Abstract
R-loop represents a prevalent and specialized chromatin structure critically involved in a wide range of biological processes. In particular, co-transcriptional R-loops, produced often due to RNA polymerase pausing or RNA biogenesis malfunction, can initiate molecular events to context-dependently regulate local gene transcription and crosstalk with chromatin modifications. Cellular “readers” of R-loops are identified, exerting crucial impacts on R-loop homeostasis and gene regulation. Mounting evidence also supports R-loop deregulation as a frequent, sometimes initiating, event during human pathologies, notably cancer and neurological disorder. The purpose of this review is to cover recent advances in understanding the fundamentals of R-loop biology, which start to unveil complex interplays of R-loops with factors involved in various biological processes such as transcription, RNA processing and epitranscriptomic modification (such as N6-methyladenosine), DNA damage sensing and repair, and epigenetic regulation.
INTRODUCTION
When RNA hybridizes with its DNA template, the RNA-DNA hybrid duplex termed R-loop is formed. Such a heteronomous three-stranded structure includes an RNA-DNA hybrid and a displaced single-stranded DNA (ssDNA) [1–5]. R-loop represents an intermediate between the RNA (A-form) and DNA (B-form) conformations, often exhibiting a higher degree of stability than double-stranded DNAs (dsDNAs) [1–5]. Although R-loop was initially considered to be a by-product of transcription, R-loop can occur genome-wide and be detected among all organisms that cover bacteria, yeast, plant and animal. R-loops are known to be directly involved in a number of specialized cellular processes, such as replication of bacterial ColE1 DNA, bacteriophage T4 and mitochondrial DNA, clustered regularly interspaced short palindromic repeats (CRISPR) RNA (crRNA)-guided dsDNA recognition, RNAi-directed heterochromatin assembly in fission yeast, class switch recombination (CSR) of immunoglobulin genes, and formation and maintenance of telomere. More broadly, R-loops occur co-transcriptionally, often near gene promoters enriched with the C/G-high content, and are involved in regulation of RNA polymerase II (Pol-II) pausing, sense versus antisense transcription, and epigenetic modifications [2–4]. R-loops generally form in cis near the sites of transcription but can also occur in trans. For the latter, Rad51p, an homologous recombination factor, was shown to promote formation of RNA-DNA hybrids in trans [6]; likewise, based on sequence complementarity, a long noncoding RNA (lncRNA) termed APOLO in Arabidopsis was reported to act in trans mediating R-loop formation at its target [7]. R-loops can also be detected among repeated elements such as LINE-1 [8].
It is generally viewed that excessive formation of R-loops induces the replication fork collision and Pol-II blocking, which are deleterious for productive transcriptional elongation and often lead to pathogenesis due to DNA double-strand breaks (DSBs) and genomic instability [2, 9, 10]. Therefore, understanding the homeostasis and regulation of R-loops shall have a far-reaching implication in biology (related to those termed “regulatory R-loops” [2, 3]) and human diseases (which, in most cases, are related to excessive production of R-loops, also termed “unscheduled or unwanted R-loops” [2, 3]). Here, we summarize the biochemical basis and molecular players that underlie the “creation”, “recognition”, “maintenance” and “resolution” of R-loops. In particular, this review will focus on the modulation of R-loops via epigenetic regulation of chromatin and RNA such as N6A methylation of DNA or RNA during the gene transcriptional and post-transcriptional processes, the newly unveiled RNA-DNA hybrid “readers”, and cancers resulting from the impaired R-loop balance.
Co-transcriptional R-loops influence gene and epigenome regulation.
Formation of R-loops is modulated by a number of factors, contributing to diverse cellular processes.
In the “thread-back” model [10–12], co-transcriptional R-loops (also known as cis R-loops) can be established genome-wide when a nascent RNA transcript reanneals with its template DNA strand at the site of transcription. However, evidence exists showing that R-loops can also form in trans [6, 11, 12], in which an RNA transcript hybridizes to the complementary strand of DNA at a location distant from the original transcription site. R-loop formation is highly supported by conformational features (Figure 1), such as negative supercoiling of DNA, the guanine (G) content and presence of G-quadruplex (G4) second structure or a single-strand DNA (ssDNA) nick [13–15]. Negative DNA superhelicity promotes R-loop formation and a recent study further suggested the important interplay between DNA base sequence and negative superhelicity in controlling R-loop stability [16]. The presence of G clusters on the RNA transcript facilitates the initiation of R-loop formation during transcription [14] due to the thermal stability of rG/dC base pairing. In accordance with thermal stability of G:C base pairing, R-loop hotspot regions were mapped to those with a feature of GC skew, including promoter and transcription termination regions exhibiting high G+C contents [17, 18]. Once the formation of R-loops is initiated, the elongation of R-loops depends on the density of G on the non-template strand of DNA [14]. In addition, R loops are favoured by nicks on the non-template DNA strand downstream of the promoter [15]. Mechanistically, the transient removal of the non-template strand after the nicked position may increase the opportunity to form RNA-DNA hybrids [15]. Additionally, a set of biological processes, which at least include RNA Pol-II pausing, antisense transcription (often producing lncRNA), RNA and DNA modifications, and availability of a free RNA end (often increased due to RNA splicing or biogenesis malfunction), all enhance R-loop occurrence. Adenosine methylation of RNA (m6A) or DNA (6mA) was also reported to modulate R-loops, potentially through recruitment of m6A “readers” (Figure 2). Conceivably, proteins with activity for binding ssDNA or R-loop may also influence R-loop formation/stabilization.
Figure 1. Biophysical basis underlying the R-loop formation.
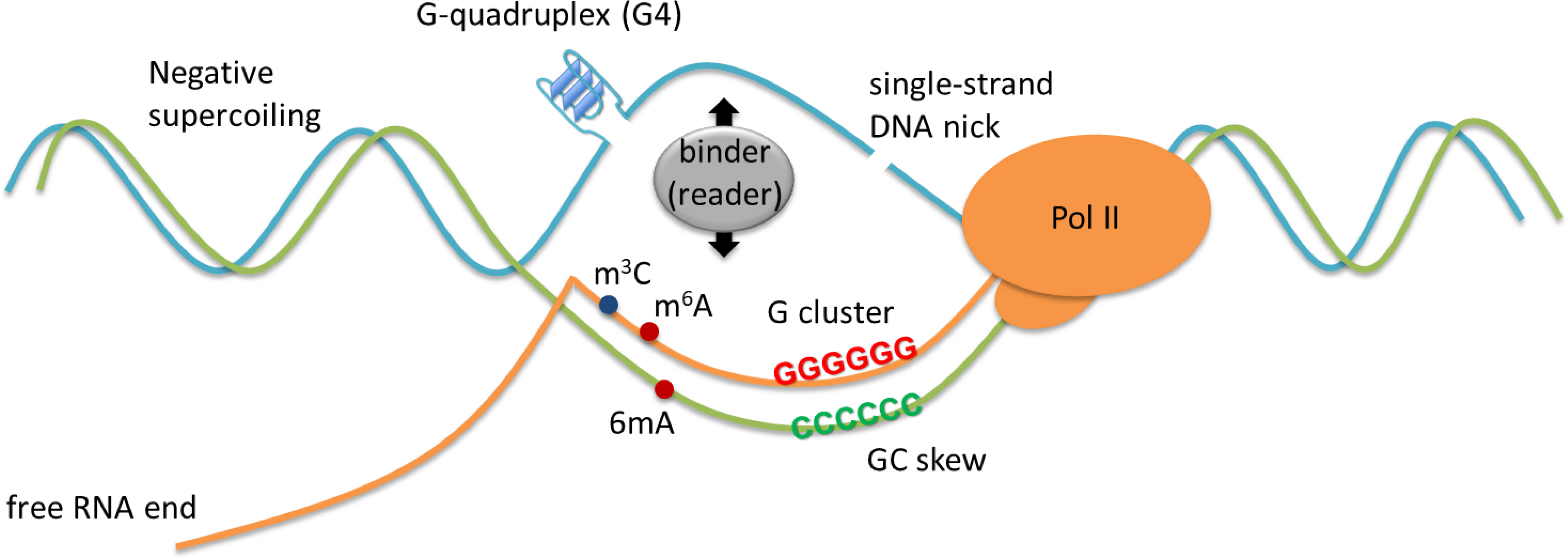
R-loop formation is achieved by conformational features such as negative supercoiling, a nick and G-quadruplex on the displaced strand of DNA, G-cluster and GC skew on the RNA transcript, and free RNA end. RNA modifications such as m3C and m6A and/or DNA modification of 6mA may also potentially modulate R-loops.
Figure 2. Adenosine Methylation of RNA and DNA.
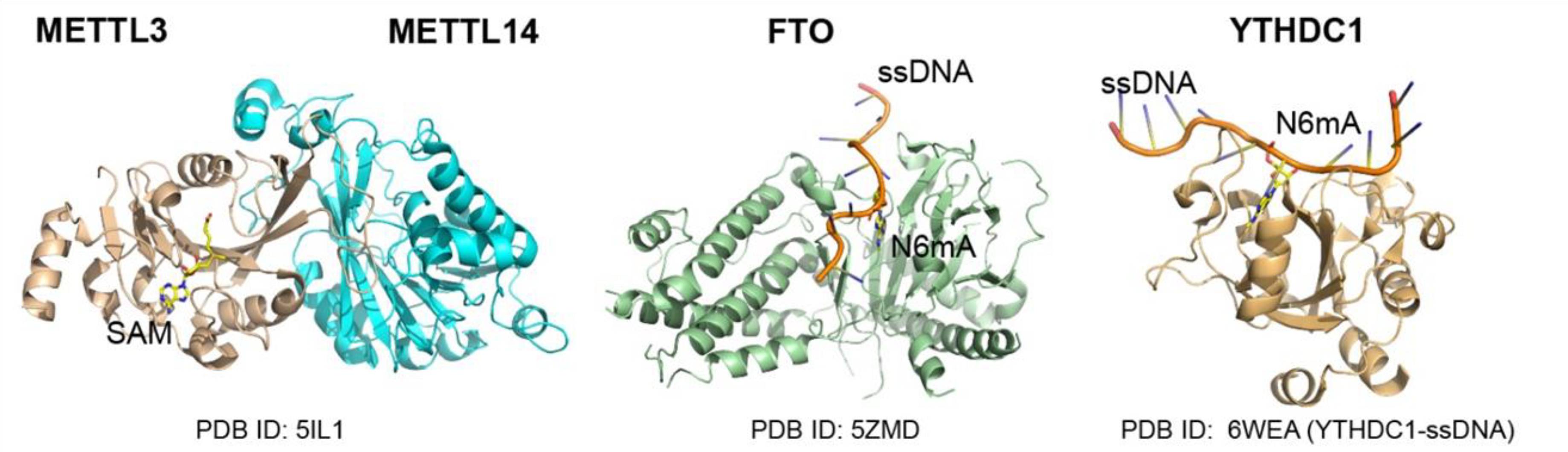
RNA m6A is installed by writers such as the METTL3/METTL14 methyltransferase, removed by erasers include the FTO demethylases, and can be recognized by readers such as YTH domain proteins. Please note that the above enzymes have promiscuous activity towards ssDNA.
While regulatory R-loops actively contribute to diverse cellular processes, excessive R-loops (i.e., “unscheduled and unwanted R-loops” [2, 3]) can be a source of a cell-intrinsic stress arising from the defects in transcription elongation, replication and DNA repair. Regarding the regulatory R-loops that are critically involved in normal physiological and biological processes, they can exert impact, not only on the RNA Pol-II behaviours but also on local chromatin structure or modifications (notably, DNA demethylation and Polycomb-related modifications). In addition, the importance of regulatory R-loops has been also demonstrated in class switch recombination (CSR) of immunoglobulin genes [19, 20]. It was reported that non-coding switch region transcripts promote formation of R-loops, which facilitate CSR, a process that also depends on DSBs, and the involvements of the origin recognition complex (ORC), MCM complex and the replicative helicase [21–23]. In the sections below, we discuss about recent advances in (epi)genomic regulation by R-loops.
