Abstract
The shift towards primary HPV-based screening has necessitated the search for a secondary triage test that provides sufficient sensitivity to detect high grade cervical intraepithelial neoplasia (CIN) and cancer, but also brings an improved specificity to avoid unnecessary clinical work and colposcopy referrals.
We evaluated the performance of the previously described DNA-methylation test (S5) in detecting CIN3 and cancers from diverse geographic settings in high, medium and low income countries, using the cut-off of 0.80 and exploratory cut-offs of 2.62 and 3.70. Assays were performed using exfoliated cervical specimens (n=808) and formalin-fixed biopsies (n=166) from women diagnosed with cytology-negative results (n=220), CIN3 (n=204) and cancer stages I (n=245), II (n=249), III (n=28) and IV (n=22).
Methylation increased proportionally with disease severity (Cuzick test for trend, p<0.0001). S5 accurately separated women with negative-histology from CIN3 or cancer (p<0.0001). At the 0.80 cut-off, 543/544 cancers were correctly identified as S5 positive (99.81%). At cut-off 3.70, S5 showed a sensitivity of 95.77% with improved specificity. The S5 odds ratios of women negative for cervical disease versus CIN3+ were significantly higher than for HPV16/18 genotyping at all cut-offs (all p<0.0001). At S5 cut-off 0.80, 96.15% of consistently hrHPV-negative cancers (tested with multiple hrHPV-genotyping assay) were positive by S5. These cancers may have been missed in current primary hrHPV-screening programmes.
The S5-test can accurately detect CIN3 and malignancy irrespective of geographic context and setting. The test can be used as a screening and triage tool. Adjustment of the S5 cut-off can be performed considering the relative importance given to sensitivity versus specificity.
Keywords: DNA methylation, triage, HPV, high-risk human papillomavirus, cervical cancer, cancer screening, molecular triage
Introduction
The implementation of cervical cancer prevention programmes by systematic cytology screening1 has contributed to a reduction in cervical cancer-associated deaths in high income countries2. Yet, cervical cancer is currently the fourth most common cancer in women and continues to increase worldwide, with 604,000 cases in 2020, accounting for 7.5% of all female cancer deaths3. To allow a further reduction in the incidence of cervical cancer, screening has shifted towards high risk human papilloma virus (hr-HPV) testing with triage of HPV positive women. Cervical cancer incidence, ranges from 5 to 50 per 100,000 women depending on setting and while hrHPV testing is highly sensitive for the detection of disease, specificity is less optimal given the benign trajectory of most infections. Triage generally relies on cytology as the preferred secondary test in HPV positive women4. However, being subjective, cytology has limitations and objective secondary triage tests are urgently needed to identify the minority of hrHPV positive women with high-grade disease4. Furthermore, triage tests that rely on molecular, rather than morphological signatures (such as cytology) remove the requirement for specialized expertise.
Methylation biomarkers can offer an accurate alternative to detect clinically significant infection and associated disease and can identify women who have the highest risk of progressing into invasive cervical cancer5,6. Aberrant DNA methylation has been reported to increase with cervical cancer disease progression7, allowing this epigenetic event to be used as a temporal biomarker, with a potential to accurately predict whether hr-HPV infection will lead to cervical intraepithelial neoplasia grade 2 or above (CIN2+) or disappear5,8.
Several methylation biomarkers tests including our S5 DNA-methylation classifier, which tests for methylation on the host tumour suppressor gene EPB41L3 and viral late genes (L1 and L2) of HPV16, HPV18, HPV31 and HPV33, can accurately separate women with CIN2/3 and cancer from those with CIN1 or normal cytology9. The S5-classifier has demonstrated improved triage performance compared to hrHPV genotyping, cytology or the combination thereof and has been validated in a HPV-positive cohort of women as part of the Canadian FOCAL clinical trial, in the FRIDA screening trial in Mexico and the Colombian ASC-US-COL trial10-12. Additionally, the S5-classifier demonstrated a potential prognostic utility, in its ability to identify women with progressive CIN28. Together, these data support the prospect of using the S5-classifier as a molecular tool to identify clinically significant cervical abnormalities and predicting their clinical course.
Validation of the S5-classifier in a large number of CIN3+ samples from both high income and low- and middle-income countries (LMICs) is required to demonstrate that the methylation test can consistently detect cervical cancers worldwide. Extensive validation of the S5 classifier will support implementation of the test in global screening and disease-management systems, especially with the rise in acceptance of screening based on self-sampling 13,14. The main objective of the present study is to analyse the performance and consistency of S5 in detecting high grade lesions and cervical cancers from diverse settings that reflect Asia, Europe, Africa and the Americas. The present study aims to complement previous works on the S5 DNA-methylation classifier6,9,11,12,15.
Materials and Methods
Study population
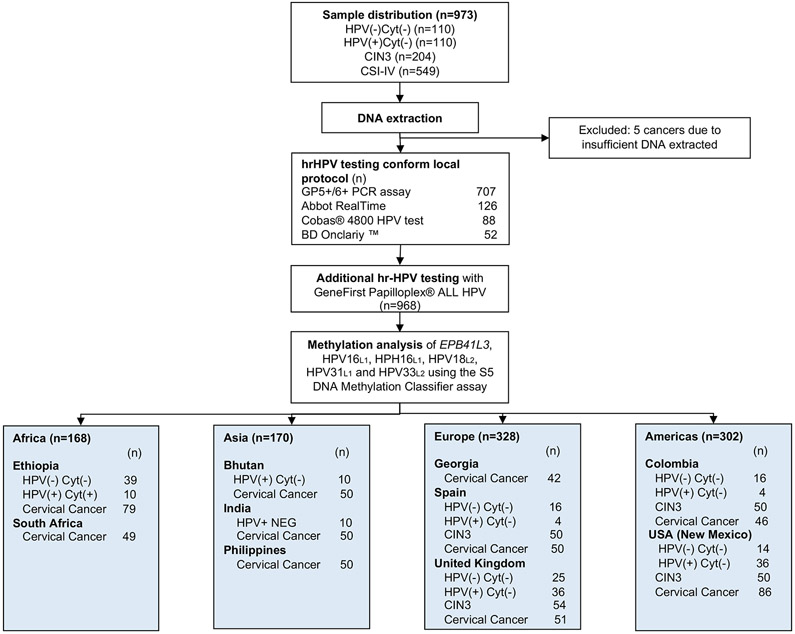
Cervical swabs and biopsies were collected from a total of 973 patients aged 21-64 as described in the referenced papers16-22. All samples included in the study were analysed by cytology or histology and had results of negative, CIN3 or invasive cervical cancer. We excluded CIN1 and CIN2 from our study because the CIN1 is a low grade lesion and CIN2 is considered increasingly, a heterogenous lesion that does not serve as a robust histological indicator of high-grade disease. Our study focussed mainly on cervical cancer stages I and II in order to have a sharper view of the epigenetic contrast between CIN3 versus early cancer and to complement previously published data on S5 performance.
Details regarding patient characteristics are described in Table 1.
Table 1.
Baseline characteristics of the 973 women from the study cohort. All women were diagnosed as histology negative (healthy women), cervical intraepithelial neoplasia grade 3 (CIN3) and cervical cancer stages I-IV (highlighted).
