhibitory concentrations (MIC) according to Clinical Laboratory Standards Institute (CLSI) breakpoints [11–13].
Genome sequencing and analyses
The 466 IPD isolates were WGS on the Illumina HiSeq platform to produce paired-end reads of 150 base pairs in length and raw data were deposited in the European Nucleotide Archive (ENA) (Supplementary Material: Data_Summary_GPS_Brazil.xlsx). WGS data were processed as previously described [14]. We derived the virulence factors (serotype [15] and pilus locus [16]) and multi-locus sequencing types (STs) [17].
The genetic structure was defined by assigning the clonal complexes (CCs) from the STs previously described by the Global Pneumococcal Sequencing Project (GPS) [14] and also by assigning Global Pneumococcal Sequence Cluster (GPSC) on each isolate using a PopPUNK [18], along with a reference list of pneumococcal isolates (n=13 454) in the GPS database (https://www.pneumogen.net/gps/assigningGPSCs.html). The STs and GPSCs described in this study were deposited in the PubMLST (https://pubmlst.org/organisms/streptococcus-pneumoniae) and GPS databases (https://www.pneumogen.net/gps/assigningGPSCs.html), respectively. Phylogenetic analysis was performed on all Brazilian isolates in this study by constructing a maximum-likelihood tree using FastTree [19]. In brief, the tree was built upon a SNP alignment after mapping reads to the reference genome of S. pneumoniae ATCC 700669 (NCBI accession number FM211187) using Burroughs Wheeler Aligner (BWA).
Capsular or serotype switching was identified in isolates with identical ST but different serotypes in this study. For each ST, we examined the genetic relatedness of isolates in lineage-specific phylogenies and place the Brazilian lineage of interest in a global context by including other GPS published isolates belonging to the same GPSC [20]. The lineage-specific tree was constructed using GUBBINS [21]. In brief, GUBBINS detects recombination regions and removes them when constructing the phylogeny. The recombination-free phylogeny created by GUBBINS was used as input for BactDating [22], an R package used to create a time-measured phylogeny performing Bayesian dating inference of the nodes on the bacterial phylogenetic tree; typically involves simultaneous Bayesian estimation of the molecular clock rate and coalescent rate as previously described [20]. The time-measured tree was used to estimate the period when capsular switching occurred.
Resistance profiles for six antibiotics, including penicillin [defined as penicillin binding protein transpeptidase amino acid sequence types (PBP types) based on pbp1A, pbp2B, pbp2X changes] [23, 24], chloramphenicol (cat), cotrimoxazole (folA and folP), erythromycin (ermB and mefA), tetracycline (tetM, tetO and tetS/M), and vancomycin (vanA, vanB, vanC, vanD, vanE and vanG) were predicted from genomic data, as previously described [16, 25]. Multidrug resistance (MDR) was defined as isolates with predicted intermediate resistance or resistance to three or more classes of antibiotics [26]. The serotype and antimicrobial resistance predictions for the 466 sampled isolates were compared to the phenotypic results generated from routine surveillance.
Statistical analyses
The pneumococcal isolates were defined as vaccine serotype (VT) when isolates belonged to predicted serotypes included in PCV10 (1, 4, 5, 6B, 7F, 9V, 14, 18C, 19F and 23F), and as non-vaccine serotypes (NVT) for the predicted serotypes non-PCV10, including the additional PCV13 serotypes 3, 6A and 19A. We defined the status of a lineage (GPSC) as VT (100 % PCV10 serotypes), NVT (100 % non-PCV10 serotypes) and GPSC with both VT and NVT isolates, based on its serotype composition detected in the whole study period. The prevalence of in silico serotypes was stratified by age groups (<5 years old and ≥5 years old).
As the population denominators are unavailable, we evaluated significant changes of VT/NVT in each GPSC lineage in proportion to all VT/NVT, respectively, using Fisher’s exact test. This calculation was performed to avoid the overestimation of the NVT increase. Overall, and by GPSC, the prevalence of antibiotic resistance between vaccine periods was also detected using Fisher’s exact test. Two-sided P-values of <0.05 were considered statistically significant. The number of samples was calculated to achieve 80 % of statistical power with a significant level of P-values. Before using the Fisher’s exact tests to compare variables (e.g. VT or penicillin resistance) before and after the PCV10 period, we calculated the number of samples that we need to achieve an 80 % statistical power with a significant level of P-value<0.05 using the R package pwr, which contains functions for basic power calculation [27]. When the variables (VT and/or antibiotic resistance) would not have sufficient statistical power to be tested, Fisher’s exact test was not performed. Multiple testing was adjusted using the Benjamin-Hochberg false discovery rate of 5 %, the statistical analysis was carried out in R version 3.5.2, and R scripts used for analyses were deposited at GitHub (https://github.com/StephanieWLo/Genomic-Surveillance).
Results
Serotype distribution
No discrepancies were observed between the 466 predicted serotypes and the Quellung results, and the frequency of the predicted serotypes in our study subset reflected the frequency of the serotypes identified in the larger 3342 isolates' collection. Figs S1 and S2 show serotype distribution by vaccine period (pre-PCV10 and post-PCV10), for age groups <5 years and ≥5 years, and for the larger and subset collections included in this WGS study.
As expected, a higher number of VT isolates was observed in the pre-PCV10 period mainly in the <5 years while NVT (including the additional PCV13, 3, 6A and 19A) were more frequent in the post-PCV10 period (Fig. S1). Before vaccination, serotype 14 was highly common in both age groups and after vaccine introduction, serotype 3 was most frequent in children aged <5 years, followed by serotype 19A, 6A, 12F and 6C (Fig. S1), and in ≥5 years serotype 12F, 3, as well the serotypes 8 and 9 N (Fig. S2).
Pneumococcal lineages
The predictions of serotype, ST and GPSC by age group and vaccine period of the 466 S . pneumoniae invasive study isolates are presented in Table S3.
A total of 159 STs were identified among 466 isolates sequenced, belonging to 54 CCs and 42 singletons. The phylogeny supported the good correlation of the CCs with the WGS-based GPSCs and, the latter typing scheme, revealed the genetic relatedness among the CCs 66, 81, 2216, 9747 plus three singletons (ST4913, 12483, 13878) in the GPSC16, one of the major GPSCs identified in the study (Fig. S3).
Overall, 65 GPSCs were identified and the most prevalent GPSCs were GPSC6 (CC156, serotype 14/9V), GPSC16 (CC66/81/2216/9747 and ST4913/12483/13878, serotypes 7C/9 N/14/15A/19F/23F/24/40), GPSC12 (CC180, serotype 3), GPSC5 (CC172, serotypes 6C/15B/15C/23A/23F) and GPSC11 (CC193, serotypes 11A/15A/15B/15C/18B/18C) (Fig. S3). Of the GPSCs identified, 36 GPSCs (55%) belonged to NVT lineages, 18 GPSCs (28 %) to VT lineages, and 11 (17 %) included both VT and NVT lineages (GPSC1, 5, 10, 11, 13, 16, 18, 23, 47, 61 and 231). Among the 466 pneumococcal isolates, 410 (88 %) were assigned to 49 GPSCs that have previously been found in the GPS reference database (last updated in April 2019, n=20 187, www.pneumogen.net\\gps\\assigningGPSCs.html) the remain 16 GPSCs (204, 231, 249, 289, 311, 341, 392-394, 571, 573-575, 577, 702, 811) included STs assigned in the international PubMLST database as mainly associated with Brazil (http://pubmlst.org, accessed: 26/05/2021) (Table S3). All eight global-spreading lineages recognized in the previous GPS study [14] were found in the current bacterial collection with an overall prevalence of 44.9 %: 17.8 % GPSC6, 9.0 % GPSC16, 5.6 % GPSC12, 3.9 % GPSC1, 3.0 % GPSC32, 2.6 % GPSC18, 1.5 % GPSC7, and 1.5 % GPSC23 (Table S3).
