Graphical abstract
Keywords: SARS-CoV-2, Graphene, Molecular dynamics, Protein adsorption, Secondary structure
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been widely spread in the world, causing more than two million deaths and seriously threatening human life. Effective protection measures are important to prevent the infection and spreading of the virus. To explore the effects of graphene on the virus adsorption and its biological properties, the adsorption process of the receptor binding domain (RBD) of SARS-CoV-2 on graphene has been investigated by molecular dynamics simulations in this paper. The results show that RBD can be quickly adsorbed onto the surface of graphene due to stacking and hydrophobic interactions. Residue PHE486 with benzene ring has stronger adsorption force and the maximum contact area with graphene. Graphene significantly affects the secondary structure of RBD area, especially on the three key sites of binding with human ACE2, GLY476, PHE486 and ASN487. The binding free energy of RBD and graphene shows that the adsorption is irreversible. Undoubtedly, these changes will inevitably affect the pathogenicity of the virus. Therefore, this study provides a theoretical basis for the application of graphene in the protection of SARS-CoV-2, and also provides a reference for the potential application of graphene in the biomedical field.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been widely spread all over the world [1], [2], [3], [4], [5]. The diameter of SARS-CoV-2 is about 50–200 nm. The inner single stranded RNA consists of 29,811 nucleotides, which is encapsulated by N (nucleocapsid) protein. E (envelope) protein and M (membrane) protein form the viral envelope, and S (spike) protein is embedded in the outermost layer of the virus [1], [2], [3]. The high affinity between the receptor binding domain (RBD) of S protein and human angiotensin converting enzyme 2 (ACE2) is the key to the strong infection of the virus [1], [6], [7]. SARS-CoV-2 has high homology with SARS-CoV appeared in 2002–2003 and MERS-Cov appeared in 2012 [8], possessing strong infection in human. It has rapidly caused the worldwide pandemic of coronavirus disease 2019 (COVID-19) [1]. According to the report of the World Health Organization (WHO), more than 100 million have been infected by COVID-19 until January 28, 2021, with 2.15 million deaths. In addition, the impact on economy and trade, industrial production and cultural exchange is enormous. Considering that SARS-CoV-2 is in nanometer size, nanotechnology may provide an effective way to solve this crisis [3].
As two-dimensional nano material with sp2 hybrid, graphene has been widely studied due to its good properties and potential application prospects [9], [10], [11]. It plays a key role in the field of medical and electronic engineering since its inception [4], [12], [13], [14], [15], [16]. In the past decades, graphene based nanomaterials have been the most attractive in biosensor design due to their high affinity, low cost and ease of manufacture [17], [18], [19]. Being a single-layer sp2 hybrid carbon atom, graphene has high surface area to volume ratio [20], [21], it can easily detect single biomolecule. Therefore, it becomes a perfect material for sensor manufacture and implantable devices [22]. The research of cytochrome on graphene revealed a wide application prospect of graphene in biosensors and green bio-photovoltaic [23], [24]. The interaction of graphene with biomolecules has become one of the research hotspots in biomedical area. Using molecular dynamics simulation, Zhao et al. found that HIV Vpr protein could be adsorbed onto graphene and the protein conformation changed significantly during the adsorption process [25]. Zou and his team used sum frequency generation(SFG)and molecular dynamics simulation to explore the interaction of cecropin P1 and MSI78 (C1) with graphene, demonstrating that molecular dynamics simulation is effective in investigating the adsorption of biomolecules on graphene [11]. Immunoglobulin G could be rapidly expanded with conformation changes in the adsorption process of graphene [26]. Peptides with larger volume were easier to be adsorbed onto graphene [27]. The properties (size, oxidation rate, curvature, etc.) of graphene and the tension of pulmonary surfactant have significant effects on the extraction kinetics [28]. In addition, researchers found that graphene and its derivatives have good surface integrity in capturing virus [4], [29], [30]. If the structural arrangement and spatial configuration of the protein are changed, its function will be lost and may lead to virus inactivation [3]. Therefore, graphene may be used to fight SARS-CoV-2.
The droplets discharged from respiratory tract of SARS-CoV-2 infected people contain virus particles. The virus in this aerosol remains active for up to 3 h in the air [31]. People inhaling this aerosol will be infected, which is also the main path of human transmission. Medical experts advocate the use of surgical masks to prevent the virus from being inhaled [32]. Some people proposed to add graphene nanofilms into the masks to more effectively block the spread of the virus. Byeongtaek Oh et al. studied the application of graphene in promoting neuronal stem cell differentiation [33]. Graphene can be made into magnetic nanocapsules, which is able to deliver oral drugs to the stomach site-selective [34]. Graphene has large surface area ratio and delocalized electronic structure. It can be used as a loading platform for chemotherapeutic drugs in cancer treatment. This strategy shows excellent effects in vitro and in vivo [35], [36]. Graphene nanocapsules (AuNR@G) were injected into mice caudal vein at certain concentration, which didn’t cause any significant damage to cell morphology and main organs [36]. Mice orally administered with appropriate concentration of magnetic graphene capsules did not cause significant damage to cell morphology and gastric mucosa, and most of the particles were excreted within 12 h [37]. These researches indicate that graphene is feasible for in vitro protection. Whether graphene is used as a sensor to detect virus or added into protective equipment, the process of virus adsorption on graphene surface should be understood.
The binding of RBD on S protein of SARS-CoV-2 with ACE2 of human [1], [6], [7] is the key for people to be infected. RBD is also the area the virus contacts outside. The adsorption process of RBD on graphene is the process of virus being captured by graphene. Thus, we investigated the whole process of RBD adsorption on graphene by molecular dynamics simulation, to excavate a significant way for graphene in virus detection and defence.
