ABSTRACT
Plant brassinosteroid hormones (BRs) regulate growth in part through altering the properties of the cell wall, the extracellular matrix of plant cells. Conversely, feedback signalling from the wall connects the state of cell wall homeostasis to the BR receptor complex and modulates BR activity. Here, we report that both pectin-triggered cell wall signalling and impaired BR signalling result in altered cell wall orientation in the Arabidopsis root meristem. Furthermore, both depletion of endogenous BRs and exogenous supply of BRs triggered these defects. Cell wall signalling-induced alterations in the orientation of newly placed walls appear to occur late during cytokinesis, after initial positioning of the cortical division zone. Tissue-specific perturbations of BR signalling revealed that the cellular malfunction is unrelated to previously described whole organ growth defects. Thus, tissue type separates the pleiotropic effects of cell wall/BR signals and highlights their importance during cell wall placement.
KEY WORDS: Brassinosteroids, Cell wall, Cell division, Arabidopsis, Root, Meristem
Summary: Both increased and reduced BR signalling strength results in altered cell wall orientation in the Arabidopsis root, uncoupled from whole-root growth control.
INTRODUCTION
The cell wall, a carbohydrate-rich structure surrounding all plant cells, fulfils numerous essential functions in growth and development; it provides mechanical support, controls cellular adhesion and forms the interface with the environment (Cosgrove, 2016; Lampugnani et al., 2018; Voxeur and Höfte, 2016). Notably, as the cell wall prevents cell migration, tight control over the plane of cell wall deposition during cytokinesis is often assumed to be important for plant patterning and morphogenesis. However, whether there is a direct relationship between cell shape, in part controlled by cell division orientations, and organ shape is not clear (Beemster et al., 2003; Kaplan and Hagemann, 1991; Torres-Ruiz and Jurgens, 1994; Traas et al., 1995). During cytokinesis, the microtubules (MT) of the phragmoplast guide secretory traffic towards the cell plate, a radially expanding disk of membrane-engulfed cell wall material that increases in diameter until it fuses with the parental walls to conclude cell division (Livanos and Müller, 2019). Cell division plane orientation is pre-determined before the cell plate actually develops. Immediately before prophase, the MT network forms a transient ring structure known as preprophase band (PPB) at the periphery of the cell, marking the future fusion site of the cell plate and the parental wall during cytokinesis (Livanos and Müller, 2019; Rasmussen and Bellinger, 2018; Rasmussen et al., 2013; Smith, 2001). This site, called the cortical division zone (CDZ) persists throughout mitosis and is populated by proteins that guide the expanding cell plate towards the parental wall. Although PPBs are at least partially dispensable for oriented cell division (Schaefer et al., 2017; Zhang et al., 2016), mutants in CDZ components such as TAN1 and POK1,2 show severely altered division angles, suggesting that these factors are important for phragmoplast guidance in the absence of these factors (Lipka et al., 2014; Stöckle et al., 2016; Walker et al., 2007). In light of the central role of cell wall biosynthesis during cytokinesis (Gu et al., 2016; Miart et al., 2014; Zuo et al., 2000) and a growing list of cell wall-mediated feedback signalling modules affecting a wide range of biological processes (Wolf, 2017), it is conceivable that cell wall state during mitosis/cytokinesis has to be monitored by cell wall surveillance systems (Rui and Dinneny, 2020; Vaahtera et al., 2019; Voxeur and Höfte, 2016).
Growth itself poses a threat to cell wall integrity and composition and physical properties of the cell wall have to be tightly monitored to ensure cell wall homeostasis and coordinated growth. For example, a compensatory response to cell wall challenge is mediated by RECEPTOR-LIKE PROTEIN 44 (RLP44), which is able to interact with the brassinosteroid (BR) receptor BRI1 (Holzwart et al., 2018) and its co-receptor BRI1-ASSOCIATED KINASE1 (BAK1) (Wolf et al., 2014) to promote BR signalling. More specifically, the degree of methylesterification (DM) of homogalacturonan (HG), a pectin component of the cell wall, has a profound impact on cell wall structure and mechanical properties: once secreted into the wall network, HG can be de-methylesterified by pectin methylesterases (PMEs), ubiquitous plant enzymes which are themselves regulated by PME inhibitor proteins (PMEIs). Reduction of PME activity through overexpression of PECTIN METHYLESTERASE INHIBITOR 5 (PMEI5) activates BR hormone signalling via RLP44, which in turn leads to a complex directional growth phenotype including organ fusion and root waving (Wolf et al., 2012, 2014).
BR-deficient mutants and plants treated with BR biosynthesis inhibitors display reduced longitudinal meristem size (Cole et al., 2014; González-García et al., 2011; Hacham et al., 2011; Vragović et al., 2015), but show an increased number of formative (periclinal) divisions, resulting in supernumerary cell files in root tissues (Holzwart et al., 2018; Kang et al., 2017). In addition to the main receptor BRI1, BRs are also perceived by two close paralogs of BRI1, BRL1 and BRL3 (Caño-Delgado et al., 2004; Zhou et al., 2004). brl1 and brl3 single and double mutants do not show growth defects and bri1 brl1 brl3 triple mutant (bri1-triple from hereon) resembles bri1. However, the absence of brl1 and brl3 enhances the vascular defects of bri1 (Caño-Delgado et al., 2004; Holzwart et al., 2018; Kang et al., 2017). BR signalling has context- and cell type-specific roles in plant growth and development (Ackerman-Lavert and Savaldi-Goldstein, 2019; Nolan et al., 2019; Planas-Riverola et al., 2019; Singh and Savaldi-Goldstein, 2015), involving cell autonomous and non-cell autonomous effects on root length and on the number of anticlinal and periclinal divisions in the meristem (Hacham et al., 2011; Vragović et al., 2015; Kang et al., 2017; Graeff et al., 2020).
Here, we dissected the phenotype induced by PMEI5-mediated reduction in PME activity on the root apical meristem and revealed that pectin-triggered cell wall signalling leads to orientation defects of newly placed walls, which are dependent on BR signalling activation. These defects are dependent on BR signalling activation, but independent of organ-level growth phenotypes, and coincide with aberrant localisation of the CDZ component POK1. Conversely, reduced BR signalling in receptor and biosynthetic mutants leads to cell wall orientation defects similar to those observed in PMEIox, but unrelated to the enhanced formative division phenotypes. These cell wall orientation defects can also be genetically separated from BR functions in root growth. Thus, we reveal a role for cell wall and BR signalling in controlling cell wall orientation.
RESULTS
Pectin-triggered cell wall signalling leads to cell division defects that are RLP44 and BRI1 dependent
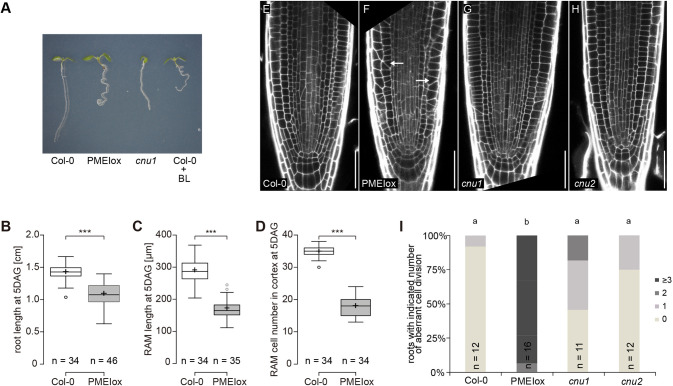
We have previously demonstrated that cell wall signalling connects the state of the cell wall to the BR signalling pathway (Wolf et al., 2012). When de-methylesterified pectin becomes limiting, such as in PMEIox plants overexpressing PMEI5, elevated BR signalling counteracts loss of cell wall integrity and leads to BRI1-dependent macroscopic growth defects such as reduced root length and root waving (Wolf et al., 2012; Fig. 1A,B). In agreement with this, external application of brassinolide (BL) also results in root waving (Lanza et al., 2012; Wolf et al., 2012) (Fig. 1A). To shed light on the cellular phenotype of PMEIox roots and to assess whether root waving is associated with meristematic defects, we analysed PMEIox root tips 5 days after germination (DAG) and found that the reduced root length of PMEIox was accompanied by a reduction in size and cell number of the root apical meristem (RAM) (Fig. 1C,D). Interestingly, we also revealed that, in contrast to the stereotypical pattern of cellular morphology and tissue organisation in wild-type root tips (Dolan et al., 1993) (Fig. 1E), PMEIox roots displayed a substantial amount of obliquely orientated transverse cell walls apparent in epidermis, cortex and endodermis (Fig. 1F). We next asked whether these cell wall orientation defects in PMEIox are also the result of elevated BR signalling mediated by RLP44 (Wolf et al., 2014). Consistent with this hypothesis, PMEIox suppressor mutants with lesions in BRI1 (cnu1; Wolf et al., 2012) and RLP44 (cnu2; Wolf et al., 2014), respectively, showed to a large extent normal cross wall orientation (Fig. 1G-I). Thus, cell wall orientation defects in PMEIox are mediated by RLP44 and BRI1, similar to the other macroscopic PMEIox phenotypes, such a root waving and organ fusion (Fig. 1A) (Wolf et al., 2012, 2014). Moreover, expression of a GFP-tagged version of RLP44 under control of the CaMV 35S promoter (RLP44-GFP) in cnu2 resulted in a significant increase of oblique cell walls, indicating complementation (Fig. S1). Together, these results suggest that cell wall signalling-triggered elevation of BR signalling is causative for the oblique cell wall phenotype in PMEIox.
