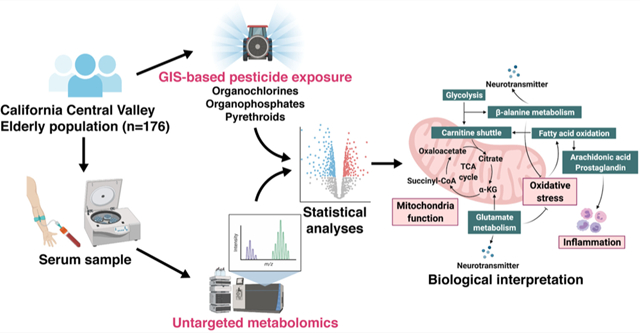
Abstract
Pesticides are widely used in the agricultural Central Valley region of California. Historically, this has included organophosphates (OPs), organochlorines (OCs), and pyrethroids (PYRs). This study aimed to identify perturbations of the serum metabolome in response to each class of pesticide and mutual associations between groups of metabolites and multiple pesticides. We conducted high-resolution metabolomic profiling of serum samples from 176 older adults living in the California Central Valley using liquid chromatography with high-resolution mass spectrometry. We estimated chronic pesticide exposure (from 1974 to year of blood draw) to OPs, OCs, and PYRs from ambient sources at homes and workplaces with a geographic information system (GIS)-based model. Based on partial least-squares regression and pathway enrichment analysis, we identified metabolites and metabolic pathways associated with one or multiple pesticide classes, including mitochondrial energy metabolism, fatty acid and lipid metabolism, and amino acid metabolism. Utilizing an integrative network approach, we found that the fatty acid β-oxidation pathway is a common pathway shared across all three pesticide classes. The disruptions of the serum metabolome suggested that chronic pesticide exposure might result in oxidative stress, inflammatory reactions, and mitochondrial dysfunction, all of which have been previously implicated in a wide variety of diseases. Overall, our findings provided a comprehensive view of the molecular mechanisms of chronic pesticide toxicity, and, for the first time, our approach informs exposome research by moving from macrolevel population exposures to microlevel biologic responses.
Graphical Abstract
1. INTRODUCTION
Agricultural pesticides are chemicals used for crop protection and combating animal pests and disease. California leads the United States in pesticide use, with a total of 205 million pounds of pesticides applied in 2017.1 In particular, the region of greatest pesticide use in California is the Central Valley area, including Fresno, Kern, and Tulare counties. Based on chemical properties, the main classes of insecticides used in California have historically included organophosphates (OPs), organochlorines (OCs), and pyrethroids (PYRs).
OPs are widely used insecticides, and they are designed to inhibit acetylcholinesterase enzyme activity, which leads to accumulation of the neurotransmitter acetylcholine and results in neurotoxic effects in the peripheral and central nervous system.2–4 OCs are a group of chlorinated compounds that are highly persistent organic pollutants (POPs) in the environment.5 The modes of action of OC pesticides include opening of sodium ion channels in neurons6 as well as binding with the GABA chloride ionophore complex producing a decreased uptake of chlorine ions in neurons.7 Due to the toxicity of OPs and the persistence of OCs, their use in agriculture has been restricted, and in recent decades, other types of insecticides, including PYRs, have been used more widely.8 The primary targets of PYRs are the membrane sodium channels, but they also act on potassium, chloride, and calcium channels.9–11 As a group they are known as axonic excitotoxins as they disturb sodium channels, leading to abnormal neural activity.7,11 Furthermore, chronic exposure of OPs, PYRs, and OCs has been shown to have a wide range of downstream secondary effects, including genotoxicity and DNA damage, oxidative stress, immunotoxicity, mitochondrial dysfunction, and regulation of neuronal apoptosis.7,12–16
Numerous epidemiological studies, as well as in vitro and in vivo experimental evidence, suggest these three pesticide groups are associated with a wide range of adverse health effects including neurodegenerative disease, such as seen in Parkinson’s disease (PD) and Alzheimer’s disease (AD), diabetes, and cancers.17 OC and PYR exposures have also been associated with neurodevelopmental impairment.18–20
As chronic pesticide exposures may have profound effects on various biological systems, characterizing metabolic response to these chemicals and how they influence human health systemically is important. High-resolution metabolomics (HRM) is a powerful analytical approach that can profile more than 10,000 endogenous and exogenous chemicals in biological specimens. Recently, HRM has been used to study the effects of environmental exposures such as metals, air pollution, tobacco smoking, and polycyclic aromatic hydro-carbons on human metabolism.21–23
A large number of metabolomic studies have employed animal models or cell lines to study the toxicity of different pesticides.8,24–29 In recent years, metabolomics has also been applied to study the effect of OP, PYR, or OC pesticide exposure. For example, a cross-sectional study of 83 pregnant women characterizing the urine metabolome found that pesticide mixtures increased oxidative stress and disturbed energy metabolism,30 while a larger study of 750 pregnant women identified mitochondrial catabolic pathways as being associated with low-level exposure to OCs.31 OP and OC metabolic profiling of 102 Chinese pregnant women found OP and OC exposures may disrupt thyroid hormone metabolism and glyceraldehyde metabolism.32 A Swedish study investigated dichlorodiphenyldichloroethylene and hexachlorobenzene exposure using serum metabolomic profiles from 1016 elderly participants and identified lipid metabolism as an essential metabolic response to OC pesticides.33 Other studies focusing on persistent organic pollutants in general indicated that POPs disturb amino acid, lipid and fatty acid, and carbohydrate metabolism.34,35
Most previous studies measured pesticide exposure levels via pesticide biomarkers in blood or in urine samples. While these biomarkers estimate exposure at the same time as the metabolome is being characterized, they may not be suitable for capturing the cumulative effects of low-level chronic past pesticide exposures or for chemicals with short biological half-lives. Also, POP exposures that occurred in the distant past may not be represented well in a recent blood draw, especially in older individuals. In addition, the majority of studies using biomarkers mostly focused on one or a few specific chemicals instead of integrating various types of pesticides. Here, we conducted an untargeted metabolomics study of exposure to three groups of pesticides (OPs, PYRs, and OCs) using metabolomes from serum samples from 176 older adults who were recruited as control subjects into a study of PD in the California Central Valley. Chronic pesticide exposure from ambient sources at homes and workplaces over decades was estimated with a geographic information system (GIS)-based model for each type of pesticide. We utilized the same liquid chromatography high-resolution mass spectrometry platform that has been used in several previous studies to examine the metabolomic response to environmental toxicants including OCs.36,37 This platform has been shown to be sensitive enough to capture metabolic perturbations associated with low-level environmental exposures, a goal of our study. Furthermore, by conducting an integrative network analysis, we aim to provide a comprehensive view of the way in which these pesticides may influence the human metabolome and how this may result in potential adverse health effects.
2. METHODS
2.1. Study Population and Samples Collection.
The Parkinson’s Environment and Gene Study (PEG) is a community-based case-control study of PD etiology in agricultural regions of the California central valley that recruited control subjects from Kern, Tulare, or Fresno counties between 2001 and 2007.38 Controls were randomly sampled either from Medicare rolls or from property tax assessor records listing residential parcels. Participation was limited to one person per household, and eligibility criteria included being 35+ years of age, having lived in one of the three counties for the past five years or more, and not having received a Parkinsonism diagnosis. More detailed information has been provided elsewhere.39
Here, we used blood samples collected at enrollment from 176 adult controls as well as demographic information, lifestyle, and medical data collected in standardized interviews. Blood samples were collected by study staff during field visits in local clinics, centrifuged, transferred on ice, and stored in a –80 °C freezer at UCLA. Serum samples were shipped frozen to Emory University on dry ice for metabolomics analyses, where they were stored at −80 °C until analyses.
2.2. Pesticide Assessment.
Pesticide exposure assessment was performed as previously described.40 Briefly, we estimated ambient pesticide exposure for each participant based on a GIS approach.41 Participants’ residential and occupational address histories were used to assess proximity to commercial agricultural pesticide applications, which are reported in the California state-mandated pesticide use reports (CA-PUR). The CA-PUR is a state-wide registry of all commercial pesticide applications since 1974, and our GIS approach linked these data to land use surveys providing locations of specific crops, and eventually to participants’ geocoded residential and occupational address histories.
Based on the California Department of Pesticide Regulation and the pesticide action network pesticide database, we identified individual chemicals that were classified as one of three types of pesticides: organophosphates (OP), pyrethroids (PYR), and organochlorines (OC). Specifically, 36 different chemicals were classified as OPs, 14 chemicals as PYRs, and 8 chemicals as OCs. We calculated the total pounds of each chemical applied within 500 m of the addresses annually from 1974 to the year of blood draw and then averaged the pounds per acre for each pesticide across residential and occupational addresses over the entire period. Exposures at both residential and occupational addresses were included, and each participant could have been exposed at both locations, only one, or neither. To calculate the total number of individual OP, PYR, and OC pesticides a participant had been exposed to, we dichotomized exposure to each of the individual chemicals based on each chemical’s median exposure level and then summed the number of chemicals that each participant was exposed to above the median, counting chemical exposures at both residences and work places.
