Abstract
Putative RNA helicases are involved in most aspects of gene expression. All previously characterized members of the DEAH-box family of putative RNA helicases are involved in pre-mRNA splicing. Here we report the analysis of two novel DEAH-box RNA helicases, Dhr1p and Dhr2p, that were found to be predominantly nucleolar. Both genes are essential for viability, and MET-regulated alleles were therefore created. Depletion of Dhr1p or Dhr2p had no detectable effect on pre-mRNA splicing in vivo or in vitro. Both Dhr1p and Dhr2p were, however, required for 18S rRNA synthesis. Depletion of Dhr2p inhibited pre-rRNA cleavage at sites A0, A1, and A2, while Dhr1p depletion inhibited cleavage at sites A1 and A2. No coprecipitation of snoRNAs was detected with ProtA-Dhr2p, but Dhr1p-ProtA was stably associated with the U3 snoRNA. Depletion of Dhr1p inhibited processing steps that require base pairing of U3 to the 5′ end of the 18S rRNA. We speculate that Dhr1p is targeted to the preribosomal particles by the U3-18S rRNA interaction and is required for the structural reorganization of the rRNA during formation of the central pseudoknot.
Putative ATP-dependent RNA helicases are ubiquitous, being present in RNA and DNA viruses, bacteria, archaea, and eukaryotes. It is generally assumed that many or all members of the family are capable of ATP-dependent RNA helicase activity, but this has been formally demonstrated in only a few cases (reviewed in references 13 and 58). These proteins are predicted to act as modulators of RNA structures (16) and are therefore expected to play key roles in all cellular processes involving structural isomerization of RNA. Putative RNA helicases have been implicated in many aspects of gene expression, including transcription, nuclear and mitochondrial RNA splicing, ribosome synthesis (pre-rRNA processing and ribosomal assembly), translation, RNA editing, mRNA export, and mRNA turnover (reviewed in references 13 and 37). However, in most cases the precise substrate, and hence the function, of these proteins is not known.
The putative RNA helicases share a conserved central core domain (ca. 290 to 360 residues in length) that consists of eight highly conserved motifs (reviewed in references 13 and 49). These include the eponymous DEAH or DEAD box (reviewed in reference 37) and are implicated in the basic functions of the proteins: binding to the ATP and RNA cofactors, ATP hydrolysis, and RNA helicase activity (reviewed in reference 37). The core region is typically flanked by poorly conserved amino- and carboxy-terminal extensions. Analysis of the DEAH-box helicase Prp16p indicated that the nonconserved extensions confer substrate specificity (70).
RNA helicases can be grouped into four distinct families based on conserved sequence elements in the core domain. The largest group, the DEAD-box proteins, includes the archetypal helicase eIF4-A that is involved in translation initiation, as well as several proteins required for pre-mRNA splicing and, remarkably, 13 proteins that are needed for different steps in ribosome synthesis. The DExH-box helicases include Ski2p, which is required during 3′ degradation of mRNA by the exosome complex (20), and Dob1p/Mtr4p, which functions as a cofactor for the exosome during nuclear RNA processing and degradation (14). The Upf1p-related family includes Upf1p itself, which is required for nonsense-mediated mRNA decay (11, 33, 44), and Sen1p which is implicated in several nuclear RNA processing activities, including tRNA splicing (47, 64, 65), and is weakly associated with several small nucleolar RNAs (snoRNAs) and snRNAs (65). The remaining group is the DEAH-box family. The four previously characterized DEAH-box proteins each function in pre-mRNA splicing. Prp2p and Prp16p are required for the first and second trans-esterification steps, respectively (36, 45, 51, 59). Prp22p and Prp43p are involved in the release of spliced mRNA products from the spliceosome and the disassembly-recycling of spliceosomal components (2, 10). Prp16p and Prp22p show ATP-dependent RNA duplex unwinding activity in vitro, indicating that the DEAH-box proteins may modify RNA structure directly (50, 68, 71).
Eukaryotic nucleoli contain large numbers of snoRNAs, which are associated with proteins in small nucleolar ribonucleoprotein (snoRNP) particles (reviewed in references 24 and 67). The bulk of the snoRNAs are involved in selection of the sites of modification in the pre-rRNAs. Sites of 2′-O-methylation and pseudouridylation are selected by the box C+D and box H+ACA families of snoRNAs, respectively (reviewed in references 26 and 57). A small number of snoRNA species are required for processing of the large pre-rRNA to the mature rRNAs (Fig. 1) (19, 34, 40, 60). The best characterized of these is the box C+D snoRNA U3, which is required for pre-rRNA cleavage in the 5′ external transcribed spacer (5′-ETS) and internal transcribed spacer 1 (ITS1) regions in yeast, in Xenopus oocytes, and for in vitro cleavage of the mouse 5′-ETS (19, 23, 41). Yeast U3 is required for cleavage at sites A0, A1, and A2 (Fig. 1) and base pairs to the pre-rRNA at two sites: in the 5′-ETS and at the 5′ end of the 18S rRNA region (5, 54). The U3–5′-ETS interaction is required for cleavage at sites A0, A1, and A2, while the U3-18S interaction is required for cleavage only at sites A1 and A2 (Fig. 1). All box C+D snoRNAs are associated with a set of common proteins specific to this class of RNA: Nop1p, Nop56p, and Nop5p/Nop58p in yeast (29, 30, 48, 74). In addition, several proteins appear to be specifically associated with the U3 snoRNP: Sof1p, Mpp10p, Imp3p, Imp4p, Lcp5p, Rcl1p, and Rrp9p (7, 15, 22, 32, 73; J. Venema, personal communication).
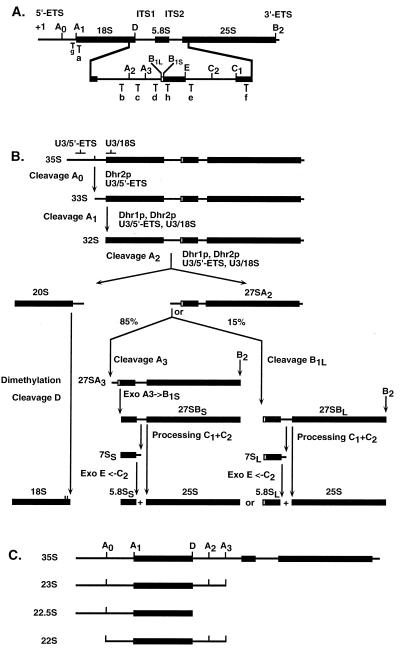
FIG. 1.