2. Computational model and methods
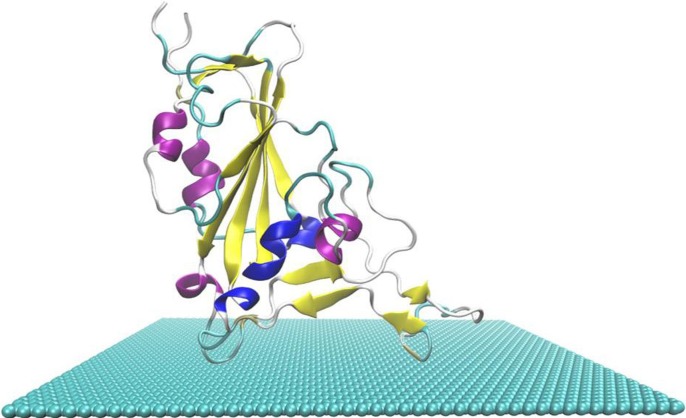
To build the simulation model, the crystal structure of RBD and ACE2 complex was downloaded from Protein Data Bank (PDB: 6M0J). RBD was separated and the missed residues were completed by homologous modeling. Then, a two-dimensional graphene with the size of 118 × 89 Å2 is constructed and placed in the X-Y plane at the bottom of the box [25], [38]. The length of the simulation system along the Z axis is 88 Å. RBD was placed on top of graphene with the outermost residue closer to graphene, which bounds to human ACE2. Residues embedded inside the virus were farther away from graphene as shown in Fig. 1 . The initial distance between them was about 9 Å. To eliminate the influence of periodicity on the system, the distance between RBD and the edge of the simulation box was at least 10 Å. Finally, 27,988 water molecules were filled into the box and solvated with SPC/E model [25], [39]. Three Cl- were added into the box and randomly distributed to keep the system electrically neutral.
Fig. 1.
Simulation model of RBD on graphene, where RBD is expressed in the form of colored Newcartoon, and the blue-green sphere represents graphene.
All simulations were exceuted with Gromacs package 5.1.4 [40], [41]. The interaction between RBD and graphene was simulated by CHARMM36, which is widely used to describe the molecular dynamics behavior of proteins [42], [43], [44], [45], [46]. The C atoms in graphene were regarded as uncharged Lennard-Jones particles. The nonbonding parameters of benzene(, )in CHARMM36 were used to describe them, which had been successfully used in the study of graphene surface adsorption [27], [42], [47]. The parameters of the nonbonding interactions between graphene, RBD and water were calculated by Lorentz-Berthelot combination rule. Periodic boundary conditions were applied in ×, y and z direction during the simulation. The cutoff values of Lennard-Jones and Coulomb interactions were 10 Å. The particle mesh Ewald (PME) method was used to calculate the electrostatic interaction [48], [49]. The bond length of hydrogen atom in the system was constrained by LINCS algorithm [50]. Firstly, the energy of the system was minimized with the steepest descent method in 50,000 steps. Then, pre equilibrium was carried out in the canonical ensemble for 1 ns to relax the position restriction of solvent molecules on protein atoms, with V-rescale temperature coupler controlling the system temperature[51]. Finally, molecular dynamics simulation was carried out in NVT ensemble of 298 K for 180 ns. There was no restriction on the protein and solvent molecules in this process, with Nose-Hoover temperature coupler [52] controlling the temperature. (On the basis of 180 ns molecular dynamics simulation, many 20 ns simulations were carried out. The results showed that RBD was stably adsorbed on the graphene surface.) The trajectory of the system was analyzed by Gromacs, and visualized by visual molecular dynamics (VMD) [53].
MM-PBSA is a method to calculate the binding free energy with the MD trajectory. It has been successfully applied into many researches in biomedical field [54], [55], [56], [57], [58], [59]. The free energy of a molecule can be expressed as
| (1) |
| (2) |
where is determined by the interaction of intramolecular bond length, bond angle and dihedral angle. and represent van der Waals and Coulomb interaction respectively. is the polar solvation energy, which can be obtained by solving Poisson Boltzmann equation. is the nonpolar solvation energy, which can be calculated according to the solvent surface area of the molecule. represents the molecular entropy. is the thermodynamic temperature. Therefore, the binding free energy of the protein and graphene can be calculated as
