Abstract
Purpose:
To investigate the editing-pulse flip angle dependence of editing efficiency and ultimately to maximize the edited signal of commonly edited MRS signals such as GABA and lactate.
Methods:
Density-matrix simulations were performed for a range of spin systems to find the editing-pulse flip angle (FA) for maximal editing efficiency. Simulations were confirmed by phantom experiments and in vivo measurements in 10 healthy participants using a 3T Philips scanner. Four MEGA-PRESS in vivo measurements targeting GABA+ and lactate were performed, comparing the conventional editing-pulse flip angle (FA = 180°) to the optimal one suggested by simulations (FA = 210°).
Results:
Simulations and phantom experiments show that edited GABA and lactate signals are maximal at FA = 210°. Compared to conventional editing (FA = 180°), in vivo signals from GABA+ and lactate signals increase on average by 8.5% and 9.3%, respectively.
Conclusion:
Increasing the flip angle of editing-pulses in the MEGA-PRESS experiment from 180° to 210° increases the edited signals from GABA+ and lactate by about 9% in vivo.
Keywords: edited MRS, flip angle, MEGA-PRESS, optimization
Introduction
Edited MRS is a non-invasive method for metabolite quantification that is used in a variety of clinical research applications.1 A number of metabolites with low in-vivo concentration, including gamma-aminobutyric acid (GABA), glutathione (GSH), lactate (Lac), and phosphorylethanolamine (PE), are better resolved with targeted edited approaches.2–5 The widely used MEGA-PRESS6–8 sequence adds two frequency-selective editing pulses to PRESS9 localization, acquiring edit-ON and edit-OFF sub-experiments in an interleaved fashion. Subtraction of these sub-experiments removes the strong overlying signals from more concentrated metabolites, yielding a difference spectrum that reveals the metabolite signal of interest.
There is extensive literature re-optimizing the MEGA-PRESS sequence for various metabolites.2,10–12 TE is generally chosen so that the detected signal is maximally negative in the edit-OFF sub-experiment due to coupling evolution. This evolution is refocused in the edit-ON scan, yielding the maximal difference-edited signal. This picture is complicated somewhat by spatially heterogeneous coupling evolution patterns that arise from limited-bandwidth slice-selective refocusing pulses.13,14 The experiment generally works as intended in the majority of the voxel, but edge effects where either coupled spin undergoes a slice-selective refocusing flip angle other than 180o lead to a loss of editing efficiency and modify the shape of the difference-edited multiplet. Increased slice-selective refocusing bandwidth can limit these losses but may come at the expense of greater sensitivity to B1 miscalibrations/inhomogeneity or increased SAR.15
Since the original proposal of J-difference editing6, editing pulses have been applied with a 180° flip angle.7,8,16 This aligns with their role of selectively refocusing coupling evolution during TE by inverting the passive spins. For this reason, the assumption that 180° is the optimal edit-ON flip angle (and 0° the optimal edit-OFF flip angle) has gone unexamined, even while the duration and frequency response profile of these pulses has received much attention. For practical purposes, it is more critical to optimize the detected SNR than it is that targeted couplings are refocused (or allowed to evolve) perfectly.
Therefore, the primary aim of this work was to investigate the flip-angle dependence of editing efficiency and ultimately to maximize the edited signal. Firstly, a range of simulations was performed in order to determine optimal editing flip angles. Secondly, phantom experiments were performed that seek to confirm the simulation results. Thirdly, editing with optimized flip angles was evaluated in healthy participants and compared with conventional MEGA-PRESS editing – i.e., with an editing-pulse flip angle of 180° – of GABA+ and Lac.
Methods
Simulations
Given the diversity of edited experiments that have been proposed, we aim to simulate a representative, rather than exhaustive set. Firstly, we consider GABA-edited MEGA-PRESS, as applied to the GABA spin system, and to co-edited spin systems of glutamine (Gln), glutamate (Glu), and lysine (Lys, as the primary component of the co-edited MM signal17). Secondly, we consider separate edited experiments of GSH, Lac, and PE.