Pre-rRNA processing pathway in yeast. (A) Structure of the rRNA gene operon and location of the oligonucleotides used in this work. The mature 18S, 5.8S, and 25S rRNAs (bold lines) are released from the 35S primary transcript following cleavages in the 5′-ETS and 3′-ETS and ITS1 and ITS2. Cleavage sites are indicated by uppercase letters (A0 to D). Oligonucleotides used for the Northern blot hybridization and primer extension experiments are indicated by lowercase letters (a to h). (B) Pre-rRNA processing pathway. The 35S pre-rRNA is successively cleaved in the 5′-ETS at site A0 (generating the 33S RNA), at site A1, the 5′ end of the mature 18S rRNA (generating the 32S RNA), and at site A2 in ITS1 (generating the 20S and 27SA2 pre-rRNAs). The 20S is dimethylated by Dim1p and cleaved at site D to generate the mature 18S rRNA. The 27SA2 is matured to the 5.8S and 25S rRNAs following two alternative pathways. Eighty-five percent of the 27SA2 population is cleaved at site A3 in ITS1 by RNase MRP; this is rapidly followed by 5′-to-3′ trimming to site B1S by Xrn1p and Rat1p. Fifteen percent of 27SA2 is cleaved at site B1L. Cleavage at site B2 occurs concomitantly with cleavage at site B1. The two forms of 27SB (27SBS and 27SBL) are matured following identical pathways involving cleavage at sites C1 and C2 and 3′-to-5′ exonucleolytic digestion to site E by the exosome. Both Dhr1p and Dhr2p are required for cleavages at sites A1 and A2; Dhr2p is also required for cleavage at site A0. Base pairing between U3 and the pre-rRNA in the 5′-ETS and 18S regions is required for cleavage at sites A1 and A2; the U3–5′-ETS interaction is also required for cleavage at site A0. (C) Structure of aberrant pre-rRNA processing intermediates detected on depletion of Dhr1p and Dhr2p. Premature cleavage of the 35S pre-rRNA at site A3 in ITS1 generates the 23S RNA. The 22.5S RNA results from cleavage of pre-rRNA molecules at site D in the absence of cleavage at sites A0 and A1. The 22S RNA is accumulated when cleavages occur at sites A0 and A3 in the absence of cleavage at sites A1 and A2.
Here we report the characterization of two new members of the DEAH family of putative RNA helicases. Despite strong homology to Prp22p and related splicing factors, Dhr1p and Dhr2p were required for pre-rRNA processing rather than for pre-mRNA splicing. Furthermore, Dhr1p was found to be stably associated with the U3 snoRNP, suggesting a function in restructuring of the pre-rRNA.
MATERIALS AND METHODS
Plasmids.
The plasmids used in this study were pRS313 (56), pTL27 (25), pTL54 (27), and pUC18-55-HA(n) (a kind gift of R. van Nues).
Yeast genetics.
All strains were constructed using one-step PCR-based technology (3, 25). The yeast strains used in this study were BMA38 (a/α trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 ura3-1/ura3-1 leu2-3,112/leu2-3,112 ade2-1/ade2-1 can1-100/can1-100) and BMA64 (a trp1Δ1 his3-11,15 ura3-1 leu2-3,112 ade2-1 can1-100) (kind gifts of B. Dujon and F. Lacroute).
Gene disruption.
The DHR1, DHR2, and YLR419w open reading frames (ORFs) were precisely deleted from the genome and replaced with the HIS3 marker. The oligonucleotides used for amplification with plasmid pRS313 were 1 and 2 (DHR1), 3 and 4 (DHR2), and 12 and 13 (YLR419w). PCR products were transformed in a diploid wild-type strain (BMA38). Transformants were selected on minimal medium lacking histidine. Correct integration was checked by PCR on yeast colonies and/or Southern blot analysis. Diploids were sporulated at 23°C.
Construction of MET-regulated alleles of DHR1 and DHR2.
Conditional alleles of DHR1 and DHR2 were constructed by placing their expression under the control of a MET-regulated promoter (pMET3). Transcription driven from MET3 promoters is strongly repressed on addition of methionine to the growth medium (9). TRP1-pMET3::HA cassettes were PCR amplified from plasmid pUC18-55-HA(n) with oligonucleotides 5 and 6 (for DHR1) and oligonucleotides 7 and 8 (for DHR2). PCR products were transformed in a wild-type haploid strain (BMA64). In addition to the fusion to pMET3, integration at the correct locus results in HA epitope tagging of the ORF. HA is a nonapeptide antigen (YPYDVPDYA) from the human influenza virus hemagglutinin protein. Transformants were selected on minimal medium lacking tryptophan and were screened by PCR on yeast colonies, methionine dependence for growth, and Western blot analysis.
Time course and RNA analysis.
MET-regulated strains were pregrown to mid-log phase in minimal medium lacking tryptophan. Depletion was achieved by the addition of methionine to a final concentration of 20 mM. Total RNA was extracted as described previously (63) and resolved on 1.2 and 2% agarose-formaldehyde gels for pre-rRNA and splicing analysis, respectively, and on 8% polyacrylamide gels for low-molecular-weight pre-rRNA and snoRNA analysis. Primer extension and in vitro splicing reactions were performed as previously described (6, 45).
Western blot analysis.
For protein extraction, cells equivalent to an optical density at 600 nm (OD600) of 10 were processed as previously described (25). Supernatants equivalent to a cell OD600 of 0.375 were loaded per lane. Samples were subjected to sodium dodecyl sulfate (SDS)–8% (Dhr1p) or 15% (Dhr2p) polyacrylamide gel electrophoresis (PAGE) and blotted in accordance with standard procedures. The following antibodies were used: PAP (peroxidase-antiperoxidase; P-2026; Sigma), anti-HA Y-11 (cat_sc-805; Santa Cruz Biotech; rabbit polyclonal immunoglobulin G [IgG]; dilution, 1:1,000), anti-Nop1p (mouse monoclonal mA66; dilution, 1:200; J. Aris, University of Florida), anti-Srp14p and anti-Srp68p (rabbit polyclonal; dilution, 1:500; J. Brown, University of Edinburgh), anti-Mpp10p (rabbit polyclonal; dilution, 1:20,000; S. Baserga, Yale School of Medicine), and anti-Rcl1p (rabbit polyclonal; dilution, 1:500; E. Billy and W. Filipowicz, Friedrich Miescher-Institut).
Immunoprecipitation of ProtA epitope-tagged Dhr1p and Dhr2p.