Interplay between RNA Pol-II and co-transcriptional R-loop profoundly influences local transcription.
It is generally viewed that, following RNA Pol-II pausing at transcription start sites (TSSs), R-loop accumulation often occurs (Figure 3A). In support, Negative Elongation Factor Complex Member B (NELFB; also known as COBRA1), an integral subunit for mediating RNA Pol-II pausing, was reported to interact physically with and functionally antagonize the Breast cancer type 1 susceptibility protein (BRCA1) [24]; due to enhanced NELFB/COBRA1 activity and Pol-II pausing, the BRCA1-deficient tumor cells are featured by excessive accumulation of R-loops [24]. On the other hand, Kim et al recently reported that the bromodomain-containing proteins BRD2 and BRD4, which are histone acetylation “readers” with transcriptional-elongation-facilitating activities, suppress R-loop formation [25]. While BRD4 recruits pTEFb for RNA Pol-II phosphorylation during transcriptional activation/elongation, a C-terminal region of BRD2 associates with DNA topoisomerase 1 (TOP1) inducing activation of the latter [25]. In consistence, a separate study also showed that depletion of BRD4 in cancer cells induces R-loop accumulation and transcription–replication collision events, leading to DNA damage and apoptosis of tumor cells [26] (Figure 3A). Likewise, TFIIS recognizes RNA Pol-II backtracking, the reversible sliding of Pol-II along DNA and RNA, induces transcript cleavage and enables Pol-II to resume transcript elongation [27]; a recent work further suggested that expression of a transcriptional-elongation-defective mutant of TFIIS (which inhibits transcript cleavage) caused Pol-II pausing, extended backtracking, and triggered cellular accumulation of anterior R-loops, which are different from posterior R-loops [28] (Figure 3A).
Figure 3. Interplay between RNA Pol-II and co-transcriptional R-loop.
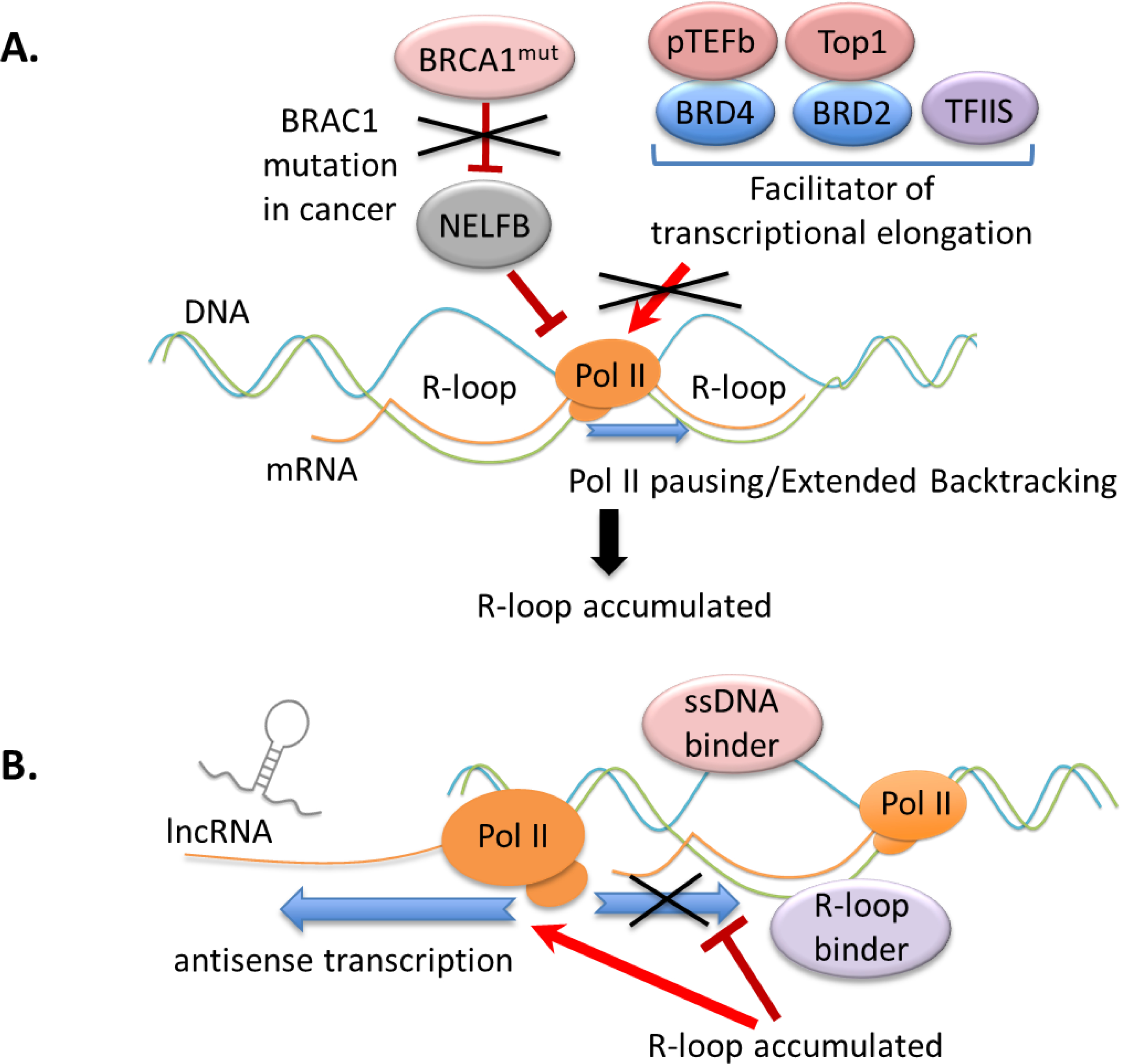
(A) The impairment of transcriptional elongation causes RNA Pol-II pausing and promotes R-loop accumulation. Depletion of BRCA1 in cancer induces Pol-II pausing and triggers excessive R-loop formation, partly through the enhanced NELFB/COBRA1 activity. Along with TFIIS, BRD4 and BRD2 recruit pTEFb and TOP1, respectively, to facilitate transcriptional elongation. A transcriptional-elongation-defective mutant of TFIIS stimulates anterior R-loop formation.
(B) Crosstalk among R-loops, Pol-II-mediated antisense transcription and various R-loop structure “sensors” such as ssDNA-specific binder and R-loop (RNA-DNA hybrid) “readers” is crucial for metabolism of R-loop and R-loop-related gene regulation. In the mammalian cells, accumulated R-loops promote antisense transcription of lncRNAs near the TSS of lncRNAs.
At promoter regions, GC skew-triggered formation of R-loop is often observed between TSS and the first exon-intron junction. Also, G-rich sequences potentially produce the G4 secondary structure on the non-template strand (Figure 1) [29–31], which further contributes to R-loop stabilization and influences transcription. In agreement, the G4-binding proteins were reported to increase R-loop establishment in cancer cells, thereby inducing DNA damage [29–31]. Meanwhile, GC skew at transcription termination sites (TTSs) also causes R-loop, leading to RNA Pol-II pausing downstream from the poly(A) signals; here, it is viewed that R-loops enable efficient transcription termination of genes within gene-dense regions [17, 32, 33], exhibiting a feature that resembles insulator or enhancer [18].
Furthermore, a free RNA end was recently appreciated to be an important initiating factor by allowing invasion of a nascent RNA into the DNA duplex [34]. Such a requirement for R-loop formation partially explains why diseased cells with mutation of RNA splicing factor or malfunction of premature transcriptional termination often display a phenotype of R-loop accumulation (will be discussed next).
Increasing evidence sheds light on reciprocal regulation involving R-loop, antisense transcription and lncRNA. Tan-Wong et al recently demonstrated that the mammalian R-loops act as intrinsic promoters for promoting antisense transcription and thus generating lncRNAs genome-wide [35] (Figure 3B); here, R-loops are located near TSSs of lncRNAs and lncRNA expression was found to be sensitive to RNase H1, suggesting that R-loops act to enhance synthesis of antisense lncRNAs [35]. Previously, it was also reported that lncRNAs often induce R-loop formation, content-dependently regulating mRNA transcription of adjacent genes, as observed for SPHK1 and vimentin in human cancer cells [36, 37] and FLOWERING LOCUS C (FLC) in Arabidopsis [38]. For the latter, AtNDX, an ssDNA-binding homeodomain protein in Arabidopsis, was identified to stabilize R-loop at the promoter of COOLAIR, which is an antisense lncRNA at the FLC locus that functions to antagonize FLC expression through epigenetic silencing mechanisms [38].
Altogether, these observations highlight a general cell stress produced by transcription and also unveil crucial crosstalk among RNA Pol-II, co-transcriptional R-loop and R-loop “sensor” (Figure 3B; such as ssDNA binder and RNA-DNA hybrid “reader” to be discussed next). Importantly, R-loop can serve as a platform for recruiting or repelling DNA/RNA-binding factor and chromatin modifier, leading to context-dependent regulation of chromatin structure and transcription control, which is elaborated below.
RNA or DNA modifications modulate homeostasis of R-loops.
In eukaryotes, methylation of adenosine within RNA (N6-methyladenosine or m6A) emerges as the most prevalent post-transcriptional modification that modulates RNA metabolism [39, 40]. RNA m6A is installed by m6A “writers” such as the METTL3/METTL14 methyltransferase and removed by m6A “erasers” such as the m6A demethylases, FTO and ALKBH5 [41] (Figure 2). Additionally, a set of m6A “readers”, including the YT521-B Homology (YTH) domain-containing proteins (such as YTHDC1, YTHDC2, YTHDF1, YTHDF2 and YTHDF3) (Figure 2), heterogeneous nuclear ribonucleoproteins (hnRNPs; such as HNRNPC, HNRNPG and HNRNPA2B1), Insulin-like Growth Factor 2 mRNA Binding Proteins 1, 2, and 3 (IGF2BP1/2/3), and FMRP [41, 42], selectively recognize and bind to m6A-containing RNAs, mediating the biological consequence of m6A [42].
A recent study uncovered that the METTL3/METTL14 complex deposits m6A in nascent RNAs, which stabilizes R-loops around TTSs of m6A+ genes (defined as genes whose transcript harbors m6A), thereby facilitating transcription termination [43] (Figure 4A, top). Such a crosstalk of METTL3/METTL14 with R-loop can be applied to cellular response to DNA double-strand break (DSB), in which METTL3 is phosphorylated by the “sensor” of DSB, ATM, and localized to DSB sites for m6A deposition [44]; in consistence, Kang et al additionally reported that TonEBP recognizes R-loops generated by DNA damaging agents (see also next section of R-loop “readers”) and also subsequently recruits METTL3 to R-loop-associated damage sites for “writing” m6A (Figure 4A, top) [45]. Then, RNA m6A recruits its “reader” YTHDC1 to enhance R-loop accumulation at DSB sites, leading to further recruitment of RAD51 and BRCA1 for homologous recombination (HR)-mediated repair and eventual resolution of R-loop [44] (Figure 4A, bottom). “Reader” of m6A can also act to suppress R-loop accumulation, thereby preventing R-loop-induced genomic instability [8]. Abakir et al recently reported that, in human pluripotent stem cells, m6A is prevalent in the RNA strands of RNA-DNA hybrids and such m6A-containing R-loops undergo turnover in a cell cycle-dependent fashion—m6A-marked R-loops are accumulated during S and G2/M phases, but depleted at G0/G1 phase [8]; here, YTHDF1, YTHDF2, HNRNPA2B1 and METTL3 all interact with R-loops, but in dividing cells, YTHDF2, an m6A “reader” that regulates degradation of cytoplasmic mRNA, associates to mitotic chromatin and binds the R-loops within the LINE-1 repeated elements and intronic regions, leading to removal of R-loops [8]. It is important because YTHDF2 depletion leads to R-loop accumulation, cell growth retardation and DNA DSBs, supporting that YTHDF2 operates to remove excessive R-loops for maintenance of genomic stability [8] (Figure 4A, bottom).
