Abstract
Mitochondria have a receptor complex in the outer membrane which recognizes and translocates mitochondrial proteins synthesized in the cytosol. We report here the identification and functional analysis of human Tom22 (hTom22). hTom22 has an N-terminal negatively charged region exposed to the cytosol, a putative transmembrane region, and a C-terminal intermembrane space region with little negative charge. Tom22 forms a complex with Tom20, and its cytosolic domain functions as an import receptor as in fungi. An import inhibition assay, using pre-ornithine transcarbamylase (pOTC) derivatives and a series of hTom22 deletion mutants, showed that the C-terminal segment of the cytosolic domain is important for presequence binding, whereas the N-terminal domain is important for binding to the mature portion of pOTC. No evidence for pOTC interaction with the Tom22 intermembrane space domain was obtained. Binding studies revealed that the presequence is critical for pOTC binding to Tom20, whereas both the presequence and mature portion are important for binding to Tom22. A cell-free immunoprecipitation assay indicated that an internal segment of the Tom22 cytosolic domain is important for interaction with Tom20.
Many nucleus-encoded mitochondrial proteins are initially synthesized on cytosolic ribosomes as larger preproteins with NH2-terminal presequences which function as mitochondrial targeting and import signals. The preproteins are then targeted to the mitochondria and imported into the organelle. An important step in this process is the interaction of the preproteins with the outer surface of the mitochondria. Genetic and biochemical studies of yeast and Neurospora crassa have identified a number of proteins in the mitochondrial outer membrane that are responsible for recognizing and translocating preproteins into the organelle (for reviews, see references 18, 20, 26, 30, and 33). They form a dynamic protein complex, termed the translocase of the outer membrane of mitochondria (Tom). Subunits of the complex include the receptor components Tom20 (22, 31), Tom22 (16, 19), and Tom70 (12, 36). An additional component, Tom37, has also been reported elsewhere for yeasts (9). The cytosolic domains of Tom22 and Tom20 are believed to form the major part of a cis site, which mediates the import of all preproteins known to use the general import machinery of mitochondria (21). The preprotein is then routed through the Tom complex translocation channel and transferred to a trans site on the intermembrane space (IMS) side of the outer membrane. Matrix-targeted proteins are further transferred to the matrix through import machinery in the inner membrane.
As the N-terminal cytosolic domain of fungal Tom22 is highly negatively charged, it has been speculated to bind the amphiphilic targeting sequences of preproteins through electrostatic interactions (16). However, it has been reported previously that the abundance of negative charges is not essential for the binding and import of preproteins and that other features in the domain are important (25). In addition to functioning as an import receptor, Tom22 forms a conducting channel with Tom40, a major component of the general insertion pore (5). Recent studies have shown that fungal Tom22 has docking sites for peripheral receptors, Tom20 and Tom70, and regulates preprotein translocation through the general insertion pore (38).
On the other hand, little is known about the import receptors of higher eukaryotes. cDNAs for human homologues of fungal Tom20 (8, 10, 35) and Tom70 (2) have been isolated, and Tom20 has been well characterized as a receptor protein (1, 14, 34, 37, 40). However, other mammalian components remain to be identified (23).
Here we report the identification and functional analysis of human Tom22 (hTom22). The cytosolic domain of hTom22 binds to preproteins. The C-terminal segment of the cytosolic domain is important for binding to the presequence of pre-ornithine transcarbamylase (pOTC), whereas the N-terminal segment is required for binding to the mature portion. hTom22 forms a complex with human Tom20 (hTom20), and the internal segment of the hTom22 cytosolic domain is important for complex formation.
MATERIALS AND METHODS
cDNA cloning and sequence analysis.
The cDNA fragments including an open reading frame sequence of hTom22 were amplified by PCR using a human cDNA library (Multi Choice cDNA; OriGene) as the template. The upstream and downstream primers were 5′-TGCTCTCTTCCGCTTCCGG-3′ and 5′-CACTGAGACAGCTCAAACAGC-3′, respectively. The amplified cDNA fragment was cloned into the HincII site of pGEM-3Zf(+), yielding pGEM-3Zf(+)-hTom22. The cDNA fragment amplified using another upstream primer, 5′-CTCTTCCGCTTCCGGTGTC-3′, was also cloned. These cDNA fragments were sequenced, and the overlapping sequences were completely identical to each other.
Construction of plasmids.
The BamHI/HindIII fragment of pGEM-3Zf(+)-hTom22 was blunt ended and cloned into the blunt-ended XhoI site of pCAGGS (28), yielding pCAGGS-hTom22. The construction of pCAGGS-hTom20, pCAGGS-pOTC, and pCAGGS-pOTC-GFP was reported previously (39).
Site-directed mutagenesis by the overlap extension method (13) was employed to produce pOTC-green fluorescent protein (GFP) mutants. The mutagenic primers to construct R23A pOTC-GFP were 5′-CACCGAAAATTGGCAACCATGAAGTTG-3′ and 5′-CTTCATGGTTGCCAATTTTCGGTGTGG-3′. The primers to construct R15/23/26A pOTC-GFP were 5′-GCAACCATGAAGTTGTGACCATTTGCAAAAGCTGCATTG-3′ and 5′-GGTCACAACTTCATGGTTGCCAATTTTGCGTGTGGACAACC-3′. The plasmid used as a template was pGEM-3Zf(+)-pOTC-GFP. The construction of pGEM-3Zf(+)-pOTC, pGEM-3Zf(+)-pOTC-GFP, and pGEM-3Zf(+)-pOTCN-GFP was described previously (39).
To construct the plasmids expressing chimeric proteins in which glutathione S-transferase (GST) was fused with hTom22 derivatives, the hTom22 gene was amplified by PCR. For constructing pGEX-2T-(1–82)hTom22, which expresses GST fused to the entire cytosolic domain of hTom22, 5′-TTTTTTGGATCCATGGCTGCCGCCGTCGC-3′ and 5′-AAAAAAGTCGACTCACCTGGAAAACCTGTACATTTTC-3′ were used as upstream and downstream primers, respectively, and the BamHI/HincII fragment of the PCR product was cloned into the BamHI/blunt-ended EcoRI site of pGEX-2T (Amersham Pharmacia Biotech). To construct the plasmids expressing GSTs fused with the C-terminally truncated cytosolic domain of hTom22, the same upstream primer was used, and the downstream primers used for each derivative were 5′-TTTTTTGAATTCTCATAGTCTCTCCGACAGGGTCTC-3′ for pGEX-2T-(1–48)hTom22, 5′-TTTTTTGAATTCTCACGCGGACCGGACCCTCTCC-3′ for pGEX-2T-(1–62)hTom22, 5′-AACGAAGAATTCTCAGAGGGAAAGATCAAAAGTGGC-3′ for pGEX-2T-(1–71)hTom22, 5′-TTTTTTGAATTCTCACTGAGCCACAAAGAGGGAAAG-3′ for pGEX-2T-(1–75)hTom22, and 5′-TTTTTTGAATTCTCACCTGTACATTTTCTGAGCCAC-3′ for pGEX-2T-(1–79)hTom22. The BamHI/EcoRI fragments of the PCR products were cloned into the same site of pGEX-2T. To construct pGEX-2T-(102–142)hTom22 expressing a GST-fused C-terminal domain of hTom22, 5′-TTTTTTGGATCCGAGACGGAGAAGTTGCAAATG-3′ and 5′-AAAAAAGTCGACTCAGATCTTTCCAGGAAGTGAGG-3′ were used, and the BamHI/HincII fragment of the PCR product was cloned into the same site of pGEX-2T. Construction of pGEX-2T-(25–145)hTom20 was previously described (40).