For the immunoprecipitation experiments, Dhr1p and Dhr2p were fused to a high-affinity tag. A repeat of the z domain of the protein A (ProtA) epitope from Staphylococcus aureus was fused either to the carboxyl (Dhr1p) or to the amino (Dhr2p) end of the proteins. For Dhr1p, a ProtA-K1URA3 cassette was generated by PCR from plasmid pTL54 with oligonucleotides 9 and 10. For Dhr2p, a HIS3-pGAL10-ProtA cassette was PCR amplified from plasmid pTL27 with oligonucleotides 3 and 11. The PCR cassettes were transformed in a haploid strain (BMA64a). Dhr1p-ProtA and ProtA-Dhr2p strains showed no growth impairment under any of the conditions tested, indicating that both fusions are functional. Transformants were screened as described above. The GAL::ProtA-DHR2 allele allowed high levels of residual expression on glucose medium and was not galactose dependent.
Immunoprecipitations were performed on IgG-agarose beads with yeast whole-cell extracts prepared as previously described (53). Lysates were made in buffer A150 (150 mM K acetate, 20 mM Tris HCl [pH 8.0], 5 mM MgCl2, 1 mM dithiothreitol, 0.2% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride), and cleared by centrifugation (12,000 × g, 4°C, 20 min). Lysates equivalent to a cell OD600 of 120 were incubated on a rotating wheel for 2 h at 4°C with 20 μl of IgG-agarose beads (A2909; Sigma), prewashed in buffer A150, in a total volume of 400 μl. Pellets were washed four times for 20 min (each time) in 1 ml of buffer A150. RNA were either analyzed by Northern blot assay or directly labeled at the 3′ end with pCp. For Northern blot analysis, each gel lane (total, supernatant, or pellet) was loaded with RNA from a fraction of the preparation equivalent to a cell OD600 of 15 (1:1:1 ratio). For 3′-end labeling, RNA from the pellet fractions equivalent to a cell OD600 of 15 was incubated overnight at 4°C with T4 RNA ligase (New England Biolabs) and pCp in accordance with the recommendations of the manufacturer. For protein analysis, pellets were submitted to four additional washes in buffer NET150 (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 0.05% NP-40, 0.5 mM phenylmethylsulfonyl fluoride), resuspended in SDS loading buffer, and boiled for 5 min with occasional vortexing. Dhr1p is a large protein (144,935 kDa) which appeared to be highly sensitive to proteolytic degradation, and in this case, pellet lanes were loaded with an excess of material (1:1:35 ratio of the total, supernatant, and pellet fractions, respectively).
Immunofluorescence analysis.
Immunofluorescence experiments were performed in accordance with standard procedures (46). HA epitope-tagged proteins were detected with a rat monoclonal antibody (3F10; Boehringer) used at a dilution of 1:200, followed by a fluorescein isothiocyanate (FITC)-coupled goat anti-rat antibody (1:200; ICN). As a nucleolar marker, Nop1p was detected with a mouse monoclonal antibody (mA66; J. Aris, University of Florida) at a dilution of 1:200, followed by an anti-mouse Cy3-coupled donkey antibody (1:2,000; Jackson Immunoresearch).
Oligonucleotides.
For the construction of yeast strains, we used oligonucleotides 1 (ATTACAACCCAATACTAATAATAGAGTGTCTTAAGAAATATAAGTACTCTTGGCCTCCTCTAG), 2 (TATGTTCTTATATAATGAAACTCTATTTCTATAATACCAGCGCATCGTTCAGAATGACACGTA), 3 (GAAAATAAAAAATTCTGCTCGATGAGATGAGATGAGGTTATGATACTCTTGGCCTCCTCTAGT), 4 (AATGCCTTTATATACAATAAAAATGCGCTAAGAGTAGGTGACTTATCGTTCAGAATGACACG), 5 (GCTATATGCTATAAAAATGCAATTACAACCCAATACTAATAATAGACTTAAATAAATACTACTC), 6 (GCCGGATCTGGCTTTTTCATTAAACCTTTTTCTGTAAGTACCCATACGAGCTCCAGCGTAATCTGGAA), 7 (TGCTGACTGAAAGATGTACTTTTCTTCTACCGTTTTTCTTCGGCGGCCTTAAATAAATACTACTC), 8 (AGAAGTATGGTTTGATGCAACTCTACTATTGCTATTTGCTGCCATACGAGCTCCAGCGTAATCTGG), 9 (AAGGGCTTCCAGACCATCACAGGTGAAGAGAAAGAAAAAAAAGGCGTGGACAACAAATTC), 10 (AAGTGGTTGCAATTATTTGATGCCCTAGATAGGAATAATGTACTGGGTAGAAGATCGGTC), 11 (AGAAGTATGGTTTGATGCAACTCTACTATTGCTATTTGCTGCCATATTCGCGTCTACTTTCGG), 12 (TCACTTTATACTTTACATCGCAATTGCTTTCATTAATTTTTGATAACTCTTGGCCTCCTCTAGT), and 13 (TGAAGATGATAATTATTATACAGGGACTACAGTAGGAACTGATTATCGTTCAGAATGACACG.
Oligonucleotides a to h, used for pre-rRNA analysis, were described in reference 29. The sequence of oligonucleotide i is GCTCTCATGCTCTTGCC. Oligonucleotides anti-U3, -U14, -U24, and -snR190 (box C+D) and oligonucleotides anti-snR10, -snR30, -snR31, -snR33, and -snR37 (box H+ACA), used for detection of snoRNA, were described in reference 29.
For detection of mRNAs, oligonucleotides anti-ACT1 (TCAATAACCAAAGCAGCAAC) and anti-CYH2 (GTGCTTTCTGTGCTTACCGATACGACCTTTACCG) were used.
RESULTS
Dhr1p and Dhr2p are essential for growth.
When these experiments were commenced, the yeast genome contained three uncharacterized, predicted ORFs that were homologous to the DEAH-box family of splicing factors (13). These were YMR128w/Ecm16p (now DHR1 for DEAH-box protein involved in ribosome synthesis), YKL078w/JA2 (now DHR2), and YLR419w. Each ORF was individually deleted from the genome of diploid wild-type strains and replaced with the HIS3 marker using a one step PCR-based strategy (see Materials and Methods). Heterozygous diploids were sporulated, and 20 tetrads of each strain were dissected. For Dhr1p and Dhr2p, all tetrads showed a 2:2 segregation for growth and no His+ spores were recovered, showing that both genes are essential, at least for spore germination. For YLR419w, all spores were viable with a 2:2 segregation for His+, showing the gene to be nonessential, as previously reported (55). YLR419w has not yet been further characterized.
