Abstract
Objective:
To study whether fluorescence lifetime imaging microscopy (FLIM) detects differences in metabolic state among cumulus. cell samples and if this metabolic state is associated with patient age, BMI and/or AMH, and maturity of the oocyte.
Design:
Prospective observational study.
Setting:
Academic laboratory.
Patients/Animals:
Cumulus cell (CC) clusters from cumulus-oocyte complexes were collected after oocyte retrieval and vitrified.
Interventions:
CC metabolism was assessed using FLIM to measure autofluorescence of NAD(P)H and FAD+, endogenous coenzymes essential for cellular respiration and glycolysis. Patient age, BMI, and AMH, and the maturity of corresponding oocytes were recorded.
Main Outcome measure(s):
Quantitative information from FLIM was obtained regarding: metabolite concentrations from fluorescence intensity; and metabolite enzyme engagement from fluorescence lifetimes. Associations were investigated between each FLIM parameter and oocyte maturity, patient age, BMI and AMH level. Variance between CC clusters within and between patients.
Results:
Of 619 CC clusters from 193 patients, 90 were associated with immature oocytes, 505 with metaphase II oocytes. FLIM enabled quantitative measurements of metabolic state of CC clusters. These parameters were significantly correlated with patient age and AMH, independently, but not BMI. CC NAD(P)H FLIM parameters and Redox Ratio were significantly associated with maturity of the enclosed oocyte.
Conclusions:
FLIM detects variations in metabolic state of CC, showing a greater variance among clusters from each patient than between patients. FLIM can detect CC metabolic associations with patient age and AMH and detects variations between mature and immature oocytes, suggesting the potential utility of this technique to help identify superior oocytes.
Keywords: Metabolism, cumulus cells, cumulus-oocyte complexes, human oocytes, fluorescence lifetime imaging microscopy, maturity
Capsule:
Metabolic imaging using fluorescence lifetime imaging microscopy (FLIM) sensitively detects variations in metabolic state of human cumulus cells. These variations are associated with maturity of the enclosed oocyte and patient clinical factors.
Introduction
Extensive evidence supports the role of bidirectional crosstalk between the oocyte and its surrounding cumulus mass for successful oocyte development (1,2). Oocyte growth is dependent on the metabolic cooperation between the oocyte and its cumulus cells (CCs), which is facilitated by the exchange of molecules through gap junctions and paracrine signals (3,4). This interaction is bidirectional: CCs provide growth factors, lipids and metabolites for oocyte maturation and development (5,6,9); oocyte secreted factors enable CCs differentiation and mucification (7,8).
Oocyte developmental potential depends on oocyte quality (9), which in turn, is reflected in nuclear and cytoplasmic maturity, both of which are essential for successful fertilization and embryo development (10). In addition, CCs help maintain the oocyte in meiotic arrest at prophase I (1). This communication is essential for the resumption of meiosis in response to the luteinizing hormone surge (11). Conversely, the meiotic status can influence the metabolism not only of the oocyte, but also of the surrounding cumulus mass. Pyruvate consumption by cumulus oocyte complexes (COCs) has been positively associated with oocyte nuclear maturation (7,12). Moreover, mitochondrial dysfunction and mitochondrial DNA (mtDNA) copy number of CCs might be directly related to oocyte maturity (13-15).
Because of the intimate interactions between the oocyte and its surrounding CC mass, measurements of the metabolic state of CCs provide a promising means to determine the developmental competence of the associated oocyte (16-19). Since cumulus masses are routinely removed for intracytoplasmic sperm injection (ICSI), and are frequently trimmed before conventional insemination, these otherwise discarded cells have the potential for use in a non-invasive assay of oocyte developmental competence (20-25). However, reliable biomarkers in CCs have yet to be identified (23).
A number of recent studies have focused on measurements of mtDNA in CCs, the results of which have been inconclusive (17,18,26). Some studies found that mtDNA copy number does not seem to be associated with oocyte maturity or fertilization (18), while other studies observed a correlation with oocyte fate (14,15). Additional studies have used mitochondrial dyes (13,27) to study mitochondrial activity in CCs. Establishing quantitative tools to measure the metabolic state of single cumulus complexes could be helpful for developing an improved understanding of the metabolic relationship between CCs and the enclosed oocyte and might enable non-invasive approaches to evaluate oocyte maturity and overall quality.
Nicotinamide adenine dinucleotide (NADH), Nicotinamide adenine phosphate dinucleotide (NADPH) and flavine adenine dinucleotide (FAD+), are central metabolic coenzymes that are naturally fluorescent (28). These molecules are electron carriers that have essential roles in metabolic pathways, such as the electron transport chain and glycolysis, hence they are ideal biomarkers of cellular metabolic state (29,30). The fluorescence spectra NADH and NADPH are almost indistinguishable (31), therefore the combined fluorescence of NADH and NADPH is often referred to as the NAD(P)H signal. Fluorescence lifetime imaging microscopy (FLIM) of NAD(P)H and FAD+ provides a means to measure the fluorescence intensity and fluorescence lifetime of these molecules. The fluorescence intensity is related to the concentrations of the coenzymes, while the fluorescence lifetime depends on the local environment of the coenzyme, varying drastically if they are bound to an enzyme or free (32).
In the present study, we used metabolic imaging via FLIM to measure metabolic state of CCs in vitro. We evaluated the sensitivity of this non-invasive metabolic imaging to investigate the associations among clinically relevant patient factors, such as maternal age, BMI and anti-mullerian hormone (AMH) level, and the metabolic state of the cumulus masses. And finally, we explored the extent to which quantitative measures of the metabolic state of CC clusters are associated with the maturity of the oocyte they enclose.
Material and Methods
Study design
Cumulus cell clusters were donated for research under consent by patients undergoing ART treatments after institutional review board (IRB) approval of the study protocol by Partners Healthcare IRB (Partners IRB # 2014P000874).
