Abstract
Germline variants might predict cancer progression. Bevacizumab improves overall survival (OS) in patients with advanced cancers. No biomarkers are available to identify patients that benefit from bevacizumab. A meta-analysis of genome-wide association studies (GWAS) was conducted in 1,520 patients from phase III trials (CALGB 80303, 40503, 80405, and ICON7), where bevacizumab was randomized to treatment without bevacizumab. We aimed to identify genes and SNPs associated with survival independently of bevacizumab treatment or through interaction with bevacizumab. A cause-specific Cox model was used to test the SNP-OS association in both arms combined (prognostic), and the effect of SNPs-bevacizumab interaction on OS (predictive) in each study. The SNP effects across studies were combined using inverse variance. Findings were tested for replication in advanced colorectal and ovarian cancer patients from The Cancer Genome Atlas (TGCA). In the GWAS meta-analysis, patients with rs680949 in PRUNE2 experienced shorter OS compared to patients without it (p=1.02×10−7, HR=1.57, 95% CI:1.33–1.86), as well as in TCGA (p=0.0219, HR=1.58, 95% CI:1.07–2.35). In the GWAS meta-analysis, patients with rs16852804 in BARD1 experienced shorter OS compared to patients without it (p=1.40×10−5, HR=1.51, 95% CI:1.25–1.82) as well as in TCGA (p=1.39×10−4, HR=3.09, 95% CI:1.73–5.51). Patients with rs3795897 in AGAP1 experienced shorter OS in the bevacizumab arm compared to the non-bevacizumab arm (p=1.43×10−5). The largest GWAS meta-analysis of bevacizumab treated patients identified PRUNE2 and BARD1 (tumor suppressor genes) as prognostic genes of colorectal and ovarian cancer, respectively, and AGAP1 as a potentially predictive gene that interacts with bevacizumab with respect to patient survival.
Keywords: Bevacizumab, overall survival, PRUNE2, BARD1, AGAP1
Introduction
Germline variants can significantly improve predictions of the survival of cancer patients compared to clinical variables alone 1. In addition to their prognostic potential, germline variants are also useful to individualize the selection of the most effective treatment 2,3. This is clearly demonstrated by the example of deficient mismatch repair of germline DNA, which creates tumors responsive to checkpoint inhibition 4. Another example is tumors carrying BRCA1 germline variants, which are more responsive to poly(ADP-ribose) polymerase (PARP) inhibitors 5. When germline variants change the biology of the tumor phenotype or its responsiveness to therapy, new targets and/or changes in the tumor microenvironment increase the efficacy of novel therapies.
There are several anti-tumor therapies lacking validated biomarkers to select patients more responsive to treatment. One of them is bevacizumab, a humanized monoclonal antibody that binds to the vascular endothelial growth factor A (VEGF-A), a proangiogenic factor that plays an important role in tumor growth. Bevacizumab is approved by the U.S. Food and Drug Administration and the European Medicines Agency for the treatment of many tumor types, including metastatic colorectal cancer (CRC), metastatic breast cancer, advanced non-squamous lung cancer, metastatic renal cell carcinoma, recurrent glioblastoma, advanced cervical and ovarian cancers, and hepatocellular carcinoma 6,7.
VEGF-A signaling is a key factor in angiogenesis, and the inhibition of the VEGF-A pathway impairs tumor angiogenesis, reducing tumor growth and distant metastatization. As angiogenesis is primarily a host-mediated process 8,9, germline variation that affects the extent of angiogenesis might interfere with the efficacy of bevacizumab.
The identification of tumor-type agnostic markers to predict outcome in cancer patients is a challenge. However, pan-cancer studies have reported germline variants associated with outcome across different tumor types 1,10. In addition, the identification of biomarkers that can indicate which patients would benefit or not from treatment with bevacizumab regardless of tumor type is even more challenging. Many studies have attempted to identify single nucleotide polymorphisms (SNPs) associated with overall survival (OS) in patients treated with bevacizumab, including a genome-wide association study (GWAS) 11 and studies focused on VEGF-pathway genes (for a review 12,13). However, for approved indications, there is no molecular marker that can identify the patients who are more or less likely to benefit from treatment with bevacizumab.
Using a GWAS meta-analysis of four phase III clinical trials of patients with different tumor types and randomized to treatment with bevacizumab, the objective of this study was to identify new genes, the genetic variation of which are associated with survival independently of treatment (prognostic). We also aimed at identifying new genes, the genetic variation of which interacts with bevacizumab with respect to patient outcome (predictive). We applied different approaches and methodologies to the newly discovered SNPs and genes, including, when available, external validation using data from The Cancer Genome Atlas (TCGA).
Materials and Methods
Patients and phase III clinical trials
The phase III clinical trials included in this study are from the Cancer and Leukemia Group B (CALGB, now part of the Alliance for Clinical Trials in Oncology, Alliance) and the Gynecologic Cancer InterGroup International Collaboration on Ovarian Neoplasms (ICON7). All studies were randomized, either bevacizumab versus placebo (CALGB 80303 and 40503), bevacizumab versus no bevacizumab (ICON7), or bevacizumab versus another targeted agent (cetuximab, CALGB 80405). All patients had either advanced or metastatic disease.
CALGB 80303 was conducted in advanced pancreatic cancer patients treated with gemcitabine and either 10 mg/kg bevacizumab or placebo. CALGB 40503 was conducted in hormone receptor-positive advanced-stage breast cancer patients treated with letrozole and either bevacizumab or placebo. CALGB 80405 was conducted in locally advanced or metastatic CRC patients treated with chemotherapy (FOLFIRI/FOLFOX) and either bevacizumab or cetuximab. ICON7 was conducted in advanced or high-risk early-stage ovarian cancer patients treated with carboplatin and paclitaxel with or without bevacizumab. Patient eligibility, characteristics, stratifications, and treatments have been previously described 14–17.
The primary endpoint of this study is OS, defined as the time from trial registration (CALGB 80303), study entry (CALGB 80405, CALGB 40503), or randomization (ICON7) until death from any cause. Patients without reported deaths were censored at their last known follow-up.
Genotyping and quality control
Germline DNA was obtained from peripheral blood. The genotyping platforms used in each study are described in Table 1, and the number of SNPs included in the analysis in each study after quality control (QC) is described in Figure S1. Additional information on the QC procedures can be found in the publications of the GWAS of CALGB 80303 18 and 80405 19. The genotyping and QC for CALGB 40503 and ICON7 have not been reported before. In CALGB 40503, 964,058 SNPs were genotyped using the Illumina Human OmniExpressExome-8 BeadChip. Based on QC criteria, specifically call rates less than 95% (51,759) and minor allele frequencies (MAF) less than 0.05 (318,063), 369,822 SNPs were excluded from the analyses. The 594,236 SNPs passing QC in CALGB 40503 were included in the association analyses (Figure S1). In ICON7, 951,117 SNPs were genotyped using the Illumina Human OmniExpressExome-8 BeadChip. Based on QC criteria, specifically call rates less than 95% (4,634), MAF less than 0.05 (338,597), SNPs in chromosome 23 (15,748), and Hardy-Weinberg equilibrium with p-value <1.00×10−8 (323), 359,302 SNPs were excluded from the analyses. The 591,815 SNPs passing QC in ICON7 were included in the association analyses (Figure S1). Each SNP that passed QC and has genotype data for at least one study was tested for association.