2-dimensional spatially resolved (21 × 21 points, 4.5 cm field of view in each direction) density-matrix simulations were performed using the MATLAB-based toolbox FID-A18 with in-house modifications. Excitation as assumed to be ideal; refocusing and editing pulse waveforms, durations and sequence timing are taken from a cross-vendor-standardized MEGA-PRESS implementation.19 The editing pulse, in particular, is available in the FID-A simulation package as ‘SampleEditPulse.pta’. The flip angle of this pulse is correctly calibrated within the simulations, such that a flip angle of 180 degrees results in full inversion of longitudinal magnetization on-resonance. The sequence parameters kept constant throughout the simulations are summarized in Table 1 for each specific spin system. For all simulated experiments, the editing-pulse flip angle was varied between 0° and 270° in steps of 10°.
Table 1.
Sequence parameters kept constant throughout the simulations for each specific spin system.
| Spin system | TE [ms] | Edit Frequency (ON/OFF) [ppm] | Edit Pulse Duration [ms] |
|---|---|---|---|
|
| |||
| GABA | 80 | 1.9/7.5 | 20 |
| Gln | 80 | 1.9/7.5 | 20 |
| Glu | 80 | 1.9/7.5 | 20 |
| Lysine | 80 | 1.9/7.5 | 20 |
| GSH | 80 | 4.56/7.5 | 20 |
| Lactate | 140 | 4.1/7.5 | 40 |
| PE | 90 | 3.98/7.5 | 20 |
Simulations were independently repeated by two co-authors (JN and BJS), one in FID-A using a different editing pulse shape (‘Gauss0_R1p0952.pta’) to ensure that the behavior investigated is not pulse-specific and one in the Vespa-Simulation application, which uses PyGAMMA to wrap the GAMMA NMR simulation library, to ensure that unexpected results are not driven by the simulation software.
Phantom Measurements
Phantom and in vivo measurements were performed on a 3T MR scanner (Philips dStream Achieva, Philips Healthcare, Best, The Netherlands) equipped with a 32-channel head coil. Phantom measurements were performed on a 10 mM GABA phantom with the voxel (24 × 24 × 24 mm3) placed in the center of the phantom. First, a short-TE PRESS measurement was performed to determine the temperature-dependent chemical shift of water (from 4.68 to 4.82 ppm), and correctly account for it in determining the editing pulse offsets in the subsequent MEGA-PRESS measurements. Note that in the MEGA-PRESS edit-OFF sub-experiment, the editing pulse was applied on-resonance for water at 4.82 ppm, providing a saturation calibration of the editing-pulse flip angle. Measurements were performed with: TE/TR = 80/2000 ms; 20-ms editing pulses; editing pulse offsets 4.82/1.9 ppm; 64 averages; 2048 datapoints sampled with 2 kHz spectral width; pencil-beam second-order shim; Philips ‘excitation’ water suppression. The editing-pulse flip angle was varied: from 20° to 100° in increments of 20°; from 110° to 170° in increments of 10°; from 175° to 210° in increments of 5°, and from 220° to 270° in increments of 10°.
In Vivo Measurements
10 healthy participants (mean age 38 ± 11.9 years, 7 females) were recruited and written informed consent was obtained. Following T1-weighted imaging, a 30 × 30 × 30 mm3 voxel was placed in the midline parietal lobe. Four separate MEGA-PRESS measurements (each 320 averages) were performed with the following parameters:
Conventional GABA: TE/TR = 80/2000 ms, ON/OFF = 1.9/7.5 ppm, edit FA = 180/180, 20 ms pulse duration
Optimized GABA: TE/TR = 80/2000 ms, ON/OFF = 1.9/7.5 ppm, edit FA = 210/210, 20 ms pulse duration
Conventional Lac: TE/TR = 140/2000 ms, ON/OFF = 4.1/7.5 ppm, edit FA = 180/180, 40 ms pulse duration
Optimized Lac: TE/TR = 140/2000 ms, ON/OFF = 4.1/7.5 ppm, edit FA = 210/210, 40 ms pulse duration
The first two measurements targeted GABA, comparing conventional 180° editing-pulse flip angles to the editing-pulse flip angle (210°) suggested by simulations to maximize the edited signal (see Results below). The latter two measurements targeted lactate and made the same comparison. A pencil-beam second-order shim routine and VAPOR20 water suppression were used for all measurements.