| (3) |
where , and represent the free energy of the protein, graphene and their complex separately.
3. Results and discussion
Molecular dynamics simulation provides a possible way to clarify the effect of graphene on RBD structure. Therefore, the adsorption process of RBD on graphene surface and resulted changes in the structure were thoroughly investigated.
3.1. Adsorption process
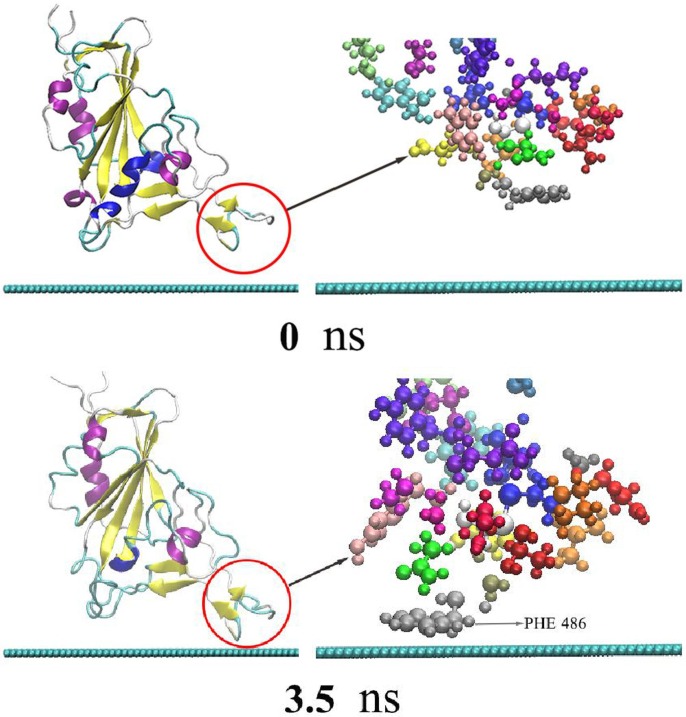
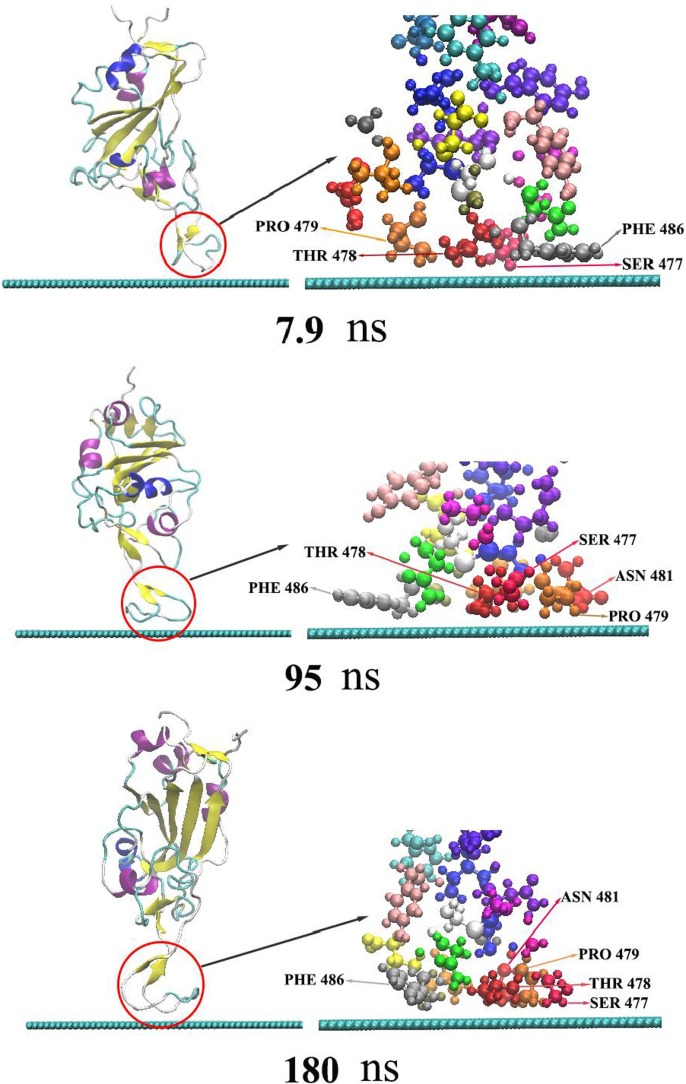
As the adsorption of RBD on graphene is time-related, the adsorption trajectory is discussed in detail. Locations of the main nodes of RBD during the simulation are shown in Fig. 2 . As shown, RBD quickly approaches graphene in the initial stage of the simulation, to a distance about 3 Å at 3.5 ns. In other words, the initial adsorption process appears in a very short time, indicating the strong adsorption effect of graphene on RBD. At the moment, residue PHE486 has been adsorbed onto the surface of graphene, and thereafter never desorbs in the whole simulation process. It illustrates that the stacking interaction between benzene ring of PHE486 and graphene plays an important role in the protein adsorption [60], which is consistent with previous research results [61], [62], [63], [64]. With the increase of simulation time, residue SER477,THR488 and PRO479 are closer on to graphene. At 7.9 ns, the three amino acid residues are also adsorbed onto graphene and never desorb thereafter. From 3.5 to 7.9 ns, the distance between protein and graphene keeps about 3 Å. As shown by Fig. 2 and Fig. 4, ASN481 begins to oscillate near graphene at 7.9 ns. The protein atoms contact the surface of graphene to the maximum extent after a long time of structural adjustment and interface rearrangement. In addition, water molecules are repulsed on the graphene surface and form a dehumidification layer to promote adsorption [65], [66]. ASN481 isn’t stably adsorbed onto graphene until 95 ns. The five adsorbed residues never desorbe in 95–180 ns. Because the charge of graphene atom is 0, there is no electrostatic interaction between the protein and graphene. Therefore, van der Waals (vdW) force is the main driving force for RBD adsorption and stabilization. Hydrophobic and stacking interactions also assist in the adsorption, which are included in vdW interaction of typical nonpolarized molecular dynamics force field [62], [67].
Fig. 2.
The adsorption process of RBD on the surface of graphene. The left is the secondary structure of RBD presented in newcartoon type, in which the red circle indicates the protein adsorption area. The right is the enlarged CPK type structure of the adsorption area. The blue-green spherical chain below is the side view of two-dimensional graphene.
Fig. 4.
Structural changes of RBD during simulation.
3.2. Energies
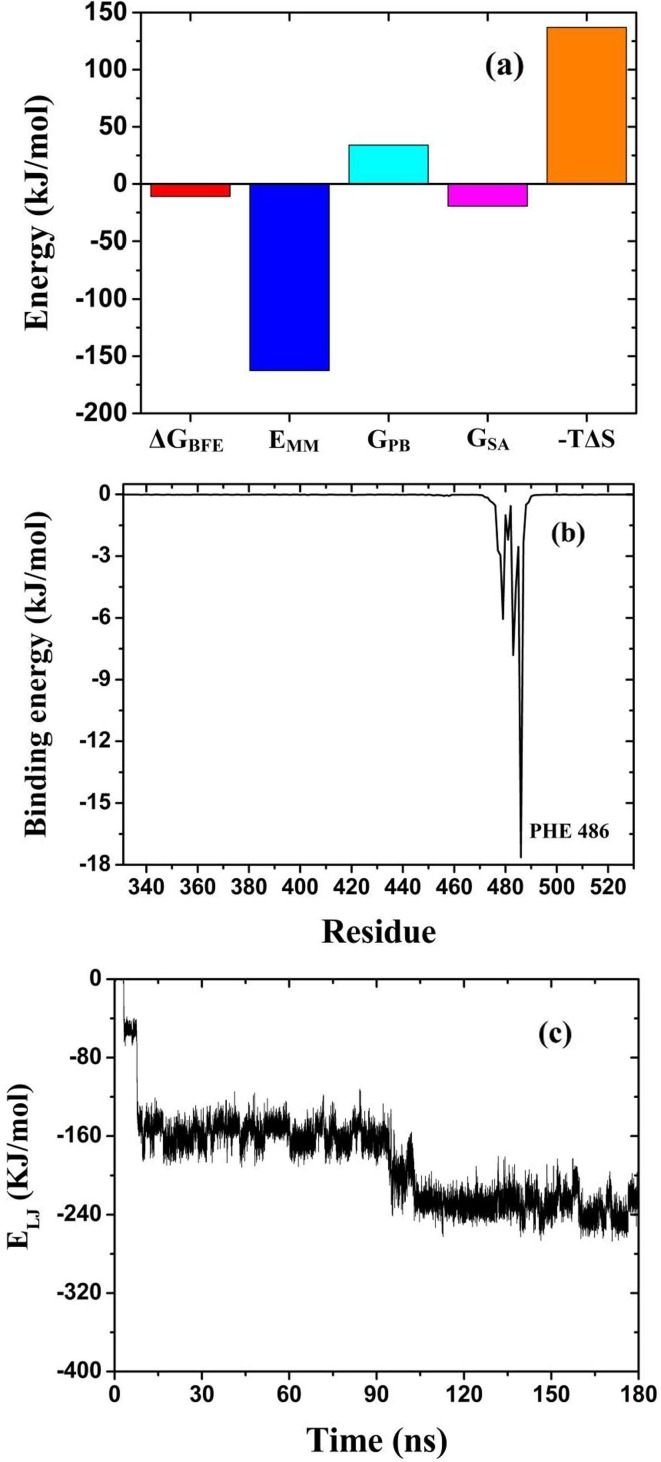
After the RBD was stably absorbed, the last 30 ns of the simulated trajectory was used to calculate the binding free energy by the shell script gmx_mmpbsa [57], [68], with one frame at 1 ns interval. The results are shown in Fig. 3 a. The binding free energy of RBD and graphene is −11 kJ/mol, the molecular mechanics energy is −163 kJ/mol, the polar solvation energy is 34 kJ/mol, the solvent accessible surface (SASA) energy is −19 kJ/mol and energy contributed by entropy is 137 kJ/mol. Negative values of energy is conducive to the binding of RBD and graphene, while the positive is not. The positive polar solvation energy and energy contributed by entropy are counterbalanced by the negative molecular mechanics energy and SASA energy. The binding free energy of RBD and graphene is negative, which means RBD will be adsorbed to graphene surface without desorbing.