After collection, vitrification and subsequent thawing, the metabolic state of each CC cluster was assessed using metabolic imaging by FLIM. Patient age, BMI and AMH within 6 months of treatment, Follicle-stimulating hormone (FSH) and luteinizing hormone (LH) levels, the number of oocytes retrieved, and the percentage of mature oocytes (number mature oocytes / total of oocytes retrieved) were recorded. The meiotic status of each oocyte was retrospectively tied to its corresponding CC cluster.
Sample preparation
COCs were retrieved, using standard clinical protocols following ovarian stimulation and ovulatory trigger, isolated and set up in individual drops of 25pl of culture medium (Global® total, Life Global Group, Cooper Surgical; Guilford, CT) overlaid with oil (Vitrolife Ovoil, Göteborg, Sweden), ) and incubated for 1-4h. CC masses of each oocyte were trimmed, and trimmed CCs were rinsed and vitrified following the Irvine vitrification protocol (90133-SO - Vit Kit-Freeze, FUJIFILM Irvine Scientific, USA). Each CC mass was then thawed, following the Irvine thawing protocol (90137-SO - Vit Kit-Thaw, FUJIFILM Irvine Scientific), and their hyaluronan matrix was disaggregated following exposure to hyaluronidase (80 IU/mL of Hyaluronidase, FUJIFILM Irvine Scientific). The disaggregated CCs were centrifuged and the pellet, which consisted of a cluster of CCs, was collected. The cluster was then placed in 5μL droplets of culture media covered with oil in a glass bottom dish (MatTek P35G-0.170-14-C, USA) designed for imaging. CC clusters were imaged in an on-stage incubation system (Ibidi GmbH, Martinsried, Germany) to maintain culture environment conditions of 37°C and 5% CO2, 5% O2, balanced with N2.
Staining protocols
For TMRM (tetramethylrhodamine, methyl ester) experiments, CC clusters were incubated with 5nM of TMRM (Thermo Fisher, USA) for 10 minutes. For DNA staining experiments, CCs were stained with 1:400 of Syto 9 (Thermo Fisher, USA) for 60 minutes. The samples were then washed 3 times through culture media (Global® total, Life Global Group, Cooper Surgical; Guilford, CT) and transferred to a glass bottom dish for imaging.
Metabolic Imaging using Fluorescence Lifetime Imaging Microscopy (FLIM)
FLIM measurements were performed on a Nikon Eclipse Ti microscope using two-photon excitation from a Ti:Sapphire pulsed laser (Mai-Tai, Spectral-Physics) with a 80MHz repetition rate and 70fs pulse width, a Galvano scanner (DCS-120, Becker and Hickl, Germany), TCSPC module (SPC-150, Becker and Hickl, Germany) and a hybrid single photon counting detector (HPM-100-40, Becker and Hickl, Germany). The wavelength of excitation was set to 750nm for NAD(P)H and 890nm for FAD+, with powers measured at the objective of 3mW for NAD(P)H and 16.8mW for FAD+. Optical bandpass filters were positioned in a filter wheel in front of the detector – 460/50 nm for NAD(P)H and 550/88nm for FAD+ and a 650nm short pass filter (Chroma technologies) was mounted on the detector. Imaging was performed with a 40X Nikon objective with 1.25 numerical aperture (NA) (CFI Apo 40 × WI, NA 1.25, Nikon). Each NAD(P)H and FAD+ FLIM image was acquired with 60 seconds of integration time. Objective piezo stage (P-725, Physik Instruments) and motorized stage (ProScan II, Prior Scientific) were used to perform multidimensional acquisition. Ten images were acquired per cumulus cluster varying x, y, and z axis. All the electronics were controlled by SPCM software (Becker and Hickl, Germany) and custom LabVIEW software.
Data Analysis
Data was analyzed using customized MATLAB (version R2019b, MathWorks, USA) code. NAD(P)H and FAD+ FLIM intensity images were subject to an intensity-based threshold to classify pixels as being in cells or background. For each cell segment, the photon arrival time histogram was modeled as a bi-exponential decay:
| (1) |
Where, A is a normalization factor, B is the background, τ1 is the short lifetime, τ2 is the long lifetime and F is the fraction of molecule with long lifetime (fraction engaged with enzymes for NAD(P)H and unengaged for FAD+). This function was convolved with a measured instrument response function, and a least square fit was used to determine the parameters. The fluorescence intensity was calculated for each cumulus cell cluster as the number of photons divided by the area of the cluster. Thus, a single FLIM measurement produced 9 parameters, 4 for NAD(P)H and 4 for FAD+ and 1 Redox Ratio (NAD(P)H fluorescence intensity / FAD+ fluorescence intensity) to characterize the metabolic state of CC clusters.
Statistical Analysis
All statistical tests were performed using Stata Statistical Software (version 16.0, LLC Stata Corp, Texas, USA) and R Studio (Version 1.3.959, R Foundation for Statistical Computing, Vienna, Austria). Our data was structured hierarchically, 1- 10 images for each cluster (i) and 1- 4 clusters (j) per patient (k). We thus used multilevel models, on the standardized FLIM parameters , to analyze this structured data. Data points that lay either 40% above or below the mean were considered outliers and removed from the analysis. We incorporated additional predictors (age, BMI, AMH levels, and oocyte maturity) for certain analysis, resulting in the multilevel model:
where β0 corresponds to the intercept; β1 is the slope; Patient Factor is age, BMI, AMH levels, or oocyte maturity (if considered); c0,k is the patient level random error; b0,jk is the cluster level random error; eijk is the image level random error (33). This modelling encodes information on the variance associated with each level: patient, CC clusters within patients and images within CC cluster. To correct for multiple comparisons, we used Benjamini – Hochberg’s false discovery rate (FDR), at a q value of 0.05. FDR p-values of <0.05 were considered significant.
