Abstract
Identification and quantification of low-frequency mutations remain challenging despite improvements in the baseline error rate of next-generation sequencing technologies. Here, we describe a method, termed SaferSeqS, that addresses these challenges by (1) efficiently introducing identical molecular barcodes in the Watson and Crick strands of template molecules and (2) enriching target sequences with strand-specific PCR. The method achieves high sensitivity and specificity and detects variants at frequencies below 1 in 100,000 DNA template molecules with a background mutation rate of <5 × 10−7 mutants per base pair (bp). We demonstrate that it can evaluate mutations in a single amplicon or simultaneously in multiple amplicons, assess limited quantities of cell-free DNA with high recovery of both strands and reduce the error rate of existing PCR-based molecular barcoding approaches by >100-fold.
Many next-generation sequencing approaches exist for the detection of rare mutations1. Such detection is critical to answer fundamental biological questions as well as to improve clinical management. Fields of use include infectious diseases2, immune repertoire profiling3, paleogenetics4, forensics5, aging6, non-invasive prenatal testing7 and cancer8. Next-generation sequencing approaches can, in principle, allow for the detection of rare mutations; but in practice, the error rate of sequencing itself is too high to allow for confident detection of mutations present at low frequencies in the original sample. One type of strategy to overcome this obstacle involves using bioinformatic analysis to calculate probabilities that an observed mutation is more likely to be due to its presence in the original sample rather than to be a technical artifact9–12. But this strategy alone is often insufficient to detect rare mutations with high confidence that is optimal for clinical use, inspiring the use of molecular barcodes to tag every original template molecule. With molecular barcoding, redundant sequencing of the PCR-generated progeny of each tagged molecule is performed, and sequencing errors are easily recognized13. For example, if all of the progeny of the barcoded template contain the same mutation, then the mutation is considered genuine (a ‘supermutant’), and if only a subset of the progeny contains the mutation of interest, then the mutation is considered an artifact.
Two types of molecular barcodes have been described: exogenous and endogenous13. Exogenous barcodes, consisting of prespecified or random nucleotides, are appended during library preparation or during PCR. Endogenous barcodes are formed by the sequences at the 5′ and 3′ ends of the template fragments. Endogenous barcodes allow for ‘duplex sequencing’, wherein each of the two strands (Watson and Crick) of the original DNA duplex can be discerned by the 5′ to 3′ directionality revealed upon sequencing6,13. Duplex sequencing reduces sequencing errors because it is extremely unlikely that both strands of DNA contain the identical mutation if that mutation was erroneously generated during library preparation or sequencing. A variety of molecular barcoding approaches based on either endogenous or exogenous barcodes, or the combination thereof, have been developed and applied to a wide range of clinical applications14–24.
A particularly clever duplex barcoding strategy was described by Schmitt and colleagues25. Their innovation was to append the identical exogenous barcode to the Watson and Crick strands of a template molecule. This allows for unambiguous determination of the identity of the two strands of a template without reference to the endogenous sequence ends. Because the method involves duplex sequencing, the error rate is minimal. Although this method has the lowest error rate of any sequencing technology described to date, two issues have limited its clinical applicability. First, it is challenging to convert a large fraction of the initial template molecules to adapter-ligated fragments with the same barcode on each strand9,25,26. This issue is particularly problematic when the amount of initial DNA is limiting, such as what is found in cell-free plasma DNA used for liquid biopsies. Second, hybridization-based capture is used to enrich for desired regions of the genome. While effective for enriching large regions of interest, hybridization capture does not scale well for small target regions27 and exhibits poor duplex recovery9,26. Sequential rounds of capture26 can partially overcome these limitations, but existing hybridization capture-based methods typically recover a minority of input molecules with sequence information from both strands26,28. When the targeted region is very small (for example, one or a few positions in the genome of particular interest) or the amount of DNA available is limited (for example <33 ng, as is often found in plasma), capture-based approaches are suboptimal. Here, we have addressed these issues by developing an approach to identically barcode both strands of templates in situ and a method for PCR-based enrichment of each strand that does not require hybridization capture.
Building upon our previously described methodology for rare variant detection13, the approach described herein, termed SaferSeqS, comprises the following three key steps: (1) library construction with in situ generation of double-stranded molecular barcodes (Fig. 1a), (2) target enrichment via anchored PCR29 (Fig. 1b) and (3) in silico reconstruction of template molecules (Fig. 1c). Bona fide mutations present in the original starting templates are identified by requiring alterations to be found on both strands of the same initial DNA molecule. This strategy should, in principle, minimize DNA damage, PCR and sequencing artifacts and permit the identification of rare mutations with high confidence.
Fig. 1 |. Overview of SaferSeqS.