Fig. 3.
The various energies of RBD and graphene. In Fig. 3a, , , , and represent binding free energy, molecular mechanics energy, polar solvation energy, solvent accessible surface energy and energy contributed by entropy, respectively.
The binding energy (BE) of each residue with graphene was also calculated using gmx_mmpbsa [57], [68], where the energy contributed by entropy was not included. The results are shown in Fig. 3b. It can be seen that the binding energies of SER477, THR488, PRO479, ASN481 and PHE486 are negative, while the binding energies of the remaining residues are basically zero. The results indicate that only these five residues are adsorbed with graphene. Among the five residues adsorbed, PHE486 has the smallest adsorption energy of −18 kJ/mol, which indicates that stacking interaction between PHE486 and graphene is strong.
Van der Waals interaction is usually calculated by Lennard-Jones (LJ) potential, as shown in Eq. (4)
| (4) |
where and are LJ parameters between atom i and j, given by the corresponding force field of GROMACS. is the distance between atom i and j. As Graphene has no charge, the energy between RBD and graphene is mainly LJ potential, whose variation with time is shown in Fig. 3c. It can be seen that the LJ potential decreases rapidly in the first 7.9 ns, corresponding to the process of RBD approaching the surface of graphene. After some residues of RBD were adsorbed onto graphene (such as SER477,THR488, PRO479 and PHE486), the energy curve shows a stable period of about 87 ns. LJ potential begins to decrease again at 95 ns, due to ASN481 contacting graphene with the change of RBD structure and interface. For another 10 ns, the adsorption of ASN481 is stable. The LJ potential between RBD and graphene keeps at −231 kJ/mol thereafter.
3.3. Sructural changes
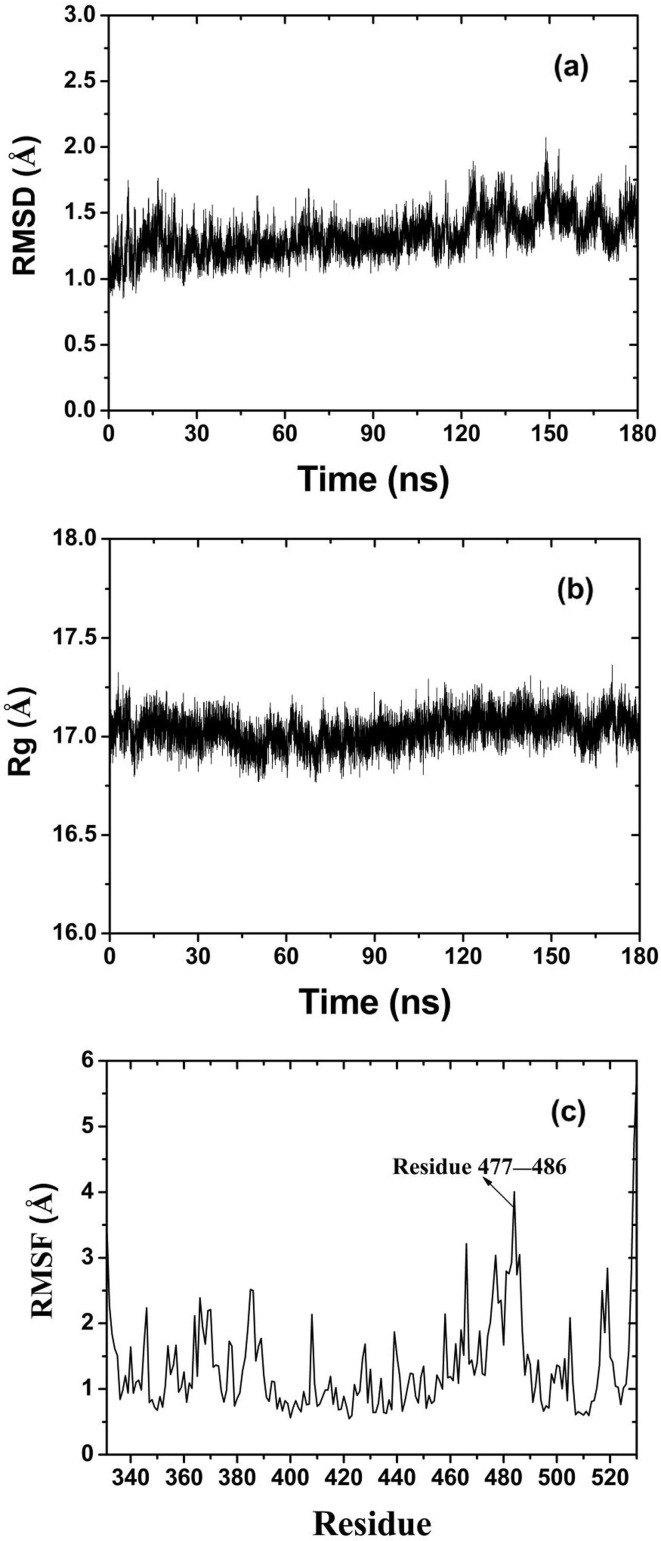
In order to reveal the structural changes of RBD during simulation, the root mean square deviation (RMSD) was calculated according to Eq. (5)
| (5) |
where is the mass of the ith atom, is the total mass of all atoms in RBD, is the total number of atoms in protein, and corresponds to the position of the ith atom at initial time and . RMSD reflects the change of protein structure during simulation. A larger RMSD indicates larger change of protein configuration. When the structure of protein is stabilized, RMSD shows little variation over a period of time. That is, RMSD converges. In Fig. 4 a, RMSD increases rapidly in the process of RBD approaching graphene at the beginning of the simulation. When the protein is
stably adsorbed, RMSD decreases for a short time and then increases slowly. Finally, RMSD converges to the average value of 1.5 Å after 130 ns.