Results
NAD(P)H autofluorescence of CCs predominantly originates from mitochondria
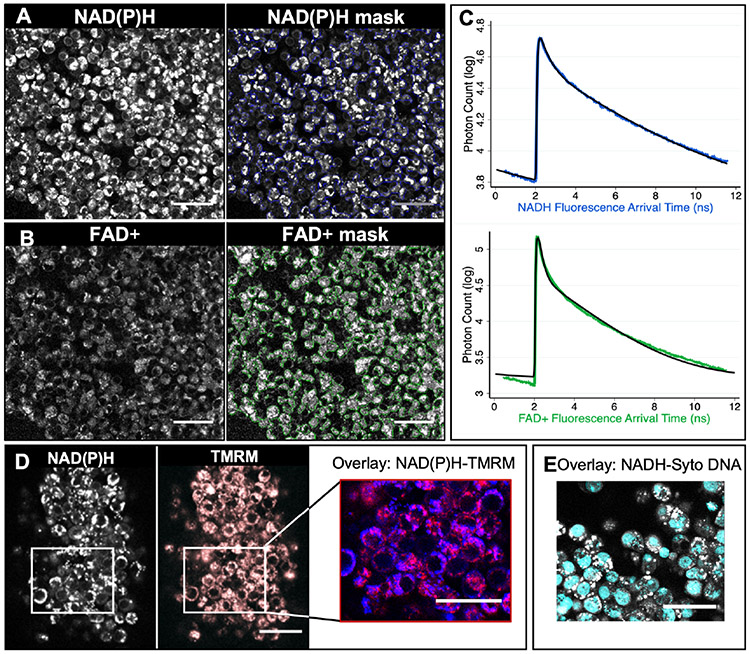
We first sought to explore the subcellular localization of NAD(P)H and FAD+ autofluorescence. After centrifuging the dissociated CCs from a cluster, the resulting pellet was moved to an onstage incubation system and imaged for NAD(P)H and FAD+ autofluorescence (Figure 1A and B, respectively) with FLIM. Each acquired image was 220 μm by 220 μm, which contains tens to hundreds of CCs (Figure 1A and B, left panels), but represented only a small region of an entire CC pellet. To quantitatively analyse the CC metabolic state in the imaged region, we used an intensity-based threshold to create masks (Figure 1A and B, right panels). We grouped all photons from each of the masked regions to create histograms of the fluorescence decay of NAD(P)H (Figure 1C, upper, blue) and FAD+ (Figure 1C, lower, green). We then fit these histograms using two-exponential decay models (equation 1) (Figure 1C, black lines), thereby obtaining a total of 6 parameters: short and long lifetimes, and the fraction of engaged molecules, for both NAD(P)H and FAD+. In addition, we computed the NAD(P)H intensity, FAD+ intensity, and the Redox Ratio, providing a total of 9 quantitative metabolic parameters per each cluster.
Figure 1. Fluorescence Lifetime Imaging Microscopy (FLIM) imaging of cumulus cells.
(A) FLIM imaging of the autofluorescence of NAD(P)H (A) of a cumulus cell mass, and FAD+(B). An intensity-based thresholding algorithm was used to create masks of CCs to integrate the fluorescence signal of NAD(P)H or FAD+ (right panels). All photon arrival times from that mask were combined to create a fluorescence decay curve for each fluorophore (NAD(P)H in blue, C top and FAD+ in green, C bottom). These curves were fit to two-exponential models (C, black curves). This approach provides 9 quantitative parameters for characterizing the metabolic state of CCs: fluorescence intensity, long and short lifetime and the fraction engaged to enzyme for molecules NAD(P)H and FAD+ and the Redox Ratio. (D) NAD(P)H autofluorescence (left) image showings a co-localization with mitochondria dye TMRM image (in red, overlay in purple). (E) Overlay image of NAD(P)H autofluorescence in grey and DNA dye Syto in Cyan. Scale bars, 40 μm.
Whereas FAD+ is highly enriched in mitochondria (29), NADH and NADPH participate in different pathways in mitochondria, the nucleus, and the cytoplasm (31,32,34). To better understand the source of the NAD(P)H signal in CCs, we compared NAD(P)H images to images with dyes for either mitochondria or DNA. We first stained a cumulus mass with TMRM, a dye known to localize to active mitochondria (Figure 1C). We segmented the mitochondria from the TMRM images using a machine-learning based segmentation software (Illastik, version 1.0,(35)), and, by comparing the overlap of the segmented TMRM and NAD(P)H images, determined that 73±5% of NAD(P)H photons come from mitochondria. We next stained the cumulus mass with Syto 9, a DNA dye (Figure 1E) and by comparing the overlap with NAD(P)H signal, we found that 16±1% of the NAD(P)H photons come from the nucleus. Hence, the remaining photons come from the cytoplasm. Thus, while the acquired NAD(P)H signal has contributions from all compartments, the majority of the signal comes from mitochondria.
CC processing for FLIM analyses
We next explored the extent to which variations in sample preparation procedures impacted the results of FLIM measurements of CC clusters. CCs that were imaged fresh gave indistinguishable FLIM parameters as compared to CCs that were first vitrified and then thawed (Figure S1). Since vitrified samples are more convenient to work with than fresh samples, we opted to vitrify CCs in subsequent experiments. We next tested the impact of hyaluronidase on FLIM parameters of CCs. CCs that were not exposed to hyaluronidase, and thus maintained their hyaluronan matrix, gave indistinguishable fraction engaged and lifetimes (Figure S2) compared to those that were dispersed with hyaluronidase. However, CCs not exposed to hyaluronidase did give significantly different intensities and Redox Ratio, which was likely caused by difficulties in successfully segmenting these far less dense cells. Thus, we proceeded to use vitrified CCs exposed to hyaluronidase in subsequent experiments.
We then explored how the location of the CCs within a cumulus cluster impacted the results of FLIM measurements. FLIM parameters of CCs that were trimmed from closer to the oocytes did not show significant differences from those of CCs that were trimmed further from the oocyte (Figure S3). Since the further the CCs are trimmed from the oocyte, the less likely any risk of damage will be introduced to the oocyte, we trimmed CCs further away from the oocytes for all subsequent sample collections. Taken together, these experiments led us to use CCs trimmed further away from the oocyte, which were vitrified and then exposed to hyaluronidase before imaging.