Fig. 1.
Cell wall signalling triggered by ubiquitous PMEI5 expression in PMEIox alters root growth and cell wall orientation. (A) PMEIox seedlings have a root waving phenotype caused by enhanced BR signalling: 5-day-old seedlings of Col-0, PMEIox, the PMEIox bri1 suppressor mutant cnu1, and Col-0 seedlings grown on plates containing 5 nM brassinolide (BL) are shown. (B-D) PMEIox plants show reduced root length (B), RAM length (C) and RAM cell number (D) 5 DAG. ***P<0.001 (Mann–Whitney U-test). Boxes indicate median, upper and lower quartile, whiskers indicate minimum and maximum except outliers beyond 1.5× interquartile range, which are indicated as dots. Cross indicates mean. (E-H) PMEIox plants show oblique cell walls in the root apical meristem, dependent on BRI1 and RLP44. Cell division defects (compared with Col-0; E) in PMEIox (F, arrows) are reduced in the PMEIox suppressor mutants cnu1, carrying a mutation in BRI1 (G), and cnu2, carrying a mutation in RLP44 (H). Cells walls of the root apical meristems are visualised using mPS-PI staining (Truernit et al., 2008). Scale bars: 50 µm. (I) Quantification of the fraction of roots with the indicated number of oblique transversal walls in cortex cells from confocal section as in E-H. Letters indicate statistically significant differences according to Dunn's test after Kruskal–Wallis analysis.
Cell wall perturbation in diverse cell types separates aberrant cell division, root waviness and root growth
Thus far, our results indicate that pectin-triggered cell wall signalling leads to root waving, root growth inhibition and aberrant wall orientation phenotypes, which are all BRI1-dependent. These pleiotropic effects could be all linked or could result from unrelated processes. As BR signalling is context-dependent, we employed a cell type-specific expression system to alter cell wall properties locally and followed the phenotypic consequences at the organ, tissue and cellular level. We selected a number of tissue-specific promoters to drive expression of the chimeric transcription factor GR-LhG4 (Craft et al., 2005; Moore et al., 1998; Schürholz et al., 2018) in the epidermis, cortex, endodermis or xylem pole pericycle cells, complemented by ubiquitous expression (Fig. S2). GR-LhG4, in turn, triggers transcription of PMEI5 under control of the synthetic pOp promoter in the presence of dexamethasone (DEX). Quantitative reverse transcription PCR (QRT-PCR) revealed that, in all lines, PMEI5 was upregulated by DEX treatment, even though absolute levels differed substantially between lines (Fig. S3). These differences might, at least in part, depend on varying strength and cell type-specificity of the respective promoters. To assess root waving of PMEI5-expressing lines, we determined the vertical growth index of 6-day-old seedlings grown on medium containing either DEX or an equal amount of DMSO (Fig. 2A). Expression of PMEI5 in hair cells, driven by the pCOBL9 promoter (pCOBL9>GR>PMEI5; Fig. 2A,B), or in all epidermal cells (pML1>GR>PMEI5; Fig. 2A,C) was sufficient to trigger root waving, similar to ubiquitous expression (pUBQ10>GR>PMEI5; Fig. 2A,D). Interestingly, expression in the xylem pole pericycle (XPP) cells of the stele (pXPP>GR>PMEI5) also led to a root waving phenotype (Fig. 2A,E). This suggests that cell wall-induced BR signalling in these cells causes organ-level responses, with the caveat that PMEI5 might be mobile in the cell wall. We then assessed the occurrence of oblique cell walls in the trans-activation lines. Notably, tissue-specific expression lines that displayed root waving, namely pUBQ10>GR>PMEI5, pML1>GR>PMEI5, pCOBL9>GR>PMEI5 and pXPP>GR>PMEI5, differed in their behaviour with respect to the cell wall orientation phenotype (Fig. 3). Whereas ubiquitous PMEI5 expression (Fig. 3A) and expression under control of the ML1 promoter (Fig. 3B) resulted in significant increase of aberrant epidermal cell wall placement, plants with hair cell (Fig. 3C) and pericycle expression of PMEI5 (Fig. 3D) were unaffected. This suggests that PMEI5-induced root waving is at least partially independent from meristematic cell wall orientation defects. Interestingly, trans-activation of PMEI5 in cortex cells (pCO2>GR>PMEI5), which did not lead to root waving (Fig. 2A), triggered PMEIox-like oblique cell walls cell autonomously in cortex cells and in neighbouring cell types such as epidermal cells (Fig. 4). Introducing a fluorescent reporter under control of the pOp6 promoter suggested that trans-activation in the pCO2>GR>PMEI5 line is indeed restricted to cortex cells (Fig. S4). However, it cannot be excluded that the observed non-cell autonomous effects were conferred by PMEI5 mobility in the apoplastic space. As the addition of fluorescent proteins or any other tags tested renders PMEI5 non-functional, this could not be experimentally addressed. In addition to the cell wall orientation defects, PMEI5 expression in cortex cells resulted in a disrupted organisation of the stem cell niche (Fig. 4B) and a reduction of meristematic cell number (Fig. S5A). In marked contrast to PMEIox (Fig. 1), the effect on cell wall orientation in pCO2>GR>PMEI5 plants was not accompanied by a significant change of root length or RAM size compared with the uninduced control (Fig. S5B,C). Taken together, these results reveal that triggering cell wall modification in either the epidermis or in XPP cells of the stele can lead to similar organ level responses and confirm that PMEI5-induced cell wall orientation alterations are independent of directional growth processes. In addition, meristematic cell wall orientation defects in pCO2>GR>PMEI5 did not affect organ-level growth.
Fig. 2.
Cell wall perturbation in diverse cell types can lead to similar organ-level responses. (A) Quantification of the vertical growth index (distance from hypocotyl junction to tip of the root divided by root length) in Col-0, PMEIox and inducible tissue-specific expression lines in the absence and presence of 30 µM dexamethasone (DEX). (B-E) Induction of PMEIox trans-activation in hair cells (B), epidermal cells (C), ubiquitously (D) and in meristematic cortex cells (E) leads to root waving. Plants were germinated and grown on plates containing 30 μM DEX or an equal volume of DMSO for 5 days. Scale bars: 1 cm. *P<0.05, ***P<0.001 (Mann–Whitney U-test). n.s., not significant. Boxes indicate median, upper and lower quartile, whiskers indicate minimum and maximum except outliers beyond 1.5× interquartile range, which are indicated as blue circles. Cross indicates mean. Individual data points (n=20) are shown to the right of the box diagrams.
Fig. 3.
Tissue-specific expression of PMEIox reveals that root waving is independent from cell wall orientation defects. (A) Ubiquitous, pUBQ10-driven trans-activation of PMEI5 recapitulates the PMEIox cell wall phenotype. (B-D) In lines with tissue-specific PMEI5 expression in epidermis, hair cells or XPP cells, cell wall orientation shows varying degrees of cell wall orientation defects. Scale bars: 50 µm. Cell walls are counterstained with mPS-PI. Bar diagrams denote fraction of roots with the indicated number of oblique transversal walls in epidermis cells (A-C) or all cells (D) from confocal sections. ***P<0.001 (Mann–Whitney U-test). n.s., not significant.