Figure 4. RNA and DNA modifications regulate R-loop homeostasis during gene transcription and cellular response to DNA damage.
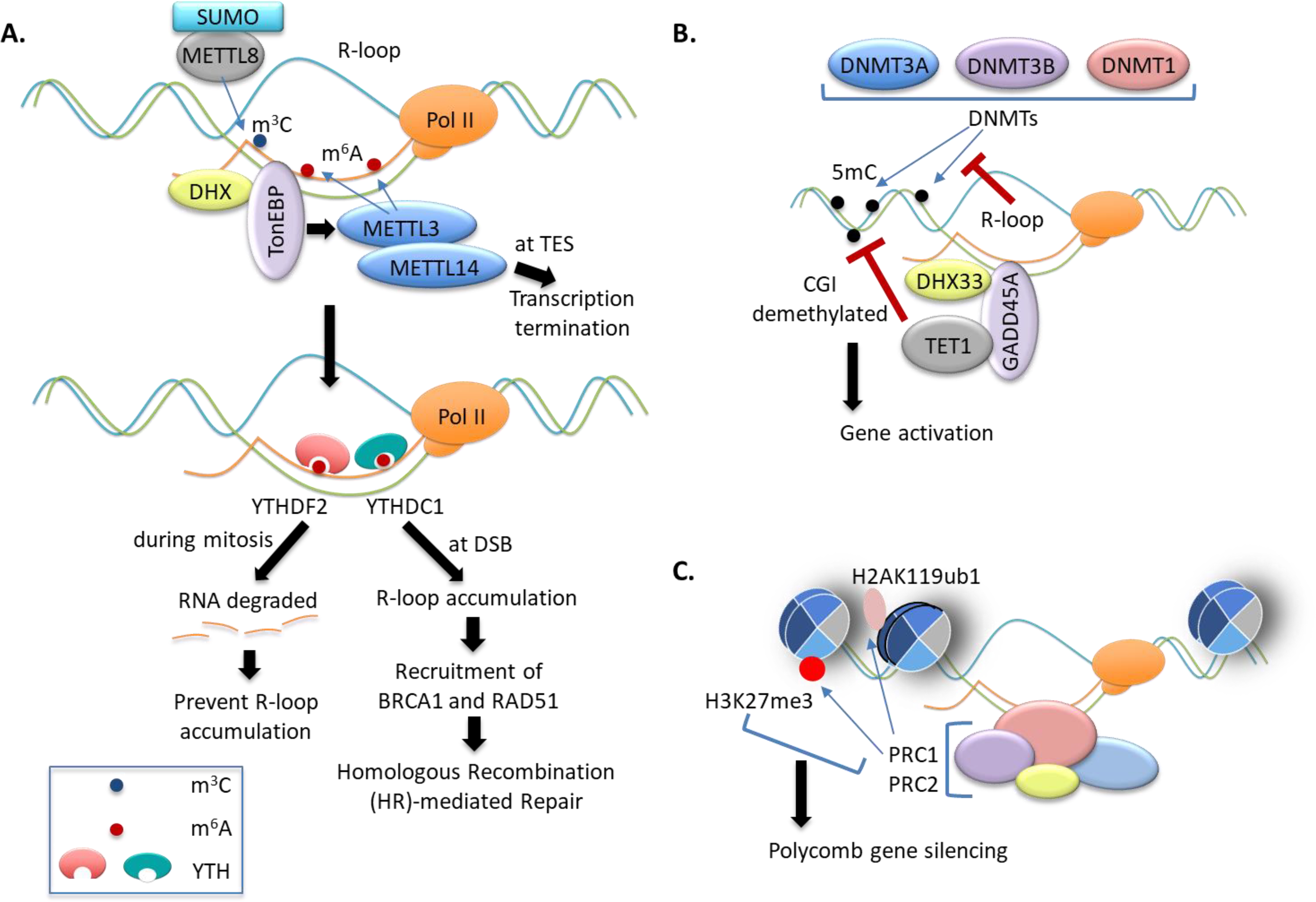
(A) The m6A methylation, installed by METTL3/METTL14 on nascent RNA, or the m3C methylation, installed by SUMOylated METTL8, promotes R-loop formation, which leads to transcription termination at TTSs. The m6A marks in the RNA strands of R-loops can be recognized by m6A “readers” such as YTHDF2 and YTHDC1. In human pluripotent stem cells, YTHDF2 binds to R-loops leading to depletion of m6A-marked R-loops during mitosis. At DSB sites, YTHDC1 increases accumulation of RNA-DNA hybrids and recruits BRCA1 and RAD51 for homologous recombination-mediated repair of DSB.
(B) R-loops are poor substrates for methylation by mammalian DNA methyltransferases (DNMTs). GADD45A binds to R-loop, an event which can be further facilitated by GADD45A interaction with the RNA helicase DHX33, further recruits TET1 to induce CGI demethylation, leading to gene activation.
(C) PcG proteins recognize R-loops and trigger local histone modifications such as H3K27me3 and H2AK119ub1, which provides a mechanism underlying PcG gene silencing.
In human cells, METTL8, another methyltransferase-like family member, was shown to induce 3-methylcytidine (m3C) within RNA, with implication in R-loop formation [46]. METTL8 interacts with various splicing and RNA-binding complexes and, in particular, a complex of METTL8-THOC2-RPA3 was suggested for R-loop association [46]. SUMOylation of METTL8 lysine 80 induces its nuclear 3 localization and METTL8’s methyltransferase activity of m C was shown to be essential for the METTL8-related R-loop formation [46] (Figure 4A, top).
In rice, m6A of DNA was reported to silence genes when present at promoters, but activate gene when present at gene bodies [47]. Furthermore, the rice genes containing R-loops contain more m6A within gene body and a short region downstream from TTS, and m6A+ genes are expressed at higher levels than those lacking m6A [48]. Though OsALKBH1 was suggested as m6A demethylase in rice [47], the causal relationship between DNA m6A and R-loop homeostasis remains to be fully determined.
Altogether, recent evidence starts to unveil context-dependent, dynamic modulation of R-loop through RNA or DNA modifications and their molecular players.
R-loop “readers” act to exert epigenetic impact for regulating gene expression.
Once formed and stabilized, R-loops can induce subsequent change of local chromatin. Initially, R-loop was shown to be a poor substrate of mammalian DNA methyltransferases (DNMTs) [49], a phenomenon corroborated in vivo by genomic profiling [50] (Figure 4B). Furthermore, Growth Arrest and DNA Damage-inducible Protein 45 alpha (GADD45A; Figure 4B), a protein essential for cell cycle arrest and genomic stability, was recently identified as RNA-DNA hybrid “readers”, exhibiting a preferential affinity in vitro towards RNA-DNA hybrid over ssDNA/RNA or dsDNA/RNA [51]. We here use R-loop reader to refer to RNA-DNA hybrid reader. In mammalian cells, GADD45A binds R-loop generated by the antisense lncRNA TARID at the TCF21 promoter where GADD45A recruits TET1 to induce demethylation of CpG island (CGI), leading to activated expression of TCF21, a tumor suppressor [51]. Furthermore, R-loop-dependent recruitment of TET1 can be applied to thousands of TSS-associated CGIs in mouse embryonic stem cells (mESCs) as demonstrated by genomic profiling following RNase H1 treatment [51]. In agreement, Feng et al reported that the RNA helicase DHX33 also recruits GADD45A leading to DNA demethylation and transcriptional activation [52] (Figure 4B); DHX33 deficiency decreases R-loop formation in the promoter of Aurora kinase B, indicative of a role for DHX33 in R-loop stabilization and/or GADD45A:R-loop-mediated gene activation [52]. As GADD45A only binds to a subset of R-loops in cells [51], its binding specificity and structure basis remain to be characterized.
Furthermore, R-loops are frequently detected at Polycomb response elements (PREs) and involved in the chromatin recruitment of Polycomb Group (PcG) Proteins in Drosophila [53] (Figure 4C). In the in vitro reconstitution system, PRC2 can mediate the formation of RNA-DNA hybrid and PcG proteins such as PRC2 and PRC1 can recognize and bind the R-loops and open DNA bubbles, indicative of an interplay between R-loop and PcG gene silencing [53]. However, a separate study using mESCs did not support the promoting effect by PcG on R-loop, as PcG depletion did not affect R-loop formation in cells [54]. Nevertheless, the role for R-loop in enhancing PcG functionality appears to be conserved among Drosophila [53], animal [54] and plant [7]. In the mESCs and Drosophila, removal of R-loop by degrading RNAs with treatment of RNase H1 reduced the chromatin binding of PcG factors, leading to reduction in PcG-induced histone modifications (namely, H3K27me3 and H2AK119ub1) and derepression of those R-loop-associated target genes, but not R-loop-negative ones [53, 54]. At the genome-wide level, the silencing effects by PcG and R-loop are additive, suggesting their cooperation in gene repression [54]. In Arabidopsis, the lncRNA APOLO acts in trans and mediates R-loop formation at its sequence-complementary targets, where R-loop subsequently recruits and decoys plant-specific Polycomb Repressive Complex 1 (PRC1) component LHP1 for controlling the local chromatin 3D conformation and gene transcription [7].
Additionally, AtALBA1 and AtALBA2, the two Arabidopsis-specific Acetylation Lowers Binding Affinity (ALBA) family proteins, were reported to be RNA-DNA hybrid “readers” [55]. In vitro, AtALBA1 and AtALBA2 bind to the RNA-DNA hybrid and ssDNA in R-loops, respectively. Colocalized in the nucleus, AtALBA1 and AtALBA2 form a heterodimer or heteropolymer and bind R-loops at genic regions to maintain genome stability [55]. The molecular underpinning of ALBA1/2-mediated actions remains to be defined.
Thus, akin to binding of the displaced ssRNA by ssRNA-specific proteins such as RPA (Replication protein A), RNA-DNA hybrid is “sensed” and engaged by their specific “readers”, causing downstream changes of (epi)genome modification and chromatin structure.
Prevention and removal of excessive R-loops
R-loops are constantly removed/resolved, with an estimated 27,000 R-loops turned over every day and about 300 R-loops at the steady state in mammalian cells [56]. When excessively formed, R-loops induce DNA damage and replication stress, resulting in genomic instability. R-loops are prone to mutagenesis induced by activation-induced cytidine deaminase (AID) [57], the DNA replication fork stalling [58] and DSBs [8, 59, 60], all of which threaten genome integrity. In addition, spontaneous deamination of the displaced ssDNA in R-loops, replication fork collisions with blocked Pol II by transcriptional R-loops and R-loops-induced histone 3 Ser10 phosphorylation (H3S10P), a mark of chromatin compaction, eventually lead to DSBs and genomic instability [9]. Moreover, aberrant R-loop accumulation activates the replicative stress “sensor”, ATR, via replication fork reversal and the MUS81 nuclease [61]. ATR then represses transcription-replication collisions and promotes DNA synthesis and a G2/M cell-cycle arrest, thereby preventing genomic instability [61].
Given the importance of regulating R-loop abundance and distribution, a wealth of mechanisms evolves in controlling excessive R-loop formation, which at least include (i) R-loop formation prevention by TOP1 and (ii) RNA-binding proteins (RBPs), (iii) R-loop removal by RNase H1/2, (iv) R-loop prevention/surveillance by various proteins involving RNA biogenesis, degradation and metabolism, such as spliceosome, RNA exosome and export complexes, and (v) R-loops resolution by helicase.