Table 1. Clinical trials and GWAS in patients of European ancestry.
CALGB Cancer and Leukemia Group B; ICON7 International Collaboration on Ovarian Neoplasms; FOLFIRI 5-fluorouracil (400 mg/m2 and 2400 mg/m2) + leucovorin (400 mg/m2) + irinotecan (180 mg/m2); mFOLFOX6 5-fluorouracil (400 mg/m2 and 2400 mg/m2) + leucovorin (400 mg/m2) + oxaliplatin (85 mg/m2); SD standard deviation; OS overall survival; CI confidence interval; NR not reached; NE not estimated.
| Clinical Trial | CALGB 80303 | CALGB 40503 | CALGB 80405 | ICON7 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Cancer type | Advanced pancreatic cancer | Hormone receptor-positive advanced-stage breast cancer | Locally advanced or metastatic colorectal cancer | Advanced or high-risk early-stage ovarian cancer | |||||
| Genotype platform | Illumina HumanHap550-Quad | Illumina Human OmniExpressExome-8 | Illumina HumanOmniExpress-12v1 and Illumina Human OmniExpressExome-8 | Illumina Human OmniExpressExome-8 | |||||
| Treatment | Gemcitabine 1,000 mg/m2 on days 1, 8, and 15 plus either placebo or bevacizumab 10 mg/kg on days 1 and 15 | Letrozole 2.5 mg orally/day plus either placebo or bevacizumab 15 mg/kg every 21 days | FOLFIRI or mFOLFOX6 every two weeks plus either cetuximab 400 mg/m2 on day 1, then 250 mg/m2 every week or bevacizumab 5 mg/kg every other week | Carboplatin AUC 5 or 6 and paclitaxel 286 mg/m2 or the same chemotherapy regimen plus bevacizumab 7.5 mg/kg every 3 weeks | |||||
|
| |||||||||
| Treatment arms | Bevacizumab (n=154) | Placebo (n=140) | Bevacizumab (n=105) | Placebo (n=100) | Bevacizumab (n=310) | Cetuximab (n=300) | Bevacizumab (n=212) | No Bevacizumab (n=199) | |
|
| |||||||||
| Age | Mean (SD) | 64.1 (10.2) | 63.6 (10.9) | 56.9 (11.7) | 58.5 (12.1) | 58.5 (12.2) | 59.3 (10.9) | 54.6 (9.6) | 57.7 (9.7) |
| Gender | Male (n, %) | 90 (58.4) | 69 (49.3) | 0 (0.0) | 0 (0.0) | 119 (38.4) | 112 (37.3) | 0 (0.0) | 0 (0.0) |
| Female (n, %) | 64 (41.6) | 71 (50.7) | 105 (100.0) | 100 (100.0) | 191 (61.6) | 188 (62.7) | 199 (100.0) | 212 (100.0) | |
| OS | Median (95% CI) | 5.8 (4.9–7.01) | 6.3 (5.2–7.9) | 49.2 (40.0-NE) | 46.0 (36.3–50.5) | 29.4 (26.0–32.6) | 30.1 (25.8–39.2) | NR (41.1-NE) | NR (38.8-NE) |
Statistical analysis
The objectives of the study were to test the association between SNPs and OS in both arms combined in each trial (prognostic effect) and the interaction between SNPs and bevacizumab treatment with respect to OS in each trial (predictive effect). A GWAS meta-analysis was performed including previously published data (CALGB 80405 19) and unpublished data (CALGB 80303, CALGB 40503, and ICON7). For the principal component analysis, the genetic ancestral origin of patients was estimated based on clustering by Eigenstrat 20. Following linkage disequilibrium (LD) pruning using an LD threshold of R2 0.2, pruned genotype data was compared to genotype data for the set of pruned variants from the 1,000 Genomes Project Phase 3 samples, including individuals from ASW, CEU, CHB, MEX, and YRI populations. Samples within each dataset that grouped with 1,000 Genomes Project CEU samples based upon the first and second principal components were categorized as of genetically determined European ancestry.
A cause-specific Cox model was fitted in each trial to test the association between SNPs and OS. The analysis was adjusted by various covariates in each study: prior radiation, extent of disease, and performance status (CALGB 80303); disease-free interval and presence of measurable disease (CALGB 40503); country group and FIGO score (ICON7); protocol chemotherapy (FOLFOX or FOLFIRI), prior radiotherapy, prior adjuvant therapy, and primary tumor location (CALGB 80405). For CALGB 80405, primary tumor location was added due to its prognostic value 19. An additive genetic model was used.
For the prognostic effect, the inverse variance formula was used to combine the SNP effect in each study to obtain the meta-analysis estimate (β) of the SNP-OS association and its standard error (SE). The exponential function was used to calculate the hazard ratio (HR) for each SNP. The SE was applied in function exp(β−1.96*(SE)) to calculate the lower bound of 95% confidence interval (CI) of HR and in the function exp(β+1.96*(SE)) to calculate the upper bound of 95% CI of HR.
For the predictive effect, the same methodology was applied, with a Cox model fitted in each study and the inverse variance formula used to combine the SNP-treatment interaction effect to obtain the meta-analysis estimate (β) of the SNP-treatment association and its SE.
The heterogeneity across studies was examined by the Cochran’s Q test, and the SNPs reported as associated with OS were those with Cochran’s Q p-value >0.20. The p-values were not corrected for multiple comparisons, as the p-values were used to prioritize associations for future analyses, including external replication in TCGA.
External replication of SNPs associated with OS (prognostic) in TCGA
We sought to replicate the prognostic associations observed in the meta-analysis using the data from cancer patients in TCGA 21 as we have done before 19. Stage IV patients with either colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) (n=80) or ovarian serous cystadenocarcinoma (OV) (n=86) were included. Patients with stage IV breast invasive carcinoma (n=20) or stage IV pancreatic adenocarcinoma (n=5) were not included due to their small sample sizes.
The ten most statistically significant prognostic SNPs associated with OS with the same direction of effect (either reduced or increased OS) in at least three out of the four trials were selected for replication. If the SNP for testing was not genotyped in TCGA, a SNP in high LD with it (R2>0.8 in Europeans, LDlink 22) was used as a proxy. The SNPs were tested for association with OS using a Cox model as described above. The analysis was adjusted by primary tumor location in COAD+READ patients. The inverse variance formula was used to combine the SNP effect in COAD+READ and OV patients to obtain the meta-analysis β estimate and SE to calculate HR and 95% CI as described above (p-value <0.05 was used as the threshold for statistical significance).