2.3. High-Resolution Metabolomics.
HRM profiling was completed according to established methods.42 Briefly, serum samples were collected during the interview and stored at –80 °C. Prior to HRM, batches of 40 serum samples were removed from storage and thawed on ice. Each sample was then thoroughly vortexed, and plasma proteins were precipitated by diluting 65 μL of serum with 130 μL of LC-MS grade acetonitrile. We previously compared extraction efficiency using different volume equivalents of acetonitrile and methanol.43 Comparison of protein removal using the Lowry method and by SDS- PAGE with Coomassie blue staining and densitometry showed a 2:1 ratio of acetonitrile removed 98% of plasma proteins, and has been used for all subsequent HRM studies. The extract was centrifuged, and the resulting supernatant was transferred to an autosampler vial containing a low-volume insert and maintained at 4 °C until analysis (<24 h). To evaluate system performance, we used two separate quality assessment methods. Our first QC sample was NIST 195044 which was analyzed at the beginning and end of the entire analytical run. The second QC sample (Q-Std) included commercially purchased plasma pooled from an unknown number of males and females. Q-Std was analyzed at the beginning, middle, and end of each batch of 40 samples for normalization and batch effect evaluation.
Sample extracts were analyzed in triplicate using a dual-column, dual-polarity approach that includes hydrophilic interaction (HILIC) chromatography with positive ESI and C18 chromatography with negative ESI (Ultimate 3000, Q-Exactive HF, Thermo Fisher, m/z range 85–1275).45 We chose HILICpos and C18neg because dual chromatography HRM with positive and negative electrospray ionization provides the optimal number of reproducible ions detected and matches to known chemicals in the Kyoto Encyclopedia of Genes and Genomes (KEGG) Human Metabolite database.46 Following a 10 μL sample injection, HILIC separation was accomplished using a 2.1 cm × 5 cm × 2.5 μm HILIC column (Waters XBridge BEH Amide XP HILIC) and acetonitrile gradient (A = water, B = acetonitrile, C = 2% formic acid) consisting of an initial 1.5 min period of 22.5% A, 75% B, and 2.5% C, followed by linear increase to 77.5% A, 20% B, and 2.5% C at 4 min and hold for 1 min. Separation by C18 was with 2.1 cm × 5 cm × 3 μm column (Higgins end-capped C18) with C = 10 mM ammonium and the following gradient: initial 0.5 min period of 60% A, 35% B, and 5% C, followed by linear increase to 0% A, 95% B, and 5% C at 1.5 min and then held for an additional 3 min. Mobile phase flow rate was held at 0.4 mL/min for 1.5 min and then increased to 0.5 mL/min. The mass spectrometer was operated using ESI mode at a resolution of 120,000 and mass-to-charge ratio (m/z) range 85–1275. High-resolution detection of m/z features was accomplished by a maximum injection time of 10 ms and an AGC target of 1 × 106. Raw data files were extracted and aligned using apLCMS47 with modifications by xMSanalyzer.48 Uniquely detected ions consisted of m/z, retention time (rt), and ion abundance, referred to as m/z features. Prior to data analysis, m/z features were batch corrected using a novel algorithm based on wavelet.49 For further analyses, we only included metabolomic features detected in >25% of all plasma samples, with median coefficients of variation among technical replicates <30% and Pearson correlation >0.7. Following quality assessment, intensities of three replicates for each feature were summarized using the median value. In addition, we conducted a log 2 transformation and autoscaling. Missing values were imputed using k-nearest neighbors (k = 10)50 imputed in the impute R package.
2.4. Metabolome-Wide A ssociation Analysis.
We present the distributions of three pesticide classes and Pearson correlations for levels of pesticide exposure. All analyses were performed using R version 4.0.1.
We conducted partial least-squares (PLS) regression to identify metabolomic features associated with each pesticide group (i.e., OPs, PYRs, and OCs). PLS regression is a supervised, multivariate analysis approach for dimensionality reduction that maximizes covariance between intensities of metabolomic features and exposures.51 To adjust for potential confounders, we regressed each feature’s intensity on potential confounding variables (age, sex, and race/ethnicity) and formed residuals as the input matrix. Features with variable importance in projection (VIP) scores >2 were selected as significant. A 10-fold cross-validation was used to assess the performance of the selected features. We further applied linear regression to test for linearity between m/z features and pesticides.
To assess the potential confounding effect of socioeconomic status, we conducted a sensitivity analysis by additionally adjusting for education.
To gain a holistic view of the relationship between pesticides and the serum metabolome, we further conducted integrated network analysis using xMWAS.52 xMWAS calculates the pairwise association scores (approximation of the correlation coefficient) between each metabolomic feature and each pesticide using partial least-squares53 and generates a multidata integrative network. Only associations with both the |association score| > 0.2 and p < 0.05 (by Student’s t test) were included in the network. Finally, a multilevel community detection algorithm54 was used to identify metabolite and exposure clusters that are tightly connected with each other, but sparsely connected with the rest of the network.
2.5. Annotation and Pathway analysis.
HRM provides accurate mass (±5 parts per million; ppm) measures of ion m/z, which can be related to chemical monoisotopic mass. Significant features selected by PLS regression were first matched to a reference database of authenticated chemical standards previously characterized in our laboratory. The list of in-house database of metabolites was confirmed using MS/MS and authentic standards; the detailed process has been published previously.42,55 The error tolerance was 5 ppm and 15 s for m/z and retention time, respectively. Additional m/z features not matching these metabolites were then annotated by xMSannotator.56 Accurate mass m/z for adducts formed under positive/negative ESI mode was matched to the Human Metabolome Database (HMDB), KEGG, and LipidMaps with a mass error threshold of 10 ppm. xMSannotator also considers the correlation of intensities and retention time and assigns confidence scores based on a multilevel scoring algorithm (0–3, a higher score representing higher-confidence result), ensuring annotation accuracy. Only results with an annotation score >2 were kept. The metabolite identification confidence levels57 were reported for all annotation results.
We conducted pathway enrichment analysis utilizing mummichog version 258 to identify perturbed metabolic pathways associated with pesticide exposure for each class of pesticides. All features previously selected by PLS regression with VIP ≥ 2 and features included in the integrated network analysis were included in this pathway enrichment analysis. Mummichog uses a permutation-based framework that accounts for the complexity of untargeted mass spectral data. Although annotation results in mummichog may include false positives, the enriched pathways inferred by the algorithm have been shown to be valid and reflect real biological activity.59 All metabolites annotated by mummichog were required to present in at least their primary adduct (M + H or M − H for positive and negative mode, respectively) to reduce the false positive match rate. A pathway was considered significant if γ adjusted p-values were smaller than 0.05. Only pathways that contained at least three discriminative metabolites were interpreted.
3. RESULTS
3.1. Study Population.
Demographics of the 176 adult controls unaffected by PD and the distribution of pesticide exposures are provided in Table 1. Lists of chemicals classified as OPs, PYRs, or OCs can be found in Table S1. Approximately half of the participants included in the study were male (51%), and a majority were white (86%) with a mean age of 66 years. Participants who did not have metabolomics data available did not differ from participants we included in terms of age, sex, ethnicity, or pesticide exposure. OP exposure counts ranged from 0 to 41, PYR from 0 to 10, and OC from 0 to 8. Exposure levels of pesticides in the three classes were moderately correlated (Figure S1).
Table 1.
Distribution of Demographics and Pesticide levels
| PEG study community controls (n = 176) | |
|---|---|
| Age | |
| min | 35 |
| max | 92 |
| mean (SD) | 66.13 (13.40) |
| Sex | |
| male (%) | 90 (51) |
| female (%) | 86 (49) |
| Race/Ethnicity | |
| white (%) | 159 (90) |
| hispanic (%) | 17 (10) |
| Education | |
| less than high school | 20 (11) |
| high school | 32 (18) |
| more than high school | 124 (71) |
| Organophosphate Count | |
| min | 0 |
| max | 41 |
| mean (SD) | 7.58 (8.46) |
| Pyrethroid Count | |
| min | 0 |
| max | 10 |
| mean (SD) | 0.94 (1.70) |
| Organochlorine Count | |
| min | 0 |
| max | 8 |
| mean (SD) | 1.23 (1.72) |
3.2. Metabolome-Wide Analysis.
We detected 12,925 metabolomic features from the HILIC column coupled with positive ionization mode (HILICpos) and 7209 metabolomic features from the C18 column coupled with negative ionization mode (C18neg). After quality control steps, a total of 16,510 metabolomic features (10,959 HILICpos and 5551 C18neg) were included in the analyses.
