Abstract
Lipoprotein lipase (LPL) is one of the most important factors in systemic lipid partitioning and metabolism. It mediates intravascular hydrolysis of triglycerides packed in lipoproteins such as chylomicrons and VLDL. Since LPL was initially discovered in the 1940s, its biology and pathophysiological significance have been well characterized and documented. Nonetheless, several studies in the past decade, with recent delineation of LPL crystal structure and the discovery of several new regulatory factors such as angiopoietin-like proteins (ANGPTL), glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1), lipase maturation factor 1 (LMF1) and Sel-1 suppressor of Lin-12-Like 1 (SEL1L), have completely transformed our understanding of LPL maturation and function.
Keywords: LPL, hyperlipidemia, LMF1, SEL1L, endoplasmic reticulum, ANGPTL, endothelium, GPIHBP1
Lipoprotein lipase (LPL): A Central Player in Systemic Lipid Metabolism
LPL was first discovered in 1943 by Dr. Paul Hanh in dogs as “heparin-activated clearing factor” [1] and renamed to “lipoprotein lipase” by Dr. Edward Korn in 1955 [2, 3]. It was later revealed that LPL is a highly conserved protein among mammalian species with a molecular mass of 50 kDa [4]. It is now well established that LPL plays a key role in lipid metabolism by catalyzing the hydrolysis of intravascular triglycerides packaged in lipoproteins such as chylomicron and very-low-density lipoprotein (VLDL) into fatty acids [5]. Highlighting the central role of LPL in triglycerides metabolism, defects in LPL function result in familial chylomicronemia (see Glossary) and ectopic lipid deposition in humans [4], and neonatal death in mice due to engorgement of chylomicrons into capillaries of the lungs [6].
LPL is highly expressed in adipocytes and myocytes as well as macrophages [7]. Its protein level and activity are tightly regulated by multiple mechanisms in response to the metabolic state and energy demands of the cell [7] (Box 1). For example, fasting, feeding, exercise and cold challenge all can alter the protein level and activity of LPL, thereby shunting fatty acids to specific cell types to meet their cellular energy demand [8, 9]. Recent years have witnessed the identification of several new factors and regulators (Table 1) involved in the regulation of LPL maturation, stability and activity, which have expanded and, in some cases, transformed our understanding of LPL biology.
BOX 1. Multiple Mechanisms in Place to Control LPL Activity in vivo.
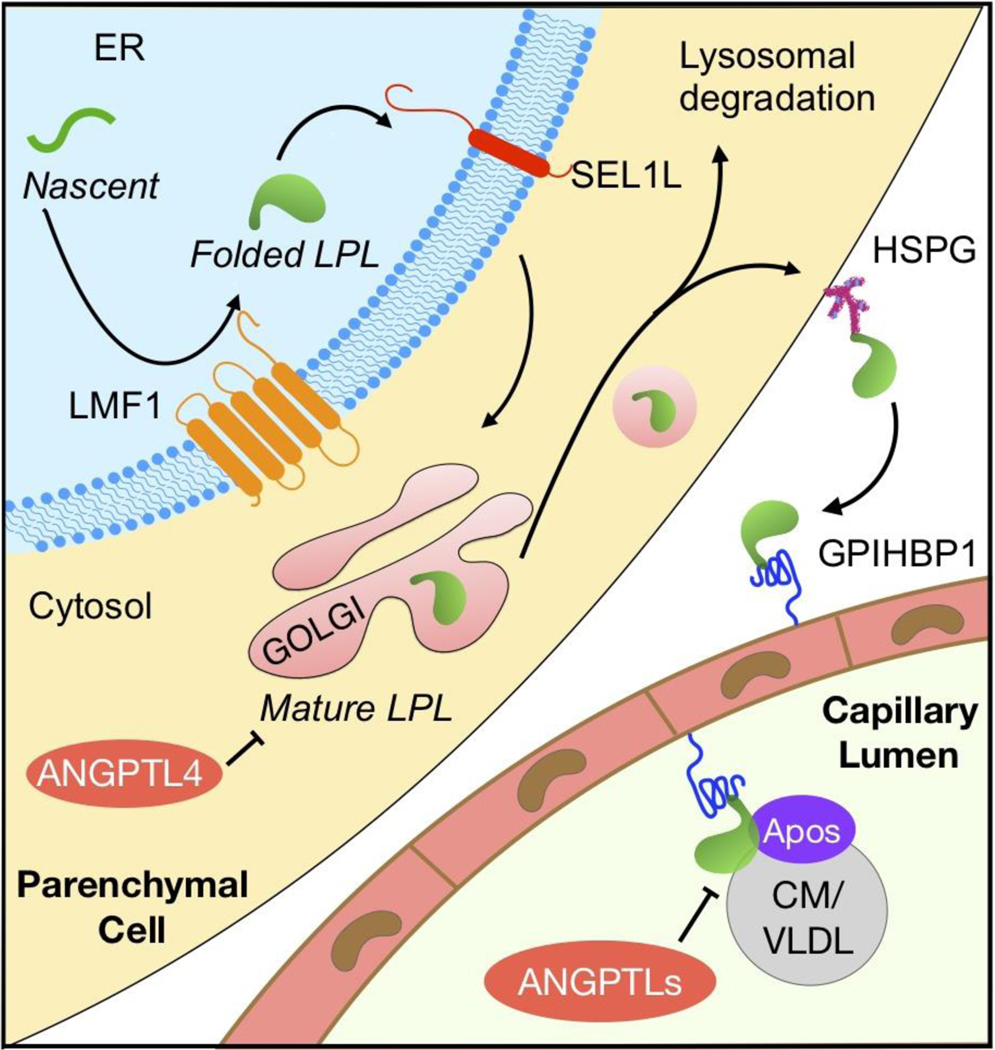
The biosynthesis and function of LPL is controlled at multiple steps (Figure I). Nascent LPL peptide is made and undergoes N-glycosylation in the ER where LMF1 and SEL1L act to ensure its maturation and ER exit [16, 22]. Once reached the Golgi apparatus, LPL is continuously modified with the addition of complex sugar groups [115]. In the trans-Golgi network, LPL binds to heparan sulfate proteoglycan (HSPG), Syndecan-1, in secretory vesicles [29] where it may be stored as oligomers [30], or is targeted for lysosomal degradation [32]. Mature LPL may attach to HSPG at the cell surface, and eventually be captured by GPIHBP1 expressed on the surface of nearby endothelial cells [38, 39]. LPL bound to GPIHBP1 is then transcytosed to the endothelial lumen [41], where LPL mediates intravascular hydrolysis of lipoproteins such as chylomicrons and VLDL. LPL is activated by apo-CII and apo-AV, but inhibited by apo-CI and apo-CIII [7]. The endogenous inhibitors of LPL, namely ANGPTL3, 4 and 8, may further contribute to the regulation of LPL activity [59, 81].