| Characteristics | HPV(−)Cyt(−) n = 110 (%) |
HPV(+)Cyt(−) n = 110 (%) |
CIN3 n = 204 (%) |
Cervical Cancer – FIGO stage |
||||
|---|---|---|---|---|---|---|---|---|
| Stage I n = 245 (%) |
Stage II n = 249 (%) |
Stage III n = 28 (%) |
Stage IV n = 22 (%) |
|||||
| Histotype of cervical cancer | Squamous cell carcinoma | 230 (93.9) | 236 (94.8) | 26 (92.8) | 19 (86.5) | |||
| Adenocarcinoma | 14 (5.7) | 10 (4.0) | 2 (7.2) | 2 (9.0) | ||||
| Adenosquamous cell carcinoma | - | 1 (0.4) | - | - | ||||
| Neuroendocrine carcinoma | 1 (0.4) | 2 (0.8) | - | 1 (4.5) | ||||
| Hr-HPV status* | Consistently Negative | 110 (100) | - | 10 (4.90) | 14 (5.7) | 10 (4.0) | 2 (7.1) | - |
| HPV16+ | - | 35 (31.8) | 120 (58.8) | 160 (65.3) | 179 (72.1) | 20 (71.4) | 19 (86.4) | |
| HPV18+ | - | 14 (12.7) | 7 (3.4) | 19 (7.7) | 15 (6.0) | 1 (3.5) | - | |
| HPV31+ | - | 6 (5.5) | 30 (14.7) | 8 (3.2) | 9 (3.6) | 2 (7.1) | 1 (4.5) | |
| HPV33+ | - | 6 (5.5) | 7 (3.4) | 9 (3.6) | 8 (3.2) | - | - | |
| Other hr-HPV+ | - | 49 (44.5) | 30 (14.7) | 35 (14.3) | 28 (10.8) | 3 (10.7) | 2 (9.1) | |
| Sample type | Cervical Scrape | 110 (100) | 110 (100) | 204 (100) | 142 (58.0) | 192 (77.0) | 28 (100) | 22 (100) |
| FFPE Tissue | - | - | - | 103 (42.0) | 57 (22.9) | - | - | |
| Age | <25 | 20 (18.18) | 12 (10.9) | 14 (6.9) | 5 (2.0) | 2 (0.8) | - | - |
| 25-29 | 27 (24.5) | 25 (22.7) | 59 (28.9) | 27 (11.0) | 10 (4.0) | - | 1 (4.5) | |
| 30-39 | 28 (25.4) | 26 (23.6) | 81 (39.7) | 83 (33.8) | 43 (17.3) | 4 (14.2) | 3 (13.6) | |
| 40-49 | 21 (19.0) | 23 (20.9) | 30 (14.7) | 56 (22.8) | 84 (33.8) | 8 (28.5) | 6 (27.4) | |
| 50-59 | 8 (7.2) | 12 (10.9) | 14 (6.9) | 46 (18.7) | 63 (25.4) | 9 (32.1) | 11 (50.0) | |
| >60 | 6 (5.5) | 12 (10.9) | 6 (2.9) | 28 (11.4) | 47 (18.5) | 7 (25.0) | 1 (4.5) | |
| Country of Origin | Bhutan | - | 10 (9.0) | - | 28 (11.4) | 22 (8.8) | - | - |
| Colombia | 16 (14.5) | 4 (3.6) | 50 (24.2) | 21 (8.5) | 25 (10.0) | - | - | |
| Ethiopia | 39 (35.5) | 10 (9.0) | - | 2 (0.8) | 18 (7.2) | 28 (100) | 22 (100) | |
| Georgia | - | - | - | 40 (16.3) | 2 (0.8) | - | - | |
| India | - | 10 (9.0) | - | 12 (4.9) | 38 (15.3) | - | - | |
| Philippines | - | - | - | 15 (6.1) | 35 (14.1) | - | - | |
| South Africa | - | - | - | 2 (0.8) | 47 (18.9) | - | - | |
| Spain | 16 (14.5) | 4 (3.6) | 50 (24.2) | 28 (11.4) | 22 (8.8) | - | - | |
| United Kingdom | 25 (22.7) | 36 (32.7) | 54 (27.1) | 48 (19.5) | 3 (1.2) | - | - | |
| USA (New Mexico) | 14 (12.7) | 36 (32.7) | 50 (24.2) | 50 (20.4) | 36 (14.5) | - | - | |
The cellular material for cervical liquid-based cytology samples was collected in PreservCyt® medium (Hologic Corporation, Marlborough MA, USA) for storage until DNA extraction. A subset of specimens from Bhutan (n=10), Ethiopia (n=49), India (n=10), Spain (n=20), UK (n=51), Colombia (n=20), USA (n=50) were selected for negative cytology results. CIN3 samples originated from Spain (n=50), UK (n=54), Colombia (n=50) and USA (n=50). Cancer samples originated from Ethiopia (n=70), South Africa (n=49), Bhutan (n=50), India (n=50), Philippines (n=50), Georgia (n=42), Spain (n=50), UK (n=51), Colombia (n=46), USA (n=86). All CIN3 and cancer samples were collected from patients showing abnormal cytology and histology results through colposcopy referral and diagnosed according to specific country recommendations. Biopsy samples were collected and stored at −70°C (IARC and Scottish HPV Archive and Addis Ababa University); formalin-fixed paraffin-embedded (FFPE) samples were stored at room temperature (Bhutan-IARC and University of New Mexico) until DNA was extracted.
S5-methylation assays
DNA was extracted using i) QIAsymphony DSP DNA Mini Kit (Qiagen, Hilden, Germany) for all samples in the IARC biobank (both PreservCyt® and FFPE specimens); ii) Abbott M2000 system (Abbott Laboratories, USA) for all samples in the Scottish HPV Archive biobank; iii) DNA from Ethiopian samples and FFPE tissue from University of New Mexico was manually extracted using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). After extraction, DNA was quantified with the Qubit Flex Fluorometer (Thermofisher Scientific, London, UK). One hundred nanograms of DNA were used in the bisulfite conversion reaction where un-methylated cytosines were converted to uracils with the EZ DNA methylation kit (Zymo Research, Irvine, USA). The converted DNA was amplified and pyrosequenced on a PyroMark Q96ID (Qiagen, Hilden, Germany) for DNA methylation on CpG islands from EPB41L3 and viral late genes (L1 and L2) of HPV16, HPV18, HPV31, and HPV33 as previously described9. Percentage methylation was taken as the mean for CpG sites involved in each case.
HPV Genotyping
The clinical samples used in the study have been initially genotyped as part of previously approved research studies. The following technologies were used: GP5+/6+ PCR assay (n= 641)17,18,20,21, Abbott RealTime assay (n=126), Roche LINEAR ARRAY® HPV Genotyping assay (n= 88), BD Onclarity™ assay (n=52). Genotyping with Papilloplex High Risk HPV test (GeneFirst, Oxford, UK) was additionally used to validate previous HPV genotyping in all samples investigated. This is a multiplexed PCR-based system that simultaneously detects 13 hr-HPV genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68 and the low risk type 66)23. Genotyping with the Papilloplex High Risk HPV test was run in house on the QuantStudio 5 Real-Time PCR System (Thermofisher Scientific, London, UK) and targeted the L1 region of all the genotypes described. Papilloplex High Risk HPV internal negative and positive controls were used as baselines and analysis was performed on the GeneFirst software.
We investigated the following HPV categories: 1) HPV16 positive, 2) HPV18 positive, 3) HPV31 positive, 4) HPV33 positive and 5) other hr-HPV positive (non-HPV16/18/31/33). We followed a hierarchical attribution of HPV genotypes, namely in cases with multiple hrHPV-infections the more prevalent genotype in cancer was attributed dominant status (e.g. a sample positive for both HPV16 and HPV18 was placed in category 1).
Statistical Analysis
We validated the performance sensitivity of the S5 classifier on CIN3 and cervical cancer samples from a global population of samples. We used the mean of methylation scores for selected CpG sites: three for EPB41L3 (438, 427, 425), two for HPV16L1 (6367, 6389), six for HPV18L2 (4256, 4261, 4265, 4269, 4275, 4282), two for HPV31L1 (6352, 6367) and three for HPV33L2 (5557, 5560, 5566) as previously described9,15,24-26. Proportion of methylation on HPV16L2 was calculated using the following three CpG sites: 4275, 4259 and 4238 as they were the most reproducible sites in this region. The S5-methylation score was calculated by using the following weighed average defined in 2016 by Lorincz A et al.9: S5 = 30.9(EPB41L3) + 13.7(HPV16L1) + 4.3(HPV16L2) + 8.4(HPV18L2) + 22.4(HPV31L1) + 20.3(HPV33L2)
The main groups being compared were HPV-negative cytology negative: HPV (−)/Cyt (−), HPV-positive cytology negative: HPV (+)/Cyt (−), CIN3 and cervical cancer including stages I (CSI), II (CSII), III (CSIII) and IV (CSIV) as per FIGO stage classification. We hypothesised that uniformity in proportions and / or levels of methylation status among the different lesion categories may imply prognostic value as a clinical marker.
