Abstract
Introduction
Prosthetic joint infections (PJIs) are challenging to treat therapeutically because the infectious agents often are resistant to antibiotics and capable of abundant growth in surface-attached biofilms. Though infection rates are low, ca. 1–2 %, the overall increase in the sheer number of joint replacement surgeries results in an increase in patients at risk.
Aims
This study investigates the consensus of microbial species comprising PJI ecology, which is currently lacking.
Methodology
In this study, PJI populations from seven patients were analysed using combined culturing and whole-genome shotgun sequencing (WGSS) to establish population profiles and compare WGSS and culture methods for detection and identification of the PJI microbiome.
Results
WGSS detected strains when culture did not, notably dormant, culture-resistant and rare microbes. The CosmosID algorithm was used to predict micro-organisms present in the PJI and discriminate contaminants. However, culturing indicated the presence of microbes falling below the WGSS algorithm threshold. In these instances, microbes cultured are believed to be minor species. The two strategies were combined to build a population profile.
Conclusion
Variability between and among PJIs showed that most infections were distinct and unique. Comparative analysis of populations revealed PJIs to form clusters that were related to, but separate from, vaginal, skin and gut microbiomes. Fungi and protists were detected by WGSS, but the role of fungi is just beginning to be understood and for protists it is unknown. These micro-organisms and their novel and strain-specific microbial interactions remain to be determined in current clinical tests.
Keywords: prosthetic joint infection, microbiology, whole-genome sequencing, microbiome, polymicrobial
Introduction
Prosthetic joint infections (PJIs) occur in 1–2 % of prosthetic joint surgery patients [1]. Even though this represents only a small percentage of individuals at risk, the infection can be devastating and even deadly. Estimates are that the number of prosthetic hip and knee replacements will increase 174 and 673 %, respectively, between 2005–2030, with concomitant increase in the incidence of infection [2].
PJIs are particularly difficult to treat, in part because biofilms tend to form and these are notoriously resistant to antibiotics [1, 3]. It has also been reported that PJIs infected with Pseudomonas spp., methicillin-resistant Staphylococcus aureus (MRSA) or Proteus spp. that have been found to be associated with biofilms, are linked to poorer patient outcomes [4]. PJIs are notoriously polymicrobial and such cases are associated with poorer outcomes compared to monomicrobial infections [5, 6]. Polymicrobial populations carry the potential for forming stable biofilms and to interact synergistically to increase resistance to both antibiotics and immune response [7]. Polymicrobial populations comprise an added complexity, because virulence, biofilm formation and antibiotic resistance are influenced by biochemical interactions specific to unique microbial combinations.
To characterize the complete PJI population, improved culturing methods can be employed to detect and identify more of the species, such as slow-growing bacteria [8–10]. Biofilms, however, sequester microbes and require additional sampling steps [11]. Furthermore, biofilms frequently contain culture-resistant microbes that thrive in a biofilm, detectable by sequencing, but frequently not by culture [12]. Several studies of PJIs have shown an advantage of non-culture-based methods for microbial detection and identification. Notably, PCR-based methods that are relatively fast and cost-effective but limited to identification of one or only a few specific agents. PCR primers are used to target specific suspected PJI microbial agents, leaving novel or unsuspected species unidentified [13, 14]. Application of 16S rRNA sequencing can be used to identify novel or unsuspected organisms to species, but not to the level of strain [15, 16]. Whole-genome shotgun sequencing (WGSS) employed in this study identifies bacteria, but also protists, fungi, viruses and detects antibiotic resistance and virulence genes, thereby providing valuable insight into virulence and related properties of species and strains comprising the PJI ecology [17, 18].
Differentiating sample contaminants from pathogens is also a challenge since some of the micro-organisms commonly isolated from nosocomial infections, including PJIs, have been recognized as pathogens only within the last few decades [19–22]. PJI diagnosis can be improved by using newly developed bioinformatics to detect and define the causative agents in PJI.
Since a PJI microbiome may comprise groups of micro-organisms not typical of the natural environment, many associated biochemical interactions need to be described. Furthermore, while in vivo studies can provide insight with respect to patient outcomes, interspecies (and even interstrain) interactions need to be determined, and understood if treatment is to be effective. The aim of this study was to characterize the ecology of PJIs, including low abundance and dormant microbes [23]. This was achieved by employing a combination of WGSS and standard clinical culturing methods. Metagenomic sequencing methods have only recently been explored for their ability to identify microbes of PJIs, finding examples when both sequencing and culturing do not detect species [24, 25]. The micro-organisms detected and identified by both methods were compared and a preliminary description of the microbial ecology of the PJI developed.
Methods
Sample processing and culturing
PJI fluid collected during prosthetic replacement and suspected PJI fluid aspirated during an office visit comprised the sample set for this study. Altogether, 50 µl of each sample was plated in triplicate on Brain Heart Infusion agar (Dot Scientific, Burton, MI, USA), Tryptic soy broth agar (Dot Scientific) with 5 % sheep’s blood (Hardy Diagnostics, Springboro, OH, USA), Columbia blood agar (5 % blood) (Acumedia, Lansing, MI, USA), Sabouraud agar (Dot Scientific), and Brucella agar (Acumedia). All plates were incubated at 37 °C for 14 days. Brucella agar was incubated anaerobically using a BD GasPak EZ Gas Generating Container Systems (BD, Franklin Lakes, NJ, USA). An uninoculated control plate was included for all media and culture conditions to identify any plate contamination. Colonies identified on day 14 were streaked to single colony purification, PCR amplified using 16S rRNA primers 8F and 1525R, and sequenced using 8F, 533F and 1525R primers. Resultant sequences were assembled and subjected to NCBI nucleotide blast search, using the non-redundant nucleotide database and Megablast. Species that were identified had >97 % identity with the query sequence. Ca. 200 µl was used to isolate genomic DNA employing QIAamp Fast DNA Tissue Kit (Qiagen, Germantown, MD, USA). The remainder of the sample served as a glycerol stock culture, prepared by combining that portion of the sample with 70 % glycerol (VWR, Radnor, PA, USA) (1 : 1) or was preserved in Zymo DNA/RNA shield (Zymo Research, Irvine, CA, USA).