Figure I. The maturation, secretion and function of LPL.
Abbreviation: ANGPTLs, angiopoietin-like proteins including angiopoietin-like proteins 3, 4, and 8. Apos: apolipoproteins including apolipoprotein-CI, -CII, -CIII, -AV and -E. CM, chylomicron. ER, endoplasmic reticulum. HSPG, heparan sulfate proteoglycan. LMF1, lipase maturation factor 1. SEL1L, Sel-1 suppressor of Lin-12-Like 1. VLDL, very low-density lipoproteins.
Table 1.
Factors and regulators involved in LPL maturation and function.
| Regulators | Tissue distribution | Major action site | Function | Potential mechanisms |
|---|---|---|---|---|
| LMF1 | Ubiquitously expressed | ER | Required for the acquisition of enzymatic activities of LPL | Serve as an ER chaperone to promote the proper LPL folding and maturation |
| SEL1L | Ubiquitously expressed | ER | Required for the ER exit of LPL | Ensure proper folding of LPL possibly by preventing LPL aggregation in the ER |
| ANGPTL4 | Adipose tissue, liver, intestine, and muscle | Likely post-ER compartments | Inhibit LPL activity in autocrine or paracrine manner | Promote LPL unfolding and intracellular cleavage; prevent the interaction between LPL and lipoproteins |
| ANGPTL3 | Liver | Mostly capillary lumen | Suppress LPL activity in an endocrinal manner during feeding | Inhibit LPL activity in a complex with ANGPTL 8 via an endocrine action |
| ANGPTL8 | Liver, white and brown adipose tissue | Capillary lumen, possibly adipose tissue | Suppress LPL activity in an endocrinal manner during feeding; may also reduce ANGPTL4 activity in adipose tissue during feeding | Work with ANGPTL3 to inhibit LPL activity via an endocrine action; may also interact and suppress ANGPTL4-mediated LPL inhibition in adipose tissue |
| GPIHBP1 | Capillary endothelium in adipose tissue, heart and skeletal muscle | Plasma membrane of capillary endothelium | Transport LPL from interstitial space to endothelial lumen, prevent LPL unfolding | Capture and stabilize LPL with high affinity, bi-directionally transcytose LPL |
| Apo-CII | Liver | Primarily capillary lumen | Cofactor required for LPL activity in endothelial lumen | May trigger LPL conformational change to allow substrate entry |
| Apo-AV | Liver | Primarily capillary lumen | Enhance LPL activity | May direct lipoprotein substrates to the active site of LPL |
| Apo-CI | Liver | Primarily capillary lumen | Reduce LPL-mediated triglyceride hydrolysis | Unclear, may displace substrates from LPL |
| Apo-CIII | Liver, small intestine | Primarily capillary lumen | Reduce LPL-mediated triglyceride hydrolysis | Unclear, may displace substrates from LPL |
| Apo-E | Liver, macrophages | Primarily capillary lumen | Reduce LPL-mediated triglyceride hydrolysis when apo-E concentration is high | Unclear |
LPL Structure
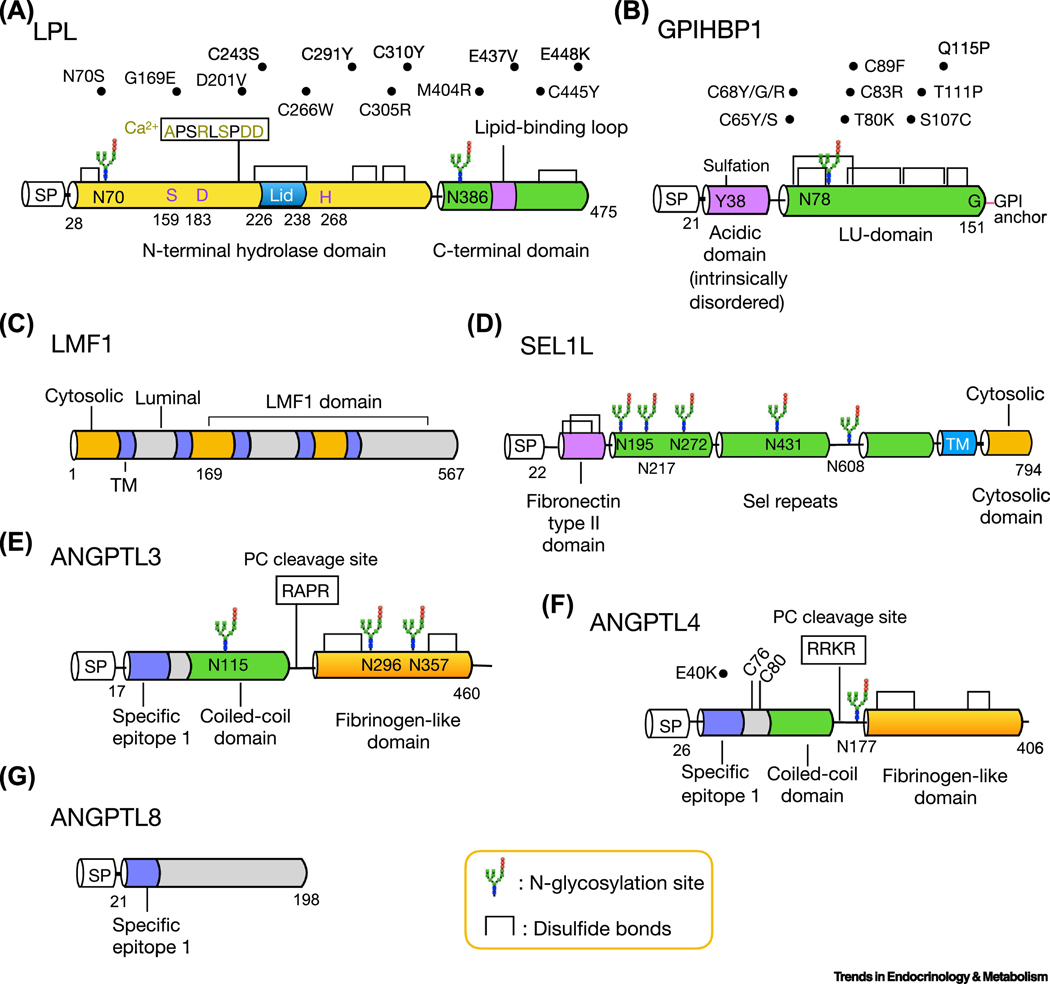
The propensity of the N-terminal domain of LPL to unfold makes structural crystallization of LPL alone impossible [10]. Recently, two groups reported the structure of the LPL protein complexed with its endothelial cofactor glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1) (more below), which provides fresh and valuable insights into the structure and functional unit of LPL [10, 11] (Box 2). Both studies revealed that LPL contains an α/β-hydrolase fold at the N-terminal domain and a flat β-barrel region at the C-terminal domain, connected by a hinge region (Figure 1a). The N-terminal domain of LPL also harbors a serine protease-like catalytic triad (Ser159, Asp183 and His268), and a lid (245–265) which presumably can adopt an open or closed conformation to regulate substrate access to the active site [11]. Several N-terminal residues (Ala194, Arg197, Ser199, Asp201 and Asp202) are involved in the coordination of a calcium ion into the LPL molecule. On the other hand, the C-terminal domain of LPL contains a tryptophan-rich lipid binding region (412–422) for substrate recognition [10, 11]. The hydrophobic residues in the N-terminal lid and C-terminal lipid binding regions of LPL create hydrophobic patches which may facilitate the entry of triglycerides to the active site [10, 11]. At the same time, a large, flat continuous basic patch spanning both N- and C-terminal domains of LPL may be involved in the interaction with the negatively-charged extracellular heparan sulphate proteoglycan and transient binding of the acidic domain of GPIHBP1 [10, 11] (Figure 1b).