We compared differences in methylation levels between groups using Kruskal-Wallis and Dunn’s multiple comparison tests and the Cuzick test for trend to assess any methylation trend with disease progression. We evaluated the S5 diagnostic potential using the pre-defined cut-off of 0.80. In addition, we explored cut-off points more suited for LMIC, using a previously defined alternative cut-off and Youden-J index12. McNemar’s test with continuity correction was used for differences in sensitivity and specificity.
We also used unconditional logistic regression to study the relationship between methylation in the invasive cervical cancer group and the covariates - stage of cancer, type, age, demographics and HPV status. Additionally, we calculated the odds ratios (ORs) for the associations between HPV16/18 positivity, S5 classifier positivity at different cut-offs and CIN3 or cervical cancer (CSI-IV, FIGO stage unknown included) diagnosis. All p-values were two sided with α ≤0.05 considered significant. Analysis was performed with GraphPad Prism v8.0 as well as R v 3.4.1 for Cuzick tests, ORs and for unconditional logistic regression analyses.
Results
Characteristics and selection criteria
We present a cross-sectional retrospective study including 973 women from ten countries to evaluate the S5 methylation classifier performance to detect CIN3 and cervical cancer. The present study aims to complement previous work on the S5 DNA-methylation classifier6,9,11,12,15. We selected 220 women cytology negative or HPV(−/+)/Cyt(−), 204 women diagnosed with CIN3 and 544 with invasive cervical cancer as described in Figure 1. Baseline characteristics of the women are presented in Table 1. The invasive cervical cancer group is further divided into squamous cell carcinomas (SCC, n = 510), adenocarcinomas (ADC, n = 29), adenosquamous cell carcinomas (ASC, n = 1) and neuroendocrine small cell carcinoma (SNEC, n = 4). The median age for women with negative cytology was 38 years (IQR, 30 – 47), for CIN3 was 31 years (IQR, 28 – 39) and for women with invasive cervical cancer was 45 years (IQR, 38 – 55).
F1 -.
Study design flow-chart
hrHPV detection
The Papilloplex High Risk HPV genotyping data was in 98.23% (95%CI 96.38-99.99%) agreement with previous genotyping methodologies used (Figure 1). The grouped prevalence of 13 types of hrHPV plus HPV66 (now regarded as a low risk HPV) was 95.09% (95%CI 92.13-97.89%) in the CIN3 group and 95.21% (95%CI 92.45-98.12%) in the cancer group. Each type of HPV in women infected by multiple HPV types was counted individually as described in Table 1 and Supplementary Figure 1. In the cancer group, 374 women were consistently positive for HPV16 (68.91%), 37 women for HPV18 (6.91%), 21 women for HPV31 (3.90%), 16 women for HPV33 (3.01%) and 68 women for other hr-HPV types (12.41%). A total of 25 (4.78%) cervical cancers tested negative for any hr-HPV type covered by the assays. HPV genotyping was in agreement with methylation data on HPV16/18/31/33 infection in 95.38% (95%CI 91.38-98.33%) of cases.
Increasing trend in the S5 methylation scores with disease severity
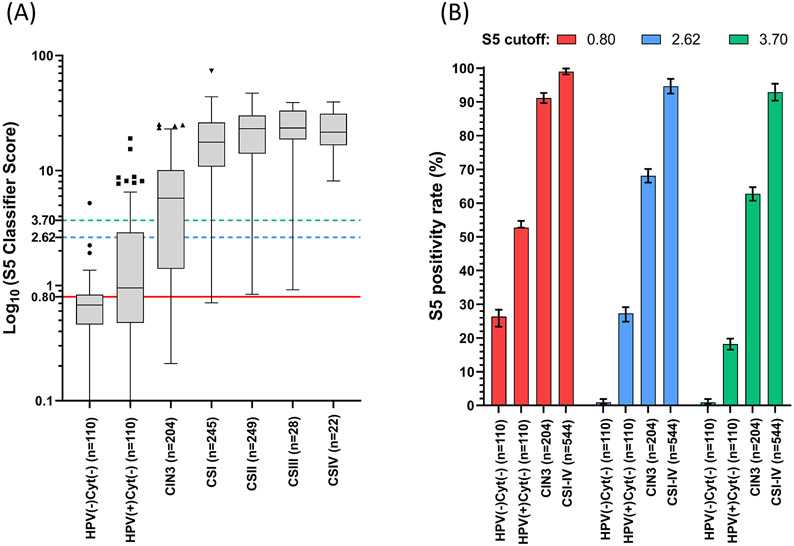
The S5 methylation scores were clustered according to severity of cervical abnormality. An outline of the methylation scores per country is provided in Supplementary Figure 2. Median methylation score was 0.66 (95%CI 0.60 – 0.78) in HPV negative and cytology negative women (HPV(−)/Cyt(−)), 0.91 (95%CI 0.86 – 0.94) in HPV positive and cytology negative women (HPV(+)/Cyt (−)), 5.64 (95%CI 3.99 – 6.88) in CIN3, 17.58 (95%CI 15.99 – 20.33) in cervical cancer stage I (CSI), 23.71 (95%CI 21.67 – 25.24) in cervical cancer stage II (CSII), 24.44 (95%CI 22.10 – 32.14) in cervical cancer stage III (CSIII) and 24.95 (95%CI 22.34 – 32.85) in cervical cancer stage IV (CSIV). The distribution of the S5 scores based on the disease diagnosis and hr-HPV status is shown in Figure 2A. Kruskal-Wallis and Dunn’s multiple comparisons tests show significant separation in the following paired comparisons: HPV(−)/Cyt(−) vs CIN3 (p<0.0001), HPV(−)/Cyt(−) vs CSI-IV (all p<0.0001), HPV(+)/Cyt(−) vs CIN3 (p<0.0001), HPV(+)/Cyt(−) vs CSI-IV (all p<0.0001), CIN3 vs CSI-IV (all p<0.0001). No other significant differences were identified. The S5-classifier scores increased significantly with the severity of lesions (Cuzick test for trend: z = 9.23, p<0.0001).
F2 -.
S5 performance in the study group: A. Distribution of the S5 scores based on the histopathological diagnostic of the patient. Data was plotted as log10 of the S5 score according to lesion grade: HPV(−)/Cyt(−), HPV(+)/Cyt(−), CIN3 and all stages of cervical cancer (CSI-IV). The S5 classifier was significantly different between the following group comparisons: HPV(−)/Cyt(−) vs CIN3 (p < 0.0001), HPV(−)/Cyt(−) vs CSI-IV (all, p < 0.0001), HPV(+)/Cyt(−)vs CIN3 (p < 0.0001), HPV(+)/Cyt(−)vs CSI-IV (all, p < 0.0001) and CIN3 vs CSI-IV (all, p < 0.0001). Other comparisons were not significant (HPV(−)/Cyt(−)vs HPV(+)/Cyt(−), among CSI-IV). The proposed cut-offs for analysis are: 0.80 (red), 2.62 (blue) and 3.70 (green). The top of box represents the upper quartile (p75), bottom the lower quartile (p25), and the line the median (p50). The upper whisker extends to the largest point of the inter-quartile range from the upper quartile. The lower whisker extends to the smallest point of the inter- quartile range from the lower quartile. The Cuzick test for trend was highly significant (p < 0.0001). B. Cumulative S5 positivity per lesion grade at the three cut-offs: 0.80 (red), 2.62 (blue) and 3.70 (green). Median values are shown by each bar and error bars show 95% CI of the median. Abbreviations: HPV(−)/Cyt(−),HPV negative and cytology negative; HPV(+)/Cyt(−), HPV positive and cytology negative; CIN, cervical intraepithelial neoplasia (of grade 3); CSI-IV, cervical cancer stages I-IV.
S5-classifier sensitivity in cervical cancers at the 0.80 predefined cut-off
The S5-classifier methylation score was successfully measured in all 544 women diagnosed with cervical cancer which were included in the study. A total of 543 out of 544 cancer patients tested positive for S5 at 0.80, yielding a sensitivity of 99.81% (95%CI 98.34-99.96). Table 2 shows the S5 sensitivity stratified per histology, FIGO stage, hrHPV status, hrHPV type, sample type, age and country of origin. At the 0.80 cut-off, cervical cancers which were consistently hrHPV-negative when tested with multiple hrHPV genotyping assays were 96.15% (95%CI 94.38-98.25) identified by the S5 classifier. The performance of the S5 classifier was uniform among all groups considered. There were no significant differences in S5 sensitivity among histology, FIGO stage, hrHPV type, sample type, age and country of origin (Fishers’ test, all p>0.05).