Whole-genome shotgun sequencing and metagenomic bioinformatics analysis
DNA was quantified using fluorometer Qubit 3.0 and Fragment libraries were constructed from 50 to 200 ng DNA, using the IonXpress Plus Fragment Library kit (ThermoFisher Scientific, USA), according to the manufacturer’s instructions. The libraries were pooled by adding an equimolar ratio of each based on concentration determined by qPCR, using TaqMan quantification Kit. The libraries were sequenced on an Ion S5XL sequencer (ThermoFisher Scientific, USA) to generate 200 bp sequence reads. Each sample was sequenced targeting 15–20M sequence read depth. A few of the samples that showed discordant results between culture and sequencing, were re-sequenced using the Nextera XT library protocol (Illumina, San Diego, CA, USA) and Illumina HiSeq v3 chemistry (Illumina, San Diego, CA, USA) targeting 50–60 M paired end 150 bp read depth. Unassembled sequencing reads were directly analysed by the CosmosID bioinformatics platform (CosmosID, Rockville, MD, USA), as described elsewhere [17, 26–28] for multi-kingdom microbiome composition analysis and quantification of relative abundances at all taxonomic levels. Briefly, the system utilizes highly curated, phylogenetically organized databases (GenBook) comprising over 150 000 genomes and gene sequences and a high-performance data-mining algorithm that rapidly disambiguates hundreds of millions of sequence reads from a metagenomic sample into discrete micro-organisms engendering the particular sequences. High performance bioinformatics enables speed and accuracy in microbial identification and the phylogenetically organized curated database helps achieve finer resolution of identification, differentiating different strains within a species, discrimination of pathogens from ‘near neighbours’, and accurate measurement of relative abundance. The pipeline has two separable comparators. The first consists of a pre-computation phase and a per-sample computation. The input to the pre-computation phase is a reference microbial database and the output is a whole-genome phylogenetic tree, together with sets of k-mer fingerprints (biomarkers) that are uniquely identified with distinct branch, nodes and leaves of the tree. The second per-sample, computational phase searches the hundreds of millions of sequence reads against the fingerprint sets in minutes. The resulting statistics are analysed to achieve fine-grain composition and relative abundance estimates at all branches, nodes and leaves of the tree. The second comparator uses edit distance-scoring techniques to compare a target sample with a reference set. Overall classification precision is maintained by aggregation statistics. The first comparator finds reads for which there is an exact match with a k-mer uniquely identified with one or a set of reference strains; the second comparator statistically scores the entire read against the reference to verify that the read is indeed uniquely identified with that set. Therefore, we can precisely attribute reads to different strains of the databases that occupy a distinct phylogenetic leaf in the tree using strain-specific (unique) k-mer sequences and differentiating them or determining their novelty, using aggregated statistics of the strain compared to its phylogenetic near neighbour. The resultant taxa abundance tables were used to calculate observed and expected species richness, alpha diversity indices and beta diversity distance matrices. Principle coordinate analysis (PCoA) was performed to cluster samples based on abundance Jaccard distance matrix (community structure).
Discrimination of samples belonging to PJI and different body sites was done using Jaccard’s distance. Jaccard’s distance was calculated from relative abundance matrix of the species between each pair of the samples using the beta-dispersion method of the Vegan package of R (https://CRAN.R-project.org/package=vegan). In order to evaluate how close the clusters are to each other, a pair-wise PERMANOVA test was performed between the PJI cluster and a cluster belonging to each of the body sites one by one using the ‘permutest’ function of the Vegan package and the ‘TukeyHSD’ function was used to assign t-scores to the distances. Lower t-scores indicate the closer the body site belonging samples to the samples of the PJI in terms of sharing the same microbial community.
In order identify subtle differences of microbial communities between different body sites and the PJI with guidance of phylogenetic relationships amongst taxa, a phylogenetic aware distance analysis was performed adopting the Unifrac approach through Python package ‘Unifrac’ [29]. The phylogenetic guidance tree was constructed from the 16S rRNA sequence derived from the GreenGene database for entire species identified in the analysis. Multiple sequence analysis (MSA) was performed on all the 16S rRNA sequences using the ClustalW tool [30] and output of the MSA was sent to construct a phylogenetic tree using the UPGMA algorithm of the BioPython packages. The file of the constructed guidance tree in newick (.nwk) format was used as the input for the Unifrac analysis and a distance matrix was obtained, which was used to construct a PCoA plot through Vegan package using the ‘Unweighted Unifrac Distance’ option and F-value was calculated based on permutation-based ANOVA (PERMANOVA) test from the Vegan package.
Growth of Cutibacterium acnes on selective media
C. acnes ATCC strains 11828, 11827 and 6919 were streaked in triplicate onto Medium 14 (Sodium Lactate 4.0 g l−1, Yeast Extract 0.5 g l−1) and incubated under oxygen-limited conditions for 14 days at 37 °C using a BD GasPak EZ Gas Generating Container System.
Targeted culturing
Glycerol stocks of original PJI samples were re-streaked using plates and media as noted in Table 2. Colonies were picked and identified by 16S rRNA sequencing as described above.
Table 2.