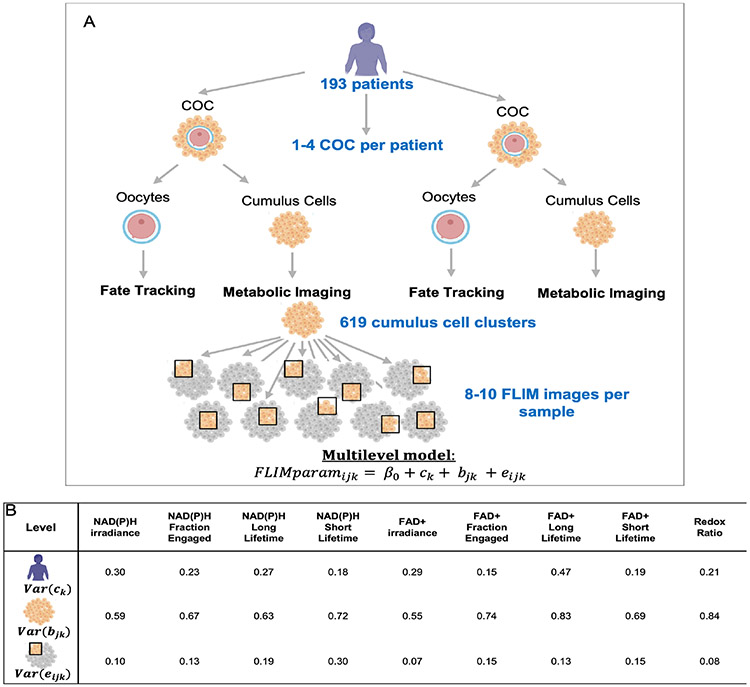
Metabolic variance between CCs clusters exceeds variance between patients and between images
A total of 619 CC samples were collected from 193 patients, 90 of which were associated with immature oocytes, 58 at the germinal vesicle (GV) stage and 32 at metaphase I (n=32), 505 were associated with mature oocytes, and 24 were associated with either degenerated or abnormal oocytes. We acquired 8-10 non-overlapping FLIM images at different locations throughout each CC pellet (Figure 2A). We used a multilevel model (33,36) to analyze this hierarchically organized data, and decomposed the variance in the FLIM parameters into three levels: variance associated with differences between patients, variance associated with differences between CC clusters from the same patient, and variance associated with differences between individual images from the same CC cluster. For all FLIM parameters, the variance associated with differences between CC clusters from one patient was substantially larger than the variance associated with differences between individual images, or the variance associated with differences among patients (Figure 2B). The large variance in FLIM parameters associated with CC clusters indicates that different clusters from the same patient have significantly different metabolic states. The small variance in FLIM parameters associated with differences between individual images suggests a relative homogeneity of metabolic state within a CC cluster and demonstrates the high robustness and reproducibility of these measurements. Despite being small, the variance in FLIM parameters associated with differences between patients indicates that patient specific factors impact the metabolism of their CCs.
Figure 2. Metabolic variance between patients, between cumulus clusters and between images.
(A) Diagram of the experimental design of this study. 1-4 COCs were collected per patient. CC masses were trimmed from the COCs and imaged using metabolic imaging. The fate of the corresponding oocyte was monitored. 8-10 FLIM metabolic images at different locations throughout the cluster were taken for each cumulus cluster. A multilevel model was applied to analyze this structured data. (B) The multilevel model encodes information on the variance of each FLIM parameter between patients, between CC clusters from the same patient, and between images from the same CC cluster. Numbers in the table correspond to the variance of each FLIM parameter associated with each level: patients, CC clusters and images.
CC metabolic parameters are associated with maternal clinical factors
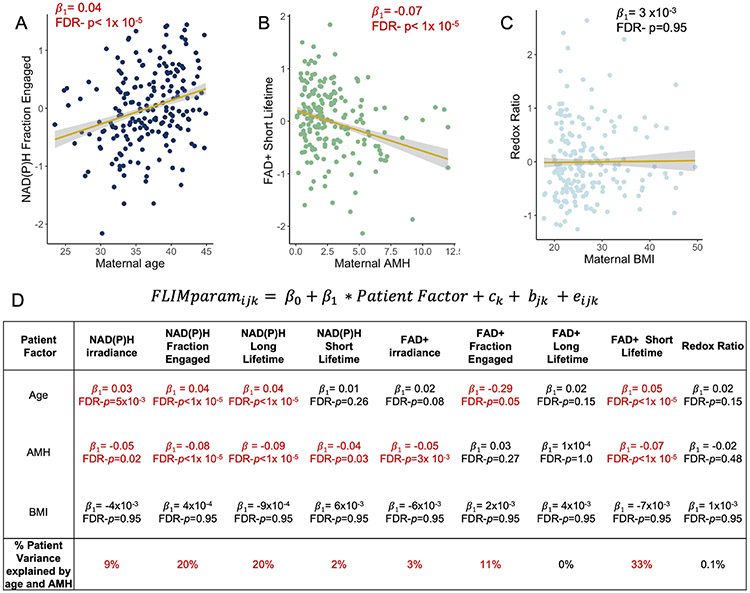
We next explored the extent to which maternal age, AMH levels and BMI independently account for the variance in FLIM parameters associated with differences between patients. These factors varied greatly across the 193 patients we studied: maternal age ranged from 23.5 to 45 years, with a mean of 36.7 years; 58.0% of the patients were less or equal than 38 years old and 42.0% were more than 38 years old. AMH levels ranged from 0.1 to 12 ng/mL, with a mean of 2.9 ng/mL; BMI ranged from 17.8 to 49.5 kg/m2, with a mean of 26.7 kg/m2. 49.8% of the patients had a BMI lower or equal than 24.9, 38.8% had a BMI between 25 and 29.9, and 11.4% had a BMI greater or equal than 30. Plotting FLIM parameters versus maternal age and AMH levels revealed apparent associations (Figure 3A, 3B, respectively). Incorporating these patient factors into the multilevel model allowed us to determine that maternal age was significantly correlated with NAD(P)H irradiance, fraction engaged and long lifetime, and FAD+ fraction engaged and short lifetime; while AMH levels were significantly correlated with NAD(P)H irradiance, fraction engaged, and both short and long lifetime, and FAD+ irradiance and short lifetime (Figure 3D). The correlations with maternal age remained after controlling for AMH levels, and correlations with AMH levels remained after controlling for maternal age. In contrast, we found no significant correlations between any FLIM parameters and maternal BMI (Figure 3C, 3D) or when stratifying the BMI by the three groups. However, the percentage of variance in CC FLIM measurements between patients was not entirely explained by maternal age and AMH levels, suggesting that other patient specific factors impact the metabolic state of their CC clusters (Figure 3D). Consistent with this, we also found that levels of both FSH and LH administered were significantly correlated with FLIM parameters (FDR p <0.05). However, these associations were dependent on both maternal age and AMH levels suggesting that the amount of administered gonadotropin might not directly cause changes in FLIM parameters. Further studies will be necessary to disentangle the interactions between patient specific factors that impact CC FLIM parameters.