Table 2 -.
S5 Classifier sensitivity at the 0.80 cut-off in a cervical cancer referral group, stratified per histotype of cervical carcinoma, FIGO stage, hrHPV status, sample type, age or country of origin.
| S5 sensitivity at cut-off 0.80 | ||||
|---|---|---|---|---|
| n / N* | % | 95% CI | P-value** | |
| Histotype of cervical carcinoma | ||||
| Squamous cell carcinoma | 509 / 510 | 99.80 | (99.10-99.95) | |
| Adenocarcinoma | 29 / 29 | 100.0 | (97.34-100.0) | 0.837 |
| Adenosquamous cell carcinoma | 1 / 1 | 100.0 | (12.8-100.0) | |
| Neuroendocrine carcinoma | 4 / 4 | 100.0 | (26.86-100.0) | |
| FIGO Stage | ||||
| Stage I | 244 / 245 | 99.60 | (99.25-99.83) | |
| Stage II | 249 / 249 | 100.0 | (99.43-100.0) | 0.687 |
| Stage III | 28 / 28 | 100.0 | (97.34-100.0) | |
| Stage IV | 22 / 22 | 100.0 | (96.45-100.0) | |
| HPV status *** | ||||
| HPV-positive | 518 / 518 | 100.0 | (99.46-100.0) | |
| HPV16 | 379 / 379 | 100.0 | (99.36-100.0) | 0.465a |
| HPV18 | 36 / 36 | 100.0 | (98.82-100.0) | 0.587b |
| HPV31 | 20 / 20 | 100.0 | (96.45-100.0) | |
| HPV33 | 17 / 17 | 100.0 | (93.12-100.0) | |
| Other hr-HPV | 66 / 66 | 100.0 | (98.93-100.0) | |
| HPV-negative | 25 / 26 | 96.15 | (94.38-98.25) | |
| Sample type | ||||
| Cervical Scrape | 383 / 384 | 99.73 | (98.34–99.96) | 0.917 |
| FFPE tissue | 160 / 160 | 100.0 | (98.76-100.0) | |
| Age | ||||
| <25 | 7 / 7 | 100.0 | (64.87-100.0) | |
| 25-29 | 38 / 38 | 100.0 | (98.62-100.0) | |
| 30-39 | 133 / 133 | 100.0 | (99.32-100.0) | 0.989 |
| 40-49 | 153 / 154 | 99.39 | (98.74-99.86) | |
| 50-59 | 129 / 129 | 100.0 | (99.22-100.0) | |
| >60 | 83 / 83 | 100.0 | (99.02-100.0) | |
| Country of Origin | ||||
| Bhutan | 50 / 50 | 100.0 | (98.89-100.0) | |
| Colombia | 46 / 46 | 100.0 | (98.87-100.0) | |
| Ethiopia | 70 / 70 | 100.0 | (98.98-100.0) | |
| Georgia | 42 / 42 | 100.0 | (98.84-100.0) | 0.892 |
| India | 50 / 50 | 100.0 | (98.89-100.0) | |
| Philippines | 50 / 50 | 100.0 | (98.89-100.0) | |
| South Africa | 49 / 49 | 100.0 | (98.88-100.0) | |
| Spain | 50 / 50 | 100.0 | (98.89-100.0) | |
| United Kingdom | 50 / 51 | 98.03 | (95.99-99.05) | |
| USA (New Mexico) | 86 / 86 | 100.0 | (99.12-100.0) | |
| Total | 543/544 | 99.81 | (98.34-99.96) | |
n = number of positive samples in a specified group; N = group total
hrHPV genotype grouping performed by hierarchical genotype attribution, as detailed in materials and methods
Determined by performing Fishers’ exact test of independence.
P-value between HPV-positive and HPV-negative subgroups
P-value among all subgroups in the HPV groups
S5 classifier cut-off adjusted per country to optimise triage capacity
Cervical cancer incidence is directly linked to the availability of screening in a particular country. Hence the importance to introduce different modalities for the implementation of a molecular triage reflecting the country clinical setting. We investigated the false positive rates in women with HPV(−)/Cyt(−), HPV(+)/Cyt(−), CIN3 and CSI-IV at the UK-predefined cut-off of 0.80, the Youden-J index cut-off based on the S5 methylation scores of cervical cancers: 2.62 and the previously explored LMIC cut-off of 3.7012. For all groups analysed, the false positive rate, decreased with the increase in cut-off (Figure 2B). The most important decrease was observed in the HPV(−)/Cyt(−) and HPV(+)/Cyt(−) groups. At a cut-off of 0.80, the false positive rate in HPV(−)/Cyt(−) women was 26.32% (95%CI 23.90 – 29.94). This may be acceptable for a country desiring very high sensitivity, where a strong health care system could accommodate the rather common false positives. The false positive rate decreased to 0.92% (95%CI 0.36 - 1.82) at both 2.62 and 3.70 cut-offs (McNemar test χ2=27.1, p<0.0001) which would better suit countries with lower screening capacity. However, the false positive rate of S5 in the HPV(+)/Cyt(−) was 52.74% (95%CI 49.71 – 55.63) at 0.80 and showed a significant decrease trend to 27.22% (95%CI 24.94 – 29.53) at cut-off 2.62 and 18.26% (95%CI 16.62 – 20.24) at cut-off 3.70 (Cuzick test for trend, p<0.0001). These possible adjustments can allow a customization of the S5 cut-off according to the country of clinical implementation.
Although we observed a significant decrease in false positive rate with the increase of the cut-off, a similar but less pronounced decrease trend was observed in S5 positivity for CIN3 and cancer detection. CIN3 sensitivity decreased from 87.26% (95%CI 84.42 – 89.93) at 0.80 to 62.74% (95%CI 60.13–65.25) at 3.70 (Cuzick test for trend, p<0.0001). Further, the S5 sensitivity for cancer decreased from 99.81% (95%CI 98.34–99.96) at 0.80 to 95.77% (95%CI 92.39- 97.40) at 3.70 (Cuzick test for trend, p=0.005).
Diagnostic potential of S5 classifier compared to HPV16/18 testing
Table 3 presents the associations between HPV16/18 and S5 classifier positivity at different cut-offs for the identification of CIN3+, compared to the HPV(+)/Cyt(−). HPV16/18 positivity was strongly associated with CIN3+ development. The univariate odds ratios (OR) of HPV16/18 positivity for CIN3 was 2.86 (95%CI 1.77 – 4.62), while for cancer the OR was approximatively two times higher: 4.80 (95%CI 3.13 – 7.96). The univariate ORs for all S5 cut-offs were higher than the univariate ORs for HPV16/18 (p<0.0001). Increased ORs were observed for the bi-variable associations of HPV16/18 and the S5 test regardless of the cut-off or the geographic location (all, p<0.0001). This indicates stronger associations between the combination of HPV16/18 positivity and S5 positivity and CIN3+ development. Although the ORs for the bi-variable analysis of HPV16/18 positivity and S5 0.80 cut-off were significantly higher than the univariate HPV16/18 ORs (p<0.0001), the highest associations for CIN3+ development were observed for S5 positivity at the 3.70 cut-off (OR: 5.63, 95%CI 3.26 – 9.73 for CIN3; and OR: 45.55, 95%CI 24.67 – 73.38 for cervical cancer).