For constructing the plasmids expressing dihydrofolate reductase (DHFR)-fused hTom22s, the hTom22 gene was amplified by PCR. The same combination of primers for constructing pGEX-2T-(1–82)hTom22 and pGEX-2T-(102–142)hTom22 was used to construct pQE40-(1–82)hTom22 and pQE40-(102–142)hTom22, which express fusion proteins in which DHFR was fused with the N-terminal and C-terminal domains of hTom22, respectively. The amplified fragments were digested with BamHI and cloned into the BglII/HincII site of pQE40 (Qiagen).
Expression and purification of GST or DHFR-fused proteins.
The plasmids encoding GST-fused proteins were transformed into TOPP 2 cells (Stratagene). Expression and purification with glutathione agarose (Amersham Pharmacia Biotech) were performed as described previously (10). The plasmids encoding DHFR-fused hTom22s were transformed into SG13009 cells (Novagen). The DHFR-fused hTom22s with histidine tags were purified by metal chelation chromatography under denaturing conditions as described in the manufacturer's instructions.
Antibodies.
The purified fusion proteins GST-(1–82)hTom22 and GST-(102–142)hTom22 were used for raising anti-T22N and anti-T22C antibodies in rabbits, respectively. Anti-T22N and anti-T22C antibodies were affinity purified using N-hydroxysuccinimide-activated Sepharose HP (Amersham Pharmacia Biotech) conjugated with DHFR-fused hTom22s. Anti-human OTC and anti-hTom20 antiserum were prepared as described previously (39). Anti-human porin (Calbiochem-Novabiochem) and anti-human Hsp60 (StressGen Biotech) antibodies were purchased commercially.
Cell culture and transfection.
COS-7 cells were cultured in growth medium (Dulbecco's modified Eagle's medium [DMEM] plus 10% fetal calf serum) at 37°C under an atmosphere of 5% CO2 and 95% air. For observation of mitochondria with a fluorescence microscope, cells were cultured on coverslips in 35-mm-diameter dishes. When cells were about 70% confluent, the cells were washed twice with serum-free DMEM, and the same medium was added. The cells were transfected with plasmids at 37°C for 4 h using TransIT LT1 polyamine (Pan Vera Corp.) and then placed in growth medium. The transfection efficiency was about 10%.
Subcellular fractionation.
COS-7 cells were harvested with phosphate-buffered saline (PBS) plus 1 mM EDTA, washed twice with PBS, and then suspended in ice-cold hypotonic buffer (10 mM Tris-HCl [pH 7.4] containing 5 mM MgCl2, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride). After sonication, the suspension was centrifuged at 500 × g for 10 min at 4°C, and the supernatant was used as whole-cell extract. The cell extract was further centrifuged at 100,000 × g for 10 min at 4°C to give the soluble and membrane fractions. The membrane fraction was extracted with 0.1 M Na2CO3 (pH 11.5) as described previously (6).
Double staining for GFP and hTom22.
COS-7 cells were transfected with pCAGGS-pOTC-GFP as described above. The cells on coverslips were fixed with 4% formaldehyde for 40 min and treated with PBS containing 1% Triton X-100. The cells were treated with anti-T22C antibody and then with goat anti-rabbit immunoglobulin G (IgG) conjugated with Cy3 (Amersham Pharmacia Biotech) as secondary antibody. The fluorescence of Cy3 and GFP was photographed with a fluorescence microscope.
Isolation of mitochondria from COS-7 cells.
COS-7 cells were harvested with PBS plus 1 mM EDTA and washed twice with PBS. The cells were suspended in the mitochondrial isolation buffer (3 mM HEPES-KOH [pH 7.4], 0.21 M mannitol, 0.07 M sucrose, 0.2 mM EGTA), homogenized with a Dounce homogenizer (Wheaton), and then centrifuged at 500 × g for 5 min at 4°C. The supernatant was further centrifuged at 8,000 × g for 5 min at 4°C, and the precipitated mitochondria were resuspended and washed twice in the same buffer.
Protease accessibility assay.
Mitochondria (20 μg) were treated with 200 μg of trypsin per ml or 200 μg of proteinase K per ml in mitochondrial isolation buffer (total, 50 μl) in the presence or absence of 0.5% Triton X-100 for 30 min on ice. The digested products were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblot analysis.
In vitro import into isolated mitochondria.
mRNAs for hTom22, human pOTC, rat pOTC (24), pig pre-aspartate aminotransferase (27), rat pre-serine:pyruvate aminotransferase (29), pOTC-GFP, R23A pOTC-GFP, R15/23/26A pOTC-GFP, and pOTCN-GFP were synthesized by in vitro transcription. For the antibody inhibition assay, COS-7 mitochondria (25 μg of protein) were incubated with affinity-purified anti-T22N or anti-T22C antibodies for 20 min at 25°C in the import mixture (total, 50 μl) (37), and then 10 μl of reticulocyte lysate containing 35S-labeled preproteins was added to start the import reaction. After incubation for the indicated times, the mitochondria were reisolated by centrifugation and subjected to SDS-PAGE. The radioactivity in the gels was visualized and quantified using a FUJIX BAS2000 analyzer (Fuji Film Co.).
Coimmunoprecipitation of Tom20 and Tom22.
COS-7 cells (0.24 g [wet weight]) were harvested and lysed in 5 ml of lysis buffer (10 mM morpholinepropanesulfonic acid [MOPS]-NaOH [pH 7.2], 0.5% digitonin, 250 mM sucrose, 1 mM EDTA, 200 mM NaCl, 3% bovine serum albumin). The cell lysate was centrifuged at 25,000 × g for 15 min at 4°C. The supernatant fraction (1 ml) was incubated with 20 μl of anti-Tom20, anti-T22C, or anti-T22N antiserum or preimmune serum for 30 min at 25°C, and then 100 μl of a 12% suspension of protein A-Sepharose (Amersham Pharmacia Biotech) was added. After mixing for 40 min at 25°C, the resin was collected by centrifugation and washed once with lysis buffer without bovine serum albumin. Proteins were extracted and subjected to SDS-PAGE and immunoblot analysis.
Pulse-chase experiments.