After adjusting for potential confounding variables (age, sex, and race/ethnicity), PLS regression selected 389 metabolomic features (254 HILICpos features and 135 C18neg features) across three components that were associated with OP exposure (Figure S2, Tables S2 and S3). Among these, 226 features (58%) were positively associated with OP exposure. Furthermore, 517 metabolomic features (331 HILICpos features and 186 C18neg features) were associated with PYR pesticide, and 233 features (45%) increased with increasing levels of PYR exposure (Figure S2, Tables S2 and S3). We also identified 485 features (311 HILICpos features and 174 C18neg features) related to OCs. Approximately half of the features (229 features, 47%) were positively associated with OCs (Figure S2, Tables S2 and S3). A large proportion of metabolomic features were uniquely associated with just one of the pesticide groups, but 72 statistically significant features were shared across all three pesticide classes (Figure 1).
Figure 1.
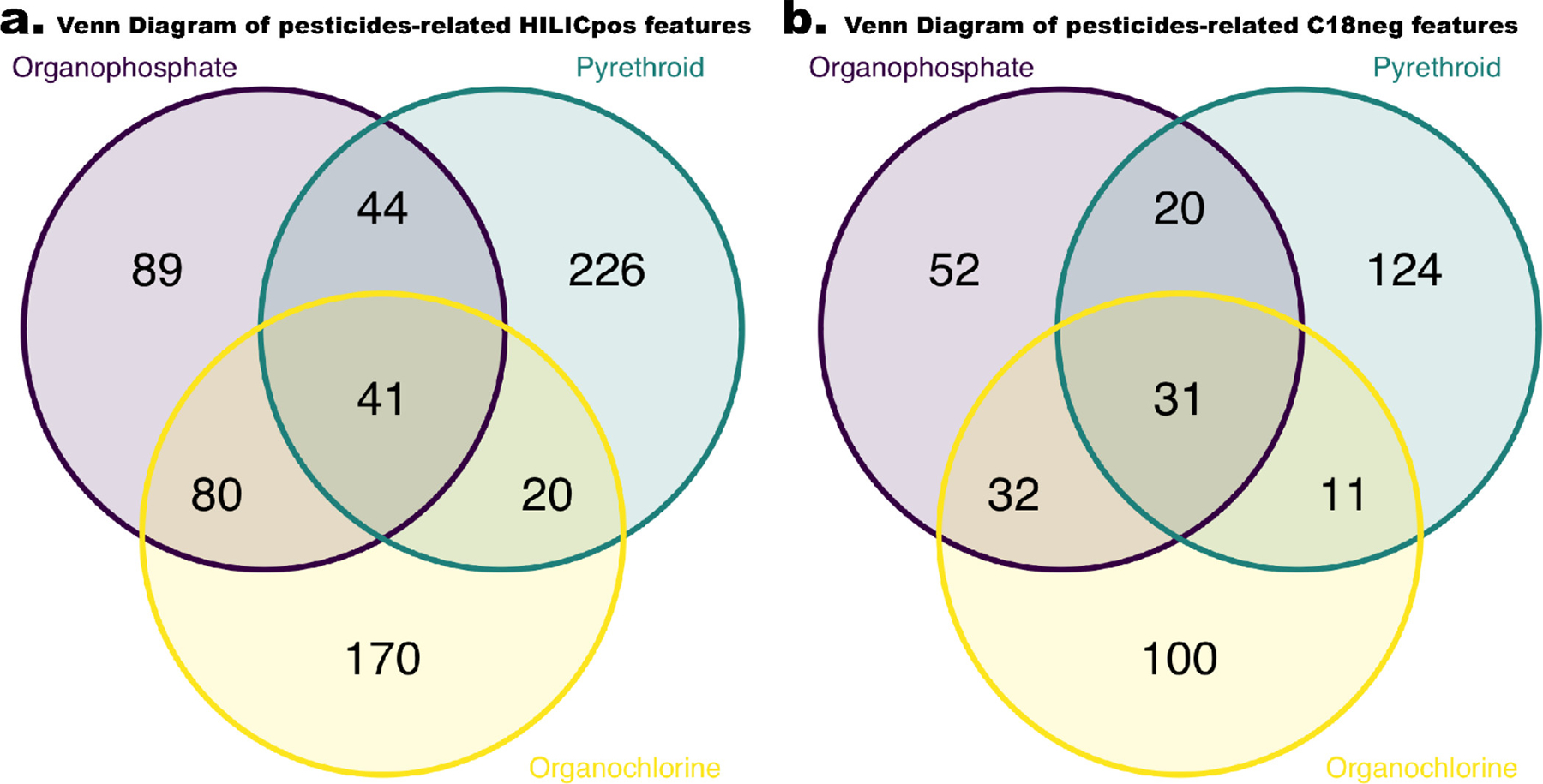
Venn diagram of pesticides-related HILICpos features (a) and C18neg features (b).
Based on xMSannotator, in total, 15.9% of the HILICpos features and 16.2% C18neg features were matched to compounds in the HMDB, KEGG, and LipidMaps databases (identification confidence level 3). We matched metabolomic features related to at least one of the pesticide groups to known metabolites based on authenticated chemical standards verified by tandem mass spectrometry (identification confidence level 1). In total, we confirmed the identities of 12 metabolites (Table 2), including metabolites involved in the carnitine shuttle pathway and β-alanine metabolism.
Table 2.
Confirmed Chemical Identity of Metabolomic Features Associated with OP, PYR, or OC Pesticide
| OP | PYR | OC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| name | KEGG ID | m/z | time | coefficient | P-value | coefficient | P-value | coefficient | P-value |
| l-histidine | C00135 | 154.0623 | 50.63 | 0.014 | 0.031 | 0.091 | 0.005 | 0.060 | 0.067 |
| lauroylcarnitine c12 | CA1406 | 344.279 | 48.7 | −0.013 | 0.070 | −0.077 | 0.027 | −0.067 | 0.055 |
| alanine | C00041 | 90.055 | 97.36 | 0.014 | 0.044 | 0.059 | 0.072 | 0.065 | 0.050 |
| octanoylcarnitine c8 | C02838 | 288.2166 | 52.25 | −0.011 | 0.114 | −0.068 | 0.037 | −0.068 | 0.039 |
| sphinganine | C00836 | 302.3049 | 46.12 | −0.018 | 0.024 | −0.044 | 0.255 | −0.122 | 0.001 |
| cytidine 2,3-cyclic monophosphate | C02354 | 306.0489 | 118.2 | −0.017 | 0.041 | −0.039 | 0.338 | −0.080 | 0.051 |
| 11-deoxycortisol | C05488 | 347.2207 | 47.92 | 0.016 | 0.078 | 0.091 | 0.043 | 0.087 | 0.054 |
| pyruvate | C00186 | 87.0087 | 38.65 | 0.019 | 0.032 | 0.064 | 0.131 | 0.065 | 0.133 |
| O-acetyl-l-carnitine | C02571 | 204.123 | 70.68 | −0.012 | 0.162 | −0.069 | 0.107 | −0.088 | 0.041 |
| l-methionine | C00073 | 148.0439 | 47.27 | 0.012 | 0.156 | 0.100 | 0.013 | 0.053 | 0.197 |
| carnosine | C00386 | 227.1136 | 110.36 | 0.012 | 0.121 | 0.095 | 0.012 | 0.034 | 0.374 |
| omega-hydroxydodecanoic acid | C08317 | 217.1798 | 20.12 | 0.022 | 0.011 | 0.057 | 0.167 | 0.028 | 0.498 |
Using mummichog, we examined whether the features that were selected by PLS regression were enriched within specific metabolic pathways. The result indicated that 33 metabolic pathways were significantly enriched with a p-value smaller than 0.05 (Figure 2, Table S4). List of putatively annotated metabolites (identification confidence level 3) within each pathway can be found in Table S5. The majority of pathways were uniquely associated with exposure to one of the pesticide groups, whereas β-alanine metabolism and glycine, serine, alanine, and threonine metabolism were linked with all three classes, that is, OPs, PYRs, and OCs. Carnitine shuttle, glutamate metabolism, glycolysis and gluconeogenesis, butanoate metabolism, and pyruvate metabolism were related to both OPs and OCs. The enriched pathways are involved in a wide range of metabolic functions, including lipid metabolism, central carbon metabolism, amino acid metabolism, neurotransmitter precursors, cofactors, and nucleotide metabolism.
Figure 2.