Figure 3. CC FLIM metabolic parameters are associated with patient clinically relevant factors.
Normalized NAD(P)H fraction engaged is significantly associated with maternal age (A), FAD+ short lifetime is significantly associated with maternal AMH levels (B) and Redox Ratio is not significantly associated with maternal BMI (C). Each dot corresponds to the mean of each patient. (D) A multilevel model was used to determine the significance (i.e., p values) extent of correlation (i.e., β1-coefficients) of CC FLIM parameters with maternal age, BMI and AMH levels. Numbers in red are statically significant after correcting for multiple comparisons using Benjamini – Hochberg’s false discovery rate (FDR p values<0.05). The last row shows the percentage of the CC FLIM parameters variance between patients that is explained by both age and AMH levels.
CC FLIM parameters are associated with the maturity of the enclosed oocyte
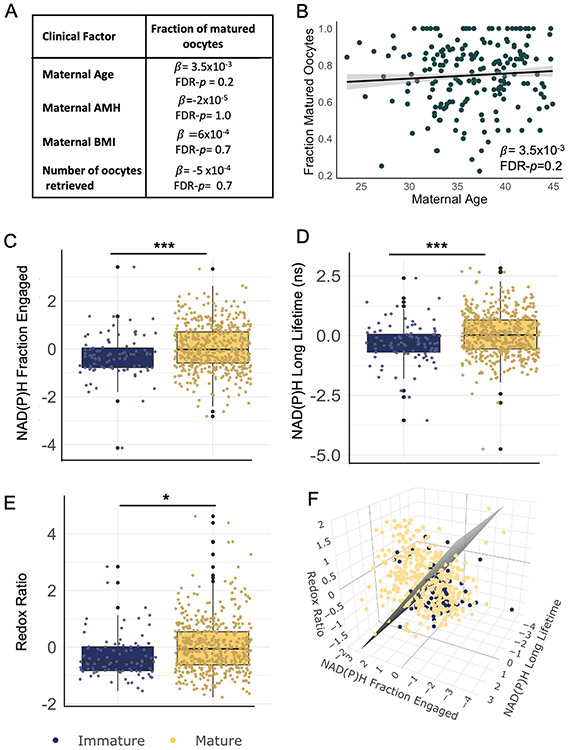
We next sought to determine if the metabolic state of CC clusters was correlated with the maturity of the oocyte with which they were associated. The percentage of mature oocytes (of total oocytes retrieved) from a patient was not associated with maternal age (p = 0.20), or BMI (p = 1.0), or AMH (p = 0.74), or the total number of oocytes retrieved from that patient (p= 0.69) (Figure 3A and b). Comparison of CC FLIM parameters from mature oocytes with those from immature oocytes revealed several interesting contrasts: NAD(P)H irradiance (FDR p=0.02), NAD(P)H fraction engaged (FDR p< 1x10−5), NAD(P)H long lifetime (FDR p< 1x10−5) and Redox Ratio (FDR p=0.02) were significantly different between mature and immature oocytes (Figure 4 C, D and E). These significant differences were upheld after controlling for maternal age and AMH levels. Plotting these three FLIM parameters for each CC cluster on a three-dimensional graph revealed that while the distributions were significantly different between CCs from mature and immature oocytes, there was also substantial overlap between these two populations (Figure 4E, blue, immature; yellow, mature; grey, support vector machine generated hyperplane that best separates the two populations).
Figure 4. CC FLIM metabolic parameters are associated with the maturity of the enclosed oocyte.
(A) β- coefficients and FDR-p values of the linear regression of maternal age, BMI, AMH levels, and number of oocytes retrieved, with the fraction of matured oocytes (number of matured oocytes / total of oocytes retrieved). There are no significant associations between these patient clinical factors and the fraction of matured oocytes. (B) Fraction of matured oocytes plotted vs maternal age indicates no significant association. However, significant differences were found in CC normalized NAD(P)H irradiance, NAD(P)H fraction bound (C), NAD(P)H long lifetime (D) and Redox Ratio(E) between CCs of immature (n=90, in blue) or mature oocytes (n=505, in yellow). (F) 3D plot of these three FLIM parameters for each CC cluster. In grey, support vector machine was applied to draw a hyperplane that best separates both groups. * signifies FDR- p<0.05, *** signifies FDR- p<0.001.
Discussion
Cumulus cells and oocyte metabolic interconnections are essential for cumulus mass expansion, oocyte maturation and development (25). Mitochondrial dysfunction in CCs affects oocyte growth and quality (37). The effects of CC metabolic state on the corresponding oocyte have attracted much attention in recent years. In view of the importance of CC mitochondrial function for successful oocyte developmental competence (14), CC metabolic state may act as a biological marker of oocyte quality.
In this study, we used FLIM of NAD(P)H and FAD+ to quantitatively determine the metabolic state of CCs. Analysis of 8-10 FLIM images acquired throughout each of 619 CC clusters from 193 patients, revealed that for all FLIM parameters, the variance associated with differences between CCs of different oocytes was larger than the variance associated with differences between patients and between individual images obtained from the same cluster. Additionally, we observed that CC metabolic parameters are associated with maternal clinical factors that are known to be key determinants of oocyte quality. Lastly, we found significant differences in NAD(P)H FLIM parameters and the Redox Ratio between CC clusters from mature and immature oocytes.