COS-7 cells were transfected as described above. After 16 h of culture, the cells were harvested with trypsinization, washed twice with methionine-free DMEM, and suspended in 1 ml of the same medium. After preincubation at 37°C for 1 h to deplete methionine, the cells were radiolabeled with 8 MBq of Pro-Mix containing l-[35S]methionine and l-[35S]cysteine (Amersham Pharmacia Biotech) for 5 min and then chased with 20 mM l-methionine in 2 ml of DMEM. At the indicated times, 0.5-ml aliquots were withdrawn and mixed with 0.5 ml of ice-cold lysis buffer (20 mM Tris-HCl [pH 7.4] containing 4 mM EDTA, 0.2% SDS, 0.2% Triton X-100, 100 μM chymostatin, 100 μM pepstatin, 100 μM leupeptin, and 100 μM antipain). Radiolabeled proteins were immunoprecipitated with 20 μl of antiserum and 200 μl of a 10% suspension of protein A-Sepharose and subjected to SDS-PAGE. The radioactivity in the gels was visualized and quantified using a FUJIX BAS2000 analyzer.
In vitro binding assay.
Purified GST-fused Tom22s or Tom20 (7 nmol) was absorbed onto glutathione-agarose in 1.25 ml of binding buffer (20 mM HEPES-KOH [pH 7.4], 50 mM KCl, 1 mM MgCl2, 0.1 mg of bovine serum albumin per ml) containing 125 μl of a 50% slurry of glutathione-agarose. The agarose beads were washed three times and resuspended in 500 μl of binding buffer. GST derivative agarose (40 μl) was suspended in 250 μl of binding buffer and then mixed with 10 μl of reticulocyte lysate containing 35S-labeled preproteins in an Ultrafree-MC centrifugal filter unit (Millipore Corp.) for 30 min at 25°C with gentle shaking. Unbound proteins were removed by centrifugation at 1,000 × g for 5 min at 4°C, and the retained beads were washed once with binding buffer. Fifty microliters of 50 mM Tris-HCl (pH 8.0) containing 15 mM glutathione was added to the wet agarose beads, and the mixture was mixed gently for 30 min at 25°C. Twenty microliters of the eluate was subjected to SDS–10% PAGE, and the radioactivity in the gels was visualized and quantified using a FUJIX BAS2000 analyzer. Ten microliters was also subjected to SDS–10% PAGE, and the proteins were stained with Coomassie brilliant blue R-250 to check the amount of eluted proteins.
Cell-free coimmunoprecipitation of hTom22 with hTom20.
Purified GST-fused hTom22s (each 0.2 nmol) were incubated with the same amount of GST-(25–145)hTom20 for 30 min at 25°C in 1 ml of lysis buffer (10 mM MOPS-NaOH [pH 7.2], 0.5% digitonin, 250 mM sucrose, 1 mM EDTA, 200 mM NaCl, 3% bovine serum albumin). After centrifugation, supernatants were incubated with anti-hTom20 antiserum (20 μl) for 30 min at 25°C, and then 80 μl of a 30% suspension of protein A-Sepharose was added. After incubation for 40 min at 25°C, the resin was collected by centrifugation and washed once with lysis buffer without bovine serum albumin. Ten percent of the extracted proteins and 10% of the input proteins were subjected to SDS-PAGE and immunoblot analysis using anti-GST antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology).
RESULTS
Cloning of human homologue of Tom22.
By searching the expressed sequence tag database with the known cDNA sequence encoding Tom22 of Neurospora crassa (16) and Saccharomyces cerevisiae (19), a mouse cDNA clone was found (GenBank accession no. AI156846) which showed a significant amino acid sequence similarity. Although human homologues were found in the database, the putative 5′ region was missing. A putative full-length cDNA for hTom22 was cloned by PCR using a primer pair, one of which was complementary to the upstream region of mouse cDNA (GenBank accession no. AI156846), and the other of which was complementary to the downstream region of the putative human cDNA fragment (EMBL accession no. Z46029).
The cDNA clone from human liver with a 426-bp open reading frame encoded a 142-amino-acid protein with a calculated molecular mass of 15.5 kDa and a pI of 4.1 (Fig. 1) (GenBank accession no. AB040119). Homology alignment showed that this human homologue had 34 and 32% similarity to N. crassa and S. cerevisiae Tom22, respectively. The similarity extended to several structural features. The proteins consist of three regions: an N-terminal negatively charged region, an internal hydrophobic region suggested to be a transmembrane region in fungi, and a C-terminal region. The N-terminal region (residues 1 to 82) of the human homologue, speculated to be important for interaction with positively charged presequences of preproteins in fungi, contains 21 negatively charged residues, whereas the corresponding regions of N. crassa and S. cerevisiae Tom22 have 19 and 24 negatively charged residues, respectively. On the other hand, the C-terminal portions of the three proteins have less similarity. The region of the human proteins has a glutamine-rich segment; a similar sequence is present in mammalian Tom20 (34). This human protein was identified as hTom22 (see below).
FIG. 1.
Comparison between human and fungal Tom22 sequences. Amino acids are designated with the single-letter code, and gaps were introduced to maximize the alignment. The identical or similar residues (I/V/L, R/K, G/A, D/E, Q/N, T/S, and F/Y) are shown in shaded blocks. Putative transmembrane regions are boxed. S.c., S. cerevisiae; N.c., N. crassa.
Mammalian Tom22 is a mitochondrial outer membrane protein.
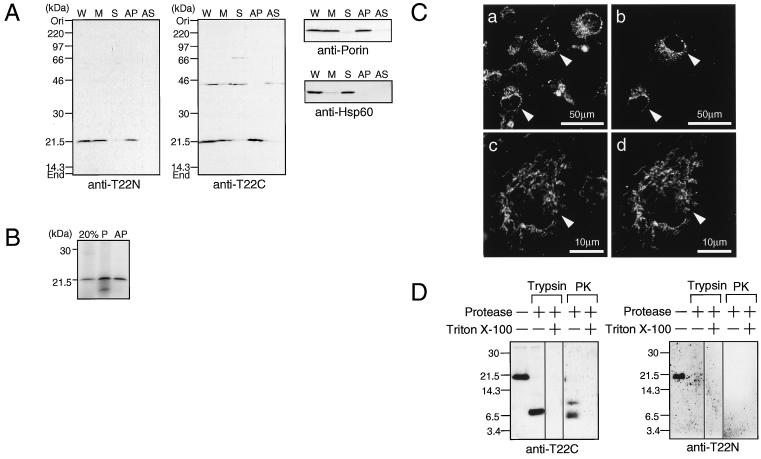
COS-7 cells were fractionated and subjected to immunoblot analysis using the affinity-purified anti-T22N antibody. A protein of 21 kDa was detected in the whole-cell extract and was recovered in the membrane fraction (Fig. 2A). This protein was not extracted with alkali, indicating that it is an integral membrane protein (6). Similar results were obtained with the anti-T22C antibody.
FIG. 2.