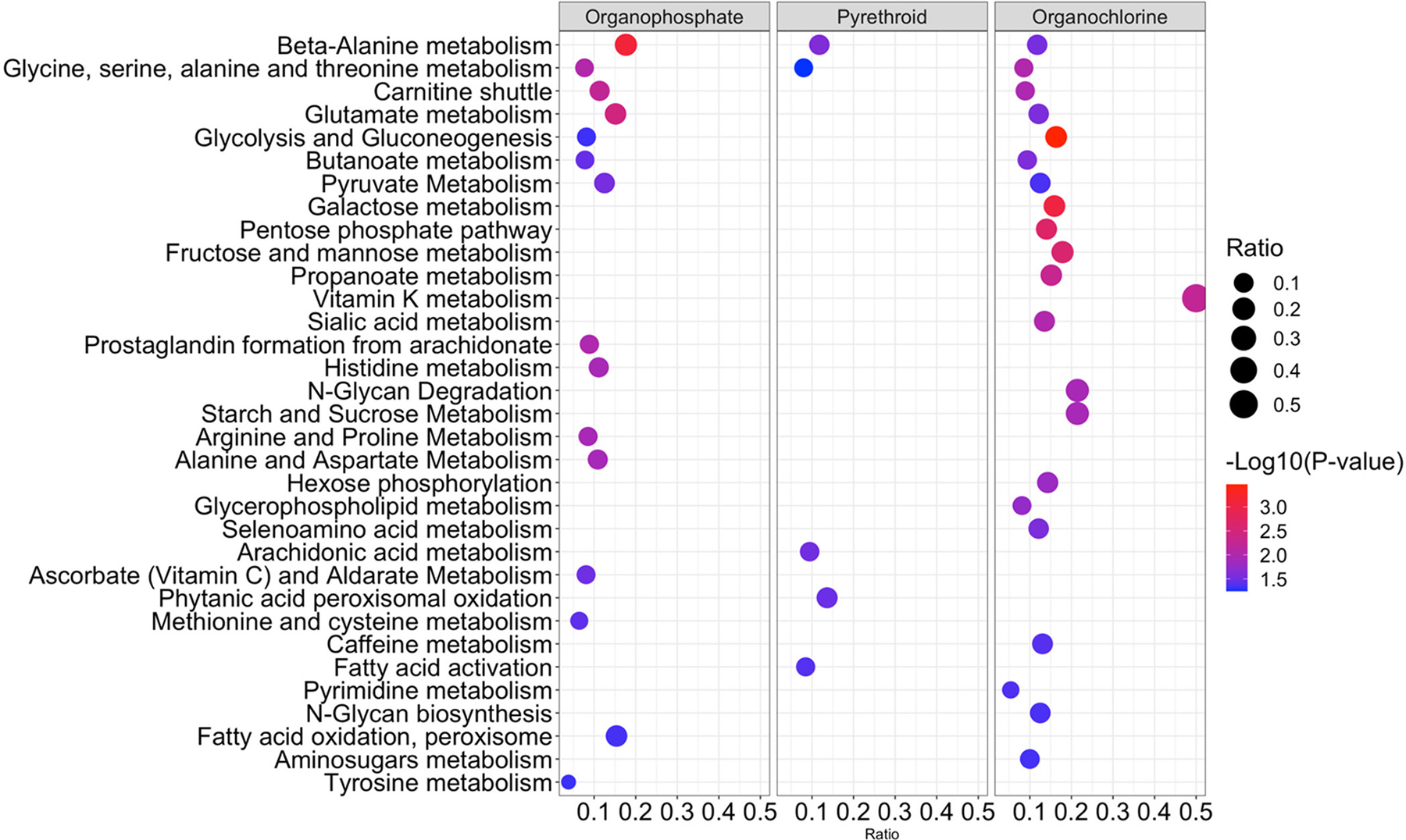
Metabolic pathways correlated with pesticides from a serum metabolome-wide association study. The vertical axis represents the pathways associated with pesticides. The circle radius is proportional to the number of correlated metabolite features within each pathway (ratio). The horizontal axis also represents the ratio. The color represents the −log 10 (p-value) of each pathway.
To assess the potential for additional confounding, we adjusted for education information (Figure S3), which can serve as a surrogate for socioeconomic status. Enriched pathways identified in the sensitivity analysis were very similar to the original results.
3.3. Integrated Network analysis.
To identify systemic metabolic alterations associated with different pesticides, we adopted a network-based approach to integrate metabolomic features with the three pesticide classes. In total, 383 features were included in the network, with an association score >0.2 and a p < 0.05 (Figure 3). Unlike the previous MWAS approach in which most of the features identified were uniquely associated with one pesticide, this network was more densely connected, that is, 187 out of 383 features within the network were correlated with all three pesticides. We identified two separate clusters using a community detection algorithm, including one cluster (blue) centered on OPs and a second cluster (yellow) that included PYRs and OCs (Figure 3). Both clusters included pathways related to fatty acid β-oxidation. In addition, the OP cluster consisted of several pathways involved in amino acid metabolism and neurotransmitter precursors (Tables S6–S8).
Figure 3.
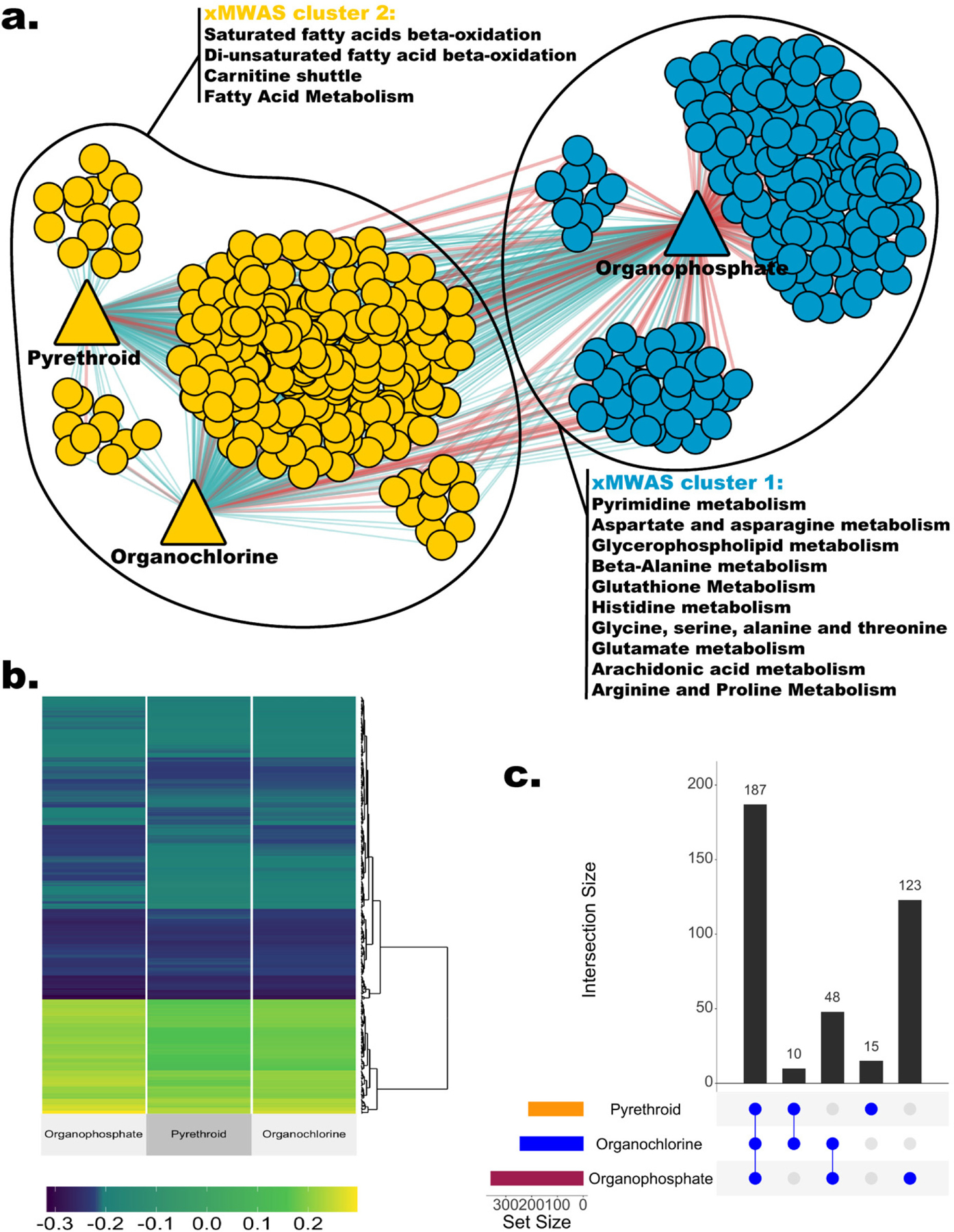
Integrative network analysis with xMWAS. (a) The metabolome-pesticide network revealed two metabolomic communities. Only metabolomic features with an association score >0.2 and p < 0.05 were included in the network. Round nodes represent metabolomic features, and triangle nodes represent three pesticides. The edges’ color represents the direction of association (red: positive, green: negative). Multilevel community detection algorithm identified two clusters, represented by different node colors. (b) Heatmap of the association scores between metabolomic features and pesticides. Only features included in the network are shown here. (c) UpSet plot shows the number of features associated with each pesticide group and the intersection size.