Table 3 –
Univariate and bivariate odds ratios (OR with 95%CI confidence intervals) for the associations between the different clinical outcomes and HPV16/18 and/or S5 cut-offs in a HPV(+)/Cyt(−)baseline.
| Variables | OR | 95% CI | Z value | P valuea | |
|---|---|---|---|---|---|
| CIN3 | HPV 16/18 | 2.86 | 1.77 - 4.62 | 4.30 | Reference |
| S5 0.80 | 4.50 | 2.71 – 7.46 | 5.83 | <0.0001 | |
| S5 2.62 | 5.63 | 3.26 – 9.73 | 6.19 | <0.0001 | |
| S5 3.70 | 6.42 | 3.67 – 11.24 | 6.52 | <0.0001 | |
| * HPV 16/18 and S5 0.80 | 3.26 | 2.01 - 5.30 | 4..79 | <0.0001 | |
| * HPV 16/18 and S5 2.62 | 3.56 | 2.12 - 5.98 | 4.80 | <0.0001 | |
| * HPV 16/18 and S5 3.70 | 5.01 | 2.82 - 8.90 | 5.49 | <0.0001 | |
| Cervical Cancer | HPV 16/18 | 4.80 | 3.13 - 7.36 | 7.19 | Reference |
| S5 0.80 | 20.94 | 7.89 - 51.71 | 7.22 | <0.0001 | |
| S5 2.62 | 36.21 | 20.9 - 62.73 | 12.80 | <0.0001 | |
| S5 3.70 | 45.55 | 24.67 – 73.38 | 13.49 | <0.0001 | |
| * HPV 16/18 and S5 0.80 | 6.32 | 4.08 - 9.80 | 8.25 | <0.0001 | |
| * HPV 16/18 and S5 2.62 | 9.87 | 6.11 - 15.96 | 9.35 | <0.0001 | |
| * HPV 16/18 and S5 3.70 | 14.90 | 8.69 - 25.56 | 9.81 | <0.0001 |
Bivariate OR for the associations between HPV16/18 and S5 cut-offs (0.80, 2.62 and 3.70) and CIN3 and cervical cancer
P value indicating an increased ORs compared to the reference HPV16/18. P value determined using Fishers’ test of independence
S5-classifier performance in detecting cervical cancers at the 3.70 cut-off
When the cut-off was increased to 3.70, the false positive rate in HPV(+)/Cyt(−) was 9.54% (95%CI 8.49 – 10.76), which was approximatively 4-fold lower than at the 0.80 cut-off (39.54%, 95%CI 37.20 – 41.86). The decrease in the false positive rate correlates to an increase in specificity of the S5 classifier. Estimates of specificity are presented in Supplementary Table 3 for cervical cancer compared to HPV(−)/Cyt(−) women or HPV(+)/Cyt(−) women (approximating a currently relevant triage population). For both cases, increasing the cut-off from 0.80 to 3.70 dramatically improved specificity at a cost of sensitivity (Supplementary Table 3). In an HPV(−)/Cyt(−) population, specificity rose from 65.12% (95%CI, 54.59 – 74.35) at 0.80 to 100% (95.19 – 100.0) at 3.70. The same trend was observed in an approximated triage population. Here, specificity increased from 50.60% (95%CI, 43.11 – 58.06) at 0.80 to 83.33% (95%CI 76.97 – 88.21) at 3.70.
A total of 521 out of 544 women with any cancer type tested positive for S5 at the 3.70 cut-off, yielding a sensitivity of 95.77% (95%CI 92.39- 97.40). A lower proportion of the hrHPV-negative cancer group tested S5 positive at a cut-off 3.70 compared to the hrHPV-positive cancer group: 73.07% (95%CI 56.85 – 86.82) versus 98.45% (95%CI 92.72 – 99.46) (Fishers’ test, p<0.0001) as described in Table 4. There were no other significant differences in S5 sensitivity among histology, FIGO stage, hrHPV type, sample type, age and country of origin. These results indicate a highly consistent performance of the S5 classifier in patients from different continents with respect to detection of cancer.
Table 4 -.
S5 Classifier sensitivity at the 3.70 cut-off in a cervical cancer referral group, stratified per histotype of cervical cancer, FIGO stage, hrHPV status, sample type, age or country of origin
| S5 sensitivity at cut-off 3.70 | ||||
|---|---|---|---|---|
| n / N* | % | 95% CI | P-value** | |
| Histotype of cervical cancer | ||||
| Squamous cell carcinoma | 491 / 510 | 96.22 | (91.32 – 98.35) | |
| Adenocarcinoma | 28 / 29 | 96.55 | (91.71 – 99.23) | |
| Adenosquamous cell carcinoma | 1 / 1 | 100.0 | (91.81 – 100.0) | 0.837 |
| Neuroendocrine carcinoma | 4 / 4 | 100.0 | (94.24 – 100.0) | |
| FIGO Stage | ||||
| Stage I | 230 / 245 | 93.87 | (89.42 – 96.91) | |
| Stage II | 242 / 249 | 97.18 | (93.62 – 99.00) | |
| Stage III | 27 / 28 | 96.42 | (90.32 – 99.42) | 0.687 |
| Stage IV | 22 / 22 | 100.0 | (97.76 – 100.0) | |
| HPV status *** | ||||
| HPV-positive | 510 / 518 | 98.45 | (92.72 – 99.46) | |
| HPV16 | 372 / 379 | 98.15 | (92.52 – 99.32) | |
| HPV18 | 34 / 36 | 94.44 | (91.60 – 98.72) | 0.465a |
| HPV31 | 19 / 20 | 95.00 | (92.31 – 96.42) | 0.587b |
| HPV33 | 16 / 17 | 94.11 | (90.22 – 95.41) | |
| Other hr-HPV | 61 / 66 | 92.42 | (81.35– 94.15) | |
| HPV-negative | 19 / 26 | 73.07 | (56.85 – 86.82) | |
| Sample type | ||||
| Cervical Scrape | 371 / 384 | 96.61 | (91.70 – 98.62) | |
| FFPE tissue | 150 / 160 | 93.75 | (90.82 – 96.62) | 0.917 |
| Age | ||||
| <25 | 6 / 7 | 85.71 | (65.55 – 90.22) | |
| 25-29 | 36 / 38 | 94.73 | (90.45 – 98.65) | |
| 30-39 | 128 / 133 | 96.24 | (91.24 – 98.75) | |
| 40-49 | 148 / 154 | 96.10 | (92.54 – 98.12) | 0.989 |
| 50-59 | 122 / 129 | 94.57 | (89.79 – 96.12) | |
| >60 | 81 / 83 | 97.59 | (94.05 – 98.92) | |
| Country of Origin | ||||
| Bhutan | 47 / 50 | 94.00 | (92.32 – 96.82) | |
| Colombia | 40 / 46 | 86.95 | (78.35 – 92.92) | |
| Ethiopia | 68 / 70 | 97.14 | (94.92 – 98.96) | |
| Georgia | 40 / 42 | 95.23 | (92.98 – 97.35) | |
| India | 48 / 50 | 96.00 | (94.12 – 97.59) | 0.892 |
| Philippines | 48 / 50 | 96.00 | (94.12 – 97.59) | |
| South Africa | 49 / 49 | 100.0 | (98.88 - 100.0) | |
| Spain | 48 / 50 | 96.00 | (94.12 – 97.59) | |
| United Kingdom | 48 / 51 | 94.11 | (93.72 – 96.59) | |
| USA (New Mexico) | 84 / 86 | 97.67 | (94.72 – 98.68) | |
| Total | 521 / 544 | 95.77 | (92.39- 97.40) | |
n = number of positive samples in a specified group; N = group total.
hrHPV genotype grouping performed by hierarchical genotype attribution, as detailed in materials and methods.
Determined by performing Fishers’ exact test of independence.
P-value among all subgroups in the HPV-positive group
P-value between HPV-positive and HPV-negative subgroups
S5 classifier components proportion vary with disease severity
On a component basis, individual EPB41L3 methylation was observed to increase in a sigmoidal curve with disease progression. The Cuzick test for trend was significant for HPV(−)/Cyt(−), CIN3, CSI and CSII (p < 0.0001). The second most important component of the S5 classifier, HPV16 methylation, showed an increasing trend with disease progression (Cuzick test for trend, both, p < 0.001) as well as a steep increase from CIN3 to CSI-IV. Additionally, both HPV18 and HPV33 methylation showed a linear increase with disease progression (Cuzick test for trend, both, p < 0.01) as described in Supplementary Figure 3.
The weight of each component of the S5-classifier was plotted for HPV(+)Cyt(−), CIN3 and the cancer CSI-IV groups in Figure 3. The Cuzick test for trend showed an increasing trend of EPB41L3 weight with disease lesion, z = 8.21 (p<0.0001). EPB41L3 weight plateaus at CSII. Unconditional logistic regression models showed the strength of the association between EPB41L3 methylation, severity of lesion, and age. The relationship between EPB41L3 methylation was stronger for severity of lesion: F = 367.50, p<0.0001 than age (F = 81.0, p<0.0001). This indicates that host EPB41L3 methylation might have a good potential to predict disease progression, independent of increasing natural epigenetic methylation levels occurring with age.