While TOP1 inhibits R-loop formation by resolving negative supercoiling behind Pol-II, RBPs, involved in RNA surveillance, interact with nascent RNAs to prevent transcription-associated R-loop formation (Figure 5A). For example, Npl3, an RNA-binding heterogeneous nuclear RNP (hnRNP), also prevents the R-loop-mediated genome instability [62]. RNases H is a class of ribonuclease that specifically degrades the RNA moiety in RNA-DNA hybrids. In eukaryotes, there are two types of RNases H, RNase H1 and RNase H2, and human RNase H1 has been adopted for genome-wide R-loop profiling. Loss of RNase H increases R-loop formation in cells. RNase H2 also binds with Breast cancer type 2 susceptibility protein (BRCA2) and manages R-loops at DSBs [63]. RNA-processing factors (Figure 5A), such as Serine/arginine-rich splicing factor (SRSF1, also known as ASF1/SF2), the THO/TREX complex and Pcf11, have been reported to prevent R-loops by promoting mRNA processing and exporting [64–66]. The absence of Trf4, a poly(A) polymerase of TRAMP (Trf4–Air2–Mtr4p polyadenylation) complex, induces R-loop accumulation [67]. RNA exosome, including exosome component 3 (EXOSC3, Rrp40 in yeast) and exosome component 10 (EXOSC10, Rrp6 in yeast), mediated degradation of non-coding RNAs prevents R-loops to maintain genome integrity [68]. R loops can be resolved by helicases, with the reported activities reported for human senataxin (SETX) or its yeast homologue Sen1, Aquarius (AQR), DDX5, DDX23, DHX9, UAP56/DDX39B and the bacterial RNA helicase Rho (Figure 5B). For instance, SETX resolves R-loops at transcriptional pause sites and enables access of XRN2, a 5’−3’ exoribonuclease that plays a role in preventing formation of R loops, at 3’ cleavage poly(A) sites, thereby promoting the termination of transcription [32, 69, 70]. AQR (also known as intron-binding protein 160) also acts as a R-loop removal factor. AQR is integrated into the spliceosome in a position-specific manner, and loss of AQR results in DSB formation and R-loop accumulation [60, 71]. This pathway depends on the transcription-coupled nucleotide excision repair (TC-NER) factor Cockayne syndrome group B (CSB), which recruits RAD52, then initiate transcription-coupled homologous recombination (TC-HR) by reactive oxygen species (ROS)-induced R-loops [60, 72, 73]. In brief, CSB recruits the nucleotide excision repair endonucleases XPF and XPG, and these endonucleases lead to the processing of R-loops into DSBs, indicating the role of AQR in R-loop repression [60, 72]. Rho, the bacterial RNA helicase involved in transcription termination, is essential for bacteria survival and Rho-dependent transcription termination is essential for R-loop prevention [74].
Figure 5. Surveillance and prevention of excessive R-loop formation.
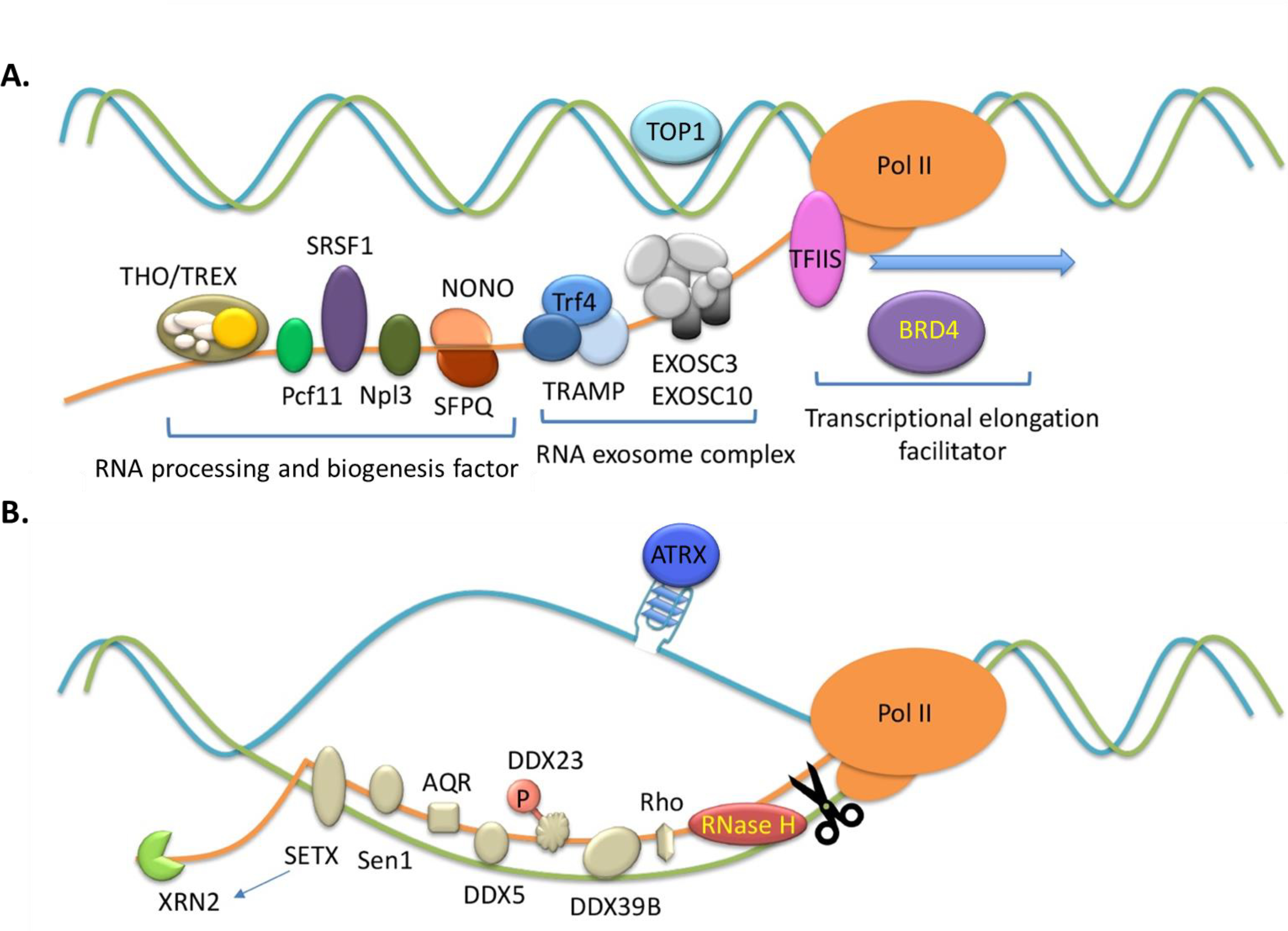
(A) R-loop formation is prevented by topoisomerase 1 (TOP1) as well as specific RNA-binding proteins (RBPs) such as SRSF1, the THO/TREX complex, Pcf11, Npl3, non-POU domain-containing octamer-binding protein (NONO) and Splicing factor proline- and glutamine-rich (SFPQ). RNA exosme complexes such as Trf4, exosome component 3 (EXOSC3) and exosome component 10 (EXOSC10), as well as transcription factor TFIIS and epigenetic reader Bromodomain-containing protein 4 (BRD4) play roles in preventing R-loops.
(B) The removal of R-loops is mediated by RNase H and helicases such as senataxin (SETX), Sen1, aquarius (AQR), DDX5, DDX23, DDX39B and Rho. 5’−3’ exoribonuclease XRN2 cooperates with SETX or DDX5 to remove R-loops. ATRX at telomere may involved in resolving R-loops by suppressing deleterious DNA secondary structures.
Additionally, recent studies highlight important crosstalk and cooperation of helicase, RNA biogenesis/splicing factor and DNA damage repair protein in regulating R-loop homeostasis and safeguarding genome integrity. For instance, DDX5, a DEAD-box RNA helicase, reduces R-loop occurrence and, when it is methylated by PRMT5 at its C-terminal RGG/RG motif, DDX5 also interacts with XRN2 for the release of Pol-II at TTSs [75]. Likewise, UAP56/DDX39B was reported as a key RNA-DNA helicase for R-loops’ resolution [76]. RNA Pol-II pausing induces the recruitment of serine/arginine protein kinase 2 (SRPK2) and DDX23 to specific R-loop-containing loci where SRPK2 phosphorylates DDX23, which then acts to prevent the R-loop-related genomic instability [77]. Mutations of DDX23 are observed in human cancer, with its homozygous deletion frequently observed in adenoid cystic carcinoma [77]. DHX9, a helicase carrying the in vitro R-loop-resolving activity, contributes to R-loop suppression and transcriptional termination in cells [78]. DHX9 interacts with poly(ADP-ribose) polymerase 1 (PARP1) and prevents R-loop-associated DNA damage in response to TOP1 inhibitor, camptothecin [78]. Another study demonstrated that, in the absence of RBPs and splicing factors, DHX9 enhances R-loop occurrence through unwinding nascent RNAs to generate the free RNA end [79], which indicates a coordinated process for R-loop prevention and RNA biogenesis. The helicase senataxin (SETX), on the other hand, is associated with AOA2/ALS4 neurodegenerative disorders. SETX interacts with BRCA1 and recruitment of the BRCA1:SETX complex to the R-loop-associated TTSs helps to restrict R-loop-induced DNA damage [80]. Transcription‐dependent recruitment of ATRX, a member of the SWI/SNF family of chromatin remodeling factors, to telomeric G-rich repeats (TTAGGG)n, is also associated with the presence of R‐loops [81]. ATRX has be shown to resolve R‐loops or recruit other enzymes that degrade R-loops [81].
R-loops and DNA repair
RNA-DNA hybrid can also be formed upon DNA damage at transcribed DNA and regulate the DNA repair process. For example, a ssDNA nick downstream of the promoter stimulates R-loop formation; and laser irradiation or radiomimetics-induced nicks in DNA trigger the formation of R-loops and RNA-DNA hybrids at DSB sites [82–84]. In addition to R-loop-mediated recruitment of RAD51 or RAD52 during the HR-mediated DSB repair (Figure 5), R-loops affect DSB repair process by HR or non-homologous end joining (NHEJ) pathway. In fission yeast, RNA-DNA hybrids in damaged regions recruit replication protein A (RPA) to ssDNA overhangs, but these hybrids should be degraded by RNase H to complete the DNA repair process [83]. Similarly, persistent R-loops in damaged sites block repair process in budding yeast [85]. On the contrary, the existence of RNA-DNA hybrids around DSB facilitates DNA repair by both HR and NHEJ pathways in human cells and these processes require Drosha, miRNA biogenesis enzyme, which manages the recruitment of repair factors [86].