Data Processing and Analysis
Phantom data were analyzed using Gannet 3.121, with default settings for analysis of GABA phantom data. The resulting ON-integrals were calculated in the ranges [4.6, 5.0] and [2.8, 3.2] ppm for water and GABA, respectively. The integrals were normalized to the maximum value for each metabolite to facilitate comparison with the simulations. Smooth models were fitted to the normalized integral curves, and the optimal flip angle was determined from the maximum model value. For the simulated data and the data from the water calibration experiment, polynomial model functions were used, while a Gaussian model function was used for the GABA phantom data.
Based on simulation results that optimal saturation of the water signal occurs at a flip angle of 180°, the prescribed flip angle of optimal water saturation FAw,opt was used to calculate the actual flip angle of the edited phantom experiments FAedit,actual from the prescribed flip angle FAedit,prescribed thus: FAedit,actual = FAedit,prescribed* 180/FAw,opt.
In vivo data were analyzed using Osprey22 with frequency-and-phase correction performed using robust spectral registration23. Eddy-current correction was performed on the averaged edit-ON and edit-OFF sub-spectra which were subsequently aligned by minimizing the water signal in the difference spectrum. Linear-combination modeling was performed on the difference spectra using TE-specific simulated basis sets that included: ascorbate, aspartate, creatine, GABA, Gln, Glu, glycerophosphocholine, GSH, H2O, myo-inositol, Lac, N-acetylaspartate, N-acetylaspartylglutamate, phosphocholine, phosphocreatine, PE, scyllo-inositol, and taurine. For the analysis of the GABA-edited data, the co-edited MM signal at 3 ppm was included in the model by setting the `coMM3` parameter to `1to1GABAsoft`, assuming a 1:1-amplitude ratio between GABA and the co-edited MM enforced as a soft constraint.24 The baseline spline knot spacing was kept constant at 0.55 ppm for all analyses. The fit range was [0.5, 4] ppm and [0.5, 3.5] ppm for GABA and Lac, respectively.25 The water-signal-normalized GABA+ and Lac signal were computed for the corresponding measurements, with corrections for relaxation but without corrections for tissue composition.22
Paired t-tests were performed in order to determine whether using an editing-pulse flip angle of 210° generated significantly (p < 0.05) higher edited signals for GABA+ and Lac than the conventional 180° editing pulse. The mean percentage signal increase was also computed and compared to the increase predicted by simulations.
Results
Simulations
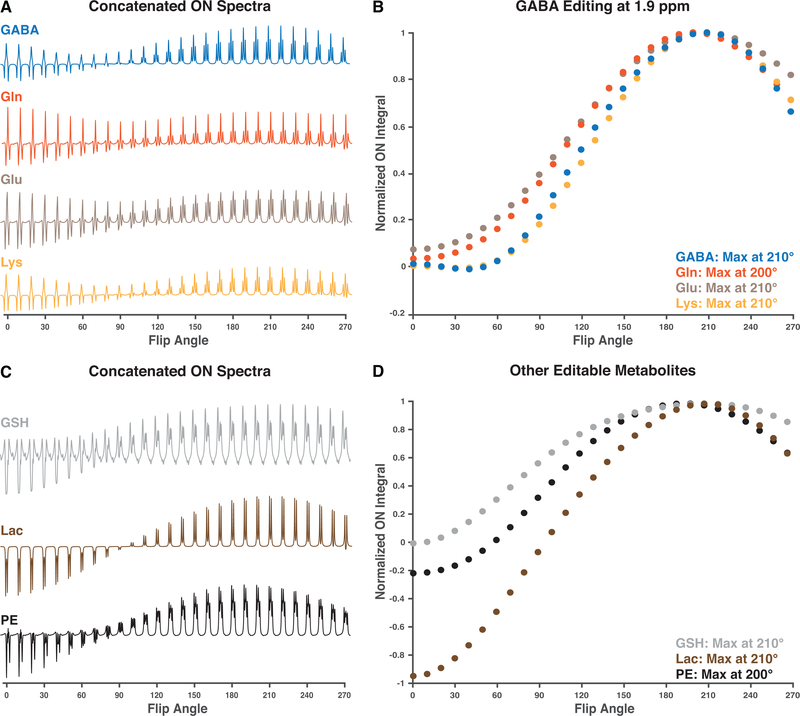
Prior understanding suggests that the flip-angle dependence of editing will be a sinusoidal function with a minimum at 0° and the first maximum at 180°. Simulated multiplets for all spin systems considered are shown in Figure 1A and Figure 1C. Both the targeted GABA spin system and the co-edited signals of Glu and Lys show maximal signal at a flip angle of 210°. Gln signal is maximal at 200° flip angle, as shown in Figure 1B. Lac and GSH show maximal signal at a flip angle of 210°, while PE signal is maximal at 200°, as shown in Figure 1D.