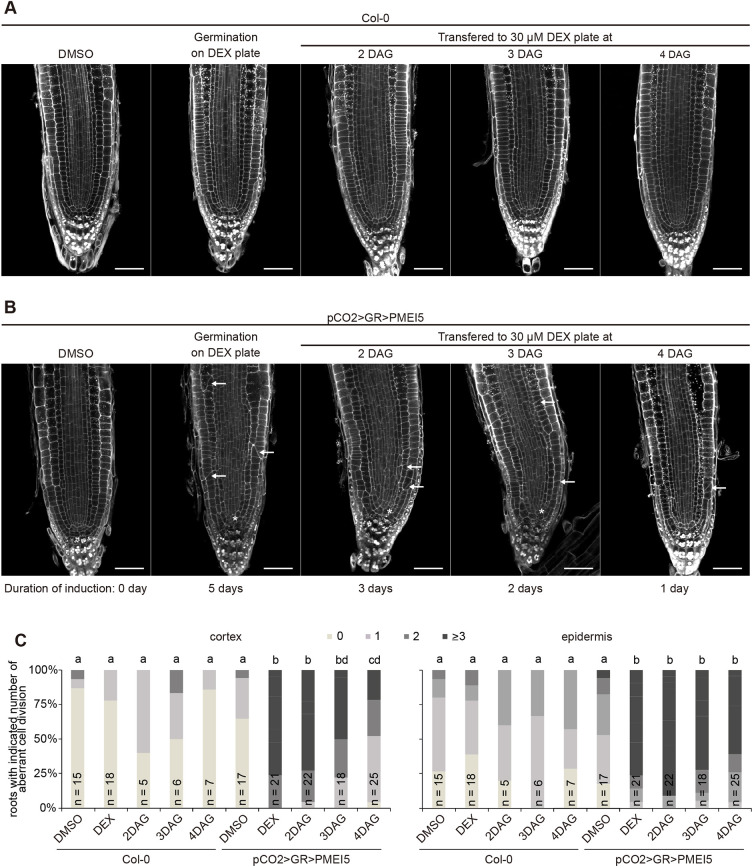
Fig. 4.
Cell wall orientation defects allow unaltered organ level growth. (A,B) Induction of PMEI5 expression in meristematic cortex cells leads to oblique cell walls in cortex and epidermis (arrows) and a disrupted stem cell region (asterisk). Plants were imaged at 5 DAG on DEX-containing medium or after germination on DMSO-containing plates and transferred to induction medium at the indicated time points. (C) Quantification of the fraction of roots with the indicated number of oblique transversal walls in cortex cells from confocal sections. Letters indicate statistically significant differences according to Dunn's test after Kruskal–Wallis analysis.
Reduced BR signalling leads to wall orientation defects that can be separated from root growth
We next characterised the cross-wall orientation in lines with reduced BR signalling and observed aberrant wall angles in bri1 that were enhanced in bri1-triple mutants. The occurrence of oblique transversal walls within the cell files of the root meristem resulted in random perturbations (Fig. 5A, asterisk), differing from the formative divisions previously observed in these mutants that result in additional cell files (Holzwart et al., 2018; Kang et al., 2017). Quantification of aberrant cell walls in cortex cells revealed that the majority of bri1 meristems had at least one oblique cortex cross wall per median confocal section, a phenotype which was enhanced in bri1-triple mutants (Fig. 5B). We recently reported that some functions of BRI1 are independent of classical BR signalling outputs (Holzwart et al., 2018, 2020). However, cell wall orientation defects in bri1 mutants appear to be caused by reduced canonical BRI1/ligand dependent signalling, as the biosynthetic mutants cpd (Szekeres et al., 1996) and det2 (Chory et al., 1991), as well as plants in which endogenous BRs were depleted by the application of brassinazole (BRZ) (Asami et al., 2000), displayed phenotypes comparable with bri1 mutants (Fig. 5B). To address whether the impact of BR activity on the cell wall orientation is tissue-specific, we used a previously reported collection of lines conferring tissue-specific BRI1 expression in bri1 and bri1-triple mutant backgrounds (Fridman et al., 2014; Hacham et al., 2011; Vragović et al., 2015). In contrast to expression of BRI1 in non-hair cells (pGL2-BRI1), expression of BRI1 from the endodermis (pSCR-BRI1) or stele (pSHR-BRI1) largely rescued cell wall orientation in the cortex of bri1 and bri1-triple mutants (Fig. 5C). Interestingly, the extent of the effect of BRI1 on the aberrant wall angles was not linked to its effect on meristem size and root length (Hacham et al., 2011; Vragović et al., 2015; Fig. 5D,E; Fig. S6). Specifically, pGL2-BRI1 roots are longer than pSCR-BRI1 and pSHR-BRI1 and can have almost wild-type length, in both bri1 and bri1-triple backgrounds (Hacham et al., 2011; Fig. 5D,E), despite the occurrence of aberrant cell walls (Fig. 5C). BRI1 activity in non-hair cells limits cell elongation in the elongation zone of the root, unless BRI1 in the neighbouring hair cells is also present (Fridman et al., 2014). High BRI1 in non-hair cells results in waviness in a mutant and wild-type background (Fridman et al., 2014; Fig. 5D). Thus, aberrant wall orientation is unlinked from root waving. Taken together, as with PMEIox, the control of cell wall orientation is separated from other growth processes controlled by BRI1. In addition, both enhanced BR signalling (PMEI5 expression) and reduced BR signalling (BR loss-of-function mutants) lead to cell wall orientation defects.
Fig. 5.
BR signalling is required for the maintenance of cell wall orientation. (A) PI-stained meristems of Col-0, bri1-triple mutants and two bri1-triple lines expressing BRI1 from cells of the epidermis (pGL2:BRI1) or stele (pSHR:BRI1). Note oblique transversal walls in epidermis, cortex and endodermis (arrows), as well as random perturbation of cell files (asterisk). (B) Quantification of the fraction of cortex cells with oblique transversal walls in cortex cells from confocal section in A for the indicated genotypes and wild-type roots in which BRs were depleted by treatment with BRZ. Numbers in the x-axis indicate independent transgenic lines. Letters indicate statistically significant differences according to Dunn's test after Kruskal–Wallis analysis. (C) Quantification of the fraction of cortex cells with altered cell wall orientation of lines expressing BRI1 from the epidermis (pGL2-BRI1), endodermis (pSCR-BRI1) and stele (pSHR-BRI1) as well as their combinations in various backgrounds. Letters indicate statistically significant differences according to Dunn's test after Kruskal–Wallis analysis. (D) Growth phenotype of 7-day-old Col-0, bri1-triple and bri1-triple plants expressing BRI1 from cells of the epidermis (pGL2:BRI1) or stele (pSHR:BRI1). (E) Quantification of root length as depicted in D. n=21-25. Letters indicate statistically significant difference after one-way ANOVA followed by Tukey's post-hoc test. Boxes indicate median, upper and lower quartile, whiskers indicate minimum and maximum except outlier beyond 1.5× interquartile range, which is indicated as red cross.
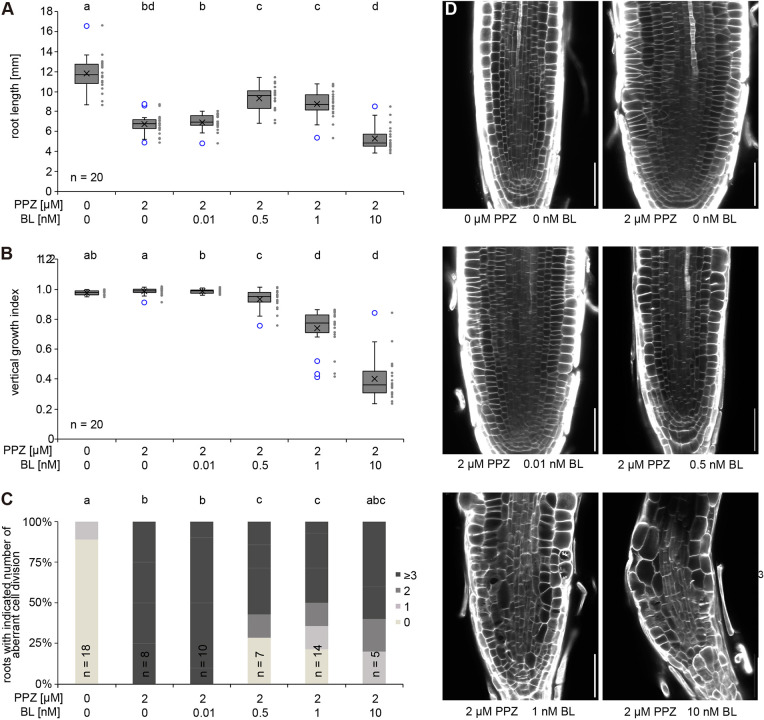
To independently test whether both growth-enhancing and growth-inhibiting doses of BRs affect the orientation of the new cell wall, plants were grown on medium containing the BR biosynthesis inhibitor propiconazole (PPZ) and supplemented with increasing concentrations of epi-BL. Treatment with PPZ inhibited root growth, which could be partially compensated by the addition of 0.5 nM epi-BL, without induction of root waving (Fig. 6A,B). Addition of 1 nM or 10 nM epi-BL to the PPZ-containing medium led to root waving and a reduction of root growth (Fig. 6A,B), demonstrating that these concentrations enhanced BR signalling. Both reduced and enhanced BR signalling strength resulted in aberrant cell wall orientation in root meristems (Fig. 6C,D), supporting our genetic evidence. Moreover, the cell wall orientation defects in pCO2>GR>PMEI5 roots (Fig. 4) were attenuated by loss of RLP44 function, and undetectable in a cross of the pCO2>GR>PMEI5 line with the hypomorphic BRI1 mutant bri1cnu4 (Holzwart et al., 2020) (Fig. S7). The bri1cnu4 mutant showed a significantly increased frequency of aberrant cell wall orientations, similar to pCO2>GR>PMEI5 (Fig. S7) and consistent with our findings on other BR loss-of-function mutants (Fig. 5). However, pCO2>GR>PMEI5 (bri1cnu4) roots showed wild type-like cell wall orientation (Fig. S7), suggesting that the reduction of BR signalling strength caused by the BRI1 mutation can counteract the PMEI5-induced activation of BR signalling. Together, optimal BR signalling strength is required for normal cell wall orientation in the Arabidopsis root.