BOX 2. LPL Functional Unit: Monomer or Dimer?
The functional LPL unit has long been thought to be a homodimer [78, 116–118]; however, this model is now being challenged. Although LPL and GPIHBP1 form dimeric 2:2 complex in the crystallography, this dimeric orientation of LPL seems to be incompatible with its function as the active site of LPL would be blocked by the lipid-binding region of another LPL molecule [10, 11]. Birrane et al. [10] proposed a lipid-binding displacement model where the lipid-binding region of LPL is flexible and can be displaced from the active site upon the binding of substrates such as lipoproteins. In contrast, Arora et al. [11] argued that this dimeric orientation of LPL is not physiologically relevant, but rather an artifact resulted from crystal packing interaction. Using size exclusion chromatography coupled with multi-angle light scattering, they observed that the molecular mass of the LPL-GPIHBP1 complex is 75 kDa, pointing to a single copy of each component in the complex [11]. In addition, Kristensen et al. [40] showed that LPL monomer is stable and functional and that ANGPTL4 inhibits LPL activity by promoting the unfolding of LPL monomer. Using density gradient ultracentrifugation, Beigneux et al. [119] showed that newly secreted LPL proteins are monomers and that the use of heparin in previous studies may be problematic as it induces its dimerization [120–122]. In conclusion, LPL may adopt different conformations in vivo depending on its subcellular localization, cell types, the nature of substrates and cofactors, and physiological settings. Additional future studies delineating LPL structure in near native conditions (i.e. cryoEM) is a key to settle the debate once and for all.
Figure 1. Functional domains, post-translational modifications and disease mutants of LPL and its regulators.
Schematic diagrams of amino acid structure of LPL (a), GPIHBP1 (b), LMF1 (c), SEL1L (d), ANGPTL3 (e), ANGPTL4 (f) and ANGPTL8 (g). The cytosolic, transmembrane and luminal domain of LMF1 are colored with orange, purple and gray, respectively (c). The two cysteine residues of ANGPTL4, C76 and C80, are required for its oligomerization (f). Several human mutations of LPL (a), GPIHBP1 (b) and ANGPTL4 (f) are listed above the structure. SP: signal peptide; TM, transmembrane domain.
Regulation of LPL Maturation in The Endoplasmic Reticulum (ER)
Nascent LPL protein undergoes folding and attains its catalytic activity in the ER, prior to entering the secretory pathway [12]. To obtain a properly folded structure, nascent LPL protein is subjected to two post-translational modifications, N-glycosylation and disulfide bond formation, in the ER [13] (Figure 1a). LPL has two N-glycosylation sites on residues Asn 70 and 386 (N70 and N386), of which glycosylation at N70 has been shown to be essential for LPL activity and intracellular trafficking [14]. In addition, LPL forms five intra-molecular disulfide bonds [10]. The formation of the three N-terminal disulfide bonds, Cys 243-Cys 266 (C243–266), C291–302 and C305–310, is essential for LPL activity [13], while the C-terminal disulfide bonds may be important for LPL and GPIHBP1 binding [10]. Below we discuss key factors involved LPL folding and maturation in the ER.