Localization of mammalian Tom22. (A) COS-7 cells were harvested and suspended in hypotonic buffer. After sonication, the suspension was centrifuged at 500 × g and the supernatant was used as whole-cell extract. The cell extract was further centrifuged at 100,000 × g to give the soluble and membrane fractions. The membrane fraction was extracted with alkali (0.1 M Na2CO3 [pH 11.5]). The whole-cell extracts (W) (44 μg of protein), soluble fractions (S) (23 μg), membrane fractions (M) (24 μg), precipitated fraction (AP) (11 μg), and soluble fraction (AS) (12 μg) after extraction with alkali were subjected to SDS–12% PAGE and immunoblot analysis using affinity-purified antibodies to the N-terminal portion (anti-T22N) and the C-terminal portion (anti-T22C) of hTom22. Mitochondrial porin (outer membrane protein) and Hsp60 (matrix protein) were also stained as controls. (B) Rabbit reticulocyte lysate (10 μl) containing 35S-labeled hTom22 was incubated with isolated mitochondria (50 μg of protein) for 40 min at 25°C. Mitochondria were precipitated and subjected to extraction with alkali. The precipitated mitochondria (P) and the alkali-insoluble fraction (AP) were subjected to SDS–14% PAGE. “20%” represents 20% of hTom22 input in the import assay. (C) COS-7 cells grown on coverslips in 35-mm-diameter culture dishes were transfected with 2 μg of pCAGGS-pOTC-GFP. After culture for 24 h, cells were subjected to immunostaining with anti-T22C antibody and secondary antibody labeled with Cy3. Fluorescence due to Cy3 (a and c) or GFP (b and d) was photographed. Arrowheads indicate the cells expressing pOTC-GFP. (D) Isolated mitochondria (20 μg) from COS-7 cells were treated with trypsin (200 μg/ml) or proteinase K (PK) (200 μg/ml) in the absence or presence of 0.5% Triton X-100 for 30 min on ice. The products were subjected to Tris-Tricine PAGE, followed by immunoblot analysis with anti-T22C or anti-T22N antibodies. Numbers at left are molecular masses in kilodaltons.
Integration of hTom22 into the mitochondrial membrane was studied (Fig. 2B). Cell-free-synthesized hTom22 migrated at the same position as that of endogenous Tom22 from COS-7 cells. When in vitro-synthesized hTom22 was incubated with isolated mitochondria, about 30% was recovered in pelleted mitochondria and was not extracted with alkali, indicating that hTom22 was integrated into the mitochondrial membrane.
COS-7 cells expressing pOTC-GFP, a fusion protein in which the presequence of human pOTC was fused to GFP, were visualized for GFP fluorescence and for immunostained endogenous Tom22 (Fig. 2C). pOTC-GFP was shown to be correctly imported into the mitochondrial matrix (39). The pattern of GFP fluorescence coincided with that of the Tom22 stain, confirming the mitochondrial localization of endogenous Tom22.
We next analyzed the sensitivity of Tom22 in intact mitochondria to trypsin and proteinase K (Fig. 2D). When the mitochondria isolated from COS-7 cells were treated with proteases, Tom22 was degradated to a major fragment of about 7 kDa, which was recognized by anti-T22C antibody but not by anti-T22N antibody. When mitochondria were treated with detergent prior to protease treatment, no fragment was recognized by these antibodies. These results indicate that the N-terminal and C-terminal portions of mammalian Tom22 are exposed to the cytosol and IMS, respectively.
Tom22 functions as an import receptor for preproteins.
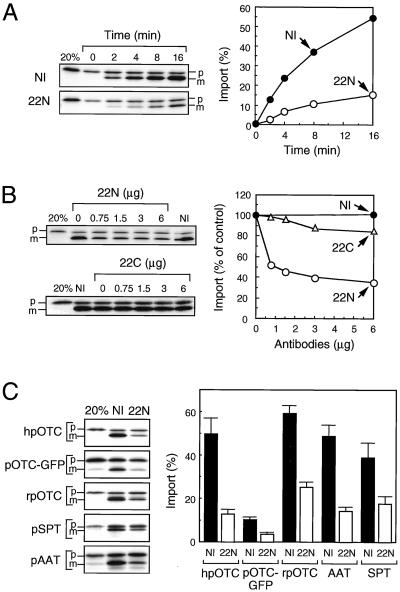
To test the involvement of Tom22 in receptor function for preproteins, we examined the effect of antibodies against hTom22 on mitochondrial import of preproteins (Fig. 3). COS-7 mitochondria were used because the antibodies against hTom22 cross-react strongly with primate Tom22 but less strongly with the rat protein. Human pOTC synthesized in rabbit reticulocyte lysates was efficiently imported into the isolated mitochondria and processed to the mature form (Fig. 3A). This import was inhibited strongly by the anti-T22N antibody but only slightly by the anti-T22C antibody (Fig. 3A and B). Import of other natural preproteins (rat pOTC, pre-serine:pyruvate aminotransferase, and pre-aspartate aminotransferase) and a chimeric protein, pOTC-GFP, was also inhibited by the anti-T22N antibody to a similar extent (Fig. 3C). These results indicate that all these preproteins are imported into the mitochondria through interaction with the cytosolic domain of hTom22.
FIG. 3.
Effect of anti-hTom22 antibodies on import of preproteins into isolated mitochondria. (A) Isolated COS-7 cell mitochondria (25 μg) were preincubated in the presence of 6 μg of nonimmune IgG (NI) or affinity-purified anti-T22N antibody (22N) for 20 min at 25°C in the import reaction mixture (50 μl). 35S-labeled reticulocyte lysate translation product containing human pOTC (10 μl) was then added to the mixture and incubated at 25°C. The import reaction was stopped at the indicated times and subjected to SDS–10% PAGE. The radioactive polypeptides were visualized by image plate analysis (left panels), and the radioactive mature OTC on the SDS-polyacrylamide gels was quantified (right panel). “20%” represents 20% of input pOTC. The percent import represents the amount of mature OTC compared with the input pOTC. (B) Import of 35S-labeled human pOTC was performed in the absence (control) or presence of indicated amounts of nonimmune IgG (NI) or affinity-purified anti-T22N (22N) or anti-T22C (22C) antibodies for 16 min at 25°C and was analyzed as described for panel A. Import without antibody was set as 100%. (C) Import of human pOTC (hpOTC), pOTC-GFP, rat pOTC (rpOTC), pre-serine:pyruvate aminotransferase (pSPT), and pre-aspartate aminotransferase (pAAT) was performed in the presence of 6 μg of nonimmune IgG (NI) or affinity-purified anti-T22N antibody (22N) for 16 min at 25°C and was analyzed as described for panel A. The percent import represents the amount of mature proteins compared with the input precursors. Values are represented by means ± standard deviations of three independent experiments. p and m, precursor and mature forms, respectively.
Tom22 forms a receptor complex with Tom20.
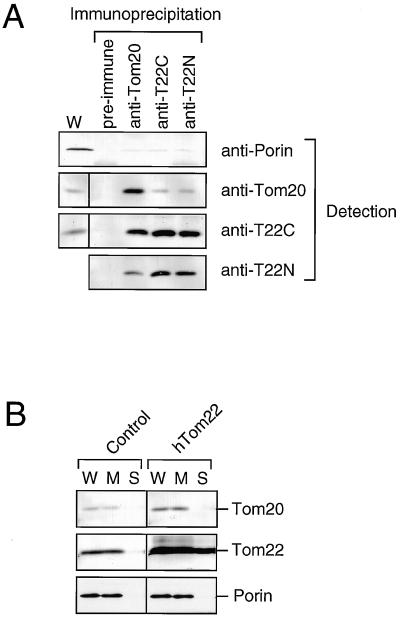
To assess whether Tom22 forms a receptor complex with Tom20, coimmunoprecipitation analysis was performed (Fig. 4A). COS-7 cells were lysed in digitonin and subjected to immunoprecipitation. When Tom20 was immunoprecipitated with anti-Tom20 antibody, Tom22 was coprecipitated. On the other hand, Tom20 was less efficiently coprecipitated with Tom22 antibodies. It should be noted that, in fungi, Tom20 is present only partly in a large complex with Tom22 and partly in a smaller subcomplex without Tom22 (5).