Fig. 6.
Optimal BR signalling strength is required for normal cell wall orientation. (A) Root length of Col-0 seedling untreated or grown in the presence of the BR biosynthesis inhibitor PPZ and the indicated concentration of epi-BL. Letters indicate statistically significant difference according to Tukey's HSD test after one-way ANOVA, n=20. (B) Vertical growth index of the same plants as in A. Letters indicate statistically significant differences according to Dunn's test after Kruskal–Wallis analysis, n=20. Boxes in A and B indicate median, upper and lower quartile, whiskers indicate minimum and maximum except outliers beyond 1.5× interquartile range, which are indicated as blue circles. Cross indicates mean. Individual data point are shown to the right of the box diagrams. (C) Quantification of the fraction of roots with the indicated number of oblique transversal walls in cortex cells from confocal sections in D. Letters indicate statistically significant differences according to Dunn's test after Kruskal–Wallis analysis. (D) Representative images of roots analysed in A-C. Cell walls were stained with SCRI 2200 in ClearSee (see Materials and Methods). Scale bars: 50 μm.
Cell wall perturbation by PMEIox leads to cytokinesis defects after specification of the CDZ
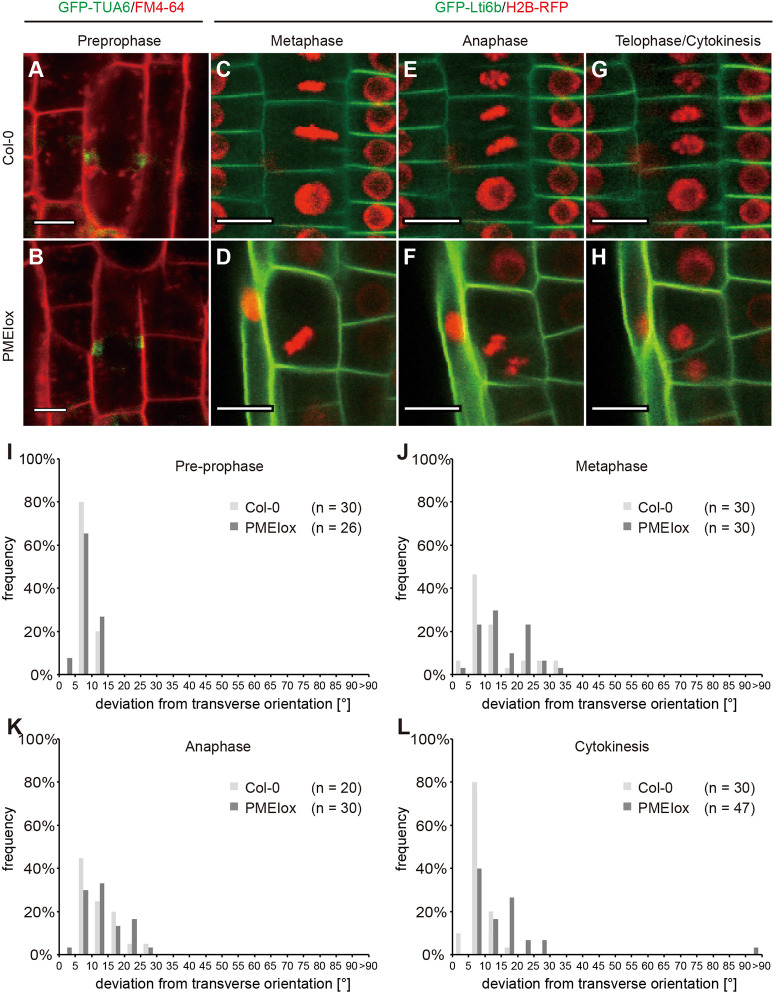
To test whether the aberrant cell wall placement in PMEIox was associated with defects in cell division plane orientation, we quantified the orientation of this plane at the different mitotic stages (Fig. 7). The PPB transiently indicates positioning of the CDZ (Rasmussen et al., 2013). Hence, to determine whether cell wall perturbations affect this positioning, we used a fluorescently labelled tubulin (GFP-TUA6) that allows visualisation of the PPB (Fig. 7A,B). Quantitative analysis of PPB orientation in wild-type cortex and epidermis cells revealed only minimal deviations from a position perpendicular to the cell long axis (Fig. 7I). PMEIox displayed a very similar distribution, indicating that division plane orientation defects are independent of PPB positioning (Fig. 7I). As the PPB disappears in pro-metaphase, we used a transgenic line expressing an RFP-fused Histone 2B to label chromatin and a GFP-fused plasma membrane protein Lti6b (Maizel et al., 2011) to visualise cell outlines and the forming cell plate. We quantified the orientation of the metaphase plate, the midline between sister chromatids, and the cell plate in metaphase, anaphase and telophase. Interestingly, the orientation of both the Col-0 wild-type and PMEIox metaphase plates deviated from a 90° angle relative to the cell long axis (Fig. 7C,D,J). A similar observation was made during anaphase (Fig. 7E,F,K). However, the forming cell plate during telophase was largely aligned with the expected division plane in Col-0, i.e. perpendicular to the cell long axis, and deviation angles appeared very similar to what was previously observed with the PPB (Fig. 7G,L). In contrast, telophase cell-plate orientation in PMEIox showed a distribution similar to what was observed in meta- and anaphase, consistent with defective re-alignment to the ∼90° angle observed in Col-0 (Fig. 7H,L). In summary, the PPB orientations in wild type and PMEIox appeared to be similar to each other, whereas both wild-type and PMEIox virtual division planes defined by the H2B marker deviated from transverse orientation during metaphase and anaphase. During telophase, wild-type cell plates largely aligned with the expected position perpendicular to the cell long axis, whereas a substantial fraction of PMEIox cell plates did not.
Fig. 7.
Cell wall perturbation by PMEIox leads to cell division defects after specification of the cortical division zone. (A,B) Orientation of pre-prophase bands labelled by GFP-TUA6 is transverse in both Col-0 (A) and PMEIox (B); see I for quantification. (C-F) Metaphase plate orientation (C,D; see J for quantification) and sister chromatid orientation during anaphase (E,F; see K for quantification) deviates from a 90°C angle in both the Col-0 wild type (C,E) and PMEIox (D,F). (G,H) Orientation of the cell plate in the Col-0 wild type stabilised at around 90°C relative to the cell long axis (G), whereas PMEIox cell plates showed a wide range of orientations (H) (see L for quantification). (I-L) Quantification of division plane orientation during mitosis and cytokinesis. Bars in histograms denote fraction of cells in bins bordered by angles indicated on the x-axis. n=30-50. Scale bars: 20 µm (A,C,E,G); 10 µm (B,D,F,H).
These observations are consistent with two hypotheses that could explain the aberrant cell wall angles in PMEIox. First, PMEIox cell plates might fail to find the CDZ initially marked by the PPB and occupied by POK1 and other components (Livanos and Müller, 2019). Second, the position of the cortical division zone might be mobile relative to the cell walls in PMEIox, although guidance is unaffected. To differentiate between the two possibilities, we introduced a fluorescently tagged version of POK1, YFP-POK1 (Lipka et al., 2014), into PMEIox and determined whether the cell plate fusion site coincided with the location of POK1 (Lipka et al., 2014; Müller et al., 2006). Cell plate fusion sites were marked by YFP-POK1, even when the cell plate fused at an angle deviating from 90° relative to the parental walls, suggesting that phragmoplast guidance towards CDZ components is unaffected in PMEIox (Fig. 8). As we did not observe aberrant PPB positioning, our results suggest that YFP-POK1, and thus potentially CDZ localisation, might deviate from the expected position during cytokinesis. Together, aberrant cell plate fusion in PMEIox coincides with POK1 localisation at positions that deviate from the cell midline. Our results show that optimal BR signalling strength is required to maintain the orientation of newly placed cell walls.