4. DISCUSSION
Our community-based study provided us with a unique opportunity to link GIS derived pesticide exposure data with a HRM approach, thus allowing the external and internal exposomes to inform on each other. Specifically, we first used a complex GIS approach to combine almost a half century of California pesticide application records spatially with home and work address histories to generate measures of long-term pesticide exposure from agricultural applications in older adults. Then we combined these exposure histories with data from HRM to interrogate links between complex patterns of pesticide exposure with metabolic responses. Our approach provided us with the opportunity to measure more than 10,000 metabolomic features in serum samples simultaneously, and we found that chronic exposure to pesticides in three classes, OPs, PYRs, or OCs, seemed to perturb a wide range of metabolic pathways. Employing MWAS for these three pesticide groups as well as an integrative network approach, we identified metabolic pathways distinctly associated with only one group of pesticides as well as metabolic response patterns shared across the pesticide classes that included fatty acid metabolism, central carbon metabolism, and amino acid metabolism. Specifically, we found that the β-alanine metabolism and glycine, serine, alanine and threonine metabolism were related to all three pesticides. Also, several fatty acid metabolic pathways were associated with both OPs and OCs. Network analysis showed that the fatty acid β-oxidation pathway was a common pathway shared by all three pesticide groups. More generally, the alterations we observed in these pathways suggested that chronic pesticide exposure may result in oxidative stress, inflammatory reactions, and mitochondrial dysfunction. These findings are informative as various pesticide-related health outcomes, such as neurodegenerative disorders, diabetes, and cancer, are related to these pathways.60–64
Previous epidemiological and experimental studies have shown that chronic exposure to OPs, PYRs, and OCs results in various secondary toxic effects, including mitochondria dysfunction and oxidative stress.7,14,64 In our study, we identified alterations in oxidative stress-induced fatty acid oxidation in peroxisomes as being associated with OP and PYR exposures and glycerophospholipid metabolism as being associated with OC pesticide exposures. Based on our network analysis, diunsaturated fatty acid β-oxidation was related to all three exposure types. This finding is consistent with previous findings that abnormal fatty acid metabolism and glycerophospholipid metabolism are correlated with persistent organic pollutant exposure.34,35 A study that focused on polybrominated and polychlorinated biphenyl POPs also found alterations in fatty acid metabolism, including fatty acid activation and glycerophospholipid metabolism.36 Moreover, the same fatty acid metabolism pathways were associated with other environmental exposures such as trichloroethylene, benzo[a]pyrene, or air pollution.65–67 Oxidative stress and disturbed fatty acid metabolism are thought to underlie many adverse health outcomes such as heart disease, diabetes, and neurodegenerative diseases including AD and PD.68–70 The pathophysiology of PD involves the depletion of dopamine neurons, which is a neuronal population that is particularly susceptible to oxidative stress owing to its low antioxidant capacity.71
Pesticide-induced oxidative stress may lead to mitochondrial dysfunction, and we observed enrichment of several pathways associated with mitochondrial energy metabolism. For example, the carnitine shuttle, short-chain fatty acid (butanoate) metabolism, glycolysis and gluconeogenesis pathway, and pyruvate metabolism were perturbed with higher OPs and OCs exposure. Fatty acid oxidation in the peroxisome was associated with both OPs and PYRs. Although the carnitine shuttle pathway was not significantly enriched with the PYR class pesticides, several acylcarnitines, including lauroylcarnitine and octanoylcarnitine, were significantly decreased when PYR exposure levels increased. The carnitine shuttle refers to the process of transporting long-chain fatty acetyl coenzyme A (acyl-CoA) into mitochondria as acylcarnitines for fatty acid oxidation.72 This process is critical for energy supply in every tissue. Studies have shown that plasma acylcarnitine levels can serve as an indicator of mitochondrial function.73,74 Mitochondrial dysfunction contributes to diabetes, cardiovascular disease, cancer, metabolic syndrome, and chronic neurodegenerative disorders such as PD and Huntington’s disease.75–77
There is also increasing evidence for immunotoxicity of certain pesticides.10,71 In our study, several inflammation-related pathways, including histidine metabolism, arachidonic acid metabolism, and prostaglandins metabolism, were associated with OPs and PYRs. Histidine has anti-inflammatory effects and is negatively associated with inflammation and oxidative stress.78 Oxidative stress can also induce the activation of phospholipase A2 and generate polyunsaturated free fatty acid, including linoleic acid and arachidonate acid.79 Arachidonic acid is an inflammatory intermediator and can be converted to prostaglandins, which has a major proinflamma-tory effect. In accordance with our results, previous studies also linked inflammation-related pathways to environmental toxicants.80
Perturbed glutamate metabolism was related to increased OPs and OC exposures, which was consistent with the results obtained in some animal models.81,82 Glutamate is a precursor for glutathione, the second major thiol redox couple for antioxidant defense. Moreover, glutamate serves as an excitatory neurotransmitter in the central nervous system. Previous studies have demonstrated a link between glutamate-mediated excitotoxicity and degeneration of dopamine neurons,83,84 and glutamate may play a role in the development of PD. Furthermore, we also observed an association between tyrosine metabolism and OP exposure, and tyrosine is a precursor of dopamine, that is, tyrosine is converted to L-dopa by tyrosine hydroxylase.36 Previous studies suggested that alterations of tyrosine hydroxylase can contribute to neurodegenerative diseases, including PD and AD because it affects the biosynthesis of dopamine.85,86 Studies also linked these pesticides to an increased risk of PD and other neurodegenerative disorders. Our data suggest that dysregulation of these neurotransmitter precursors are tentatively associated with OPs or OCs.
By utilizing an integrative network analysis, we obtained a holistic view of interactions between the metabolome and various pesticides. Even though there were distinct mechanisms of action for each pesticide class, most metabolomic connections were shared across pesticide groups. A similar overlap of metabolomic responses has been described before for dichlorodiphenyltrichloroethane, poly and perfluoroalkyl substances, oxychlordane, hexachlorobenzenes, and several polychlorinated biphenyl compounds; specifically, the metabolome-exposure network converged on oxidative stress, fatty acid metabolism, and mitochondrial energy metabolism.37 Our findings, and those of previous studies, might suggest general metabolic mechanisms an organism employs in response to the stress induced by various environmental toxicants.
Two amino acid metabolism pathways, β-alanine metabolism and glycine, serine, alanine, and threonine metabolism, were found to be associated with all three pesticide groups. β-Alanine is a nonessential amino acid the human body makes by converting pyruvate.82 Alanine has various biological functions in different tissues. For example, β-alanine functions as a neurotransmitter or a neuromodulator in the brain, while in skeletal muscle, it is a major energy source and a component of carnosine.87,88 Using authentic standards, we were able to identify both alanine and carnosine within the β-alanine metabolism pathway. Alanine increased with higher levels of OPs and OCs, and carnosine was significantly and positively associated with OPs and PYRs. Similarly, increased alanine levels have been found in the brain of goldfish after exposure to PYRs.82 Rats exposed to OP pesticides expressed an abnormal level of alanine aminotransferase due to disturbances in the oxidative–reductive hepatocyte system.89 A human study found elevated urine alanine levels in pesticide applicators compared to nonapplicators.90 Carnosine can function as an antioxidant that scavenges ROS.91,92 Similarly, glycine and serine also act as a cytoprotective agent.90 Therefore, the increased activity in alanine, glycine, and serine metabolism may indicate the activation of antioxidative defense mechanism in response to pesticide-induced oxidative stress, as has been shown in previous metabolomics studies of pesticide mixtures.30 Vitamin C is a water-soluble antioxidant,93 and disturbances of vitamin C metabolism may also indicate responses to oxidative damage caused by OP exposure. Finally, there are several other amino acid metabolism and cofactor pathways related to OP pesticide exposure that are involved in oxidative stress and inflammation. Methionine is an essential amino acid that promotes ROS production, in line with chronic OP exposure inducing oxidative stress.
Our study has some limitations. Our GIS model-based chronic pesticide exposure measure allowed us to investigate low-level exposure that occurred over decades; however, it is challenging to estimate the absolute exposure levels for each chemical we investigated. In addition, our pesticide measure does not necessarily translate into total pesticide exposure levels as other sources of pesticide exposures may also contribute. Despite the inherent challenges in GIS model-based exposure estimation, our pesticide measurements have been associated with a number of prior health outcomes such as PD, autism spectrum disorders, and neural tube defects.94–96 Also, our previous epigenomic studies of OPs and PYRs identified critical genetic pathways consistent with the pathophysiology of each pesticide type of action, in support of the validity of our exposure assessment model. Diet may also explain differences in metabolites, and we did not have information on the participants’ dietary intake; however, in order for dietary differences to have confounded our analyses, they would have had to be related to agricultural pesticide exposure, which would be more likely for dietary pesticide sources rather than ambient exposures due to agricultural applications near homes and workplaces. By controlling for age, sex, and race/ethnicity, we hope to have at least partially addressed potential confounding. Another limitation is the annotation of metabolomic features detected using untargeted analysis. Adopting a pathway and network analysis approach, we were able to improve annotation results, but there may still be incorrect matches that could have influenced the interpretation of our results. It is recommended to improve metabolite identification using either tandem MS or internal standards in future studies.