FIG. 4.
Interaction between Tom22 and Tom20. (A) COS-7 cells were lysed in 10 mM MOPS-NaOH (pH 7.2) containing 0.5% digitonin, 250 mM sucrose, 1 mM EDTA, 200 mM NaCl, and 3% bovine serum albumin. The cell lysates were centrifuged at 25,000 × g for 15 min at 4°C, and the soluble fractions (340 μg of protein) were subjected to immunoprecipitation using anti-Tom20, anti-hT22C, or anti-hT22N antiserum or preimmune serum as described in Materials and Methods. The immunoprecipitates were subjected to SDS–12% PAGE and immunoblot analysis. W, COS-7 cell extracts (38 μg of protein). (B) COS-7 cells were transfected with 10 μg of pCAGGS (control) or pCAGGS-hTom22. After culture for 20 h, cells were harvested and fractionated as described in Materials and Methods. Whole-cell extracts (W) (30 μg), soluble fractions (S) (18 μg), and membrane fractions (M) (15 μg) were subjected to SDS–12% PAGE and immunoblot analysis for the indicated proteins.
When hTom22 was transiently overexpressed in COS-7 cells, the amount of endogenous Tom20 was also increased whereas that of porin was not affected (Fig. 4B). This observation is in agreement with the finding that depletion of Tom20 in fungi decreases the level of Tom22 (19). These results point to an association of Tom22 and Tom20. In contrast, overexpression of hTom20 did not result in Tom22 accumulation (see Fig. 5A).
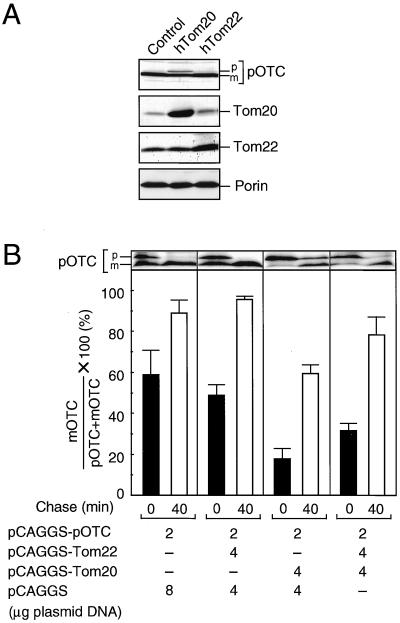
FIG. 5.
Cooperation of hTom22 and hTom20 on preprotein import. COS-7 cells were cultured in 10-cm-diameter dishes. (A) Two micrograms of pCAGGS-pOTC was cotransfected with 4 μg of pCAGGS (control), pCAGGS-hTom20, or pCAGGS-hTom22. After culture for 20 h, cells were harvested, and cell extracts (45 μg of protein) were subjected to SDS–12% PAGE and immunoblot analysis for the indicated proteins. (B) Two micrograms of pCAGGS-pOTC was cotransfected with indicated amounts of plasmids. After 16 h of culture, cells were harvested and subjected to pulse (5-min)-chase (40-min) experiments as described in Materials and Methods. The radioactive pOTC and mature OTC (mOTC) on the SDS-polyacrylamide gel were quantified by image plate analysis, and percentages of mOTC versus pOTC plus mOTC are shown. Values are represented by means ± standard deviations of three independent experiments. p and m, precursor and mature forms, respectively.
Cooperation of Tom22 with Tom20 on preprotein import.
When human pOTC is transiently expressed in COS-7 cells, it is imported efficiently into the mitochondria and processed to the mature form, as revealed by cell fractionation and immunoblot analysis (37). We analyzed whether overexpression of hTom22 had any effect on pOTC import in cultured cells (Fig. 5A). When hTom20 was coexpressed with pOTC, its mitochondrial import and processing were inhibited and unprocessed pOTC accumulated (see also references 37 and 40). When hTom22 was coexpressed, mitochondrial import and processing of pOTC were little affected.
To examine the effect of overexpression of Tom22 in detail, pulse-chase experiments were performed (Fig. 5B). When the COS-7 cells expressing human pOTC alone were labeled for 5 min with [35S]methionine, 60% of the newly synthesized pOTC was converted to the mature form. When the cells were then chased with cold methionine, the labeled pOTC was converted to the mature form almost completely. When hTom22 was coexpressed with pOTC, no significant change of pOTC import was observed. In contrast, when hTom20 was coexpressed with pOTC, only about 20% of newly synthesized pOTC was processed in a 5-min pulse and a moderate amount of pOTC remained after the chase. When hTom22 was coexpressed with pOTC and hTom20, inhibition of pOTC import by Tom20 was partly canceled. These results indicate cooperation of Tom22 and Tom20 in preprotein import.
Differential domains of hTom22 contribute to interaction with presequence and mature portion of preprotein.
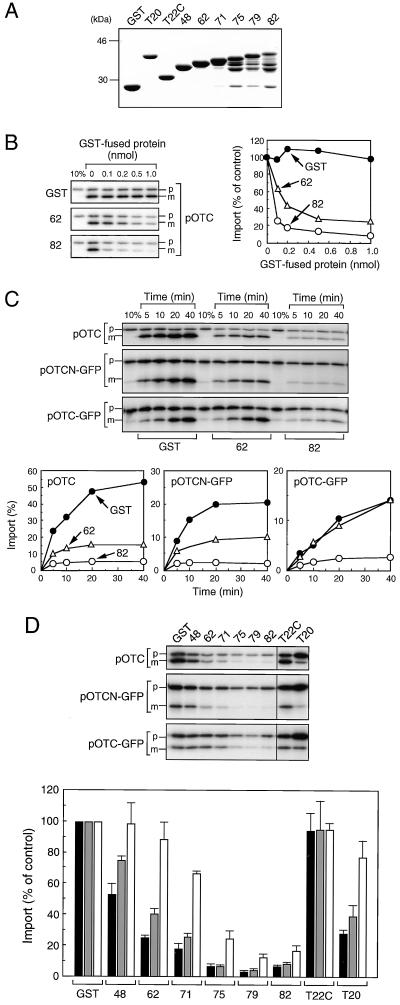
To analyze the roles of Tom22 in mitochondrial protein import in vitro, we expressed and purified a GST fusion protein containing the entire N-terminal cytosolic domains of hTom22 [GST-(1–82)hTom22] and five containing deletions [GST-(1–48)hTom22, GST-(1–62)hTom22, GST-(1–71)hTom22, GST-(1–75)hTom22, and GST-(1–79)hTom22] (Fig. 6A). Unfortunately, the three longer fusion proteins were partly degraded, and attempts to obtain the intact proteins were unsuccessful. GST fusion proteins containing the C-terminal domain of hTom22 facing the IMS [GST-(102–142)hTom22] and the cytosolic domain of hTom20 [GST-(25–145)hTom20] were also purified.