Lipase maturation factor 1 (LMF1)
Combined lipase deficiency (Cld) is a naturally occurring mutation in mouse chromosome 17 discovered in 1983 [15]. Mice carrying the cld mutation in both alleles (cld/cld) develop severe hypertriglyceridemia and die within 1 to 2 days postnatally, due to the accumulation of inactive LPL and hepatic lipase (HL) in the ER (Table 2) [15]. In 2007, using positional cloning technique, Péterfy et al. identified the pathogenic gene, Tmem112, in cld/cld mice and re-named it as lipase maturation factor 1 (LMF1) [16]. LMF1 has been predicted to be an ER-resident protein containing five transmembrane domains and a conserved C-terminal LMF1 domain [16] (Figure 1c). Mice with global LMF1 deficiency phenocopies cld/cld mice with severe hypertriglyceridemia, while expression of wildtype LMF1 restores the function of LPL and HL in cld/cld cells (Table 2) [16, 17], demonstrating the requirement and necessity of LMF1 in lipase maturation.
Table 2.
Mouse models of LPL dysfunction and hyperlipidemia.
| Mouse model | Description | The effect on LPL | Phenotype | Reference(s) |
|---|---|---|---|---|
| cld/cld | Autosomal recessive mutation in mouse chromosome 17, a naturally occurring mutation | The loss of LPL (and hepatic lipase) activity | Develop severe hyperlipidemia, milky serum, ischemia, cyanosis soon after birth, and died within 36–48 hours after birth. | [15] |
| Lmf1 −/− | Global LMF1 deficiency by gene trap mutagenesis | The loss of LPL (and hepatic lipase) activity | Recapitulating the phenotypes of cld/cld mice. | [17] |
| Sel1LAdipoq-cre | Adipocyte-specific SEL1L deficiency using adiponectin promoter-driven Cre | ER retention of LPL aggregates, and reduced LPL secretion | Postprandial hyperlipidemia and resistant to HFD-induced obesity with ectopic lipid accumulation in the liver upon HFD. | [22] |
| Angptl4-Tg | Whole body overexpression of mouse ANGPTL4 | Significantly reduced postheparin LPL activity | Severe hypertriglyceridemia and chylomicronemia in fasted state; retention of chylomicrons in plasma after 24-hour fasting; reduced fat mass; increased lipolysis in adipose tissue; mild insulin resistance | [83, 84] |
| Hepatocyte Angptl4-Tg | Hepatocytespecific overexpression of human ANGPTL4 driven by the ApoE promoter | Mildly reduced post-heparin LPL activity | Increased plasma triglycerides and cholesterol under fed condition. | [85] |
| Angptl3 and Angptl8 | Overexpression of mouse ANGPTL3 by adenoviral infection or human ANGPTL8 by hydrodynamic DNA delivery | Not tested | Increased plasma triglyceride levels | [93], [92] |
| Gpihbp1 −/− | Global deficiency in GPIHBP1 | Mis-localization of LPL in the interstitial space | Early onset of severe chylomicronemia within 4–14 weeks of age | [34, 38] |
| Apoa5−/−; Apoa5 knockdown | Global knockout of Apo-AV; 60% knockdown of hepatic Apo-AV protein using targeted antisense oligonucleotide treatment | Decreased plasma LPL activity | Significantly increased plasma triglyceride levels; reduced plasma lipid clearance rate | [49, 50] |
| Apoc1 overexpression | Hemizygous expression of the human apoc1 gene; | Not tested | Markedly increased plasma triglyceride levels; mildly increased plasma cholesterol levels | [55] |
| Apoc3 overexpression | Overexpression of human apo-CIII | Not tested | Markedly increased plasma triglyceride levels; reduced clearance of VLDL triglycerides | [54] |
Abbreviation: HFD: high-fat diet; Tg: Transgenic.
The mechanism underlying the role of LMF1 in LPL maturation remains largely unclear. To add further confusion, a series of LMF1 papers were retracted. LMF1 may stabilize LPL in the ER by promoting its dimerization [18] (Box 2), or interact with ER-resident chaperones such as ERp72, ERp44, ERdj5 and thioredoxin to promote disulfide bond formation of LPL [19]. Hence, LMF1 is likely involved either directly or indirectly in the folding and maturation of nascent LPL in the ER.
Sel-1 suppressor of Lin-12-Like 1 (SEL1L)
SEL1L is a type-I transmembrane protein with a large N-terminal luminal domain, a transmembrane and short cytosolic domain [20] (Figure 1d). It is an obligatory cofactor for the E3 ligase HRD1 in ER-associated degradation (ERAD), a principal mechanism that specifically targets ER proteins for proteasomal degradation in the cytosol [21]. In 2014, while characterizing adipocyte-specific Sel1L-deficient mice (Table 2), we found that these mice develop postprandial hypertriglyceridemia, as a result of LPL retention in the ER [22]. In the absence of SEL1L, LPL forms high-molecular-weight complexes or aggregates in the ER. While at the time we proposed that SEL1L may act alone in helping LPL maturation in the ER, whether SEL1L acts through HRD1 ERAD or not and whether nascent LPL is a misfolding prone protein remain exciting open questions. Indeed, our recent studies demonstrated that SEL1L-HRD1 ERAD controls many physiological processes such as food and water intake, and energy metabolism via controlling the maturation and hence function of specific substrates, such as proopiomelanocortin, vasopressin prohormone, and cAMP-responsive element-binding protein 3-like protein 3 (CREBH), respectively [23–26]. In light of these new data, we now speculate that nascent LPL may be misfolding-prone and that clearance of misfolded LPL by SEL1L-HRD1 ERAD may play a critical role in LPL maturation and subsequent acquisition of its enzymatic activity.