To further investigate the variation of RBD configuration in simulation, its radius of gyration (Rg) was calculated according to Eq. (6)
| (6) |
where is the mass of the ith atom, is the position of the ith atom relative to the molecular centroid. Rg can roughly reflect the structural compactness of protein. A higher Rg means larger volume and smaller density of the protein. In Fig. 4b, Rg firstly decreases slowly until 30 ns, and then begins to increase slowly to the maximum at about 130 ns. During 130–180 ns, Rg keeps at an average value of 17.1 Å. This shows that in the initial stage of the adsorption, the molecular volume first decreases due to the inertia effect. When the adsorption is stable, the molecular volume begins to recover and increase slowly under the interacton of Coulomb and van der Waals forces. After 130 ns, the configuration and volume of the protein tend to be stable.
What’s more, the root mean square fluctuation (RMSF) of each residue was calculated according to Eq. (7)
| (7) |
where is the position of conformation at time t, is the position of the reference conformation, and is the total simulation time. RMSF reflects the average fluctuation of residues relative to the reference position in the simulation process. In Fig. 4c, there are two parts of residues fluctuated greatly in the whole simulation process. One part is the residues at the end of protein peptide chain (residue 331 and 530). As only one end of the these residues is connected with others, the other end can fluctuate freely. The other part is residues 477–486, which is the adsorption area of RBD on the surface of graphene. It indicates that the hydrophobic interface of graphene has a strong effect on these residues and changes their structure obviously. This domain is also the main binding region between RBD and ACE2.
3.4. Contact area
The contact areas are calculated according to Eq. (8)
| (8) |
where is the solvent accessible surface (SASA) of monomer X, is the SASA of monomer Y, is the SASA of the adsorbed complex, and is the contact area of monomer X and Y. The probe radius for SASA calculatation is 1.4 Å.
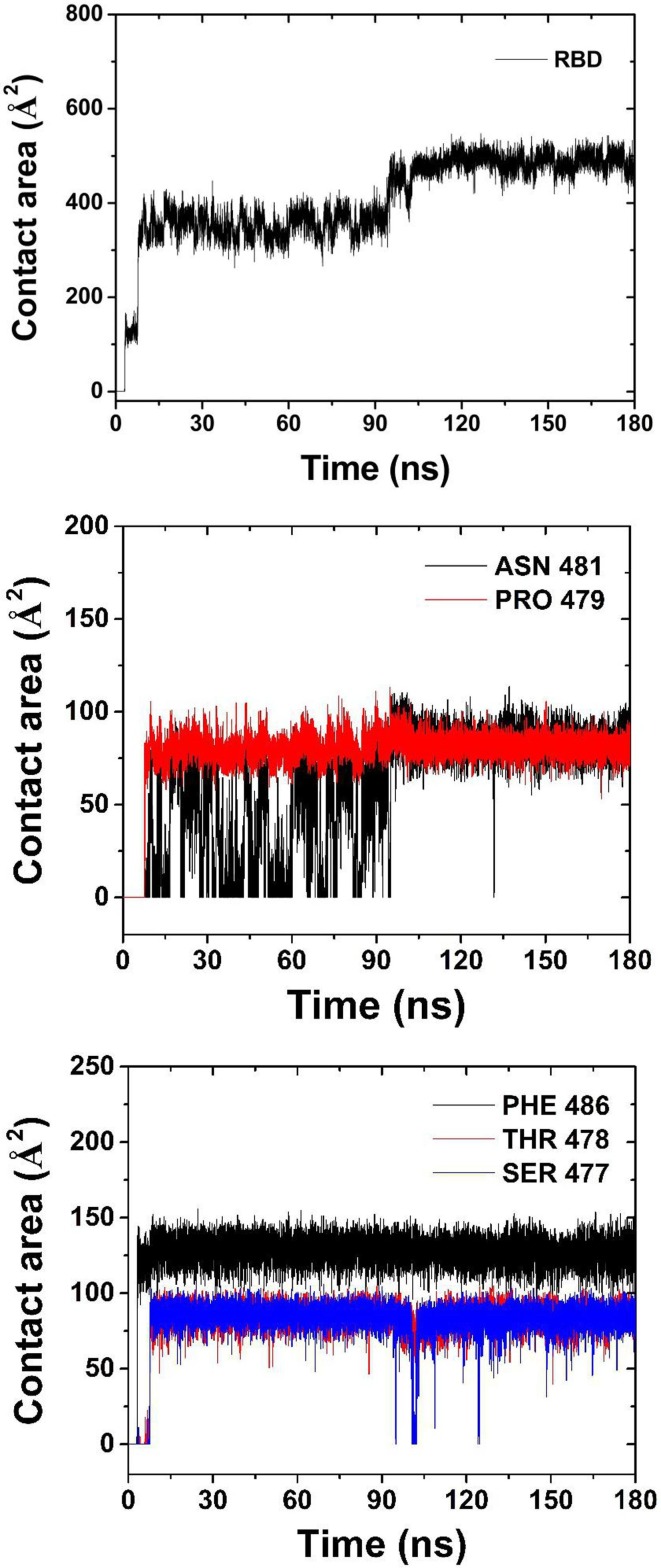
The contact area of RBD and some of its residues with graphene is shown in Fig. 5 . The average contact area of RBD with graphene is 129 Å2 in 3.5–7.9 ns, and 354 Å2 in 7.9 ns-95 ns. After 95 ns, the average contact area rests around 488 Å2. As seen from Fig. 2 and Fig. 5, the variation in contact area of RBD with graphene results from the adsorption process of different residues onto graphene. There is only PHE486 contacting graphene in 3.5–7.9 ns, while SER477, THR488 and PRO479 also adsorbed onto graphene in 7.9–95 ns. And, ASN481 residue doesn’t get a stable adsorption until 95 ns. The average contact areas of SER477,THR488,PRO479 and PHE486 in stable adsorption with graphene were 84 Å2, 85 Å2, 81 Å2 and 129 Å2, respectively. ASN481 doesn’t get stable adsorption area of 84 Å2 until 95 ns, as it flops near graphene in 7.9 ns-95 ns. This indicates that the hydrophobicity of ASN481 is weaker than that of the other four residues, while PHE486 with aromatic ring has the strongest adsorption capacity. It is worth noting that PHE486 is also one of the main residues of RBD binding to human ACE2 [6], [8].
Fig. 5.
Contact area of RBD and some residues with graphene.