Genome-wide R-loop profiling methodologies
Techniques for profiling R-loop distribution genome-wide were developed, including (i) bisulfite-based footprinting, (ii) S9.6 antibody-based DNA:RNA Immunoprecipitation coupled with sequencing (DRIP-seq) and (iii) RNase H-based approach (Table 1). Sodium bisulfite treatment induces the deamination of unmethylated cytosines to uracils in ssDNA [87]. Under non-denaturing conditions, bisulfite conversion can be used as marks for R-loop footprinting which detects RNA-DNA hybrid-dependent ssDNA footprints in genomic DNA [50]. The most widely adopted strategy for profiling R-loops has been based on the intrinsic specificity of the monoclonal S9.6 antibody for the RNA-DNA hybrid structure [88]. DNA:RNA Immunoprecipitation (DRIP), which displays a specificity for RNA-DNA hybrids, has been coupled with high-throughput sequencing, termed DRIP-seq, for mapping R-loops [50, 89]. Based on the robustness of DRIP-seq, various modified methodologies have been established such as RDIP-seq (RNA:DNA immunoprecipitation, followed by sequencing) [90], S1-DRIP-seq (S1 nuclease DNA:RNA immunoprecipitation, followed by deep sequencing) [91], DRIPc-seq (DNA:RNA immunoprecipitation, followed by cDNA conversion coupled to high-throughput sequencing) [18, 89], ssDRIP-seq (single-strand DNA ligation-based library construction from RNA-DNA hybrid immunoprecipitation, followed by sequencing) [92] and bisDRIP-seq (bisulfite-DNA:RNA immunoprecipitation, followed by sequencing) [93]. Recently, a modified DRIP-seq technology termed qDRIP-seq (quantitative differential DNA:RNA immunoprecipitation followed by sequencing) uses synthetic RNA-DNA hybrid internal standards as spike-in control, which enables absolute quantification of R-loop content with high resolution [56]. Alternatively, DRIP-chip (DNA:RNA immunoprecipitation followed by hybridization on tiling microarrays) [94] and S9.6 ChIP-seq (chromatin immunoprecipitation with the S9.6 antibody, followed by deep sequencing) [95] have been also proposed as S9.6-based methods for the identification of R-loop structures. Distinctive from S9.6 antibody-dependent R-loop detection, other mapping techniques rely on RNase H, an enzyme that degrades the RNA strand in RNA-DNA hybrids in the 3’→5’ direction. In DRIVE-seq (DNA:RNA in vitro Enrichment, followed by sequencing), a catalytically-dead human RNase H1, which binds to RNA-DNA hybrids but not resolve them, is used to affinity pull-down to capture in vitro R-loops [50]. R-ChIP (R-loop chromatin immunoprecipitation) is also based on a catalytically-dead human RNase H1, fused with a nuclear localization signal (NLS) at the N terminus and a V5 tag at the C terminus, for in vivo R-loop profiling [34, 96]. A R-loop-capturing method, termed MapR, employs CUT&RUN strategy to overcome certain disadvantages of previous RNase H-based approaches [97, 98]. In MapR, R-loop regions are recognized by a catalytically-dead RNase H1, cleaved by micrococcal nuclease (MNase) digestion, and released from chromatin for sequencing [97, 98]. More recently, a R-loop imaging technique for detecting the RNA-DNA hybrids by immunofluorescence has been reported [99]. Using catalytically inactive human RNase H1 tagged with GFP (GFP-dRNH1), this method allows imaging and quantifying R-loops [99]. Please note that both S9.6 antibody- and RNase H1-based mapping techniques exhibit intrinsic target bias [56, 100], off-target effects (e.g. S9.6 for dsRNA) or high background (for RNase H1). Further optimized strategies for accurately identifying R-loops genome-wide are required, due to recently appreciated relevance of R-loops in human diseases.
Table 1.
Techniques for profiling genome-wide R-loop distribution.
| Technique | Innovation | Experimental workflow | Strengths | Weaknesses | Ref. | |
|---|---|---|---|---|---|---|
| DRIP-seq and its derivatives | DRIP-seq | Using the intrinsic specificity of the S9.6 antibody for RNA-DNA hybrid | 1. DNA extraction 2. Restriction enzyme digestion 3. Immunoprecipitation with S9.6 antibody 4. ds DNA sequencing |
Convenient, robust signal | Low resolution, higher background, not strand specific, S9.6 antibody’s bias in sequence recognition [100] | [50, 89] |
| RDIP-seq | Pre-treatment with RNase I, use of sonication, directional sequencing | 1. Whole cell nucleic acid sonication 2. RNase I pre-treatment 3. Immunoprecipitation with S9.6 antibody 4. RNA sequencing |
Reduced background, Strand-specific | A sonication step for nucleic acid fragmentation, which has been shown to reduce the number of genomic R-loops, off target affinity of the S9.6 antibody for dsRNA | [90] | |
| S1-DRIP-seq | Using optimized levels of S1 nuclease to preserve RNA-DNA hybrid during sonication |
1. DNA extraction 2. S1 nuclease digestion 3. Sonication 4. Immunoprecipitation with S9.6 antibody 5. dsDNA sequencing |
Quantitative recovery of R loops, High-resolution | Not strand specific, S9.6 antibody’s bias in sequence recognition | [91] | |
| DRIPc-seq | Further digestion by DNase I, cDNA conversion | 1. DNA extraction 2. Restriction enzyme digestion 3. Immunoprecipitation with S9.6 antibody 4. DNase I treatment 5. RNA recovery and reverse transcription to cDNA 6. RNA sequencing |
High resolution, strand specific | Off target affinity of the S9.6 antibody for dsRNA | [18, 89] | |
| ssDRIP-seq | Distinguish specific DNA strands with fewer steps for library construction than DRIPc-seq | 1. DNA extraction 2. Sonication 3. Immunoprecipitation with S9.6 antibody 4. ssDNA sequencing |
Strand specific | S9.6 antibody’s bias in sequence recognition | [92] | |
| bisDRIP-seq | In vivo R-loop profiling, combining the use of the S9.6 antibody with sodium bisulfite treatment | 1. Cell lysis in the presence of bisulfite 2. DNA extraction 3. Restriction enzyme digestion 4. Immunoprecipitation with S9.6 antibody 5. Bisulfite-modified dsDNA sequencing |
Discriminate between the R-loop sequence and the surrounding non-R-loop sequence, high resolution, Strand specific | High sequencing depth is needed, background conversions in ds DNA by bisulfite, S9.6 antibody’s bias in sequence recognition | [93] | |
| qDRIP-seq | Combining synthetic RNA-DNA hybrid internal standards (spike-in) | 1. Adding spike-in to cell lysate 2. Sonication 3. Immunoprecipitation with S9.6 antibody 4. ssDNA sequencing |
Accurate cross-condition normalization, absolute quantitation, sensitive, high resolution, strand-specific | S9.6 antibody’s bias in sequence recognition | [56] | |
| Other S9.6 based approaches | DRIP-chip | DRIP followed by hybridization on tiling microarray | 1. Crosslinking with formaldehyde 2. Sonication 3. Immunoprecipitation with S9.6 antibody 4. T7 RNA polymerase amplification 5. Biotin labeling 6. Microarray |
S9.6 antibody’s bias in sequence recognition | [94] | |
| S9.6 ChIP-seq (chromatin immunoprecipitation with antibody S9.6, followed by deep sequencing of immunopurified DNA fragments) | Application of ChIP-seq to mapping R loops | 1. Crosslinking with formaldehyde 2. Sonication 3. Immunoprecipitation with S9.6 antibody 4. Reverse crosslinking 5. dsDNA sequencing |
High-resolution | Formaldehyde crosslinking could affect results, not strand specific | [95] | |
| RNase H based approaches | DRIVE-seq (DNA:RNA in vitro Enrichment) | Using specificity of catalytically dead RNase H1 for RNA-DNA hybrid |
1. Genomic DNA extraction 2. Restriction enzyme digestion 3. Catalytically dead RNase H1 incubation 4. Pull-down 5. dsDNA sequencing |
Enable the specific and near quantitative recovery of R loop molecules in complexnucleic acid mixture exquisite specificity of RNase H | Low capture efficiency, not strand specific, substantial RNase H-resistant regions on the genome | [50] |
| R-ChIP | In vivo R-loop profiling using catalytically dead RNase H1 | 1. Introduce V5-tagged catalytically dead mutant RNase H1 into cells 2. Sonication 3. Immunoprecipitation with anti-V5 antibody 4. ssDNA sequencing |
exquisite specificity of RNase H, strand-specific | substantial RNase H- resistant regions on the genome, require the generation of stable cell lines (time-consuming) | [34, 96] | |
| MapR | Combining the specificity of RNase H for RNA-DNA hybrid with CUT&RUN approach | 1. Immobilize cells on beads 2. Incubate with GST-RNaseHΔcat-MNase 3. R-loop recognition by RNaseHΔcat 4. MNase mediated R-loop cleavage 5. DNA sequencing |
Antibody-independent, does not require the generation of stable cell lines (fast and convenient), high sensitivity with low input material | substantial RNase H-resistant regions on the genome | [97, 98] | |
| R-loop imaging | Imaging and quantifying R-loops using GFP-catalytically dead RNase H1 (dRNH1) | 1. Expression and purification of GFP-dRNH 2. Transfection in fixed cells 3. Imaging |
Using purified GFP-dRNH1 protein, bypassing the need for cell line engineering | GFP-dRNH1 has its own limitations (non-specific binding, binding preference to G-rich) | [99] |
R-Loops and human disease
Previous evidence has demonstrated an intimate relationship between R-loop and pathogenesis of neurodegenerative disease, motor neuron disorder and autoimmune disease; for these topics, reviewers shall refer to other reviews [101–103]. We focus this section on the role for R-loops in oncogenesis. R-loop-induced DNA damage causes genomic instability, which can drive cancer initiation/progression [104]. In support, the tumor suppressors BRCA1 and BRCA2 participate in repair of R-loop-induced DNA damage and depletion of BRCA1/2 increased R-loop accumulation [63, 80, 105]. In ER-responsive breast cancer, estrogen can stimulate R-loop formation leading to DNA damage and genome instability, suggesting that estrogen-induced R-loops to be tumorigenic [106].
R-loop accumulation underlying cancer with spliceosome mutation.
Myelodysplastic syndromes (MDS) are clonal hematopoietic diseases, featured with ineffective hematopoiesis and morphological dysplasia leading to cytopenias and a higher risk of development to acute myeloid leukemia (AML) [107]. Somatic mutations of splicing factors, including SF3B1, U2AF1 (also known as U2AF35), SRSF2 and ZRSR2, are prevalent in MDS patients [107, 108]. SF3B1, a component of the core spliceosome U2 complex, is the most commonly mutated spliceosomal gene in MDS [109]. SF3B1 mutations were typically found in MDS with increased ring sideroblasts, including refractory anaemia with ring sideroblasts (RARS) and refractory cytopenia with multilineage dysplasia with ring sideroblasts (RCMD-RS). In bone marrow stem/progenitor cells from MDS and AML patients with SF3B1 mutation, there is a significant increase of R-loops, which cause DNA damage and ATR-Chk1 activation [110] (Figure 6A). Similar effects of U2AF1 and SRSF2 mutations were also observed [104, 111, 112]. Mechanistically, SRSF2 mutation interferes with p-TEFb translocation from the 7SK complex to Pol-II resulting in transcriptional elongation defection, which may explain the contribution of this splicing factor mutation to R-loop elevation [111]. Accumulating evidence also revealed that splicing factor mutations are mutually exclusive in MDS patients [113] and that each mutation exhibits own specific/selective effect on R-loop formation. Indeed, SRSF2 mutation induces R-loops at TSSs and U2AF1 mutation enhances R-loops in both promoter and non-promoter regions [111], indicating dynamic characteristics of R-loop formation. Importantly, splicing factor mutation-caused perturbations of RNA splicing and R-loop accumulation occur independently of one another [111, 112], indicative of cooperation between the two changes during MDS pathogenesis (Figure 6A).
Figure 6. R-loop aberration is prevalent, sometimes causal, during development of human cancer.