Regulation of LPL in Post-ER compartments
After exiting the ER, LPL moves to the Golgi apparatus for additional trimming and modifications of its glycans. In the trans-Golgi network (TGN), LPL is sorted to different vesicles for secretion via distinct mechanisms, including a constitutive secretory pathway via a non-specific “bulk-flow” mechanism or a regulated secretory pathway requiring certain sorting signals, as demonstrated in in vitro studies [27, 28]. Recently, Sundberg et al. [29] showed that LPL in TGN may enter regulated secretion following binding to the heparan sulfate chain of Syndecan-1, an integral membrane heparan sulfate proteoglycan enriched in sphingolipid sphingomyelin in exocytic vesicles. LPL may be stored in intracellular vesicles in an inactive helical oligomeric form, possibly through the binding with Syndecan-1, which move to the cell surface and is released upon nutritional signaling [30]. However, the physiological relevance of these findings remains to be demonstrated.
Mature LPL protein may also be degraded by the lysosome-mediated mechanism [28]. A study using cardiomyocyte-specific unc-51 like autophagy activating kinase 1 (ULK1) knockout mice showed that LPL is degraded by macroautophagy [31]. SorLA, a family member of vacuolar protein sorting 10 (VPS10) involved in TGN-to-endosome sorting, may mediate LPL trafficking from the Golgi to endosomes and lysosomes [32]. Future studies are required to delineate the physiological significance and molecular mechanism underlying autophagy-mediated LPL degradation.
Regulation of LPL Function at The Endothelium
GPIHBP1
GPIHBP1 was cloned in a screen for HDL binding factors in 2003 [33]. GPIHBP1, as suggested by its name, is a capillary endothelial protein anchored to the cell surface via a glycosylphosphatidylinositol (GPI-) anchor that can be cleaved by phosphatidylinositol-specific phospholipase C [33]. The tissue distribution pattern of GPIHBP1 resembles that of LPL, with the exception of capillaries of the lung where GPIHBP1 is expressed but LPL is not [34]. The structure of human GPIHBP1 is highly asymmetric containing an intrinsically disordered N-terminal acidic region and a Ly6/uPAR (LU)-domain composed of three-fingered loop mediated by disulfide bonds [35] (Figure 1b). N-glycosylation at Asn78 of GPIHBP1 is required for its intracellular trafficking as replacing Asn78 to Gln leads to ER retention [36]. Like other GPI-anchored proteins, GPIHBP1 contains a hydrophobic GPI signal peptide at the C-terminal Gly151, which is replaced by a GPI moiety following proteolytic cleavage [4].
LPL interacts with GPIHBP1 at 1:1 ratio via a dual binding mechanism: the basic residues in the C-terminal domain of LPL first interact with the acidic domain of GPIHBP1 to form an “encounter complex”, which then guides the complex maturation by forming a large interface between the C-terminal domain of LPL and the LU-domain of GPIHBP1 [35, 37]. This high affinity interaction allows GPIHBP1 to quickly capture heparan sulfate proteoglycan-bound LPL in the subendothelial spaces and then shuttle it to the capillary lumen [38, 39]. In addition, GPIHBP1 may prevent LPL unfolding that occurs either spontaneously or induced by endogenous inhibitors such as angiopoietin-like proteins (ANGPTLs, more below) [4, 37, 40]. Moreover, the Tyr38 of GPIHBP1 is modified by O-sulfate (Figure 1b), which has been shown to enhance the GPIHBP1-LPL binding and promote LPL stabilization [35].
GPIHBP1 is also required for LPL function. Using confocal and electron microscopy, Davies et al. [41] reported that the movement of LPL by GPIHBP1 across the endothelial cells is through transcytosis, independent of caveolin-1. Indeed, mice with GPIHBP1 deficiency (Gpihbp1−/−) develop severe chylomicronemia under chow diet due to the retention of LPL in the interstitial space surrounding adipocytes and myocytes [34, 38] (Table 2). Moreover, GPIHBP1 serves as a platform for LPL and lipoproteins. Goulbourne et al. [42] demonstrated that the physical association of triglyceride-rich lipoproteins along the capillary endothelium depends on the presence of the GPIHBP1-LPL complex. In either Gpihbp1−/− or LPL-scarce capillaries, lipoproteins are unable to bind to the endothelial wall.
Apolipoproteins
The obligatory lipoprotein-derived cofactor for LPL activation was identified in 1970 as apolipoprotein-CII (apo-CII) in HDL [43]. The first loss-of-function mutation in human apo-CII was identified in 1978. Deficiency of apo-CII leads to failure to clear plasma triglycerides and subsequent development of severe hyperlipidemia [44]. Apo-CII is primarily produced by the liver and incorporated into chylomicrons, VLDL and HDL [45]. The binding of apo-CII to LPL may trigger a conformational change in LPL to allow subsequent substrate entry to the active site [45]. The activity of apo-CII may be further increased by the binding of apolipoprotein-AV (apo-AV), a low abundance plasma apolipoprotein produced by the liver [46]. It has been shown that overexpression of apo-AV in mice promotes postprandial triglycerides clearance and lipoprotein-derived fatty acid uptake by heart, skeletal muscle and adipose tissue [47, 48], while mice with apo-A5 deficiency develop hyperlipidemia with reduced post-heparin plasma LPL activity [49, 50] (Table 2 and 3). Although the underlying molecular mechanism remains obscure, it has been suggested that apo-AV may help direct lipoproteins to the site where hydrolysis occurs [51].
Table 3.