Targeted culturing of suspected PJI strains
|
Sample |
Medium |
Species identification sought |
Species cultured |
|---|---|---|---|
|
A |
Modified FAB |
None |
|
|
A |
MSA |
(RA <1) |
E. faecalis (previously identified) |
|
C |
MSA |
S. epidermidis, S. hominis |
S. aureus (previously identified) |
|
D |
MSA |
None |
|
|
A |
Brucella Blood (anaerobic) |
E. faecalis (previously identified) |
|
|
C |
Brucella Blood (anaerobic) |
S. aureus (previously identified) |
|
|
B,D,1,15 |
Brucella Blood (anaerobic) |
C. acnes / |
None |
|
A,C,1,15 |
LB (Lysogeny broth) (anaerobic) |
C. acnes/ |
None |
|
A,C,1,15 |
Medium 14 (anaerobic) |
C. acnes/ |
S. aureus (sample C, previously identified) E. faecalis (sample A, previously identified) |
Results
Bacterial populations of PJIs
Fluids subjected to study were from five PJIs collected at the time of surgical intervention and two suspected infections confirmed by physicians during office visits. PJI diagnosis is made with a combination of signs and tests. Clinical suspicion comes with a picture of increased pain, swelling or redness with drainage of purulent material being highly indicative of PJI. Suspicion of infection leads to laboratory tests of Westergreen sedimentation rate and C reactive protein. Elevation of these tests and a clinical suspicion prompts joint aspiration. Elevated white count in the acute or late postoperative joint aspirations and/or positive cultures is indicative of infection. Additionally, sensitive tests such as alpha defensin and esterases can indicate PJI.
In the laboratory, each sample was split into two fractions. Genomic DNA was isolated from one fraction and subjected to whole-genome shotgun sequencing. The second fraction served as inoculum for culturing on and enrichment in multiple media types (see Methods) under aerobic and anaerobic conditions. Plates were incubated for 14 days to allow growth of slow-growing micro-organisms. Colonies were picked, cultured in LB broth, and analysed by 16S rRNA sequencing. Whole-genome shotgun metagenomics coupled with the CosmosID algorithm was used to predict organisms present in the infection versus contaminant. Culturing results, however, suggest the presence of some microbes that fall below the algorithm threshold. Additionally, a negative control of microbial DNA-free water indicates known microbes unique to this study. Combining this information with identified phages and virulence genes as well as past studies we comment on infection composition and diversity.
Shotgun metagenomics offers the opportunity to sequence the entire genome of an organism allowing characterization of all core and accessory genes and genetic elements associated with an organism, hence if a given bacteriophage that infects certain genera and species is detected by metagenomic sequencing then it can be used as additional evidence of the presence of their bacterial host. As an example, two groups of bacteriophages that infect E. coli, somatic and F-specific coliphages, have been used as both faecal bacterial and viral indicators in academia for many years [31–34], Additionally, bacteriophages of B. fragilis have also been used as indicators of human faecal bacterial contamination [35]. However, such evidence should not be used in isolation as the degree of host specificity could be wide-ranging and depends on the bacteriophage.
Micro-organisms determined to be of relative abundance of 1 % or greater and species identified by culturing are listed in Table 1, with a complete listing in Table S1 (available in the online version of this article), and sequences available at NCBI Short Read Achieve (https://www.ncbi.nlm.nih.gov/sra). Results of whole-genome shotgun sequencing to identify bacteria, as well as phages and virulence factors are provided in Table 1. Phages and virulence factors can serve as markers to support the presence of a microbe within the sample [36], while Qiagen microbial DNA free water was analysed to identify contaminants originating from laboratory procedures, which have been shown to be variable based on the DNA preparation kits used. Common contaminants were also often the same species found in PJIs [22].
Table 1.
PJI bacterial species identified by WGSS and culturing
|
Sample |
Bacterial species identified |
Relative abundance (%) |
Cultured |
Phage present |
Virulence factor present |
Present in negative control |
|---|---|---|---|---|---|---|
|
A |
Pseudomonas aeruginosa* |
65.11 |
x |
x |
||
|
|
Streptococcus australis* |
12.34 |
x |
|||
|
|
Streptococcus ssp* |
5.376 |
x |
|||
|
|
Propionibacterium (Cutibacterium) acnes* |
4.487 |
x |
|||
|
|
Klebsiella pneumonia* |
3.872 |
x |
|||
|
|
Enterobacter ssp. |
2.259 |
x |
|||
|
|
2.125 |
x |
x |
x |
||
|
|
0.04511 |
x |
||||
|
B |
Bacteroides fragilis* |
81.37 |
x |
|||
|
|
Lactobacillus ssp. |
3.508 |
x |
|||
|
|
Bacteroides ssp. |
1.735 |
x |
|||
|
|
2.705 |
x |
||||
|
|
Bifidobacterium Longum subsp. Infantis |
2.619 |
||||
|
|
1.688 |
x |
||||
|
|
Clostridiales order |
1.621 |
||||
|
|
1.125 |
x |
||||
|
C |
47.14 |
|||||
|
|
27.27 |
x |
x |
|||
|
|
18.86 |
|||||
|
|
6.729 |
|||||
|
D |
32.22 |
|||||
|
|
25.49 |
|||||
|
|
Candidate division TM7 |
18.83 |
||||
|
|
14.25 |
|||||
|
|
11.3 |
x |
||||
|
E |
100 |
x |
||||
|
|
|
(74 reads/ 54 million) |
||||
|
|
|
(6 reads/ 54 million) |
||||
|
|
|
(2 reads/ 54 million) |
||||
|
1 |
Propionibacterium ssp.* |
57.04 |
x |
|||
|
|
Clostridium autoethanogenum* |
10.81 |
||||
|
|
Clostridium Ijungdahlii* |
10.78 |
||||
|
|
Lachnospiraceae bacterium* |
6.602 |
||||
|
|
1.6 |
|||||
|
|
|
(9 reads/ 46 million) |
||||
|
15 |
Propionibacterium ssp.* |
48.59 |
x |
|||
|
|
Propionibacterium acnes* |
43.83 |
x |
|||
|
|
Staphylococcus epidermidis* |
2.433 |
x |
|||
|
|
|
(6 reads/ 44 million) |
||||
|
S. aureus control |
99.85 |
n/a |
x |
x |
||
|
P. aeruginosa control |
P. aeruginosa strain AZPAE14876 |
91.61 |
n/a |
x |
x |
x |
|
|
5.78 |
n/a |
x |
|||
|
|
Pseudomonas ssp. |
2.229 |
n/a |
x |
*Denotes bacteria likely to be present in the sample, based on the CosmosID algorithm.