F3 -.
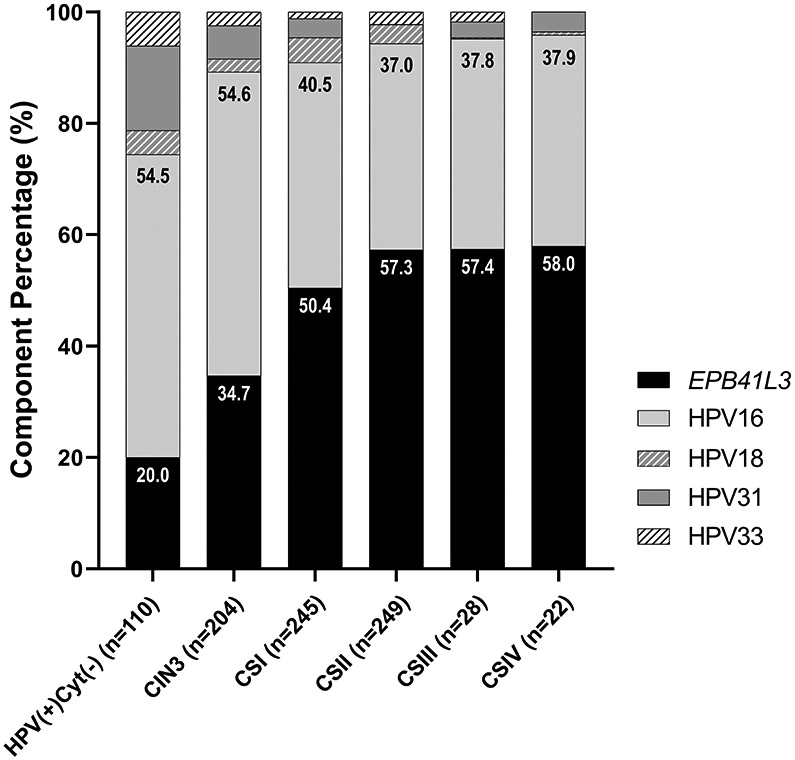
S5 component breakdown in HPV+ NEG, CIN3 and CSI-IV samples. The weight of the methylation on the S5 components: EPB41L3, HPV16, HPV18, HPV31 and HPV33 has been calculated for each group. Percentages of EPB41L3 and HPV16 weights in the classifier are noted at the top of the corresponding bars. HPV methylation becomes less important versus EPB41L3 as the lesions become more advanced, however EPB41L3 weight in the classifier does not change after stage II cancer.
Interestingly, the relative proportion of the HPV components of the S5-classifier decreased slightly with severity of lesion (Cuzick test for trend, z = −6.52, p<0.0001). HPV16 had the highest weight out of all viral components, however this was 1.8 times lower than the weight of EPB41L3 in CSII+ specimens.
Discussion
To our knowledge, our study represents one of the most comprehensive assessment of viral and host cell DNA methylation data in invasive cervical cancer to date, particularly given its multi-site dimension(s) and number of cases of high grade and invasive disease27. We show that there is a strong increasing trend of S5 DNA methylation score with cervical disease severity in our global collection of samples. Our results also show a very high S5 sensitivity for CIN3+ at the predefined cut-off of 0.80, while there was high to moderate sensitivity at the exploratory cut-offs of 2.62 and 3.70 respectively.
We found an S5 test sensitivity of 91.18% (186/204) for CIN3 and 99.81% (543/544) for cervical cancer detection at the predefined cut-off of 0.809. At this cut-off S5 sensitivity for cervical cancer was higher than the sensitivity of HPV DNA testing: 95.21% (518/544) (McNemar χ2=5.08, p=0.032). Additionally, at the S5 cut-off of 3.70, previously demonstrated useful in LMIC settings12, we found a sensitivity of 62.74% (128/204) for CIN3 and 95.77% (521/544) for cervical cancer.
This study complements previous results in Colombia10, Canada11 and Mexico12, by including data on cancers from additional countries and thus, describing a larger study for cervical cancer identification. A recent Dutch study of 519 cervical cancer samples, FAM19A4/miR124-2 methylation analysis yielded a sensitivity of 98.30% (510/519)27. Additionally, the four-gene methylation marker panel comprising of the host genes JAM3, EPB41L3, TERT and C13ORF18 identified 94.20% (65/69) of cervical cancers28. S5 demonstrated a slightly higher sensitivity of 99.81% (543/544) compared to the above mentioned tests (p=0.047 and p=0.029, respectively). Most importantly, S5 detected 25 out of 26 hrHPV-negative cancers which were not explored in other studies.
We explored the performance of the S5 classifier at three cut-offs: 0.80, 2.62 and 3.70. At cut-off 0.80, 26.32% of HPV(−)Cyt(−) women tested positive for S5, which indicates either a potential specificity issue of the tool in our selected group of HPV(−)Cyt(−) women or a higher than expected prevalence of occult CIN in these women. The lowest false positive rate was observed at the 3.70 cut-off (0.92%), a cut-off at which 95.77% (521/544) of cancer cases were still identified. A similar trend was observed with CIN3 cases (Supplementary Table 2). Due to the referral nature of the samples included, exact specificity values could not be calculated, however estimated values are provided in Supplementary Table 3.
Our data on S5 sensitivity combined with our earlier results from studies in the UK, Canada, Mexico and Colombia suggest that the prevalence of HPV infection as well as the difference in screening capacity and performance of populations can affect disease prevalence, thereby arguably the optimal cut-off of S5 could be made ‘setting specific’9-12. An increased cut-off with a lower number of false positives rate would maximise the detection of cancer, which is favourable in countries with minimal screening resources such as in LMICs. Indeed, in settings with no or patchy screening programmes, an increasedcut-off might be justified11,12,29.
The major strengths of our study are its size, the incorporation of sample set that reflects diverse settings from ten different countries spanning five continents. Our study highlights the general trend of increasing DNA methylation with disease progression.
Although HPV infection is an important co-factor in cervical cancer development, a small proportion of cervical cancer samples in the study tested hr-HPV negative as confirmed by HPV testing with multiple assays. Though rare, these cases represent a challenge for detection in the current primary hr-HPV screening programme. Our data shows that nearly all of these cancers (25/26) were identified by the S5 classifier at a cut-off of 0.80. Regardless of the cut-off examined, performance of the S5 classifier was uniform among the stratified groups: histology, FIGO stage, hrHPV status, hrHPV type, sample type, age and country of origin.
A limitation to our study is that all CIN3 and cervical cancer cases come from referral populations and do not accurately represent those that may be apparent in the screening population or those that do not present to clinics. The proportion of rare histological subtypes in our study was also small, so this element would benefit from further investigation. Moreover, much more emphasis was placed on cancers FIGO stage I and II as previously published data indicates that aberrant methylation is an early event in cervical carcinogenesis14,29. An intentional limitation of our study is that we excluded CIN1 and CIN2 which would be present in a real-world setting. Addition of these samples to our study in realistic proportions would likely lower the sensitivity and specificity of the S5 test. There is a further limitation in our selection of the cytology negative women who were presumed to have no disease on the basis of cytological testing. Although we divided these women into HPV+ and HPV− groups these women may not be representative of the routine screening populations in many geographic locations including in Europe and the USA. Therefore, the aim of our present work was to assess a larger panel of CIN3+ samples to confirm the sensitivity and robustness of the assay for the detection of significant disease.
The present findings highlight the major contribution of host EPB41L3 methylation in the S5 score. We showed that the relationship between EPB41L3 methylation was approximatively 4.5 times stronger for severity of lesion than age (p<0.0001). This indicates that host EPB41L3 methylation might have a strong potential to predict disease progression, independent of increasing natural epigenetic methylation levels occurring with age. This is in line with previously published data on the S5 where it better identified women with CIN2 that were more likely to progress to higher stages of the disease8. Additionally, the weight of EPB41L3 methylation shows an increasing trend (p<0.0001, Cuzick test for trend), up to cervical cancer FIGO stage II, where it plateaus. However, the strength of this observation is limited by the decreased number of cervical cancer samples of FIGO stage III and IV, included in the study.