3.5. Secondary structure
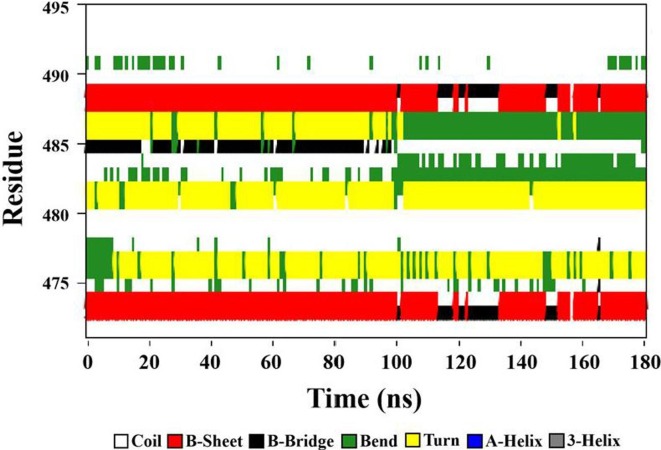
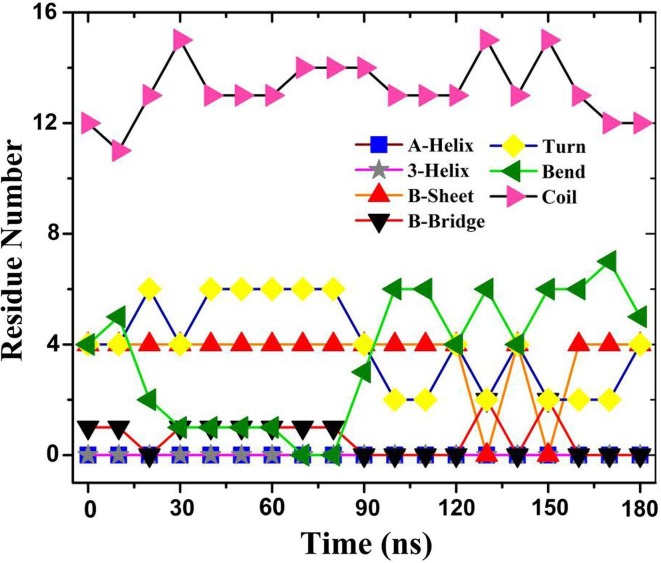
To further study the effect of graphene on RBD, the secondary structure of RBD adsorption region was calculated using Define Secondary Structure of Proteins (DSSP) [44], [69], as shown in Fig. 6, Fig. 7 . The adsorption region initially contains two B-sheets, one B-bridge, two Turns and two Bends. The B-sheets are respectively composed of residues 473–474 and 488–489 , while the B-bridge is composed of residue 485. The Turns are respectively composed of residues 481–482 and 486–487, as well as the Bends correspondingly of residues 476–478 and 491. The remaining 12 residues form five Coil. No A-helix and 3-helix appear in the adsorption region. The secondary structure of RBD adsorption region changes obviously after the simulation of 180 ns. Two B-sheets evolves into B-bridge and Coil in some time and finally forms B-sheet in the simulation process. The B-bridge formed by residue 485 becomes Coil in some time and finally evolves into Bend. The Turn formed by residues 486–487 evolves into Bend. Bend formed by residues 476–478 evolves into a Turn and a Coil. It is worth noting that GLY476, PHE486 and ASN487 are the key sites for SARS-CoV-2 virus binding to human ACE2 [6], [8]. These changes in the secondary structure are bound to influence the biological properties of the virus, and even make it lose the pathogenicity.
Fig. 6.
Evolution of secondary structure in RBD adsorption region.
Fig. 7.
Number of residues in various secondary structures of RBD adsorption region.
4. Conclusions
In this paper, the adsorption process of the receptor binding domain (RBD) of SARS-CoV-2 S protein with graphene was studied by molecular dynamics simulations. The results show that graphene has strong adsorption on RBD. Within 3.5 ns, RBD can be approach to and contact graphene. Five amino acid residues are adsorbed onto the surface of graphene. Compared to the other four residues, PHE486 gets stronger adsorption and more contact area, because of the stacking interaction between benzene ring and graphene. Due to the adsorption of graphene, the secondary structure of RBD adsorption region changes significantly, with Turn formed by residues 486–487 becoming Bend, and the Bend formed by residues 476–478 becoming a Turn and a Coil. Among them, GLY476, PHE486 and ASN487 are the key sites for SARS-CoV-2 binding to ACE2. These changes of the secondary structure will inevitably affect the pathogenicity of the virus. The simulations after stable adsorption show that RBD will not desorb. The binding free energy of −11 kJ/mol also shows that the adsorption is irreversible. This is very important to prevent the spread of SARS-CoV-2. Therefore, it provides a theoretical basis for the application of graphene in defending SARS-CoV-2 and preventing its spread, as well as in the biomedical field.
CRediT authorship contribution statement
Jianbin Du: Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. Chunmei Yang: Visualization, Investigation. Xiangyun Ma: Software. Qifeng Li: .
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Key Research and Development Program of China (2017YFC0803600),National Natural Science Foundation of China (NSFC22174098) and Key Projects in the Science & Technology Pillar Program of Tianjin (20YFZCSN00530).