By analyzing our hierarchically organized data using multilevel modelling (33,36) we obtained the variances in the FLIM parameters within the three levels: variance associated with differences between patients, variance associated with differences between CC clusters from the same patient, and variance associated with differences between individual images from the same cluster. We found a large variance in all FLIM metabolic parameters between CC clusters, indicating that CCs associated with individual oocytes from the same patient have significantly different metabolic states.
The fact that there are measurable differences between CCs associated with individual oocytes highlight the potential use of this approach to determine oocyte specific characteristics. Additionally, the small variance in FLIM parameters associated with the differences between images of the same CC cluster, indicates the high reproducibility of these measurements. However, this variance was not null, which might either be due to minor errors in the FLIM measurements or a true heterogeneity of metabolic state within a CC cluster. The latter is unlikely as our validation studies showed only minor deviations in FLIM measurements among samples from the same cumulus clusters. The variance in FLIM parameters associated with differences between patients indicates that patient specific factors impact the metabolism of their CCs. Patient specific effects on CC mtDNA measures have been previously reported (18).
Patient factors such as age, BMI and measures of ovarian reserve, such as AMH, have long been associated with oocyte quality (38,39). Here, we found significant associations of CC metabolic state, as measured by FLIM of NAD(P)H and FAD+, and maternal age. The influence of maternal ageing on CC metabolism has been demonstrated in several studies (21,40-42). Both proteomic and genomic analyses of CCs in woman with advanced age differ from those of younger women (40,41). CCs from older woman have been shown to contain higher number of damaged mitochondria and mtDNA alterations (42,43). In addition, we observed a significant association between CC FLIM parameters and AMH levels, even after controlling for maternal age. This observation is consistent with previous results that found a correlation between CC mitochondrial membrane potential and measures of ovarian reserve (13). The levels of gonadotropins administered were significantly correlated with several FLIM parameters. Since these associations depend on both maternal age and AMH levels they are unlikely to be caused by direct interactions. Future studies will be required to clarify the interplay between patient specific factors that impact CC FLIM parameters.
Our results did not indicate an association between CC metabolic state and maternal BMI. This contrasts with a study by Gorshinova et al., where they observed a reduction in active mitochondria with increasing maternal BMI in a group of 13 patients (27). However, only a small portion of the patient level variance in FLIM parameters can be explained by age and AMH levels. This argues that CC metabolic state is also influenced by other patient specific factors, such as the presence of metabolic diseases, genetic predisposition, diet, or smoking. Future studies will disentangle the nature of those additional factors. It has been demonstrated that in mitochondria of mouse oocytes, a simultaneous increase in NAD(P)H intensity and fraction engaged correspond to an increase in metabolic flux through the electron transport chain (44). If the same held for human CCs, this would imply that mitochondrial metabolic fluxes in CCs increase with patient age and decreases with ovarian reserve. However, this conclusion must be treated with caution because of the extensive metabolic differences between CCs and oocytes. Additional experiments will be necessary to definitively determine the biological basis of the trends in FLIM parameters in CCs we observed in this study.
Cumulus cell-oocyte crosstalk is a key determinant for acquisition of oocyte developmental competence and CC maturation (4,7,11,12). Oocyte quality depends on successful nuclear and cytoplasmic maturation (9,10) as well as maternal clinical factors (38,39). The results of our study indicate that the fraction of mature oocytes at retrieval did not correlate with any patient clinical characteristic such as age, BMI or AMH levels, or even the number of oocytes retrieved. However, variations in FLIM metabolic parameters were observed between those CC clusters associated with mature oocytes and those associated with oocytes that were immature. CC and oocyte metabolic communication is essential not only for the resumption of meiosis (1,11), but also for acquisition of developmental competence (7,12). Interestingly, we found that CCs from mature oocytes showed a higher NAD(P)H fraction Bound, NAD(P)H long lifetime and Redox Ratio (NAD(P)H intensity: FAD+ intensity) than cumulus masses from immature oocytes. These findings are in line with previous studies demonstrating a low mitochondrial membrane potential in CCs from immature oocytes compared to those from matured oocytes (13), as well as those demonstrating differences in mtDNA copy number(14,15). We therefore conclude that FLIM imaging can detect metabolic variations of cumulus masses from immature oocytes compared to mature oocytes. However, we also observed large variations in FLIM parameters across CC clusters from mature oocytes and large variations in FLIM parameters across CCs from immature oocytes. These findings highlight the difficulty in developing a prediction model to assess oocyte maturity.
Conclusions
In summary, metabolic imaging via FLIM of human CCs is a promising approach for detecting variations of metabolic state of these cells. FLIM sensitively detects variations in CC metabolism and shows a greater variance in FLIM parameters among oocytes than between patients. CC metabolic parameters are associated with clinically relevant factors such as age and AMH levels, but not with BMI. Moreover, metabolic imaging detects significant differences between CC metabolic state of immature oocytes and those that matured. A greater understanding of the metabolic underpinnings of human cumulus-oocyte interactions could offer new opportunities for improving fertility interventions, in particular in vitro maturation protocols.
Supplementary Material
Supplementary Figure 1. CCs that were imaged fresh show no significant differences in FLIM parameter from CCs that were first vitrified and then thawed (A) Mean FLIM measurements of both fresh and vitrified cumulus clusters (n=9), the FDR p-value of the t-test performed in the ratio between fresh/ vitrified compared to 1. (B) Box plots showing the ratios of each FLIM measurement.
Supplementary Figure 2. CCs that were not exposed to hyaluronidase show significant differences in intensities and redox ratio from those exposed to hyaluronidase (A, left) NAD(P)H autofluorescent intensity image of CCs exposed to hyaluronidase enzyme to dissolve its matrix and (A, right) CCs not exposed with hyaluronidase. (B) Mean of FLIM measurements of CCs exposed and not exposed to hyaluronidase (n=9), the p-values obtained from the comparison of the ratio between CCs not exposed to hyaluronidase / CCs exposed with hyaluronidase enzyme to 1 and FDR p-values. (C) Box plots showing these ratios of each FLIM measurement.