FIG. 6.
Effect of GST-fused hTom22s on mitochondrial import of pOTC, pOTCN-GFP, and pOTC-GFP. (A) GST or GST-fused hTom22s and Tom20 were expressed and purified as described in Materials and Methods. Purified proteins (5 μg) were subjected to SDS–10% PAGE. T20, GST-(25–145)hTom20; T22C, GST-(102–142)hTom22; 48, GST-(1–48)hTom22; 62, GST-(1–62) hTom22; 71, GST-(1–71)hTom22; 75, GST-(1–75)hTom22; 79, GST-(1–79) hTom22; 82, GST-(1–82)hTom22. (B) Import of 35S-labeled human pOTC translated in reticulocyte lysate (10 μl) into isolated mitochondria (25 μg) was performed in the absence (control) or presence of indicated amounts of purified GST or GST-fused hTom22 derivatives at 25°C for 16 min in the import reaction mixture (50 μl) and was analyzed as described for Fig. 3. Ten percent of input preprotein was placed in the first lane. (C) Mitochondrial import of 35S-labeled human pOTC, pOTCN-GFP, and pOTC-GFP was performed in the presence of 1 nmol of GST or GST-fused hTom22 derivatives at 25°C. At the indicated times, the import reaction was stopped and analyzed as described for Fig. 3. (D) Mitochondrial import was performed in the presence of 1 nmol of GST (control), GST-fused hTom22 derivatives, and GST-fused hTom20 at 25°C for 16 min for human pOTC, 20 min for pOTCN-GFP, or 40 min for pOTC-GFP. The import reaction was stopped and analyzed as described for Fig. 3. Import was expressed as percentage of controls in which GST was included. Values are represented by means ± standard deviations of three independent experiments. Solid bars, pOTC; shaded bars, pOTCN-GFP; open bars; pOTC-GFP. p and m, precursor and mature forms, respectively.
When in vitro-synthesized pOTC was incubated with isolated mitochondria in the presence of the GST fusion proteins, pOTC import was markedly inhibited by increasing amounts of GST-(1–62)hTom22 and more strongly by GST-(1–82)hTom22 (Fig. 6B). A nearly complete inhibition was observed with 0.2 nmol of the longer fusion protein.
The effect of these hTom22 fusion proteins on the import of pOTCN-GFP and pOTC-GFP as well as pOTC was examined (Fig. 6C). pOTC-GFP is a fusion protein in which the presequence of pOTC was fused with GFP, and the GFP domain appears to be folded immediately after translation (40). pOTCN-GFP is another fusion protein, in which the presequence plus 58 residues in the mature portion of pOTC was fused to GFP, and the GFP domain appears to remain unfolded (39). pOTC was also shown to remain unfolded after translation (40). Import of pOTC was strongly inhibited by both GST-(1–82)hTom22 and GST-(1–62)hTom22. Similar results were obtained for pOTCN-GFP. In contrast, import of pOTC-GFP was inhibited by GST-(1–82)hTom22 but not by GST-(1–62)hTom22. These results suggest that the hTom22 segment of residues 63 to 82 is important for binding to all preproteins, presumably to the presequence, and that the N-terminal segment (residues 1 to 62) is involved in binding to the unfolded mature portions.
The effect of a series of fusion proteins on preprotein import was tested (Fig. 6D). The inhibition of pOTC and pOTCN-GFP import was progressively decreased with the increasing length of a C-terminal truncation, whereas the import inhibition of pOTC-GFP was decreased with a much smaller truncation. The inhibition of import of pAAT, another authentic preprotein, was very similar to that of pOTC (data not shown). These results suggest that the C-terminal segment of the cytosolic domain of hTom22 (residues 63 to 82) is important for presequence binding, whereas the N-terminal domain is important for binding to the unfolded mature portions of pOTC and pOTCN-GFP. Importantly, the C-terminal IMS domain did not inhibit import, suggesting that it does not bind to preproteins. The import of pOTC and pOTCN-GFP was inhibited by the hTom20 fusion, but that of pOTC-GFP was inhibited much less strongly, suggesting the involvement of unfolded mature portions in binding to Tom20 (40). To be noted is that pOTC-GFP import was much more strongly inhibited by the cytosolic domain of Tom22 than by that of Tom20.
Contribution of presequence and mature portion of preproteins to receptor binding differs between Tom22 and Tom20.
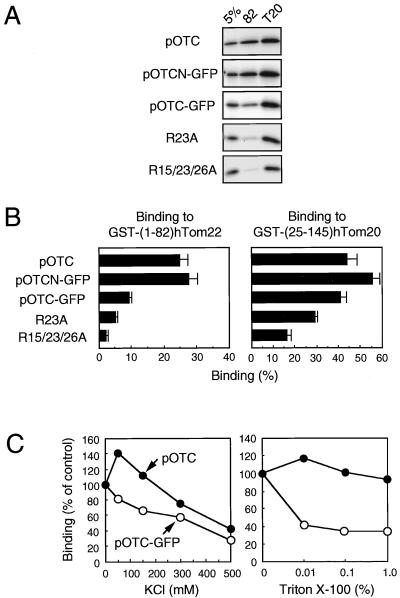
We next examined the direct interaction between the two receptors and preproteins by using the GST-fused Tom proteins (Fig. 7). Preproteins synthesized in vitro were incubated with glutathione-agarose beads prebound with GST-fused hTom22 or hTom20 proteins, and the preproteins and GST fusions were then eluted with reduced glutathione. Almost 100% of GST fusions that were applied to the binding assay were recovered in the eluate (data not shown). About 25% of applied pOTC and pOTCN-GFP were bound to the entire cytosolic domain of hTom22, whereas only 10% of pOTC-GFP was bound (Fig. 7A and B), suggesting that both the presequence and the mature portion of preproteins are important for binding to Tom22. In contrast, all three preproteins were bound to hTom20 to the same degree, suggesting that the presequence is critical for preprotein binding to Tom20. When pOTC-GFP mutants, in which one (R23A) or three (R15/23/26A) Arg residues in the presequence of pOTC-GFP were replaced by Ala, were used, preprotein binding was decreased partly and stepwisely as the number of replacements increased. Considering that pOTC has only four positive charges in the presequence, these results indicate that positive charges are important but not sufficient for receptor binding. Hydrophobic interaction between the presequence and receptors may also be important, as shown previously for Tom20 (1). GFP did not bind to the receptors (data not shown), indicating that pOTC-GFP interacts with hTom22 only via the presequence. The C-terminal IMS domain of hTom22 did not interact with preproteins (data not shown).
FIG. 7.