Figure 1:
Results from the simulations. A. Concatenated edit-ON spectra for GABA and co-edited metabolites. B. The normalized edit-ON integrals computed for GABA, and the co-edited metabolites Gln, Glu and Lys, simulated for GABA editing with the editing pulse at 1.9 ppm. C. Concatenated edit-ON spectra for some other editable metabolites GSH, Lac, and PE. D. The normalized edit-ON integrals computed for the other editable metabolites. For all investigated spin-systems, the maximal integral was achieved for flip angles between 200° and 210°.
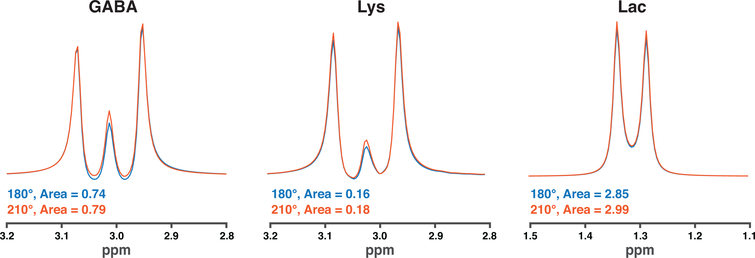
Figure 2 overlays the simulated GABA, Lys and Lac difference multiplets for a 210° editing-pulse flip angle compared to the conventional 180° flip angle. A 210° flip angle resulted in integral areas that were 0.79, 0.18, and 2.99, while the resulting integral areas using a 180° flip angle were 0.74, 0.16, and 2.85 for GABA, Lys and Lac, respectively. This corresponded to 7.0%, 7.6%, and 4.9% increase for GABA, Lys, and Lac, respectively.
Figure 2:
Simulated difference spectra computed for GABA, lysine, and lactate, comparing an edit flip angle of 180° to 210°. The calculated integral areas were larger using a 210° flip angle.
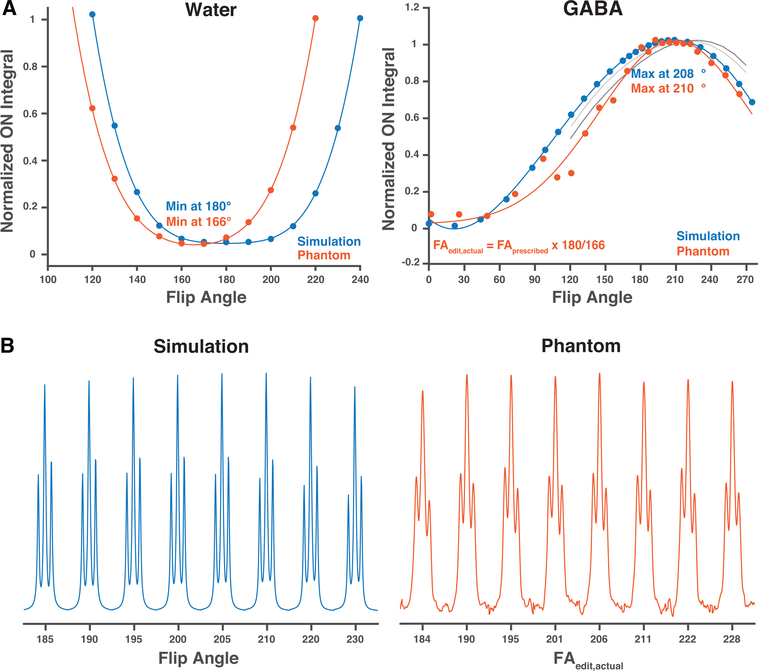
Prior understanding would predict optimal editing and optimal on-resonance water suppression at a flip angle of 180°. These simulations show clear divergence – whereas optimal water suppression occurs at a flip angle of 180 degrees as expected, and as shown in Figure 3, optimal editing occurs at a higher flip angle. This in itself is strong evidence that the effect under discussion is not simply an artefact for poor pulse flip-angle specification.