Bioinformatics and downstream analyses
To test the hypothesis that germline variants associated with outcome might act as expression quantitative trait loci (eQTL) for tumor infiltrating immune cells, we investigated selected SNPs using the Database of Immune Cell eQTLs (DICE) 23. Moreover, because germline variants might change gene expression in non-tumoral tissues that interact with cancer cells, for example aorta, blood cells, and fibroblasts 24, we have also used the data from The Genotype-Tissue Expression project (GTEx v8) 25. In order to postulate the mechanisms by which the SNPs may be exerting their effect, SNPs were referenced in the RegulomeDB 26 for functional inference. For those SNPs with experimentally-determined binding site motifs according to RegulomeDB 26, AtSNP was used to quantify the impact of SNPs on transcription factor binding 27.
The cBioPortal 28 tool was used to obtain mRNA expression data of patients from TCGA. In TCGA, mRNA gene expression data was available for 51 stage IV COAD+READ patients and for 38 stage IV OV patients. The association between prognostic SNPs and mRNA expression (log2 RNA Seq V2) in patients with stage IV COAD+READ and OV from TCGA was tested by the Jonckheere-Terpstra test (patients having homozygous reference versus one or more alternate alleles, p-value < 0.05 was used for statistical significance).
Results
A total of 1,520 cancer patients of genetically determined European ancestry were genotyped and included in this study. The characteristics and demographics of patients are in Table 1. The Q-Q plots for the association between SNPs and OS show no evident deviation in distribution (Figure S2).
SNPs associated with OS in both arms (prognostic)
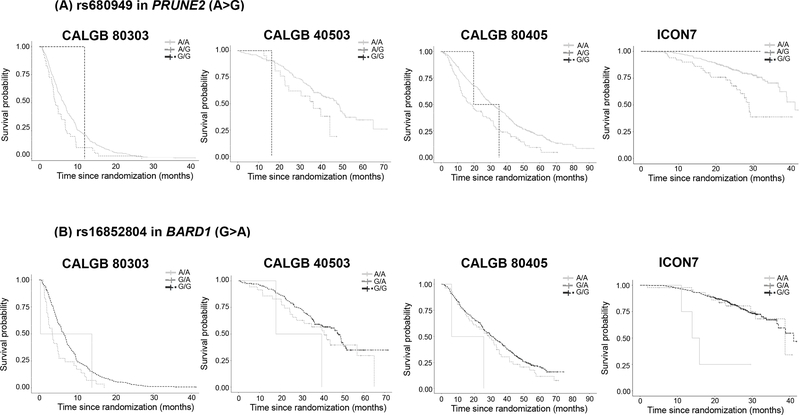
The Manhattan and Q-Q plots are shown in Figure S2. The ten most statistically significant SNPs that had the same direction of effect (either reduced or increased OS) in at least three out of the four studies are shown in Table 2. Variant rs680949 (A>G) in the intron 9 of PRUNE2 had the strongest evidence for association (p=1.02×10−7, HR=1.57, 95% CI 1.33–1.86) with a p-value <0.05 in each of the four studies (results in each study are shown in Table S1). Patients with the AG genotype of rs680949 (A>G) in PRUNE2 experienced significantly shorter OS than those with AA genotype (Figure 1A). The median (95% CI) OS (months) for the AG and AA genotypes in each CALGB study, respectively, was 4.9 (3.7–6.9) and 6.4 (5.7–7.2) in CALGB 80303, 36.3 (24.5-not estimated, NE) and 48.7 (42.2–51.3) in CALGB 40503, and 18.8 (12.9–27.6) and 31.1 (28.1–34.5) in CALGB 80405. Because the majority of genotypes did not reach median OS in ICON7, we report the 75th percentile (95% CI) OS (months) for the AG and AA genotypes, which was 23.9 (17.1–29.2) and 33.1 (29.9–41.1), respectively. Due to the very small number of GG patients, their median or 75th percentile (95% CI) OS (months) in each study is reported in Table 3.
Table 2. Results of the association between SNPs and OS as a prognostic effect and as a predictive effect. Ten most statistically significant SNPs associated with OS with same direction of effect (either reduced or increased OS), in at least three out of four trials.
Rows in bold are those with p-value<0.05 in at least three out of four trials. Ch chromosome, NA Intergenic SNP, MAF minor allele frequencies in CALGB 80303, 40503, 80405, and ICON7, respectively, HR hazard ratio, CI confidence interval. Direction of effect increasing (−) or decreasing (+) OS in CALGB 80303, 40503, 80405, and ICON7, respectively. “_” SNP not present in the genotyping platform of the trial.
| Prognostic effect | |||||||||||
|
| |||||||||||
| SNP | Ch | Gene | Feature | 5’ flanking | 3’ flanking | Base change | MAF | Direction of effect | Effect size (β) | HR (95% CI) | p-value |
|
| |||||||||||
| rs680949 | 9 | PRUNE2 | intron | LOC392352 | LOC392352 | A>G | 0.07/0.06/0.07/0.06 | ++++ | 0.45 | 1.57 (1.33– 1.86) | 1.02x10 −7 |
| rs218527 | 18 | NA | NA | LOC100129774 | LOC100128360 | G>A | 0.09/0.10/0.10/0.11 | ++++ | 0.39 | 1.48 (1.28–1.71) | 1.83x10 −7 |
| rs2795492 | 9 | CORO2A | intron | TRIM14 | TBC1D2 | G>A | 0.41/0.41/0.42/0.42 | ---- | −0.22 | 0.81 (0.73–0.88) | 5.41x10−6 |
| rs476612 | 9 | LOC100129762 | intron | LOC392352 | PRUNE2 | A>C | _/0.06/0.07/0.06 | _+++ | 0.52 | 1.68 (1.34–2.12) | 8.45x10 −6 |
| rs7615734 | 3 | NA | NA | TPRG1 | TP63 | A>G | 0.16/0.18/0.18/0.21 | ++++ | 0.26 | 1.29 (1.15–1.45) | 9.58x10−6 |
| rs6712389 | 2 | NA | NA | KIAA1604 | LOC729009 | G>A | _/0.31/0.29/0.27 | _--- | −0.28 | 0.76 (0.67–0.86) | 1.16x10−5 |