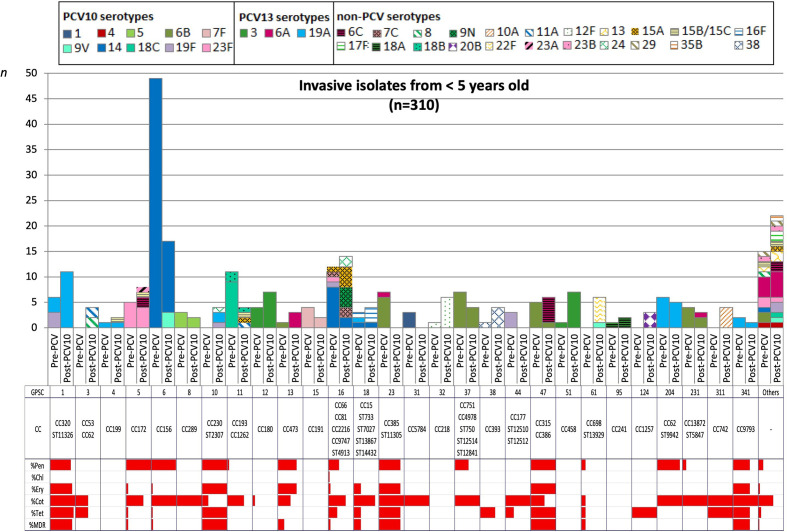
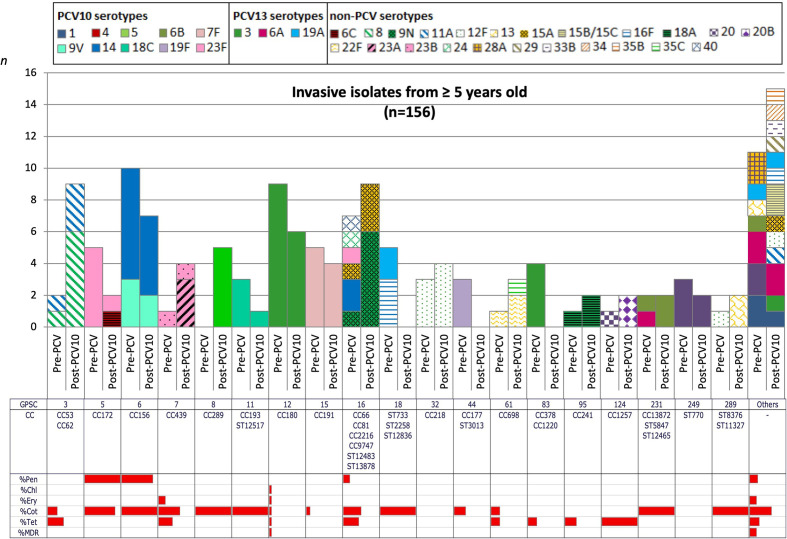
In comparison with the pre-PCV10 period, we detected any significant changes in the frequency of VT and NVT within GPSC, but we do not have sufficient statistical power to detect changes in each serotype. However, isolates associated with VT GPSCs decreased from 48–20 % (P<0.0001) and 32–24 % (P=0.2871) in the age groups <5 and ≥5 years old respectively, while isolates belonging to NVT GPSCs increased from 17–39 % (P<0.0001) and 34–52 % (P=0.0246). The five most frequent GPSCs by age groups are listed in Tables 1 and 2. GPSC6 and GPSC16 were among the top lineages in both age groups, pre-PCV10 and post-PCV10 periods. Though GPSC6, composed of CC156 and VT 9V and 14, remained a predominant lineage during the whole period of study and showed a decreasing trend among the <5 years old group (Fig. S4). In contrast, GPSC16 lineage persisted with the NVT components 9 N and 15A frequently observed in the post-PCV10 period. In the post-PCV10 NVT lineages were mainly associated with children aged <5 years; GPSC1 (CC320, serotype 19A), GPSC12 (CC180, serotype 3) and GPSC51 (CC458, serotype 3) (Fig. 1, Table 1). In the ≥5 years old group, GPSC3 expressing serotypes 8 (CC53) and 11A (CC62) became the predominant lineage 2–3 years after PCV10 introduction, though it was not in the top five lineages before vaccine roll-out (Fig. 2, Table 2).
Table 1.
The five most frequent lineages associated with serotypes of IPD isolates in the age group <5 years old (N=310) in the pre-PCV10 (2008–2009) and the post-PCV10 periods (2012–2013), Brazil
|
Rank |
Pre-PCV10 period (n=155) |
Post-PCV10 period (n=155) |
||||
|---|---|---|---|---|---|---|
|
GPSC (CC) |
N (%) |
Associated serotypes (n) a |
GPSC (CC) |
N (%) |
Associated serotypes (n) a |
|
|
First |
GPSC6 (CC156) |
49 (32 %) |
14 (49) |
GPSC6 (CC156) |
17 (11 %) |
14 (14), 9V (3) |
|
Second |
GPSC16 (CC66, CC81) |
12 (8 %) |
14 (8), 7 C (1), 15A (1), 19F (1), 23F (1) |
GPSC16 (CC66, ST4913, CC2216, CC9747) |
14 (9 %) |
9 N (4), 15A (4), 7 C (2), 14 (2), 24 (2) |
|
Third |
GPSC11 (CC193) |
11 (7 %) |
18C (9), 18B (2) |
GPSC1 (CC320) |
11 (7 %) |
19A (11) |
|
Fourth |
GPSC23 (CC385, ST11305) |
7 (5 %) |
6B (6), 6A (1) |
GPSC5 (CC172) |
8 (5 %) |
23F (4), 6 C (2), 15B/ 15 C (1), 23A (1) |
|
GPSC37 (CC751, CC4978, ST750, ST12514, ST12841) |
7 (5 %) |
6B (7) |
||||
|
Fifth |
GPSC1 (CC320, ST11326) |
6 (4 %) |
19A (3), 19F (3) |
GPSC12 (CC180) |
7 (4,5 %) |
3 (7) |
|
GPSC204 (CC62, ST9942) |
6 (4 %) |
19A (6) |
GPSC51 (CC458) |
7 (4,5 %) |
3 (3) |
|
a, Black, VT or PCV10 serotypes; blue, NVT additional PCV13 serotypes; and red, NVT non-PCV serotypes.
Table 2.
The five most frequent lineages associated with serotypes of IPD isolates in the age group ≥5 years old (N=156) in the pre-PCV10 (2008–2009) and the post-PCV10 periods (2012–2013), Brazil
|
Rank |
Pre-PCV10 period (n=77) |
Post-PCV10 period (n=79) |
||||
|---|---|---|---|---|---|---|
|
GPSC (CC) |
N (%) |
Associated serotypes (n) a |
GPSC (CC) |
N (%) |
Associated serotypes (n) a |
|
|
First |
GPSC6 (CC156) |
10 (13 %) |
14 (7), 9V (3) |
GPSC3 (CC53, CC62) |
9 (11 %) |
8 (6), 11A (3) |
|
GPSC16 (CC66, CC2216, ST13878) |
9 (11 %) |
9 N (6), 15A (3) |
||||
|
Second |
GPSC12 (CC180) |
9 (12 %) |
3 (9) |
GPSC6 (CC156) |
7 (9 %) |
14 (5), 9V (2) |
|
Third |
GPSC16 (CC66, CC2216, CC81, CC9747, ST12483) |
7 (9 %) |
14 (2), 9N (1), 15A (1), 23F (1), 24 (1), 40 (1) |
GPSC12 (CC180) |
6 (8 %) |
3 (6) |
|
Fourth |
GPSC5 (CC172) |
5 (6 %) |
23F (5) |
GPSC8 (CC289) |
5 (6 %) |
5 (5) |
|
|
GPSC15 (CC191) |
5 (6 %) |
7F (5) |
|||
|
|
GPSC18 (ST2258, ST12836, ST733) |
5 (6 %) |
16 F (3), 19A (2) |
|||
|
Fifth |
GPSC83 (CC378, CC1220) |
4 (5 %) |
3 (4) |
GPSC7 (CC439) |
4 (5 %) |
23A (3), 23B (1) |
|
GPSC15 (CC191) |
4 (5 %) |
7F (4) |
||||
|
GPSC32 (CC218) |
4 (5 %) |
12 F (4) |
||||
a, Black, VT or PCV10 serotypes; blue, NVT additional PCV13 serotypes; and red, NVT non-PCV serotypes.