Figure 3:
Phantom measurements compared to the simulations. A. The normalized edit-ON integrals and fitted curves for the water calibration experiment and the GABA measurements adjusted based on the water calibration. B. The resulting concatenated GABA edit-ON spectra for the simulations and GABA phantom data. The actual editing flip angle was calculated using the results from the water calibration experiment. The gray lines plot the results of confirmatory simulations, for different editing pulse shapes and sequence timings, performed in FID-A (light) and Vespa-Simulation (dark), illustrating maximal edit-ON integral above flip angle 180°.
Phantom Measurements
Phantom results are compared to the simulations in Figure 3. The model fitted to the water-saturation calibration experiment resulted in strongest water suppression for a 166° flip angle, while the model fitted to the simulated data resulted in strongest water suppression for a 180° flip angle. For the GABA-edited phantom data, the model indicated maximal signal for a prescribed flip angle of 194°, while the model fitted to the simulated data generated maximal signal for a 208° flip angle. After applying the flip-angle calibration from the water saturation experiment, the maximal actual flip angle was shown to be 208°, in close agreement with simulations. This experimental design, using water suppression as an internal calibration, again demonstrates that water suppression behavior diverges from the editing behavior.
In Vivo Results
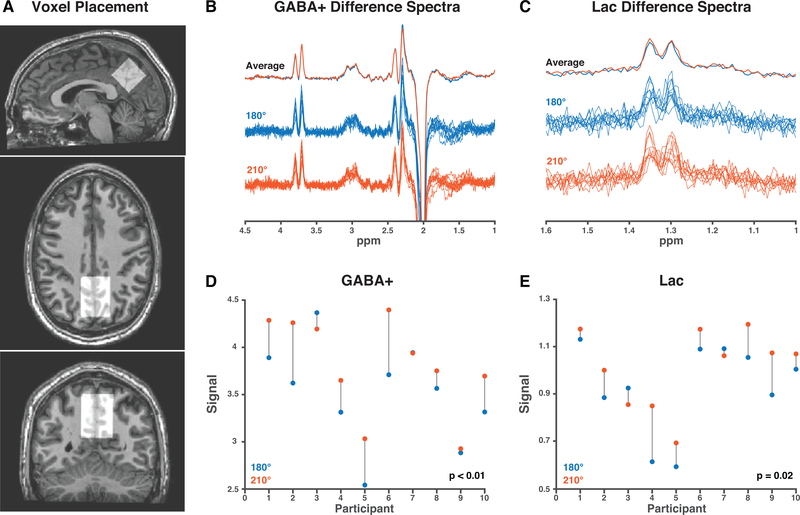
A summary of the in vivo data is shown in Figure 4. Comparing data acquired with an editing-pulse flip angle of 210° to 180°, a small increase can be seen in averaged GABA+ and Lac signal. The modeled signals were higher for 8/10 datasets using the 210° flip angle for both GABA+ and Lac. The ratio of the signal acquired at the two flip angles (S210/S180) is 1.09 ± 0.08 and 1.11 ± 0.13 for GABA+ and Lac, respectively, and the paired t-tests (comparing S210 and S180) were significant for both GABA+ (p < 0.01) and Lac (p = 0.02).
Figure 4:
In vivo comparison of 180° and 210° flip angle. A. The voxel (30 × 30 × 30 mm3) was placed in the medial parietal lobe. B. GABA+-edited difference spectra. C. Ladc-edited difference spectra. D. The water-referenced GABA+ signal is higher for 210° FA than 180° FA in 8/10 participants. E. The water-referenced Lac signals is also higher in 8/10 participants.
Discussion
Edited MRS allows the selective quantification of low-concentration metabolites. It is an SNR-limited technique, requiring larger voxels and longer experiments than would ideally be chosen. It is therefore critical to investigate and optimize all aspects of the experiment – even relatively modest signal improvements can meaningfully reduce scan-time because of the square-root relationship between scan time and SNR.