| rs16852804 | 2 | BARD1 | intron | LOC100128203 | ABCA12 | G>A | 0.06/0.10/0.07/0.06 | ++++ | 0.41 | 1.51 (1.25–1.82) | 1.40x10−5 |
| rs10950639 | 7 | NA | NA | AGR3 | LOC100131425 | A>G | _/0.17/0.21/0.20 | _+++ | 0.29 | 1.33 (1.17–1.52) | 1.40x10−5 |
| rs4807493 | 19 | PIP5K1C | UTR-3’ | C19orf29 | TJP3 | G>A | 0.21/0.17/0.18/0.20 | -+-- | −0.25 | 0.78 (0.70–0.88) | 2.26x10−5 |
| rs2103445 | 8 | NA | NA | CALB1 | TMEM64 | G>A | _/0.30/0.29/0.31 | _+++ | 0.25 | 1.29 (1.15–1.45) | 2.29x10−5 |
|
| |||||||||||
| Predictive effect | |||||||||||
|
| |||||||||||
| SNP | Ch | Gene | Feature | 5’ flanking | 3’ flanking | Base change | MAF | Direction of effect | Effect size (β) | p-value | |
|
| |||||||||||
| rs4969481 | 17 | DCXR | near 3’ | RAC3 | DCXR | G>A | 0.13/0.16/0.14/0.14 | ---- | −0.70 | 1.18x10−7 | |
| rs11895736 | 2 | LTBP1 | intron | TTC27 | RASGRP3 | A>G | 0.12/0.14/0.14/0.14 | ---- | −0.68 | 6.85x10−7 | |
| rs448960 | 20 | LOC100129869 | near 3’ | PMEPA1 | LOC100129869 | A>G | _/0.49/0.46/0.47 | _--- | −0.58 | 7.50x10−6 | |
| rs10763269 | 10 | NA | NA | LOC389970 | ZWINT | G>A | 0.27/0.24/0.22/0.21 | ++++ | 0.48 | 1.09x10−5 | |
| rs13392750 | 2 | NA | NA | ANKRD44 | LOC729342 | A>G | _/0.46/0.46/0.43 | _+++ | 0.58 | 1.23x10−5 | |
| rs3795897 | 2 | AGAP1 | intron | LOC642692 | LOC100130154 | G>A | 0.13/0.11/0.17/0.13 | ++++ | 0.68 | 1.43x10 −5 | |
| rs1150743 | 6 | NA | NA | HLA-J | ETF1P1 | G>A | 0.11/0.11/0.10/0.10 | ++++ | 0.66 | 1.83x10−5 | |
| rs10915428 | 1 | NA | NA | hCG_2036596 | LOC644357 | A>C | 0.46/0.44/0.51/0.48 | ---- | −0.39 | 1.86x10−5 | |
| rs11860804 | 16 | NA | NA | LOC729847 | DYNLRB2 | G>A | _/0.49/0.50/0.49 | _--- | −0.49 | 1.87x10−5 | |
| rs6453031 | 5 | RGNEF | intron | UTP15 | ENC1 | A>C | _/0.15/0.16/0.16 | _--- | −0.63 | 1.88x10−5 | |
Fig. 1. Kaplan-Meier estimates of OS for (A) rs680949 in PRUNE2 (A>G, MAF 0.06–0.07) and (B) rs16852804 in BARD1 (G>A, MAF 0.06–0.10), prognostic effect.
CALGB Cancer and Leukemia Group B, ICON7 International Collaboration on Ovarian Neoplasms.
Table 3.
Median (95% CI) OS (months) – CALGB 80303, 40503, 80405 – and 75th percentile (95% CI) OS (months) – ICON7 – for rs680949 in PRUNE2 (A>G, MAF 0.06–0.07) and rs16852804 in BARD1 (G>A, MAF 0.06–0.10), prognostic effect, and for rs3795897 in AGAP1 (G>A, MAF 0.11–0.17), predictive effect.
| Prognostic effect: rs680949 in PRUNE2 (A>G, MAF 0.06–0.07) | ||||||||
|
| ||||||||
| Genotype |
CALGB 80303
Median OS (95% CI) - n |
CALGB 40503
Median OS (95% CI) - n |
CALGB 80405
Median OS (95% CI) - n |
ICON7 75th percentile OS (95% CI) - n |
||||
|
| ||||||||
| AA | 6.4 (5.7–7.2) - 253 | 48.7 (42.2–51.3) - 183 | 31.1 (28.1–34.5) −527 | 33.1 (29.9–41.1) - 360 | ||||
| AG | 4.9 (3.7–6.9) - 40 | 36.3 (24.5- NE) – 21 | 18.8 (12.9–27.6) - 81 | 23.9 (17.1–29.2) - 50 | ||||
| GG | 12.0 (NE-NE) - 1 | 16.3 (NE-NE) - 1 | 32.2 (19.8-NE) – 2 | NR (NE-NE) - 1 | ||||
|
| ||||||||
| Prognostic effect: rs16852804 in BARD1 (G>A, MAF 0.06–0.10) | ||||||||
|
| ||||||||
| Genotype |
CALGB 80303
Median OS (95% CI) - n |
CALGB 40503
Median OS (95% CI) - n |
CALGB 80405
Median OS (95% CI) - n |
ICON7 75th percentile OS (95% CI) - n |
||||
|
| ||||||||
| GG | 6.5 (5.8–7.2) - 263 | 48.7 (40.4–51.3) - 168 | 29.6 (26.7–33.0) - 537 | 32.3 (27.6–38.9) - 362 | ||||
| GA | 3.8 (2.7–6.8) - 29 | 42.2 (35.0-NE) - 35 | 29.0 (19.3–36.4) - 70 | 38.8 (22.9-NE) - 45 | ||||
| AA | 13.9 (0.4-NE) - 2 | 39.9 (17.9-NE) - 2 | 15.9 (6.11-NE) - 2 | NR (11.1-NE) - 4 | ||||
|
| ||||||||
| Predictive effect: rs3795897 in AGAP1 (G>A, MAF 0.11–0.17) | ||||||||
|
| ||||||||
|
CALGB 80303
Median OS (95% CI) - n |
CALGB 40503
Median OS (95% CI) - n |
CALGB 80405*
Median OS (95% CI) - n |
ICON7 75th percentile OS (95% CI) - n |
|||||
|
| ||||||||
| Genotype | Placebo | Bevacizumab | Placebo | Bevacizumab | Cetuximab | Bevacizumab | No bevacizumab | Bevacizumab |
|
| ||||||||
| GG | 6.31 (5.22–7.88) −107 | 6.54 (4.90–7.33) - 118 | 46.6 (36.4-NE) −75 | 49.2 (40.4-NE)- 87 | 23.2 (18.7–28.8) - 66 | 31.7 (25.2–41.0) - 71 | 27.6 (24.0-NE) - 160 | 37.0 (33.1-NE) −157 |
| GA | 5.45 (3.78–9.66) - 30 | 5.06 (3.71–6.64) - 33 | 40.1 (35.8-NE) - 22 | 33.4 (22.2-NE) −16 | 41.1 (22.5–60.9) - 35 | 30.1 (11.2–39.7) −20 | NR (1.91-NE) - 36 | 26.1 (21.1-NE) - 49 |
| AA | 6.08 (4.99–7.88) - 3 | 7.23 (2.83-NE) - 3 | NR (35.5-NE) - 2 | NR (22.7-NE) −2 | 45.9 (45.9-NE) - 4 | 22.0 (NE-NE) - 3 | NR (NE-NE) - 3 | 17.4 (16.3-NE) - 6 |
genotyped only in patients from the GWAS-Batch 1 (n=199, Fig. S1). CALGB Cancer and Leukemia Group B, ICON7 International Collaboration on Ovarian Neoplasms; OS overall survival; CI confidence interval; NR not reached; NE not estimated.