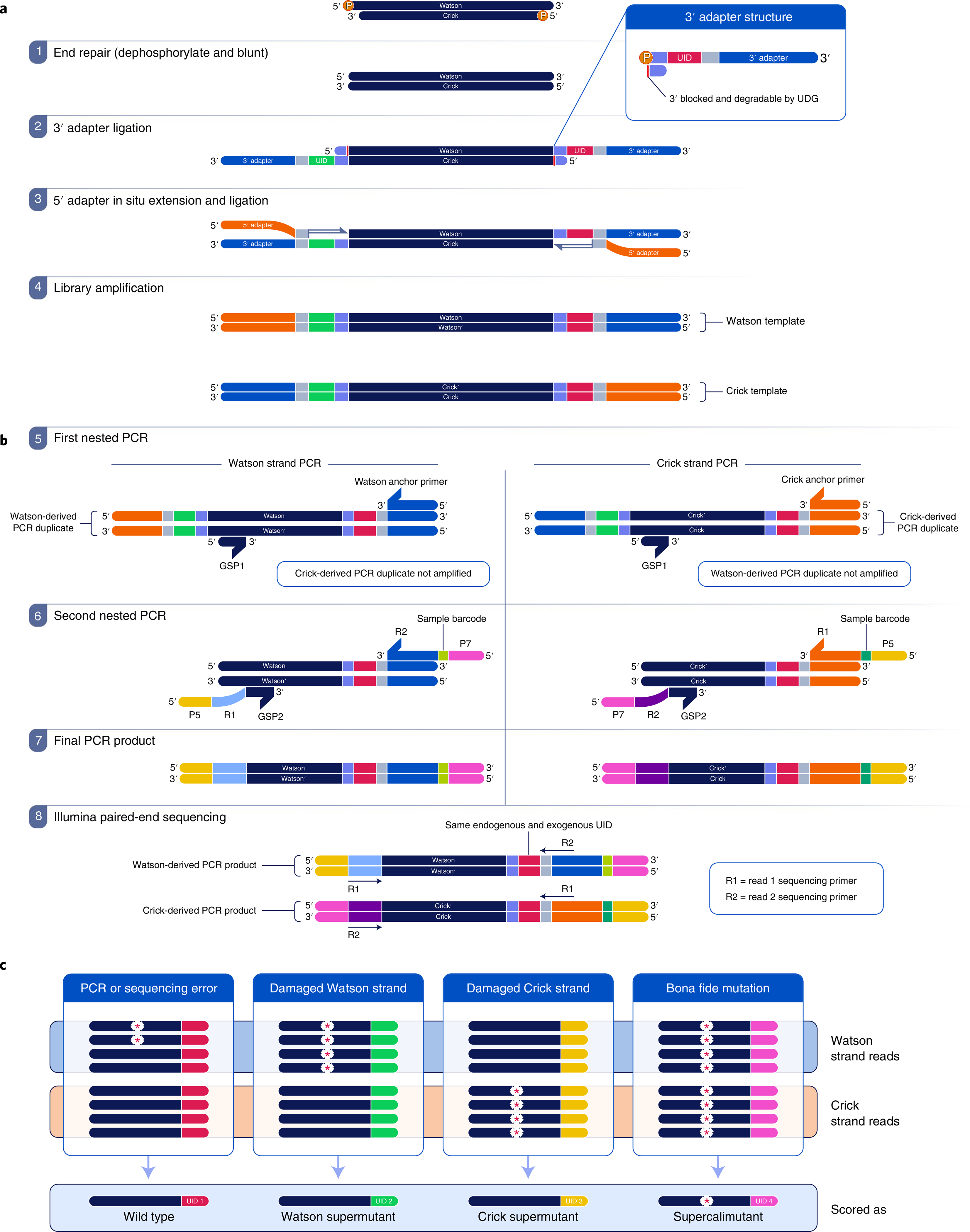
a, Library preparation begins with end repair (step 1), in which DNA template molecules are dephosphorylated and blunted. Next, a 3′ adapter (blue) containing a unique identifier (UID) sequence (red or green) is ligated to the 3′ fragment ends (step 2). The UID sequences are converted into double-stranded barcodes upon extension and ligation of the 5′ adapter (step 3). Finally, redundant PCR copies of each original template molecule are generated during library amplification (step 4). UDG, uracil-DNA glycosylase. b, Target enrichment is achieved with strand-specific hemi-nested PCRs. The amplified library is partitioned into Watson- and Crick-specific reactions (step 5), which selectively amplify products derived from one of the DNA strands. Additional on-target specificity and incorporation of sample barcodes are achieved with a second nested PCR (step 6). The final PCR products (step 7) are subjected to paired-end sequencing (step 8). The endogenous barcode represents the end of the template fragment before library construction. GSP, gene-specific primer; P5 and P7, Illumina P5 and P7 graft sequences. c, Following sequencing, reads are determined to be derived from the Watson or Crick strand. Because each strand of the original template is tagged with the same exogenous barcode and has the same endogenous barcode, reads derived from each of the two strands of the same parental DNA duplex can be grouped together into a duplex family. The red, green, yellow and pink colors at the right ends of the strands represent different barcodes. Bona fide mutations, represented by the asterisks within the pink family, are present in both parental strands of a DNA duplex and are therefore found in both Watson and Crick families. By contrast, PCR or sequencing errors, represented by asterisks within the red family, are limited to a subset of reads from one of the two strands. Watson strand-specific (asterisks within the green family) and Crick strand-specific (asterisks within the yellow family) mutations, such as those due to pre-existing, unrepaired, DNA damage, are found in all copies of the Watson or Crick family, but not in both.
To address inefficiencies and introduced errors typically associated with library construction, we designed a strategy that relies on the sequential ligation of adapter sequences to the 3′ and 5′ DNA fragment ends30 and the generation of double-stranded molecular barcodes in situ (Fig. 1a). The in situ generation of molecular barcodes is the key innovation of our library preparation method. The enzymes used for the in situ generation of double-stranded molecular barcodes uniquely barcoded each DNA fragment and obviated the need to enzymatically prepare duplex adapters, which has been noted to adversely affect input DNA recovery9 (Fig. 1a, steps 2 and 3). Following adapter ligation, the fragments were subjected to a limited number of PCR cycles to create redundant copies of the two original DNA strands (Fig. 1a, step 4).
Another innovation in our protocol is the use of a hemi-nested PCR-based approach for enrichment. Although hemi-nested PCR has previously been used for target enrichment29,31, major changes were required to apply it to duplex sequencing with high efficiency. Previous descriptions of hemi-nested PCR either do not retain the requisite strand information to reconstruct the original duplex molecule29 or do not recover a high enough fraction of template molecules to permit the detection of variants present at frequencies below 0.1% within limited quantities of DNA31. The hemi-nested approach described herein used two separate PCRs, one for the Watson strand and one for the Crick strand (Fig. 1b).
Following sequencing, reads corresponding to each strand of the original DNA duplex were grouped into Watson and Crick families. Each family member had the identical endogenous barcode representing the sequence at one end of the initial template fragment and the identical exogenous barcode introduced in situ during library construction. Mutations present in a Watson strand family were called ‘Watson supermutants’. Mutations present in a Crick strand family were called ‘Crick supermutants’. Those present in both the Watson and Crick families with the same molecular barcode (a ‘duplex family’) were called ‘supercalifragilisticexpialidocious mutants’, hereinafter referred to as ‘supercalimutants’ (Fig. 1c).
As an initial proof-of-principle demonstration of SaferSeqS, we conducted a mixing experiment in which DNA with a known mutation was spiked into DNA from a healthy individual’s leukocytes at ratios varying from approximately 8% to 0%. The fraction of on-target reads (that is, reads comprised of the intended amplicon) was 88%, considerably higher than achievable with a single round of hybrid capture27. Moreover, a strong correlation between the expected and observed allele frequencies was demonstrated across five orders of magnitude (Supplementary Fig. 1; Pearson’s r > 0.999, P = 9.89 × 10−12). Not a single mutant corresponding to the prespecified admixed variant was observed in DNA from the healthy individual, indicating very high specificity for the mutation of interest. Specificity was also determined for any base within the amplicon rather than just the queried base. Across a total of 37,747,670 bases queried among all DNA samples, only six supercalimutants were observed, representing a mutation frequency of 1.59 × 10−7 supercalimutants per bp (Supplementary Table 1).