The COVID-19 pandemic points towards a shift to self-sampling for hrHPV primary screening to reduce the burdens on the healthcare professionals and access women who do not respond to screening invitations. Having the possibility to triage hrHPV positive women from the same self-collected specimen would bring many advantages including a reduction in logistical issues associated to systematic screening as well as reducing the subjectivity of cytology30-34. A pilot study tested the accuracy of S5 classifier in cervical self-samples. S5 showed a statistically significant separation between <CIN2 and CIN2+ samples for both urine and cervical self-samples (p≤0.0001). At the predefined cut-off of 0.80, the sensitivity for cervical self-samples was 71% and specificity 68% and for urine samples was 66% and specificity 72%33.
In conclusion, our study shows that the S5 classifier at a cut-off of 0.80 identifies more than 90% CIN3 cases and almost 100% of cervical cancers, independent of histology, FIGO stage hrHPV status, hrHPV genotype, sample type and geographical origin. Adjustment of the cut-off leads to an increase in specificity with only a small decrease in sensitivity. The 3.70 cut-off could allow for a better triage modality for LIMC where screening is not performed as systematically as in higher income countries. Additionally, high methylation levels on the host gene component of the S5 classifier, EPB41L3 is associated with higher severity of the disease, indicating prognostic potential. Thus, considering the growing acceptability of self-sampling, our results support the utility of the S5 classifier as a credible tool for enhanced risk stratification of women in cancer screening programmes.
Data Availability Statement
The data that supports the findings of this study are available from the corresponding author upon reasonable request
Supplementary Material
Novelty and Impact.
Here we confirm that the S5 DNA-methylation test can accurately detect and predict CIN3 and cervical cancer. A fundamental strength of our test is its ability to prioritise women for clinical attention. The S5 test-score is directly proportional to disease severity. Therefore, a high S5 score calls for prompt visualisation of the cervix at colposcopy to inform timely treatment decisions. The S5 test threshold may be adjusted according to context. In a low to medium income setting, an increased threshold with a lower number of false-positives would maximise detection of cancer. Implementation of DNA-methylation biomarkers in clinical settings, ideally requires translation into point-of-care tests, which is challenging but achievable with recent technology developments. Additionally, the COVID-19 pandemic could disrupt efforts to reach cervical cancer elimination targets outlined by WHO. An objective biomarker test, like S5 coupled with minimally invasive self-sampling strategies has the potential to improve the utilisation of healthcare resources and save lives globally.
Acknowledgements
We acknowledge Dr Daniel Seifu from the Department of Biochemistry, Global Health Equity, Kigali, Rwanda, Dr Yirgu Gebrehiwot from the Department of Obstretics and Gynecology, Addis Ababa University, Addis Abbaba, Ethiopia, Dr Vanessa Tenet from the International Agency for Research on Cancer (IARC), World Health Organization. Lyon France for their contribution in collecting the samples included in the study. We also acknowledge Cancer Research UK who funded part of this study (Cancer Prevention programme Grant C569/A10404/CRUK_/CancerResearchUK/UnitedKingdom.
WHO disclaimer:
Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Funding:
Cancer Research UK funded part of this study (grant: Cancer Prevention programme C569/A10404/CRUK). The authors gratefully recognize the patients who participated in this study. The study sponsors CRUK had no role in the study design, collection, analysis and interpretation of data, in the writing of the manuscript and in the decision to submit the manuscript for publication.
Abbreviations:
- CI
confidence interval
- CIN
cervical intraepithelial neoplasia
- HPV
human papillomavirus
- hrHPV
high risk human papillomavirus
- IQR
interquartile range
- LBC
liquid based cytology
- PPV
positive predictive value
- NPV
negative predictive value
- OR
Odd ratio
Footnotes
Conflict Interest
KC reports her employer having received research funding or gratis consumables to support research from the following in the last 3 years: Cepheid, Furoimmun, Genefirst, Selfscreen, Hiantis, Seegene, Roche, Abbott, Hologic. CMW reports receiving the following outside the submitted work: cooperative agreements and grants from the US National Institutes of Health for research on cancer prevention and sexually transmitted infections, reagents and equipment from Roche Molecular Systems, Roche/Ventana Medical Systems, Hologic and Genera Biosystems, research funding from Hologic and Becton Dickinson (BD) and personal fees from BD. The other authors declare no conflict of interest.
Ethics Statement
The samples used for this study have received ethical and research approvals from: i) The International Agency for Research on Cancer (IARC) of the World Health Organization (WHO) for all samples from Bhutan, Colombia, Georgia, India and Philippines, South Africa and Spain; ii) The Scottish HPV Archive, MRC Centre for Reproductive Health, The University of Edinburgh for all samples from the United Kingdom; iii) Institutional Review Board of College of Health, Addis Ababa University for all samples from Ethiopia; iv) University of New Mexico Human Research Review Committee for all samples from USA – New Mexico. The study protocol has been approved by Queen Mary University of London’s Ethical Committee (MTA-2018-ICE-0426, MTA-2018-ICE-1353, 11/06/2018).
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record.
References
- 1.Allen-Leigh B, Uribe-Zúñiga P, León-Maldonado L, Brown BJ, Lörincz A, Salmeron J, Lazcano-Ponce E. Barriers to HPV self-sampling and cytology among low-income indigenous women in rural areas of a middle-income setting: a qualitative study. BMC Cancer. 2017. November 9;17(1):734. doi: 10.1186/s12885-017-3723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catarino R, Petignat P, Dongui G, Vassilakos P. Cervical cancer screening in developing countries at a crossroad: Emerging technologies and policy choices. World J Clin Oncol. 2015. December 10;6(6):281–90. doi: 10.5306/wjco.v6.i6.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021. February 4. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Cuschieri K, Ronco G, Lorincz A, Smith L, Ogilvie G, Mirabello L, Carozzi F, Cubie H, Wentzensen N, Snijders P, Arbyn M, Monsonego J, Franceschi S. Eurogin roadmap 2017: Triage strategies for the management of HPV-positive women in cervical screening programs. Int J Cancer. 2018. August 15;143(4):735–745. doi: 10.1002/ijc.31261. [DOI] [PubMed] [Google Scholar]
- 5.Lorincz AT. The Promise and the Problems of Epigenetics Biomarkers in Cancer. Expert Opin Med Diagn. 2011. September 1;5(5):375–379. doi: 10.1517/17530059.2011.590129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nedjai B, Reuter C, Ahmad A, Banwait R, Warman R, Carton J, Boer S, Cuzick J, Lorincz AT. Molecular progression to cervical precancer, epigenetic switch or sequential model? Int J Cancer. 2018. October 1;143(7):1720–1730. doi: 10.1002/ijc.31549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorincz AT. Virtues and Weaknesses of DNA Methylation as a Test for Cervical Cancer Prevention. Acta Cytol. 2016;60(6):501–512. doi: 10.1159/000450595. [DOI] [PubMed] [Google Scholar]
- 8.Louvanto K, Aro K, Nedjai B, Bützow R, Jakobsson M, Kalliala I, Dillner J, Nieminen P, Lorincz A. Methylation in Predicting Progression of Untreated High-grade Cervical Intraepithelial Neoplasia. Clin Infect Dis. 2020. June 10;70(12):2582–2590. doi: 10.1093/cid/ciz677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorincz AT, Brentnall AR, Scibior-Bentkowska D, Reuter C, Banwait R, Cadman L, Austin J, Cuzick J, Vasiljević N. Validation of a DNA methylation HPV triage classifier in a screening sample. Int J Cancer. 2016. June 1;138(11):2745–51. doi: 10.1002/ijc.30008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramírez AT, Sánchez GI, Nedjai B, Agudelo MC, Brentnall AR, Cuschieri K, Castañeda KM, Cuzick J, Lorincz AT; ASC-US-COL Trial Group. Effective methylation triage of HPV positive women with abnormal cytology in a middle-income country. Int J Cancer. 2021. March 15;148(6): 1383–1393. doi: 10.1002/ijc.33314. [DOI] [PubMed] [Google Scholar]