Simulations clearly indicate that the edited signal is larger at a flip angle of 210° than 180°. This is a very surprising result. Simulations conducted with ideal editing pulses (perfect inversion of only the passive spins) have the expected optimal flip angle at 180°, so the effect relates to some non-ideality of pulses. One potential mechanism that we considered is that increasing the flip-angle of the selective editing pulse might result in fuller inversion of the complete target multiplet, but this is not the case – all lines of the target multiplet are less efficiently inverted with the increased flip angle. Comparing the simulated propagators for shaped pulses (at flip angles 180° and 210°) to the ideal propagator (perfect inversion of editing-target spins and no effect on the other spins), it can be seen that they differ not only in their treatment of the editing-target spins (i.e. real pulses have less-than-ideal inversion) but also in their treatment of the detected spins. Thus, we suggest that the increased flip angle counteracts the unintended effect of the editing pulse on the detected spins, resulting in a net increase in edited signal.
Acquiring phantom experiments to validate a set of simulations for flip-angle dependence is challenging, because the local amplitude of B1 fields are themselves poorly defined, even after pre-scan calibration. In this work, we used the saturating effect of on-resonance pulses to calibrate the editing pulse flip-angle. The measured optimal water suppression was achieved for a 166° prescribed flip angle, indicating an 8% adjustment to the prescan calibration. Agreement between re-calibrated phantom data and simulations is relatively good, but not perfect, and certainly much better after applying this calibration than before. This further emphasizes the importance of accurate, localized prescan B1 calibration, although it is likely that scanner routines are more effective for in vivo calibrations than for one-liter cylindrical phantoms.
Simulations predict a 7.0% increase in GABA signal and 7.6% increase in Lys signal for the higher editing-pulse flip angle, which agrees relatively well with the mean in vivo GABA+ increase of 8.5%. It is worth noting that GABA and co-edited MM signals increase at very similar rates, and the higher flip angle does not result in a meaningfully larger contribution of MM signal to the GABA+ signal. Simulations for Lac predict a 4.9% increase, while the in vivo measurements indicated 9.3% increase. Since both values represent the ratio of two low-SNR signals, this discrepancy is not overly concerning.
It can be seen both in the mean difference spectra and the modeled signals that GABA+ and Lac signals were increased by using an editing-pulse flip angle of 210° rather than 180°. While this difference was small, the effect was statistically significant and seen in 8/10 participants. It is interesting to note that the same two participants (3 and 7, shown in Figure 4) showed the opposite effect for both GABA+ and Lac, potentially due to an over-calibration of the RF in those subjects; B1 calibration was not repeated between GABA+ and Lac experiments.
In conclusion, we have demonstrated that increasing the flip angle of editing pulses in the MEGA-PRESS experiment from 180° to 210° increases the resulting GABA+ and lactate difference signals by about 9% in vivo. This observation was supported by simulations and phantom measurements. Although modest, such an increase may afford 16% scan time reduction with constant SNR - a meaningful impact, and the broader project of point-by-point optimization yields compounding benefits.
Acknowledgements
This work was supported by NIH grants R01 EB016089, R01 EB023963, R00 AG062230, K99 DA051315, P41 EB015909, S10 OD021648, and P41 EB031771.