We then sought to determine whether SaferSeqS could be applied to clinical samples in which the quantity of DNA was limiting. For example, as little as 33 ng of DNA is often present in 10 ml of cell-free plasma DNA samples used for liquid biopsies14–18,20,22–24. The vast majority of DNA template molecules in these samples are wild type, with as few as 1 or 2 mutant templates among the 10,000 wild-type templates present in samples from individuals with low tumor burdens14–17. To sensitively detect this exceedingly small number of mutant templates, the assay must efficiently recover the starting molecules.
To assess SaferSeqS in such a challenging context, we mixed cell-free plasma DNA from individuals with cancer with cell-free plasma DNA from healthy individuals to mimic mutation frequencies that are typically observed in clinical samples. In these experiments, 33 ng of each sample was assayed for one of three different mutations in TP53. The median number of duplex families (that is, both Watson and Crick strands containing the same endogenous and exogenous barcodes) was 89% (range, 65% to 102%) of the number of original template molecules (Supplementary Fig. 2a). The median fraction of on-target reads across the 27 experimental conditions (3 TP53 amplicons × 3 samples × 3 aliquots per sample) was 80% (range, 72% to 91%) (Supplementary Fig. 2b). Moreover, in all six admixed samples, the supercalimutant of interest was identified at the expected frequency (Fig. 2b,d,f and Supplementary Table 2). Mutations at this expected frequency were also identified in these same samples using a previously described molecular barcoding method13 (‘SafeSeqS’ rather than ‘SaferSeqS’) (Fig. 2a,c,e and Supplementary Table 2). The key advantage of SaferSeqS was its specificity. There were a total of 1,406 additional supermutants representing 153 distinct mutations observed with the previously described method, reflecting an average error rate of 9.39 × 10−6 supermutants per bp (Fig. 2a,c,e and Supplementary Table 2). The vast majority of these mutations were presumably polymerase errors that arose during early barcoding cycles in only one of the two strands. Similarly, if only Watson supermutants or Crick supermutants (that is, those observed in one but not both of the two strands; Fig. 1c) were considered in the SaferSeqS-generated libraries, an error rate of 6.03 × 10−6 supermutants per bp was observed (Supplementary Fig. 3a,c,e and Supplementary Table 3). By contrast, when only supercalimutants were considered (that is, those observed in both strands of the same starting template), zero additional mutations were found among 3,573,481 bases queried, reflecting an error rate of <2.80 × 10−7 supercalimutants per bp (Fig. 2b,d,f, Supplementary Fig. 3b,d,f and Supplementary Tables 2 and 3). These differences in specificity between SaferSeqS and previously described molecular barcoding methods (that is, those using direct PCR or adapter ligation to incorporate strand-agnostic molecular barcodes before sequencing) were highly significant (P < 7.0 × 10−6, two-sided Z-test for proportions comparing SaferSeqS with each of the other methods).
Fig. 2 |. Detection of mutations in liquid biopsy samples.
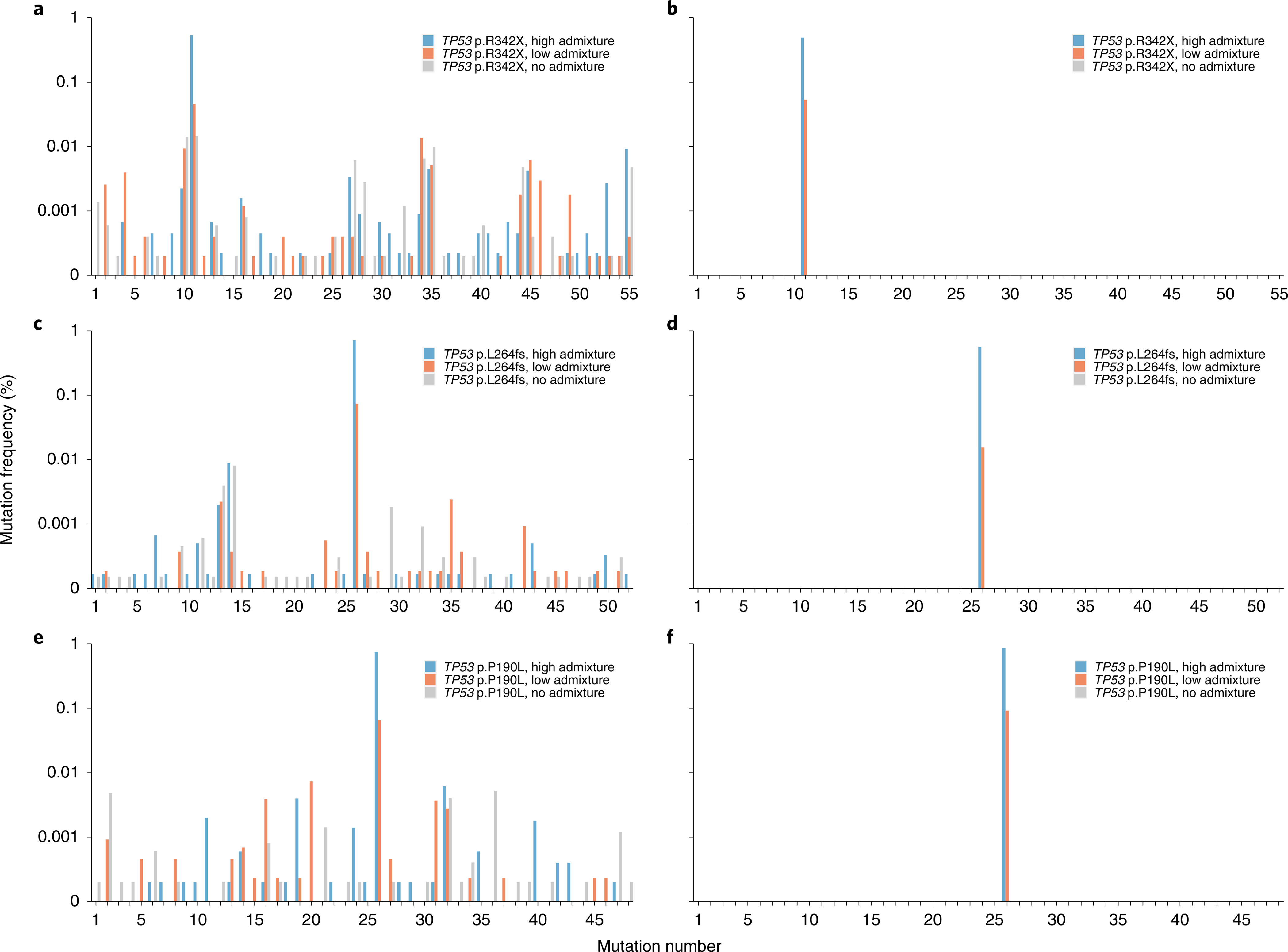
Analysis of 33 ng of plasma cell-free DNA from healthy individuals admixed with cell-free plasma DNA from an individual with cancer. Mixtures were created to generate a high frequency (~0.5–1%) of mutation (blue bars), low frequency (~0.01–0.1%) of mutation (orange bars) or no mutation (gray bars). The admixed TP53 p.R342X sample was assayed with SafeSeqs (a) and SaferSeqS (b). Similarly, the admixed TP53 p.L264fs sample was assayed with SafeSeqs (c) and SaferSeqS (d), and the admixed TP53 p.P190L sample was assayed with SafeSeqs (e) and SaferSeqS (f). Mutation numbers represent each of the 153 distinct mutations observed with SafeSeqS defined in Supplementary Table 2.