Fig. 1.
Dynamics of Global Pneumococcal Sequence Clusters (GPSCs) among invasive isolates from children aged <5 years old over vaccine periods in Brazil. The number of invasive pneumococcal isolates, coloured by serotypes, is plotted by GPSC with stratification into two vaccine periods (pre-PCV10 and post-PCV10) and MLST CC. Solid fill represented the VT and NVT additional PCV13 serotypes while hatched patterns represented the NVT non-PCV serotypes. The antibiotic resistance pattern to penicillin, chloramphenicol, erythromycin, cotrimoxazole, tetracycline and MDR are present for each GPSC and in the entire period studied.
Fig. 2.
Dynamics of Global Pneumococcal Sequence Clusters (GPSCs) among invasive isolates from children aged ≥5 years old over vaccine periods in Brazil. The number of invasive pneumococcal isolates, coloured by serotypes, is plotted by GPSC with stratification into two vaccine periods (pre-PC10 and post-PCV10) and MLST CC. Solid fill represented the VT and NVT additional PCV13 serotypes while hatched patterns represented the NVT non-PCV serotypes. The antibiotic resistance pattern to penicillin, chloramphenicol, erythromycin, cotrimoxazole, tetracycline and MDR are present for each GPSC and in the entire period studied.
Capsular switch variants
Among the STs identified, nine occurred in more than one serotype and are suggestive of capsular switching events: STs 66, 156, 193, 199, 338 and 386; four are NVT switches and were observed in the post-PCV10 period (ST66 serotype 9 N, ST 199 serotypes 19A/15B/15C, ST338 serotype 15B/15C, and ST386 serotype 6C).
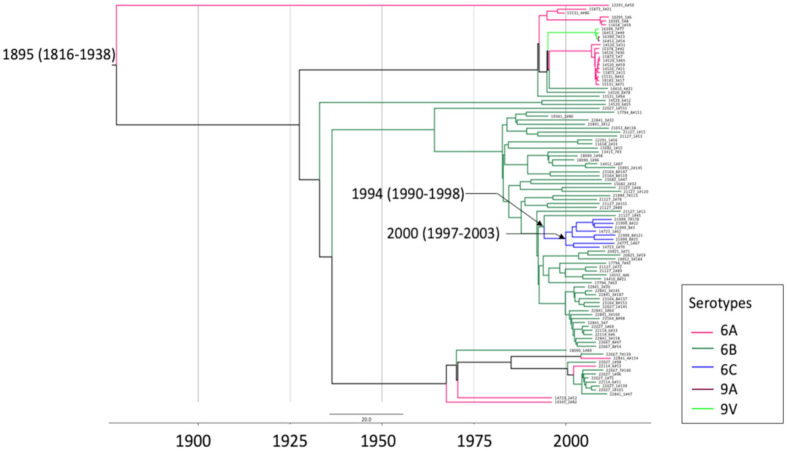
The ST386 (GPSC47) was represented by serotypes 6B in the pre-PCV10 period and 6C in the post-PCV10 period (Fig. 1) and a time-measured phylogeny of GPSC47 using isolates from the GPS database and including the ST386 of this study, showed serotype 6C grouped separately from the ST386 serotype 6B isolates (Fig. 3). Using BactDating software we estimated a possible capsular switch may have occurred between serotypes 6B and 6C isolates around 1994 (95 % confidence interval: 1990–1997). This suggests the capsular switch event occurred in the pre-PCV era leading to the selection of the NVT lineage GPSC47 (ST386, serotype 6C) over the VT lineage GPSC47 (ST386, serotype 6B) following PCV introduction.
Fig. 3.
A timed-measured phylogeny of GPSC47 (ST386) isolates from Brazil and the other 15 countries from Gladstone et al. [14]. The phylogeny is built using BactDating [22] with 100 000 000 generations on a recombination-free SNP alignment generated by GUBBINs [21]. The most recent common ancestor (tMRCA) of the serotype 6C clade (all isolates are ST386) is estimated to emerge in around 2000 (95 % confidence interval: 1997–2003), and the capsular switching occurred among 1990–2003. The phylogeny and metadata can be interactively visualized at https://microreactorg/project/tmxT66fLq56uZb5VfA1BYC.
Antimicrobial resistance
The antimicrobial susceptibility testing performed at IAL showed good correlation (susceptible, intermediate or resistant category) with the in silico prediction, presenting a 100 % of the agreement for erythromycin, chloramphenicol and vancomycin; 98.9 % (461/466) for tetracycline, 98.7 %(460/466) for penicillin and 93.6 %(436/466) for cotrimoxazole. Due to the high level of concordance between phenotype and genotype results, we used the WGS predicted antimicrobial resistance in the following analyses.
The antimicrobial non-susceptibility patterns between pre-and post-PCV10 periods showed a significant increase of tetracycline resistance (P=0.0019) and a decrease of penicillin (P=0.0123) and cotrimoxazole resistance (P<0.0001) among isolates from children aged <5 years after vaccination. No significant difference (P≥0.05) was observed in the frequency of predicted non-susceptibility to chloramphenicol, erythromycin and MDR isolates from <5 years, as well for all studied antibiotics in the isolates from ≥5 years (Table 3). All isolates were predicted as vancomycin susceptible.
Table 3.
Proportions of IPD isolates with antibiotic non-susceptibility in pre-PCV10 and post-PCV10 periods (N=466), Brazil
|
Antibiotics a |
Age <5 years old |
Age ≥5 years old |
||||
|---|---|---|---|---|---|---|
|
Pre-PCV10 (N=155) |
Post-PCV10 (N=155) |
P-value b |
Pre-PCV10 (N=77) |
Post-PCV10 (N=79) |
P-value b |
|
|
Penicillin |
88 (57 %) |
66 (42 %) |
0.0123 |
18 (23 %) |
17 (22 %) |
0.8488 |
|
Chloramphenicol |
1 (0.6 %) |
0 |
1.0000 |
1 (1 %) |
1 (1 %) |
1.0000 |
|
Cotrimoxazole |
133 (86 %) |
81 (52 %) |
<0.0001 |
42 (55 %) |
37 (47 %) |
0.3426 |
|
Erythromycin |
19 (12 %) |
32 (21 %) |
0.0653 |
3 (4 %) |
5 (6 %) |
0.7195 |
|
Tetracycline |
24 (15 %) |
48 (31 %) |
0.0019 |
11 (14 %) |
17 (22 %) |
0.2982 |
|
MDR c |
19 (12 %) |
31 (20 %) |
0.0887 |
3 (4 %) |
4 (5 %) |
1.0000 |
a, CLSI, breakpoints: penicillin, susceptible ≤0.06 mg l−1 and resistant ≥0.125 mg l−1; chloramphenicol, susceptible ≤4 mg l−1 and resistant ≥8 mg l−1; cotrimoxazole, susceptible ≤0.5/9.5 mg l−1, intermediate 1/19-2/38 mg l−1 and resistant ≥4/76 mg l−1; erythromycin, susceptible ≤0.25 mg l−1, intermediate 0.5 mg l−1 and resistant ≥1 mg l−1; and tetracycline, susceptible ≤1 mg l−1, intermediate 2 mg l−1, and resistant ≥4 mg l−1.
b, Fisher´s two-tailed test.
c, MDR, multidrug resistance; intermediate or resistant isolates to three or more classes of antibiotic.