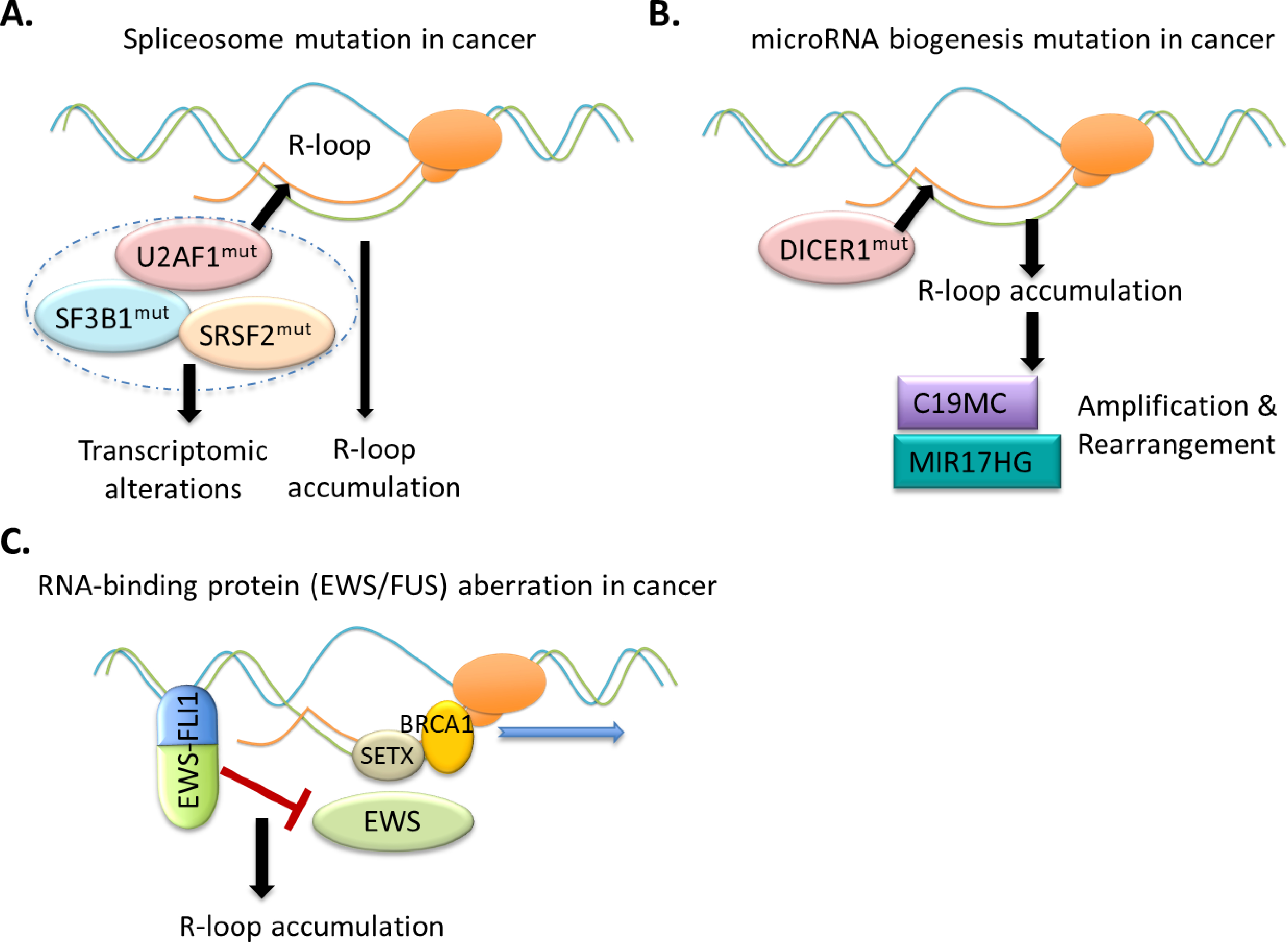
(A) Splicing factor mutations induce Pol II pausing and increase R-loops, which lead to DNA damage and ATR-Chk1 activation, influencing clonal hematopoiesis and/or leukemic transformation.
(B) During clonal evolution of ETMRs, R-loops are enriched around C19MC or MIR17HG, likely due to the loss-of-function mutation of the microRNA-processing enzyme DICER1, which induces the chromosomal breakage and rearrangement, such as the TTYH1-C19MC fusion and somatic amplification of C19MC or MIR17HG as recurrently detected in ETMRs.
(C) In Ewing sarcoma, the oncogenic EWS–FLI1 chimera promotes the accumulation of R-loops. The N-terminal protein-protein interaction domain of EWS–FLI1 is essential for R-loop accumulation, which likely acts to interfere with functionality of wild-type EWS; and a functional sequestration and impairment of BRCA1, due to R-loop accumulation and DNA damage management, might underlie the genomic instability and therapeutic response to PARP inhibitor.
R-loop accumulation underlying cancer with microRNA-processing mutation.
R-loop is potentially involved in generating chromosomal breakpoints and rearrangement during tumorigenesis. Embryonal tumors with multilayered rosettes (ETMRs) are highly aggressive pediatric CNS malignancies, including embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma (EBL) and medulloepithelioma (MEPL) [114]. A proposed driver mutation of ETMRs is genomic alteration of the chromosome 19 (19q13.42) microRNA cluster (C19MC) [114–116], either amplification of C19MC or aberrant fusion of TTYH1-C19MC, which leads to high expression of C19MC and an embryonic brain-specific DNMT3B isoform. However, the molecular mechanism underlying C19MC abnormality was previously unclear. Lambo et al. recently reported that R-loops accumulated in ETMRs are enriched around C19MC, which likely induced chromosomal instability and breakage leading to C19MC amplification and/or rearrangement in ETMRs [117]. Interestingly, a subset of ETMRs is featured with damaging lesion of DICER1, an miRNA-processing endoribonuclease, with somatic or germline mutations found clustered at its helicase and RNASE functional domains [117]. DICER1 malfunction, likely acting as a cancer susceptibility mutation, potentially interferes with biogenesis of microRNAs being transcribed at the C19MC locus, indicative of a casual mechanism underlying the R-loop-associated instability/breakage of C19MC [117]. In support, somatic amplification of the miR-17–92 miRNA cluster on chromosome 13 (also known as MIR17HG) also exists in ETMRs, regardless of C19MC amplification [117] (Figure 6B).
RNA-binding factor aberration resulting in R-loop accumulation in sarcoma.
Ewing sarcoma, a highly aggressive cancer that occurs primarily in the bone or soft tissue, is characterized by chromosomal translocation of t(11;22)(q12;q24), which generates a chimeric oncoprotein EWS–FLI1. Recently, Gorthi et al reported that wild-type EWS exhibits a multifaceted function in binding RNA, slowing transcriptional elongation and decreasing R-loop formation; on the other hand, EWS–FLI1 facilitates transcription and enhances the R-loop accumulation, resulting in the increased replication stress in Ewing sarcoma [118]. EWS–FLI1 forms interaction with EWS through its N-terminal region and the R-loop-inducing effect by EWS–FLI1 is likely due to EWS interference [118] (Figure 6C). Interestingly, Ewing sarcoma also displays BRCA1-deficient tumor like phenotypes due to association/sequestration of BRCA1 (and associated helicase SETX [80]) with R-loop and stalled transcriptional machinery, suggesting a potential mechanism underlying oncogenesis [118]. Importantly, BRCA1 deficiency rendered a hypersensitivity of Ewing sarcoma to chemotherapy such as PARP1 inhibitor [118].
CONCLUDING REMARKS
Intensive studies during recent years have allowed important insights into R-loop biology. R-loop is now appreciated to be relevant in a wide range of biological processes. As mentioned above, their roles can be beneficial (so-called “regulatory R-loops”) or detrimental (so-called “unscheduled or unwanted R-loops” when too many of them are excessively generated and not resolved), which depends on cellular context.
On one hand, regulatory roles of R-loops are important in normal physiology. Besides the above-mentioned functions of regulatory R-loops in chromatin regulation (DNA methylation and histone modifications), transcription and a range of specialized processes such as CRIPSR and class switch recombination, other documented examples include prevention of telomere shortening and premature cellular senescence by R-loops [119]; also, the oncogene c-MYC is activated by the interaction of Tudor domain containing protein 3 (TDRD3) and topoisomerase IIIB (TOP3B), which decreases negative supercoiled DNA and transcription-generated R-loops [120], indicating a regulatory role of R-loops. On the other hand, when R-loops cannot be efficiently resolved, unwanted, excessively accumulated R-loops elicit signals of DNA damage repair and responses, which can shape and/or initiate the development of human diseases.
R-loop’s function and regulation are context-dependent and complicated, with questions and issues to be answered. First, further research needs to be directed at developing optimized techniques for more accurate genome-wide R-loop detection and mapping strategies, partly due to a prevalent involvement of R-loop in human pathologies such as cancer. Second, a set of outstanding questions related to R-loop biology and regulation remain unaddressed. For example, while most of the produced R-loops are either removed or resolved, the subset of R-loops retained in the genome, so what determines the fate of R-loops? In other words, what differentiates a minority of ‘regulatory’ R-loops from a majority of those ‘unscheduled or unwanted’ ones to be removed? Also, once R-loop is formed and retained, what are the molecular determinants as of context-dependent functions of R-loops? Because it has been reported that some of R-loops are related to gene activation and some others associated with repression. Furthermore, m6A modifications occur frequently in the RNA strands of RNA-DNA hybrids, involving m6A “writer” (i.e., the METTL3-METTL14 heterodimer) and “reader” (YTH domain proteins). Then, what is the causal and sequential relationship among these events, which include the R-loop formation and removal and m6A “writing” and “reading”? In general, does m6A serve as a cellular ‘sensing’ mechanism for R-loop detection and subsequent removal? Moreover, as deregulations of RNA splicing and processing processes and alteration/mutation of RNA-binding factors are frequent in a wide range of human cancers. Does this mean that excessive R-loop accumulation so far observed in a few of cancer types can be generalized to many others tumors.
Last, can the field leverage upon current understanding of R-loops for the development of new therapeutic strategies? One important therapeutic implication from studies of cancers exhibiting R-loop accumulation is that these cancers generally have a high demand for DNA damage management and repair, which thereby renders a relative hypersensitivity to therapy with PARP and/or TOP1 inhibitors as reported in ETMRs [117], Ewing sarcoma [118] and other tumor models [26]. Future investigation is warranted to expand such ideas to other tumor types exhibiting similar R-loop accumulation, since several PARP inhibitors are now FDA-approved for treatment of breast, ovarian and prostate cancer patients carrying the mutation of DNA damage repair gene (BRCA1/2, ATM or ATR). A better understanding of R-loop homeostasis and the underlying molecular basis shall continue to impact on biology and medicine.
Highlights.
Regulatory R-loops are crucially involved in numerous physiological processes.
Unscheduled R-loop accumulation is a source of genomic instability.
RNA/DNA modifications (m6A/6mA) emerge to be crucial for R-loop modulation.
R-loop “readers” regulate chromatin states and gene transcription.
R-loop accumulation is prevalent, and sometimes causal, in oncogenesis.
ACKNOWLEDGEMENT
We graciously thank S Zhao and the Wang Laboratory members for providing illustration and helpful discussion. We also apologize to authors whose works could not be covered due to the space limit.
FUNDING
This work was supported by NIH grants (R01-CA215284 and R01-CA218600 to G.G.W.). G.G.W. is an American Cancer Society (ACS) Research Scholar and a Leukemia and Lymphoma Society (LLS) Scholar.