Mouse models with LPL gain-of-function and hypolipidemia.
| Mouse model | Description | The effect on LPL function | Phenotype | Reference(s) |
|---|---|---|---|---|
| Angptl4 −/− | Global deficiency in ANGPTL4 | Significantly increased postheparin LPL activity in both feeding and fasting conditions | May increase the mass of white adipose tissue; reduced plasma levels of triglycerides in both feeding and fasting conditions; reduced survival when fed saturated fat-rich diet due to the development of ascites and peritonitis | [85, 123, 124] |
| Adipocytespecific Angptl4−/− | Adipocyte-specific ANGPTL4 deficiency using adiponectin promoter-driven Cre | Increased LPL activity in both adipose tissue and plasma | Reduced plasma triglyceride levels when fed both chow- and highfat-diet; increased triglyceride clearance from the blood; increased lipid uptake and lipolysis in adipose tissue | [86] |
| Angptl3 −/− | Global deficiency in ANGPTL3 | Increased postheparin plasma activity of LPL (and hepatic lipase) | Low plasma lipid concentration; increased triglyceride clearance; reduced weight of white adipose tissue under a HFD | [102, 125] |
| Angptl8 −/− | Global deficiency in ANGPTL8 | Increased postheparin plasma activity of LPL | Markedly reduced plasma triglyceride levels in the fed state; accelerated clearance of dietary lipid from the blood; reduced white adipose tissue size; reduced postprandial uptake of triglycerides from VLDL by white adipose tissue | [102] |
| Hepatocyte- specific Angptl8−/− | Hepatocytespecific ANGPTL8 deficiency using albumin promoterdriven Cre | Significantly increased postheparin plasma LPL activity | Reduced plasma triglycerides levels in fed status | [104] |
| Ad-Apoa5 or hApoa5-Tg | Adenoviral- mediated Apo-AV overexpression; Transgenic mice with overexpression of human Apo-AV | Increased postheparin plasma LPL activity | Accelerated clearance of VLDL-triglycerides; Reduced plasma triglyceride levels | [49, 126, 127] |
| Apoc1 −/− | Global knockout of apo-CI | not tested | Reduced plasma triglycerides levels; accelerated clearance of dietary lipids from the blood; increased fatty acids uptake by white adipose tissue; reduced hepatic production of VLDL | [53] |
| Apoc3 −/− | Global knockout of apo-CIII | not tested | Marked reduction in VLDL cholesterol and triglycerides; Accelerated clearance of triglyceride-labeled emulsion particles | [52] |
Abbreviation: Ad, adenoviral; HFD: high-fat diet;
In contrast to apo-CII and AV, apo-CI and CIII have been implicated as negative regulators of LPL activity [52–55]. Mice with overexpression of human apo-CI or apo-CIII exhibited hypertriglyceridemia and delayed clearance of plasma lipid [54–56] (Table 2), whereas the deletion of either apo-CI or apo-CIII has the opposite effect [52, 53] (Table 3). Similarly, apo-E, an apolipoprotein required for hepatic uptake of the remanent of chylomicron and VLDL, may also be able to inhibit LPL activity, especially at high concentration [57, 58]. However, the mechanism by which they inhibit LPL activity and their physiological relevance remain unclear.
Tissue-Specific LPL Regulators
Angiopoietin-like proteins (ANGPTLs)
ANGPTL3, 4 and 8 are endogenous LPL antagonists and contribute to tissue-specific regulation of LPL under diverse physiological cues [59]. While ANGPTL3 is exclusively expressed in the liver, ANGPTL4 and 8 are abundant in the liver and adipose tissue. The loss-of-function variants of ANGPTL3, 4 and 8 have been identified in humans with reduced plasma lipid levels [60–62], implicating them as potential targets treating cardiovascular and metabolic diseases [59]. All ANGPTL proteins, except ANGPTL8, share a similar molecular structure with an N-terminal coiled-coil and a C-terminal fibrinogen-like domain connected by a linker region [63–65] (Figure 1e-g). Both ANGPTL3 and 4 are cleaved by proprotein convertases (PC) in the linker region between two functional domains [66, 67]. A conserved region, LAXGLLXLGXGL, designated as specific epitope 1 (SE1) shared by the N-terminal domain of ANGPTL3 and ANGPTL4, is required for LPL binding and inhibition [68]. ANGPTL8 structurally resembles the coiled-coil domain of ANGPTL3 and 4, which also contains the conserved motif for LPL binding [69]. Hence, ANGPTL8 is an atypical member of the ANGPTL family [69].
ANGPTL4
Angptl4, originally named as fasting-induced adipose factor (Fiaf), was reported independently by three groups in 2000 in screens for novel targets of peroxisome proliferator-activated receptors (PPARs) [70–72]. ANGPTL4 is evolutionarily conserved between human, mouse and rat [73], and human ANGPTL4 is 405 amino acids in length with a molecular weight of 50 kDa. ANGPTL4 alone is a potent inhibitor of LPL; however, the underlying mechanisms remain controversial. It may be through different, but not necessarily mutually exclusive, mechanisms: 1) ANGPTL4 may promote intracellular cleavage of LPL in post-ER compartment via PC subtilisin/kexin type 3 (PCSK3) (a.k.a. furin), leading to its subsequent intracellular degradation [74, 75]; 2) ANGPTL4 may reversibly bind to LPL, possibly at the lid domain, to block its interaction with substrates [76, 77]; 3) ANGPTL4 may convert LPL homodimers to monomers [78] and 4) ANGPTL4 may promote the unfolding of the hydrolase domain of LPL monomer irreversibly [40]. In vitro, the estimated t1/2 for ANGPTL4-mediated unfolding of LPL monomer is about 45–60 min [40]. Moreover, it has been suggested that the inhibitory effect of ANGPTL4 on LPL requires its oligomerization through disulfide bonds formed between Cys 76 and Cys 80 [64, 73, 79]. However, a more recent study reported that ANGPTL4 with reduced and alkylated disulfide bonds has a similar capacity to inhibit LPL and that the covalent oligomerization of ANGPTL4 may be more important for its stability rather than activity [80].
