ABSTRACT
Motile cilia have a ‘9+2’ structure containing nine doublet microtubules and a central apparatus (CA) composed of two singlet microtubules with associated projections. The CA plays crucial roles in regulating ciliary motility. Defects in CA assembly or function usually result in motility-impaired or paralyzed cilia, which in humans causes disease. Despite their importance, the protein composition and functions of most CA projections remain largely unknown. Here, we combined genetic, proteomic and cryo-electron tomographic approaches to compare the CA of wild-type Chlamydomonas reinhardtii with those of three CA mutants. Our results show that two proteins, FAP42 and FAP246, are localized to the L-shaped C1b projection of the CA, where they interact with the candidate CA protein FAP413. FAP42 is a large protein that forms the peripheral ‘beam’ of the C1b projection, and the FAP246–FAP413 subcomplex serves as the ‘bracket’ between the beam (FAP42) and the C1b ‘pillar’ that attaches the projection to the C1 microtubule. The FAP246–FAP413–FAP42 complex is essential for stable assembly of the C1b, C1f and C2b projections, and loss of these proteins leads to ciliary motility defects.
KEY WORDS: Central pair complex, Axoneme, Flagella, Cryo-electron tomography, Subtomogram averaging
Summary: We provide insight into the subunit organization and 3D structure of the C1b projection of the ciliary central apparatus and the mechanism by which it regulates dynein activity and ciliary beating.
INTRODUCTION
Cilia are highly conserved organelles in eukaryotes, where they play a variety of roles in cells, including motility, generating fluid flow and sensing extracellular signals. A wide range of human diseases, known as ciliopathies, are associated with cilia dysfunction, leading to symptoms including chronic respiratory infections, laterality abnormalities and infertility (Afzelius, 2004; Braun and Hildebrandt, 2017; Brown and Witman, 2014). The core component of the motile cilium is the ‘9+2’ axoneme, which consists of nine outer doublet microtubules (DMTs) and a central apparatus (CA) composed of two singlet microtubules and associated projections. Each DMT consists of many copies of 96-nm-long units that repeat along the axoneme; each repeat includes major substructures such as outer and inner dynein arms (ODAs and IDAs, respectively), radial spokes (RSs), and nexin–dynein regulatory complexes (N-DRCs) (Fig. 1A). Together with the CA, these substructures produce and regulate the cilium's motility (Dymek and Smith, 2007; Gui et al., 2019; Kikkawa, 2013; Lin and Nicastro, 2018; Loreng and Smith, 2017; Nicastro et al., 2006; Roberts et al., 2013; Viswanadha et al., 2017).
Fig. 1.
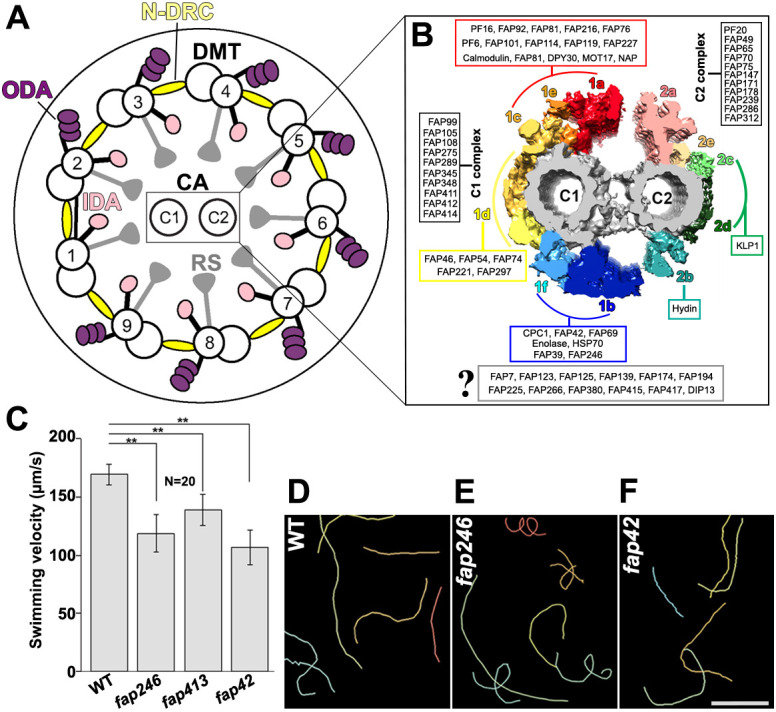
Known structure and proteome of the CA, and motility phenotypes of Chlamydomonas CA mutants. (A) Schematic of a Chlamydomonas flagellum in cross-sectional view, showing the DMTs with IDAs and ODAs, N-DRCs, and RSs that project toward the CA. The cross-section is viewed from the proximal end (flagellar base). Box indicates the CA region shown in B. (B) Isosurface rendering of the averaged Chlamydomonas CA in cross-sectional view (from the proximal end) and predicted locations of CA proteins based on previous proteomic studies (Zhao et al., 2019). Naming and coloring of CA projections are adapted from Carbajal-Gonzalez et al. (2013) and Fu et al. (2019) using our data. (C) Average swimming velocities of WT Chlamydomonas and the three CA mutants fap246, fap413 and fap42. Mean±s.d. of n=20 cells. **P<0.01 (two-tailed, paired Student's t-test). (D–F) Swimming trajectories of Chlamydomonas WT and CA mutants fap246 and fap42. Each colored line represents the swimming path of one cell recorded for 2 s. Scale bar: 100 µm.
The CA is the largest ciliary regulatory complex. The two singlet microtubules, termed C1 and C2, are both canonical 13-protofilament microtubules; however, they differ in stability and associated projections. Recent cryo-electron tomography (cryo-ET) studies of the Chlamydomonas reinhardtii axoneme have identified at least 11 protein projections with a total molecular mass of greater than 14 MDa that form 16- or 32-nm repeating units along the C1 and C2 microtubules (Carbajal-González et al., 2013). Among the 11 projections, six (C1a–C1f) are associated with the C1 microtubule and five (C2a–C2e) are associated with the C2 microtubule (Fig. 1B; Movie 1). The projections consist of many proteins that connect the C1 and C2 microtubules and/or extend toward the RS heads. Presumably the CA regulates ciliary motility through the CA–RS–N-DRC–IDA signaling pathway (Wirschell et al., 2009); however, the molecular details are still elusive. The CA is essential for normal ciliary motility from Chlamydomonas to mammals, and mutations affecting the CA often lead to impaired or paralyzed cilia (Dutcher et al., 1984; Smith and Lefebvre, 1996; Smith and Yang, 2004; Witman et al., 1978). In mice and humans, defects in CA proteins are associated with the ciliopathy primary ciliary dyskinesia (PCD) (Loreng and Smith, 2017; Poprzeczko et al., 2019; Teves et al., 2016). For instance, defects in the C1d proteins FAP221 (also known as PCDP1 or CFAP221) or FAP54 (also known as CFAP54) lead to typical PCD symptoms in mice, including accumulation of mucus in the lungs, male infertility and hydrocephalus (Lee et al., 2008; McKenzie et al., 2015). In humans, mutations in the CA protein PF20 (human SPAG16L) can cause male infertility, and recessive mutations in the C2b protein HYDIN cause PCD (Olbrich et al., 2012; Zhang et al., 2007). Therefore, it is important to establish a detailed understanding of CA structures and the mechanisms by which the CA regulates ciliary motility.
Despite the importance of the CA for ciliary motility, the specific locations of most of its proteins remain unknown, which makes the CA the least understood axonemal structure to date. Several recent proteomic and structural studies have shed light on the protein composition and 3D structure of the CA (Carbajal-González et al., 2013; Dai et al., 2020; Fu et al., 2019; Zhao et al., 2019). For example, proteomic comparisons of CAs from wild-type (WT) and CA-mutant Chlamydomonas have identified 37 and 44 new candidate CA proteins, respectively (Dai et al., 2020; Zhao et al., 2019), many of which have been assigned to the C1 or C2 microtubule and some to specific projections (Fig. 1B). Our previous cryo-ET study visualized the CA structure with up to 2.3 nm resolution and provided details about the molecular architecture of the PF16-dependent C1a–e–c supercomplex, which contains at least 16 proteins (Fu et al., 2019). However, due to the high connectivity within and between CA projections, the assignment of CA proteins to certain projections based on biochemical and mass spectrometry (MS) data can lead to ambiguous or even erroneous conclusions. For instance, the two CA proteins FAP76 and FAP81 were unambiguously shown to be components of the C1a–e–c supercomplex by cryo-ET studies (Fu et al., 2019), but were originally mistakenly assigned to the C1d projection in a preprint version of a proteomic study (Dai et al., 2019 preprint; see Dai et al., 2020 for final results). Given this caveat, it is important to complement MS studies with structural studies by, for example, using cryo-ET of mutants or tagged candidate proteins, to localize specific subunits within the CA.
The C1b projection is one of the largest and most complex CA projections. Pioneering studies on this projection in Chlamydomonas have shown that it physically interacts with the C2b projection of the C2 microtubule, and that the cpc1 mutant, which swims slowly due to reduced ciliary beat frequency, lacks the entire C1b projection, as assessed using conventional electron microscopy (EM), suggesting that the C1b projection has a role in the control of beat frequency (Mitchell and Sale, 1999). Biochemical analyses have found that the cpc1 axonemes lack three proteins that co-sediment as part of a larger complex, termed the CPC1 complex, which contains at least six proteins (Mitchell and Sale, 1999). Further analysis has identified five of these proteins as CPC1, FAP42, FAP69, enolase and HSP70A (Fig. 1B) (Mitchell et al., 2005). The first cryo-ET study of the CA (Carbajal-González et al., 2013) revealed that the C1b projection is more architecturally complex than previously known, and that its estimated mass is nearly 2 MDa – twice the sum of the masses of the known C1b subunits (883 kDa). Cryo-ET has also shown that C1b interacts with a previously undescribed projection, C1f, which is located between the C1b and C1d projections (Carbajal-González et al., 2013). In our previous analyses of the CA proteome, we identified two novel CA candidate proteins, FAP39 and FAP246, which were assigned to the C1 microtubule and co-immunoprecipitated with known C1b proteins, indicating that they likely are C1b or C1f proteins. We also found that axonemes of the mutant fap246-1 had greatly reduced amounts of both FAP246 and another candidate CA protein, FAP413, which was predicted to be a C1 protein, but could not be assigned to a specific projection (Zhao et al., 2019). However, none of these proteins have been localized precisely within the CA projections.