Fig. 8.
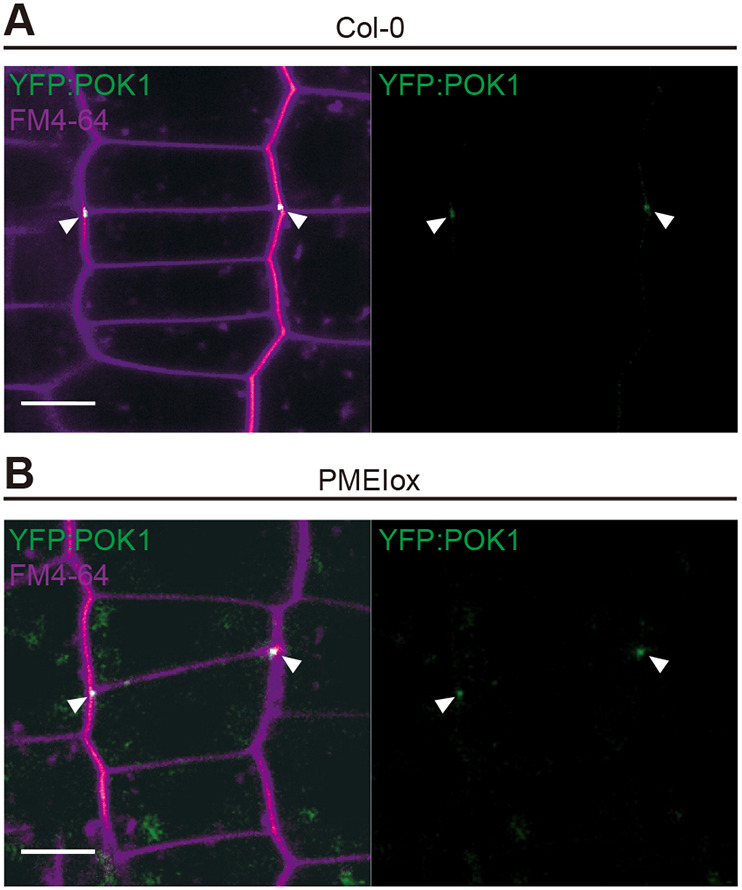
Cell plate fusion in PMEIox coincides with POK1 localisation at aberrant positions. (A,B) Cell plate fusion site position and YFP-POK1 localisation at the end of cytokinesis in Col-0 (A) and PMEIox (B). Note that the cell plate fusion at POK1-positive sites in the parental walls deviates from the normal position in PMEIox. White arrowheads indicate YPF-POK1-marked cell plate fusion sites. Membranes are labelled with FM4-64. Scale bars: 20 μm.
DISCUSSION
Here, we report that cell wall integrity and BR signalling are involved in the control of cell wall orientation in the Arabidopsis root. Both pectin-triggered cell wall signalling, as in PMEIox plants, and impaired BR signalling resulted in aberrant cross walls in the Arabidopsis root meristem.
In PMEIox plants, pectin modification triggers an RLP44-mediated activation of BR signalling which, in turn, prevents loss of cell wall integrity, but results in a wide variety of growth-related phenotypes (Holzwart et al., 2018; Wolf et al., 2012, 2014; this study). Aberrant cell wall orientation due to an altered cell wall in PMEI5-expressing plants appears to occur downstream of BR signalling, as mutations in BRI1 (cnu1, bri1cnu4) or RLP44 (cnu2) mutants, largely suppressed the oblique cell wall phenotype of PMEIox. Analysis of PMEIox cell divisions at the subcellular level revealed that the virtual cell division plane between meta- and telophase, marked by the midline between chromatin structures (metaphase plate and segregating sister chromatids), showed deviations from the 90° angle in both wild-type and PMEIox cells. Wild-type cell plates later displayed the expected transversal orientation, whereas PMEIox cell plates frequently did not. Our use of metaphase plate and sister chromatids as a read-out for the ‘virtual’ cell division plane can be questioned as, for example, mitotic features such as spindle orientation do not always correlate with division plane orientation (Cleary and Smith, 1998; Galatis et al., 1984; Marcus et al., 2003; Oud and Nanninga, 1992; Rasmussen et al., 2011, 2013). However, that wild-type cell division plane orientations show considerable variation and noise, but are later harmonised by interaction with CDZ components, is in agreement with the phenotype of CDZ mutants such as tan1, pok1 pok2 and phgap1 phgap2 (Lipka et al., 2014; Müller et al., 2006; Smith et al., 1996; Stöckle et al., 2016; Walker et al., 2007). In these mutants, oblique cell walls are presumably the result of a lack of phragmoplast guidance. Our analysis suggests that PMEIox and Col-0 behave similarly with respect to all aspects analysed up until the very last steps of cell division, during which a substantial fraction of PMEIox cell plates fail to show orientation perpendicular to the cell long axis. Notably, it is conceivable that oblique cell walls in PMEIox occur through a mechanism different from that in the aforementioned CDZ mutants, as the CDZ in some PMEIox cells appears to have shifted from the expected position, based on YFP-POK1 localisation. This raises the issue of how the relative position of the CDZ is maintained in wild-type cells and whether this could involve interactions with the cell wall as previously suggested (Smertenko et al., 2017). Supporting an involvement of the cell wall in CDZ maintenance is the observation that four-way junctions comprising cross walls of adjacent cells fusing at a similar position of their shared longitudinal wall are actively avoided: PPBs are placed with an offset from the predicted division plane if a cross wall or PPB from an adjacent cell is found at this position and it is conceivable that the necessary signal to trigger this response involves the cell wall (Flanders et al., 1990; Martinez et al., 2018). In addition, cell wall attachment of the CDZ is a plausible way to maintain its relative position in the plasma membrane, in line with the observation that the cell wall is required for the maintenance of PIN polarity (Feraru et al., 2011) and acts to restrict the mobility of plasma membrane proteins in general (Martiniere et al., 2012; McKenna et al., 2019).
Whether there is a direct relationship between cell shape, in part controlled by cell division orientation, and organ shape is a long-standing question (Beemster et al., 2003; Hong et al., 2018; Kaplan and Hagemann, 1991; Martinez et al., 2017). Both continuous pectin-triggered cell wall signalling activation and exogenous application of BRs result in root growth alterations such as waviness and a reduction in root length (Lanza et al., 2012; Wolf et al., 2012; this study). We observed that cell type-specific induction of cell wall signalling separated root growth, its directionality and cell wall orientation defects based on which cell type expressed PMEI5. Furthermore, pectin-triggered cell wall signalling and reduced BR signalling in bri1 mutants led to similar cell wall orientation defects in the meristem, but only PMEIox and plants treated with high BR concentrations displayed root waviness. In addition, plants expressing PMEI5 in cortex cells showed pronounced cell division orientation defects without influencing organ morphology, growth, meristem size or meristematic cell number, whereas epidermal expression of BRI1 in bri1-triple mutants rescued organ level growth but enhanced cell division orientation defects, likely because of high BR signalling. Although cell wall orientation defects in BR loss-of-function mutants and in plants with enhanced BR signalling might occur through different mechanisms, these results demonstrate that aberrant placement of new walls is compatible with normal organ growth.
Reminiscent of previous findings demonstrating non-cell autonomous effects of BR signalling, cortical PMEI5 expression led to oblique cell divisions in both epidermal and cortex cells. We cannot determine whether the non-cell autonomous effect is directly linked to PMEI5 or to downstream signalling components. However, it is noteworthy that expression of PMEI5 in two spatially separated cell types, hair cells in the epidermis and the XPP cells, affects whole organ level responses (root waving). Future work needs to address how these organ-level responses are connected to cellular effects of BR signalling and how the cell wall is connected to cell division orientation maintenance, taking into account potential mechanical feedbacks and the contribution of cell geometry and developmental signalling (Besson and Dumais, 2011, 2014; Chakrabortty et al., 2018; Lintilhac and Vesecky, 1984; Louveaux et al., 2016; Martinez et al., 2018; Moukhtar et al., 2019; Yoshida et al., 2014).