As a further demonstration of the clinical applicability of SaferSeqS, we evaluated five individuals with cancer with minimal tumor burden. In each case, mutations in the primary tissues (rather than the plasma) were identified, as described previously20. We then divided the plasma from these individuals into two equal aliquots and evaluated one aliquot with a previously described molecular barcoding method13 and the other with SaferSeqS. In both cases, primers targeting the mutations of interest were designed. Evaluation with the previously described barcoding method revealed that the plasma samples harbored in aggregate eight mutations that were originally identified. The frequencies of these mutations in the plasma varied from 0.01% to 0.1% (Supplementary Fig. 4 and Supplementary Table 4). In addition to the 8 known mutations, the previously described method identified 334 distinct mutations present at frequencies up to 0.013%, none of which were found in the primary tumors of these individuals. These 334 mutations comprised 10,347 supermutants, reflecting an average error rate of 1.23 × 10−5 supermutants per bp (Supplementary Fig. 4 and Supplementary Table 4). With SaferSeqS, the eight expected mutations were detected in all five individuals at frequencies similar to those found with the previously described method (Supplementary Fig. 4 and Supplementary Table 4). However, among the 6,138,524 queried bases, no additional supercalimutants (rather than 334 mutations) were identified with SaferSeqS, representing an average error rate of <1.63 × 10−7 supercalimutants per bp (Supplementary Table 4). This >100-fold improvement in specificity over the previously described molecular barcoding method was highly significant (P < 2.2 × 10−16, two-sided Z-test for proportions).
To evaluate the multiplexing capabilities of SaferSeqS, we designed 48 primers to query regions of driver genes that are commonly mutated in cancer32 (Supplementary Table 5). These primers were combined in two reactions, one targeting 25 regions and the other targeting 23 regions. Each of the 48 primer pairs specifically amplified their intended targets (Supplementary Fig. 5); the median on-target rate for Watson-derived reads was 95% (range, 34% to 97%), and the median on-target rate for Crick-derived reads was 94% (range, 34% to 96%). The targets demonstrated relatively uniform recovery of the input molecules, with a coefficient of variation of only 14% (Fig. 3a). The duplex recoveries for each amplicon were invariant beyond a threshold level of sequencing (Fig. 3b), suggesting that they were not artificially inflated due to polymerase or sequencing errors in the exogenous barcode sequences. Finally, the lengths of the amplicons sequenced (median, 77 bp; interquartile range, 71–83 bp) were similar in all amplicons and consistent with expectations given that the initial size of cell-free plasma DNA is ~167 bp ± 10.4 bp (ref. 33) (Fig. 3a and Supplementary Note).
Fig. 3 |. Multiplex panel for the detection of cancer driver gene mutations.
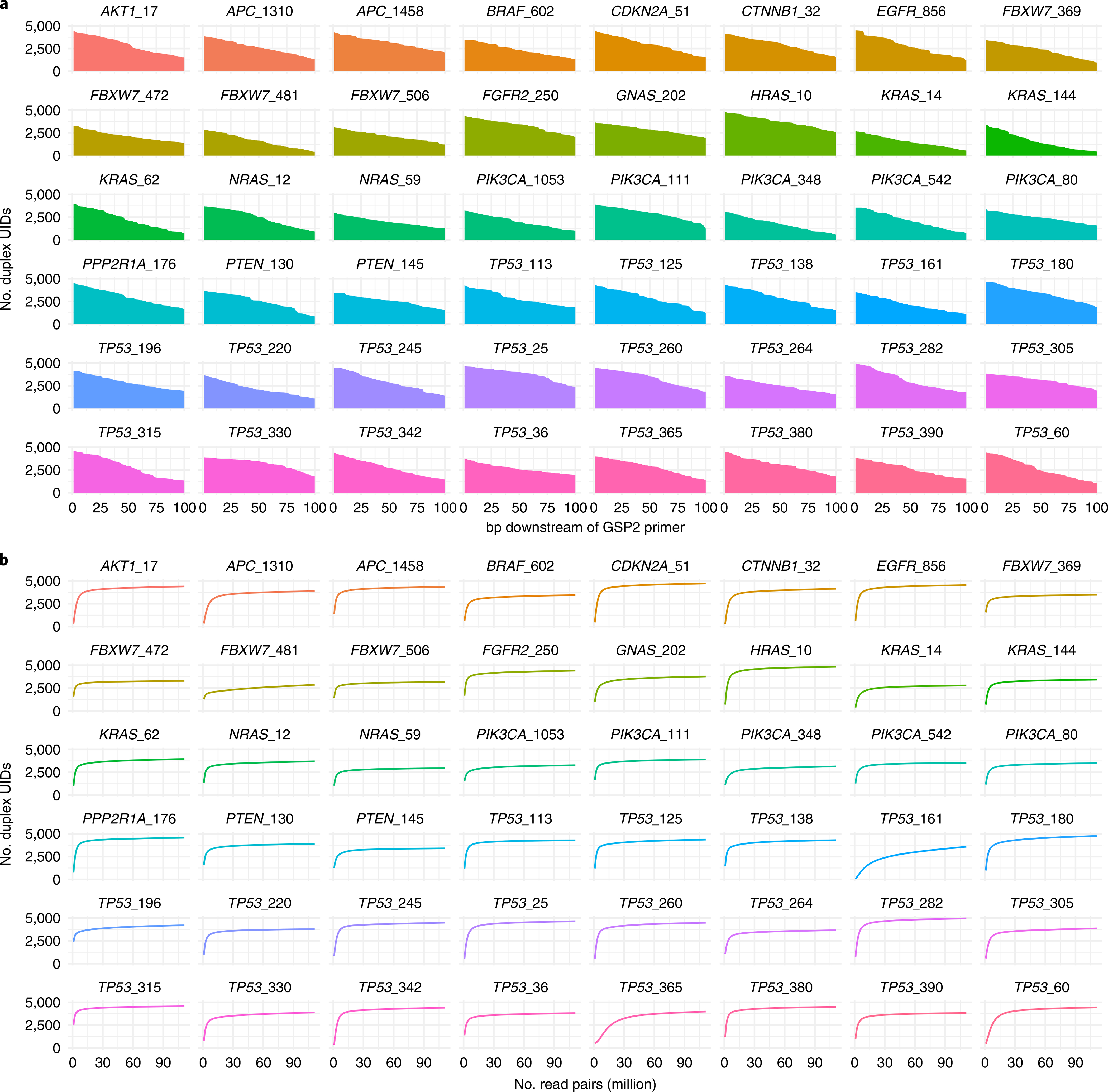
a, Recovery and coverage of the 48 amplicons within the multiplex panel. The horizontal axis displays the position downstream of the 3′ end of the second gene-specific primer (GSP2). The gradual decline in coverage with increasing distance from the 3′ primer end is a consequence of the input DNA fragmentation pattern. Details regarding the theoretical recovery of reads with specific amplicon lengths are discussed in the Supplementary Note. b, Duplex recoveries for each of the 48 amplicons with varying levels of sequencing depth. Recoveries were invariant beyond a threshold level of sequencing, suggesting that they are not artificially inflated due to polymerase or sequencing errors in the UID sequences.