For sample A, more than 30 species of bacteria were identified (Table S1), with seven having relative abundance greater than 1.0 % (Table 1). Of these seven, five were identified by the CosmosID algorithm as likely to be present (noted by asterisk). Streptococcus , Enterobacter and Klebsiella pneumoniae appear in the microbial DNA-free water sample specific to this study. Pseudomonas aeruginosa , the most abundant bacterium detected by sequencing, was not able to be cultured. This species was also identified in the microbial DNA-free negative control, but of a different strain. This level of identification is not possible with other methods, notably culture, with the exception of extensive employment of PCR. Additionally, phages and virulence factors related to P. aeruginosa were identified in the sample. Linking genera-specific factors is a tool to support the conclusion that P. aeruginosa was present in sample A [36]. Initial and directed attempts at culturing (as discussed below) were unsuccessful. This may be a result of the sample collection method employed since biofilms are difficult to disrupt. Also, P. aeruginosa is known to become culture-resistant following biofilm growth [12].
P. (C.) acnes is a common contaminant [37], but was identified in sample A at a higher relative abundance compared to microbial DNA-free water control, but falling below the CosmosID algorithm. Additionally, a Propionibacterium phage was identified in high relative abundance, supporting the presence of this organism in the sample. Similar to P. aeruginosa , this bacterial species was not detected in initial or directed culturing and has been shown by other investigators to be culture-resistant [38].
Cultured microbes included E. faecalis and C. striatum (0.04511 RA), both of which were identified by WGSS and have been previously found in PJIs [24, 39], but below the CosmosID algorithm threshold, suggesting these may be low incidence species within the population.
Sample B was culture negative but WGSS detected B. fragilis , not commonly associated with PJI, however, reported cases of infection with B. fragilis are associated with concomitant conditions, suggesting an haematogenous origin [40]. Ivy et al. suggest that metagenomics shotgun sequencing may be a useful tool for culture-negative samples [25].
Interestingly, additional species identified (though falling below the CosmosID algorithm threshold) included Lactobacillus , Bifidobacteria longum, Bacteroides stercoris , Bacteroides caccae and Clostridiales [41–43], all associated with the human gut microbiome. Because these species fall below the algorithm and were present in the negative control, it is concluded they were contaminants.
Bacteria detected by WGSS in sample C were at very low levels. However, amongst these S. aureus was also detected by culture and the presence of this bacterium in the infection was supported by identification of multiple associated virulence factors by WGSS analysis. A similar study of PJIs using metagenomics sequencing also reports instances of S. aureus identified by culture, but not by sequencing [24]. Slow-growing Propionibacterium , found in PJIs [8], was indicated by WGSS to be a predominant species in this PJI. Anaerobic culturing was carried out for 14 days in all cases, as slow-growing species require longer incubation time [8], but Propionibacterium could not be cultured, even when targeted methods were used. While its presence in the negative control and lack of culturing could be interpreted as the Propionibacterium being a contaminating species, its abundance identified within the sample by WGSS supports its presence in the original sample. Both C. acnes (formerly Propionibacterium ) and S. aureus have been shown to be culture-impaired during biofilm growth [38].
Sample D was culture negative, with an interesting mix of bacterial species detected by WGSS, but at very low concentrations. Georgenia , a genus found in the environment, has not been previously reported to occur in infections. S. epidermidis , a common skin bacterium often identified in PJIs [16], was identified by WGSS but not cultured by initial or directed culturing. Candidate division TM7, also identified by sequencing, has been identified in several human microbiomes, but was only recently successfully cultured. TM7 is a potential pathogen and because it is described as an epibiont, identification is not possible using traditional culturing methods [44, 45]. Veillonella and Bacteroides stercoris are both gut microbiome species, with only Veillonella previously identified in PJIs [46].
Sample E revealed only Faecalibacterium , also identified in the negative control. Culture detected S. lugdenensis, S. epidermidis, C. jeikeium, with WGSS also detecting these species with a low number of reads in a second analysis.
Samples 1 and 15 were from suspected infections determined during office visits. Both samples yielded Microbacterium foliorum in culture and also by WGSS. M. foliorum has been identified in clinical specimens, but not reported previously to occur in PJIs [47]. WGSS analysis of sample 15 showed a Propionibacterium phage to be present, supporting the occurrence of C. acnes , a bacterium known to produce infections characterized by milder symptoms [48]. Sample 1 also contained Clostridium autoethanogenum and Clostridium ljungdahlii , not previously reported in PJIs.