Fasting increases ANGPTL4 levels in adipose tissue, downregulating adipose LPL activity and routing triglycerides towards oxidative tissues [59, 81, 82]. Similarly, physical exercise has been shown to induce ANGPTL4 specifically in non-exercising muscles to shunt fatty acids to exercising muscles [8]. Mice with ANGPTL4 overexpression developed hypertriglyceridemia in both fed and fasting conditions [83–85], whereas ANGPTL4-deficient mouse exhibited accelerated blood triglyceride clearance and hypotriglyceridemia [85–88] (Tables 2 and 3). Several genome-wide association (GWAS) studies have identified a loss-of-function variant of human ANGPTL4 with Glu40-to-Lys substitution (E40K) in the coiled-coil domain which is associated with lower serum triglycerides and reduced risk of coronary artery disease (Figure 1f) [60].
ANGPTL3
ANGPTL3 was initially identified in 2002 as angiopoietin-like lipoprotein modulator 1 (Allm1) in KK/San mice that exhibit abnormally low plasma triglyceride levels [89]. The inhibitory effect of ANGPTL3 towards LPL may be activated by PC-mediated proteolytic cleavage to generate a potent N-terminal peptide [90] – a process that may be negatively modulated by the O-glycosylation at Thr 220 immediately adjacent to the PC cleavage site [91]. Unlike ANGPTL4, ANGPTL3 itself has low potency towards LPL. Although overexpression of ANGPTL3 has been shown to reduce LPL activity and increase plasma triglyceride levels in the absence of ANGPTL8 [92], ANGPTL3 at a physiological-relevant concentration has little effect on plasma triglyceride levels, unless interacted with ANGPTL8 [93–95] (Table 2, more below).
ANGPTL8
In 2012, three groups independently identified ANGPTL8, which was originally named as refeeding-induced fat and liver (RIFL) or lipase inhibition (lipasin), as a novel regulator of lipid metabolism [95–97]. The expression of ANGPTL8 in liver and adipose tissue is highly induced by feeding [98]. It has been shown that ANGPTL8 is activated upon binding to ANGPTL3 [92, 99], which in turn enhances the potency of ANGPTL3 towards LPL [93, 95, 99]. The formation of the ANGPTL3–8 complex during feeding has been proposed to direct dietary fatty acids away from muscle and heart, which may facilitate the energy storage in white adipose tissue [59]. In keeping with this model, LPL activity is enhanced in cardiac and skeletal myocytes during feeding in Angptl3- or Angptl8-deficient mice, leading to increased fatty acids uptake by heart and muscle and reduced plasma triglyceride levels [100–102].
Both ANGPTL4 and 8 are expressed in adipose tissue; however, how they interact remains poorly understood. Recent studies using purified ANGPTL4 and 8 proteins showed that ANGPTL8 can interact and suppress the ANGPTL4 activity in a dose-dependent manner [94, 103]. In line with this finding, Oldoni et al. [104] recently reported that deletion of ANGPTL8 in adipocytes leads to increased postprandial plasma triglyceride levels in mice, and that ANGPTL4-mediated inhibition of LPL activity can be attenuated by co-expression of ANGPTL8. Hence, these studies suggest that ANGPTL8 inhibits ANGPTL4 activity to promote LPL-mediated hydrolysis and fatty acids uptake in adipose tissue during feeding [104]. Future studies are required to further establish the (patho-)physiological importance of this model and delineate the underlying mechanism.
ANGPTLs vs. GPIHBP1
Although GPIHBP1 has been shown to protect LPL from the inhibitory effects of ANGPTLs at the endothelium [4, 37, 40], several in vitro experiments suggest that ANGPTLs still retain substantial capacity to bind and inhibit the activity of GPIHBP1-bound LPL [105, 106]. Using a real-time system to monitor the direct interaction between LPL and GPIHBP1 in vitro, Shetty et al. [106] reported that both ANGPTL4 and ANGPTL3–8 complex may dissociate and hence inactivate LPL from GPIHBP1 at a slow but steady rate. However, it should be noted that, unlike the ANGPTL3–8 complex, the primary physiological locations where ANGPTL4 inhibits LPL, either in an intracellular, subendothelial, or intravascular compartments, remain unclear and may vary according to different physiological conditions [74].
Human Disease Mutants
To date, more than 100 mutations in the LPL gene have been reported in patients with familial chylomicronemia. In most cases, LPL deficiency is diagnosed during infancy or childhood [107]. Several mutations may interfere with LPL maturation and function by disrupting N-glycosylation [108] and disulfide bonds formation [109] (Figure 1a). In addition, LPL Asp201Val [110] abolishes the binding of calcium to LPL, leading to LPL misfolding and secretion defect [10]. Substitution of glycine with glutamic acid at residue 169increases lysosomal degradation of LPL, leading to LPL deficiency [111]. Several C-terminal mutants, such as Met404Arg, Cys445Tyr and Glu448Lys, have been shown to interfere with the interaction between LPL and GPIHBP1 [10, 112]. The Glu437Val mutation increases the susceptibility of LPL to endoproteolytic cleavage, while having little-to-no effect on LPL-GPIHBP1 interaction [113]. Nevertheless, the underlying molecular mechanisms for other LPL mutants remain obscure.
Loss-of-function disease mutations leading to LPL deficiency and hyperlipidemia have also been identified in LPL regulators such as LMF1, apo-CII, apo-AV and GPIHBP1. Many mutations in the LU-domain of GPIHBP1 interfere the formation of disulfide bonds in the LU-domain, which causes multimerization of GPIHBP1 protein on the cell surface and abolishes LPL-GPIHBP1 interaction [4] (Figure 1b). Other GPIHBP1 mutations, such as Thr80Lys and Gly175Arg that interfere with N-glycosylation and GPI-anchor of GPIHBP1, respectively, may alter intracellular trafficking of GPIHBP1 [114]. Unlike these GPIHBP1 mutants, how the mutations of LMF1, apo-CII and apo-AV affect LPL function requires further investigation.