WGS analysis identified 127 PBP types allele combinations, 51 of them predicted non-susceptibility to penicillin (MIC ≥0.125 mg l−1). Independent of age group, the PBP allele combinations 13-11-16 (n=12, MIC=4 mg l−1), 15-12-18 (n=11, MIC=2 mg l−1) and 45-12-63 (n=8, MIC=2 mg l−1) were predominant and associated with specific international antimicrobial-resistant lineages, GPSC1 (CC320, serotype 19A), GPSC6 (CC156, serotypes 9V) and GPSC6 (CC156, serotype 14), respectively. Despite the other PBP profiles that confer penicillin resistance in the post-PCV10 period, we highlight the third most frequent PBP profile in the <5 years old group the profile 2-53-77 (n=6, MIC=0.125 mg l−1) observed in the lineage GPSC47 (ST386, serotype 6C and CC315, serotype 6B) since it is associated with an important NVT and capsular switching described in our study (Table S4).
The frequency of the gene combination ermB and mef increased from 2 (1 %) to 11 (7 %) isolates in the pre-PCV10 vs post-PCV10 period in the <5 years old and was associated with GPSC1 (CC320, serotype 19A). Full resistance to cotrimoxazole (MIC ≥4 mg l−1) was characterized for the presence of alterations in genes folA (all isolates have the I100L substitution) and folP, which were detected in a large proportion of the isolates (n=210, 46 %). As previously shown [16], a mutation within folA or folP alone conferred intermediate cotrimoxazole resistance, while mutations within both folA (I100L) and folP (1–2 codon insertions) conferred full resistance. For tetracycline, the most frequent resistance gene detected was tetM (n=97, 21 %), mainly identified in the post-PCV10 period (n=62/234, 26 %) and associated with GPSC1 (CC320, serotype 19A) and GPSC16 (CC66, serotype 9 N). Additionally, we observed lower frequencies of tetO and the combination of the tetS and tetM genes also predicting tetracycline resistance. The cat gene conferring chloramphenicol resistance, substitutions in the rpoB gene (P15A, H21N, or K22N) that predict rifampicin resistance (MIC >2 mg l−1) and substitutions in the parC gene (S79C, S29F, or S79Y) predicting fluoroquinolone resistance were identified in only a few isolates (Table 4).
Table 4.
Non-penicillin resistance gene determinants from IPD isolates by age groups in pre-PCV10 and post-PCV10 periods, Brazil
|
Antibiotic |
Resistance genes |
Age <5 years old (N=310) |
Age≥5 years old (N=156) |
Total (N=466) |
||
|---|---|---|---|---|---|---|
|
Pre-PCV10 (n=155) |
Post-PCV10 (n=155) |
Pre-PCV10 (n=77) |
Post-PCV10 (n=79) |
|||
|
N (%) |
N (%) |
N (%) |
N (%) |
N (%) |
||
|
Macrolide a |
mef |
2 (1) |
6 (4) |
1 (1) |
1 (1) |
10 (2) |
|
ermB |
15 (10) |
15 (10) |
2 (3) |
3 (4) |
35 (8) |
|
|
ermB +mef |
2 (1) |
11 (7) |
0 (0) |
1 (1) |
14 (3) |
|
|
rplD2 b |
0 (0) |
1 (1) |
0 (0) |
0 (0) |
1 (0.2) |
|
|
Cotrimoxazole c |
folA only |
6 (4) |
3 (2) |
3 (4) |
2 (3) |
14 (3) |
|
folP only |
24 (15) |
19 (12) |
12 (16) |
10 (13) |
65 (14) |
|
|
folA +folP |
104 (67) |
59 (38) |
27 (35) |
24 (30) |
214 (46) |
|
|
Tetracycline |
tetO |
0 (0) |
0 (0) |
0 (0) |
2 (3) |
2 (0.4) |
|
tetM |
24 (15) |
47 (30) |
11 (14) |
15 (19) |
97 (21) |
|
|
tetS +tetM |
0 (0) |
1 (1) |
0 (0) |
0 (0) |
1 (0.2) |
|
|
Chloramphenicol d |
cat |
1 (1) |
0 (0) |
1 (1) |
1 (1) |
3 (1) |
|
Rifampin e |
rpoB1 |
0 (0) |
1 (1) |
2 (3) |
0 (0) |
3 (1) |
|
Fluoroquinolone f |
parC |
1 (1) |
0 (0) |
1 (1) |
1 (1) |
3 (1) |
a, mef, macrolide efflux pumps gene resistance; ermB, macrolide erythromycin methylation.
b, One rare substitution (E7K) within rplD2 gene core genome mutations conferred erythromycin resistance.
c, One to five substitutions within the folA gene (Q1H, D2N, V3I, D12T, E14D or I20L) or one to two codon insertions within the folP gene (at nucleotides 169, 174, 176, 177, 178, 180, 182, 185, 186, 188, 189 or 195) result in an intermediate phenotype (MIC 1–2 mg l−1) against cotrimoxazole. The folA substitutions (Q1H, Q1Y, D2N, V3I, V6A, Q11H, D12G, E14D or I20L) combined with folP insertions (at the nucleotides 169, 175, 176, 177, 178, 179, 180, 182, 187, 186, 189 or 195) result in a resistant phenotype (MIC ≥4 mg l−1).
d, cat, Chloramphenicol acetyltransferase.
e, One substitution within the rpoB1 gene (P15A, H21N or K22N) in each isolated.
f, One substitution within the parC gene (S79C, S29F, or S79Y) in each isolated.
This study observed MDR in 57 (12.2 %) pneumococcal isolates, 22 (9.5 %) in the pre-PCV10, and 35 (14.9 %) in the post-PCV10 period (Table 3). Figs 1 and 2 illustrated the overall resistance among the GPSCs in each age group. The <5 years old group presented higher levels of MDR associated mainly with GPSCs 1, 10, 23, 47 and 341. Focusing on the post-PCV10 period, 12 isolates were associated with the lineage GPSC1 (CC320, serotype 19A) with a profile of high resistance to penicillin (MIC=4 mg l−1) plus resistance to cotrimoxazole, erythromycin, and tetracycline; and five related to GPSC47 (ST386, serotype 6C) with a profile of lower resistance to penicillin (MIC=0.125 mg l−1) and resistance to erythromycin and tetracycline.
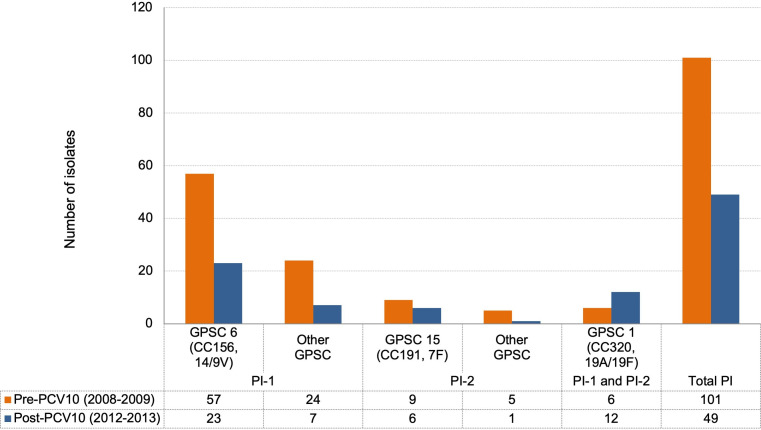
Pilus islets
During the study period, the presence of pilus islets (PIs) was observed in 32 % (n=150/466) of isolates (pre-PCV10: n=81 PI-1, n=14 PI-2, and n=6 PI-1 and PI-2; and post-PCV10: n=30 PI-1, n=7 PI-2, and n=12 PI-1 and PI-2). The PI-1 type was related to GPSC6 (CC156, serotypes 14/9V) lineage. The combination of P-1 and P-2 was observed only in the lineage GPSC1 (CC320, serotypes 19A/19F) (Fig. 4). In 59 % (n=89/150) of isolates with a pilus we observed high level penicillin resistance, 75 isolates presented PI-1 [MIC=2 mg l−1; GPSC6 (CC156, serotype 14)] and 14 isolates presenting PI-1and PI-2 (MIC=4 mg l−1; GPSC1 (CC320, serotype 19A), with 13 of them also MDR (cotrimoxazole, erythromycin and tetracycline).