Controls were run using Qiagen microbial DNA-free water and P. aeruginosa and S. aureus overnight cultures. Microbial DNA-free water analysed as a negative control yielded a variety of potential contaminants, ostensibly from laboratory and sequencing operations. Species identified were also detected in the PJI samples, the most abundant of which included Bacteroides , Faecalibacterium, Ruminococcus, Alistipes and Parabacteroides . Controls using overnight cultures suggested the isolation/sequencing method introduced minimal contamination. However, contaminants may play an interfering role in samples containing very low quantities of DNA, especially culture-negative samples, where infecting agents are present in very low numbers detectible using WGSS.
Targeted culturing of bacteria
Several samples contained species detected by WGSS but not by culture. Once the results of sequencing were available, directed culture for specific organisms, including P. aeruginosa , multiple Staphylococcus species, B. fragilis and C. acnes , was done. Glycerol stocks from the original sample were grown under conditions favouring the species of interest, as indicated in Table 2. In every case, culturing was unsuccessful, though species previously identified by culture were often again, but not always, obtained in culture, supporting the hypothesis that bacteria in PJIs may not be cultured following biofilm transition.
Interestingly, medium 14, a lactate rich medium, did not support growth of the C. acnes control strains. While this medium was designed to isolate lactic acid-fermenting Propionibacterium (e.g. from Emmentaler cheese), it did not support growth of (ATCC strains used as control) C. acnes (formerly P. acnes ). The recent re-definition of Cutibacterium from Propionibacterium is based on genetic discrepancies accumulated from adaptation on the skin environment [49]. In retrospect, due to the difference in habitat, it might not be surprising that growth of C. acnes growth was not supported by medium 14, reinforcing re-classification of skin adapted C utibacterium as separate from Propionibacterium .
Diversity of PJI populations
Comparison with negative controls and with other microbiomes
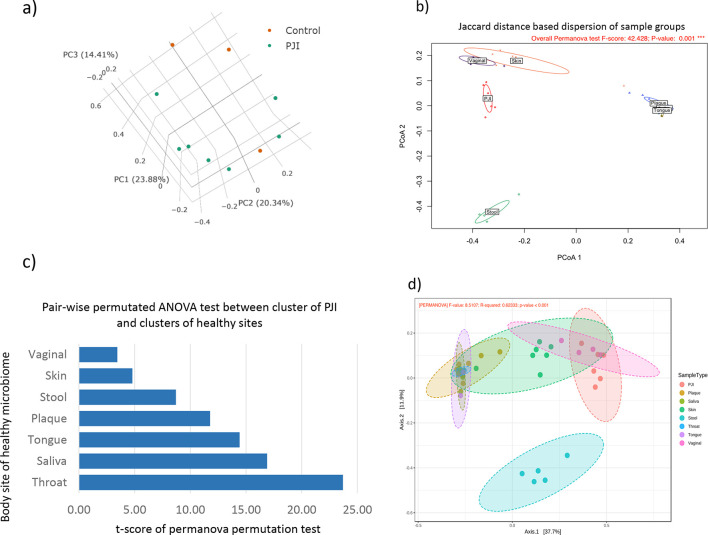
Comparative analysis of all PJI samples included in this study with controls indicated that addition of P. aeruginosa and S. aureus in the controls showed higher abundance than the cluster of strains detected in PJI samples, while the control containing microbial DNA-free water located within the cluster, i.e. grouping with the PJI populations (Fig. 1a). This clustering of samples with the DNA-free water control, suggested that bacteria from similar sources may play a role in PJIs and, perhaps also during sample processing. To determine potential sources of bacteria involved in PJI, microbiome profiles of PJI samples were compared to that of healthy human skin, saliva, stool, gingival plaque, throat, tongue and vagina datasets collected from human microbiome project (HMP) (https://portal.hmpdacc.org/) (Fig. 1b and Table S2). PJI populations clustered together, spanning vaginal, skin and gut microbiomes (Fig. 1b). In order to evaluate how close the PJI cluster is to samples of HMP body sites, a pair-wise distance comparison was carried out using Jaccard distances and t-scores were derived through PERMANOVA test (Fig. 1c). Lower t-scores indicate greater similarity between populations with skin, vaginal and stool microbiomes being most similar to PJIs (Fig. 1c). To evaluate what extent the distance between PJI and a body site is contributed by closely related species, a phylogenetic tree guidance-based discrimination of the samples was performed. The results (Fig. 1d) show that PJIs remained closer to the vaginal, skin and stool samples compared to other body sites indicating that the closer distance is significantly contributed by close phylogenetically related microbial community.
Fig. 1.
Jaccard PCoA comparative analysis showing PJI populations, positive and negative controls (a). Jaccard PCoA comparative analysis showing the PJI population compared to body sites of a healthy population (b). Distribution of t-scores derived from PERMANOVA test on the distance of each cluster between PJI and the body site. Lower t-scores indicate the abundance of microbial populations are more similar (c). A Unifrac analysis reflecting a phylogenetically aware distance between PJI and the HMP samples (d).
Additional micro-organisms identified in PJI samples
As WGSS analysis includes all micro-organisms and not just bacteria, it can be used to detect and identify viruses, fungi and protists in samples. The presence of viruses related to microbes identified is noted in Table 1, supporting detection and identification of the micro-organism in the infection. Protists and fungi identified in the PJI samples included in this study are listed in Table 3. Fungi have been found to cause or contribute to ca. 1 % of PJIs, particularly Candida spp. accounting for >80 % of fungi identified in infections [50]. The importance of fungal PJIs is just beginning to be understood, while the significance of protists within these communities is yet to be determined. It is noteworthy that none were identified in the negative control, and hence should be considered as potential pathogens.
Table 3.