Supplementary Figure 3. CCs closed to the oocyte show no significant differences in FLIM parameters from CCs that were located further from the oocyte (A) Mean FLIM parameter values of both the CCs closer to the oocytes and those further from the oocytes (n=7), the p-value after a t-test performed in the ratio between CCs further from the oocytes / CCs closer to the oocytes compared to 1 and FDR p-values. (C) Box plots showing these ratios of each FLIM measurement.
Acknowledgements
Authors thank Becker and Hickl for contributing a single-photon-counting detector and TCSPC electronics to this research. Authors would also like to acknowledge Brian Leahy for useful advice, as well as all members of the embryology team at Brigham and Women's Hospital for their assistance in the collection of the cumulus samples for this research. Images Created with BioRender.com
Funding
NIH R01HD092550-03
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eppig JJ. Intercommunication between mammalian oocytes and companion somatic cells. Bioessays 1991;13:569–74. [DOI] [PubMed] [Google Scholar]
- 2.Albertini DF, Combelles CM, Benecchi E, Carabatsos MJ. Cellular basis for paracrine regulation of ovarian follicle development. Reproduction 2001;121:647–53. [DOI] [PubMed] [Google Scholar]
- 3.Anderson E, Albertini DF. Gap junctions between the oocyte and companion follicle cells in the mammalian ovary. J Cell Biol 1976;71:680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction 2001;122:829–38. [DOI] [PubMed] [Google Scholar]
- 5.Seli E, Babayev E, Collins SC, Nemeth G, Horvath TL. Minireview: Metabolism of Female Reproduction: Regulatory Mechanisms and Clinical Implications. Mol Endocrinol 2014;28:790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumesic DA, Meldrum DR, Katz-Jaffe MG, Krisher RL, Schoolcraft WB. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertility and Sterility 2015;103:303–16. [DOI] [PubMed] [Google Scholar]
- 7.Sutton ML, Cetica PD, Beconi MT, Kind KL, Gilchrist RB, Thompson JG. Influence of oocyte-secreted factors and culture duration on the metabolic activity of bovine cumulus cell complexes. Reproduction 2003;126:27–34. [DOI] [PubMed] [Google Scholar]
- 8.Gilchrist RB, Lane M, Thompson JG. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update 2008;14:159–77. [DOI] [PubMed] [Google Scholar]
- 9.Keefe D, Kumar M, Kalmbach K. Oocyte competency is the key to embryo potential. Fertil Steril 2015;103:317–22. [DOI] [PubMed] [Google Scholar]
- 10.Watson AJ. Oocyte cytoplasmic maturation: a key mediator of oocyte and embryo developmental competence. J Anim Sci 2007;85:E1–3. [DOI] [PubMed] [Google Scholar]
- 11.Funsho Fagbohun C, Downs SM. Metabolic Coupling and Ligand-Stimulated Meiotic Maturation in the Mouse Oocyte-Cumulus Cell Complex1 Biology of Reproduction 1991;45:851–9. [DOI] [PubMed] [Google Scholar]
- 12.Downs SM, Humpherson PG, Leese HJ. Pyruvate utilization by mouse oocytes is influenced by meiotic status and the cumulus oophorus. Mol Reprod Dev 2002;62:113–23. [DOI] [PubMed] [Google Scholar]
- 13.Anderson SH, Glassner MJ, Melnikov A, Friedman G, Orynbayeva Z. Respirometric reserve capacity of cumulus cell mitochondria correlates with oocyte maturity. J Assist Reprod Genet 2018;35:1821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucret L, Bris C, Seegers V, Goudenège D, Desquiret-Dumas V, Domin-Bernhard M, et al. Deep sequencing shows that oocytes are not prone to accumulate mtDNA heteroplasmic mutations during ovarian ageing. Hum Reprod 2017;32:2101–9. [DOI] [PubMed] [Google Scholar]
- 15.Lan Y, Zhang S, Gong F, Lu C, Lin G, Hu L. The mitochondrial DNA copy number of cumulus granulosa cells may be related to the maturity of oocyte cytoplasm. Hum Reprod 2020;35:1120–9. [DOI] [PubMed] [Google Scholar]
- 16.Fontana J, Martínková S, Petr J, Žalmanová T, Trnka J. Metabolic cooperation in the ovarian follicle. Physiol Res 2020;69:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino M, Tsubamoto H, Sakata K, Oohama N, Hayakawa H, Kojima T, et al. Mitochondrial DNA copy number in cumulus cells is a strong predictor of obtaining good-quality embryos after IVF. J Assist Reprod Genet 2016;33:367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desquiret-Dumas V, Clèment A, Seegers V, Boucret L, Ferrè-L’Hotellier V, Bouet PE, et al. The mitochondrial DNA content of cumulus granulosa cells is linked to embryo quality. Hum Reprod 2017;32:607–14. [DOI] [PubMed] [Google Scholar]
- 19.Cinco R, Digman MA, Gratton E, Luderer U. Spatial Characterization of Bioenergetics and Metabolism of Primordial to Preovulatory Follicles in Whole Ex Vivo Murine Ovary. Biol Reprod 2016;95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Assou S, Haouzi D, Mahmoud K, Aouacheria A, Guillemin Y, Pantesco V, et al. A non-invasive test for assessing embryo potential by gene expression profiles of human cumulus cells: a proof of concept study. Molecular Human Reproduction 2008;14:711–9. [DOI] [PubMed] [Google Scholar]
- 21.Boucret L, Chao de la Barca JM, Morinière C, Desquiret V, Ferrè-L’Hôtellier V, Descamps P, et al. Relationship between diminished ovarian reserve and mitochondrial biogenesis in cumulus cells. Hum Reprod 2015;30:1653–64. [DOI] [PubMed] [Google Scholar]
- 22.Gebhardt KM, Feil DK, Dunning KR, Lane M, Russell DL. Human cumulus cell gene expression as a biomarker of pregnancy outcome after single embryo transfer. Fertility and Sterility 2011;96:47–52. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z, Wells D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod 2010;16:715–25. [DOI] [PubMed] [Google Scholar]
- 24.McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, et al. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod 2004;19:2869–74. [DOI] [PubMed] [Google Scholar]