The defects in cell wall orientation described here for BR receptor mutants appear to be random and it is unclear whether these cell division defects are related to previously described disturbed cell files and altered tissue organisation in rice bri1 mutants (Nakamura et al., 2006). The use of a tissue-specific approach to perturb the BR signalling was an instrumental tool to disentangle the pleiotropic effects of the BR pathway. Hence, despite a ubiquitous expression of BRI1, it enabled the discovery of tissue-specific effects on shoot growth, root meristem size, final cell size, root length and gene expression that were otherwise masked by alternative overexpression or loss-of-function studies (Singh and Savaldi-Goldstein, 2015). Here, we show that aberrant cell wall orientation can be largely rescued in both bri1 and bri1-triple mutant backgrounds while other growth parameters are poorly rescued, and vice versa. For example, pGL2-BRI1 in bri1-triple has long root meristems and almost wild type-like root length and shows an increased number of anticlinal divisions (Hacham et al., 2011; Vragović et al., 2015) but harbours severe wall orientation defects. This could be the result of high local BR signalling intensity (Fridman et al., 2014; Vragović et al., 2015), as in pML1>GR>PMEI5. However, pSHR-BRI1 has a shorter root and wide meristem (Vragović et al., 2015; Kang et al., 2017) that largely rescued these wall orientation defects. A tissue-specific approach recently also assisted in separating BRI1 control of phloem differentiation from that of growth (Graeff et al., 2020). As BRI1 is not expressed in the phloem in the lines analysed here, restoration of BR signalling in diverse cell types is sufficient to control root length and orientation of transversal walls.
Taken together, our results demonstrate that cell wall integrity and optimal BR signalling levels are required for a correct cell wall placement. This BR signalling-mediated control of cell wall orientation occurs both cell autonomously and non-cell autonomously and is uncoupled from organ-level growth control.
MATERIALS AND METHODS
Plant material
All genotypes used in this study were in the Col-0 background and are listed in Table S1. For PMEox-related experiments, seeds were sterilised with 1.3% (v/v) sodium hypochlorite (NaOCl) diluted in 70% ethanol for 3 min, then washed twice with 100% ethanol and dried in laminar flow hood. Seeds were sowed out on plate with growth medium containing half-strength (1⁄2) Murashige & Skroog (MS) medium (Duchefa), 1% D-sucrose (Car Roth) and 0.9% phytoagar (Duchefa) with pH adjusted to 5.8 with KOH. After 2 days stratification at 4°C in darkness, plates were placed vertically in long-day conditions (16 h light/8 h dark cycles) with equal light conditions (∼100 μE m-2s-1) for 5 days. All analyses have been carried out on 5-day-old seedlings. For DEX induction on plate, DEX (Sigma-Aldrich, D4902) was added to the growth medium, and an equal volume of dimethyl sulfoxide (DMSO) was added to the control plate. For DEX induction on soil, 30 μM DEX was used to spray the aerial part of the plant and to water every other day starting from 3 days after transfer of seedling on soil. All plants were grown on soil under long-day conditions (16 h light/8 h dark cycles) at 23°C with 65% humidity. For BRI1-related experiments (i.e. Fig. 5), seeds were sterilised and grown as described in Fridman et al. (2014).
Microscopy
Microscopic analyses were carried out using Zeiss LSM 510 Meta, a Zeiss LSM710, and Leica TCS SP5 laser scanning confocal microscopes. For mTurquoise2, an excitation wavelength of 458 nm was used and emission was collected between 460 and 520 nm. GFP was excited with a 488 nm laser line and fluorescence was collected between 490 and 530 nm. mVenus was excited with a 514 nm laser line and fluorescence was collected between 520 and 560 nm. For propidium iodide (PI) fluorescence, an excitation wavelength of 488 nm was used, whereas emission was collected between 600 and 670 nm. RFP and FM4-64 fluorescence was excited at 561 nm and emission was collected between 560 and 620 nm (RFP) or between 675 and 790 nm (FM4-64). SCRI Renaissance 2200 was excited with the 405 nm laser and fluorescence was collected between 425 and 475 nm. Images were analysed with ImageJ/Fiji (https://imagej.net/software/fiji/).
Plasmid construction
All constructs were produced using GreenGate cloning (Lampropoulos et al., 2013) with modules and primers listed in Table S2. PCR products were generated using Q5® High-Fidelity DNA Polymerase (NEB) and column-purified by using GeneJET PCR Purification Kit (Thermo Fisher Scientific), followed by restriction digest with Eco31I FD restriction enzyme (Thermo Fisher Scientific) at 37°C for 15 min. Products were column-purified as described above. Empty entry vectors (pGGA000, pGGC000; Lampropoulos et al., 2013) were digested and purified separately. Digested and purified insert and vector were ligated using Instant Sticky-end Ligase Master Mix (New England Biolabs) following the manufacturer's instructions. The ligation products were then used to transform chemically competent Escherichia coli strain DH5α or XL1-blue and cultivated in LB medium supplied with Ampicillin. Plasmid sequences were verified by Sanger sequencing. Confirmed entry modules are ligated into intermediate vectors by using a GreenGate reaction (Lampropoulos et al., 2013). The generation of expression plasmids involved the creation of two intermediate constructs, one in pGGM000 carrying the GR-LhG4 expression cassette, and one in pGGN000 carrying the PMEI5 coding sequence under control of the pOp6 promoter. The assembly of two expression cassettes each carried by one intermediate vector was achieved by running the same GreenGate reaction and replacing the entry module and empty intermediate vector by an intermediate module and an empty pGGZ001 destination vector, respectively. Final plasmids were verified by colony PCR and restriction digest, and then used to transform Agrobacterium tumefaciens strain ASE (pSOUP+) carrying resistance to chloramphenicol, kanamycin and tetracycline. All constructs were transformed by the floral dip method as previously described (Clough and Bent, 1998; Zhang et al., 2006).
Root staining and analysis
Staining of cell outlines with PI (Sigma-Aldrich, P4170) using a modified pseudo-PI staining method has been performed as described in Truernit et al. (2008), with modifications. Seedlings at 5 DAG were fixed in solution containing 50% methanol and 10% acetic acid for 3 days at 4°C. Samples were then washed twice with H2O and incubated in 1% periodic acid (Sigma-Aldrich, P0430) at room temperature for 40 min. Samples were washed twice with H2O and then stained with 100 μg/ml PI freshly diluted in Schiff's reagent (100 mM sodium metabisulphite, 75 mM HCl). Stained samples were transferred onto microscope slides covered by chloral hydrate solution (4 g chloral hydrate, 1 ml glycerol, 2 ml H2O) and incubated overnight at room temperature in a closed environment. Excess of chloral hydrate solution was removed and several drops of Hoyer's solution (3 g gum arabic, 20 g chloral hydrate, 2 g glycerol, 5 ml H2O) was added to the samples, which were covered gently by cover slips and kept at room temperature for 3 days before imaging. Alternatively (Fig. S1), roots were fixed in paraformaldehyde and stained with SCRI Renaissance 2200 (Renaissance Chemicals) as previously described (Musielak et al., 2015) and cleared in ClearSee (Kurihara et al., 2015). Samples were mounted in ClearSee using imaging chambers.
Orientation of mitotic structures, RAM length and meristematic cell number were measured using ImageJ. The meristem was defined as the region between the quiescent centre and the first cortex cells with twice the length of the previous cell. Directional growth of the roots was analysed by determining the vertical growth index, i.e. the distance of base of the root to the tip divided by root length. Quantification of aberrant cell wall orientation per root was performed by counting the number of cross walls with >15° deviance from transverse orientation in median confocal sections. If not stated otherwise, only cortex cell files were considered. In diagrams, stacked bars denoting more than three aberrant cell walls per root are assigned the same colour for clarity, but statistics were performed with the actual numerical and un-binned values.
QRT-PCR analysis
Seedling roots were harvested at 5 DAG and directly frozen in liquid nitrogen. After tissue homogenisation, RNA extraction was carried out using the GeneMATRIX Universal RNA Purification Kit (EURx, 3598), following the manufacturer's instructions. cDNA synthesis and QRT-PCR analysis was performed as previously described (Holzwart et al., 2020). PMEI5 cDNA was amplified with primers 5′-ACGTGCTTTTAATTGCGATAACG-3′ and 5′-GAAGTCCAAGTTCCCAAGCTG-3. All experiments have been repeated at least twice, with identical results.
Supplementary Material
Acknowledgements
The authors thank Sabine Müller for the YFP-POK1 line.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Z.L., H.H., S.S.-G., S.W.; Validation: L.G.; Formal analysis: Z.L., A.S., Y.F., L.G., S.S.-G., S.W.; Investigation: Z.L., A.S., Y.F.; Writing - original draft: S.S.-G., S.W.; Writing - review & editing: S.S.-G., S.W.; Visualization: Z.L., A.S., Y.F.; Supervision: H.H., S.S.-G., S.W.; Project administration: S.S.-G., S.W.; Funding acquisition: S.S.-G., S.W.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through grant WO 1660/6-1 and through the DFG's Emmy Noether Programme (WO 1660/2 to S.W.). This work in the Savaldi-Goldstein lab was supported by the Israel Science Foundation (grant 1725/18). Deposited in PMC for immediate release.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199504.