Despite strong conservation in their core domains, Dhr1p and Dhr2p differ substantially in size (1,267 amino acids for Dhr1p and 735 amino acids for Dhr2p) and diverge outside the core region. Overexpression of Dhr2p failed to suppress the growth defect in a strain depleted of Dhr1p (data not shown).
Dhr1p and Dhr2p are not required for pre-mRNA splicing.
To test the requirements for Dhr1p and Dhr2p in pre-mRNA splicing, conditional alleles were constructed. A one-step PCR technique was used to replace the chromosomal DHR1 or DHR2 promoter with a pMET::HA cassette (see Materials and Methods). Correct integration resulted in expression of fusion constructs with the HA nonapeptide from the human influenza virus hemagglutinin, under the control of the MET promoter, which is strongly repressed by addition of methionine to the growth medium (9).
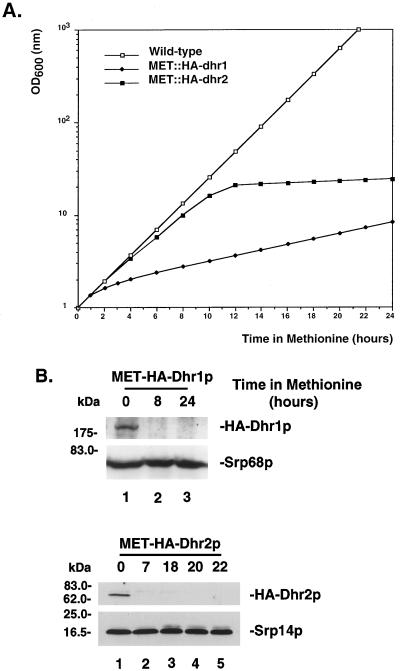
MET::HA-dhr1 and MET::HA-dhr2 strains were pregrown in minimal medium lacking methionine. At mid-log phase, methionine was added to the medium to a final concentration of 20 mM and the OD600 was measured at regular intervals (Fig. 2A). Cells were maintained in exponential growth by dilution with prewarmed medium. Following addition of methionine to the medium, the growth rate of both the MET::HA-dhr1 and MET::HA-dhr2 strains was severely affected. For Dhr1p, the effects of methionine addition were rapid and growth was clearly slowed within 2 h. For Dhr2p, a major increase in doubling time occurred between 10 and 12 h after addition of methionine (Fig. 2A).
FIG. 2.
Construction of MET-regulated alleles of DHR1 and DHR2. (A) Growth rates of MET::HA-dhr1 (closed diamonds), MET::HA-dhr2 (closed squares), and wild-type (open squares) strains following addition of methionine to the medium. Values are corrected for dilution. (B) Western blot analysis of strains described in panel A. Total protein was extracted from similar amounts of cells (according to OD600), separated on SDS-polyacrylamide gels (8 and 15% PAGE for Dhr1p and Dhr2p, respectively), and transferred to nitrocellulose. Membranes were decorated with anti-HA antibodies. Two protein components of the signal recognition particle, Srp14p (16.4 kDa) and Srp68p (69.0 kDa), were decorated with specific antibodies as loading controls.
Depletion of Dhr1p and Dhr2p was followed by Western blot analysis (Fig. 2B). Total protein was extracted at several time points after methionine addition. Equivalent amounts of total protein were resolved by SDS-PAGE and transferred to nitrocellulose membranes that were decorated with anti-HA antibodies. Both HA-Dhr1p and HA-Dhr2p were readily detected under permissive conditions (Fig. 2B, lanes 1). At the first time point inspected after methionine addition (8 h for Dhr1p and 7 h for Dhr2p), no residual HA-tagged protein was detected. As loading controls, Western blots were decorated with anti-Srp14p and anti-Srp68p specific antibodies, directed against protein subunits of the signal recognition particle.
The effects of Dhr1p and Dhr2p depletion on pre-mRNA splicing were tested both in vivo and in vitro. Total RNA was extracted from MET::dhr1 and MET::dhr2 strains grown in the absence of methionine or in the presence of methionine for up to 20 h. Primer extension analysis revealed no effects of depletion of Dhr1p or Dhr2p on the levels of the ACT1 mRNA or pre-mRNA (data not shown). Northern hybridization showed no defects in splicing of the CYH2 pre-mRNA (data not shown) or the U3 snoRNA, which is processed from an intron-containing precursor (see Fig. 9).
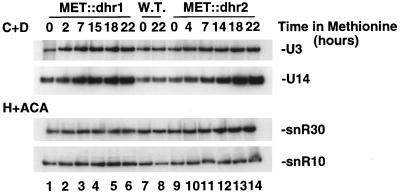
FIG. 9.
Dhr1p and Dhr2p are not required for accumulation of snoRNAs. Total RNA was extracted from MET::dhr1, MET::dhr2, and otherwise isogenic wild-type (W.T.) strains grown in selective minimal medium lacking methionine (0-h time points) and following addition of methionine for the times indicated. Total RNA was resolved in an 8% polyacrylamide gel, transferred to a nylon membrane, and hybridized with oligonucleotides specific for the box C+D snoRNAs (U3 and U14) and the H+ACA snoRNAs (snR10 and snR30).
Silent mutations in vivo can have deleterious effects under the more stringent and suboptimal in vitro conditions. This has been reported for both pre-mRNA splicing (21, 52) and translation (28). Splicing extracts were therefore prepared from MET::dhr1 and MET::dhr2 strains 15 h after methionine addition. Extracts were incubated with in vitro-transcribed ACT1 pre-mRNA, and the products of the reactions were separated on polyacrylamide gels. No clear differences were observed between the wild-type extract and the extracts from the Dhr1p- or Dhr2p-depleted strains (data not shown).
We conclude that neither Dhr1p nor Dhr2p is required for pre-mRNA splicing.
Dhr1p and Dhr2p are localized to the nucleolus.