Preprotein binding to hTom22 and hTom20. (A) 35S-labeled translation product (10 μl) was incubated for 30 min at 25°C with glutathione-agarose prebound with about 0.56 nmol of GST-(1–82)hTom22 or GST-(25–145)hTom20. After washing, GST derivatives were eluted with 15 mM reduced glutathione, and 40% of the eluted protein was subjected to SDS–10% PAGE and fluorography using a FUJIX BAS2000 analyzer as described in Materials and Methods. Five percent of input preproteins were put in the first lanes. R23A, R23A pOTC-GFP; R15/23/26A, R15/23/26A pOTC-GFP. (B) The radioactive preproteins eluted were quantified by image plate analysis. The binding was expressed as percentage of input precursors. Values are represented as means ± standard deviations of three independent experiments. (C) The binding assay was performed as described for panel A except that the indicated concentrations of KCl or Triton X-100 were added to binding and washing buffers. Binding was expressed as percentage of controls without KCl and Triton X-100.
Sensitivity of the interaction between preproteins and hTom22 to salt and detergent was also examined (Fig. 7C). The binding of pOTC-GFP to Tom22 decreased gradually along with increasing concentrations of KCl, suggesting hydrophilic interaction between the presequence and Tom22. In contrast, the binding of pOTC increased at 50 mM KCl and then decreased gradually. This increased binding may reflect increased binding of the unfolded mature portion of pOTC to the receptor. The binding of pOTC-GFP to Tom22 was decreased in the presence of Triton X-100, whereas the binding of pOTC was little affected. This suggests that hydrophobic interaction between the presequence and Tom22 exists and that the binding of the mature portion of pOTC to Tom22 is stable in the presence of the detergent.
Internal segment in cytosolic domain of hTom22 is important for interaction with hTom20.
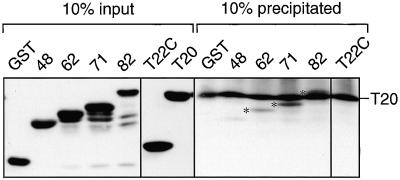
Interaction between Tom22 and Tom20 was analyzed by using a cell-free binding assay (Fig. 8). Purified GST-fused hTom20 was incubated with a series of purified GST-fused hTom22s used in Fig. 6. When GST-fused hTom20 was immunoprecipitated with an antiserum to hTom20, a fusion protein with the whole cytosolic domain of hTom22 was coprecipitated, showing the direct interaction between hTom22 and hTom20. Binding was similar for GST-(1–71)hTom22, decreased for GST-(1–62)hTom22, and was lost for GST-(1–48)hTom22. These results indicate that an internal segment of the cytosolic domain of hTom22 (residues 49 to 71) is important for interaction with hTom20. No binding was observed for the IMS domain of hTom22.
FIG. 8.
Analysis of sites of interaction of hTom22 with hTom20. Purified GST-fused hTom22s (each 0.2 nmol) were incubated with the same amount of GST-(25–145)hTom20 (T20) for 30 min at 25°C in the lysis buffer used for the coimmunoprecipitation assay. After centrifugation, immunoprecipitation using anti-Tom20 antiserum was performed as described in Materials and Methods. Ten percent of the immunoprecipitated proteins and 10% of the input proteins were subjected to SDS–12% PAGE and immunoblot analysis using an anti-GST antibody conjugated with horseradish peroxidase. Asterisks indicate the coprecipitated Tom22 proteins.
DISCUSSION
We have identified a protein import receptor from human mitochondria of 15.5 kDa (hTom22) and demonstrated it to be the human ortholog of fungal Tom22 based on the following findings: (i) hTom22 and fungal Tom22s show significant sequence homology and have similar domains, (ii) hTom22 is an integral mitochondrial outer membrane protein with the N-terminal portion exposed to the cytosol and the C-terminal portion exposed to the IMS, (iii) the cytosolic domain is involved in preprotein binding, and (iv) hTom22 forms a complex with Tom20.
We have shown that the C-terminal region (residues 63 to 82) of the hTom22 cytosolic domain is important for presequence binding, whereas the N-terminal region is required for binding to the mature portion of the mitochondrial preprotein, pOTC. This C-terminal region is predicted to form a β-sheet or turn structure and contains only one negative charge. We found that the positive charges in the presequence were important but not essential for binding to Tom22. Taken together, the interaction between Tom22 and presequence appeared to be mediated by both hydrophobic and hydrophilic interactions. Recent structural analysis of hTom20 by nuclear magnetic resonance analysis revealed that presequence binding to this receptor is mediated mainly by hydrophobic rather than by ionic interaction (1). Besides binding to presequences, receptors also appear to interact with the mature parts of preprotein, although the contribution of this interaction in total binding may differ among receptors. We found that hTom20 bound mainly to the presequence of pOTC and partly to the mature portion (40). Brix et al. (3), using a peptide scan method for analyzing the binding of preprotein to yeast receptors, found that, whereas CoxVI-derived peptides corresponding to its presequence bound mainly to Tom20, peptides corresponding to the mature protein also bound. Indeed, the mature peptides bound more strongly to Tom22, which is in agreement with the findings we report here that hTom20 binds mainly to the presequence of pOTC, whereas hTom22 binds mainly to the mature portion. Stronger inhibition of pOTC-GFP import by the cytosolic domain of Tom22 than by that of Tom20 (Fig. 6D) suggests that Tom22 has a higher affinity for the presequence than does Tom20.
Although the cytosolic domains of hTom22 and fungal Tom22s have a significant homology, the IMS domains are much less similar. The IMS domain of hTom22 has few negative charges and contains a unique segment rich in glutamine residues. A similar segment, called the Q-rich motif, is also present in hTom20. This segment was shown previously to be important for binding to preproteins and for preprotein import (34, 40). The involvement of the IMS domain of fungal Tom22s in preprotein import has been debated. Studies using mitochondria lacking the IMS domain suggested that the domain is required for promoting the transfer of presequence to the machinery in the inner membrane (4, 15). Chemical and photo-cross-linking experiments suggested that the presequence binds mainly to Tom40 in the trans site after entry of preprotein into the translocation pore (15, 32). In contrast, direct binding of preprotein to the IMS domain of Tom22 was reported by Komiya et al. (17). In the present study, we could not detect preprotein binding to the IMS domain of hTom22.
Haucke et al. (11) have suggested that fungal Tom20 and Tom70 interact with each other through tetratricopeptide repeat motifs, which are thought to mediate protein-protein interaction (7). However, recently, van Wilpe et al. (38) reported that the interaction of the receptors Tom20 and Tom70 with the translocation pore, Tom40, is through the cytosolic domain of Tom22 in yeast. Since neither fungal Tom22 nor hTom22 has an apparent tetratricopeptide repeat motif, some other interaction must be involved. In this study, we found that the internal segment of the cytosolic domain of hTom22 (residues 49 to 71) is important for interaction with hTom20. This remains to be confirmed by structural analysis.
ACKNOWLEDGMENTS
We thank J. Miyazaki (Osaka University, Japan) for pCAGGS, colleagues of this laboratory (Kumamoto University) for discussions, and M. Imoto for secretarial services.
This work was supported by a grant for a JSPS Research Fellow (to M.Y.); grants-in-aid 08457040 and 0725321 (to M.M.) from the Ministry of Education, Science, Sports and Culture of Japan; and a grant from the Australian Research Council (to N.H.).