References
- Ackerman-Lavert, M. and Savaldi-Goldstein, S. (2019). Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 53, 90-97. 10.1016/j.pbi.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Asami, T., Min, Y. K., Nagata, N., Yamagishi, K., Takatsuto, S., Fujioka, S., Murofushi, N., Yamaguchi, I. and Yoshida, S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123, 93-100. 10.1104/pp.123.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster, G. T. S., Fiorani, F. and Inzé, D. (2003). Cell cycle: the key to plant growth control? Trends Plant Sci. 8, 154-158. 10.1016/S1360-1385(03)00046-3 [DOI] [PubMed] [Google Scholar]
- Besson, S. and Dumais, J. (2011). Universal rule for the symmetric division of plant cells. Proc. Natl. Acad. Sci. USA 108, 6294-6299. 10.1073/pnas.1011866108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson, S. and Dumais, J. (2014). Stochasticity in the symmetric division of plant cells: when the exceptions are the rule. Front. Plant Sci. 5, 538. 10.3389/fpls.2014.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado, A., Yin, Y., Yu, C., Vafeados, D., Mora-García, S., Cheng, J.-C., Nam, K. H., Li, J. and Chory, J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131, 5341-5351. 10.1242/dev.01403 [DOI] [PubMed] [Google Scholar]
- Chakrabortty, B., Willemsen, V., de Zeeuw, T., Liao, C.-Y., Weijers, D., Mulder, B. and Scheres, B. (2018). A plausible microtubule-based mechanism for cell division orientation in plant embryogenesis. Curr. Biol. 28, 3031-3043.e3032. 10.1016/j.cub.2018.07.025 [DOI] [PubMed] [Google Scholar]
- Chory, J., Nagpal, P. and Peto, C. A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in arabidopsis. Plant Cell 3, 445-459. 10.2307/3869351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, A. L. and Smith, L. G. (1998). The Tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 10, 1875-1888. 10.1105/tpc.10.11.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J. and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cole, R. A., McInally, S. A. and Fowler, J. E. (2014). Developmentally distinct activities of the exocyst enable rapid cell elongation and determine meristem size during primary root growth in Arabidopsis. BMC Plant Biol. 14, 386. 10.1186/s12870-014-0386-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D. J. (2016). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 67, 463-476. 10.1093/jxb/erv511 [DOI] [PubMed] [Google Scholar]
- Craft, J., Samalova, M., Baroux, C., Townley, H., Martinez, A., Jepson, I., Tsiantis, M. and Moore, I. (2005). New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41, 899-918. 10.1111/j.1365-313X.2005.02342.x [DOI] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K. and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71-84. 10.1242/dev.119.1.71 [DOI] [PubMed] [Google Scholar]
- Feraru, E., Feraru, M. I., Kleine-Vehn, J., Martinière, A., Mouille, G., Vanneste, S., Vernhettes, S., Runions, J. and Friml, J. (2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21, 338-343. 10.1016/j.cub.2011.01.036 [DOI] [PubMed] [Google Scholar]
- Flanders, D. J., Rawlins, D. J., Shaw, P. J. and Lloyd, C. W. (1990). Nucleus-associated microtubules help determine the division plane of plant epidermal cells: avoidance of four-way junctions and the role of cell geometry. J. Cell Biol. 110, 1111-1122. 10.1083/jcb.110.4.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, Y., Elkouby, L., Holland, N., Vragović, K., Elbaum, R. and Savaldi-Goldstein, S. (2014). Root growth is modulated by differential hormonal sensitivity in neighboring cells. Genes Dev. 28, 912-920. 10.1101/gad.239335.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatis, B., Apostolakos, P. and Katsaros, C. (1984). Positional inconsistency between preprophase microtubule band and final cell plate arrangement during triangular subsidiary cell and atypical hair cell formation in two Triticum species. Can. J. Bot. 62, 343-359. 10.1139/b84-053 [DOI] [Google Scholar]
- González-García, M. P., Vilarrasa-Blasi, J., Zhiponova, M., Divol, F., Mora-Garcia, S., Russinova, E. and Cano-Delgado, A. I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849-859. 10.1242/dev.057331 [DOI] [PubMed] [Google Scholar]
- Graeff, M., Rana, S., Marhava, P., Moret, B. and Hardtke, C. S. (2020). Local and systemic effects of brassinosteroid perception in developing phloem. Curr. Biol. 30, 1626-1638.e1623. 10.1016/j.cub.2020.02.029 [DOI] [PubMed] [Google Scholar]
- Gu, F., Bringmann, M., Combs, J. R., Yang, J., Bergmann, D. C. and Nielsen, E. (2016). Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell 28, 1722-1737. 10.1105/tpc.16.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham, Y., Holland, N., Butterfield, C., Ubeda-Tomas, S., Bennett, M. J., Chory, J. and Savaldi-Goldstein, S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138, 839-848. 10.1242/dev.061804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzwart, E., Huerta, A. I., Glockner, N., Garnelo Gomez, B., Wanke, F., Augustin, S., Askani, J. C., Schurholz, A. K., Harter, K. and Wolf, S. (2018). BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc. Natl. Acad. Sci. USA 115, 11838-11843. doi:10.1073/pnas.1814434115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzwart, E., Wanke, F., Glockner, N., Hofte, H., Harter, K. and Wolf, S. (2020). A mutant allele uncouples the brassinosteroid-dependent and independent functions of BRASSINOSTEROID INSENSITIVE 1. Plant Physiol. 182, 669-678. 10.1104/pp.19.00448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Dumond, M., Zhu, M., Tsugawa, S., Li, C.-B., Boudaoud, A., Hamant, O. and Roeder, A. H. K. (2018). Heterogeneity and robustness in plant morphogenesis: from cells to organs. Annu. Rev. Plant Biol. 69, 469-495. 10.1146/annurev-arplant-042817-040517 [DOI] [PubMed] [Google Scholar]
- Kang, Y. H., Breda, A. and Hardtke, C. S. (2017). Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development 144, 272-280. 10.1242/dev.145623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, D. R. and Hagemann, W. (1991). The relationship of cell and organism in vascular plants. Bioscience 41, 693-703. 10.2307/1311764 [DOI] [Google Scholar]
- Kurihara, D., Mizuta, Y., Sato, Y. and Higashiyama, T. (2015). ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168-4179. 10.1242/dev.127613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos, A., Sutikovic, Z., Wenzl, C., Maegele, I., Lohmann, J. U. and Forner, J. (2013). GreenGate---a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8, e83043. 10.1371/journal.pone.0083043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani, E. R., Khan, G. A., Somssich, M. and Persson, S. (2018). Building a plant cell wall at a glance. J. Cell Sci. 131, jcs207373. 10.1242/jcs.207373 [DOI] [PubMed] [Google Scholar]
- Lanza, M., Garcia-Ponce, B., Castrillo, G., Catarecha, P., Sauer, M., Rodriguez-Serrano, M., Paez-Garcia, A., Sanchez-Bermejo, E., TC, M., Leo del Puerto, Y.et al. (2012). Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 22, 1275-1285. 10.1016/j.devcel.2012.04.008 [DOI] [PubMed] [Google Scholar]
- Lintilhac, P. M. and Vesecky, T. B. (1984). Stress-induced alignment of division plane in plant tissues grown in vitro. Nature 307, 363-364. 10.1038/307363a0 [DOI] [Google Scholar]
- Lipka, E., Gadeyne, A., Stockle, D., Zimmermann, S., De Jaeger, G., Ehrhardt, D. W., Kirik, V., Van Damme, D. and Muller, S. (2014). The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26, 2617-2632. 10.1105/tpc.114.124933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livanos, P. and Müller, S. (2019). Division plane establishment and cytokinesis. Annu. Rev. Plant Biol. 70, 239-267. 10.1146/annurev-arplant-050718-100444 [DOI] [PubMed] [Google Scholar]
- Louveaux, M., Julien, J.-D., Mirabet, V., Boudaoud, A. and Hamant, O. (2016). Cell division plane orientation based on tensile stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 113, E4294-E4303. 10.1073/pnas.1600677113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel, A., von Wangenheim, D., Federici, F., Haseloff, J. and Stelzer, E. H. K. (2011). High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J. 68, 377-385. 10.1111/j.1365-313X.2011.04692.x [DOI] [PubMed] [Google Scholar]