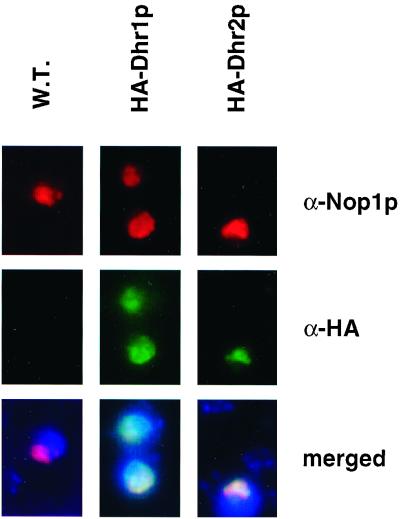
To gain an insight into the function of Dhr1p and Dhr2p, their subcellular localization was analyzed by indirect immunofluorescence microscopy. Strains expressing either the HA-Dhr1p or HA-Dhr2p fusion were grown to mid-log phase and processed for immunofluorescence (see Materials and Methods). Spheroplasts were incubated with a rat monoclonal anti-HA antibody, followed by FITC staining (Fig. 3, green channel). As a nucleolar marker, Nop1p was decorated with a mouse monoclonal antibody in combination with Cy3 staining (red channel). The signal for HA-Dhr1p and HA-Dhr2p was exclusively nuclear with strong enrichment in a crescent-shaped region which colocalized with Nop1p (yellow in merged panels). In yeast, the nucleolus normally occupies a crescent-shaped region covering around a third of the nuclear space and is largely resolved from the chromosomal DNA (stained in blue with 4′,6′-diamidino-2-phenylindole [DAPI], merged panels).
FIG. 3.
Dhr1p and Dhr2p localize to the nucleolus. Indirect immunofluorescence on cells expressing an HA epitope fusion of either Dhr1p or Dhr2p. The Dhr fusions were detected with a rat anti-HA (α-HA) monoclonal antibody, followed by FITC staining. For Nop1p detection, a mouse anti-Nop1p (α-Nop1p) monoclonal antibody was used in combination with Cy3 staining. Both HA fusions showed an intranuclear staining (green channel) which colocalized with Nop1p (red channel) and revealed a classical nucleolar crescent-like shape structure (yellow in merged images). DNA was stained with DAPI (blue in merged images). W.T., wild type.
We conclude that both Dhr1p and Dhr2p localize predominantly to the nucleolus, the site of ribosome synthesis.
Dhr1p and Dhr2p are required for 18S rRNA synthesis.
The nucleolar localization of Dhr1p and Dhr2p prompted us to analyze the requirement for these proteins in pre-rRNA processing.
In eukaryotes, three of the four rRNAs (the 18S, 5.8S, and 25S rRNAs) are produced from a single high-molecular-weight precursor (35S pre-rRNA in yeast) by a complex processing pathway involving both endonuclease and exonuclease digestion (reviewed in references 24 and 67). The legend to Fig. 1 contains a complete description of the pathway.
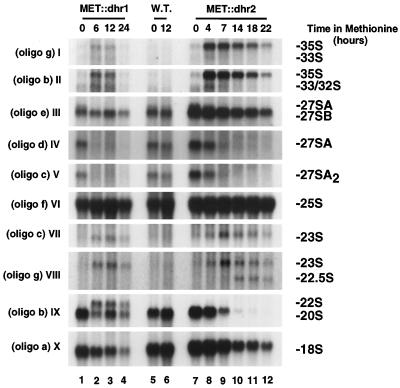
Pre-rRNA processing was analyzed by Northern blot hybridization and primer extension using the MET-regulated alleles. Total RNA was extracted from MET::dhr1 and MET::dhr2 strains grown in medium lacking methionine (0-h samples) and at time points after methionine addition. An isogenic wild-type strain was used as a control. Hybridization with oligonucleotides specific for the mature rRNA species (oligonucleotides a, f, and h) revealed that depletion of either Dhr1p or Dhr2p led to reduced levels of 18S rRNA (Fig. 4X). The steady-state levels of the 25S (Fig. 4VI) and 5.8S (Fig. 5II) rRNAs were mostly unaffected (see below).
FIG. 4.
Dhr1p and Dhr2p are required for 18S rRNA synthesis. Total RNA was extracted from MET::dhr1, MET::dhr2, and otherwise isogenic wild-type (W.T.) strains grown in selective minimal medium lacking methionine (0-h time points) and following addition of methionine for the times indicated. RNA was resolved in a 1.2% agarose-formaldehyde gel, transferred to nylon membrane and hybridized with the oligonucleotides indicated. Oligonucleotide (oligo) b does not distinguish between the 33S and 32S pre-rRNAs, while oligonucleotides d and e do not distinguish between the 27SA2 and 27SA3 pre-rRNAs. However, the 33S and 27SA3 pre-rRNAs are very low in abundance and the signals observed are predominantly due to the 32S and 27SA2 pre-rRNAs.
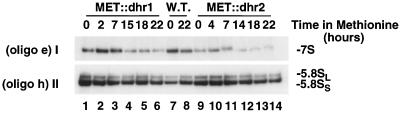
FIG. 5.
Dhr1p and Dhr2p are not involved in the maturation of the 7S pre-rRNA. Total RNA was extracted from MET::dhr1, MET::dhr2, and isogenic wild-type (W.T.) strains grown in selective minimal medium lacking methionine (0-h time points) and following addition of methionine for the times indicated. RNA was resolved in an 8% polyacrylamide gel, transferred to nylon membrane, and hybridized with oligonucleotide (oligo) e (I) or h (II).
Although both Dhr1p and Dhr2p were required for 18S rRNA accumulation, the pre-rRNA processing defects observed on depletion were significantly different. In the Dhr2p-depleted strain, the 35S pre-rRNA was strongly accumulated (Fig. 4I and II), demonstrating the inhibition of cleavage at site A0. The products of cleavage at sites A1 and A2, the 32S, 27SA2, and 20S pre-rRNAs, were all strongly codepleted, showing that processing at these sites was also inhibited (Fig. 4II, V, and IX). Two aberrant processing intermediates were detected in the Dhr2p-depleted strain; the 23S and 22.5S RNAs (Fig. 4VII and VIII and 1C). The 23S RNA is the product of cleavage of the 35S pre-rRNA at site A3 in the absence of prior cleavage at sites A0, A1, and A2 (Fig. 1B). The 22.5S RNA is the product of processing of the 35S and/or 23S RNA at site D, the 3′ end of the mature 18S rRNA. Together, these data indicate that depletion of Dhr2p strongly inhibits processing at sites A0, A1, and A2.
Depletion of Dhr1p resulted in only mild accumulation of the 35S pre-rRNA (Fig. 4I) but strongly reduced the levels of the 27SA2 and 20S pre-rRNAs (Fig. 4V and IX). These results indicated that the inhibition of cleavage was greater at sites A1 and A2 than at site A0. Consistent with this, an aberrant 22S processing intermediate that extends from site A0 to site A3 was detected in the Dhr1p-depleted strain (Fig. 4IX and 1C). The 23S RNA was detected at similar levels in strains depleted of Dhr1p or Dhr2p (Fig. 4VIII). However, the 23S RNA is normally rapidly degraded by the exosome complex of 3′→5′ exonucleases (1) and the level of this intermediate may not well reflect the relative flux through the pathway in different strains.