References
- 1.Harris AD, Saleh MG, Edden RAE. Edited 1H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn Reson Med. 2017;77(4):1377–1389. doi: 10.1002/mrm.26619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi I-Y, Andronesi OC, Barker P, et al. Spectral editing in (1) H magnetic resonance spectroscopy: Experts’ consensus recommendations. NMR Biomed. 2021;34(5):e4411. doi: 10.1002/nbm.4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikkelsen M, Barker PB, Bhattacharyya PK, et al. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage. 2017;159:32–45. doi: 10.1016/j.neuroimage.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikkelsen M, Rimbault DL, Barker PB, et al. Big GABA II: Water-referenced edited MR spectroscopy at 25 research sites. Neuroimage. 2019;191:537–548. doi: 10.1016/j.neuroimage.2019.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edden RA, Harris AD, Murphy K, et al. Edited MRS is sensitive to changes in lactate concentration during inspiratory hypoxia. J Magn Reson Imaging. 2010;32(2):320–325. doi: 10.1002/jmri.22233 [DOI] [PubMed] [Google Scholar]
- 6.Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90(12):5662–5666. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8516315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mescher M, Tannus A, Johnson MO, Garwood M. Solvent Suppression Using Selective Echo Dephasing. J Magn Reson Ser A. 1996;123(2):226–229. doi: 10.1006/JMRA.1996.0242 [DOI] [Google Scholar]
- 8.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272. doi: [pii] [DOI] [PubMed] [Google Scholar]
- 9.Bottomley PA. Spatial Localization in NMR Spectroscopy in Vivo. Ann N Y Acad Sci. 1987;508(1):333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen M, Loo RS, Puts NAJ, Edden RAE, Harris AD. Designing GABA-edited magnetic resonance spectroscopy studies: Considerations of scan duration, signal-to-noise ratio and sample size. J Neurosci Methods. 2018;303:86–94. doi: 10.1016/j.jneumeth.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan KL, Puts NAJ, Snoussi K, Harris AD, Barker PB, Edden RAE. Echo time optimization for J-difference editing of glutathione at 3T. Magnetic Resonance in Medicine. 2017:498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prisciandaro JJ, Mikkelsen M, Saleh MG, Edden RAE. An evaluation of the reproducibility of 1H-MRS GABA and GSH levels acquired in healthy volunteers with J-difference editing sequences at varying echo times. Magn Reson Imaging. 2020;65:109–113. doi: 10.1016/j.mri.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. 2008;21(1):22–32. doi: 10.1002/nbm.1150 [DOI] [PubMed] [Google Scholar]
- 14.Edden RAE, Barker PB. Spatial effects in the detection of gamma-aminobutyric acid: Improved sensitivity at high fields using inner volume saturation. Magn Reson Med. 2007;58(6):1276–1282. doi: 10.1002/mrm.21383 [DOI] [PubMed] [Google Scholar]
- 15.Puts NAJ, Schär M, Zhu H, Barker PB, Edden RAE. B1-dependence of single-voxel MRS sequences: STEAM, PRESS and MEGA-PRESS. In: Proc. Intl. Soc. Magn. Reson. Med.; 2012:4460. [Google Scholar]
- 16.Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. 1998;133(2):243–254. doi: 10.1006/jmre.1998.1458 [DOI] [PubMed] [Google Scholar]
- 17.Deelchand DK, Marjańska M, Henry P-G, Terpstra M. MEGA-PRESS of GABA+: Influences of acquisition parameters. NMR Biomed. 2019;n/a(n/a):e4199. doi: 10.1002/nbm.4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)—An open source, MATLAB-based toolkit. Magn Reson Med. 2017;77(1):23–33. doi: 10.1002/mrm.26091 [DOI] [PubMed] [Google Scholar]
- 19.Saleh MG, Rimbault D, Mikkelsen M, et al. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. Neuroimage. 2019;189:425–431. doi: 10.1016/j.neuroimage.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tkáč I, Starčuk Z, Choi I-Y, Gruetter R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn Reson Med. 1999;41(4):649–656. doi: [DOI] [PubMed] [Google Scholar]
- 21.Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans CJ. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid–edited MR spectroscopy spectra. J Magn Reson imaging JMRI. 2014;40(6):1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oeltzschner G, Zöllner HJ, Hui SCN, et al. Osprey: Open-Source Processing, Reconstruction & Estimation of Magnetic Resonance Spectroscopy Data. J Neurosci Methods. 2020;September 1(343):108827. doi: 10.1016/j.jneumeth.2020.108827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikkelsen M, Tapper S, Near J, Mostofsky SH, Puts NAJ, Edden RAE. Correcting frequency and phase offsets in MRS data using robust spectral registration. NMR Biomed. 2020. doi: 10.1002/nbm.4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murdoch, James B, Dydak U. Modeling MEGA-PRESS Macromolecules for a Better Grasp of GABA. Proc 19th Annu Meet ISMRM, Montr Canada. 2011;Number 139. [Google Scholar]
- 25.Zöllner HJ, Tapper S, Hui SCN, Barker PB, Edden RAE, Oeltzschner G. Comparison of linear combination modeling strategies for GABA-edited MRS at 3T. bioRxiv. January 2021:2021.05.26.445817. doi: 10.1101/2021.05.26.445817 [DOI] [Google Scholar]