- Marcus, A. I., Li, W., Ma, H. and Cyr, R. J. (2003). A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 14, 1717-1726. 10.1091/mbc.e02-09-0586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, P., Luo, A., Sylvester, A. and Rasmussen, C. G. (2017). Proper division plane orientation and mitotic progression together allow normal growth of maize. Proc. Natl. Acad. Sci. USA 114, 2759-2764. 10.1073/pnas.1619252114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, P., Allsman, L. A., Brakke, K. A., Hoyt, C., Hayes, J., Liang, H., Neher, W., Rui, Y., Roberts, A. M., Moradifam, A.et al. (2018). Predicting division planes of three-dimensional cells by soap-film minimization. Plant Cell 30, 2255-2266. 10.1105/tpc.18.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniere, A., Lavagi, I., Nageswaran, G., Rolfe, D. J., Maneta-Peyret, L., Luu, D.-T., Botchway, S. W., Webb, S. E. D., Mongrand, S., Maurel, C.et al. (2012). Cell wall constrains lateral diffusion of plant plasma-membrane proteins. Proc. Natl. Acad. Sci. USA 109, 12805-12810. 10.1073/pnas.1202040109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, J. F., Rolfe, D. J., Webb, S. E. D., Tolmie, A. F., Botchway, S. W., Martin-Fernandez, M. L., Hawes, C. and Runions, J. (2019). The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc. Natl. Acad. Sci. USA 116, 12857-12862. 10.1073/pnas.1819077116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miart, F., Desprez, T., Biot, E., Morin, H., Belcram, K., Höfte, H., Gonneau, M. and Vernhettes, S. (2014). Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J. 77, 71-84. 10.1111/tpj.12362 [DOI] [PubMed] [Google Scholar]
- Moore, I., Galweiler, L., Grosskopf, D., Schell, J. and Palme, K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95, 376-381. 10.1073/pnas.95.1.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukhtar, J., Trubuil, A., Belcram, K., Legland, D., Khadir, Z., Urbain, A., Palauqui, J.-C. and Andrey, P. (2019). Cell geometry determines symmetric and asymmetric division plane selection in Arabidopsis early embryos. PLoS Comput. Biol. 15, e1006771. 10.1371/journal.pcbi.1006771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, S., Han, S. and Smith, L. G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16, 888-894. 10.1016/j.cub.2006.03.034 [DOI] [PubMed] [Google Scholar]
- Musielak, T. J., Schenkel, L., Kolb, M., Henschen, A. and Bayer, M. (2015). A simple and versatile cell wall staining protocol to study plant reproduction. Plant Reprod. 28, 161-169. 10.1007/s00497-015-0267-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., Fujioka, S., Sunohara, H., Kamiya, N., Hong, Z., Inukai, Y., Miura, K., Takatsuto, S., Yoshida, S., Ueguchi-Tanaka, M.et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140, 580-590. 10.1104/pp.105.072330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, T., Vukasinovic, N., Liu, D., Russinova, E. and Yin, Y. (2019). Brassinosteroids: multi dimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295-318. 10.1105/tpc.19.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oud, J. L. and Nanninga, N. (1992). Cell shape, chromosome orientation and the position of the plane of division in Vicia faba root cortex cells. J. Cell Sci. 103, 847. 10.1242/jcs.103.3.847 [DOI] [Google Scholar]
- Planas-Riverola, A., Gupta, A., Betegon-Putze, I., Bosch, N., Ibanes, M. and Cano-Delgado, A. I. (2019). Brassinosteroid signaling in plant development and adaptation to stress. Development 146, dev151894. 10.1242/dev.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, C. G. and Bellinger, M. (2018). An overview of plant division-plane orientation. New Phytol. 219, 505-512. 10.1111/nph.15183 [DOI] [PubMed] [Google Scholar]
- Rasmussen, C. G., Sun, B. and Smith, L. G. (2011). Tangled localization at the cortical division site of plant cells occurs by several mechanisms. J. Cell Sci. 124, 270-279. 10.1242/jcs.073676 [DOI] [PubMed] [Google Scholar]
- Rasmussen, C. G., Wright, A. J. and Müller, S. (2013). The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J. 75, 258-269. 10.1111/tpj.12177 [DOI] [PubMed] [Google Scholar]
- Rui, Y. and Dinneny, J. R. (2020). A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428-1439. 10.1111/nph.16166 [DOI] [PubMed] [Google Scholar]
- Schaefer, E., Belcram, K., Uyttewaal, M., Duroc, Y., Goussot, M., Legland, D., Laruelle, E., de Tauzia-Moreau, M.-L., Pastuglia, M. and Bouchez, D. (2017). The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356, 186-189. 10.1126/science.aal3016 [DOI] [PubMed] [Google Scholar]
- Schürholz, A.-K., López-Salmerón, V., Li, Z., Forner, J., Wenzl, C., Gaillochet, C., Augustin, S., Barro, A. V., Fuchs, M., Gebert, M.et al. (2018). A comprehensive toolkit for inducible, cell type-specific gene expression in arabidopsis. Plant Physiol. 178, 40-53. 10.1104/pp.18.00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. P. and Savaldi-Goldstein, S. (2015). Growth control: brassinosteroid activity gets context. J. Exp. Bot. 66, 1123-1132. 10.1093/jxb/erv026 [DOI] [PubMed] [Google Scholar]
- Smertenko, A., Assaad, F., Baluška, F., Bezanilla, M., Buschmann, H., Drakakaki, G., Hauser, M.-T., Janson, M., Mineyuki, Y., Moore, I.et al. (2017). Plant cytokinesis: terminology for structures and processes. Trends Cell Biol. 27, 885-894. 10.1016/j.tcb.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Smith, L. G. (2001). Plant cell division: building walls in the right places. Nat. Rev. Mol. Cell Biol. 2, 33-39. 10.1038/35048050 [DOI] [PubMed] [Google Scholar]
- Smith, L. G., Hake, S. and Sylvester, A. W. (1996). The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122, 481-489. 10.1242/dev.122.2.481 [DOI] [PubMed] [Google Scholar]
- Stöckle, D., Herrmann, A., Lipka, E., Lauster, T., Gavidia, R., Zimmermann, S. and Müller, S. (2016). Putative RopGAPs impact division plane selection and interact with kinesin-12 POK1. Nat. Plants 2, 16120. 10.1038/nplants.2016.120 [DOI] [PubMed] [Google Scholar]
- Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., Rédei, G. P., Nagy, F., Schell, J. and Koncz, C. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171-182. 10.1016/S0092-8674(00)81094-6 [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz, R. A. and Jurgens, G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967-2978. 10.1242/dev.120.10.2967 [DOI] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D. and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676-677. 10.1038/375676a0 [DOI] [Google Scholar]
- Truernit, E., Bauby, H., Dubreucq, B., Grandjean, O., Runions, J., Barthélémy, J. and Palauqui, J.-C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. Plant Cell 20, 1494-1503. 10.1105/tpc.107.056069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtera, L., Schulz, J. and Hamann, T. (2019). Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924-932. 10.1038/s41477-019-0502-0 [DOI] [PubMed] [Google Scholar]
- Voxeur, A. and Höfte, H. (2016). Cell wall integrity signaling in plants: “To grow or not to grow that's the question”. Glycobiology 26, 950-960. 10.1093/glycob/cww029 [DOI] [PubMed] [Google Scholar]
- Vragović, K., Sela, A., Friedlander-Shani, L., Fridman, Y., Hacham, Y., Holland, N., Bartom, E., Mockler, T. C. and Savaldi-Goldstein, S. (2015). Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. USA 112, 923-928. 10.1073/pnas.1417947112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, K. L., Müller, S., Moss, D., Ehrhardt, D. W. and Smith, L. G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17, 1827-1836. 10.1016/j.cub.2007.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, S. (2017). Plant cell wall signalling and receptor-like kinases. Biochem. J. 474, 471-492. 10.1042/BCJ20160238 [DOI] [PubMed] [Google Scholar]
- Wolf, S., Mravec, J., Greiner, S., Mouille, G. and Höfte, H. (2012). Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr. Biol. 22, 1732-1737. 10.1016/j.cub.2012.07.036 [DOI] [PubMed] [Google Scholar]
- Wolf, S., van der Does, D., Ladwig, F., Sticht, C., Kolbeck, A., Schurholz, A.-K., Augustin, S., Keinath, N., Rausch, T., Greiner, S.et al. (2014). A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA 111, 15261-15266. 10.1073/pnas.1322979111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Barbier de Reuille, P., Lane, B., Bassel, G. W., Prusinkiewicz, P., Smith, R. S. and Weijers, D. (2014). Genetic control of plant development by overriding a geometric division rule. Dev. Cell 29, 75-87. 10.1016/j.devcel.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Henriques, R., Lin, S.-S., Niu, Q.-W. and Chua, N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641-646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Iakovidis, M. and Costa, S. (2016). Control of patterns of symmetric cell division in the epidermal and cortical tissues of the Arabidopsis root. Development 143, 978-982. 10.1242/dev.129502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, A., Wang, H., Walker, J. C. and Li, J. (2004). BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 40, 399-409. 10.1111/j.1365-313X.2004.02214.x [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.-W., Nishizawa, N., Wu, Y., Kost, B. and Chua, N.-H. (2000). KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12, 1137-1152. 10.1105/tpc.12.7.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.