Here, we integrated genetic and quantitative proteomic approaches with cryo-ET and subtomogram averaging to compare CAs from WT Chlamydomonas and mutants for FAP42, FAP246 and FAP413. The mutants showed impaired motility compared to WT, indicating the importance of the mutated proteins for the regulation of ciliary beating. Our data allowed localization of FAP42, FAP246 and FAP413 within the C1b projection, as well as refined predictions for the locations of FAP39 and six additional CA proteins. 3D classification of the CA structures of the mutants revealed that the FAP246–FAP413–FAP42 subcomplex is critical for the stability of neighboring structures and may play a role in mechanosignaling from the CA to the RS and dyneins to regulate ciliary beating.
RESULTS
Chlamydomonas fap246, fap42 and fap413 mutants have motility defects
Chlamydomonas FAP246 (Cre14.g618750.t1.1) is a 120 kDa protein that is predicted to contain conserved leucine-rich repeat (LRR), papain-like and EF-hand domains (Fig. S1A,B). We first re-examined the phenotype of the fap246-1 (hereafter fap246) mutant that was partially characterized in our previous study (Zhao et al., 2019). Nearly all fap246 cells were motile, but they swam ∼30% slower than WT cells (Fig. 1C), a reduction in swimming velocity slightly greater than we previously reported (possibly reflecting culture in liquid TAP medium here versus culture in modified M medium previously). Compared to WT cells, many fap246 cells also had an altered swimming pattern in which cells frequently changed swimming direction, resulting in abnormally curved and spiraling swimming paths (Fig. 1D,E). This suggests that FAP246 plays a role in controlling flagellar motility. The reduction of the swimming velocity of fap246 cells was similar to that of the previously described cpc1 mutant, which lacks the entire C1b projection (Mitchell and Sale, 1999). The previously reported reduction of swimming velocity in the cpc1 mutant, to about two-thirds that of WT, has been shown to be due to reduced beat frequency (Mitchell and Sale, 1999). To determine whether this is also the case for fap246, we directly compared cpc1 and fap246 cells and found that their beat frequencies were reduced to 0.56 and 0.72 of that of WT, respectively (Fig. S2D). This reduction is proportional to the reduction in swimming speed of the two mutants and suggests that for both cpc1 and fap246 cells the reduced swimming velocity is mainly due to reduced beat frequency. In addition, cpc1 cells – like fap246 cells – had abnormally curved swimming paths with transient events of cell body waggle that occurred more frequently than in WT cells (data not shown). These waggles, in which the cell body rocked back and forth in place, likely were caused by momentary asynchrony of the two flagella (Wan et al., 2014). As with flagellar beat frequency, this phenotype was more severe in cpc1 than in fap246.
FAP42 (Cre12.g519950) is an ∼270 kDa protein that is predicted to contain a highly conserved papain-like domain and four guanylate kinase domains (Fig. S1C,D). We obtained a fap42 mutant (CLiP ID: LMJ.RY0402.205930) from the Chlamydomonas Library Project (CLiP; Li et al., 2016) and, using PCR, confirmed disruption of the FAP42 gene by the insertion of the cassette in a genetically homogenous clone (Fig. S2A). Compared to WT cells, fap42 cells swam ∼37% slower (Fig. 1C). Cells of fap42 swam slightly slower than fap246-mutant cells, but their swimming paths were less curved (Fig. 1F). The results suggest that FAP42 is also important for the regulation of flagellar motility.
As detailed in the following subsection, our MS analyses suggested that FAP246 and FAP42 are associated with the algae-specific FAP413 (Cre08.g364000.t1.1), which is a 215 kDa protein predicted to have a large WD40-repeat domain at the C terminus (Fig. S1E) (Zhao et al., 2020). We obtained a fap413 mutant (CLiP ID: LMJ.RY0402.087215) from CLiP (Li et al., 2016) and, using PCR, confirmed disruption of the FAP413 gene by the insertion of the cassette (Fig. S2B). The swimming velocity of fap413 mutant cells showed only ∼10% decrease compared to that of WT cells (Fig. 1C), which is the least disrupted motility phenotype of the mutants analyzed here.
Proteomic studies of fap246, fap42 and fap413 axonemes
We used quantitative MS to compare the axonemal proteome of WT cells with those of the fap246, fap42, and fap413 mutants. Label-free quantitation of proteins across samples was performed using SINQ normalized spectral index software (Trudgian et al., 2011) (Table 1; Table S1). The MS results confirmed that the protein level of FAP246 is greatly reduced in the fap246 axoneme compared to levels in WT cells (fap246/WT ratio=0.05) (Table 1). Only a few unique FAP246 N-terminal peptides were identified in the mutant, suggesting that a small amount of C-terminally truncated FAP246 is assembled into fap246 axonemes (Table S2). The results also confirmed the previous finding that levels of FAP413, a candidate C1 protein, are reduced in fap246 axonemes (fap246/WT ratio=0.01) (Table 1; Table S2) (Zhao et al., 2019). No other known or predicted CA proteins had similarly reduced levels in fap246 axonemes.
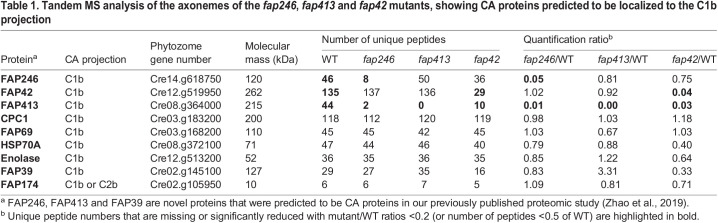
Table 1.
Tandem MS analysis of the axonemes of the fap246, fap413 and fap42 mutants, showing CA proteins predicted to be localized to the C1b projection
The MS analysis of fap413 axonemes showed that FAP413 was absent or greatly reduced compared to levels in WT cells (fap413/WT ratio=0) (Table 1). No other known or predicted CA protein, including FAP246 and FAP42, had greatly decreased levels in fap413 axonemes (Table 1; Table S1).
The MS analyses of fap42 axonemes revealed that the protein level of FAP42 was greatly reduced in the mutant axonemes compared to levels in WT cells (fap42/WT ratio=0.04) (Table 1). The few peptides detected in the mutant originated from throughout the FAP42 sequence (Table S3), suggesting that a small amount of FAP42 is assembled into the fap42 mutant axoneme. The only other protein that had greatly reduced levels in the fap42 axoneme was FAP413 (fap42/WT ratio=0.03) (Table 1; Table S3). The level of FAP246 in fap42 axonemes was mildly decreased (fap42/WT ratio=0.75) (Table 1).
Taken together, the MS results indicate that FAP413 is dependent on both FAP42 and FAP246 for its assembly into the axoneme, but not vice versa. FAP246 is moderately dependent on FAP42 for its stable assembly into the axoneme. The results strongly suggest that FAP413 interacts with FAP42 and FAP246, and that the three proteins are part of the same complex.
Several other proteins have been shown or predicted to be in the C1b projection, including CPC1, FAP39, FAP69, the chaperone HSP70A, the glycolytic enzyme enolase and, with less confidence, FAP174 and FAP380 (Fig. 1B) (Mitchell et al., 2005; Zhang and Mitchell, 2004; Zhao et al., 2019). Our MS analyses showed that the levels of these proteins were not greatly affected in fap246 or fap413 axonemes. In fap42 axonemes, the levels of FAP69 and CPC1 were also not affected, but the levels of HSP70A, enolase, FAP39 and FAP380 were reduced to 40%, 64%, 33% and 40% of the WT level, respectively (Table 1; Table S1). HSP70A and enolase are known to be located not only to the C1b projection but also elsewhere in the flagellum or axoneme (Mitchell et al., 2005; Zhao et al., 2019). It is not possible from these results alone to know whether the amounts of HSP70A and enolase remaining in fap42 axonemes represent only the non-C1b pool, or the non-C1b pool plus a fraction of the C1b pool. In either case, the amounts of these proteins lost from the C1b projection of fap42 axonemes are likely to be greater than the percentage lost from the entire sample as reported by MS. None of the known proteins in the C1a–e–c supercomplex or C1d projection were significantly affected in any of the three mutants (fap246, fap42 and fap413) (Table 1; Table S1).
FAP246 and FAP413 form a complex that is localized to the C1b projection
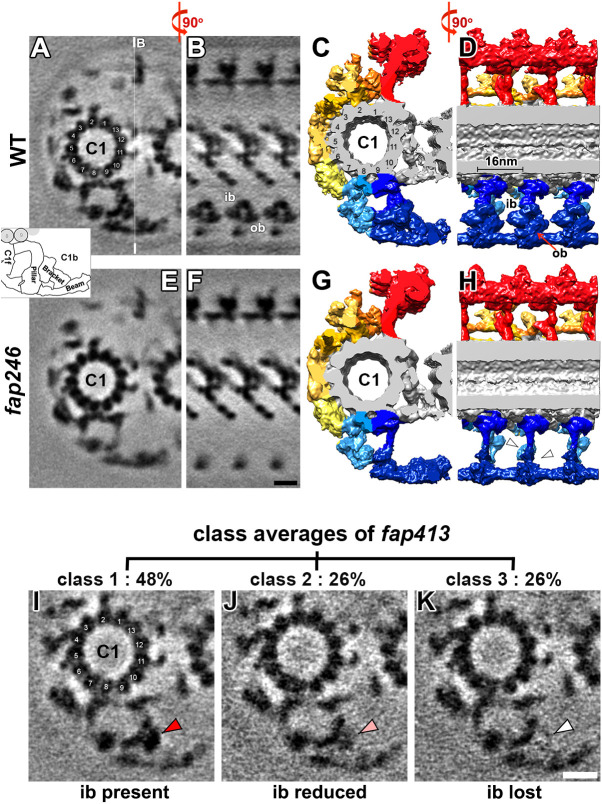
We next carried out cryo-ET and subtomogram averaging to investigate the CA structure of the fap246 mutant and compare it with that of the WT CA. Our subtomogram averages revealed the CA structures of WT and fap246 axonemes with 2.3 nm and 2.2 nm resolution, respectively [Fourier shell correlation (FSC) 0.5 criterion] (Fig. S2C), similar to what we reported previously (Fu et al., 2019). In WT axonemes, C1b is one of the largest CA projections (∼1.3 MDa). In cross-sectional view, C1b has a roughly triangular shape. We named the three sides of the triangle (1) the ‘pillar’, which projects 17 nm outwards from C1 protofilament 9; (2) the ‘beam’, a long component perpendicular to the pillar at the periphery of the projection; and (3) the ‘bracket’, which connects diagonally between pillar and beam (Fig. 2A–D and inset). Morphologically, the bracket density consists of two subareas: the slightly more distal ‘outer bracket’ spans the entire diagonal and connects the beam and pillar, whereas the more proximal ‘inner bracket’ density attaches to the outer bracket and forms a small connection to the neighboring C1b repeat (Fig. 2B,D). The beam structure extends longitudinally and forms a large connection with the beam structures of the neighboring repeats along the axoneme length. The tip of the beam also connects with the tip of the C2b projection (Movie 1). All C1b structures have a 16 nm periodicity and form multiple connections to neighboring CA structures.