Fig. 4.
Distribution of pilus islets genes in the main GPSCs by vaccine period studied (n=150).
Discussion
Our study analysed the genomic characteristics of a select subset of invasive pneumococcus strains obtained through national laboratory-based surveillance in Brazil. The distribution of serotypes predicted by our WGS study represented as closely as possible the national serotype distribution in the pre- (2008–2009) and post-PCV10 (2012–2013) periods. As expected, a lower prevalence of VT in the post-PCV10 period was due to the substantial PCV10 impact on IPD in the country [28, 29] suggesting that even in a short period after the introduction of the vaccine, it was possible to observe the phenomenon of herd immunity since this VT reduction was also observed in the group of ≥5 years old age, who are not targeted for PCV10 vaccination. However, some increase in NVTs was also documented. A previous Brazilian study [9] analysed a large collection (n=8971) of IPD isolates and compared the prevalence of VT and NVT over a longer period, pre-PCV10 (2005–2009) and post-PCV10 (2010–2015), and showed a large IPD VT reduction among children and adult population, documenting a direct and indirect vaccine effect. They also showed a change to NVT as the main cause of the IPD in the post-PCV10 period and concluded that in Brazil there is evidence of cross-protection between serotypes 6B/6A, a fact not observed among the serotypes 6B/6C and 19F/19A. In the post-PCV10 period, our study observed the NVT 3, 19A, 6A, 12F and 6C in the <5 years old age group, and serotypes 12F, 3, 8 and 9 N in the ≥5 years old age group; suggesting, as observed by Brandileone et al. [9], an absence of cross-protection between the serotypes 6B and 19F present in the PCV10 composition and the serotypes 6C and 19A, respectively, and revealing that the burden of pneumococcal disease could be further reduced in the country with the introduction in the national childhood immunization programme of PCV13 or other new generation of PCVs (new-PCV10, PCV15 and PCV20), which include the serotypes 3, 6A (with cross-protection to serotype 6C), and 19A in their composition.
Bacterial molecular typing is essential in the surveillance of infectious diseases [20]. This study characterized the baseline population pneumococcal structure for continued vaccine impact monitoring using whole-genome sequencing. Genome data not only allow us to extract public health-relevant data (e.g. serotype and antibiotic resistance profile) from a single experiment but also delineates genetic lineages using both whole-genome clustering method (GPSC) and multi-locus sequencing typing (MLST). Our findings showed a good concordance between these two typing methods. The whole-genome clustering method has further revealed the relationships between strains over a longer timescale by accounting for genetic variations across the whole genome [14]. The GPSCs characterization of the 466 IPD isolates presented a similar genetic structure to the globally GPSC described by the GPS project [14] with the majority of the isolates belonging to these previously described GPSCs. The study collection showed the majority of the GPSC lineages belonged to NVT lineages and a smaller proportion of lineages expressing both VT and NVT serotypes. The post-PCV10 period was marked by the increase in NVT lineages for both the <5 and ≥5 years old age groups. We observed the emergence of NVT lineages, GPSC1 (CC320, serotype 19A), GPSC12 (CC180, serotype 3) and GPSC51 (CC458, serotype 3) in <5 years old and GPSC3 (CC53, serotype 8 and CC62, serotype 11A) in ≥5 years old age groups, highlighting the importance of expanding the PCV coverage for a higher valence vaccine such as PCV13 or others that are still in development (new-PCV10, PCV15 and PCV20) would be useful to further reduce pneumococcal diseases in Brazil.
The pneumococcal population presents several strategies that allow the maintenance of a lineage in the face of pressures imposed through the use of vaccines or antimicrobials: (i) the replacement of a VT by NVT lineage through capsular switching to avoid vaccine-induced immunity, (ii) the acquisition of resistance genes and (iii) the expansion of an existing NVT lineage by filling the open niche left by VT lineage [3, 4]. In our study, we showed the lineage GPSC6 (CC156, serotypes 14–9V) associated with penicillin resistance (PBP 15-12-18 and 45-12-63) was frequent in both vaccine periods and age groups studied. This lineage, well documented in Brazil and globally, is often MDR [30–34], and as expected for a VT lineage [9, 35, 36], showed a decreasing trend, primarily in the <5 years old. A few lineages associated with NVTs in this study have also been documented globally in the post-pneumococcal vaccine periods [14, 37], for example, GPSC1 (CC320, serotype 19A), GPSC3 (CC53, serotype 8), GPSC12 (CC180, serotype 3) and GPSC16 (CC66, serotype 9 N). CC320 serotype 19A was one of the predominant emerging lineages in PCV7 and PCV10 countries [38–43]. Its expansion has been associated with capsular switch events from serotype 19F to 19A and the association with MDR has further allowed its selection in the post-PCV period [41, 43–48]. This lineage was been detected in Brazil since the pre-PCV10 period, but it increased after the PCV10 introduction independent of the age group [49–51]. Another important lineage disseminated worldwide is GPSC12 (CC180, serotype 3), recently associated with MDR [52–54]. In Brazil, this lineage has expanded as a predominant cause of serotype 3 invasive disease in the post-PCV10 era among adults [9, 36]. In this study, GPSC12 was associated with antimicrobial susceptibility, but at IAL has rarely been identified in some isolates as MDR in chloramphenicol, erythromycin, clindamycin and tetracycline (IAL unpublished data). The serotype 8 GPSC3 (CC53) lineage is widely distributed [37, 53, 55, 56] and has been observed in Brazil before PCV introduction [36, 57, 58]. In many countries, the GPSC16 (CC66) is primarily a serotype 9 N lineage associated with carriage [56] but has also been reported with different serotypes [56]. In Brazil and our study, this lineage is mostly invasive isolates expressing the capsular VTs 14 and 19F [31, 33, 36] and rarely are found as serotype 9 N.
The molecular characterization of isolates enabled the identification of several possible capsular switching events from VT to NVT. Initial molecular studies in the serotype 6C reported its origin related to independent recombination events involved isolates from serotype 6A [59], but after that other studies reported possible recombinant events from other serotypes like 6B [60, 61] suggesting multiple genetic origins for serotype 6C. In our study, we estimated a switch from serotype 6B to 6C occurred in the GPSC47 (ST386) before vaccine implementation with an expansion of the serotype 6C clone in the post-PCV10. This clonal expansion correlates with previous data from Lo et al. [37] that suggests serotype replacement is mostly mediated by expansion of NVT within VT lineages following vaccine implementation.
The use of PCVs in routine immunization has resulted in a significant effect on the prevalence of antimicrobial resistance, as their formulations include serotypes mostly associated with penicillin and multidrug resistance [62–64]. Beta-lactams are widely used and generally effective for the treatment of pneumococcal infections. Penicillin is recommended to treat non-meningitis pneumococcal infection caused by strains with penicillin MIC <8 mg l−1, instead of broad-spectrum antimicrobials such as the third-generation cephalosporin [65]. In Brazil, the use of third-generation cephalosporins is the standard choice for the empiric treatment of meningitis independent of the antimicrobial susceptibility testing results and the combination of cephalosporin and vancomycin has been used in cases of failure to respond to initial treatment. In the present study, we observed significant reductions in penicillin and cotrimoxazole resistance rates and increases in the frequency of tetracycline resistance in the post-PCV10 period for the <5 years old group. We identified resistance determinants by WGS commonly conferring resistance to penicillin, macrolides, cotrimoxazole, tetracycline and chloramphenicol [45]. In concordance, a recent Brazilian study [2] observed a reduction of isolates expressing penicillin MIC ≥0.125 mg l−1 in the first 3 years of post-PCV10 introduction (2011 to 2013), with high rates of cotrimoxazole non-susceptibility found during the study years (2007 to 2019), but showing a declining trend after PCV10 implementation, and a gradual increase of non-susceptibility to erythromycin and tetracycline over the study, reaching high rates in the years 2017–2019. We demonstrated that the most frequent lineages related to MDR in the post-PCV10 were NVT GPSC1 (CC320, serotype 19A) with high resistance to penicillin (MIC=4 mg l−1), cotrimoxazole, erythromycin and tetracycline, and the single lineage with the presence of the pilus islet PI-1 and PI-2; and the GPSC47 (ST386, serotype 6C) with lower resistance to penicillin (MIC=0.125 mg l−1) and resistance to erythromycin and tetracycline. We recommend continued genomic surveillance for long-term monitoring following data presented by Brandileone et al. [2] showing how the early impact of PCV10 in reducing non-susceptibility to beta-lactam antibiotics was eroded by increases in penicillin resistance, mainly associated with NVT S. pneumoniae , and reaching the highest rates in the years 2017–2019.