To demonstrate the performance of SaferSeqS with this multiplex panel in a clinically relevant context, we evaluated 86 primary tumor-derived mutations from a cohort of 74 plasma samples obtained from individuals with cancer. Aliquots of these same plasma samples were previously evaluated with a multiplex PCR-based, single-stranded molecular barcoding method16, but the circulating tumor DNA levels observed in these individuals failed to reach the requisite degree of statistical significance at high specificity (Methods). All 74 individuals had surgically resectable disease at the time of diagnosis (stage 1 to 3) and a median of 5.5 ml (range, 2 to 10 ml) of residual plasma available for reanalysis with the SaferSeqS multiplex panel described above (Supplementary Table 6).
When assayed with multiplex SaferSeqS assays, a total of 43 (58%) of these individuals harbored a detectable tumor-derived mutation in their plasma. In accordance with the relatively minimal disease burden present in these individuals, both the proportion of samples detectable (Fig. 4a) and the absolute number of supercalimutants (Fig. 4b) increased with plasma volume. The efficiency of library construction and target enrichment in these samples was high, with a median of 69% of the original starting templates recovered with sequence information from both strands (Fig. 4c). Furthermore, to orthogonally validate the sensitivity of the multiplex panel, we assayed the SaferSeqS libraries generated from these plasma samples for the participant-specific mutations of interest in separate, individual PCR reactions. Because SaferSeqS libraries can be partitioned into multiple PCR reactions without adversely impacting sample recovery, this analysis afforded a direct comparison of the performance of these two approaches. Both the supercalimutant allele frequencies (Fig. 4d) and the absolute number of supercalimutants (Supplementary Table 6) were highly concordant between the multiplex and single amplicon assays (Pearson’s r = 0.98, P < 2.2 × 10−16 for supercalimutant allele frequencies; Pearson’s r = 0.92, P < 2.2 × 10−16 for supercalimutant counts). In comparison to the multiplex assays, an identical number of individuals (43 of 74, 58%) harbored detectable tumor-derived mutations when assessed with the single amplicon assays. Specifically, of the 43 individuals detectable with the multiplex assay, 42 were detectable with the single amplicon assay; of the 43 individuals detectable with the single amplicon assays, 42 were detectable with the multiplex assay (Supplementary Fig. 6). In accordance with stochastic sampling effects, the two discordant plasma samples each harbored one supercalimutant when evaluated with either assay (Supplementary Table 6).
Fig. 4 |. Clinical application of the SaferSeqS multiplex panel.
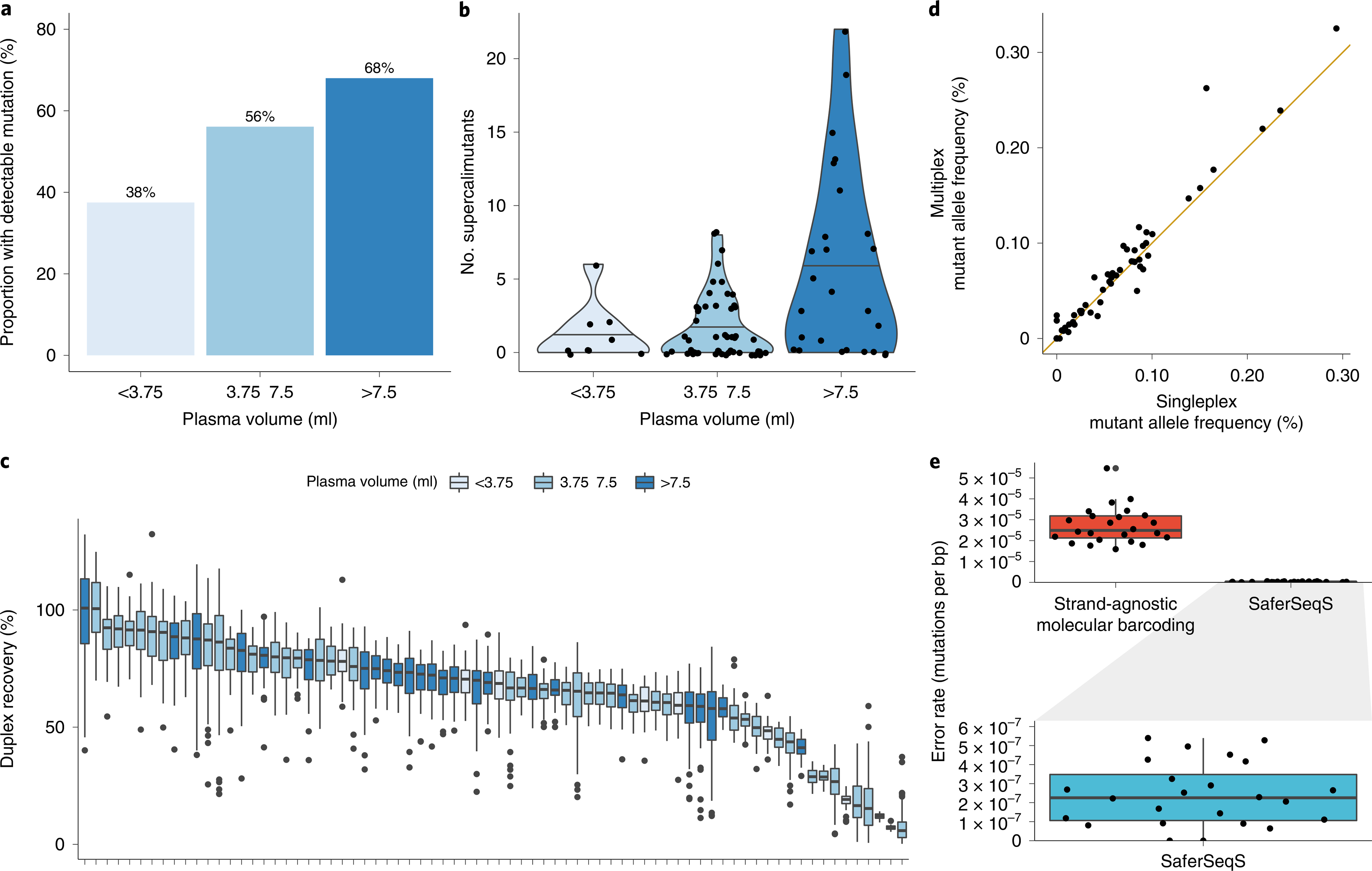
a, Proportion of plasma samples with a detectable primary tumor-concordant mutation as a function of plasma volume assayed (<3.75 ml, 3.75 to 7.5 ml and >7.5 ml). b, Absolute number of supercalimutants (that is, high-confidence mutant DNA molecules) detected stratified by plasma volume (<3.75 ml, 3.75 to 7.5 ml and >7.5 ml). c, Duplex recoveries for the multiplex panel in each of the 74 plasma samples obtained from individuals with cancer. Each plasma sample is represented by an individual box plot showing the distribution of duplex recoveries across the 48 amplicons assayed within the multiplex panel. Lower and upper edges correspond to the 25th and 75th percentile, whiskers extend to 1.5× the interquartile range and values outside of this range are plotted as individual points. d, Reproducibility between supercalimutant allele frequencies measured by singleplex and multiplex SaferSeqS assays. A solid 45° line passing through the origin is plotted for reference. e, Evaluation of the specificity of the multiplex panel in a cohort of 24 plasma samples obtained from healthy donors. Background error rates of the multiplex panel when evaluated by strand-agnostic molecular barcoding (that is, scoring mutations observed in only one of the two strands) in comparison to SaferSeqS are shown. Expanded view of the background error rate of SaferSeqS is shown in the box plot below. Note the change in scale by two orders of magnitude. Lower and upper edges correspond to the 25th and 75th percentile, and whiskers extend to 1.5× the interquartile range. Individual data points are overlaid with random scatter.