Processing of pre-rRNAs on the pathway of 5.8S and 25S rRNA synthesis appeared to be mostly unaffected by depletion of Dhr1p or Dhr2p. At late time points, some reduction in the levels of the 27SB (Fig. 4III) and 7S (Fig. 5I) pre-rRNAs was seen. However, the 35S rRNA was also reduced at these times and this is probably a consequence of the reduced growth of the strains. No alteration was seen in the 5.8SL:5.8SS rRNA ratio (Fig. 5II), indicating that relative processing at sites B1S and B1L was unaffected.
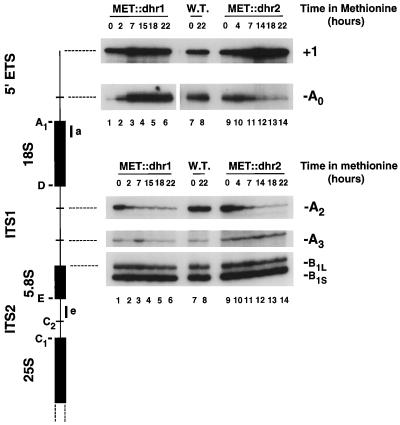
Cleavage in the 5′-ETS and ITS1 was also analyzed by primer extension (Fig. 6). Primer extension from oligonucleotide e, complementary to the 5′ end of ITS2, revealed that depletion of either Dhr1p or Dhr2p strongly inhibited cleavage at site A2 while not affecting processing at sites A3 and B1L or B1S. Primer extension from oligonucleotide a, complementary to the 5′ end of the 18S rRNA, showed an increase in the stop at +1, the 5′ end of the 35S pre-rRNA, that was in good agreement with the results of Northern hybridization, with stronger accumulation on Dhr2p than on Dhr1p depletion. The primer extension stop at A0, the 5′ end of the 33S pre-rRNA, was strongly reduced on depletion of Dhr2p, indicating a greatly reduced level of the 33S pre-rRNA, which is not readily detected by Northern hybridization. In contrast, the primer extension stop at site A0 was greatly increased on depletion of Dhr1p (Fig. 6; in the panel showing the stop at site A0, lanes 1 to 6 were exposed sixfold less than the other lanes). Much of this increase is likely due to the appearance of the 22S RNA (Fig. 4IX) that extends from A0 to A3 (Fig. 1C). In keeping with the onset of growth inhibition, pre-rRNA processing was clearly inhibited in the MET::dhr1 strain 2 h after methionine addition, as shown by the decreased primer extension stop at site A2 and the increase at site A0, whereas a slower onset was seen in the MET::dhr2 strain.
FIG. 6.
Dhr1p and Dhr2p are required for cleavages in 5′-ETS and ITS1. Primer extension analysis through 5′-ETS and ITS1 was performed from oligonucleotides a and e (diagram on left). Total RNA was extracted from MET::dhr1, MET::dhr2, and isogenic wild-type (W.T.) strains grown in selective minimal medium lacking methionine (0-h time points) and following addition of methionine for the times indicated. On Dhr1p depletion, the primer extension stop at site A0 is substantially elevated and the corresponding panel (lanes 1 to 6) is from a sixfold shorter exposure than lanes 7 to 14.
We conclude that both Dhr1p and Dhr2p are required for pre-rRNA cleavage at sites A1 and A2. Impairment of cleavage at these sites leads to strong inhibition of 18S rRNA synthesis that most likely underlies the lethality observed on deletion or genetic depletion of Dhr1p or Dhr2p. The requirements for Dhr1p and Dhr2p in processing at site A0 are different; Dhr2p is strictly required for cleavage at site A0, while cleavage at this site is only mildly delayed in Dhr1p-depleted strains.
Dhr1p is associated with the U3 snoRNA.
The pre-rRNA processing defects reported on Dhr1p and Dhr2p depletion resemble phenotypes associated with depletion and conditional inactivation of several snoRNP components (24, 67). This prompted us to test for physical interactions between Dhr1p or Dhr2p and the snoRNAs and to test for their requirements in snoRNA accumulation.
Interactions between the putative helicases and snoRNAs were addressed by immunoprecipitation. For this, Dhr1p and Dhr2p were each fused to two copies of the z domain of S. aureus ProtA using a one-step PCR transformation procedure in haploid strains (see Materials and Methods). In each case, the ProtA fusion construct was the only copy of the gene and supported wild-type growth, showing the fusion proteins to be fully functional.
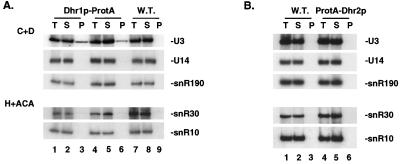
Lysates were prepared from strains expressing Dhr1p-ProtA or ProtA-Dhr2p, and the fusion proteins were bound to IgG-agarose beads. Two independently isolated Dhr1p-ProtA strains were used. Coprecipitating RNAs were extracted from the total, supernatant, and pellet fractions, and cell-equivalent amounts of each fraction were separated on 8% polyacrylamide gels. Hybridization with oligonucleotides specific to several box C+D and box H+ACA snoRNAs revealed that box C+D snoRNA U3 specifically coprecipitated with the Dhr1p-ProtA fusion (Fig. 7A, compare lanes 3 and 6 with lane 9). The efficiency of precipitation (∼30%) was in the same range as that observed for the core box C+D snoRNP protein Nop56p (30). No coprecipitation with Dhr1p-ProtA was observed for the other snoRNAs tested: U14 and snR190 (box C+D), snR10, snR30, snR31, snR33, and snR37 (box H+ACA) (Fig. 7A and data not shown). No coprecipitation of these snoRNAs was detected with ProtA-Dhr2p (Fig. 7B) or with the otherwise isogenic wild-type control strains (Fig. 7A, lanes 7 to 9, and B, lanes 1 to 3). Immunoprecipitation of the fusion proteins was analyzed in parallel by Western blot assay. The two fusion proteins were detected with similar efficiencies in the pellet fractions, indicating that the apparent lack of snoRNA association with ProtA-Dhr2p is not due to a failure in precipitation (data not shown).
FIG. 7.
Dhr1p-ProtA is specifically associated with the U3 snoRNA. Coprecipitation experiments with IgG-agarose beads were performed with lysates from cells expressing ProtA epitope-tagged versions of Dhr1p and Dhr2p. For Dhr1p-ProtA, two independently isolated strains were used. (A and B) Northern blot analyses. RNA was extracted from equivalent amounts of the total (T), supernatant (S), and pellet (P) fractions and loaded in a 1:1:1 ratio. Membranes were hybridized with oligonucleotides specific for the box C+D snoRNAs (U3, U14, and snR190) and the H+ACA snoRNAs (snR10 and snR30).