- 11.Cook DA, Krajden M, Brentnall AR, Gondara L, Chan T, Law JH, Smith LW, van Niekerk DJ, Ogilvie GS, Coldman AJ, Warman R, Reuter C, Cuzick J, Lorincz AT. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int J Cancer. 2019. May 15;144(10):2587–2595. doi: 10.1002/ijc.31976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernández-López R, Lorincz AT, Torres-Ibarra L, Reuter C, Scibior-Bentkowska D, Warman R, Nedjai B, Mendiola-Pastrana I, León-Maldonado L, Rivera-Paredez B, Ramírez-Palacios P, Lazcano-Ponce E, Cuzick J, Salmerón J; FRIDA Study Group. Methylation estimates the risk of precancer in HPV-infected women with discrepant results between cytology and HPV16/18 genotyping. Clin Epigenetics. 2019. October 12;11(1):140. doi: 10.1186/s13148-019-0743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aarnio R, Östensson E, Olovsson M, Gustavsson I, Gyllensten U. Cost-effectiveness analysis of repeated self-sampling for HPV testing in primary cervical screening: a randomized study. BMC Cancer. 2020. July 13;20(1):645. doi: 10.1186/s12885-020-07085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenzi NPC, Termini L, Longatto Filho A, Tacla M, de Aguiar LM, Beldi MC, Ferreira-Filho ES, Baracat EC, Soares-Júnior JM. Age-related acceptability of vaginal self-sampling in cervical cancer screening at two university hospitals: a pilot cross-sectional study. BMC Public Health. 2019. July 18;19(1):963. doi: 10.1186/s12889-019-7292-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorincz AT, Brentnall AR, Vasiljević N, Scibior-Bentkowska D, Castanon A, Fiander A, Powell N, Tristram A, Cuzick J, Sasieni P. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int J Cancer. 2013. August 1;133(3):637–44. doi: 10.1002/ijc.28050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alibegashvili T, Clifford GM, Vaccarella S, Baidoshvili A, Gogiashvili L, Tsagareli Z, Kureli I, Snijders PJ, Heideman DA, van Kemenade FJ, Meijer CJ, Kordzaia D, Franceschi S. Human papillomavirus infection in women with and without cervical cancer in Tbilisi, Georgia. Cancer Epidemiol. 2011. October;35(5):465–70. doi: 10.1016/j.canep.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Muñoz N, Bosch FX, de Sanjosé S, Vergara A, del Moral A, Muñoz MT, Tafur L, Gili M, Izarzugaza I, Viladiu P, et al. Risk factors for cervical intraepithelial neoplasia grade III/carcinoma in situ in Spain and Colombia. Cancer Epidemiol Biomarkers Prev. 1993. Sep-Oct;2(5):423–31. [PubMed] [Google Scholar]
- 18.De Vuyst H, Ndirangu G, Moodley M, Tenet V, Estambale B, Meijer CJ, Snijders PJ, Clifford G, Franceschi S. Prevalence of human papillomavirus in women with invasive cervical carcinoma by HIV status in Kenya and South Africa. Int J Cancer. 2012. August 15;131(4):949–55. doi: 10.1002/ijc.26470. [DOI] [PubMed] [Google Scholar]
- 19.Clifford GM, Rana RK, Franceschi S, Smith JS, Gough G, Pimenta JM. Human papillomavirus genotype distribution in low-grade cervical lesions: comparison by geographic region and with cervical cancer. Cancer Epidemiol Biomarkers Prev. 2005. May;14(5):1157–64. doi: 10.1158/1055-9965.EPI-04-0812. [DOI] [PubMed] [Google Scholar]
- 20.Bosch FX, Manos MM, Muñoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995. June 7;87(11):796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 21.Ngelangel C, Muñoz N, Bosch FX, Limson GM, Festin MR, Deacon J, Jacobs MV, Santamaria M, Meijer CJ, Walboomers JM. Causes of cervical cancer in the Philippines: a case-control study. J Natl Cancer Inst. 1998. January 7;90(1):43–9. doi: 10.1093/jnci/90.1.43. [DOI] [PubMed] [Google Scholar]
- 22.Tshomo U, Franceschi S, Dorji D, Baussano I, Tenet V, Snijders PJ, Meijer CJ, Bleeker MC, Gheit T, Tommasino M, Clifford GM. Human papillomavirus infection in Bhutan at the moment of implementation of a national HPV vaccination programme. BMC Infect Dis. 2014. July 22;14:408. doi: 10.1186/1471-2334-14-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatia R, Serrano I, Wennington H, Graham C, Cubie H, Boland E, Fu G, Cuschieri K. Evaluation of a Novel Single-Tube Method for Extended Genotyping of Human Papillomavirus. J Clin Microbiol. 2018. February 22;56(3):e01687–17. doi: 10.1128/JCM.01687-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn. 2014. April;14(3):293–305. doi: 10.1586/14737159.2014.897610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirabello L, Sun C, Ghosh A, Rodriguez AC, Schiffman M, Wentzensen N, Hildesheim A, Herrero R, Wacholder S, Lorincz A, Burk RD. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J Natl Cancer Inst. 2012. April 4;104(7):556–65. doi: 10.1093/jnci/djs135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirabello L, Schiffman M, Ghosh A, Rodriguez AC, Vasiljevic N, Wentzensen N, Herrero R, Hildesheim A, Wacholder S, Scibior-Bentkowska D, Burk RD, Lorincz AT. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J Cancer. 2013. March 15;132(6):1412–22. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vink FJ, Meijer CJLM, Clifford GM, Poljak M, Oštrbenk A, Petry KU, Rothe B, Bonde J, Pedersen H, de Sanjosé S, Torres M, Del Pino M, Quint WGV, Cuschieri K, Boada EA, van Trommel NE, Lissenberg-Witte BI, Floore AN, Hesselink AT, Steenbergen RDM, Bleeker MCG, Heideman DAM. FAM19A4/miR124-2 methylation in invasive cervical cancer: A retrospective cross-sectional worldwide study. Int J Cancer. 2020. August 15;147(4):1215–1221. doi: 10.1002/ijc.32614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eijsink JJ, Lendvai Á, Deregowski V, Klip HG, Verpooten G, Dehaspe L, de Bock GH, Hollema H, van Criekinge W, Schuuring E, van der Zee AG, Wisman GB. A four-gene methylation marker panel as triage test in high-risk human papillomavirus positive patients. Int J Cancer. 2012. April 15;130(8):1861–9. doi: 10.1002/ijc.26326. [DOI] [PubMed] [Google Scholar]
- 29.Cuschieri K, Lorincz AT, Nedjai B. Human Papillomavirus Research: Where Should We Place Our Bets? Acta Cytol. 2019;63(2):85–96. doi: 10.1159/000493800. [DOI] [PubMed] [Google Scholar]
- 30.Stanczuk G, Baxter G, Currie H, et al. Clinical validation of hrHPV testing on vaginal and urine self-samples in primary cervical screening ( cross-sectional results from the Papillomavirus Dumfries and Galloway — PaVDaG study ). 2016. doi: 10.1136/bmjopen-2015-010660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snoek BC, Splunter Van AP, Bleeker MCG, et al. Cervical cancer detection by DNA methylation analysis in urine. 2019:1–9. doi: 10.1038/s41598-019-39275-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verlaat W, Snoek BC, Wilting SM, et al. Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin Cancer Res. 2018. doi: 10.1158/1078-0432.CCR-17-3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cadman L, Reuter C, Jitlal M, Kleeman M, Austin J, Hollingworth T, Parberry AL, Ashdown-Barr L, Patel D, Nedjai B, Lorincz AT, & Cuzick J (2021). A randomised comparison of different vaginal self-sampling devices and urine for human papillomavirus testing - Predictors 5.1. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, cebp.1226.2020. Advance online publication. 10.1158/1055-9965.EPI-20-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernández-López R, Hermosillo L, León-Maldonado L, Velázquez-Cruz R, Torres-Ibarra L, Lazcano-Ponce E, Lörincz A, Wheeler CM, Bosch FX, Cuzick J, Rivera-Paredez B, Nedjai B, & Salmerón J (2021). Performance of an affordable urine self-sampling method for human papillomavirus detection in Mexican women. PloS one, 16(7), e0254946. 10.1371/journal.pone.0254946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the findings of this study are available from the corresponding author upon reasonable request