As a final demonstration of the specificity of SaferSeqS, we evaluated a cohort of 24 plasma samples obtained from healthy donors with the 48-amplicon multiplex panel described above (Fig. 3). Consistent with error rates of strand-agnostic molecular barcoding methods (that is, those that score mutations present in one of the two strands), the median mutation frequency was 2.49 × 10−5 supermutants per bp (range, 1.59 × 10−5 to 5.46 × 10−5). By contrast, the median mutation frequency was reduced to 2.25 × 10−7 supercalimutants per bp (range, 0.00 × 10−7 to 5.40 × 10−7) with SaferSeqS (Fig. 4e). This greater than two orders-of-magnitude improvement in specificity was highly significant (P < 2.2 × 10−16, two-sided Z-test for proportions).
The results presented above demonstrate that SaferSeqS can detect rare mutations with extremely high specificity. The technique is highly scalable, cost effective and amenable to high-throughput automation. SaferSeqS achieved up to a 5- to 75-fold improvement in input recovery over existing duplex sequencing techniques9,25,26,28,31,34,35, can be applied to limited amounts of starting material and resulted in a >100-fold improvement in error correction over standard PCR and ligation-based approaches using strand-agnostic molecular barcodes13 (Fig. 2, Supplementary Figs. 3 and 4 and Supplementary Tables 2–4).
Limitations of SaferSeqS should be noted. PCR-based approaches have an advantage over those using hybridization-based capture in that relatively small regions of the genome (for example, a single amplicon) can be assessed. One limitation of PCR-based approaches such as SaferSeqS, however, is that primers must be designed for each region of interest, and this poses challenges for regions that are duplicated within the genome or are difficult to amplify. To date, we have attempted to design primers to 182 regions, and our first attempt at primer design was successful in 163 (90%) of these regions (Supplementary Fig. 7). We expect that further refinements to the primer sequences and concentrations will result in even greater uniformity and on-target performance. A second limitation is that the anchored, hemi-nested PCR approach described herein is particularly effective for the analysis of relatively small genomic regions. For many clinical and research applications, only a small number of base pairs are of interest (for example, one to several thousand), and the technology described here is able to accomplish this feat while essentially eliminating PCR errors. For other applications requiring the assessment of larger regions, such as those encompassing >10,000 bp, hybrid capture may be the preferred alternative27. However, the duplex barcoding and library preparation components of SaferSeqS could be used for hybrid capture and should allow for the preservation of a considerably higher fraction of initial template molecules, thereby enhancing the efficiency of hybrid capture-based approaches for duplex sequencing, such as those described previously9,26,34.
By permitting efficient detection and quantification of rare genetic alterations, we envision that SaferSeqS will enable the development of highly sensitive and specific DNA-based molecular diagnostics as well as help to answer a variety of basic scientific questions.
Methods
Plasma and peripheral blood DNA samples.
This study was approved by the Institutional Review Boards for Human Research at participating institutions in compliance with the Health Insurance Portability and Accountability Act. Informed consent was obtained from all individuals. DNA was purified from plasma using a cfPure MAX Cell-Free DNA Extraction Kit (BioChain, K5011625MA), as specified by the manufacturer’s instructions. DNA from peripheral white blood cells (WBCs) was purified with the QIAsymphony DSP DNA Midi Kit (Qiagen, 937255) according to manufacturer’s instructions. Purified DNA from all samples was quantified with qPCR36 using the following primers: 5′-CACACAGGAAACAGCTATGACCATGGGTAACAGCTTTATCTATTGACATTATGC-3′ and 5′-CGACGTAAAACGACGGCCAGTNNNNNNNNNNNNNNAAACTTCATGCTTCATCTAGTCAGC-3′. For the evaluation of previously assayed plasma samples from individuals with cancer16, samples were selected if they (1) had residual volume available for purification and library construction and (2) were found to contain some evidence of a missed primary tumor-derived mutation (that is, mutant allele frequency >0) by a multiplex PCR-based assay, yet (3) were previously deemed undetectable at high specificity (defined as having an Ω score <1.6). This statistical significance threshold was chosen because it previously yielded a specificity of >99% in a large cohort of healthy donors37.
Library preparation.