5. CONCLUSION
In summary, we utilized a HRM approach to identify perturbations in the serum metabolome of older adults in response to long-term ambient OP, PYR, and OC exposures measured via a complex GIS approach and pesticide application records. We identified disturbances in metabolic pathways related to oxidative stress, inflammation, lipid and fatty acid metabolism, mitochondrial energy metabolism, and neurotransmitter precursors. Furthermore, by adopting integrative network analysis, we illustrated that different pesticides might share similar biochemical fingerprints at the metabolomic response level. Collectively, our study points to potential common molecular mechanisms of chronic pesticide toxicity and thus may also shed light on the physiology these exposures affect that eventually can lead to various adverse pesticide-related health outcomes. Most importantly, our approach for the first time informs the field of exposome research by moving from macrolevel population exposures to microlevel biologic responses, thereby generating hypotheses about toxicological mechanisms that can be studied in vivo.
Supplementary Material
Funding
This work was supported by the National Institute of Environmental Health Sciences grants R01ES010544 and R21ES030175
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of funding bodies such as the National Institutes of Health or National Institute of Environmental Health Sciences.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.0c00523.
Heatmap of pairwise correlations of pesticide counts; identification of metabolomic features associated with pesticide exposure; enriched pathways identified from the sensitivity analysis by additionally adjusting for education; list of chemicals within OP, PYR, or OC groups; significant HILICpos features; significant C18neg features; annotated metabolites within each enriched pathway based on mummichog; enriched pathways associated with pesticide exposures; enriched metabolic pathways associated with xMWAS cluster 1 (OP); enriched metabolic pathways associated with xMWAS cluster 2 (PYR, OC); pathway enrichment analysis for features associated with all three pesticides in xMWAS (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.chemrestox.0c00523
The authors declare no competing financial interest.
Contributor Information
Qi Yan, Department of Epidemiology, UCLA Fielding School of Public Health, Los Angeles, California 90095, United States.
Kimberly C. Paul, Department of Epidemiology, UCLA Fielding School of Public Health, Los Angeles, California 90095, United States
Douglas I. Walker, Department of Environmental Medicine and Public Health, Icahn School of Medicine at Mount Sinai, New York 10019, United States.
Melissa A. Furlong, Department of Community, Environment, and Policy, University of Arizona Mel and Enid Zuckerman College of Public Health, Tucson, Arizona 85724, United States
Irish Del Rosario, Department of Epidemiology, UCLA Fielding School of Public Health, Los Angeles, California 90095, United States.
Yu Yu, Department of Environmental Health Science, UCLA Fielding School of Public Health, Los Angeles, California 90095, United States.
Keren Zhang, Department of Epidemiology, UCLA Fielding School of Public Health, Los Angeles, California 90095, United States.
Myles G. Cockburn, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California 90089, United States
Dean P. Jones, Clinical Biomarkers Laboratory, Division of Pulmonary, Allergy, and Critical Care Medicine, School of Medicine and Department of Medicine, Emory University, Atlanta, Georgia 30322, United States
Beate R. Ritz, Department of Epidemiology, UCLA Fielding School of Public Health, Los Angeles, California 90095, United States; Department of Neurology, UCLA School of Medicine, Los Angeles, California 90095, United States.
REFERENCES
- (1).(2017) Summary of Pesticide Use Report Data - 2017. California Department of Pesticide Regulation, California Environmental Protection Agency, Sacramento, CA. [Google Scholar]
- (2).Terry AV Jr (2012) Functional consequences of repeated organophosphate exposure: potential non-cholinergic mechanisms. Pharmacol. Ther 134 (3), 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).van der Plaat DA, de Jong K, de Vries M, van Diemen CC, Nedeljković I, Amin N, Kromhout H, Vermeulen R, Postma DS, van Duijn CM, et al. (2018) Occupational exposure to pesticides is associated with differential DNA methylation. Occup. Environ. Med 75 (6), 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Paul KC, Chuang YH, Cockburn M, Bronstein JM, Horvath S, and Ritz B (2018) Organophosphate pesticide exposure and differential genome-wide DNA methylation. Sci. Total Environ 645, 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Jayaraj R, Megha P, and Sreedev P (2016) Organochlorine pesticides, their toxic effects on living organisms and their fate in the environment. Interdiscip. Toxicol 9 (3–4), 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dong K (2007) Insect sodium channels and insecticide resistance. Invertebr. Neurosci 7 (1), 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zuluaga M, Robledo S, Osorio-Zuluaga GA, Yathe L, Gonzalez D, and Taborda G (2016) Metabolomics and pesticides: systematic literature review using graph theory for analysis of references. Nova 14, 121–138. [Google Scholar]
- (8).Jellali R, Gilard F, Pandolfi V, Legendre A, Fleury MJ, Paullier P, Legallais C, and Leclerc E (2018) Metabolomics-on-a-chip approach to study hepatotoxicity of DDT, permethrin and their mixtures. J. Appl. Toxicol 38 (8), 1121–1134. [DOI] [PubMed] [Google Scholar]
- (9).Castellanos A, Andres A, Bernal L, Callejo G, Comes N, Gual A, Giblin JP, Roza C, and Gasull X (2018) Pyrethroids inhibit K2P channels and activate sensory neurons: basis of insecticide-induced paraesthesias. Pain 159 (1), 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Furlong MA, Paul KC, Yan Q, Chuang YH, Cockburn MG, Bronstein JM, Horvath S, and Ritz B (2020) An epigenome-wide association study of ambient pyrethroid pesticide exposures in California’s central valley. Int. J. Hyg. Environ. Health 229, 113569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bradberry SM, Cage SA, Proudfoot AT, and Vale JA (2005) Poisoning due to pyrethroids. Toxicol. Rev 24 (2), 93–106. [DOI] [PubMed] [Google Scholar]
- (12).Banerjee BD, Seth V, and Ahmed RS (2001) Pesticide-induced oxidative stress: perspectives and trends. Rev. Environ. Health 16 (1), 1–40. [DOI] [PubMed] [Google Scholar]
- (13).Costa LG (2006) Current issues in organophosphate toxicology. Clin. Chim. Acta 366 (1–2), 1–13. [DOI] [PubMed] [Google Scholar]
- (14).Karami-Mohajeri S, and Abdollahi M (2011) Toxic influence of organophosphate, carbamate, and organochlorine pesticides on cellular metabolism of lipids, proteins, and carbohydrates: a systematic review. Hum. Exp. Toxicol 30 (9), 1119–40. [DOI] [PubMed] [Google Scholar]
- (15).Patel S, Pandey AK, Bajpayee M, Parmar D, and Dhawan A (2006) Cypermethrin-induced DNA damage in organs and tissues of the mouse: evidence from the comet assay. Mutat. Res., Genet. Toxicol. Environ. Mutagen 607 (2), 176–183. [DOI] [PubMed] [Google Scholar]
- (16).Hossain MM, and Richardson JR (2011) Mechanism of pyrethroid pesticide-induced apoptosis: role of Calpain and the ER stress pathway. Toxicol. Sci 122 (2), 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Blair A, Ritz B, Wesseling C, and Freeman LB (2015) Pesticides and human health. Occup. Environ. Med 72 (2), 81–82. [DOI] [PubMed] [Google Scholar]
- (18).Ferreira JD, Couto AC, Pombo-de-Oliveira MS, and Koifman S (2013) In utero pesticide exposure and leukemia in Brazilian children < 2 years of age. Environ. Health Perspect 121 (2), 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Furlong MA, Barr DB, Wolff MS, and Engel SM (2017) Prenatal exposure to pyrethroid pesticides and childhood behavior and executive functioning. NeuroToxicology 62, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Weisskopf M, Knekt P, O’reilly E, Lyytinen J, Reunanen A, Laden F, Altshul L, and Ascherio A (2010) Persistent organochlorine pesticides in serum and risk of Parkinson disease. Neurology 74 (13), 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Garcia-Sevillano MA, Garcia-Barrera T, and Gomez-Ariza JL (2015) Environmental metabolomics: Biological markers for metal toxicity. Electrophoresis 36 (18), 2348–2365. [DOI] [PubMed] [Google Scholar]