Replication in TCGA of the SNPs associated with OS in both arms (prognostic)
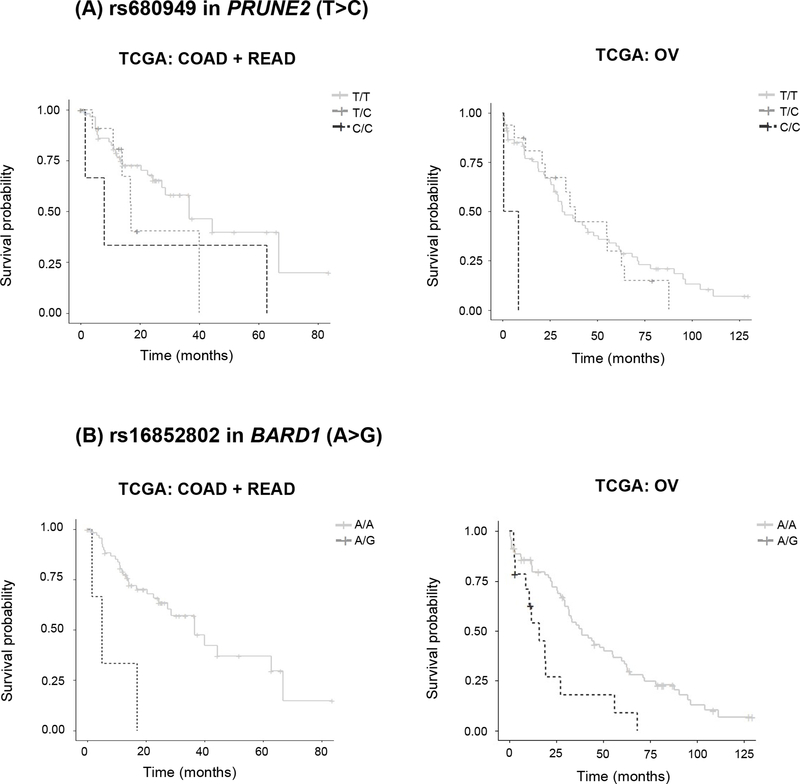
Seven out of the ten SNPs in Table 2 had genotype data (same SNP or a proxy) in TCGA and were tested for replication in COAD+READ and OV patients (Table S2). Out of the seven SNPs, rs680949 in PRUNE2 and rs16852802 in BARD1 showed a p-value <0.05 in the TCGA meta-analysis (Table S2). Patients with the CC and TC genotypes in rs680949 in PRUNE2, genotyped in the opposite strand in TCGA (T>C in TCGA corresponding to A>G in the GWAS), experienced shorter OS than patients with the TT genotype (p=0.0219, HR=1.58, 95% CI 1.07–2.35) (Figure 2A). The median (95% CI) OS (months) for the CC, TC, and TT genotypes, respectively, was 8.07 (1.63-NE), 17.0 (14.1-NE) and 36.5 (27.4-NE) in COAD+READ and 4.67 (0.8-NE), 38.53 (22.7-NE) and 31.73 (27.4–56.2) in OV.
Fig. 2. Kaplan-Meier estimates of OS for (A) rs680949 in PRUNE2 (T>C, MAF 0.11–0.12) and (B) rs16852802 in BARD1 (A>G, MAF 0.02–0.08, complete LD, R2=1.0, with rs16852804) in TCGA patients.
It should be noted that the frequency of the AG genotype of rs16852802 in BARD1 (A>G) in COAD+READ patients is low (n=2). TCGA The Cancer Genome Atlas, COAD colon adenocarcinoma, READ rectum adenocarcinoma, OV ovarian serous cystadenocarcinoma.
Patients with the AG genotype of rs16852802 (A>G) in BARD1 experienced shorter OS than patients with the AA genotype (p=1.39×10−4, HR=3.09, 95% CI 1.73–5.51) (Figure 2B). The median (95% CI) OS (months) for the AG and AA genotypes, respectively, was 5.1 (1.63-NE) and 36.5 (27.4-NE) in COAD+READ and 16.0 (10.6-NE) and 38.5 (31.4–60.0) in OV. No patients with the GG genotype were found. Variant rs16852802 (A>G) in BARD1 is in complete LD (R2=1.0) with rs16852804 (G>A) genotyped in the GWAS, and A>G of rs16852802 corresponds to G>A of rs16852804. In the GWAS, patients with the AA and GA genotypes of rs16852804 (G>A) in BARD1 experienced significantly shorter OS than those with GG genotype (p=1.40×10−5, HR=1.51, CI 1.25–1.82) (Figure 1B, Table 3).
The association between prognostic SNPs in PRUNE2 and BARD1 and their tumor mRNA expression in TCGA was tested, and only the AG genotype of rs16852802 (A>G) in BARD1 was associated with lower BARD1 mRNA expression in OV patients compared to the AA genotype (p=0.0169, Figure S3).
Bioinformatic functional analyses of the rs680949 in PRUNE2 and rs16852804 in BARD1
These analyses show that rs680949 (T>C corresponding to A>G in the GWAS) in PRUNE2 is located within a binding motif for Myc and Max transcription factors in human embryonic stem cell lines, where T>C creates a binding site for the Myc transcription factor (p=0.006) 27. The Myc transcription factor binds preferentially to the C than the T allele (p=0.0038, log-likelihood=−23.7 for the C allele, and p=0.094, log-likelihood=−46.7 for the T allele) (Figure S4) 27.
The A allele of rs16852804 (G>A) decreases BARD1 expression in whole blood in GTEx (p=1.90×10−5, normalized effect size, NES=−0.25) 25. The A allele of rs368962482 in BARD1 (G>A, complete LD, R2=1.0, with rs16852804) decreases BARD1 expression in non-classical monocytes in DICE (p=0.013, effect size=−1.3) (Figure S5) 23.
SNP-bevacizumab interactions associated with OS (predictive)
The Manhattan and Q-Q plots are shown in Figure S2. The ten SNPs with the smallest interaction p-values that had the same direction of effect (conferring either reduced or increased OS in the bevacizumab arm versus the non-bevacizumab arm) in at least three out of the four studies are shown in Table 2. The only SNP in Table 2 with p-value <0.05 in three out of the four studies was rs3795897 (G>A, MAF 0.11–0.17) in AGAP1 (p=1.43×10−5) (details on the results of each study are shown in Table S1). The genotype-bevacizumab interaction shows that patients with the GA genotype of rs3795897 in AGAP1 tended to experience shorter OS in the bevacizumab arm compared to the non-bevacizumab arm. In contrast, patients with the GG genotype of rs3795897 in AGAP1 tended to experience longer OS in the bevacizumab arm compared to the non-bevacizumab arm. This is more evident in ICON7, where the 3-year survival for the GA and GG genotypes, respectively, was 57.1% and 81.5% in the bevacizumab arm, and 96.7% and 64.3% in the non-bevacizumab arm. The effect of the AA genotype is more difficult to detect due to its small sample size (Figure S6, Table 3).