Fig. 2.
Cryo-ET and subtomogram averaging of the fap246 CA reveals defects in the C1b projection, and 3D classification of the fap413 CA reveals partial loss of the bracket density. (A–H) Tomographic slices (A,B,E and F) and isosurface renderings (C,D,G and H; color-coded as in Fig. 1B) of the averaged CA repeats of WT (A–D) and fap246 (E–H) in cross-sectional (A,C,E and G; viewed from the proximal end) and longitudinal (B,D,F and H) views show that the C1b inner and outer bracket (ib and ob, respectively) densities are present in WT but missing in fap246 (white arrowheads). The thin white line in A indicates the position of the slices shown in B and F. In longitudinal slices, the proximal side is on the left. Inset, schematic of the C1b projection shows pillar, bracket and beam regions. Scale bar: 10 nm (shown in F, valid for A,B,E and F). (I–K) 3D classification focused on the bracket region of the fap413 CA average revealed three distinct classes (percentages of repeats are indicated for each class). Cross-sectional views (viewed from the proximal end) of the class averages show the presence (red arrowhead in I), reduction (pink arrowhead in J) and loss (white arrowhead in K) of the inner bracket density. Scale bar: 10 nm (shown in K, valid for I–K). In A,C,I, numbers indicate the protofilament numbers of C1.
In the fap246-mutant structure, the averaged CA repeat lacked the entire bracket density of the C1b projection, confirming that FAP246 is a C1b protein (Fig. 2E–H; Fig. S4C,D). The molecular mass of the FAP246-dependent bracket density was estimated to be ∼350 kDa based on the volume difference between the mutant and WT CA average (for details see Materials and Methods). This suggested that the bracket either contains three copies of FAP246 (120 kDa) or additional C1b subunit(s). The only protein besides FAP246 with significantly reduced levels in the MS analyses of fap246 axonemes was FAP413 (215 kDa). Previously, FAP413 has been predicted to be a C1 protein but could not be assigned to a specific projection (Zhao et al., 2019). Our molecular mass estimates for the densities missing in the fap246 CA are in good agreement with the combined predicted masses of FAP246 and FAP413, suggesting that they form the bracket subcomplex, with FAP246 being required for stable assembly of FAP413. The inner bracket is estimated to be slightly larger (∼180 kDa) than the outer bracket (∼150 kDa) density in the WT structure. However, without knowing the 3D structures of FAP413 and FAP246, we cannot distinguish whether the proteins are globular and arranged next to each other in the inner and outer bracket densities, respectively, or whether their arrangement is more complex, with both proteins being present in the inner and outer bracket structure.
Compared to the WT CA, the density of the pillar and beam appeared to be weakened and blurred in the fap246 CA average (compare Fig. S3E and F). This blurring suggests either partial reduction of the structures (i.e. not all repeats contain them) or positional flexibility. Therefore, we applied 3D classification focused on the blurred densities (Heumann et al., 2011). Classification of the WT CA structure in this region revealed one structurally homogenous class. In contrast, the C1b structure of fap246 was separated into four structurally different classes (Fig. S3G–J, Movie 2): in classes 1–3 (Fig. S3G–I) the pillar was present but differently tilted, and in 7% of the repeats the distal half of the pillar was missing (Fig. S3J, class 4). The 3D classification of the blurred beam density also revealed four classes with a comparable distribution as the pillar classes, which is consistent with the pillar and beam forming a unit that moves together. These results showed both positional flexibility and a small reduction of the pillar and beam densities in fap246, strongly suggesting that the FAP246–FAP413 bracket plays a role in stabilizing the neighboring C1b pillar and beam. In addition, 3D classification of neighboring CA projections in fap246 revealed an apparent reduction of the C2b projection in 25% of the repeats (Fig. S3K,L).
We next performed cryo-ET and subtomogram averaging of the fap413 mutant and visualized its CA with 2.5 nm resolution (FSC 0.5 criterion) (Fig. S2C). Compared to that of the WT structure, the density of the C1b outer bracket was noticeably weakened in fap413 (compare Fig. S3A,B with Fig. S3C,D). 3D classification focused on the outer bracket density revealed one structurally homogenous class in WT, but three classes in fap413 (Fig. 2I–K). Specifically, in 48% of the fap413 CA averages, the inner bracket density was similar to that of WT (class 1, Fig. 2I), in 26% the inner bracket density was weakly visible (class 2, Fig. 2J) and in the remaining 26% the inner bracket density was completely missing (class 3, Fig. 2K). We closely investigated other structures associated with the averaged CA and DMTs, but the only other axonemal structure partially reduced in the fap413 mutant was the C2b projection, which was lacking or blurred in 47% of the repeats (Fig. S3M,N). Based on the MS results, FAP413 assembly showed a strong dependence on the presence of FAP246, suggesting that FAP413 localizes to the bracket – possibly the inner bracket – of C1b. Possible reasons for why the inner bracket density showed only partial loss in the CA averages of fap413, which appears to lack FAP413 based on our MS results, include possible structural rearrangements or stabilization of otherwise flexible domains from neighboring proteins (e.g. FAP246), or functional redundancy of another protein with FAP413, which is a WD40-repeat protein (Fig. S1E). WD40 repeats are one of the most abundant domains in the eukaryotic genome, and the WD40-repeat protein family is very large (Jain and Pandey, 2018). There are many WD40-repeat domain proteins encoded in the Chlamydomonas genome and, because of the shared domain organization, some of these have high sequence similarity with FAP413. Redundancy is not unprecedented in the Chlamydomonas axoneme; for example, the WD40-rich protein FAP244 has been shown to substitute for another WD40-rich protein, FAP43, resulting in only partial loss of the I1 dynein tether and tether head complex in the corresponding mutants (Fu et al., 2018; Kubo et al., 2018).
FAP42 forms the beam of the C1b projection
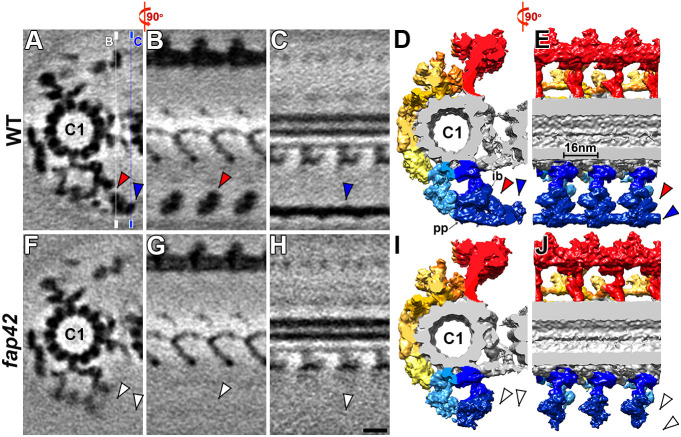
FAP42 was one of the five originally identified C1b proteins in the CPC1 complex (Mitchell et al., 2005), but so far it has not been localized within the C1b projection. Using cryo-ET and subtomogram averaging, we visualized the fap42 CA with 2.5 nm resolution (FSC 0.5 criterion) (Fig. S2C). Compared to the WT CA, the averaged fap42 CA repeats lacked the entire C1b beam and large parts of the outer bracket density that usually connects to the beam (Fig. 3; Fig. S4E,F). The mass of the entire FAP42-dependent density was estimated to be ∼400 kDa: ∼300 kDa for the beam and ∼100 kDa for the missing part of the outer bracket structure. This suggests that FAP42 (∼270 kDa protein) forms the beam structure and that stable assembly of the bracket depends on FAP42.
Fig. 3.
Cryo-ET and subtomogram averaging localizes FAP42 to the C1b projection. (A–J) Tomographic slices (A–C and F–H) and isosurface renderings (D,E,I and J; color-coded as in Fig. 1B) of the averaged CA repeats of WT (A–E) and fap42 (F–J) in cross-sectional (A,D,F and I; viewed from the proximal end) and longitudinal views (B,C,E,G,H and J) show that the beam and outer bracket are present in the WT CA repeats (blue and red arrowheads, respectively), but are missing in fap42 (white arrowheads). The thin lines in A indicate the position of the slices shown in B and G (white line) and in C and H (blue line). In longitudinal slices, the proximal side is on the left. ib, inner bracket; pp, peripheral part of the pillar. Scale bar: 10 nm (shown in H, valid for all EM images).
In the WT CA, the beam is highly connected to the bracket, the peripheral tip of the C1b pillar, the distal tips of neighboring beams and the tip of the C2b projection (Fig. 3A,D,E; Fig. S4A,B; Movie 1). Therefore, it is not surprising that several structures in the C1b, C1f and C2b projections that interact with the FAP42-dependent complex were weakened or blurred in the fap42 structure compared to the WT structure (Fig. 4A–H). We applied automatic 3D classification (Heumann et al., 2011) focusing on these weakened densities, revealing only one structurally homogenous class in the WT structures. In contrast, for fap42, two distinct classes were identified for the inner bracket and the peripheral part of the C1b pillar: in class 1 (54%) both bracket densities were present, as in WT (Fig. 4I–K), whereas in class 2 (46%) the entire bracket complex was missing (Fig. 4L–N). For the C1f projection, three classes were identified in fap42: in class 1 (50%) the C1f density connecting with the C1b pillar was missing (Fig. 4O,P); in classes 2 and 3 (26 and 24%, respectively) the C1f density was present but showed positional flexibility (Fig. 4Q–T; Movie 3). Similar to fap246 and fap413, the 3D classification of fap42 revealed that the density of the C2b projection was reduced in 56% of the CA repeats (Fig. S3O,P). In summary, the loss of FAP42 resulted in both reduced occupancy and/or positional flexibility of the C1b bracket and pillar, and of the neighboring C1f and C2b projections. Therefore, FAP42 is essential for the stable assembly and positioning of neighboring structures in the C1b, C1f and C2b projections.
Fig. 4.