In addition to the capsular polysaccharide associated with nasopharyngeal colonization, studies show that S. pneumoniae has pilus structure that is involved in the adhesion and invasion of the bacteria in human respiratory epithelial cells [66, 67]. Some studies demonstrated the association of antimicrobial resistance and pili presence, suggesting the pili structure may have a role in the spread of these antimicrobial-resistant lineages [66, 68, 69]. One-third of our isolates had target sequences for the pilus with the majority PI-1 type (74%) and associated with penicillin resistance GPSC6 (CC156, serotype 14) lineage. A recent review [66] analysing the role of the pilus islet in S. pneumoniae showed similar overall rates of pili with the predominance of PI-1, presence of PI-1and PI-2 in CC320 serotype 19A lineage, and the association of these genes with antimicrobial and MDR. The presence of the pilus islets PI-1and PI-2 in the MDR GPSC1 (CC320, serotype 19A) lineage may provide an additional advantage for these isolates as they are thought to enhance adherence and colonization [66]. The fact that this lineage is also primarily MDR could explain the success in the establishment of this NVT lineage in the post-PCV10 period.
Despite the limitation that our WGS study was only performed on a subset of the invasive isolates from the pneumococcal laboratory-based surveillance system in Brazil, we did show that the sampling was representative of the overall collection of isolates. We also provided serotype data available on the entire collection and used the subset to identify major antibiotic resistance mechanisms and important genetic lineages in the post-PCV10 period, highlighting the importance of specific NVT genetic lineages in the post-PCV10 period.
Even with the global widespread use of the PCVs, S. pneumoniae remains a major bacterial cause of community-acquired pneumonia [70] and one of the main bacterial agents associated with viral co-infections. Since the first great influenza pandemic in 1918 [71], followed almost a century later in 2009 by H1N1 [72] and currently the worldwide COVID-19 pandemic [73] highlights continued surveillance and monitoring of S. pneumoniae as a priority. This study provides detailed genomic data of invasive pneumococcal isolates from national surveillance in Brazil, generating a baseline that can help for the creation of long-term surveillance to monitor the vaccine impact and public health strategies.
Supplementary Data
Funding information
This study was co-funded by the Bill and Melinda Gates Foundation (grant code OPP1034556), the Wellcome Sanger Institute (core Wellcome grants 098051 and 206194), and the USA Centres for Disease Control and Prevention. The funding sources had no role in isolate selection, analysis, or data interpretation.
Acknowledgements
We would like to thank all microbiologists from the laboratories in the Brazilian States for sending the invasive isolates of S. pneumoniae to the IAL and for supporting IPD surveillance in the country. At the IAL, our thanks to the laboratory staff: Lincoln S. do Prado, Maria Luiza L. S. Guerra, Rosemeire C. Almendros, and Ueslei J. Dias for carrying out the phenotypic characterization of the isolates. We would like to thank all members of the Global Pneumococcal Sequencing Consortium (www.pneumogen.net/gps/partners.html) for creating a rich database, the team at CDC for DNA extraction, the Wellcome Sanger Institute sequencing facility for whole-genome sequencing, and the Pathogen Informatics Team at the Wellcome Sanger Institute for technical support of bioinformatic analyses. The corresponding author had full access to the data and is responsible for the final decision to submit for publication.
Author contributions
S.C.G.A.: conceptualization, data curation, investigation, visualisation, writing (original draft preparation); S.W.L.: conceptualization, methodology, software, project administration, data curation, investigation, formal analysis, visualization, writing (review and editing); P.A.H.: methodology, project administration, data curation, formal analysis, visualization, writing (review and editing); R.A.G.: methodology, software, project administration, data curation, formal analysis, visualization, writing (review and editing); A.P.C.: investigation, writing (review and editing); M.C.C.B.: investigation, writing (review and editing); K.P.K.: conceptualization, funding; R.F.B.: conceptualization, funding; S.D.B.: conceptualization, funding, project administration; L.M.: conceptualization, resources, funding, methodology, project administration, data curation, investigation, formal analysis, writing (review and editing). All authors contributed to the final version of the manuscript and approved it.
Conflicts of interest
S.C.G.A. reports a travel grant from Pfizer, outside the submitted work. R.A.G. reports a Ph.D. studentship from Pfizer, outside the submitted work. M.C.C.B. reports personal fees from Pfizer and Merck, outside the submitted work. S.D.B. reports personal fees from Pfizer and Merck, outside the submitted work. All other authors declare no conflicts of interest.
Ethical statement
Isolates for this study were selected from a retrospective bacterial collection from Institute Adolfo Lutz. The patient data were fully anonymized and obtained as part of the routine clinical care procedures. No tissue material or other biological material was obtained from humans. The study was approved by the Technical and Scientific Council (CTC70H-2015) and Ethics Committee (CAAE: 54891516.4.0000.0059) of Institute Adolfo Lutz (São Paulo, Brazil). The findings and conclusions in this manuscript are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Abbreviations: CC, clonal complex; CLSI, Clinical Laboratory Standards Institute; ENA, European Nucleotide Archive; GPS, Global Pneumococcal Sequencing Project; GPSC, Global Pneumococcal Sequence Cluster; IAL, Institute Adolfo Lutz; IPD, invasive pneumococcal disease; MDR, Multidrug resistance; MIC, minimal inhibitory concentration; MLST, Multi-locus sequencing typing; NVT, non-vaccine serotype; PBP, Penicillin Binding Protein; PCV10, 10-valent pneumococcal conjugate vaccine; PI, pilus islets; SNP, single nucleotide polymorphisms; ST, sequence types; VT, vaccine serotype; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.Andam CP, Hanage WP. Mechanisms of genome evolution of Streptococcus . Infect Genet Evol. 2015;33:334–342. doi: 10.1016/j.meegid.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandileone MC, Almeida SCG, Bokermann S, Minamisava R, Berezin EN, et al. Dynamics of antimicrobial resistance of Streptococcus pneumoniae following PCV10 introduction in Brazil: Nationwide surveillance from 2007 to 2019. Vaccine. 2021;39:3207–3215. doi: 10.1016/j.vaccine.2021.02.063. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011;378:1962–1973. doi: 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewnard JA, Hanage WP. Making sense of differences in pneumococcal serotype replacement. Lancet Infect Dis. 2019;29:1–8. doi: 10.1016/S1473-3099(18)30660-1. [DOI] [PubMed] [Google Scholar]
- 5.MS/Brasil - Ministério da Saúde Informe Técnico da vacina Pneumocócica 10-valente Conjugada. Ministério da Saúde. 2010. http://www.sgc.goias.gov.br/upload/links/arq_723_infotec.pdf
- 6.MS/Brasil - Ministério da Saúde Nota Informativa 149/2015. 2015. http://www.aids.gov.br/pt-br/legislacao/nota-informativa-no-1492015