Bioinformatics functional analysis of rs3795897 in AGAP1
Variant rs3795897 (G>A) is an eQTL for AGAP1-IT1, with the A allele significantly increasing gene expression in many different cell types in GTEx, the strongest of these effects being in cultured fibroblasts (p=7.8×10−14, NES=0.59) 25.
Discussion
This GWAS meta-analysis of 1,520 cancer patients treated with bevacizumab-containing regimens has selected PRUNE2 and BARD1 as genes with potential prognostic significance. These findings were supported by validation in stage IV cancer patients from TCGA. Moreover, AGAP1 was selected as a potentially novel gene with predictive significance, interacting with bevacizumab with respect to OS. To the best of our knowledge, this is the largest GWAS meta-analysis of patients treated with bevacizumab. Here, we discuss the main findings from our studies in the context of available knowledge of the biology and genetics of these genes in oncology.
PRUNE2, prune homolog 2 with BCH domain, acts as a tumor suppressor gene through the suppression of Ras homolog family member A activity 29,30. Variant rs680949 (A>G) in PRUNE2 was identified in this study as being prognostic of OS, with a p-value close to the cut-off of genome-wide significance (p-value=1.02×10−7) and a p-value <0.05 in each of the four studies. The AG genotype of rs680949 (A>G) in PRUNE2 was associated with shorter OS and was replicated in the meta-analysis of COAD+READ and OV patients from TCGA. However, considering COAD+READ and OV independently, the effect is not present in OV. Bioinformatics functional analysis showed that the G allele of rs680949 (A>G) in PRUNE2 increases the likelihood of Myc binding to PRUNE2 compared to the A allele 27. Myc amplifies the expression of oncogenes 31,32 and decreases the expression of tumor suppressor genes 33. We then hypothesize that the G allele of rs680949 (A>G) in PRUNE2 might decrease the expression of PRUNE2 by increasing Myc binding. The reduced expression of PRUNE2 and its tumor suppressor activity might lead to reduced survival in stage IV CRC patients with the G allele of rs680949 (A>G) in PRUNE2.
This study also proposes BARD1 as a prognostic gene in stage IV ovarian cancer. BARD1, BRCA1-associated RING domain 1, acts as a tumor suppressor gene and its interaction with BRCA1 is essential for the tumor suppression activity of BRCA1 34. In this study, the AA and AG genotypes of rs16852804 (G>A) in BARD1 were associated with shorter OS. Although the p-value did not reach the cut-off of genome-wide significance (p-value=1.40×10−5), the association was also observed in COAD+READ and OV patients from TCGA. The AG genotype of rs16852802 (A>G, G allele equivalent to the A allele of rs16852804, G>A) in BARD1 was associated with lower BARD1 mRNA expression in OV but not in COAD+READ tumors. This finding is consistent with the tumor suppressor activity of BARD1, with a more prominent effect in ovarian cancer, where higher mRNA expression leads to a higher tumor suppressor activity and hence longer OS. Consistently, the GA genotype of rs16852804 (G>A) in BARD1 might be decreasing BARD1 expression in the tumor of patients in our study, which leads to a reduced tumor suppressor activity, contributing to the poor prognosis. Our analysis also showed that the A allele of rs16852804 (G>A) decreases the expression of BARD1 in whole blood, and the A allele of rs368962482 (G>A, in LD with rs16852804, R2=1.0) decreases the expression of BARD1 in non-classical monocytes. These findings indicate that genetic variation in BARD1 might be exerting its effect also through indirect, non-tumoral mechanisms. Non-classical monocytes are essential to maintain vascular homeostasis in endothelial cells 35. When BRCA1 was knocked out in mice, an impaired endothelial function was observed 36, suggesting an important role of BRCA1 and its regulation by BARD1 in the normal function of vascular endothelial cells. A dysfunctional endothelium promotes inflammatory signaling and aggressive properties of cancer cells 37. Thus, we postulate an additional mechanism where the A allele of rs16852804 decreases the expression of BARD1 in non-classical monocytes and leads to a higher endothelial dysfunction and higher tumor aggressiveness, resulting in poor prognosis.
BARD1 germline mutations have been linked to the risk of ovarian cancer 38–40. Most of them are exonic, but some intronic germline variants have been reported in BRCA1/2 negative high-risk breast and /or ovarian cancer patients, including rs71579844 (T>-, in complete LD with rs16852804, R2=1.0) and rs5031010 (T>G, in very high LD with rs16852804, R2=0.9) 38. Germline variation of risk of sporadic cancer can be associated with worse patient survival 41. Moreover, the BARD1 coding region is sequenced in several mutation panels, such as Foundation Medicine (FoundationOne® CDX) and MSK-IMPACT. Because rs16852804 in BARD1 is germline and intronic, BARD1 could be fully sequenced by including intronic regions in these panels. Orthogonal confirmation of the rs16852804 genotype in each patient will aid the collection of outcome data and response to therapy in larger cohorts in the future.
The discovery of new SNPs and genes that interact with a drug with respect to patient survival is much more challenging than the discovery of SNPs and genes with prognostic significance. We have provided evidence of AGAP1 as a potentially novel predictive candidate gene interacting with bevacizumab with respect to OS. In this study, patients with the GA genotype of rs3795897 (G>A) in AGAP1 experienced shorter OS in the bevacizumab arm compared to the non-bevacizumab arm, and patients with the GG genotype experienced longer OS in the bevacizumab arm compared to the non-bevacizumab arm (Tables 3, Figure S6). AGAP1 encodes a member of an ADP-ribosylation factor GTPase-activating protein family, and its role in the bevacizumab interaction with respect to patient survival needs to be further explored.
This study has some limitations. None of the SNPs reached the cut-off of genome-wide significance; however, the test for replication of the effect in TCGA patients and the concordance of effects in three out of the four studies potentiate the validity of these associations. The independent relative contribution of the effect of each variant in each tumor type cannot be ascertained in the TCGA datasets due to the small sample sizes and the relatively low frequency of these variants. Our study included patients with different tumor types and treatments, and the heterogeneity tests and the selection of SNPs based on a Cochran’s Q p-value >0.20 might mitigate the contribution of the confounding effects of these differences across studies. Only two out of the four studies were randomized to placebo, limiting the detection of predictive effects of SNPs that are specific to bevacizumab. Also, tumor molecular data are not always available to confirm some of our hypotheses. Still, we have applied different approaches using mRNA data from TCGA to circumvent this limitation. We did not perform imputation to these data, and this approach will be further applied to larger future studies aimed at discovering additional variants. Lastly, the mechanistic hypotheses proposed in this study should be evaluated in experimental models.