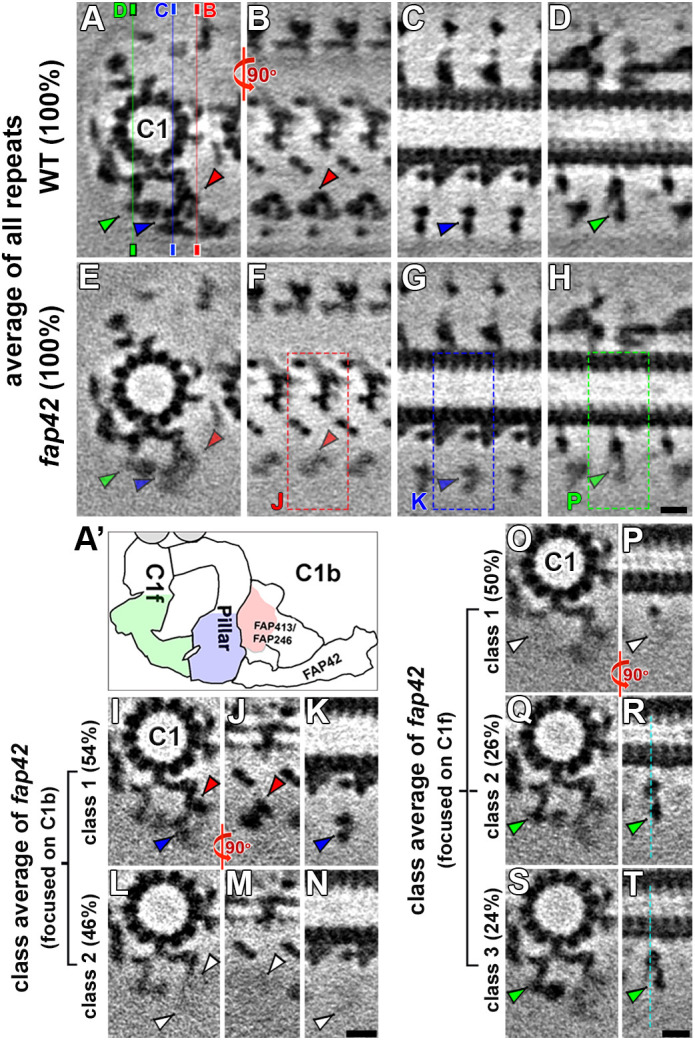
3D classification reveals structural heterogeneity for the C1b and C1f projections in fap42 but not in WT. (A–H) Tomographic slices of averaged CA repeats of WT (A–D) and fap42 (E–H) axonemes viewed in cross-sectional (A,E; viewed from the proximal end) and longitudinal orientations (B–D and F–H) show that, in addition to lacking the beam and outer bracket density, several C1b and C1f densities, including the inner bracket (red arrowheads), the peripheral part of the pillar (blue arrowheads) and the peripheral part of C1f (green arrowheads) are blurred in the averages of all CA repeats of fap42 axonemes compared to those of the WT. The thin lines in A indicate the position of the slices shown in B and F (red line), C and G (blue line), and D and H (green line). In longitudinal slices, the proximal side is on the left. Dashed boxes in F,G and H indicate regions shown as separate classes in J,M; K,N; and P,R,T, respectively. (A′) Schematic of the C1b (with pillar, FAP246–FAP413 bracket and FAP42 beam) and C1f projections. The red, blue and green colored regions indicate regions that are blurred in the average of all fap42 CA repeats, as shown in E–H. (I–T) 3D classification focused on the peripheral regions of C1b (I–N) and on C1f (O–T) reveals distinct classes (percentages of repeats are indicated for each class). Cross-sectional (I,L,O,Q and S; viewed from the proximal end) and longitudinal views (J,K,M,N,P,R and T). In longitudinal slices, the proximal side is on the left. The red, blue and green arrowheads indicate presence of densities, whereas the white arrowheads indicate missing densities. The turquoise dotted lines in R and T serve as references to show the positional differences of structures in different classes. Scale bars: 10 nm (in H, valid for A–H; in N, valid for I–N; in T, valid for O–T).
C1b pillar interacts with several structures in the C1b and C1f projections
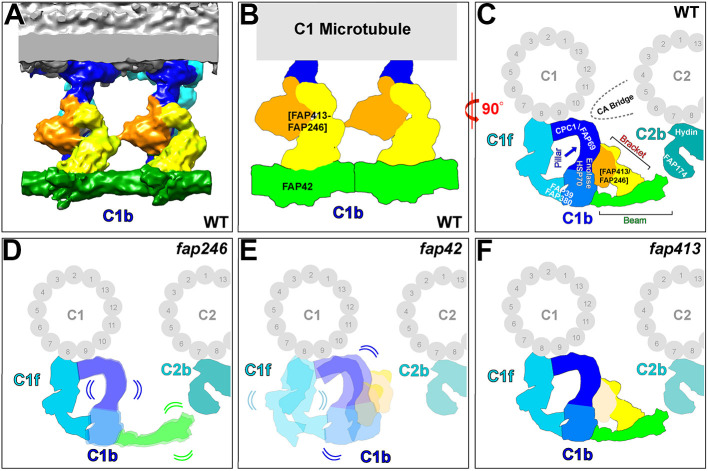
Our cryo-ET results showed that the C1b pillar is highly connected with neighboring CA structures through six interfaces (Fig. 2A,C; Fig. 5). Specifically, we observed two connections from the pillar within the C1b projection: to the FAP42 beam (Fig. 5A–C, green) and to the FAP246–FAP413 bracket (Fig. 5A–C, yellow and orange). Additional interfaces of the C1b pillar are the attachment site to protofilament 9 of the C1 microtubule, two connections with the C1f projection (Fig. 5C, cyan) and a connection to part of the CA bridge that links the C1 and C2 microtubules (Fig. 5C, gray).
Fig. 5.
Summary model depicting the structural organization of the C1b projection and showing the effects of partial loss of the FAP246–FAP413–FAP42 subcomplex on the CA structure. (A–C) The isosurface rendering (A) and schematic drawings (B,C) show part of the 3D structure of the averaged Chlamydomonas WT CA repeat viewed in longitudinal orientation (A,B; two 16 nm repeats are shown, with the proximal end to the left) and cross-sectional orientation (C; viewed from the proximal end). The views highlight the subunit organization of the C1b projection, including the FAP246–FAP413 bracket, with the longer ‘outer bracket’ (yellow) and the attached ‘inner bracket’ density (orange), and the FAP42 beam (green). In C, the pillar base and peripheral density are shown in dark and medium blue, respectively. The predicted locations of neighboring candidate proteins in the C1b, C1f and C2b projections are indicated in C, along with the C1 and C2 microtubules (numbers indicate protofilament numbers of C1 and C2). Dashed line in C indicates the position of the CA bridge. (D–F) Schematic drawings of the averaged 16 nm CA repeat of fap246 (D), fap42 (E) and fap413 (F) in cross-sectional view (viewed from the proximal end), showing the observed structural CA defects in the mutants compared to the WT CA. In fap246 the FAP246–FAP413 bracket is missing, and the pillar (base and peripheral density colored dark and medium blue, respectively) and the beam (green) densities show positional flexibility. In fap42 the beam (FAP42) and partial bracket (FAP413) are missing, and neighboring C1b and C1f densities show positional flexibility and partial reduction. In fap413 the bracket density (FAP413) is reduced. Compared to the WT structure, the C2b density (turquoise) in all three C1b mutants was reduced, suggesting partial protein loss.
The protein composition of the pillar is unknown and cannot be precisely dissected using the CA mutants studied here because the pillar structure remained mostly present in all three mutants. However, previous studies have predicted five additional C1b proteins, CPC1 (205 kDa), FAP69 (114 kDa), enolase (51 kDa), HSP70A (71 kDa) and FAP39 (127 kDa) (Mitchell et al., 2005; Zhao et al., 2019), which we did not find in the C1b beam and bracket structures. Therefore, the C1b pillar likely contains most or all of these proteins, which in sum would be ∼570 kDa, slightly larger than the estimated total molecular mass of ∼450 kDa for the pillar based on the cryo-ET averages.
DISCUSSION
Protein composition and structural organization of the C1b projection
The CA is highly conserved and plays essential roles in regulating ciliary beating, likely through the CA–RS–IDA–N-DRC–dynein signaling pathway (Wirschell et al., 2009). Single protein mutations often cause CA defects that result in impaired or paralyzed cilia and are associated with various ciliopathies (Loreng and Smith, 2017; Poprzeczko et al., 2019; Teves et al., 2016). Our previous CA study, which combined structural and biochemical data, revealed a 2 MDa C1a–e–c supercomplex (Fu et al., 2019). In the same study, we identified several key proteins in this supercomplex, including PF16, FAP76, FAP81, FAP92 and FAP216, and precisely localized them using cryo-ET (Fu et al., 2019). Our study also highlighted that structural data are often critical for guiding the interpretation of biochemical data. For example, FAP81 has been found to immunoprecipitate with C1a and C1e proteins, but could not be definitively assigned to either of these projections (Zhao et al., 2019); however, our previous cryo-ET study revealed that FAP81 localizes to the C1c projection within the interconnected C1a–e–c supercomplex (Fu et al., 2019). For most of the remaining CA projections, subunits have so far only been characterized biochemically, including for the C1d projection (FAP46, FAP54, FAP74, FAP221 and FAP297) and for the C2b, C2c and C2d projections (Brown et al., 2012; DiPetrillo and Smith, 2010, 2011; Lechtreck and Witman, 2007; Zhao et al., 2019).
The protein composition of the C1b projection has previously been predicted based on co-sedimentation in sucrose gradients, proteomics studies of CA mutants and co-immunoprecipitation assays. The C1b candidate proteins reported previously include CPC1, FAP39, FAP42, FAP69, FAP246, HSP70A, enolase, and – with less confidence – FAP174 and FAP380 (Dai et al., 2020; Mitchell et al., 2005; Zhang and Mitchell, 2004; Zhao et al., 2019). In the present study, we compared axonemes of WT cells and three mutants, fap42, fap246 and fap413, using a combination of MS and cryo-ET. We localized FAP42 and FAP246 within the C1b projection, and identified and localized a new C1b subunit, FAP413, which was previously predicted to be a C1 protein (Zhao et al., 2019). Finally, we described the structural organization of the subcomplex formed by these three subunits (Fig. 5; Movie 4).