We developed a custom library preparation workflow that can efficiently recover input DNA fragments and simultaneously incorporate double-stranded molecular barcodes. Conceptual and practical details of this strategy are discussed in the Supplementary Note. In brief, duplex sequencing libraries were prepared with cell-free DNA or peripheral WBC DNA using an Accel-NGS 2S DNA Library Kit (Swift Biosciences, 21024) with the following critical modifications: (1) DNA was pretreated with 3 U of USER enzyme (New England BioLabs, M5505L) for 15 min at 37 °C to excise uracil bases; (2) the SPRI bead/PEG NaCl ratios used after each reaction were 2.0×, 1.8×, 1.2× and 1.05× for end repair 1, end repair 2, ligation 1 and ligation 2, respectively; (3) a custom 50 μM 3′ adapter (Supplementary Table 7) was substituted for reagent Y2 and (4) a custom 42 μM 5′ adapter (Supplementary Table 7) was substituted for reagent B2. Libraries were subsequently PCR amplified in 50-μl reactions using primers targeting the ligated adapters (Supplementary Table 7). The following reaction conditions were used: 1× NEBNext Ultra II Q5 Master Mix (New England BioLabs, M0544L), 2 μM universal forward primer and 2 μM universal reverse primer (Supplementary Table 7). Libraries were amplified with 5, 7 or 11 cycles of PCR, depending on how many experiments were planned, according to the following protocol: 98 °C for 30 s, cycles of 98 °C for 10 s, 65 °C for 75 s and hold at 4 °C. If five or seven cycles were used, the libraries were amplified in single 50-μl reactions. If 11 cycles were used, the libraries were divided into eight aliquots and amplified in eight 50-μl reactions, each supplemented with an additional 0.5 U of Q5 Hot Start High-Fidelity DNA Polymerase (New England BioLabs, M0493L), 1 μl of 10 mM dNTPs (New England BioLabs, N0447L) and 0.4 μl of 25 mM MgCl2 solution (New England BioLabs, B9021S). The products were purified with 1.8× SPRI beads (Beckman Coulter, B23317) and eluted in EB buffer (Qiagen).
Anchored hemi-nested PCR.
Target enrichment of the regions of interest was achieved using critical modifications of anchored hemi-nested PCR29 that were necessary for duplex sequencing. Two separate PCRs were designed to selectively enrich the Watson or Crick strand. Both PCRs used the same gene-specific primer, but each used a different anchoring primer. PCR duplicates derived from each strand could be distinguished by the orientation of the insert relative to the exogenous UID. During the development of this custom stand-specific assay, we optimized various reaction conditions, including the number of cycles, the primer concentrations and the polymerase formulation. Our final optimized protocol included a first round of PCR performed in a 50-μl reaction with 1× NEBNext Ultra II Q5 Master Mix (New England BioLabs, M0544L), 2 μM GSP1 primer and 2 μM P7 short anchor primer for amplification of the Watson strand. The GSP1 primer was specific for each amplicon, and the P7 short anchor primer was used as the anchor primer for the Watson strand of all amplicons (Supplementary Tables 5 and 7). The Crick strand was amplified the same way in a separate well, with the exception that the P5 short anchor primer was substituted for the P7 short anchor primer. Note that the GSP1 primer used for amplification of the Watson strand was identical to the GSP1 primer used for the Crick strand; the only difference between the Watson and Crick strand PCRs was the anchor primer. Both reactions (Watson and Crick strands) were amplified with 19 cycles according to the thermocycling protocol described above.
For the Watson strand, a second round of PCR was performed in 50-μl reactions using the identical reaction conditions used for the first round of PCR. The differences were (1) the template, where 1% of product from the first anchored Watson strand PCR was used as the template instead of the library used as template for the first PCR, and (2) the primers, where the GSP2 gene-specific primers were substituted for the GSP1 gene-specific primers and the anchor P5 indexing primer was substituted for the P7 short anchor primer. The second round of PCR for the Crick strand was performed identically except for (1) the template, where the first Crick strand PCR was used as the template, and (2) the primers, where the anchor P7 indexing primer was substituted for the anchor P5 indexing primer. Both reactions (Watson and Crick strands) were amplified with 17 cycles according to the thermocycling protocol described previously. Sequences of the primers used for the second round of PCR are listed in Supplementary Table 7. The products of the second round of PCR were pooled and purified with 1.8× SPRI beads before sequencing.
For experiments in which multiple targets were simultaneously amplified within a single reaction, the PCR conditions were identical to those described above except (1) each gene-specific primer was included at the final concentration listed in Supplementary Table 5, and (2) the anchor primer was included at a final concentration equal to the total concentration of the gene-specific primer set (for example, at a final concentration of 17.6 μM in the Watson strand PCR if 25 targets were coamplified).
Sequencing.
Library concentrations were determined using a KAPA Library Quantification Kit (KAPA Biosystems, KK4824) according to the manufacturer’s instructions. Paired-end sequencing was performed with eight-base dual indexing on an Illumina MiSeq, HiSeq 2500 or HiSeq 4000 instrument. A dual-indexed PhiX control library (SeqMatic, TM-502-ND) was spiked in at 25% of the total templates to ensure base diversity across all cycles. Custom read 1, index and read 2 sequencing primers (Supplementary Table 7) were combined with standard Illumina sequencing primers at a final concentration of 1 μM.
Mutation calling and SaferSeqS analysis pipeline.
Analysis of SafeSeqS data was performed as previously described13 using custom Python scripts. Sequencing reads underwent initial processing by extracting the first 14 nucleotides as the exogenous barcode (that is, UID) sequence and masking adapter sequences using Picard’s IlluminaBasecallsToSam (http://broadinstitute.github.io/picard). Reads were then mapped to the hg19 reference genome using BWA-MEM (version 0.7.17)38 and sorted by UID sequence using SAMtools39. We arbitrarily defined the hg19 reference sequence as the Watson strand and its reverse complement as the Crick strand. UID families were scored if they consisted of two or more reads and if >90% of the reads mapped to the reference genome with the expected primer sequences. Supermutants were identified as mutations that were present in >95% of the mapped reads and had an average Phred score greater than 20.
A custom analysis pipeline was developed for the analysis of SaferSeqS. Details are discussed in the Supplementary Note. In brief, reads were demultiplexed, and the strand from which the reads were derived was identified using the index sequences. For clarity and succinctness, reads derived from the Watson strand are referred to as ‘Watson reads’, and reads derived from the Crick strand are referred to as ‘Crick reads.’ For the Watson reads, the first 14 bases of read 1 were extracted as the UID sequence. Because the orientation of the insert is reversed for the Crick strand, the first 14 bases of read 2 were extracted as the UID sequence for the Crick reads. Adapter sequences were masked using Picard’s IlluminaBasecallsToSam (http://broadinstitute.github.io/picard), and the resulting template-specific portions of the reads were mapped to the hg19 reference genome using BWA-MEM (version 0.7.17)38. Following alignment, the mapped Watson and Crick reads were merged and sorted using SAMtools39.