REFERENCES
- 1.Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez D M, Gonzalez M M, Valencia A, Zenke M, Bernal J, Munoz A. Identification of a mammalian homologue of the fungal Tom70 mitochondrial precursor protein import receptor as a thyroid hormone-regulated gene in specific brain regions. J Neurochem. 1999;73:2240–2249. doi: 10.1046/j.1471-4159.1999.0732240.x. [DOI] [PubMed] [Google Scholar]
- 3.Brix J, Rudiger S, Bukau B, Schneider M J, Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J Biol Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- 4.Court D A, Nargang F E, Steiner H, Hodges R S, Neupert W, Lill R. Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol Cell Biol. 1996;16:4035–4042. doi: 10.1128/mcb.16.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker P J, Ryan M T, Brix J, Muller H, Honlinger A, Pfanner N. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiki Y, Hubbard A L, Fowler S, Lazarow P B. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 8.Goping I S, Millar D G, Shore G C. Identification of the human mitochondrial protein import receptor, huMas20p. FEBS Lett. 1995;373:45–50. doi: 10.1016/0014-5793(95)01010-c. [DOI] [PubMed] [Google Scholar]
- 9.Gratzer S, Lithgow T, Bauer R E, Lamping E, Paltauf F, Kohlwein S D, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson B, Nuttal S, Hoogenraad N. A receptor for the import of proteins into human mitochondria. Eur J Biochem. 1996;235:750–753. doi: 10.1111/j.1432-1033.1996.t01-1-00750.x. [DOI] [PubMed] [Google Scholar]
- 11.Haucke V, Horst M, Schatz G, Lithgow T. The Mas20p and Mas70p subunits of the protein import receptor of yeast mitochondria interact via the tetratricopeptide repeat motif in Mas20p: evidence for a single hetero-oligomeric receptor. EMBO J. 1996;15:1231–1237. [PMC free article] [PubMed] [Google Scholar]
- 12.Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 14.Iwahashi J, Yamazaki S, Komiya T, Nomura N, Nishikawa S, Endo T, Mihara K. Analysis of the functional domain of the rat liver mitochondrial import receptor Tom20. J Biol Chem. 1997;272:18467–18472. doi: 10.1074/jbc.272.29.18467. [DOI] [PubMed] [Google Scholar]
- 15.Kanamori T, Nishikawa S, Nakai M, Shin I, Schultz P G, Endo T. Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc Natl Acad Sci USA. 1999;96:3634–3639. doi: 10.1073/pnas.96.7.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiebler M, Keil P, Schneider H, van der Klei I J, Pfanner N, Neupert W. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- 17.Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acid chain’ hypothesis. EMBO J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lill R, Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Biochem Sci. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- 19.Lithgow T, Junne T, Suda K, Gratzer S, Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc Natl Acad Sci USA. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lithgow T, Glick B S, Schatz G. The protein import receptor of mitochondria. Trends Biochem Sci. 1995;20:98–101. doi: 10.1016/s0968-0004(00)88972-0. [DOI] [PubMed] [Google Scholar]
- 21.Mayer A, Nargang F E, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moczko M, Ehmann B, Gartner F, Honlinger A, Schafer E, Pfanner N. Deletion of the receptor MOM19 strongly impairs import of cleavable preproteins into Saccharomyces cerevisiae mitochondria. J Biol Chem. 1994;269:9045–9051. [PubMed] [Google Scholar]
- 23.Mori M, Terada K. Mitochondrial protein import in animals. Biochim Biophys Acta. 1998;1403:12–27. doi: 10.1016/s0167-4889(98)00021-4. [DOI] [PubMed] [Google Scholar]
- 24.Murakami K, Amaya Y, Takiguchi M, Ebina Y, Mori M. Reconstitution of mitochondrial protein transport with purified ornithine carbamoyltransferase precursor expressed in Escherichia coli. J Biol Chem. 1988;263:18437–18442. [PubMed] [Google Scholar]
- 25.Nargang F E, Rapaport D, Ritzel R G, Neupert W, Lill R. Role of the negative charges in the cytosolic domain of TOM22 in the import of precursor proteins into mitochondria. Mol Cell Biol. 1998;18:3173–3181. doi: 10.1128/mcb.18.6.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 27.Nishi T, Nagashima F, Tanase S, Fukumoto Y, Joh T, Shimada K, Matsukado Y, Ushio Y, Morino Y. Import and processing of precursor to mitochondrial aspartate aminotransferase. Structure-function relationships of the presequence. J Biol Chem. 1989;264:6044–6051. [PubMed] [Google Scholar]
- 28.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 29.Oda T, Funai T, Ichiyama A. Generation from a single gene of two mRNAs that encode the mitochondrial and peroxisomal serine:pyruvate aminotransferase of rat liver. J Biol Chem. 1990;265:7513–7519. [PubMed] [Google Scholar]
- 30.Pfanner N, Craig E A, Meijer M. The protein import machinery of the mitochondrial inner membrane. Trends Biochem Sci. 1994;19:368–372. doi: 10.1016/0968-0004(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 31.Ramage L, Junne T, Hahne K, Lithgow T, Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapaport D, Neupert W, Lill R. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- 33.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 34.Schleiff E, Shore G C, Goping I S. Interactions of the human mitochondrial protein import receptor, hTom20, with precursor proteins in vitro reveal pleiotropic specificities and different receptor domain requirements. J Biol Chem. 1997;272:17784–17789. doi: 10.1074/jbc.272.28.17784. [DOI] [PubMed] [Google Scholar]
- 35.Seki N, Moczko M, Nagase T, Zufall N. A human homolog of the mitochondrial protein import receptor Mom19 can assemble with the yeast mitochondrial receptor complex. FEBS Lett. 1995;375:307–310. doi: 10.1016/0014-5793(95)01229-8. [DOI] [PubMed] [Google Scholar]
- 36.Sollner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- 37.Terada K, Kanazawa M, Yano M, Hanson B, Hoogenraad N, Mori M. Participation of the import receptor Tom20 in protein import into mammalian mitochondria: analyses in vitro and in cultured cells. FEBS Lett. 1997;403:309–312. doi: 10.1016/s0014-5793(97)00070-7. [DOI] [PubMed] [Google Scholar]
- 38.van Wilpe S, Ryan M T, Hill K, Maarse A C, Meisinger C, Brix J, Dekker P J, Moczko M, Wagner R, Meijer M, Guiard B, Honlinger A, Pfanner N. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- 39.Yano M, Kanazawa M, Terada K, Namchai C, Yamaizumi M, Hanson B, Hoogenraad N, Mori M. Visualization of mitochondrial protein import in cultured mammalian cells with green fluorescent protein and effects of overexpression of the human import receptor Tom20. J Biol Chem. 1997;272:8459–8465. doi: 10.1074/jbc.272.13.8459. [DOI] [PubMed] [Google Scholar]
- 40.Yano M, Kanazawa M, Terada K, Takeya M, Hoogenraad N, Mori M. Functional analysis of human mitochondrial receptor Tom20 for protein import into mitochondria. J Biol Chem. 1998;273:26844–26851. doi: 10.1074/jbc.273.41.26844. [DOI] [PubMed] [Google Scholar]