The C1b projection can be divided into three major structural parts (Fig. 5; Movie 4): (1) FAP42 (Fig. 5, green) forms repeating beams at the interface between the CA and RS heads; (2) the pillar (Fig. 5, blue) attaches the C1b projection to protofilament 9 of the C1 microtubule and is a major interaction hub with five additional interfaces with neighboring CA structures, including C1f and the C1–C2 bridge; and (3) FAP246 and FAP413 (Fig. 5, yellow and orange) form a bracket between the pillar and beam. We named the FAP246–FAP413 complex the ‘bracket’ because the C1b projection architecture resembles a pillar–beam construction for roofs, and our 3D classification revealed that disruption of the bracket structure in fap246 and fap42 mutants resulted in structural destabilization (positional flexibility and reduction) of the remaining C1b and C1f structures. The C1b projection has a 16 nm periodicity along the C1 microtubule, whereas the C1f projection contains structural features that repeat with 32 nm periodicity, similar to the neighboring C1d projection (Movie 1) (Carbajal-González et al., 2013). The bracket structure is not present in the CA of sea urchin sperm flagella (Carbajal-González et al., 2013), consistent with previous phylogenetic analyses showing that FAP413 is limited to green algae and that FAP246 belongs to a sizable group of CA proteins that are members of large protein families or contain common conserved motifs, but may not have true orthologs in metazoans (Zhao et al., 2020). Surprisingly, the beam structure is present in the sea urchin (Carbajal-González et al., 2013), despite FAP42 likely not having a true ortholog in metazoans (Carbajal-González et al., 2013; Zhao et al., 2020). FAP42 is a member of a large protein family (transferases), and it is possible that another family member substitutes for it in metazoans.
A previous study has shown that FAP42 co-sediments with CPC1, FAP69, HSP70A and enolase (Mitchell et al., 2005), and that FAP39 co-immunoprecipitates with HA-tagged FAP246 (Zhao et al., 2019). Abundance of the five previously identified candidate C1b proteins was not greatly reduced in the axonemal proteomes of the three mutants of the C1b beam–bracket (FAP246–FAP413–FAP42) subcomplex studied here (Table 1), suggesting that they might form the C1b pillar, or part of the C1f or C2b projections. Previous classical EM studies have observed that the entire C1b projection is missing from Chlamydomonas cpc1 mutant flagella (Mitchell and Sale, 1999; Zhang and Mitchell, 2004). Therefore, CPC1 likely is located at the base of the C1b pillar and serves as a scaffold or docking protein that stabilizes the attachment of C1b to the C1 microtubule, possibly by interacting also on two sides with C1f and/or the C1–C2 bridge (Fig. 5). In contrast to CPC1 and FAP69, which were completely unaffected in our MS of fap42 and fap246 axonemes, the levels of enolase, HSP70A and FAP39 were reduced, with a reduction to 33–64% of the WT levels in fap42 axonemes (Table 1). Because enolase and HSP70A are known to locate not only to the C1b projection but also elsewhere in the axoneme (Mitchell et al., 2005; Zhao et al., 2019), our MS results likely underestimate the loss of these two proteins from the C1b projection. Especially in fap42, enolase and HSP70A could be greatly reduced if not entirely lost from the C1b complex. Overall, this suggests that enolase, HSP70A and FAP39 localize to the peripheral half of the C1b pillar (Fig. 5, medium blue) that connects to FAP42, FAP246 and FAP413, and that was missing in ∼46% of the fap42 repeats (Fig. 4I–N, class 2) and in 7% of the fap246 repeats (Fig. S3J, class 4). The estimated size of the pillar region by cryo-ET is ∼450 kDa, which is slightly smaller than the ∼570 kDa sum of the predicted pillar proteins CPC1 (205 kDa), FAP39 (127 kDa), FAP69 (114 kDa), HSP70 (71 kDa) and enolase (51 kDa). However, mass estimations based on cryo-ET averages could be underestimates, because if some of the complex subunits have intrinsically disordered domains or exhibit positional flexibility, these densities would not be well-represented in averages. Alternatively, some of these proteins could belong to the neighboring C1f projection (Fig. 5), which would be consistent with our finding that part of the C1f projection also showed missing density in 50% of the repeats of the fap42 CA (Fig. 4O,P), similar to the peripheral pillar density.
To date, no C1f proteins have been identified. Our present proteomic analyses of fap42 axonemes revealed that another candidate CA protein, FAP380 (20 kDa) (Zhao et al., 2019), was reduced to 40% of WT level (Table S1). Given the partial loss of the C1b pillar and C1f projection in the fap42 mutant, FAP380 may be a subunit of the C1b pillar or C1f projection. None of the known proteins of the C1a–e–c supercomplex or the C1d projection had substantially decreased levels in the C1b mutants studied here (Table 1).
The candidate CA protein FAP174 (10 kDa), which has previously been assigned to C2 (Rao et al., 2016), has also been observed to co-immunoprecipitate with FAP246–HA, and thus is suggested to be a C1b or C2b protein (Zhao et al., 2019). Levels of FAP174 were mildly reduced in our MS analyses of fap42 and fap413 axonemes, but were not reduced in fap246 axonemes. By cryo-ET, the C2b projection was reduced in all C1b mutants studied here, in particular in the fap42 mutant when the beam structure was missing (Fig. S3P, 56%), suggesting that FAP174 could localize to C2b. Based on our MS results, the proposed C2b protein hydin was still present in the mutant axonemes. Possible explanations could be that hydin – which contains multiple proposed microtubule-binding ASH domains – may detach from C2, but rebind elsewhere on the axoneme (Zhao et al., 2020), or it could become positionally flexible, blurring out in the cryo-ET averages.
C1b proteins contribute to the regulation of ciliary beating
FAP246 contains highly conserved LRR, papain-like and EF-hand domains (Fig. S1A). The mammalian homolog of FAP246, LRGUK (NP_653249) (Fig. S1B), has previously been observed in the mouse spermatid manchette, sperm basal body and flagellum, and seems to be required for sperm assembly (Liu et al., 2015). FAP42 contains four guanylate kinase domains, of which one is highly conserved (Fig. S1C) with similarity to guanylate kinase isoform b (GUK1; NP_000849.1) (Fig. S1D). The latter catalyzes the transfer of a phosphate group from ATP to GMP to form GDP and is a potential cancer chemotherapy target (Khan et al., 2019). Guanylate kinase activity – possibly provided by both FAP42 and FAP246 – has not been studied in cilia, but maintenance of flagellar GTP concentrations may be important for both guanylate cyclase activity and tubulin polymerization (Mitchell et al., 2005). CPC1 contains an adenylate kinase domain, indicating that it might be involved in maintaining stable intraciliary ATP levels during ciliary beating (Zhang and Mitchell, 2004). Another C1b protein, enolase, might also have a role in ATP production in the cilium. In fact, it was previously proposed that reduced flagellar enolase levels in the cpc1 mutant could cause a reduced ATP concentration in the ciliary matrix, resulting in reduced beat frequency (Mitchell et al., 2005). Although we cannot exclude that reduced enolase activity contributes to the motility defects in C1b mutants, it is likely not the sole factor, because the enolase level was substantially reduced in fap42, but only mildly reduced in fap246, yet both mutants showed similarly reduced swimming speeds.
All C1b mutants studied so far showed reduced swimming velocity, with the severity ranging from medium in cpc1, to mild in fap246 and fap42, and very mild in fap413. This is in contrast to mutations of some other CA proteins (such as PF16 and FAP216 from the C1a–e–c supercomplex, and FAP46 and FAP74 from the C1d projection) that cause severely impaired motility or paralyzed cilia (Brown et al., 2012; Dutcher et al., 1984; Fu et al., 2019; Smith and Lefebvre, 1996). This likely means that the C1b projection, including the FAP246–FAP413–FAP42 subcomplex, is not essential for transmission of the signal that regulates dynein activity and thus ciliary beating. However, swimming cpc1, fap246 and, to a lesser extent, fap42 cells exhibited an abnormally high frequency of cell body waggles. This is typically observed when the two Chlamydomonas cilia beat asynchronously, suggesting that the C1b projection has some role in coordinating and/or timing the oscillatory switching of dynein activity to maintain synchrony between the two beating cilia (Polin et al., 2009).
It likely is relevant that hydin knockdown strains, which are observed to lack the C2b projection when assessed by conventional EM, exhibit an unusual form of paralysis that strongly suggests a defect in switching dynein on and off at necessary times in the flagellar beat cycle. Biochemical studies have shown that hydin interacts with the CPC1 complex, and that the attachment of hydin to the C2 microtubule is destabilized in the cpc1 mutant (Brown et al., 2012; DiPetrillo and Smith, 2011; Lechtreck and Witman, 2007; Mitchell and Sale, 1999; Rupp et al., 2001; Yokoyama et al., 2004; Zhao et al., 2019). These previous findings are consistent with our current cryo-ET results that the C2b density is reduced in the C1b mutants analyzed here. Taken together, the results suggest that the C1b and C2b projections function in concert to regulate the timing of dynein activity.
The FAP246–FAP413–FAP42 subcomplex is part of a large interconnected CA network
A recurring feature of CA projections and subunits is their high connectivity within the projection and with neighboring CA structures, resulting in a massive CA network. For example, we previously showed that the C1a–e–c supercomplex forms multiple internal connections and at least three connections with the neighboring C1d projection (Fu et al., 2019). Here, we found that the C1b projection has four internal connections and seven connections with neighboring structures, including with C1f, the C1–C2 bridge and C2b (Fig. 5). Furthermore, 3D classification revealed that loss of any protein of the FAP246–FAP413–FAP42 subcomplex caused positional flexibility and/or partial reduction of the neighboring C1b pillar structure and/or the C1f and C2b projections. The C1f projection is not in direct contact with the FAP246–FAP413–FAP42 subcomplex, and thus is likely affected through its connections with the C1b pillar, suggesting a C1b–f supercomplex. Previous findings have shown that the C1f projection interacts with the C1d projection, because knockdown or mutation of the C1d proteins FAP74 and FAP46, respectively, led to loss of the C1d and C1f projections (Brown et al., 2012). The C1d projection itself interacts with the C1c protein FAP76 from the C1a–e–c supercomplex (Fu et al., 2019). In addition, in our cryo-ET averages of the WT CA, the tip of the C1b beam (FAP42) appears to connect with corresponding structures of the C2b projection, and in all the C1b mutants studied here, the density of C2b was partially reduced. Previous studies have suggested that the C1b and C2b projections physically interact; for example, the C1b and C1f projections (identifiable as the ‘sheath’ when observed using conventional EM) remain attached to the C2 microtubule when the C1 microtubule is lost from axonemes of the pf16 mutant, and isolated cpc1 axonemes lacking the C1b projection also frequently lack the C2b projection (Mitchell and Sale, 1999). Conversely, biochemical studies have shown that the C2b protein hydin remains associated with the C1 microtubule when the C2 microtubule is solubilized by high-salt extraction from WT axonemes (Lechtreck and Witman, 2007). Taking all these data together, a picture of a highly interconnected network emerges that includes at least the C1a,e,c,d,f,b and C2b projections. This also explains why biochemical analyses (such as MS and co-immunoprecipitation) alone may not be sufficient to assign subunits to specific CA projections.