Fungal and protist species present
|
Sample |
Protist (%RA) |
Fungi (%RA) |
|---|---|---|
|
A |
Sarcocystis neurona strain SN1 (74.58) |
|
|
Plasmodium gaboni strain Pkg (19.62) | ||
|
Toxoplasma gondii GT1 (5.798) | ||
|
B |
Sarcocystis neurona strain SN1 (96.2) |
|
|
Toxoplasma gondii CtCo5 (3.799) | ||
|
1 |
Plasmodium yoelii (81.18) |
Malassezia restricta CBS 7877* (100) |
|
Plasmodium berghei ANKA (18.82) | ||
|
15 |
|
Malassezia restricta CBS 7877* (100) |
Note: Samples C, D and E had none present. *Denotes species that fall below the CosmosID algorithm identifying species likely to be present.
Conclusions
There is a clear need for improved methods to assess polymicrobial PJI populations. Improved culturing methods of PJIs has increased sensitivity of culture, however dormant and culture-resistant microbes will not be detected and/or identified by culture, as was observed in this study, when directed culture is done. There are limitations in this study to consider when interpreting this data. The accuracy of algorithms to discern minority species from contaminant is not clear. Culture identified multiple species that fell below the metagenomic sequencing-based detection as profiled by the CosmosID algorithm, suggesting these species may exist as a minority organism. Also, well-established standards for quantification of metagenomic sequencing depth necessary to ensure a lower level of detection for PJI do not exist and biases occur due to factors such as sampling technique, DNA preparation, proportion of host DNA as background and sequenceability. This complicates the identification of contaminants from negative controls, as common contaminants are often the same organisms found in PJIs. Here we have designated organisms identified by metagenomic sequencing coupled with the CosmosID algorithm as likely present to be contaminants. Though this and other recent studies shed light on PJI population make-up, it is clear that the current understanding of PJI communities is still shrouded in mystery. In this study, combining the metagenomic sequencing coupled with the CosmosID algorithm, culture results and negative control we hope to better distinguish minority species from contaminant.
Detection and identification of the epibiont candidate division TM7 by WGSS highlights a potential shortfall of culturing methods. Certainly culturing as a method of detection and identification is well recognized to have limitations. In addition to viable but not culturable (VBNC) micro-organisms, some culturable microbes can also become culture-resistant following biofilm formation [15, 38]. Other potentially culturable microbes may require specific nutrients unknown a priori when culturing media are employed. Not surprisingly, in this study WGSS identified several bacterial species not cultured on laboratory media, even when additional targeted culturing was attempted. WGSS offers the advantage of providing an untargeted range of information as well as the presence of associated virulence factors and phages that can serve as internal validation. An interesting disconnect between WGSS and culturing is that cultured species do not always correspond to highest relative abundance species and may represent rapid growers and not the most abundant in the initial population. Indeed, culturable C. striatum was found by WGSS to be of very low relative abundance (0.04511 %) while a high abundance organism, P. aeruginosa (65.11 %), was not cultured, even in directed culture. While a higher relative abundance of DNA determined by WGSS suggests a greater proportion of the given species to be present, it is yet to be determined what can be inferred with respect to their role in the infection. Nevertheless, WGSS is an exciting technology with great promise in analysis of complex infections.
In this study, the most prominent bacterial species identified by WGSS and confirmed by culture and/or presence of an associated phage/virulence factor included S. aureus , C. acnes , P. aeruginosa , E. faecalis , S. epidermidis and C. striatum . In sample B, B. fragilis could not be confirmed, but it is possible this obligate anaerobe was not able to be cultured from the sample. Interestingly, PJI reports note this species is often the only one detected by culture [40]. WGSS identified this species, when present, to be at relatively high concentration and dominant. In contrast, other PJI populations appear to be polymicrobial. E. faecalis is more often found in polymicrobial infections [51], as was observed for sample A, which harboured E. faecalis , P. aeruginosa , C. acnes and C. striatum. C. acnes is often identified with Staphylococcal species on prosthetics [38], as was observed for samples C and 15. While variability within these infections was observed, a given microbial player may play a key role in directing community formation along a finite number of paths.
Controls were vital to this study. The DNA-free water control in WGSS sample analysis allowed identification of potential DNA contaminants and, possibly, an ultra-low level of contaminating DNA in the DNA-free water control. In any case, the control was helpful in deciphering pathogen from contaminant. Several of the antibiotic-resistant species detected and identified in PJIs were considered only a few decades ago to be harmless contaminants and not potential pathogens. Reference cultures employed as positive controls indicated that the sequencing protocol was, indeed, able to identify predominant species accurately. WGSS also provided internal confirmation of species present by the identification of associated phages and virulence genes.
Comparative analyses showed PJI populations identified by WGSS, using Jaccard PCoA, were similar to inhabitants of the human body. Potential sources of micro-organisms comprising a PJI were most closely related to vaginal, skin and gut microbiome populations. Because the combinations of micro-organisms comprising PJIs are not normally found, as such, in nature, it is important to begin to observe these unique microbial interactions more closely.
Since biochemical interactions of micro-organisms can influence factors such as virulence, biofilm formation and antibiotic resistance, studies of interactions shaping a polymicrobial PJI are needed. Studies of pathogenic versus commensal strains of bacteria have found that there are differences, pathogenic strains having been shaped by prior exposure to antibiotics [19–21], for example. Microbial interactions in infections are not likely to be simply species specific, but also strain specific, highlighting the necessity for study of PJIs employing clinical strain isolates. Additionally, since strains may become culture-resistant, as shown by other investigators, culture must be recognized as an important limiting factor in understanding PJIs.
Finally, much remains to be learned about PJIs and those factors promoting PJI and limiting therapeutic efficacy. In this study, WGSS detected and identified protist and fungal micro-organisms in some of the PJI samples. While it is known that humans are carriers of protists, the presence of these micro-organisms can be problematic and not only for the immunocompromised [52]. These protists may play an important role in PJI onset and persistence and also in post-surgical infections.