- (22).Jones DP (2016) Sequencing the exposome: A call to action. Toxicol Rep 3, 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gu F, Derkach A, Freedman ND, Landi MT, Albanes D, Weinstein SJ, Mondul AM, Matthews CE, Guertin KA, Xiao Q, Zheng W, Shu XO, Sampson JN, Moore SC, and Caporaso NE (2016) Cigarette smoking behaviour and blood metabolomics. Int. J. Epidemiol 45 (5), 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yang J, Sun X, Feng Z, Hao D, Wang M, Zhao X, and Sun C (2011) Metabolomic analysis of the toxic effects of chronic exposure to low-level dichlorvos on rats using ultra-performance liquid chromatography-mass spectrometry. Toxicol. Lett 206 (3), 306–13. [DOI] [PubMed] [Google Scholar]
- (25).Gu J, Su F, Hong P, Zhang Q, and Zhao M (2019) (1)H NMR-based metabolomic analysis of nine organophosphate flame retardants metabolic disturbance in Hep G2 cell line. Sci. Total Environ 665, 162–170. [DOI] [PubMed] [Google Scholar]
- (26).Li F, Wang L, Ji C, Wu H, Zhao J, and Tang J (2017) Toxicological effects of tris (2-chloropropyl) phosphate in human hepatic cells. Chemosphere 187, 88–96. [DOI] [PubMed] [Google Scholar]
- (27).Xu MY, Wang P, Sun YJ, and Wu YJ (2019) Disruption of Kidney Metabolism in Rats after Subchronic Combined Exposure to Low-Dose Cadmium and Chlorpyrifos. Chem. Res. Toxicol 32 (1), 122–129. [DOI] [PubMed] [Google Scholar]
- (28).Yang J, Wang H, Xu W, Hao D, Du L, Zhao X, and Sun C (2013) Metabolomic analysis of rat plasma following chronic low-dose exposure to dichlorvos. Hum. Exp. Toxicol 32 (2), 196–205. [DOI] [PubMed] [Google Scholar]
- (29).Du L, Li S, Qi L, Hou Y, Zeng Y, Xu W, Wang H, Zhao X, and Sun C (2014) Metabonomic analysis of the joint toxic action of long-term low-level exposure to a mixture of four organophosphate pesticides in rat plasma. Mol. BioSyst 10 (5), 1153–61. [DOI] [PubMed] [Google Scholar]
- (30).Bonvallot N, Tremblay-Franco M, Chevrier C, Canlet C, Warembourg C, Cravedi JP, and Cordier S (2013) Metabolomics tools for describing complex pesticide exposure in pregnant women in Brittany (France). PLoS One 8 (5), No. e64433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Maitre L, Robinson O, Martinez D, Toledano MB, Ibarluzea J, Marina LS, Sunyer J, Villanueva CM, Keun HC, Vrijheid M, and Coen M (2018) Urine Metabolic Signatures of Multiple Environmental Pollutants in Pregnant Women: An Exposome Approach. Environ. Sci. Technol 52 (22), 13469–13480. [DOI] [PubMed] [Google Scholar]
- (32).Yang X, Zhang M, Lu T, Chen S, Sun X, Guan Y, Zhang Y, Zhang T, Sun R, Hang B, Wang X, Chen M, Chen Y, and Xia Y (2020) Metabolomics study and meta-analysis on the association between maternal pesticide exposome and birth outcomes. Environ. Res 182, 109087. [DOI] [PubMed] [Google Scholar]
- (33).Salihovic S, Ganna A, Fall T, Broeckling CD, Prenni JE, van Bavel B, Lind PM, Ingelsson E, and Lind L (2016) The metabolic fingerprint of p, p’-DDE and HCB exposure in humans. Environ. Int 88, 60–66. [DOI] [PubMed] [Google Scholar]
- (34).Valvi D, Walker DI, Inge T, Bartell SM, Jenkins T, Helmrath M, Ziegler TR, La Merrill MA, Eckel SP, Conti D, Liang Y, Jones DP, McConnell R, and Chatzi L (2020) Environmental chemical burden in metabolic tissues and systemic biological pathways in adolescent bariatric surgery patients: A pilot untargeted metabolomic approach. Environ. Int 143, 105957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Carrizo D, Chevallier OP, Woodside JV, Brennan SF, Cantwell MM, Cuskelly G, and Elliott CT (2017) Untargeted metabolomic analysis of human serum samples associated with exposure levels of Persistent organic pollutants indicate important perturbations in Sphingolipids and Glycerophospholipids levels. Chemosphere 168, 731–738. [DOI] [PubMed] [Google Scholar]
- (36).Walker DI, Marder ME, Yano Y, Terrell M, Liang Y, Barr DB, Miller GW, Jones DP, Marcus M, and Pennell KD (2019) Multigenerational metabolic profiling in the Michigan PBB registry. Environ. Res 172, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Li S, Cirillo P, Hu X, Tran V, Krigbaum N, Yu S, Jones DP, and Cohn B (2020) Understanding mixed environmental exposures using metabolomics via a hierarchical community network model in a cohort of California women in 1960’s. Reprod. Toxicol 92, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ritz BR, Paul KC, and Bronstein JM (2016) Of Pesticides and Men: a California Story of Genes and Environment in Parkinson’s Disease. Curr. Environ. Health Rep 3 (1), 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kang GA, Bronstein JM, Masterman DL, Redelings M, Crum JA, and Ritz B (2005) Clinical characteristics in early Parkinson’s disease in a central California population-based study. Mov. Disord 20 (9), 1133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Paul KC, Sinsheimer JS, Rhodes SL, Cockburn M, Bronstein J, and Ritz B (2016) Organophosphate Pesticide Exposures, Nitric Oxide Synthase Gene Variants, and Gene-Pesticide Interactions in a Case-Control Study of Parkinson’s Disease, California (USA). Environ. Health Perspect 124 (5), 570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Cockburn M, Mills P, Zhang X, Zadnick J, Goldberg D, and Ritz B (2011) Prostate cancer and ambient pesticide exposure in agriculturally intensive areas in California. Am. J. Epidemiol 173 (11), 1280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Liu KH, Nellis M, Uppal K, Ma C, Tran V, Liang Y, Walker DI, and Jones DP (2020) Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem 92 (13), 8836–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Johnson JM, Yu T, Strobel FH, and Jones DP (2010) A practical approach to detect unique metabolic patterns for personalized medicine. Analyst 135 (11), 2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Simon-Manso Y, Lowenthal MS, Kilpatrick LE, Sampson ML, Telu KH, Rudnick PA, Mallard WG, Bearden DW, Schock TB, Tchekhovskoi DV, Blonder N, Yan X, Liang Y, Zheng Y, Wallace WE, Neta P, Phinney KW, Remaley AT, and Stein SE (2013) Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal. Chem 85 (24), 11725–31. [DOI] [PubMed] [Google Scholar]
- (45).Walker DI, Lane KJ, Liu K, Uppal K, Patton AP, Durant JL, et al. (2019) Metabolomic assessment of exposure to near-highway ultrafine particles. J. Exposure Sci. Environ. Epidemiol 29 (4), 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Liu KH, Walker DI, Uppal K, Tran V, Rohrbeck P, Mallon TM, and Jones DP (2016) High-Resolution Metabolomics Assessment of Military Personnel: Evaluating Analytical Strategies for Chemical Detection. J. Occup. Environ. Med 58, S53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yu T, Park Y, Johnson JM, and Jones DP (2009) apLCMS–adaptive processing of high-resolution LC/MS data. Bioinformatics 25 (15), 1930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, and Jones DP (2013) xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinf. 14, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Deng K, Zhang F, Tan Q, Huang Y, Song W, Rong Z, Zhu Z-J, Li Z, and Li K (2019) WaveICA: A novel algorithm to remove batch effects for large-scale untargeted metabolomics data based on wavelet analysis. Anal. Chim. Acta 1061, 60. [DOI] [PubMed] [Google Scholar]
- (50).Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, and Altman RB (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17 (6), 520–5. [DOI] [PubMed] [Google Scholar]
- (51).Wold S, Sjostrom M, and Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemom. Intell. Lab. Syst 58 (2), 109–130. [Google Scholar]
- (52).Uppal K, Ma C, Go Y-M, and Jones DP (2018) xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 34 (4), 701–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Le Cao KA, Rossouw D, Robert-Granie C, and Besse P (2008) A sparse PLS for variable selection when integrating omics data. Stat. Appl. Genet. Mol. Biol 7 (1), 35 DOI: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- (54).Blondel VD, Guillaume J-L, Lambiotte R, and Lefebvre E (2008) Fast unfolding of communities in large networks. J. Stat. Mech.: Theory Exp 2008 (10), P10008. [Google Scholar]
- (55).Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, Strobel F, Quyyumi AA, Ziegler TR, Pennell KD, Miller GW, and Jones DP (2015) Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol. Sci 148 (2), 531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Uppal K, Walker DI, and Jones DP (2017) xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal. Chem 89 (2), 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, and McLean JA (2016) Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom 27 (12), 1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Bonvallot N, Tremblay-Franco M, Chevrier C, Canlet C, Warembourg C, Cravedi J-P, and Cordier S (2013) Metabolomics Tools for Describing Complex Pesticide Exposure in Pregnant Women in Brittany (France). PLoS One 8 (5), No. e64433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Uppal K, Walker DI, Liu K, Li S, Go YM, and Jones DP (2016) Computational Metabolomics: A Framework for the Million Metabolome. Chem. Res. Toxicol 29 (12), 1956–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Jenner P (2003) Oxidative stress in Parkinson’s disease. Ann. Neurol 53 (S3), S26–S38. [DOI] [PubMed] [Google Scholar]