Footnotes
Conflict of Interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Roberts RW, Crothers DM, Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition, Science, 258 (1992) 1463–1466. [DOI] [PubMed] [Google Scholar]
- [2].Garcia-Muse T, Aguilera A, R Loops: From Physiological to Pathological Roles, Cell, 179 (2019) 604–618. [DOI] [PubMed] [Google Scholar]
- [3].Niehrs C, Luke B, Regulatory R-loops as facilitators of gene expression and genome stability, Nat Rev Mol Cell Biol, 21 (2020) 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sollier J, Cimprich KA, Breaking bad: R-loops and genome integrity, Trends Cell Biol, 25 (2015) 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chedin F, Nascent Connections: R-Loops and Chromatin Patterning, Trends Genet, 32 (2016) 828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wahba L, Gore SK, Koshland D, The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability, Elife, 2 (2013) e00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ariel F, Lucero L, Christ A, Mammarella MF, Jegu T, Veluchamy A, Mariappan K, Latrasse D, Blein T, Liu C, Benhamed M, Crespi M, R-Loop Mediated trans Action of the APOLO Long Noncoding RNA, Mol Cell, 77 (2020) 1055–1065 e1054. [DOI] [PubMed] [Google Scholar]
- [8].Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, Rubio-Roldan A, Li M, Eleftheriou M, Crutchley J, Flatt L, Young L, Gaffney DJ, Denning C, Dalhus B, Emes RD, Gackowski D, Correa IR Jr., Garcia-Perez JL, Klungland A, Gromak N, Ruzov A, N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells, Nat Genet, 52 (2020) 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Skourti-Stathaki K, Proudfoot NJ, A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression, Genes Dev, 28 (2014) 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Aguilera A, Garcia-Muse T, R loops: from transcription byproducts to threats to genome stability, Mol Cell, 46 (2012) 115–124. [DOI] [PubMed] [Google Scholar]
- [11].Kasahara M, Clikeman JA, Bates DB, Kogoma T, RecA protein-dependent R-loop formation in vitro, Genes Dev, 14 (2000) 360–365. [PMC free article] [PubMed] [Google Scholar]
- [12].Zaitsev EN, Kowalczykowski SC, A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange, Genes Dev, 14 (2000) 740–749. [PMC free article] [PubMed] [Google Scholar]
- [13].Broccoli S, Rallu F, Sanscartier P, Cerritelli SM, Crouch RJ, Drolet M, Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation, Mol Microbiol, 52 (2004) 1769–1779. [DOI] [PubMed] [Google Scholar]
- [14].Roy D, Lieber MR, G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter, Mol Cell Biol, 29 (2009) 3124–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR, Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site, Mol Cell Biol, 30 (2010) 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Stolz R, Sulthana S, Hartono SR, Malig M, Benham CJ, Chedin F, Interplay between DNA sequence and negative superhelicity drives R-loop structures, Proc Natl Acad Sci U S A, 116 (2019) 6260–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ginno PA, Lim YW, Lott PL, Korf I, Chedin F, GC skew at the 5’ and 3’ ends of human genes links R-loop formation to epigenetic regulation and transcription termination, Genome Res, 23 (2013) 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, Chedin F, Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals, Mol Cell, 63 (2016) 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pavri R, R Loops in the Regulation of Antibody Gene Diversification, Genes (Basel), 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yu K, Lieber MR, Current insights into the mechanism of mammalian immunoglobulin class switch recombination, Crit Rev Biochem Mol Biol, 54 (2019) 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Roy D, Yu K, Lieber MR, Mechanism of R-loop formation at immunoglobulin class switch sequences, Mol Cell Biol, 28 (2008) 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wiedemann EM, Peycheva M, Pavri R, DNA Replication Origins in Immunoglobulin Switch Regions Regulate Class Switch Recombination in an R-Loop-Dependent Manner, Cell Rep, 17 (2016) 2927–2942. [DOI] [PubMed] [Google Scholar]
- [23].Zhang ZZ, Pannunzio NR, Han L, Hsieh CL, Yu K, Lieber MR, The strength of an Ig switch region is determined by its ability to drive R loop formation and its number of WGCW sites, Cell Rep, 8 (2014) 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang X, Chiang HC, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M, Oliver B, Wang H, Petit A, Soler T, Brunet J, Mateo F, Angel Pujana M, Poggi E, Chaldekas K, Isaacs C, Peshkin BN, Ochoa O, Chedin F, Theoharis C, Sun LZ, Curiel TJ, Elledge R, Jin VX, Hu Y, Li R, Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis, Nat Commun, 8 (2017) 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, Refvik ST, Chiang CM, Xhemalce B, Paull TT, Brodbelt JS, Marcotte EM, Miller KM, Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity, Genes Dev, 33 (2019) 1751–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lam FC, Kong YW, Huang Q, Vu Han TL, Maffa AD, Kasper EM, Yaffe MB, BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage, Nat Commun, 11 (2020) 4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nudler E, RNA polymerase backtracking in gene regulation and genome instability, Cell, 149 (2012) 1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zatreanu D, Han Z, Mitter R, Tumini E, Williams H, Gregersen L, Dirac-Svejstrup AB, Roma S, Stewart A, Aguilera A, Svejstrup JQ, Elongation Factor TFIIS Prevents Transcription Stress and R-Loop Accumulation to Maintain Genome Stability, Mol Cell, 76 (2019) 57–69 e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].De Magis A, Manzo SG, Russo M, Marinello J, Morigi R, Sordet O, Capranico G, DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells, Proc Natl Acad Sci U S A, 116 (2019) 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee CY, McNerney C, Ma K, Zhao W, Wang A, Myong S, R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation, Nat Commun, 11 (2020) 3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Miglietta G, Russo M, Capranico G, G-quadruplex-R-loop interactions and the mechanism of anticancer G-quadruplex binders, Nucleic Acids Res, 48 (2020) 11942–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Skourti-Stathaki K, Proudfoot NJ, Gromak N, Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination, Mol Cell, 42 (2011) 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, Vedadi M, Min J, Pawson TJ, Blencowe BJ, Greenblatt JF, SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination, Nature, 529 (2016) 48–53. [DOI] [PubMed] [Google Scholar]
- [34].Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H, Zhou Y, Zhang DE, Fu XD, R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters, Mol Cell, 68 (2017) 745–757 e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tan-Wong SM, Dhir S, Proudfoot NJ, R-Loops Promote Antisense Transcription across the Mammalian Genome, Mol Cell, 76 (2019) 600–616 e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, Villanueva A, Esteller M, Guil S, Head-to-head antisense transcription and R-loop formation promotes transcriptional activation, Proc Natl Acad Sci U S A, 112 (2015) 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I, LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure, Mol Cell, 60 (2015) 626–636. [DOI] [PubMed] [Google Scholar]
- [38].Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C, R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus, Science, 340 (2013) 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C, N6-methyladenosine-dependent regulation of messenger RNA stability, Nature, 505 (2014) 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Roundtree IA, Evans ME, Pan T, He C, Dynamic RNA Modifications in Gene Expression Regulation, Cell, 169 (2017) 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zaccara S, Ries RJ, Jaffrey SR, Reading, writing and erasing mRNA methylation, Nat Rev Mol Cell Biol, 20 (2019) 608–624. [DOI] [PubMed] [Google Scholar]
- [42].Zhou KI, Pan T, An additional class of m(6)A readers, Nat Cell Biol, 20 (2018) 230–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yang X, Liu QL, Xu W, Zhang YC, Yang Y, Ju LF, Chen J, Chen YS, Li K, Ren J, Sun Q, Yang YG, m(6)A promotes R-loop formation to facilitate transcription termination, Cell Res, 29 (2019) 1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, Xie C, Zhou H, Luo X, Liu H, Chen L, Ren J, Wang W, Zhao Y, METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation, Mol Cell, 79 (2020) 425–442 e427. [DOI] [PubMed] [Google Scholar]
- [45].Kang HJ, Cheon NY, Park H, Jeong GW, Ye BJ, Yoo EJ, Lee JH, Hur JH, Lee EA, Kim H, Lee KY, Choi SY, Lee-Kwon W, Myung K, Lee JY, Kwon HM, TonEBP recognizes R-loops and initiates m6A RNA methylation for R-loop resolution, Nucleic Acids Res, 49 (2021) 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang LH, Zhang XY, Hu T, Chen XY, Li JJ, Raida M, Sun N, Luo Y, Gao X, The SUMOylated METTL8 Induces R-loop and Tumorigenesis via m3C, iScience, 23 (2020) 100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhou C, Wang C, Liu H, Zhou Q, Liu Q, Guo Y, Peng T, Song J, Zhang J, Chen L, Zhao Y, Zeng Z, Zhou DX, Identification and analysis of adenine N(6)-methylation sites in the rice genome, Nat Plants, 4 (2018) 554–563. [DOI] [PubMed] [Google Scholar]
- [48].Fang Y, Chen L, Lin K, Feng Y, Zhang P, Pan X, Sanders J, Wu Y, Wang XE, Su Z, Chen C, Wei H, Zhang W, Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice, Genome Res, 29 (2019) 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ross JP, Suetake I, Tajima S, Molloy PL, Recombinant mammalian DNA methyltransferase activity on model transcriptional gene silencing short RNA-DNA heteroduplex substrates, Biochem J, 432 (2010) 323–332. [DOI] [PubMed] [Google Scholar]
- [50].Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F, R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters, Mol Cell, 45 (2012) 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Arab K, Karaulanov E, Musheev M, Trnka P, Schafer A, Grummt I, Niehrs C, GADD45A binds R-loops and recruits TET1 to CpG island promoters, Nat Genet, 51 (2019) 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Feng W, Chen S, Wang J, Wang X, Chen H, Ning W, Zhang Y, DHX33 Recruits Gadd45a To Cause DNA Demethylation and Regulates a Subset of Gene Transcription, Mol Cell Biol, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Alecki C, Chiwara V, Sanz LA, Grau D, Arias Perez O, Boulier EL, Armache KJ, Chedin F, Francis NJ, RNA-DNA strand exchange by the Drosophila Polycomb complex PRC2, Nat Commun, 11 (2020) 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Skourti-Stathaki K, Torlai Triglia E, Warburton M, Voigt P, Bird A, Pombo A, R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes, Mol Cell, 73 (2019) 930–945 e934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yuan W, Zhou J, Tong J, Zhuo W, Wang L, Li Y, Sun Q, Qian W, ALBA protein complex reads genic R-loops to maintain genome stability in Arabidopsis, Sci Adv, 5 (2019) eaav9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Crossley MP, Bocek MJ, Hamperl S, Swigut T, Cimprich KA, qDRIP: a method to quantitatively assess RNA-DNA hybrid formation genome-wide, Nucleic Acids Res, 48 (2020) e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Petersen-Mahrt SK, Harris RS, Neuberger MS, AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification, Nature, 418 (2002) 99–103. [DOI] [PubMed] [Google Scholar]
- [58].Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X, R-loop-mediated genomic instability is caused by impairment of replication fork progression, Genes Dev, 25 (2011) 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cristini A, Ricci G, Britton S, Salimbeni S, Huang SN, Marinello J, Calsou P, Pommier Y, Favre G, Capranico G, Gromak N, Sordet O, Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks, Cell Rep, 28 (2019) 3167–3181 e3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA, Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability, Mol Cell, 56 (2014) 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Matos DA, Zhang JM, Ouyang J, Nguyen HD, Genois MM, Zou L, ATR Protects the Genome against R Loops through a MUS81-Triggered Feedback Loop, Mol Cell, 77 (2020) 514–527 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Santos-Pereira JM, Herrero AB, Garcia-Rubio ML, Marin A, Moreno S, Aguilera A, The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability, Genes Dev, 27 (2013) 2445–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].D’Alessandro G, Whelan DR, Howard SM, Vitelli V, Renaudin X, Adamowicz M, Iannelli F, Jones-Weinert CW, Lee M, Matti V, Lee WTC, Morten MJ, Venkitaraman AR, Cejka P, Rothenberg E, d’Adda di Fagagna F, BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment, Nat Commun, 9 (2018) 5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li X, Manley JL, Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability, Cell, 122 (2005) 365–378. [DOI] [PubMed] [Google Scholar]