Supplementary Data
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
This research was made possible in part by an Indiana CTSI Fellowship to A.A.W. through NIH NCATS Grant TL1TR001107 (A. Shekhar, PI) and NIH NIAID Grant R01AI113219 to J.D.S.
Author contributions
A.A.W., N.H., M.K., R.R.C. and J.D.S. were involved in conceptualization and methodology. M.K. provided samples. A.A.W. carried out laboratory investigations and formal analysis. R.R.C., N.H., H.K. and CosmosID carried out genomic sequencing and formal analysis. A.A.W., N.H., M.K., R.R.C. and J.D.S. wrote the manuscript. This research has IRB approval from The University of Notre Dame and Elkhart General Hospital with all patients involved completing an informed consent.
Conflicts of interest
The following authors are affiliated with CosmosID; R.R.C., N.H. and H.K. A.A.W., J.D.S. and M.K. declare no conflict of interest.
Footnotes
Abbreviations: DNA, deoxyribonucleic acid; FAB, fastidious anaerobe broth; HMP, human microbiome project; MSA, mannitol salt agar; PCR, polymerase chain reaction; PJI, prosthetic joint infection; qPCR, quantitative polymerase chain reaction; VBNC, viable but not culturable; WGSS, whole genome shotgun sequencing.
Two supplementary tables are available with the online version of this article.
References
- 1.Zimmerli W, Trampuz A, Ochsner PE, Infections P-J. Prosthetic-joint infections. N Engl J Med. 2004;351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 2.Kurtz S, Ong K, Lau E, Mowat F, Halpern M. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J Bone Jt Surg. 2007;89-A:780–785. doi: 10.2106/JBJS.F.00222. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham DJ, Kavolus JJ, Bolognesi MP, Wellman SS, Seyler TM. Specific infectious organisms associated with poor outcomes in treatment for hip periprosthetic infection. J Arthroplasty. 2017;32:1984–1990. doi: 10.1016/j.arth.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimmer MD, Friedrich MJ, Randau TM, Ploeger MM, Schmolders J, et al. Polymicrobial infections reduce the cure rate in prosthetic joint infections: outcome analysis with two-stage exchange and follow-up ≥two years. Int Orthop. 2016;40:1367–1373. doi: 10.1007/s00264-015-2871-y. [DOI] [PubMed] [Google Scholar]
- 6.Guren E, Figved W, Frihagen F, Watne LO, Westberg M. Prosthetic joint infection-a devastating complication of hemiarthroplasty for hip fracture. Acta Orthop. 2017;88:383–389. doi: 10.1080/17453674.2017.1301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elias S, Banin E. Multi-Species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 8.Butler-Wu SM, Burns EM, Pottinger PS, Magaret AS, Rakeman JL, et al. Optimization of periprosthetic culture for diagnosis of Propionibacterium acnes prosthetic joint infection. J Clin Microbiol. 2011;49:2490–2495. doi: 10.1128/JCM.00450-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peel TN, Dylla BL, Hughes JG, Lynch DT, Greenwood-Quaintance KE, et al. Improved diagnosis of prosthetic joint infection by culturing periprosthetic tissue specimens in blood culture bottles. MBio. 2016;7:1–8. doi: 10.1128/mBio.01776-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen LH, Lange J, Xu Y, Schønheyder HC. Optimizing culture methods for diagnosis of prosthetic joint infections: a summary of modifications and improvements reported since 1995. J Med Microbiol. 2012;61:309–316. doi: 10.1099/jmm.0.035303-0. [DOI] [PubMed] [Google Scholar]
- 11.Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med. 2007;357:654–663. doi: 10.1056/NEJMoa061588. [DOI] [PubMed] [Google Scholar]
- 12.Penterman J, Nguyen D, Anderson E, Staudinger BJ, Greenberg EP, et al. Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Rep. 2014;6:293–300. doi: 10.1016/j.celrep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Karau MJ, Schmidt SM, et al. Rapid molecular microbiologic diagnosis of prosthetic joint infection. J Clin Microbiol. 2013;51:2280–2287. doi: 10.1128/JCM.00335-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgenstern C, Cabric S, Perka C, Trampuz A, Renz N. Synovial fluid multiplex PCR is superior to culture for detection of low-virulent pathogens causing periprosthetic joint infection. Diagn Microbiol Infect Dis. 2018;90:115–119. doi: 10.1016/j.diagmicrobio.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Swearingen MC, DiBartola AC, Dusane D, Granger J, Stoodley P. 16S rRNA analysis provides evidence of biofilms on all components of three infected periprosthetic knees including permanent braided suture. Pathog Dis. 2016;74:ftw083–11. doi: 10.1093/femspd/ftw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, Rudkjøbing VB, Simonsen O, Pedersen C, Lorenzen J, et al. Bacterial diversity in suspected prosthetic joint infections: an exploratory study using 16S rRNA gene analysis. FEMS Immunol Med Microbiol. 2012;65:291–304. doi: 10.1111/j.1574-695X.2012.00949.x. [DOI] [PubMed] [Google Scholar]
- 17.Ponnusamy D, Kozlova EV, Sha J, Erova TE, Azar SR, et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc Natl Acad Sci U S A. 2016;113:722–727. doi: 10.1073/pnas.1523817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Q, Wi M, Thoendel MJ, Raval YS, Greenwood-quaintance KE, et al. Evaluation of the CosmosID bioinformatics platform for prosthetic Joint-Associated sonicate fluid shotgun metagenomic data analysis. J Clin Microbiol. 2019;57:1–13. doi: 10.1128/JCM.01182-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMullen AR, Anderson N, Wallace MA, Shupe A, Burnham C-AD. When good bugs go bad: epidemiology and antimicrobial resistance profiles of Corynebacterium striatum, an emerging multidrug-resistant, opportunistic pathogen. Antimicrob Agents Chemother. 