- 25.Uyar A, Torrealday S, Seli E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertility and Sterility 2013;99:979–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taugourdeau A, Desquiret-Dumas V, Hamel JF, Chupin S, Boucret L, Ferré-L’Hotellier V, et al. The mitochondrial DNA content of cumulus cells may help predict embryo implantation. J Assist Reprod Genet 2019;36:223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorshinova VK, Tsvirkun DV, Sukhanova IA, Tarasova NV, Volodina MA, Marey MV, et al. Cumulus cell mitochondrial activity in relation to body mass index in women undergoing assisted reproductive therapy. BBA Clin 2017;7:141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heikal AA. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark Med 2010;4:241–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumollard R, Marangos P, Fitzharris G, Swann K, Duchen M, Carroll J. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 2004;131:3057–67. [DOI] [PubMed] [Google Scholar]
- 30.Klaidman LK, Leung AC, Adams JD. High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal Biochem 1995;228:312–7. [DOI] [PubMed] [Google Scholar]
- 31.Ghukasyan VV, Heikal AA. Natural Biomarkers for Cellular Metabolism: Biology, Techniques, and Applications. CRC Press; 2014. [Google Scholar]
- 32.Becker W Fluorescence lifetime imaging--techniques and applications. J Microsc 2012;247:119–36. [DOI] [PubMed] [Google Scholar]
- 33.Snijders TAB, Bosker RJ. Multilevel Analysis: An Introduction to Basic and Advanced Multilevel Modeling. SAGE; 2011. [Google Scholar]
- 34.Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 2012;23:420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, et al. ilastik: interactive machine learning for (bio)image analysis. Nat Methods 2019;16:1226–32. [DOI] [PubMed] [Google Scholar]
- 36.Lorah J Effect size measures for multilevel models: definition, interpretation, and TIMSS example. Large-scale Assessments in Education 2018;6:8. [Google Scholar]
- 37.Dumesic DA, Guedikian AA, Madrigal VK, Phan JD, Hill DL, Alvarez JP, et al. Cumulus Cell Mitochondrial Resistance to Stress In Vitro Predicts Oocyte Development During Assisted Reproduction. J Clin Endocrinol Metab 2016;101:2235–45. [DOI] [PubMed] [Google Scholar]
- 38.Cimadomo D, Fabozzi G, Vaiarelli A, Ubaldi N, Ubaldi FM, Rienzi L. Impact of Maternal Age on Oocyte and Embryo Competence. Front Endocrinol (Lausanne) [Internet] 2018. [cited 2021 Mar 26];9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6033961/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J-J, Liu X, Chen L, Zhang S, Zhang X, Hao C, et al. Advanced maternal age alters expression of maternal effect genes that are essential for human oocyte quality. Aging (Albany NY) 2020;12(4):3950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.May-Panloup P, Boucret L, Chao de la Barca J-M, Desquiret-Dumas V, Ferré-L’Hotellier V, Morinière C, et al. Ovarian ageing: the role of mitochondria in oocytes and follicles. Human Reproduction Update 2016;22(6):725–43. [DOI] [PubMed] [Google Scholar]
- 41.McReynolds S, Dzieciatkowska M, McCallie BR, Mitchell SD, Stevens J, Hansen K, et al. Impact of maternal aging on the molecular signature of human cumulus cells. Fertil Steril 2012;98(6):1574–1580.e5. [DOI] [PubMed] [Google Scholar]
- 42.Tatone C, Carbone MC, Falone S, Aimola P, Giardinelli A, Caserta D, et al. Age-dependent changes in the expression of superoxide dismutases and catalase are associated with ultrastructural modifications in human granulosa cells. Mol Hum Reprod 2006;12(11):655–60. [DOI] [PubMed] [Google Scholar]
- 43.Seifer DB, DeJesus V, Hubbard K. Mitochondrial deletions in luteinized granulosa cells as a function of age in women undergoing in vitro fertilization. Fertility and Sterility 2002;78(5):1046–8. [DOI] [PubMed] [Google Scholar]
- 44.Yang X, Needleman DJ. Coarse-grained model of mitochondrial metabolism enables subcellular flux inference from fluorescence lifetime imaging of NADH [Internet]. Biophysics; 2020. [cited 2021 Apr 13]. Available from: http://biorxiv.org/lookup/doi/10.1101/2020.11.20.392225 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. CCs that were imaged fresh show no significant differences in FLIM parameter from CCs that were first vitrified and then thawed (A) Mean FLIM measurements of both fresh and vitrified cumulus clusters (n=9), the FDR p-value of the t-test performed in the ratio between fresh/ vitrified compared to 1. (B) Box plots showing the ratios of each FLIM measurement.
Supplementary Figure 2. CCs that were not exposed to hyaluronidase show significant differences in intensities and redox ratio from those exposed to hyaluronidase (A, left) NAD(P)H autofluorescent intensity image of CCs exposed to hyaluronidase enzyme to dissolve its matrix and (A, right) CCs not exposed with hyaluronidase. (B) Mean of FLIM measurements of CCs exposed and not exposed to hyaluronidase (n=9), the p-values obtained from the comparison of the ratio between CCs not exposed to hyaluronidase / CCs exposed with hyaluronidase enzyme to 1 and FDR p-values. (C) Box plots showing these ratios of each FLIM measurement.
Supplementary Figure 3. CCs closed to the oocyte show no significant differences in FLIM parameters from CCs that were located further from the oocyte (A) Mean FLIM parameter values of both the CCs closer to the oocytes and those further from the oocytes (n=7), the p-value after a t-test performed in the ratio between CCs further from the oocytes / CCs closer to the oocytes compared to 1 and FDR p-values. (C) Box plots showing these ratios of each FLIM measurement.