- 7.Chaguza C, Cornick JE, Andam CP, Gladstone RA, Alaerts M, et al. Population genetic structure, antibiotic resistance, capsule switching, and evolution of invasive pneumococci before conjugate vaccination in Malawi. Vaccine. 2017;35:4594–4602. doi: 10.1016/j.vaccine.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaguza C, Heinsbroek E, Gladstone RA, Tafatatha T, Alaerts M, et al. Early signals of vaccine-driven perturbation seen in pneumococcal carriage population genomic data. Clin Infect Dis. 2020;70:1294–1303. doi: 10.1093/cid/ciz404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandileone MCC, Almeida SCG, Minamisava R, Andrade AL. Distribution of invasive Streptococcus pneumoniae serotypes before and 5 years after the introduction of a 10-valent pneumococcal conjugate vaccine in Brazil. Vaccine. 2018;36:2559–2566. doi: 10.1016/j.vaccine.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 10.WHO Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae . 2011. https://apps.who.int/iris/handle/10665/70765
- 11.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk Susceptibility Tests. 13th edn. Wayne, PA: Clinical and Laboratory Standard Institute; 2018. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute (CLSI) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 11th edn. Wayne, PA: Clinical and Laboratory Standard Institute; 2018. [Google Scholar]
- 13.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. 29th edn. Wayne, PA: Clinical and Laboratory Standard Institute; 2019. [Google Scholar]
- 14.Gladstone RA, Lo SW, Lees JA, Croucher NJ, van Tonder AJ, et al. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine. 2019;43:338–346. doi: 10.1016/j.ebiom.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epping L, van Tonder AJ, Gladstone RA, Bentley SD, Page AJ, et al. SeroBA: rapid high-throughput serotyping of Streptococcus pneumoniae from whole-genome sequence data. Microb Genomics. 2018;4:1–6. doi: 10.1099/mgen.0.000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metcalf BJ, Gertz RE, Jr, Gladstone RA, Walker H, Sherwood LK, et al. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect. 2016;22:60. doi: 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page AJ, Taylor B, Keane JA. Multilocus sequence typing by blast from de novo assemblies against PubMLST. J Open Source Softw. 2016;1:10–11. doi: 10.21105/joss.00118. [DOI] [Google Scholar]
- 18.Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price MN, Dehal PS, Arkin AP. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:1–10. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gladstone RA, Lo SW, Goater R, Yeats C, Taylor B, et al. Visualizing variation within Global Pneumococcal Sequence Clusters (GPSCs) and country population snapshots to contextualize pneumococcal isolates. Microb Genomics. 2020;6:1–13. doi: 10.1099/mgen.0.000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46:e134. doi: 10.1093/nar/gky783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting-lactam resistance levels in Streptococcus pneumoniae . mBio. 2016;7:1–9. doi: 10.1128/mBio.00756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, et al. Validation of β-lactam minimum inhibitory concentration predictions for pneumococcal isolates with newly encountered penicillin-binding protein (PBP) sequences. BMC Genomics. 2017;18:1–10. doi: 10.1186/s12864-017-4017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: Rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genomics. 2017;3:10. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/J.1469-0691.2011.03570.X. [DOI] [PubMed] [Google Scholar]
- 27.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. New York: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 28.Domingues C, Verani JR, Renoiner EIM, Brandileone MCC, Flannery B, et al. Effectiveness of ten-valent pneumococcal conjugate vaccine against invasive pneumococcal disease in Brazil: A matched case-control study. Lancet Respir Med. 2014;2:1–8. doi: 10.1016/S2213-2600(14)70060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrade AL, Minamisava R, Policena G, Cristo EB, Domingues CMS, et al. Evaluating the impact of PCV-10 on the invasive pneumococcal disease in Brazil: A time-series analysis. Hum Vaccines Immunother. 2016;12:285–292. doi: 10.1080/21645515.2015.1117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto TCA, Neves FPG, Souza ARV, Oliveira LMA, Costa NS, et al. Evolution of penicillin non-susceptibility among Streptococcus pneumoniae isolates recovered from asymptomatic carriage and invasive disease over 25 years in Brazil, 1990-2014. Front Microbiol. 2019;10:486. doi: 10.3389/fmicb.2019.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida SCG, Hawkins PA, Cassiolato AP, Dias UJ, Gladstone RA, et al. Molecular characterization of Streptococcus pneumoniae strains isolated from the invasive disease in the pre- and post-PCV10 periods in Brazil, 2005 to 2015. 11th International Symposium on Pneumococci & Pneumococcal Diseases, April 15-19, 2018. Melbourne, Australia: 2018. [Google Scholar]
- 32.Kim L, Mcgee L, Tomczyk S, Beall BW. Biological and epidemiological features of antibiotic-resistant Streptococcus pneumoniae in pre- and post-conjugate vaccine eras: A United States perspective. Clin Microbiol Rev. 2016;29:525–552. doi: 10.1128/CMR.00058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.dos Santos MS, Azevedo J, de Oliveria Menezes AP, Cordeiro SM, Escobar EC, et al. Temporal trends and clonal diversity of penicillin non-susceptible pneumococci from meningitis cases from 1996 to 2012, in Salvador, Brazil. BMC Infect Dis. 2015;15:1–10. doi: 10.1186/s12879-015-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caierão J, Hawkins P, Sant’anna FH, da Cunha GR, D’Azevedo PA, et al. Serotypes and genotypes of invasive Streptococcus pneumoniae before and after PCV10 implementation in southern Brazil. PLoS One. 2014;9:1–20. doi: 10.1371/journal.pone.0111129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandileone MCC, Zanella RC, Almeida SCG, Cassiolato AP, Lemos APS, et al. Long-term effect of 10-valent pneumococcal conjugate vaccine on nasopharyngeal carriage of Streptococcus pneumoniae in children in Brazil. Vaccine. 2019;37:5357–5363. doi: 10.1016/j.vaccine.2019.07.043. [DOI] [PubMed] [Google Scholar]
- 36.Almeida SCG, Cassiolato AP, Dias UJ, Guerra MLL e S, Prado LS do, et al. Molecular characterization of invasive Streptococcus pneumoniae isolated in Pre (2005-2009) and Post (2011-2015) 10-valent pneumococcal conjugate vaccine introduction in Brazil. 37th Annual Meeting of the European Society for Paediatric Infectious Diseases, May 5 – 11, 2019. Ljubljana, Slovenia: 2019. abstracts (ESPID19-0518):1164. https://2019.espidmeeting.org/espid-abstracts/ [Google Scholar]
- 37.Lo SW, Gladstone RA, van Tonder AJ, Lees JA, du Plessis M, et al. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis. 2019;3099:1–11. doi: 10.1016/S1473-3099(19)30297-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wouters I, Desmet S, Van Heirstraeten L, Blaizot S, Verhaegen J, et al. Follow-up of serotype distribution and antimicrobial susceptibility of Streptococcus pneumoniae in child carriage after a PCV13-to-PCV10 vaccine switch in Belgium. Vaccine. 2019;37:1080–1086. doi: 10.1016/j.vaccine.2018.12.068. [DOI] [PubMed] [Google Scholar]
- 39.Rinta-Kokko H, Palmu AA, Auranen K, Nuorti JP, Toropainen M, et al. Long-term impact of 10-valent pneumococcal conjugate vaccination on invasive pneumococcal disease among children in Finland. Vaccine. 2018;36:1934–1940. doi: 10.1016/j.vaccine.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Desmet S, Verhaegen J, Van Ranst M, Peetermans W, Lagrou K. Switch in a childhood pneumococcal vaccination programme from PCV13 to PCV10: a defendable approach? Lancet Infect Dis. 2018;18:830–831. doi: 10.1016/S1473-3099(18)30346-3. [DOI] [PubMed] [Google Scholar]
- 41.Isturiz R, Sings HL, Hilton B, Arguedas A, Reinert R-R, et al. Streptococcus pneumoniae serotype 19A: worldwide epidemiology. Expert Rev Vaccines. 2017;16:1007–1027. doi: 10.1080/14760584.2017.1362339. [DOI] [PubMed] [Google Scholar]
- 42.Potin M, Fica A, Wilhem J, Cerda J, Contreras L, et al. Opinión del Comité Consultivo de Inmunizaciones Sociedad Chilena de Infectología: vacuna neumocóccica conjugada en niños y la emergencia de serotipo 19A. Rev Chil Infectología. 2016;33:304–306. doi: 10.4067/S0716-10182016000300009. [DOI] [PubMed] [Google Scholar]