Coprecipitated RNAs were also directly labeled at the 3′ end with T4 RNA ligase and pCp and separated on 6% polyacrylamide gels. For Dhr1p-ProtA strains, a singly labeled RNA with a gel mobility compatible with the size of U3 was specifically recovered in the pellet (data not shown). No RNA was detectably recovered with the ProtA-Dhr2p fusion (data not shown).
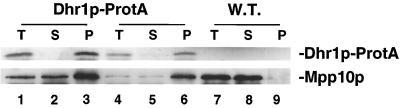
Several proteins are known to interact with U3 (see introduction), and two of these, Mpp10p and Rcl1p (7, 15), were tested for coprecipitation with the Dhr1p-ProtA fusion. Both proteins were efficiently recovered in the pellet fractions (shown for Mpp10p in Fig. 8, compare lanes 3 and 6 with lane 9). We conclude that Dhr1p-ProtA, but not ProtA-Dhr2p, is stably and specifically associated with box C+D snoRNA U3. Attempts to further demonstrate the association of Dhr1p and U3 by density gradient centrifugation were unsuccessful due to the high sensitivity of the ProtA fusion construct to proteolytic degradation (data not shown).
FIG. 8.
Dhr1p-ProtA is specifically associated with Mpp10p. Coprecipitation experiments with IgG-agarose beads were performed with two independently isolated strains expressing a Dhr1p-ProtA fusion. Proteins from the total (T), supernatant (S), and pellet (P) fractions (in a 1:1:35 ratio; see Materials and Methods) were resolved by SDS-PAGE and transferred to nitrocellulose. Membranes were decorated with PAP or anti-Mpp10p antibodies.
The requirement for Dhr1p and Dhr2p in snoRNA accumulation was addressed by Northern blot hybridization (Fig. 9). Total RNA was extracted from cells grown under permissive conditions and after addition of methionine. Hybridization with probes specific to the snoRNAs U3, U14, and U24 (box C+D) and snR10, snR30, snR31, snR33, and snR37 (box H+ACA) revealed no clear reduction on depletion of Dhr1p or Dhr2p (shown for U3, U14, snR10, and snR30 in Fig. 9). Indeed, mild snoRNA accumulation was observed (most evident for U14). The gels were loaded with equal amounts of total RNA, and depletion of the rRNAs leads to an increased signal for unaffected species.
We conclude that neither Dhr1p nor Dhr2p was required for the accumulation of any snoRNA tested, including U3.
DISCUSSION
Here we report the functional characterization of Dhr1p and Dhr2p, two novel members of the DEAH subfamily of putative RNA helicases. Both proteins were initially predicted to be required for pre-mRNA splicing, based on strong homology to splicing factors Prp2p, Prp16p, Prp22p, and Prp43p. However, genetic depletion of neither Dhr1p nor Dhr2p detectably inhibited pre-mRNA splicing in vivo or in vitro.
HA epitope-tagged Dhr1p and Dhr2p fusions were found to colocalize with the nucleolar antigen Nop1p, and consistent with this subcellular localization, both proteins were required for ribosome synthesis. Depletion of either Dhr1p or Dhr2p inhibited pre-rRNA processing at sites A1 and A2 on the pathway of 18S rRNA synthesis but had little, if any, effect on the synthesis of the 5.8S and 25S rRNAs. Interestingly, the requirements for Dhr1p and Dhr2p in cleavage at site A0 were markedly different. A0 cleavage showed little requirement for Dhr1p but was strongly dependent on Dhr2p.
Pre-rRNA processing at sites A1 and A2 also requires the U3, U14, snR10, and snR30 snoRNAs (19, 34, 40, 61). Interaction with the snoRNAs was therefore tested by coprecipitation experiments with ProtA-tagged fusion proteins. The U3 snoRNA was specifically and efficiently (∼30%) coprecipitated with Dhr1p-ProtA, whereas no snoRNA was detected in association with ProtA-Dhr2p. Dhr1p is the first putative RNA helicase to be found stably associated with a snoRNA. The DEAD-box helicases Dpb4p and Rok1p interact genetically with snoRNAs but are not detectably in physical association (35, 66). In contrast, Sen1p interacts weakly (with ∼1% efficiency) with several snoRNA species, possibly reflecting transient interactions at some stages in their biosynthetic pathways (47, 65), and this may also be the case for the nucleoplasmic helicases p50 and p55 (42). Depletion of Dhr1p or Dhr2p did not affect the accumulation of U3 or the other snoRNA species tested, including U14, snR10, and snR30.
In addition to the common box C+D snoRNP proteins Nop1p, Nop56p, and Nop5p/Nop58p (29, 30, 48, 74), several proteins have been detected in specific association with U3 (7, 15, 22, 32, 73; J. Venema, personal communication; this work). Two of these, Mpp10p and Rcl1p (7, 15), were shown to efficiently coprecipitate with Dhr1p-ProtA. Genetic depletion of the U3 snoRNA or any of the previously characterized U3 snoRNP components inhibited cleavage at site A0 in addition to sites A1 and A2 (7, 15, 19, 22, 32, 73). In contrast, depletion of Dhr1p had only a mild effect on A0 cleavage while strongly inhibiting A1 and A2 cleavage. This led to strong accumulation of the 22S pre-rRNA that extends from site A0 to site A3. A similar phenotype was previously seen in strains carrying a partial C-terminal truncation of Mpp10p (31), suggesting that this domain functions together with Dhr1p.
Several sites of interaction between U3 and the pre-rRNAs have been mapped by chemical cross-linking experiments and/or inferred from sequences with potential complementarity and phylogenetically conserved substitutions (6, 8, 17, 18, 38, 39). In yeast, U3 makes two functionally distinct interactions with the pre-rRNAs, at site +470 (in the 5′-ETS) and with the loop of a stem structure located at the 5′ end of the 18S rRNA (5, 54) (Fig. 1B). Analysis of compensatory mutations demonstrated that both of these interactions involve the formation of several consecutive Watson-Crick base pairs and both interactions are required for pre-rRNA cleavage at sites A1 and A2. However, cleavage at site A0 requires the interaction of U3 with the 5′-ETS but not the interaction with the 18S rRNA region (4, 54) (Fig. 1B). This suggested that the U3 snoRNP carries out distinct functions in ribosome synthesis by base pairing to these two sites. This model was supported by the uncoupling of these cleavages on truncation of Mpp10p (31). Since Dhr1p was required for cleavage at sites A1 and A2 but not at site A0, it is likely to mediate the function of the U3 snoRNP at the 5′ end of 18S rRNA rather than the 5′-ETS.