2017;61:AAC.01111–.01117. doi: 10.1128/AAC.01111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmark B, Söderquist B, Unemo M, Nilsdotter-Augustinsson Åsa. Comparison of Staphylococcus epidermidis isolated from prosthetic joint infections and commensal isolates in regard to antibiotic susceptibility, agr type, biofilm production, and epidemiology. Int J Med Microbiol. 2013;303:32–39. doi: 10.1016/j.ijmm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Thoendel M, Jeraldo P, Greenwood-Quaintance KE, Yao J, Chia N, et al. Impact of contaminating DNA in whole-genome amplification kits used for metagenomic shotgun sequencing for infection diagnosis. J Clin Microbiol. 2017;55:1789–1801. doi: 10.1128/JCM.02402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis K, Cells P. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 24.Street TL, Sanderson ND, Atkins BL, Brent AJ, Cole K, et al. Molecular diagnosis of Orthopedic-Device-Related infection directly from sonication fluid by metagenomic sequencing. J Clin Microbiol. 2017;55:2334–2347. doi: 10.1128/JCM.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivy MI, Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Hanssen AD, et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J Clin Microbiol. 2018;56:JCM.00402–.00418. doi: 10.1128/JCM.00402-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottesen A, Ramachandran P, Reed E, White JR, Hasan N, et al. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 2016;16:1–11. doi: 10.1186/s12866-016-0894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasan NA, Young BA, Minard-Smith AT, Saeed K, Li H, et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One. 2014;9:e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Q, Luan Y, Sun F. Variance adjusted weighted UniFrac: a powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinformatics. 2011;12:118–14. doi: 10.1186/1471-2105-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Havelaar AH, Van OM, Drost YC. F-Specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. 1993;59:2956–2962. doi: 10.1128/aem.59.9.2956-2962.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armon R, Kott Y. Bacteriophages as indicators of pollution. Crit Rev Environ Sci Technol. 1996;26:299–335. doi: 10.1080/10643389609388494. [DOI] [Google Scholar]
- 33.Jofre J. In: Human Viruses in Water. Bosch A, editor. Elsevier B.V; 2007. Indicators of waterborne enteric viruses; pp. 227–249. editor. [Google Scholar]
- 34.Grabow W. Bacteriophages : Update on application as models for viruses in water. Water Res. 2001;27:251–268. [Google Scholar]
- 35.Mesquita MM, Emelko MB. In: Kurtboke Ipek., editor. Bacteriophages InTech; 2012. Bacteriophages as surrogates for the fate and transport of pathogens in source water and in drinking water treatment processes; p. ISBN:978-953-51-0272-4. editor. p. [Google Scholar]
- 36.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 37.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:1–12. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyner H, Patel R. Propionibacterium acnes biofilm - a sanctuary for Staphylococcus aureus? Anaerobe. 2016;40:63–67. doi: 10.1016/j.anaerobe.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 39.Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Patel R. Corynebacterium prosthetic joint infection. J Clin Microbiol. 2012;50:1518–1523. doi: 10.1128/JCM.06439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah NB, Tande AJ, Patel R, Berbari EF. Anaerobic prosthetic joint infection. Anaerobe. 2015;36:1–8. doi: 10.1016/j.anaerobe.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Johnson JL, Moore WEC, Moore LVH. Bacteroides caccae sp. nov., Bacteroides merdae sp. nov., and Bacteroides stercoris sp. nov. isolated from human feces. Int J Syst Bacteriol. 1986;36:499–501. doi: 10.1099/00207713-36-4-499. [DOI] [Google Scholar]
- 42.Hong P-Y, Wu J-H, Liu W-T. Relative abundance of Bacteroides spp. in stools and wastewaters as determined by hierarchical oligonucleotide primer extension. Appl Environ Microbiol. 2008;74:2882–2893. doi: 10.1128/AEM.02568-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol. 2012;2:1–11. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, et al. Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He X, McLean JS, Edlund A, Yooseph S, Hall AP, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 2015;112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchandin H, Jean-Pierre H, Carrière C, Canovas F, Darbas H, et al. Prosthetic joint infection due to Veillonella dispar. EJCMID. 2001;20:340–342. doi: 10.1007/PL00011273. [DOI] [PubMed] [Google Scholar]
- 47.Gneiding K, Frodl R, Funke G. Identities of Microbacterium spp. encountered in human clinical specimens. J Clin Microbiol. 2008;46:3646–3652. doi: 10.1128/JCM.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boisrenoult P. Cutibacterium acnes prosthetic joint infection: diagnosis and treatment. Orthopaedics & Traumatology: Surgery & Research. 2018;104:S19–S24. doi: 10.1016/j.otsr.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 49.Scholz CFP, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol. 2016;66:4422–4432. doi: 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 50.Brown TS, Petis SM, Osmon DR, Mabry TM, Berry DJ, et al. Periprosthetic joint infection with fungal pathogens. J Arthroplasty. 2018;33:2605–2612. doi: 10.1016/j.arth.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 51.Tornero E, Senneville E, Euba G, Petersdorf S, Rodriguez-Pardo D, et al. Characteristics of prosthetic joint infections due to Enterococcus sp. and predictors of failure: a multi-national study. Clin Microbiol Infect. 2014;20:1219–1224. doi: 10.1111/1469-0691.12721. [DOI] [PubMed] [Google Scholar]
- 52.Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. Int J Parasitol. 2000;30:1217–1258. doi: 10.1016/S0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.