The FAP246–FAP413–FAP42 subcomplex provides mechanical support
In straight flagella, the nine DMTs are cylindrically arranged, and the RSs, which project towards the center of the axoneme, end with their heads on a virtual cylinder with ∼40-nm radius. The two parallel CA microtubules themselves cannot interact with all RS heads at the same time, but the different CA projections, which vary considerably in length, angle and shape, form a dense cylindrical CA network with ∼40-nm radius that interacts with the RSs from all nine DMTs (Nicastro et al., 2005). At the current cryo-ET resolution, there appears to be an ∼5-nm gap between the periphery of the CA projections and the RS heads in straight (inactive) axonemes (Oda et al., 2014). In both forward and backward swimming Chlamydomonas, the diameter of the axoneme is greater in the plane of bending of bent regions than in straight regions of the flagellum, presumably due to transverse stress acting across the axoneme during bending (Fig. S5) (Lindemann and Mitchell, 2007). In the bent regions, the two CA microtubules, which in Chlamydomonas rotate during flagellar beating, are both in the plane of bending, with the C1 microtubule (predominantly) facing DMT-1, at least in principal bends (Mitchell, 2003). The lateral compression of the axoneme in bent regions implies that the RSs mechanically push against the CA projections in a direction perpendicular to the bending plane. Therefore, the C1a–C2a and C1b–C2b projections would be subject to the greatest compression force by the RSs in the bent region (Fig. S5).
Recent cryo-EM studies have shown that the surfaces of the RS heads facing the CA are negatively charged; therefore, if CA projections are also negatively charged, this could prevent sticking of the projections to the RS heads during CA rotation (Grossman-Haham et al., 2021). FAP42, which comprises the beam and is the part of C1b that would directly interact with the RS heads, is, in fact, a moderately acidic protein (pI=6.1). It will be very interesting to determine the distribution of FAP42 acidic (negatively charged) residues relative to the architecture of the beam.
Unlike, for example, the C1c and C1d projections, which are short and closely associated with the C1 microtubule, the C1b beam, which would be closest to the RS heads, is ∼20 nm away from the C1–C2 bridge without being directly supported by the C1 microtubule. However, the structure of C1b appears to be ideally suited to resist compressive force: the shape of the projection resembles that of an architectural pillar–beam construction that transmits load imposed on a horizontal beam (e.g. by a roof) to a vertical pillar and floor that can resist the compression force. In case of the C1b projection, the compression force imposed by the RSs onto the FAP42 beam could be transmitted through the connection with the pillar and the FAP246–FAP413 bracket onto the pillar and ultimately the C1 microtubule – both of which should provide more resistance to inwardly directed compressive force. In addition, neighboring beams are connected to each other (Fig. 5A,B; Movie 4), which could also distribute force across a sheet-like beam network. Defects in the FAP246–FAP413–FAP42 subcomplex resulted in destabilization of the remaining C1b structures, and of the C1f and C2b projections, even in isolated inactive axonemes (i.e. without compression force imposed by bending), suggesting that the FAP246–FAP413–FAP42 subcomplex does indeed play a role in providing mechanical strength and support. The motility defects seen in cpc1, fap42 and fap246 mutants may, in part, reflect an abnormal but transient compression-induced partial collapse of the C1f–C1b–C2b network in bent regions of the axoneme during each beat cycle.
Defects in C1b also may affect the regulation of dynein arm activity. Previous studies have shown that mechanical interactions between the CA and RS heads are critical for ciliary motility. For example, Oda and colleagues have shown that in the paralyzed Chlamydomonas mutant pf6, which lacks most of the C1a–e complex, extending the RS heads by addition of a non-ciliary protein of suitable size (e.g. a BCCP tag) is sufficient to restore flagellar motility (Oda et al., 2014). Oda and colleagues have proposed that the C1a projection interacts mechanically and non-specifically with the RSs of DMTs 2, 3 and 4 to transmit signals to the dynein arms. In the fap42 and fap246 mutants, loss of the C1b bracket and beam structures could allow the entire CA to move away from the RSs of DMTs 2, 3, and 4 in bent regions, partially impairing CA–RS mechanosignaling required to activate or inactivate the dynein arms in those regions. The positions of specific projections relative to the DMTs undergoing active sliding in bent regions could explain why loss of different CA projections has different effects on flagellar motility (Brown et al., 2012; DiPetrillo and Smith, 2011; Hou et al., 2021; Lechtreck and Witman, 2007; Mitchell and Sale, 1999; Rupp et al., 2001; Yokoyama et al., 2004; Zhao et al., 2019). Thus, the C1b projection might not only support structural integrity and stability of the CA and axoneme, but the mechanical resistance that it provides might also be necessary for proper mechanosignaling between the CA and RSs to regulate dynein activity and thus ciliary beating.
MATERIALS AND METHODS
Bioinformatics
Domain predictions of FAP246, FAP42 and FAP413 were performed using PROSITE webserver (http://prosite.expasy.org/) (Sigrist et al., 2013). Sequence alignments and BLAST analyses were performed using the BLAST tool from NCBI (Altschul et al., 1990).
Strains and culture conditions
Chlamydomonas reinhardtii WT strains used were g1 (nit1, agg1, mt+; CC-5415, Chlamydomonas Resource Center; https://www.chlamycollection.org) and A54-e18 (CC-2929). A fap246 mutant was originally obtained from the CLiP collection (CLiP ID: LMJ.RY0402.135524) via the Chlamydomonas Resource Center and then crossed to g1; a progeny (135524-8A) was named fap246-1 and used here, as in our previous study (Zhao et al., 2019). Another CLiP mutant (LMJ.RY0402.2059300) with an insertion in exon 7 of the fap42 gene (Li et al., 2016) was also obtained from the Chlamydomonas Resource Center; analysis by PCR (Fig. S2) revealed that the cells originally supplied were genetically heterogeneous, so they were cloned, and one clone harboring the insertion was named fap42-1 (referred to here as fap42) and used in subsequent experiments. The fap413 mutant was obtained from the CLiP collection (LMJ.RY0402.087215); loss of FAP413 was confirmed by PCR analysis (Fig. S2).
As previously described (Fu et al., 2018), Chlamydomonas cells were maintained in solid Tris-acetate-phosphate (TAP) plates (supplied with 7.5 μg/ml paromomycin for CLiP mutants) and cultured in liquid TAP medium or modified M medium (Witman, 1986) under a 12:12 h light:dark cycle at 23°C with filtered air bubbling into the growth culture. Insertion sites of CLiP mutant fap42 were confirmed by PCR using the primers listed in Table S4.
Axoneme preparation
Axonemes of Chlamydomonas cells were purified by the pH-shock method as previously described (Song et al., 2015; Witman, 1986). Briefly, cells were cultured in liquid TAP medium, harvested by centrifugation (1100 g for 5 min) and washed twice with fresh M-N/5 minimal medium (Iomini et al., 2009). The cell pellet was resuspended in pH-shock buffer (10 mM HEPES, 1 mM SrCl2, 4% sucrose and 1 mM DTT, pH 7.4), and 0.5 M acetic acid was added to the buffer to adjust the pH to 4.3. After 80 s, 1 M KOH was added to increase the pH to 7.2. The pH shock treatment was performed on ice. After pH shock, 5 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA and 100 μl protease inhibitor PMSF (Sigma-Aldrich) were added to the solution. The solution was centrifuged (1800 g for 10 min, 4°C) to separate the detached flagella from the cell bodies. To further purify the flagella, the flagella-containing supernatant was centrifuged twice with a 20% sucrose cushion (2400 g for 10 min, 4°C). After centrifugation, 1% IGEPAL CA-630 (Sigma-Aldrich) was added to the supernatant for 20 min at 4°C to demembranate the flagella. Axonemes were collected by centrifugation (10,000 g for 10 min, 4°C), and the freshly isolated axonemes were resuspended in HMEEK buffer (30 mM HEPES, 25 mM KCl, 5 mM MgSO4, 0.1 mM EDTA and 1 mM EGTA, pH 7.2). Axonemal samples were either plunge frozen for cryo-ET or stored at −80°C for biochemical assays or MS.
Analysis of motility phenotypes
All observations and recordings were performed on cells grown in TAP or modified M medium at room temperature. To analyze swimming speed, 100 μl of Chlamydomonas cell culture in TAP medium was transferred to a plastic chamber (0.127-mm-deep Fisherbrand UriSystem DeciSlide; Thermo Fisher Scientific). Cells were imaged using a Nikon ECLIPSE LV-N microscope equipped with an Andor Zyla sCMOS Camera. The objective lens used was a Nikon CFI S Plan Fluor ELWD 20× NA 0.45 with a working distance of 8.2–6.9 mm. Movies were recorded at 100 frames/s using NIS-Elements AR (Nikon) software. Swimming speeds were determined using the measurement tools in NIS-Elements AR software. The TrackMate plugin (Tinevez et al., 2017) in Fiji ImageJ was used to trace the swimming trajectory of the cells.
For the flagella beat frequency analyses, cells were grown in 120 ml of modified M medium aerated with 5% CO2 under a 14:10 h light:dark cycle at 23°C. 180–200 µl aliquots of mid-log phase cells were transferred to an ∼0.6-mm-deep chamber made from two 18×18 mm glass coverslips. Movies were recorded at 478 frames/s using a Nikon DIAPHOT 200 inverted microscope with a Nikon Fluor 10× Ph2 DL objective (NA 0.5) and a ZWO AS1174MM-Cool camera with SharpCap software (Version 3.1.5220.0). Beat frequency was calculated by counting the number of ‘back and forth’ cell body movements (Kamiya, 2000) in 400 frames when the movies were played at 15–20 frames/s, and then dividing that number by the duration of the movie segment analyzed. At least 50 cells were analyzed for each strain.