References
- 1.Han Y., Kral P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J.a., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruiz‐Hitzky E., Darder M., Wicklein B., Ruiz‐Garcia C., Martín‐Sampedro R., del Real G., Aranda P. Nanotechnology responses to COVID-19. Adv. Healthcare Mater. 2020;9(19):2000979. doi: 10.1002/adhm.202000979. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A., Sharma K., Dixit A.R. Role of graphene in biosensor and protective textile against viruses. Medical Hypotheses. 2020;144:110253. doi: 10.1016/j.mehy.2020.110253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasrollahzadeh M., Sajjadi M., Soufi G.J., Iravani S., Varma R.S. Nanomaterials and nanotechnology-associated innovations against viral infections with a focus on coronaviruses. Nanomaterials. 2020;10(6):1072. doi: 10.3390/nano10061072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 7.Huang H., Fan C., Li M., Nie H.L., Wang F.B., Wang H., Wang R., Xia J., Zheng X., Zuo X., Huang J. COVID-19: a call for physical scientists and engineers. ACS Nano. 2020;14:3747–3754. doi: 10.1021/acsnano.0c02618. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro Neto A.H., Guinea F., Peres N.M.R., Novoselov K.S., Geim A.K. The electronic properties of graphene. Rev. Modern Phys. 2009;81:109–162. [Google Scholar]
- 10.Geim A.K. Graphene: status and prospects. Science. 2009;324:1530–1534. doi: 10.1126/science.1158877. [DOI] [PubMed] [Google Scholar]
- 11.Zou X., Wei S., Jasensky J., Xiao M., Wang Q., Brooks Iii C.L., Chen Z. Molecular interactions between graphene and biological molecules. J. Am. Chem. Soc. 2017;139:1928–1936. doi: 10.1021/jacs.6b11226. [DOI] [PubMed] [Google Scholar]
- 12.Singh M., Holzinger M., Tabrizian M., Winters S., Berner N.C., Cosnier S., Duesberg G.S. Noncovalently functionalized monolayer graphene for sensitivity enhancement of surface plasmon resonance immunosensors. JACS. 2015;137:2800–2803. doi: 10.1021/ja511512m. [DOI] [PubMed] [Google Scholar]
- 13.Ohno Y., Maehashi K., Matsumoto K. Label-free biosensors based on aptamer-modified graphene field-effect transistors. JACS. 2010;132:18012–18013. doi: 10.1021/ja108127r. [DOI] [PubMed] [Google Scholar]
- 14.Chung C., Kim Y.-K., Shin D., Ryoo S.-R., Hong B.H., Min D.-H. Biomedical applications of graphene and graphene oxide. Acc. Chem. Res. 2013;46:2211–2224. doi: 10.1021/ar300159f. [DOI] [PubMed] [Google Scholar]
- 15.Ye S., Shao K., Li Z., Guo N., Zuo Y., Li Q., Lu Z., Chen L., He Q., Han H. Antiviral activity of graphene oxide: how sharp edged structure and charge matter. ACS Appl. Mater. Interfaces. 2015;7(38):21571–21579. doi: 10.1021/acsami.5b06876. [DOI] [PubMed] [Google Scholar]
- 16.Chou S.S., De M., Luo J., Rotello V.M., Huang J., Dravid V.P. Nanoscale graphene oxide (nGO) as artificial receptors: implications for biomolecular interactions and sensing. JACS. 2012;134:16725–16733. doi: 10.1021/ja306767y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Ma L., Zhou M., Li Y., Xia Y., Fan X., Cheng C., Luo H. New opportunities for emerging 2D materials in bioelectronics and biosensors. Curr. Opin. Biomed. Eng. 2020;13:32–41. [Google Scholar]
- 18.Jiang Z., Feng B., Xu J., Qing T., Zhang P., Qing Z. Graphene biosensors for bacterial and viral pathogens. Biosens. Bioelectron. 2020;166:112471. doi: 10.1016/j.bios.2020.112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermisoglou E., Panacek D., Jayaramulu K., Pykal M., Frebort I., Kolar M., Hajduch M., Zboril R., Otyepka M. Human virus detection with graphene-based materials. Biosens. Bioelectron. 2020;166:112436. doi: 10.1016/j.bios.2020.112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A., Sharma K., Dixit A.R. Carbon nanotube- and graphene-reinforced multiphase polymeric composites: review on their properties and applications. J. Mater. Sci. 2019;55:2682–2724. [Google Scholar]
- 21.Kumar A., Sharma K., Dixit A.R. A review of the mechanical and thermal properties of graphene and its hybrid polymer nanocomposites for structural applications. J. Mater. Sci. 2019;54(8):5992–6026. [Google Scholar]
- 22.Nag A., Mitra A., Mukhopadhyay S.C. Graphene and its sensor-based applications: a review. Sens. Actuators, A. 2018;270:177–194. [Google Scholar]
- 23.Kowalska A.M., Trzaskowski B., Osella S. Assessing the charge transfer at the cytochrome c553/graphene interface: a multiscale investigation. J. Phys. Chem. C. 2018;122(51):29405–29413. [Google Scholar]
- 24.Kiliszek M., Harputlu E., Szalkowski M., Kowalska D., Unlu C.G., Haniewicz P., Abram M., Wiwatowski K., Niedziółka-Jönsson J., Maćkowski S., Ocakoglu K., Kargul J. Orientation of photosystem I on graphene through cytochrome c553 leads to improvement in photocurrent generation. J. Mater. Chem. A. 2018;6(38):18615–18626. [Google Scholar]
- 25.Zhao D., Li L., He D., Zhou J. Molecular dynamics simulations of conformation changes of HIV-1 regulatory protein on graphene. Appl. Surf. Sci. 2016;377:324–334. [Google Scholar]
- 26.Ortega M., Vilhena J.G., Rubio-Pereda P., Serena P.A., Perez R. Assessing the accuracy of different solvation models to describe protein adsorption. J. Chem. Theory Comput. 2019;15:2548–2560. doi: 10.1021/acs.jctc.8b01060. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Wang M., Wei Q., Yang X., Yang Y., Cui B., Yang X., Xu Z. Modulation of solid-water-peptide interfacial properties towards surface adsorption/bioresistance. Appl. Surf. Sci. 2019;483:373–382. [Google Scholar]
- 28.Luo Z., Li S., Xu Y., Ren H., Zhang X., Hu G., Huang F., Yue T. Extracting pulmonary surfactants to form inverse micelles on suspended graphene nanosheets. Environ. Sci. Nano. 2018;5:130–140. [Google Scholar]
- 29.Innocenzi P., Stagi L. Carbon-based antiviral nanomaterials: graphene, C-dots, and fullerenes. a perspective. Chem. Sci. 2020;11:6606–6622. doi: 10.1039/d0sc02658a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar Raghav P., Mohanty S. Are graphene and graphene-derived products capable of preventing COVID-19 infection? Med. Hypotheses. 2020;144:110031. doi: 10.1016/j.mehy.2020.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doremalen N.v., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Lloyd-Smith J.O., Wit E.d., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382 doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Morawska, D.K. Milton, It Is Time to Address Airborne Transmission of Coronavirus Disease 2019 (COVID-19), Clinical infectious diseases : an official publication of the Infectious Diseases Society of America, 71 (2020) 2311-2313. [DOI] [PMC free article] [PubMed]