- (61).Lin MT, and Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443 (7113), 787–795. [DOI] [PubMed] [Google Scholar]
- (62).Barnham KJ, Masters CL, and Bush AI (2004) Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discovery 3 (3), 205–214. [DOI] [PubMed] [Google Scholar]
- (63).McGeer PL, and McGeer EG (2004) Inflammation and neurodegeneration in Parkinson’s disease. Parkinsonism & related disorders 10, S3–S7. [DOI] [PubMed] [Google Scholar]
- (64).Parrón T, Requena M, Hernández AF, and Alarcón R (2011) Association between environmental exposure to pesticides and neurodegenerative diseases. Toxicol. Appl. Pharmacol 256 (3), 379–385. [DOI] [PubMed] [Google Scholar]
- (65).Walker DI, Pennell KD, Uppal K, Xia X, Hopke PK, Utell MJ, Phipps RP, Sime PJ, Rohrbeck P, Mallon CT, and Jones DP (2016) Pilot Metabolome-Wide Association Study of Benzo(a)pyrene in Serum From Military Personnel. J. Occup. Environ. Med 58, S44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Walker DI, Uppal K, Zhang L, Vermeulen R, Smith M, Hu W, Purdue MP, Tang X, Reiss B, Kim S, Li L, Huang H, Pennell KD, Jones DP, Rothman N, and Lan Q (2016) High-resolution metabolomics of occupational exposure to trichloroethylene. Int. J. Epidemiol 45 (5), 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Yan Q, Liew Z, Uppal K, Cui X, Ling C, Heck JE, von Ehrenstein OS, Wu J, Walker DI, Jones DP, and Ritz B (2019) Maternal serum metabolome and traffic-related air pollution exposure in pregnancy. Environ. Int 130, 104872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Foley JE (1992) Rationale and application of fatty acid oxidation inhibitors in treatment of diabetes mellitus. Diabetes Care 15 (6), 773–784. [DOI] [PubMed] [Google Scholar]
- (69).Fillmore N, Mori J, and Lopaschuk G (2014) Mitochondrial fatty acid oxidation alterations in heart failure, ischaemic heart disease and diabetic cardiomyopathy. British journal of pharmacology 171 (8), 2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Lane RK, Hilsabeck T, and Rea SL (2015) The role of mitochondrial dysfunction in age-related diseases. Biochim. Biophys. Acta, Bioenerg 1847 (11), 1387–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Gangemi S, Gofita E, Costa C, Teodoro M, Briguglio G, Nikitovic D, Tzanakakis G, Tsatsakis AM, Wilks MF, Spandidos DA, and Fenga C (2016) Occupational and environmental exposure to pesticides and cytokine pathways in chronic diseases (Review). Int. J. Mol. Med 38 (4), 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Reuter SE, and Evans AM (2012) Carnitine and acylcarnitines. Clin. Pharmacokinet 51 (9), 553–572. [DOI] [PubMed] [Google Scholar]
- (73).Peng K-Y, Watt MJ, Rensen S, Greve JW, Huynh K, Jayawardana KS, Meikle PJ, and Meex RC (2018) Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J. Lipid Res 59 (10), 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Jarrell ZR, Smith MR, Hu X, Orr M, Liu KH, Quyyumi AA, Jones DP, and Go Y-M (2020) Plasma acylcarnitine levels increase with healthy aging. Aging 12 (13), 13555–13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Nicolson GL (2007) Metabolic syndrome and mitochondrial function: molecular replacement and antioxidant supplements to prevent membrane peroxidation and restore mitochondrial function. J. Cell. Biochem 100 (6), 1352–1369. [DOI] [PubMed] [Google Scholar]
- (76).Swerdlow RH (2011) Brain aging, Alzheimer’s disease, and mitochondria. Biochim. Biophys. Acta, Mol. Basis Dis 1812 (12), 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Soane L, Kahraman S, Kristian T, and Fiskum G (2007) Mechanisms of impaired mitochondrial energy metabolism in acute and chronic neurodegenerative disorders. J. Neurosci. Res 85 (15), 3407–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Liang D, Moutinho JL, Golan R, Yu T, Ladva CN, Niedzwiecki M, Walker DI, Sarnat SE, Chang HH, Greenwald R, Jones DP, Russell AG, and Sarnat JA (2018) Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environ. Int 120, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Anthonymuthu TS, Kenny EM, Lamade AM, Kagan VE, and Bayir H (2018) Oxidized phospholipid signaling in traumatic brain injury. Free Radical Biol. Med 124, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Banks CN, and Lein PJ (2012) A review of experimental evidence linking neurotoxic organophosphorus compounds and inflammation. NeuroToxicology 33 (3), 575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Torres-Altoro MI, Mathur BN, Drerup JM, Thomas R, Lovinger DM, O’Callaghan JP, and Bibb JA (2011) Organophosphates dysregulate dopamine signaling, glutamatergic neurotransmission, and induce neuronal injury markers in striatum. J. Neurochem 119 (2), 303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Li M, Wang J, Lu Z, Wei D, Yang M, and Kong L (2014) NMR-based metabolomics approach to study the toxicity of lambda-cyhalothrin to goldfish (Carassius auratus). Aquat. Toxicol 146, 82–92. [DOI] [PubMed] [Google Scholar]
- (83).Meredith G, Totterdell S, Beales M, and Meshul C (2009) Impaired glutamate homeostasis and programmed cell death in a chronic MPTP mouse model of Parkinson’s disease. Exp. Neurol 219 (1), 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Sonsalla P, Albers D, and Zeevalk G (1998) Role of glutamate in neurodegeneration of dopamine neurons in several animal models of parkinsonism. Amino Acids 14 (1–3), 69–74. [DOI] [PubMed] [Google Scholar]
- (85).Tabrez S, Jabir NR, Shakil S, Greig NH, Alam Q, Abuzenadah AM, Damanhouri GA, and A Kamal M (2012) A synopsis on the role of tyrosine hydroxylase in Parkinson’s disease. CNS Neurol. Disord.: Drug Targets 11 (4), 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Priyadarshini M, Kamal MA, Greig NH, Realef M, Abuzenadah AM, Chaudhary AGA, and Damanhouri GA (2012) Alzheimer’s disease and type 2 diabetes: exploring the association to obesity and tyrosine hydroxylase. CNS Neurol. Disord.: Drug Targets 11 (4), 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Shetewy A, Shimada-Takaura K, Warner D, Jong CJ, Mehdi A-BA, Alexeyev M, Takahashi K, and Schaffer SW (2016) Mitochondrial defects associated with β-alanine toxicity: relevance to hyper-β-alaninemia. Mol. Cell. Biochem 416 (1–2), 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Tiedje KE, Stevens K, Barnes S, and Weaver DF (2010) Beta-alanine as a small molecule neurotransmitter. Neurochem. Int 57 (3), 177–88. [DOI] [PubMed] [Google Scholar]
- (89).Lukaszewicz-Hussain A, and Moniuszko-Jakoniuk J (2005) A Low Dose of Chlorfenvinphos Affects Hepatic Enzymes in Serum and Antioxidant Enzymes in Erythrocytes and Liver of Rats. Pol. J. Environ. Stud 14 (2), 199–202. [Google Scholar]
- (90).Ch R, Singh AK, Pathak MK, Singh A, Kesavachandran CN, Bihari V, and Mudiam MKR (2019) Saliva and urine metabolic profiling reveals altered amino acid and energy metabolism in male farmers exposed to pesticides in Madhya Pradesh State, India. Chemosphere 226, 636–644. [DOI] [PubMed] [Google Scholar]
- (91).Boldyrev AA, Dupin AM, Aya B, Babizhaev M, and Severin SE (1987) The antioxidative properties of carnosine, a natural histidine containing dipeptide. Biochem. Int 15 (6), 1105–1113. [PubMed] [Google Scholar]
- (92).Baraniuk JN, El-Amin S, Corey R, Rayhan R, and Timbol C (2013) Carnosine treatment for gulf war illness: a randomized controlled trial. Glob J. Health Sci 5 (3), 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Padayatty SJ, Katz A, Wang Y, Eck P, Kwon O, Lee JH, Chen S, Corpe C, Dutta A, Dutta SK, and Levine M (2003) Vitamin C as an antioxidant: evaluation of its role in disease prevention. J. Am. Coll. Nutr 22 (1), 18–35. [DOI] [PubMed] [Google Scholar]
- (94).Wang A, Costello S, Cockburn M, Zhang X, Bronstein J, and Ritz B (2011) Parkinson’s disease risk from ambient exposure to pesticides. Eur. J. Epidemiol 26 (7), 547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Shelton JF, Geraghty EM, Tancredi DJ, Delwiche LD, Schmidt RJ, Ritz B, Hansen RL, and Hertz-Picciotto I (2014) Neurodevelopmental disorders and prenatal residential proximity to agricultural pesticides: the CHARGE study. Environ. Health Perspect 122 (10), 1103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Rull RP, Ritz B, and Shaw GM (2006) Neural tube defects and maternal residential proximity to agricultural pesticide applications. Am. J. Epidemiol 163 (8), 743–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