- 43.Moore MR, Gertz RE, Woodbury RL, Barkocy‐Gallagher GA, Schaffner W, et al. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J Infect Dis. 2008;197:1016–1027. doi: 10.1086/528996. [DOI] [PubMed] [Google Scholar]
- 44.Chen YY, Wang JT, Lin TL, Gong YN, Li TH, et al. Prophage excision in Streptococcus pneumoniae serotype 19A ST320 promote colonization: Insight into its evolution from the ancestral clone Taiwan 19F-14 (ST236. Front Microbiol. 2019;10:1–10. doi: 10.3389/fmicb.2019.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metcalf BJ, Chochua S, Gertz RE, Li Z, Walker H, et al. Using whole-genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for the year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect. 2016;22:e1-1002.e8. doi: 10.1016/J.CMI.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Dagan R, Givon‐Lavi N, Leibovitz E, Greenberg D, Porat N. Introduction and proliferation of multidrug‐resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. J Infect Dis. 2009;199:776–785. doi: 10.1086/597044. [DOI] [PubMed] [Google Scholar]
- 47.Beall B, Gertz RE, Hulkower RL, Whitney CG, Moore MR, et al. Shifting genetic structure of invasive serotype 19A pneumococci in the United States. J Infect Dis. 2011;203:1360–1368. doi: 10.1093/infdis/jir052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi EH, Kim SH, Eun BW, Kim SJ, Kim NH, et al. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg Infect Dis. 2008;14:275–281. doi: 10.3201/eid1402.070807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mott M, Caierão J, Cunha GR, Del Maschi MM, Pizzuti K, et al. Emergence of serotype 19A Streptococcus pneumoniae after PCV10 associated with an ST320 in adult population, in Porto Alegre. Epidemiol Infect. 2019;147:1–7. doi: 10.1017/s0950268819000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassiolato AP, Almeida SCG, Andrade AL, Minamisava R, Brandileone MCC. Expansion of the multidrug-resistant clonal complex 320 among invasive Streptococcus pneumoniae serotype 19A after the introduction of a ten-valent pneumococcal conjugate vaccine in Brazil. PLoS One. 2018;13:1–13. doi: 10.1371/journal.pone.0208211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christophe BL, Mott M, da Cunha G, Caierão J, D’Azevedo PA, et al. Characterisation of Streptococcus pneumoniae isolates from the invasive disease in adults following the introduction of PCV10 in Brazil. J Med Microbiol. 2018;67:687–694. doi: 10.1099/jmm.0.000717. [DOI] [PubMed] [Google Scholar]
- 52.Azarian T, Mitchell PK, Georgieva M, Thompson CM, Ghouila A, et al. Global emergence and population dynamics of divergent serotype 3 CC180 pneumococci. PLoS Pathog. 2018;14:e1007438. doi: 10.1371/journal.ppat.1007438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Golden AR, Adam HJ, Karlowsky JA, Baxter MR, Nichol KA, et al. Molecular characterization of predominant Streptococcus pneumoniae serotypes causing invasive infections in Canada: the SAVE study, 2011–15. J Antimicrob Chemother. 2018;73:vii20–31. doi: 10.1093/jac/dky157. [DOI] [PubMed] [Google Scholar]
- 54.Setchanova L, Alexandrova A, Pencheva D, Sirakov I, Mihova K, et al. Rise of multidrug-resistant Streptococcus pneumoniae clones expressing nonvaccine serotypes among children after introduction of the 10-valent pneumococcal conjugate vaccine in Bulgaria. J Glob Antimicrob Resist. 2018;15:6–11. doi: 10.1016/j.jgar.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 55.Del Amo E, Esteva C, Hernandez-Bou S, Galles C, Navarro M, et al. Serotypes and clonal diversity of Streptococcus pneumoniae causing invasive disease in the Era of PCV13 in Catalonia, Spain. PLoS One. 2016;11:1–12. doi: 10.1371/journal.pone.0151125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;24:124. doi: 10.12688/wellcomeopenres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neves FPG, Cardoso NT, Souza AR, Snyder RE, Marlow MM, et al. Population structure of Streptococcus pneumoniae colonizing children before and after universal use of pneumococcal conjugate vaccines in Brazil: emergence and expansion of the MDR serotype 6C-CC386 lineage. J Antimicrob Chemother. 2018;73:1206–1212. doi: 10.1093/jac/dky001. [DOI] [PubMed] [Google Scholar]
- 58.Pinto TCA, Kegele FCO, Dias CAG, Barros RR, Peralta JM, et al. Streptococcus pneumoniae serotypes 9 and 14 circulating in Brazil over 23 years before the introduction of the 10-Valent pneumococcal conjugate vaccine: role of international clones in the evolution of antimicrobial resistance and description of a novel genotype. Antimicrob Agents Chemother. 2016;60:6664–6672. doi: 10.1128/AAC.00673-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carvalho MDG, Pimenta FC, Gertz RE, Joshi HH, Trujillo AA, et al. PCR-based quantitation and clonal diversity of the current prevalent invasive serogroup 6 pneumococcal serotype, 6C, in the United States in 1999 and 2006 to 2007. J Clin Microbiol. 2009;47:554–559. doi: 10.1128/JCM.01919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lambertsen L, Kerrn MB. Test of a novel Streptococcus pneumoniae serotype 6C type specific polyclonal antiserum (Factor Antiserum 6d) and characterisation of serotype 6C isolates in Denmark. BMC Infect Dis. 2010;10 doi: 10.1186/1471-2334-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janoir C, Cohen R, Levy C, Bingen E, Lepoutre A, et al. Clonal expansion of the macrolide resistant ST386 within pneumococcal serotype 6C in France. PLoS One. 2014;9:1–11. doi: 10.1371/journal.pone.0090935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Domínguez-Alegría AR, Pintado V, Barbolla I. Treatment and prevention of invasive pneumococcal disease. Rev Clínica Española. 2018;218:244–252. doi: 10.1016/S2213-2600(14)70060-8. [DOI] [PubMed] [Google Scholar]
- 63.Cohen R, Biscardi S, Levy C. The multifaceted impact of pneumococcal conjugate vaccine implementation in children in France between 2001 to 2014. Hum Vaccin Immunother. 2016;12:277–284. doi: 10.1080/21645515.2015.1116654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cornick JE, Bentley SD. Streptococcus pneumoniae: the evolution of antimicrobial resistance to beta-lactams, fluoroquinolones, and macrolides. Microbes Infect. 2012;14:573–583. doi: 10.1016/J.MICINF.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 65.Yildirim I, Shea KM, Pelton SI. Pneumococcal disease in the era of pneumococcal conjugate vaccine. Infect Dis Clin North Am. 2015;29:679–697. doi: 10.1016/j.idc.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dzaraly ND, Muthanna AR, Desa NNM, Taib NM, Masri SN, et al. Pilus islets and the clonal spread of piliated Streptococcus pneumoniae: A review. Int J Med Microbiol. 2020;310:151499. doi: 10.1016/j.ijmm.2020.151449. [DOI] [PubMed] [Google Scholar]
- 67.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zemlickova H, Jakubu V, Fridrichova M, Malisova L, Musilek M, et al. The association of pili with the emergence and replacement of the major antibiotic resistant pneumococcal clones. J Microbiol Immunol Infect. 2020;53:690–695. doi: 10.1016/j.jmii.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 69.Aguiar SI, Serrano I, Pinto FR, Melo-Cristino J, Ramirez M. The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol. 2008;8:1–11. doi: 10.1186/1471-2180-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Heal. 2018;6:e744–57. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis. 2006;6:303–312. doi: 10.1016/S1473-3099(06)70466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacIntyre CR, Chughtai AA, Barnes M, Ridda I, Seale H, et al. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza an (H1N1) pdm09. BMC Infect Dis. 2018;18:1–20. doi: 10.1186/s12879-018-3548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu X, Ge Y, Wu T, Zhao K, Chen Y, et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.