- 33.Oh B., Wu Y.W., Swaminathan V., Lam V., Ding J., George P.M. Modulating the electrical and mechanical microenvironment to guide neuronal stem cell differentiation. Adv. Sci. 2021;8:2002112. doi: 10.1002/advs.202002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai X., Xu Y., Zhao L., Xu J., Li S., Wen C., Xia X., Dong Q., Hu X., Wang X., Chen L., Chen Z., Tan W. In situ pepsin-assisted needle assembly of magnetic-graphitic-nanocapsules for enhanced gastric retention and mucus penetration. Nano Today. 2021;36:101032. [Google Scholar]
- 35.Xu M.L., Guan L.Y., Li S.K., Chen L., Chen Z. Stable gold graphitic nanocapsule doped hydrogels for efficient photothermal antibacterial applications. Chem. Commun. 2019;55:5359–5362. doi: 10.1039/c9cc01933b. [DOI] [PubMed] [Google Scholar]
- 36.Dong Q., Wang X., Hu X., Xiao L., Zhang L., Song L., Xu M., Zou Y., Chen L., Chen Z., Tan W. Simultaneous application of photothermal therapy and an anti-inflammatory prodrug using pyrene-aspirin-loaded gold nanorod graphitic nanocapsules. Angew. Chem. 2018;57:177–181. doi: 10.1002/anie.201709648. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Hu X., Ding D., Zou Y., Xu Y., Wang X., Zhang Y., Chen L., Chen Z., Tan W. In situ targeted MRI detection of Helicobacter pylori with stable magnetic graphitic nanocapsules. Nat. Commun. 2017;8:15653. doi: 10.1038/ncomms15653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao D., Peng C., Zhou J. Lipase adsorption on different nanomaterials: a multi-scale simulation study. Phys. Chem. Chem. Phys. 2015;17:840–850. doi: 10.1039/c4cp04696j. [DOI] [PubMed] [Google Scholar]
- 39.Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 40.Yoon T., Baek I., Lee S., Choi H., Yoon S., Lee H., Ung Kim S., Na S. Immobilization of laccase on a graphene interface: direct electron transfer and molecular dynamics study. Appl. Surf. Sci. 2020;521:146378. [Google Scholar]
- 41.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 42.Wang X., Yang X., Chen H., Yang X., Xu Z. Entropy-enthalpy compensation in peptide adsorption on solid surfaces: dependence on surface hydration. Langmuir : ACS J. Surf. Colloids. 2020;36:10822–10829. doi: 10.1021/acs.langmuir.0c01845. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z., Yang X., Wei Q., Zhao W., Cui B., Yang X., Sahai N. Quantitatively identifying the roles of interfacial water and solid surface in governing peptide adsorption. Langmuir : ACS J. Surf. Colloids. 2018;34:7932–7941. doi: 10.1021/acs.langmuir.8b01189. [DOI] [PubMed] [Google Scholar]
- 44.Li B., Bell D.R., Gu Z., Li W., Zhou R. Protein WW domain denaturation on defective graphene reveals the significance of nanomaterial defects in nanotoxicity. Carbon. 2019;146:257–264. [Google Scholar]
- 45.Mucksch C., Urbassek H.M. Accelerated molecular dynamics study of the effects of surface hydrophilicity on protein adsorption. Langmuir : ACS J. Surf. Colloids. 2016;32:9156–9162. doi: 10.1021/acs.langmuir.6b02229. [DOI] [PubMed] [Google Scholar]
- 46.Hughes Z.E., Walsh T.R. Probing nano-patterned peptide self-organisation at the aqueous graphene interface. Nanoscale. 2017;10:302–311. doi: 10.1039/c7nr06441a. [DOI] [PubMed] [Google Scholar]
- 47.Utesch T., Daminelli G., Mroginski M.A. Molecular dynamics simulations of the adsorption of bone morphogenetic protein-2 on surfaces with medical relevance. Langmuir : ACS J. Surf. Colloids. 2011;27:13144–13153. doi: 10.1021/la202489w. [DOI] [PubMed] [Google Scholar]
- 48.Darden T., York D., Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 49.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 50.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver formolecular simulations. J Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 51.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 52.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 53.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020;60:3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R. Kumari, R. Kumar, C. Open Source Drug Discovery, A. Lynn, g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations, Journal of chemical information and modeling, 54 (2014) 1951-1962. [DOI] [PubMed]
- 56.Miller B.R., 3rd, McGee T.D., Jr., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 57.Ding H.-M., Yin Y.-W., Ni S.-D., Sheng Y.-J., Ma Y.-Q. Accurate evaluation on the interactions of SARS-CoV-2 with its receptor ACE2 and antibodies CR3022/CB6*. Chin. Phys. Lett. 2021;38:018701. [Google Scholar]
- 58.Wang C., Greene D., Xiao L., Qi R., Luo R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2017;4:87. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma J., Kumar Bhardwaj V., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346:128933. doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge C., Du J., Zhao Y., Zhao L., Chai Z., Wang L., Liu Y., Li D., Yang Y., Zhou R., Chen C. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. PNAS. 2011;108:16968–16973. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Z., Zhou J. Probing carbon nanotube–amino acid interactions in aqueous solution with molecular dynamics simulations. Carbon. 2014;78:500–509. [Google Scholar]
- 62.Sherrill C.D., Sumpter B.G., Sinnokrot M.O., Marshall M.S., Hohenstein E.G., Walker R.C., Gould I.R. Assessment of standard force field models against high-quality ab initio potential curves for prototypes of pi-pi, CH/pi, and SH/pi interactions. J. Comput. Chem. 2009;30:2187–2193. doi: 10.1002/jcc.21226. [DOI] [PubMed] [Google Scholar]
- 63.Tomásio S.M., Walsh T.R. modeling the binding affinity of peptides for graphitic surfaces. influences of aromatic content and interfacial shape. J. Phys. Chem. C. 2009;113:8778–8785. [Google Scholar]
- 64.Umadevi D., Panigrahi S., Sastry G.N. Noncovalent interaction of carbon nanostructures. Acc. Chem. Res. 2014;47:2574–2581. doi: 10.1021/ar500168b. [DOI] [PubMed] [Google Scholar]
- 65.Jana A.K., Sengupta N. Surface induced collapse of Abeta1-42 with the F19A replacement following adsorption on a single walled carbon nanotube. Biophys. Chem. 2013;184:108–115. doi: 10.1016/j.bpc.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 66.Liu J., Yang Z., Li H., Gu Z., Garate J.A., Zhou R. Dewetting transition assisted clearance of (NFGAILS) amyloid fibrils from cell membranes by graphene. J. Chem. Phys. 2014;141:22D520. doi: 10.1063/1.4901113. [DOI] [PubMed] [Google Scholar]
- 67.Hunter C.A., Meah M.N., Sanders J.K.M. DABCO-metalloporphyrin binding: ternary complexes, host-guest chemistry, and the measurement of π-π interactions. J. Am. Chem. Soc. 1990;112:5773–5780. [Google Scholar]
- 68.https://jerkwin.github.io/gmxtool.
- 69.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]