Liquid chromatography–mass spectrometry
Axonemal proteins (40 μg) of the WT, fap42 and fap246 strains were separated on a 4–12% gradient SDS–polyacrylamide gel (Genscript Biotech). After the dye front migrated on the gel for 3.0–3.5 cm, the gel was stained with Coomassie Brilliant Blue for 30 min and destained until the background was clear. Each gel lane was cut into four slices, and each slice was further excised into pieces of 1 mm3. In-gel trypsin digestion, liquid chromatography–mass spectrometry (LC-MS) and peptide identification were conducted by the proteomics core facility at the University of Texas Southwestern Medical Center. Briefly, protein gel pieces were digested overnight using trypsin (Pierce) following reduction and alkylation with DTT and iodoacetamide (Sigma-Aldrich). The samples then underwent solid-phase extraction cleanup with an Oasis HLB µElution plate (Waters), and the resulting samples were analyzed by LC-MS, using an Orbitrap Fusion Lumos mass spectrometer (Thermo Electron) coupled to an Ultimate 3000 RSLC-Nano liquid chromatography system (Dionex). Samples were injected onto a 75 μm internal diameter, 75 cm long EasySpray column (Thermo Electron) and were eluted with a gradient from 0–28% buffer B over 90 min. Buffer A contained 2% (v/v) acetonitrile and 0.1% formic acid in water, and buffer B contained 80% (v/v) acetonitrile, 10% (v/v) trifluoroethanol and 0.1% formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 1.8 kV and an ion transfer tube temperature of 275°C. MS scans were acquired at 120,000 resolution in the Orbitrap and up to 10 MS spectra were obtained in the ion trap for each full spectrum acquired using higher-energy collisional dissociation (HCD) for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion was selected for fragmentation. Raw MS data files were converted to a peak list format and analyzed using the central proteomics facilities pipeline (CPFP), version 2.0.3 (Trudgian and Mirzaei, 2012; Trudgian et al., 2010). Peptide identification was performed using the X!Tandem (Craig and Beavis, 2004) and Open MS Search Algorithm (OMSSA) (Geer et al., 2004) search engines against the Chlamydomonas reinhardtii protein database, with common contaminants and reversed decoy sequences appended (Elias and Gygi, 2007). Another round of peptide identification search was conducted using a more updated Chlamydomonas reinhardtii protein database generated from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). Fragment and precursor tolerances of 10 ppm and 0.5 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification and oxidation of Met was set as a variable modification. Label-free quantitation of proteins across samples was performed using SINQ normalized spectral index software (Trudgian et al., 2011).
Cryo-ET
Freshly prepared axonemal samples (30 μl) were gently mixed with 10 μl of 10-fold-concentrated, BSA-coated 10-nm gold solution (Sigma-Aldrich). A 4 μl volume of the solution was applied to a glow-discharged (30 s at 35 mA) copper R2/2 holey carbon grid (Quantifoil Micro Tools). After removing excess liquid by blotting the grid from the back side with Whatman filter paper for 2 s, the grid was plunge frozen into liquid ethane using a homemade plunge freezer. Grids were then stored in liquid nitrogen until use.
Grids were loaded into a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV. The microscope control software SerialEM (Mastronarde, 2005) was used to acquire tilt series images in low-dose mode from −60° to 60° with 2° increments using a dose-symmetric tilting scheme (Hagen et al., 2017). For WT axonemes, the images were recorded with a 4000×4000 K2 DDD camera (Gatan) in counting mode (15 frames, 0.4 s exposure time per frame, dose rate of 8 electrons/pixel/s for each tilt image). For fap246, fap42 and fap413 axonemes, the images were recorded with a 5760×4092 K3 DDD camera (Gatan) in counting mode (10 frames, 0.05 s exposure time per frame, dose rate of 26 electrons/pixel/s for each tilt image). For all strains, the post-column energy filter (Gatan) was operated in zero-loss mode (20 eV slit width), and a VPP (Danev et al., 2014) was used with −0.5 μm defocus. The magnification was set to 26,000, with an effective pixel size of 3.2 Å (K3 camera) or 5.5 Å (K2 camera). The total electron dose per tilt series was limited to ∼100 e/Å2.
Image processing
In brief, motion correction of the frames was performed using MotionCorr2 (Zheng et al., 2017). IMOD software (Kremer et al., 1996) was used to align the tilt serial images using the 10 nm gold particles as fiducial markers and to reconstruct the tomograms by the weighted back-projection (WBP) approach. For subtomogram averaging, either CA or DMT repeats were picked from the raw tomograms, and alignment and missing-wedge compensated averaging were performed using PEET software (Nicastro et al., 2006), which is integrated in the IMOD software package. The axoneme and CA orientation (proximal to distal) was determined for each tomogram based on both DMT orientation in the axoneme cross-section and initial averages that included only the CA repeats within each tomogram. After the CA polarity and the same center of the repeat were determined for each tomogram, a second alignment was performed combining the CA repeats from all tomograms in the correct orientation and periodicity register. After global alignment of all CA repeats, the alignment of each individual CA microtubule was refined by local alignment of each microtubule and its associated projections separately, while the other microtubule was masked as described previously (Carbajal-González et al., 2013; Fu et al., 2019). Visualization of the 3D structures of the averaged CA repeats was done with the UCSF Chimera package software (Pettersen et al., 2004). For generating isosurface renderings, the same isosurface threshold was applied to the WT and mutant averages. Mass estimations of the protein complexes and subvolumes were calculated using the average density of 1.43 g/cm3 for proteins (Quillin and Matthews, 2000) and after normalizing the isosurface-rendering threshold to the mass of microtubules in Chimera. Classification analyses used a principal component analysis (PCA) clustering method incorporated in the PEET software (Heumann et al., 2011). The number of tomograms and CA repeats, as well as the estimated resolution of the averages (using FSC 0.5 criterion), are summarized for each strain in Table S5.
Supplementary Material
Acknowledgements
We thank Dr Andrew Lemoff and the proteomics core facility at the University of Texas Southwestern Medical Center (UTSW) for the liquid chromatography-MS analyses. We thank Dr Christina Baer and the Sanderson Center for Optical Experimentation at the University of Massachusetts Medical School for help with the flagellar beat frequency analysis. We are grateful to Dr Daniel Stoddard for management of the UTSW cryo-electron microscope facility, which is funded in part by a Cancer Prevention and Research Institute of Texas Core Facility Award (RP170644). This research was supported in part by the computational resources provided by the BioHPC supercomputing facility located in the Lyda Hill Department of Bioinformatics, UT Southwestern Medical Center.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.N.; Methodology: K.C., Y.Z., L.Z., N.P., D.N.; Validation: K.C., G.B.W., D.N.; Formal analysis: K.C., Y.Z., L.Z., Y.H., X.C., G.B.W., D.N.; Investigation: K.C., Y.Z., L.Z., N.P., Y.H., X.C.; Data curation: K.C.; Writing - original draft: K.C.; Writing - review & editing: K.C., G.B.W., D.N.; Visualization: K.C., Y.Z.; Supervision: D.N.; Funding acquisition: G.W., D.N.
Funding
This study was supported by National Institutes of Health grants R01 GM083122 to D.N. and R35 GM122574 to G.B.W., by a Cancer Prevention and Research Institute of Texas grant RR140082 to D.N., and by the R.W.B. Endowment at the University of Massachusetts Medical School to G.B.W. Deposited in PMC for release after 12 months.
Data availability
The averaged 3D structures of the CA focused on the C1b projection from different strains have been deposited in the Electron Microscopy Data Bank under accession codes EMD-22648 (WT), EMD-22649 (fap246), EMD-22651 (fap42) and EMD-24489 (class average 3 of fap413).
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.254227.
References
- Afzelius, B. A. (2004). Cilia-related diseases. J. Pathol. 204, 470-477. 10.1002/path.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403-410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Braun, D. A. and Hildebrandt, F. (2017). Ciliopathies. Cold Spring Harb. Perspect. Biol. 9, a028191. 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. M. and Witman, G. B. (2014). Cilia and Diseases. Bioscience 64, 1126-1137. 10.1093/biosci/biu174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J. M., Dipetrillo, C. G., Smith, E. F. and Witman, G. B. (2012). A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell Sci. 125, 3904-3913. 10.1242/jcs.107151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajal-González, B. I., Heuser, T., Fu, X., Lin, J., Smith, B. W., Mitchell, D. R. and Nicastro, D. (2013). Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken) 70, 101-120. 10.1002/cm.21094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, R. and Beavis, R. C. (2004). TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466-1467. 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- Dai, D., Ichikawa, M., Peri, K., Rebinsky, R. and Bui, K. (2019). Identification and mapping of central pair proteins by proteomic analysis (version 1). bioRxiv, 739383. 10.1101/739383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, D., Ichikawa, M., Peri, K., Rebinsky, R. and Bui, K. (2020). Identification and mapping of central pair proteins by proteomic analysis. Biophys. Physicobiol. 17, 71-85. 10.2142/biophysico.BSJ-2019048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. and Baumeister, W. (2014). Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA 111, 15635-15640. 10.1073/pnas.1418377111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo, C. G. and Smith, E. F. (2010). Pcdp1 is a central apparatus protein that binds Ca(2+)-calmodulin and regulates ciliary motility. J. Cell Biol. 189, 601-612. 10.1083/jcb.200912009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo, C. G. and Smith, E. F. (2011). The Pcdp1 complex coordinates the activity of dynein isoforms to produce wild-type ciliary motility. Mol. Biol. Cell 22, 4527-4538. 10.1091/mbc.e11-08-0739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S. K., Huang, B. and Luck, D. J. (1984). Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 98, 229-236. 10.1083/jcb.98.1.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek, E. E. and Smith, E. F. (2007). A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J. Cell Biol. 179, 515-526. 10.1083/jcb.200703107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias, J. E. and Gygi, S. P. (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207-214. 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- Fu, G., Wang, Q., Phan, N., Urbanska, P., Joachimiak, E., Lin, J., Wloga, D. and Nicastro, D. (2018). The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol. Biol. Cell 29, 1048-1059. 10.1091/mbc.E18-02-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G., Zhao, L., Dymek, E., Hou, Y., Song, K., Phan, N., Shang, Z., Smith, E. F., Witman, G. B. and Nicastro, D. (2019). Structural organization of the C1a-e-c supercomplex within the ciliary central apparatus. J. Cell Biol. 218, 4236-4251. 10.1083/jcb.201906006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer, L. Y., Markey, S. P., Kowalak, J. A., Wagner, L., Xu, M., Maynard, D. M., Yang, X., Shi, W. and Bryant, S. H. (2004). Open mass spectrometry search algorithm. J. Proteome Res. 3, 958-964. 10.1021/pr0499491 [DOI] [PubMed] [Google Scholar]