In conclusion, we have identified genes and new germline variants associated with prognosis in cancer patients. Two germline variants were also associated with prognosis in cancer patients from TCGA, rs680949 (A>G) in PRUNE2 in CRC cancer patients and rs16852804 (G>A) in BARD1 in ovarian cancer patients. Furthermore, we have also identified rs3795897 (G>A) in AGAP1 as a potentially new predictive germline variant that should be further evaluated as a marker affecting the response of patients to bevacizumab. Prospective testing of the association of these germline variants with survival should be performed to translate these findings to clinical practice.
Supplementary Material
Novelty and Impact.
We have provided evidence of potentially prognostic germline variants in PRUNE2 and BARD1 (tumor suppressor genes) in advanced colorectal and ovarian cancer, respectively. We have also identified a potentially predictive germline variant in AGAP1 that should be further evaluated as a marker affecting the response of patients to bevacizumab.
Acknowledgments
Funding
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882, and U24CA196171 (Alliance for Clinical Trials in Oncology), UG1CA233180, UG1CA233253, UG1CA233290, UG1CA233327, UG1CA233373, U10CA180863 (CCTG), and U10CA180888 (SWOG). https://acknowledgments.alliancefound.org. JCFQ was supported by the São Paulo Research Foundation-FAPESP (2018/04491-2). Also supported in part by Genentech; Bristol-Myers Squibb and Pfizer (CALGB 80405). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- CALGB
Cancer and Leukemia Group B
- CI
confidence interval
- COAD
colon adenocarcinoma
- CRC
colorectal cancer
- DICE
Database of Immune Cell eQTLs
- eQTL
expression quantitative trait loci
- GTEx
Genotype-Tissue Expression project
- GWAS
genome-wide association study
- HR
hazard ratio
- ICON7
International Collaboration on Ovarian Neoplasms
- LD
linkage disequilibrium
- NE
not estimated
- NES
normalized effect size
- OS
overall survival
- OV
ovarian serous cystadenocarcinoma
- PARP
poly(ADP-ribose) polymerase
- QC
quality control
- READ
rectum adenocarcinoma
- SE
standard error
- SNPs
single nucleotide polymorphisms
- TCGA
The Cancer Genome Atlas
- VEGF-A
vascular endothelial growth factor A
Footnotes
Conflict of interest
HJL is an advisor for BMS, Merck, Merck KG, Genentech, and Bayer. MND is presently employed by Genentech, since July 2021.
Ethics statement
All trials were conducted in accordance with recognized ethical guidelines. The study was performed in accordance with the Declaration of Helsinki and was approved by the local IRB. All participants provided written informed consent for sample collection and analysis.
ClinicalTrials.gov Identifier: NCT00601900 (CALGB 40503); NCT00088894 (CALGB 80303); NCT00265850 (CALGB 80405), and NCT00483782 (ICON7).
Data availability statement
The GWAS summary statistics are available through the NHGRI-EBI GWAS Catalog under study accession numbers GCST90020034 and GCST90020035. Further details and other data that support the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1.Chatrath A, Przanowska R, Kiran S, Su Z, Saha S, Wilson B, Tsunematsu T, Ahn JH, Lee KY, Paulsen T, Sobierajska E, Kiran M, Tang X, Li T, Kumar P, Ratan A, Dutta A. The pan-cancer landscape of prognostic germline variants in 10,582 patients. Genome Med 2020; 12(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Mattia E, Roncato R, Palazzari E, Toffoli G, Cecchin E. Germline and Somatic Pharmacogenomics to Refine Rectal Cancer Patients Selection for Neo-Adjuvant Chemoradiotherapy. Front Pharmacol 2020; 11:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, Ng PK, Jeong KJ, Cao S, Wang Z, Gao J, Gao Q, Wang F, Liu EM, Mularoni L, Rubio-Perez C, Nagarajan N, Cortés-Ciriano I, Zhou DC, Liang WW, Hess JM, Yellapantula VD, Tamborero D, Gonzalez-Perez A, Suphavilai C, Ko JY, Khurana E, Park PJ, Van Allen EM, Liang H; MC3 Working Group; Cancer Genome Atlas Research Network, Lawrence MS, Godzik A, Lopez-Bigas N, Stuart J, Wheeler D, Getz G, Chen K, Lazar AJ, Mills GB, Karchin R, Ding L. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018; 173(2):371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee V, Murphy A, Le DT, Diaz LA Jr. Mismatch Repair Deficiency and Response to Immune Checkpoint Blockade. Oncologist 2016; 21(10):1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faraoni I, Graziani G. Role of BRCA Mutations in Cancer Treatment with Poly(ADP-ribose) Polymerase (PARP) Inhibitors. Cancers (Basel) 2018; 10(12):487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EMA Summary of Product Characteristics. Available online: https://www.ema.europa.eu/en/documents/product-information/avastin-epar-product-information_en.pdf (accessed on 05 October 2020).
- 7.FDA AVASTIN®Prescribing Information: Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/125085s332lbl.pdf (accessed on 05 October 2020).
- 8.Crona DJ, Skol AD, Leppänen VM, Glubb DM, Etheridge AS, Hilliard E, Peña CE, Peterson YK, Klauber-DeMore N, Alitalo KK, Innocenti F. Genetic Variants of VEGFA and FLT4 Are Determinants of Survival in Renal Cell Carcinoma Patients Treated with Sorafenib. Cancer Res 2019; 79(1):231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glubb DM, Paré-Brunet L, Jantus-Lewintre E, Jiang C, Crona D, Etheridge AS, Mirza O, Zhang W, Seiser EL, Rzyman W, Jassem J, Auman T, Hirsch FR, Owzar K, Camps C, Dziadziuszko R, Innocenti F. Functional FLT1 Genetic Variation is a Prognostic Factor for Recurrence in Stage I-III Non-Small-Cell Lung Cancer. J Thorac Oncol 2015; 10(7):1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, Paczkowska M, Reynolds S, Wyczalkowski MA, Oak N, Scott AD, Krassowski M, Cherniack AD, Houlahan KE, Jayasinghe R, Wang LB, Zhou DC, Liu D, Cao S, Kim YW, Koire A, McMichael JF, Hucthagowder V, Kim TB, Hahn A, Wang C, McLellan MD, Al-Mulla F, Johnson KJ; Cancer Genome Atlas Research Network, Lichtarge O, Boutros PC, Raphael B, Lazar AJ, Zhang W, Wendl MC, Govindan R, Jain S, Wheeler D, Kulkarni S, Dipersio JF, Reimand J, Meric-Bernstam F, Chen K, Shmulevich I, Plon SE, Chen F, Ding L. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018; 173(2):355–370.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingle JN, Kalari KR, Wickerham DL, von Minckwitz G, Fasching PA, Furukawa Y, Mushiroda T, Goetz MP, Barman P, Carlson EE, Rastogi P, Costantino JP, Cairns J, Paik S, Bear HD, Kubo M, Wang L, Wolmark N, Weinshilboum RM. Germline genome-wide association studies in women receiving neoadjuvant chemotherapy with or without bevacizumab Pharmacogenet Genomics 2018; 28(6):147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider BP, Shen F, Miller KD. Pharmacogenetic biomarkers for the prediction of response to antiangiogenic treatment. Lancet Oncol 2012; 13(10):e427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papachristos A, Sivolapenko GB. Pharmacogenomics, Pharmacokinetics and Circulating Proteins as Biomarkers for Bevacizumab Treatment Optimization in Patients with Cancer: A Review. J Pers Med 2020; 10(3):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kindler HL, Niedzwiecki D, Hollis D, Sutherland S, Schrag D, Hurwitz H, Innocenti F, Mulcahy MF, O’Reilly E, Wozniak TF, Picus J, Bhargava P, Mayer RJ, Schilsky RL, Goldberg RM. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303). J Clin Oncol 2010; 28(22):3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickler MN, Barry WT, Cirrincione CT, Ellis MJ, Moynahan ME, Innocenti F, Hurria A, Rugo HS, Lake DE, Hahn O, Schneider BP, Tripathy D, Carey LA, Winer EP, Hudis CA. Phase III Trial Evaluating Letrozole As First-Line Endocrine Therapy With or Without Bevacizumab for the Treatment of Postmenopausal Women With Hormone Receptor-Positive Advanced-Stage Breast Cancer: CALGB 40503 (Alliance). J Clin Oncol 2016; 34(22):2602–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA, Schrag D, Greene C, O’Neil BH, Atkins JN, Berry S, Polite BN, O’Reilly EM, Goldberg RM, Hochster HS, Schilsky RL, Bertagnolli MM, El-Khoueiry AB, Watson P, Benson AB 3rd, Mulkerin DL, Mayer RJ, Blanke C. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA 2017; 317(23):2392–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, Carey MS, Beale P, Cervantes A, Kurzeder C, du Bois A, Sehouli J, Kimmig R, Stähle A, Collinson F, Essapen S, Gourley C, Lortholary A, Selle F, Mirza MR, Leminen A, Plante M, Stark D, Qian W, Parmar MK, Oza AM; ICON7 Investigators. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med 2011; 365(26):2484–2496. [DOI] [PubMed] [Google Scholar]
- 18.Innocenti F, Owzar K, Cox NL, Evans P, Kubo M, Zembutsu H, Jiang C, Hollis D, Mushiroda T, Li L, Friedman P, Wang L, Glubb D, Hurwitz H, Giacomini KM, McLeod HL, Goldberg RM, Schilsky RL, Kindler HL, Nakamura Y, Ratain MJ. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res 2012; 18(2):577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, Mahajan S, Goldberg RM, Bertagnolli MM, Blanke CD, Sanoff H, Atkins J, Polite B, Venook AP, Lenz HJ, Kabbarah O. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol 2019; 37(14):1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V; Cancer Genome Atlas Research Network, Hu H. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018; 173(2):400–416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015; 31(21):3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, Seumois G, Rao A, Kronenberg M, Peters B, Vijayanand P. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018; 175(6):1701–1715.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 25.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015; 348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuo C, Shin S, Keleş S. atSNP: transcription factor binding affinity testing for regulatory SNP detection. Bioinformatics 2015; 31(20):3353–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu W, McPherson JR, Stevenson M, van Eijk R, Heng HL, Newey P, Gan A, Ruano D, Huang D, Poon SL, Ong CK, van Wezel T, Cavaco B, Rozen SG, Tan P, Teh BT, Thakker RV, Morreau H. Whole-exome sequencing studies of parathyroid carcinomas reveal novel PRUNE2 mutations, distinctive mutational spectra related to APOBEC-catalyzed DNA mutagenesis and mutational enrichment in kinases associated with cell migration and invasion. J Clin Endocrinol Metab 2015; 100(2):E360–E364. [DOI] [PubMed] [Google Scholar]
- 30.Salameh A, Lee AK, Cardó-Vila M, Nunes DN, Efstathiou E, Staquicini FI, Dobroff AS, Marchiò S, Navone NM, Hosoya H, Lauer RC, Wen S, Salmeron CC, Hoang A, Newsham I, Lima LA, Carraro DM, Oliviero S, Kolonin MG, Sidman RL, Do KA, Troncoso P, Logothetis CJ, Brentani RR, Calin GA, Cavenee WK, Dias-Neto E, Pasqualini R, Arap W. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A 2015; 112(27):8403–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012; 151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012; 151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tikhonenko AT, Black DJ, Linial ML. Viral Myc oncoproteins in infected fibroblasts down-modulate thrombospondin-1, a possible tumor suppressor gene. J Biol Chem 1996; 271(48):30741–30747. [DOI] [PubMed] [Google Scholar]
- 34.Watters AK, Seltzer ES, MacKenzie D Jr, Young M, Muratori J, Hussein R, Sodoma AM, To J, Singh M, Zhang D. The Effects of Genetic and Epigenetic Alterations of BARD1 on the Development of Non-Breast and Non-Gynecological Cancers. Genes (Basel) 2020; 11(7):829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas G, Tacke R, Hedrick CC, Hanna RN. Nonclassical patrolling monocyte function in the vasculature. Arterioscler Thromb Vasc Biol 2015; 35(6):1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh KK, Shukla PC, Quan A, Al-Omran M, Lovren F, Pan Y, Brezden-Masley C, Ingram AJ, Stanford WL, Teoh H, Verma S. BRCA1 is a novel target to improve endothelial dysfunction and retard atherosclerosis. J Thorac Cardiovasc Surg 2013; 146(4):949–960.e4. [DOI] [PubMed] [Google Scholar]
- 37.Franses JW, Drosu NC, Gibson WJ, Chitalia VC, Edelman ER. Dysfunctional endothelial cells directly stimulate cancer inflammation and metastasis. Int J Cancer 2013; 133(6):1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ratajska M, Antoszewska E, Piskorz A, Brozek I, Borg Å, Kusmierek H, Biernat W, Limon J. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res Treat 2012; 131(1):89–97. [DOI] [PubMed] [Google Scholar]
- 39.Sauer MK, Andrulis IL. Identification and characterization of missense alterations in the BRCA1 associated RING domain (BARD1) gene in breast and ovarian cancer. J Med Genet 2005; 42(8):633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, Roeb W, Agnew KJ, Stray SM, Wickramanayake A, Norquist B, Pennington KP, Garcia RL, King MC, Swisher EM. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 2011; 108(44):18032–18037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Y, Ek WE, Whiteman D, Vaughan TL, Spurdle AB, Easton DF, Pharoah PD, Thompson DJ, Dunning AM, Hayward NK, Chenevix-Trench G; Q-MEGA and AMFS Investigators; ANECS-SEARCH; UKOPS-SEARCH; BEACON Consortium, Macgregor S. Most common ‘sporadic’ cancers have a significant germline genetic component. Hum Mol Genet 2014; 23(22):6112–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GWAS summary statistics are available through the NHGRI-EBI GWAS Catalog under study accession numbers GCST90020034 and GCST90020035. Further details and other data that support the findings of this study are available from the corresponding author upon request.