Python scripts were used for subsequent reconstruction of the duplex families and identification of Watson supermutants, Crick supermutants and supercalimutants. After correcting for PCR and sequencing errors within the molecular barcode sequences40, Watson and Crick reads belonging to the same duplex family were grouped together to reconstruct the sequence of the original template molecule. To exclude artifacts stemming from the end repair step of library construction35, bases fewer than ten bases from the 3′ adapter sequence were not considered for mutation analysis of sheared leukocyte DNA. In the case of cell-free DNA, which can contain single-stranded overhangs resulting from in vivo fragmentation41, positions fewer than 30 bases from the 3′ adapter sequence were excluded. These excluded positions were not counted in the total number of queried bases for the calculation of mutation background rates. Watson and Crick supermutants were defined as mutations present in >80% of the Watson or Crick reads of a duplex family, respectively. Supercalimutants were defined as mutations present in >80% of both the Watson and Crick families with the same UID. Variation in this parameter did not significantly alter estimates of background error rates (Supplementary Fig. 8).
Statistical analyses.
Continuous variables were reported as medians and range, while categorical variables were reported as whole numbers and percentages. All statistical tests were conducted using R’s stats package (version 4.0.3).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank the individuals who participated in this study for their courage and generosity. We also thank M. Hoang, S. Sur, A. Mattox, A. Pearlman and members of the Ludwig Center at Johns Hopkins for insightful and helpful scientific discussions. We are grateful to C. Blair and K. Judge for expert technical and administrative assistance and to E. Cook for illustrative assistance. This work was supported by The Lustgarten Foundation for Pancreatic Cancer Research, The Marcus Foundation, The Virginia and D.K. Ludwig Fund for Cancer Research, The Conrad N. Hilton Foundation, The John Templeton Foundation, Medical Research Future Fund Investigator Grant (APP1194970) and National Institutes of Health grants (T32 GM007309, U01 CA230691-01, P50 CA228991, U01 CA200469, R37 CA230400-01, and U01 CA152753).
Footnotes
Competing interests
B.V., K.W.K. and N.P. are founders of Thrive and Personal Genome Diagnostics and own equity in Exact Sciences and Personal Genome Diagnostics. K.W.K. and N.P. are consultants to Thrive. K.W.K. and B.V. are consultants to Sysmex and Eisai, and K.W.K., B.V. and N.P. are advisors to CAGE Pharma. B.V. is also a consultant to Catalio, and K.W.K., B.V. and N.P. are consultants to Neophore. C.D. is a consultant to Thrive and is compensated with income and equity. The companies named above, as well as other companies, have licensed previously described technologies related to the work described in this paper from Johns Hopkins University. J.D.C., C.D., B.V., K.W.K., C.T. and N.P. are inventors on some of these technologies. Licenses to these technologies are or will be associated with equity or royalty payments to the inventors as well as to Johns Hopkins University. Additional patent applications on the work described in this paper are being filed by Johns Hopkins University. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. The remaining authors declare no competing interests.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41587-021-00900-z.
Code availability
The SaferSeqS bioinformatics pipeline is implemented in Python. The source code is available in a Zenodo repository (https://doi.org/10.5281/zenodo.4588264).
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-021-00900-z.
Peer review information Nature Biotechnology thanks Paul Spellman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The sequencing data generated in this study can be obtained from the European Genome–phenome Archive (accession number EGAS00001005048).
References
- 1.Shendure J et al. DNA sequencing at 40: past, present and future. Nature 550, 345–353 (2017). [DOI] [PubMed] [Google Scholar]
- 2.McMahon MA et al. The HBV drug entecavir—effects on HIV-1 replication and resistance. N. Engl. J. Med. 356, 2614–2621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robins HS et al. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood 114, 4099–4107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller W et al. Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456, 387–390 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Bruijns B, Tiggelaar R & Gardeniers H Massively parallel sequencing techniques for forensics: a review. Electrophoresis 39, 2642–2654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoang ML et al. Genome-wide quantification of rare somatic mutations in normal human tissues using massively parallel sequencing. Proc. Natl Acad. Sci. USA 113, 9846–9851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu RW et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl Acad. Sci. USA 105, 20458–20463 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattox AK et al. Applications of liquid biopsies for cancer. Sci. Transl. Med 11, eaay1984 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Newman AM et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou Y et al. Accurate detection of mosaic variants in sequencing data without matched controls. Nat. Biotechnol. 38, 314–319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Razavi P et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meynert AM, Bicknell LS, Hurles ME, Jackson AP & Taylor MS Quantifying single nucleotide variant detection sensitivity in exome sequencing. BMC Bioinformatics 14, 195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinde I, Wu J, Papadopoulos N, Kinzler KW & Vogelstein B Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettegowda C et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6, 224ra224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen JD et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl Acad. Sci. USA 114, 10202–10207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen JD et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phallen J et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springer S et al. A multimodality test to guide the management of patients with a pancreatic cyst. Sci. Transl. Med 11, eaav4772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Springer S et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 149, 1501–1510 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tie J et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra392 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 7, 293ra104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl Acad. Sci. USA 112, 9704–9709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y et al. Diagnostic potential of tumor DNA from ovarian cyst fluid. eLife 5, e15175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Springer SU et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. eLife 7, e32143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt MW et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt MW et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat. Methods 12, 423–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samorodnitsky E et al. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum. Mutat. 36, 903–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabon JJ et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Z et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Makarov V & Laliberte J Enhanced adapter ligation. US patent 10,208,338B2 (2019).
- 31.Peng Q et al. Targeted single primer enrichment sequencing with single end duplex-UMI. Sci. Rep. 9, 4810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder MW, Kircher M, Hill AJ, Daza RM & Shendure J Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachmanson D et al. Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR–DS). Genome Res. 28, 1589–1599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy SR et al. Detecting ultralow-frequency mutations by duplex sequencing. Nat. Protoc. 9, 2586–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rago C et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 67, 9364–9370 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Lennon AM et al. Feasibility of blood testing combined with PET–CT to screen for cancer and guide intervention. Science 369, eabb9601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
- 39.Li H et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith T, Heger A & Sudbery I UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang P et al. Detection and characterization of jagged ends of double-stranded DNA in plasma. Genome Res. 30, 1144–1153 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study can be obtained from the European Genome–phenome Archive (accession number EGAS00001005048).