- Grossman-Haham, I., Coudray, N., Yu, Z., Wang, F., Zhang, N., Bhabha, G. and Vale, R. D. (2021). Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating. Nat. Struct. Mol. Biol. 28, 20-28. 10.1038/s41594-020-00519-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, L., Song, K., Tritschler, D., Bower, R., Si, Y., Dai, A., Augspurger, K., Sakizadeh, J., Grzemska, M., Ni, T.et al. (2019). Scaffold subunits support associated subunit assembly in the Chlamydomonas ciliary nexin-dynein regulatory complex. Proc. Natl. Acad. Sci. USA 116, 23152-23162. 10.1073/pnas.1910960116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, W. J. H., Wan, W. and Briggs, J. A. G. (2017). Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191-198. 10.1016/j.jsb.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann, J. M., Hoenger, A. and Mastronarde, D. N. (2011). Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 175, 288-299. 10.1016/j.jsb.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Zhao, L., Kubo, T., Cheng, X., McNeill, N., Oda, T. and Witman, G. B. (2021). Chlamydomonas FAP70 is a component of the previously uncharacterized ciliary central apparatus projection C2a. J. Cell Sci. 134, jcs258540. 10.1242/jcs.258540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini, C., Till, J. E. and Dutcher, S. K. (2009). Genetic and phenotypic analysis of flagellar assembly mutants in Chlamydomonas reinhardtii. Methods Cell Biol. 93, 121-143. 10.1016/S0091-679X(08)93007-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, B. P. and Pandey, S. (2018). WD40 repeat proteins: signalling scaffold with diverse functions. Protein J. 37, 391-406. 10.1007/s10930-018-9785-7 [DOI] [PubMed] [Google Scholar]
- Kamiya, R. (2000). Analysis of cell vibration for assessing axonemal motility in Chlamydomonas. Methods 22, 383-387. 10.1006/meth.2000.1090 [DOI] [PubMed] [Google Scholar]
- Khan, N., Shah, P. P., Ban, D., Trigo-Mouriño, P., Carneiro, M. G., DeLeeuw, L., Dean, W. L., Trent, J. O., Beverly, L. J., Konrad, M.et al. (2019). Solution structure and functional investigation of human guanylate kinase reveals allosteric networking and a crucial role for the enzyme in cancer. J. Biol. Chem. 294, 11920-11933. 10.1074/jbc.RA119.009251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa, M. (2013). Big steps toward understanding dynein. J. Cell Biol. 202, 15-23. 10.1083/jcb.201304099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J. R., Mastronarde, D. N. and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kubo, T., Hou, Y., Cochran, D. A., Witman, G. B. and Oda, T. (2018). A microtubule-dynein tethering complex regulates the axonemal inner dynein f (I1). Mol. Biol. Cell 29, 1060-1074. 10.1091/mbc.E17-11-0689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.-F. and Witman, G. B. (2007). Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176, 473-482. 10.1083/jcb.200611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L., Campagna, D. R., Pinkus, J. L., Mulhern, H., Wyatt, T. A., Sisson, J. H., Pavlik, J. A., Pinkus, G. S. and Fleming, M. D. (2008). Primary ciliary dyskinesia in mice lacking the novel ciliary protein Pcdp1. Mol. Cell. Biol. 28, 949-957. 10.1128/MCB.00354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhang, R., Patena, W., Gang, S. S., Blum, S. R., Ivanova, N., Yue, R., Robertson, J. M., Lefebvre, P. A., Fitz-Gibbon, S. T.et al. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367-387. 10.1105/tpc.15.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. and Nicastro, D. (2018). Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science 360, eaar1968. 10.1126/science.aar1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann, C. B. and Mitchell, D. R. (2007). Evidence for axonemal distortion during the flagellar beat of Chlamydomonas. Cell Motil. Cytoskeleton 64, 580-589. 10.1002/cm.20205 [DOI] [PubMed] [Google Scholar]
- Liu, Y., DeBoer, K., de Kretser, D. M., O'Donnell, L., O'Connor, A. E., Merriner, D. J., Okuda, H., Whittle, B., Jans, D. A., Efthymiadis, A.et al. (2015). LRGUK-1 is required for basal body and manchette function during spermatogenesis and male fertility. PLoS Genet. 11, e1005090. 10.1371/journal.pgen.1005090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreng, T. D. and Smith, E. F. (2017). The central apparatus of cilia and eukaryotic flagella. Cold Spring Harb. Perspect. Biol. 9, a028118. 10.1101/cshperspect.a028118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D. N. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36-51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- McKenzie, C. W., Craige, B., Kroeger, T. V., Finn, R., Wyatt, T. A., Sisson, J. H., Pavlik, J. A., Strittmatter, L., Hendricks, G. M., Witman, G. B.et al. (2015). CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Mol. Biol. Cell 26, 3140-3149. 10.1091/mbc.e15-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D. R. (2003). Reconstruction of the projection periodicity and surface architecture of the flagellar central pair complex. Cell Motil. Cytoskeleton 55, 188-199. 10.1002/cm.10121 [DOI] [PubMed] [Google Scholar]
- Mitchell, D. R. and Sale, W. S. (1999). Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144, 293-304. 10.1083/jcb.144.2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, B. F., Pedersen, L. B., Feely, M., Rosenbaum, J. L. and Mitchell, D. R. (2005). ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell 16, 4509-4518. 10.1091/mbc.e05-04-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro, D., McIntosh, J. R. and Baumeister, W. (2005). 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. USA 102, 15889-15894. 10.1073/pnas.0508274102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro, D., Schwartz, C., Pierson, J., Gaudette, R., Porter, M. E. and McIntosh, J. R. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944-948. 10.1126/science.1128618 [DOI] [PubMed] [Google Scholar]
- Oda, T., Yanagisawa, H., Yagi, T. and Kikkawa, M. (2014). Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J. Cell Biol. 204, 807-819. 10.1083/jcb.201312014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich, H., Schmidts, M., Werner, C., Onoufriadis, A., Loges, N. T., Raidt, J., Banki, N. F., Shoemark, A., Burgoyne, T., Al Turki, S.et al. (2012). Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am. J. Hum. Genet. 91, 672-684. 10.1016/j.ajhg.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. and Ferrin, T. E. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605-1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Polin, M., Tuval, I., Drescher, K., Gollub, J. P. and Goldstein, R. E. (2009). Chlamydomonas swims with two “gears” in a eukaryotic version of run-and-tumble locomotion. Science 325, 487-490. 10.1126/science.1172667 [DOI] [PubMed] [Google Scholar]
- Poprzeczko, M., Bicka, M., Farahat, H., Bazan, R., Osinka, A., Fabczak, H., Joachimiak, E. and Wloga, D. (2019). Rare human diseases: model organisms in deciphering the molecular basis of primary ciliary dyskinesia. Cells 8, 1614. 10.3390/cells8121614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin, M. L. and Matthews, B. W. (2000). Accurate calculation of the density of proteins. Acta Crystallogr. D Biol. Crystallogr. 56, 791-794. 10.1107/S090744490000679X [DOI] [PubMed] [Google Scholar]
- Rao, V. G., Sarafdar, R. B., Chowdhury, T. S., Sivadas, P., Yang, P., Dongre, P. M. and D'Souza, J. S. (2016). Myc-binding protein orthologue interacts with AKAP240 in the central pair apparatus of the Chlamydomonas flagella. BMC Cell Biol. 17, 24. 10.1186/s12860-016-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, A. J., Kon, T., Knight, P. J., Sutoh, K. and Burgess, S. A. (2013). Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 14, 713-726. 10.1038/nrm3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, G., O'Toole, E. and Porter, M. E. (2001). The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 12, 739-751. 10.1091/mbc.12.3.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, C. J. A., de Castro, E., Cerutti, L., Cuche, B. A., Hulo, N., Bridge, A., Bougueleret, L. and Xenarios, I. (2013). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344-D347. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. F. and Lefebvre, P. A. (1996). PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 132, 359-370. 10.1083/jcb.132.3.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. F. and Yang, P. (2004). The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton 57, 8-17. 10.1002/cm.10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K., Awata, J., Tritschler, D., Bower, R., Witman, G. B., Porter, M. E. and Nicastro, D. (2015). In situ localization of N and C termini of subunits of the flagellar nexin-dynein regulatory complex (N-DRC) using SNAP tag and cryo-electron tomography. J. Biol. Chem. 290, 5341-5353. 10.1074/jbc.M114.626556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves, M. E., Nagarkatti-Gude, D. R., Zhang, Z. and Strauss, J. F.III. (2016). Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton (Hoboken) 73, 3-22. 10.1002/cm.21271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., Bednarek, S. Y., Shorte, S. L. and Eliceiri, K. W. (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80-90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Trudgian, D. C. and Mirzaei, H. (2012). Cloud CPFP: a shotgun proteomics data analysis pipeline using cloud and high performance computing. J. Proteome Res. 11, 6282-6290. 10.1021/pr300694b [DOI] [PubMed] [Google Scholar]
- Trudgian, D. C., Thomas, B., McGowan, S. J., Kessler, B. M., Salek, M. and Acuto, O. (2010). CPFP: a central proteomics facilities pipeline. Bioinformatics 26, 1131-1132. 10.1093/bioinformatics/btq081 [DOI] [PubMed] [Google Scholar]
- Trudgian, D. C., Ridlova, G., Fischer, R., Mackeen, M. M., Ternette, N., Acuto, O., Kessler, B. M. and Thomas, B. (2011). Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics 11, 2790-2797. 10.1002/pmic.201000800 [DOI] [PubMed] [Google Scholar]
- Viswanadha, R., Sale, W. S. and Porter, M. E. (2017). Ciliary motility: regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol. 9, a018325. 10.1101/cshperspect.a018325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, K. Y., Leptos, K. C. and Goldstein, R. E. (2014). Lag, lock, sync, slip: the many ‘phases’ of coupled flagella. J. R Soc. Interface 11, 20131160. 10.1098/rsif.2013.1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell, M., Nicastro, D., Porter, M. E. and Sale, W. S. (2009). The regulation of axonemal bending. In The Chlamydomonas Sourcebook, 2nd edn. Vol. 3 (ed. Harris E. H., Stern D. B. and Witman G. B.), pp. 253-282: Elsevier Inc. [Google Scholar]
- Witman, G. B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280-290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Witman, G. B., Plummer, J. and Sander, G. (1978). Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76, 729-747. 10.1083/jcb.76.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, R., O'Toole, E., Ghosh, S. and Mitchell, D. R. (2004). Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA 101, 17398-17403. 10.1073/pnas.0406817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. and Mitchell, D. R. (2004). Cpc1, a Chlamydomonas central pair protein with an adenylate kinase domain. J. Cell Sci. 117, 4179-4188. 10.1242/jcs.01297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Zariwala, M. A., Mahadevan, M. M., Caballero-Campo, P., Shen, X., Escudier, E., Duriez, B., Bridoux, A.-M., Leigh, M., Gerton, G. L.et al. (2007). A heterozygous mutation disrupting the SPAG16 gene results in biochemical instability of central apparatus components of the human sperm axoneme. Biol. Reprod. 77, 864-871. 10.1095/biolreprod.107.063206 [DOI] [PubMed] [Google Scholar]
- Zhao, L., Hou, Y., Picariello, T., Craige, B. and Witman, G. B. (2019). Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol. 218, 2051-2070. 10.1083/jcb.201902017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L., Hou, Y., McNeill, N. A. and Witman, G. B. (2020). The unity and diversity of the ciliary central apparatus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190164. 10.1098/rstb.2019.0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S. Q., Palovcak, E., Armache, J.-P., Verba, K. A., Cheng, Y. and Agard, D. A. (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331-332. 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.