The interaction of U3 with the stem-loop structure at the 5′ end of the 18S rRNA region is mutually exclusive with the formation of the central pseudoknot (18, 39, 54). This is a highly conserved, long-range interaction within the mature small-subunit rRNA that is a key feature in the overall folding of the molecule. We speculate that the U3-18S interaction targets Dhr1p to the pre-rRNA, where it functions in the formation of the pseudoknot structure. Formation of the central pseudoknot in the Escherichia coli rRNA precursor is also prevented by a base-paired interaction, which occurs in cis between the 5′ end of the 16S rRNA and the 5′-ETS. A mutation which blocks the isomerization of this structure prevents processing of the 5′ end of the 16S rRNA, showing the two events to be coupled (12). In both yeast and E. coli, these base-paired interactions are likely to delay the formation of the central pseudoknot, a potentially irreversible step in ribosome assembly. This may be important to allow sufficient time for the binding of ribosomal proteins and other trans-acting factors. We speculate that in strains lacking Dhr1p, this structural rearrangement does not take place, leading to the inhibition of cleavage at sites A1 and A2.
There is an obvious analogy between the model we present here for the role of U3 in targeting an RNA helicase to its site of action and the functions of the bulk of the snoRNAs as modification guides. Other box C+D snoRNAs base pair with the pre-rRNAs to select sites of 2′-O-methylation, which is likely to be carried out by the snoRNP protein Nop1p (43, 62, 69). Similarly the H+ACA snoRNAs select sites for pseudouridine formation by the snoRNA-associated pseudouridine synthase Cbf5p (27, 72, 75). In each case, the snoRNA not only carries the rRNA-modifying enzyme to the rRNA but, by specific base pairing, creates the enzyme recognition site. Similarly, we envisage that the U3-18S base pairing delivers Dhr1p to the site of formation of the central pseudoknot, where it catalyzes the structural isomerization of the rRNA.
ACKNOWLEDGMENTS
We thank J. Aris (University of Florida), S. Baserga (Yale School of Medicine), J. Brown (University of Edinburgh), E. Billy, and W. Filipowicz (Friedrich Miescher-Institut) for their generous gift of antibodies and R. van Nues for plasmid pUC18-55-HA(n).
This work was supported by the European commission (grant Ct95-0009 to the TAPIR project) (A.C. and J.D.B.), the Royal Society (J.D.B.), and the Wellcome Trust (D.L.J.L. and D.T.). D.L.J.L. is on leave from the FNRS.
REFERENCES
- 1.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenas J E, Abelson J N. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billy E, Wegierski T, Nasr F, Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000;19:2115–2126. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvet J P, Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981;26:363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- 9.Cherest H, Kerjan P, Surdin-Kerjan Y. The Saccharomyces cerevisiae MET3 gene: nucleotide sequence and relationship of the 5′ non-coding region to that of MET25. Mol Gen Genet. 1987;210:307–313. doi: 10.1007/BF00325699. [DOI] [PubMed] [Google Scholar]
- 10.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 11.Czaplinski K, Weng Y, Hagan K W, Peltz S W. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 12.Dammel C S, Noller H F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 14.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuller-Pace F V. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 17.Hartshorne T. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 1998;26:2541–2553. doi: 10.1093/nar/26.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes J M. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 19.Hughes J M, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs J S, Anderson A R, Parker R P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacquier A, Rodriguez J R, Rosbash M. A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985;43:423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- 22.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 24.Kressler D, Linder P, de la Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3471. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafontaine D L, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 27.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafontaine D L J, Preiss T, Tollervey D. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol. 1998;18:2360–2370. doi: 10.1128/mcb.18.4.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafontaine D L J, Tollervey D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol. 2000;20:2650–2659. doi: 10.1128/mcb.20.8.2650-2659.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S J, Baserga S J. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc Natl Acad Sci USA. 1997;94:13536–13541. doi: 10.1073/pnas.94.25.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S J, Baserga S J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leeds P, Wood J M, Lee B-S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H V, Zagorski J, Fournier M J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang W-Q, Clark J A, Fournier M J. The rRNA-processing function of the yeast U14 small nucleolar RNA can be rescued by a conserved RNA helicase-like protein. Mol Cell Biol. 1997;17:4124–4132. doi: 10.1128/mcb.17.7.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin R J, Lustig A J, Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987;1:7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- 37.Luking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol. 1998;33:259–296. doi: 10.1080/10409239891204233. [DOI] [PubMed] [Google Scholar]
- 38.Maser R L, Calvet J P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5′ external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci USA. 1989;86:6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 40.Morrissey J P, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mougey E B, Pape L K, Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5′ external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993;13:5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman D R, Kuhn J F, Shanab G M, Maxwell E S. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae: identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 44.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 45.Plumpton M, McGarvey M, Beggs J D. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pringle J R, Adams A E, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen T P, Culbertson M R. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid S R, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 50.Schwer B, Gross C H. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 52.Seraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 54.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiratori A, Shibata T, Arisawa M, Hanaoka F, Murakami Y, Eki T. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast. 1999;15:219–253. doi: 10.1002/(SICI)1097-0061(199902)15:3<219::AID-YEA349>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 56.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 58.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 59.Teigelkamp S, McGarvey M, Plumpton M, Beggs J D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 1994;13:888–897. doi: 10.1002/j.1460-2075.1994.tb06332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tollervey D, Guthrie C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985;4:3873–3878. doi: 10.1002/j.1460-2075.1985.tb04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 63.Tollervey D, Mattaj I W. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ursic D, DeMarini D J, Culbertson M R. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol Gen Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- 65.Ursic D, Himmel K L, Gurley K A, Webb F, Culbertson M R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Venema J, Bousquet-Antonelli C, Gelugne J P, Caizergues-Ferrer M, Tollervey D. Rok1p is a putative RNA helicase required for rRNA processing. Mol Cell Biol. 1997;17:3398–3407. doi: 10.1128/mcb.17.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 68.Wagner J D, Jankowsky E, Company M, Pyle A M, Abelson J N. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Boisvert D, Kim K K, Kim R, Kim S H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Wagner J, Guthrie C. The DEAH-box splicing factor Prp16 unwinds RNA duplexes. Curr Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- 72.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiederkehr T, Pretot R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zebarjadian Y, King T, Fournier M J, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]