- [65].Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A, Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles, EMBO J, 30 (2011) 3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Grzechnik P, Gdula MR, Proudfoot NJ, Pcf11 orchestrates transcription termination pathways in yeast, Genes Dev, 29 (2015) 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gavalda S, Gallardo M, Luna R, Aguilera A, R-loop mediated transcription-associated recombination in trf4Delta mutants reveals new links between RNA surveillance and genome integrity, PLoS One, 8 (2013) e65541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, Economides AN, Bradner JE, Rabadan R, Basu U, RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity, Cell, 161 (2015) 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V, Pasero P, Canitrot Y, Legube G, Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations, Nat Commun, 9 (2018) 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, Boothman DA, XRN2 Links Transcription Termination to DNA Damage and Replication Stress, PLoS Genet, 12 (2016) e1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].De I, Bessonov S, Hofele R, dos Santos K, Will CL, Urlaub H, Luhrmann R, Pena V, The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes, Nat Struct Mol Biol, 22 (2015) 138–144. [DOI] [PubMed] [Google Scholar]
- [72].Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, Meyer T, Cimprich KA, A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability, Mol Cell, 35 (2009) 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Teng Y, Yadav T, Duan M, Tan J, Xiang Y, Gao B, Xu J, Liang Z, Liu Y, Nakajima S, Shi Y, Levine AS, Zou L, Lan L, ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB, Nat Commun, 9 (2018) 4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Leela JK, Syeda AH, Anupama K, Gowrishankar J, Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli, Proc Natl Acad Sci U S A, 110 (2013) 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mersaoui SY, Yu Z, Coulombe Y, Karam M, Busatto FF, Masson JY, Richard S, Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids, EMBO J, 38 (2019) e100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Perez-Calero C, Bayona-Feliu A, Xue X, Barroso SI, Munoz S, Gonzalez-Basallote VM, Sung P, Aguilera A, UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide, Genes Dev, 34 (2020) 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sridhara SC, Carvalho S, Grosso AR, Gallego-Paez LM, Carmo-Fonseca M, de Almeida SF, Transcription Dynamics Prevent RNA-Mediated Genomic Instability through SRPK2-Dependent DDX23 Phosphorylation, Cell Rep, 18 (2017) 334–343. [DOI] [PubMed] [Google Scholar]
- [78].Cristini A, Groh M, Kristiansen MS, Gromak N, RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage, Cell Rep, 23 (2018) 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chakraborty P, Huang JTJ, Hiom K, DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing, Nat Commun, 9 (2018) 4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, Kellis M, Hill SJ, Parmigiani G, Proudfoot NJ, Livingston DM, BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair, Mol Cell, 57 (2015) 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nguyen DT, Voon HPJ, Xella B, Scott C, Clynes D, Babbs C, Ayyub H, Kerry J, Sharpe JA, Sloane-Stanley JA, Butler S, Fisher CA, Gray NE, Jenuwein T, Higgs DR, Gibbons RJ, The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats, EMBO Rep, 18 (2017) 914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Aguilera A, Gomez-Gonzalez B, DNA-RNA hybrids: the risks of DNA breakage during transcription, Nat Struct Mol Biol, 24 (2017) 439–443. [DOI] [PubMed] [Google Scholar]
- [83].Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T, Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair, Cell, 167 (2016) 1001–1013 e1007. [DOI] [PubMed] [Google Scholar]
- [84].Crossley MP, Bocek M, Cimprich KA, R-Loops as Cellular Regulators and Genomic Threats, Mol Cell, 73 (2019) 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Costantino L, Koshland D, Genome-wide Map of R-Loop-Induced Damage Reveals How a Subset of R-Loops Contributes to Genomic Instability, Mol Cell, 71 (2018) 487–497 e483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lu WT, Hawley BR, Skalka GL, Baldock RA, Smith EM, Bader AS, Malewicz M, Watts FZ, Wilczynska A, Bushell M, Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair, Nat Commun, 9 (2018) 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hayatsu H, Wataya Y, Kazushige K, The addition of sodium bisulfite to uracil and to cytosine, J Am Chem Soc, 92 (1970) 724–726. [DOI] [PubMed] [Google Scholar]
- [88].Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ, Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids, J Immunol Methods, 89 (1986) 123–130. [DOI] [PubMed] [Google Scholar]
- [89].Sanz LA, Chedin F, High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing, Nat Protoc, 14 (2019) 1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, Sato OBP,H, Zhang Z, Jeddeloh J, Montagna C, Golden A, Seoighe C, Greally JM, RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships, Epigenetics Chromatin, 8 (2015) 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D, S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation, Genes Dev, 30 (2016) 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Xu W, Xu H, Li K, Fan Y, Liu Y, Yang X, Sun Q, The R-loop is a common chromatin feature of the Arabidopsis genome, Nat Plants, 3 (2017) 704–714. [DOI] [PubMed] [Google Scholar]
- [93].Dumelie JG, Jaffrey SR, Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chan YA, Aristizabal MJ, Lu PY, Luo Z, Hamza A, Kobor MS, Stirling PC, Hieter P, Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip, PLoS Genet, 10 (2014) e1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].El Hage A, Webb S, Kerr A, Tollervey D, Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria, PLoS Genet, 10 (2014) e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chen JY, Zhang X, Fu XD, Chen L, R-ChIP for genome-wide mapping of R-loops by using catalytically inactive RNASEH1, Nat Protoc, 14 (2019) 1661–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yan Q, Sarma K, MapR: A Method for Identifying Native R-Loops Genome Wide, Curr Protoc Mol Biol, 130 (2020) e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Yan Q, Shields EJ, Bonasio R, Sarma K, Mapping Native R-Loops Genome-wide Using a Targeted Nuclease Approach, Cell Rep, 29 (2019) 1369–1380 e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Crossley MP, Brickner JR, Song C, Zar SMT, Maw SS, Chedin F, Tsai MS, Cimprich KA, Catalytically inactive, purified RNase H1: A specific and sensitive probe for RNA-DNA hybrid imaging, J Cell Biol, 220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Konig F, Schubert T, Langst G, The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences, PLoS One, 12 (2017) e0178875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Richard P, Manley JL, R Loops and Links to Human Disease, J Mol Biol, 429 (2017) 3168–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Groh M, Gromak N, Out of balance: R-loops in human disease, PLoS Genet, 10 (2014) e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Perego MGL, Taiana M, Bresolin N, Comi GP, Corti S, R-Loops in Motor Neuron Diseases, Mol Neurobiol, 56 (2019) 2579–2589. [DOI] [PubMed] [Google Scholar]
- [104].Gaillard H, Garcia-Muse T, Aguilera A, Replication stress and cancer, Nat Rev Cancer, 15 (2015) 276–289. [DOI] [PubMed] [Google Scholar]
- [105].Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A, BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2, Nature, 511 (2014) 362–365. [DOI] [PubMed] [Google Scholar]
- [106].Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chedin F, Swigut T, Cimprich KA, Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage, Elife, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Sperling AS, Gibson CJ, Ebert BL, The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia, Nat Rev Cancer, 17 (2017) 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S, Frequent pathway mutations of splicing machinery in myelodysplasia, Nature, 478 (2011) 64–69. [DOI] [PubMed] [Google Scholar]
- [109].Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, Bowen DT, Campbell PJ, Ebert BL, Fenaux P, Haferlach T, Heuser M, Jansen JH, Komrokji RS, Maciejewski JP, Walter MJ, Fontenay M, Garcia-Manero G, Graubert TA, Karsan A, Meggendorfer M, Pellagatti A, Sallman DA, Savona MR, Sekeres MA, Steensma DP, Tauro S, Thol F, Vyas P, Van de Loosdrecht AA, Haase D, Tuchler H, Greenberg PL, Ogawa S, Hellstrom-Lindberg E, Cazzola M, SF3B1-mutant MDS as a distinct disease subtype: a proposal from the International Working Group for the Prognosis of MDS, Blood, 136 (2020) 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Singh S, Ahmed D, Dolatshad H, Tatwavedi D, Schulze U, Sanchi A, Ryley S, Dhir A, Carpenter L, Watt SM, Roberts DJ, Abdel-Aal AM, Sayed SK, Mohamed SA, Schuh A, Vyas P, Killick S, Kotini AG, Papapetrou EP, Wiseman DH, Pellagatti A, Boultwood J, SF3B1 mutations induce R-loop accumulation and DNA damage in MDS and leukemia cells with therapeutic implications, Leukemia, 34 (2020) 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chen L, Chen JY, Huang YJ, Gu Y, Qiu J, Qian H, Shao C, Zhang X, Hu J, Li H, He S, Zhou Y, Abdel-Wahab O, Zhang DE, Fu XD, The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations, Mol Cell, 69 (2018) 412–425 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Nguyen HD, Leong WY, Li W, Reddy PNG, Sullivan JD, Walter MJ, Zou L, Graubert TA, Spliceosome Mutations Induce R Loop-Associated Sensitivity to ATR Inhibition in Myelodysplastic Syndromes, Cancer Res, 78 (2018) 5363–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Dvinge H, Kim E, Abdel-Wahab O, Bradley RK, RNA splicing factors as oncoproteins and tumour suppressors, Nat Rev Cancer, 16 (2016) 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Korshunov A, Sturm D, Ryzhova M, Hovestadt V, Gessi M, Jones DT, Remke M, Northcott P, Perry A, Picard D, Rosenblum M, Antonelli M, Aronica E, Schuller U, Hasselblatt M, Woehrer A, Zheludkova O, Kumirova E, Puget S, Taylor MD, Giangaspero F, Peter Collins V, von Deimling A, Lichter P, Huang A, Pietsch T, Pfister SM, Kool M, Embryonal tumor with abundant neuropil and true rosettes (ETANTR), ependymoblastoma, and medulloepithelioma share molecular similarity and comprise a single clinicopathological entity, Acta Neuropathol, 128 (2014) 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pfister S, Remke M, Castoldi M, Bai AH, Muckenthaler MU, Kulozik A, von Deimling A, Pscherer A, Lichter P, Korshunov A, Novel genomic amplification targeting the microRNA cluster at 19q13.42 in a pediatric embryonal tumor with abundant neuropil and true rosettes, Acta Neuropathol, 117 (2009) 457–464. [DOI] [PubMed] [Google Scholar]
- [116].Kleinman CL, Gerges N, Papillon-Cavanagh S, Sin-Chan P, Pramatarova A, Quang DA, Adoue V, Busche S, Caron M, Djambazian H, Bemmo A, Fontebasso AM, Spence T, Schwartzentruber J, Albrecht S, Hauser P, Garami M, Klekner A, Bognar L, Montes JL, Staffa A, Montpetit A, Berube P, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel PM, Duchaine T, Perotti C, Fleming A, Faury D, Remke M, Gallo M, Dirks P, Taylor MD, Sladek R, Pastinen T, Chan JA, Huang A, Majewski J, Jabado N, Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR, Nat Genet, 46 (2014) 39–44. [DOI] [PubMed] [Google Scholar]
- [117].Lambo S, Grobner SN, Rausch T, Waszak SM, Schmidt C, Gorthi A, Romero JC, Mauermann M, Brabetz S, Krausert S, Buchhalter I, Koster J, Zwijnenburg DA, Sill M, Hubner JM, Mack N, Schwalm B, Ryzhova M, Hovestadt V, Papillon-Cavanagh S, Chan JA, Landgraf P, Ho B, Milde T, Witt O, Ecker J, Sahm F, Sumerauer D, Ellison DW, Orr BA, Darabi A, Haberler C, Figarella-Branger D, Wesseling P, Schittenhelm J, Remke M, Taylor MD, Gil-da-Costa MJ, Lastowska M, Grajkowska W, Hasselblatt M, Hauser P, Pietsch T, Uro-Coste E, Bourdeaut F, Masliah-Planchon J, Rigau V, Alexandrescu S, Wolf S, Li XN, Schuller U, Snuderl M, Karajannis MA, Giangaspero F, Jabado N, von Deimling A, Jones DTW, Korbel JO, von Hoff K, Lichter P, Huang A, Bishop AJR, Pfister SM, Korshunov A, Kool M, The molecular landscape of ETMR at diagnosis and relapse, Nature, 576 (2019) 274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, Iniguez AB, Bernard X, Masamsetti VP, Roston S, Lawlor ER, Toretsky JA, Stegmaier K, Lessnick SL, Chen Y, Bishop AJR, EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma, Nature, 555 (2018) 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Perez-Martinez L, Ozturk M, Butter F, Luke B, Npl3 stabilizes R-loops at telomeres to prevent accelerated replicative senescence, EMBO Rep, 21 (2020) e49087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT, Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation, Mol Cell, 53 (2014) 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]