Conclusion Remarks and Future Perspectives
As a central player in systemic triglycerides partitioning and metabolism, LPL has been intensively studied since its first discovery around 80 years ago. Today, tremendous efforts are continuously being invested to understand LPL regulation. Recent studies have indeed revealed that LPL function is meticulously regulated at multiple stages during its biosynthetic process (Box 1). These mechanisms as described above may work together to ensure a more precise lipid partitioning based on the energy needs of the cell [7].
However, despite these advances, many questions remain (summarized in Outstanding Questions). With the identification of new factors or pathways, these future studies will certainly help paint a more comprehensive picture for LPL regulation and function, which will help design more accurate approaches to target LPL in vivo.
Outstanding Questions.
What is the structure of LPL in near native conditions? Does LPL adopt different conformations in vivo depending on its subcellular localization, cell types, the nature of substrates and cofactors, and physiological settings?
How does nascent LPL protein undergo maturation in the ER? Whether nascent LPL protein is misfolding prone? If so, how is misfolded LPL cleared from the ER?
Is the folding efficiency of LPL in the ER regulated by physiological signals such as fasting and feeding in different cell types?
Does misfolded LPL form aggregates if not cleared from the ER? Whether autophagy is involved in the clearance of LPL aggregates?
How is GPIHBP1-mediated transcytosis of LPL across the endothelial cells regulated? How is the activity of GPIHBP1 regulated? Does apolipoproteins regulate LPL activity by influencing the function of GPIHBP1 and/or LPL-GPIHBP1 complex?
How are the expression and activity of ANGPTLs regulated under physiological and pathological conditions? How do they interact with each other to regulate LPL activity? How can they be targeted therapeutically in the treatment of hyperlipidemia?
Disease mutants of LPL and its regulators may regulate its biogenesis and activities, but underlying molecular mechanism remains unclear.
Highlights.
LPL is a central player in triglyceride metabolism by catalyzing intravascular triglyceride hydrolysis. Loss-of-function Lpl causes familial chylomicronemia syndrome in humans and mice.
The functional unit of LPL was thought to be a dimer, but recent studies have demonstrated that it may be a monomer.
Given its pathophysiological importance in lipid partitioning, the maturation and activity of LPL is tightly controlled.
In the ER, the maturation and folding of nascent LPL protein requires LMF1 and SEL1L; and in the post-ER compartments, the acitivity of LPL is inhibited by ANGPTL3, 4 and 8 in a tissue-specific manner; in the endothelial lumen, LPL is associated with GPIHBP1 and regulated by various apolipoproteins.
Acknowledgement
We apologize to colleagues whose works were not cited due to the space limitations. The work in the Qi laboratory is supported by NIH R35DM130292, R01DK111174, R01DK120047, R01DK120330, R01DK117639, University of Michigan Protein Folding Diseases Initiative, Juvenile Diabetes Research Foundation (JDRF) 2-SRA-2018-539-AB and American Diabetes Association (ADA) 1-19-IBS-235.
GLOSSARY
- Angiopoietin-like (ANGPTL) proteins
A family of proteins structurally resembling endothelial growth factors, angiopoietins. Among them, ANGPTL3, 4 and 8 are involved in the regulation of plasma triglyceride levels by inhibiting LPL activity.
- Disulfide bond
A posttranslational protein modification occurred in the ER, in which a covalent bond formed between the thiol groups of two cysteine residues either within a protein (intramolecularly) or between the proteins (intermolecularly).
- ER-associated protein degradation (ERAD)
A principal quality-control mechanism in the ER responsible for the recruitment and retrotranslocation of misfolded ER proteins for proteasomal degradation in the cytosol.
- Familial chylomicronemia
Disease condition characterized by severe fasting hypertriglyceridemia and the accumulation of chylomicrons in the plasma, often caused by mutations in Lpl and its cofactors such as Gpihbp1, Apoc2 and Apoa5. These patients have to restrict dietary fat intake.
- Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1)
An essential capillary protein is required for LPL transcytosis from the interstitial space to endothelial lumen, as well as for LPL stability and function.
- Heparan sulfate proteoglycan
Protein found at the cell surface, extracellular matrix and vesicles with the attachment of two or three chains of heparan sulfate. LPL bound to heparan sulfate proteoglycans is released by heparin.
- Lipase maturation factor 1 (LMF1)
An ER-resident protein required for the folding of nascent LPL in the ER.
- Macroautophagy
A major type of autophagy, in which cellular protein aggregates or damaged organelles are engulfed by a double-membrane structure, autophagosome, and degraded by lysosomal hydrolases.
- N-glycosylation
A post-translational protein modification initiated in the ER, in which oligosaccharide chains are attached to the nitrogen atom of asparagine (Asn) at the consensus sequence, Asn–X–Ser/Thr.
- Tyrosine O-sulfation
A post-translational modification occurred in trans-Golgi, in which a sulfate group is added to a tyrosine residue of a protein.
- SEL1L-HRD1 protein complex
The most conserved and best characterized branch of ER-resident mammalian ERAD machinery, with SEL1L being the obligatory cofactor for the E3 ligase HRD1. SEL1L is a single-span transmembrane protein with large luminal domain.
- Transcytosis
A vesicle-mediated transportation that move macromolecules from one side of a cell to the other.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hahn PF, Abolishment of alimentary lipemia following injection of heparin. science, 1943. 98(2531): p. 19–20. [DOI] [PubMed] [Google Scholar]
- 2.Korn ED, Clearing factor, a heparin-activated lipoprotein lipase I. Isolation and characterization of the enzyme from normal rat heart. Journal of Biological Chemistry, 1955. 215(1): p. 1–14. [PubMed] [Google Scholar]
- 3.Korn ED, Clearing factor, a heparin-activated lipoprotein lipase II. Substrate specificity and activation of coconut oil. Journal of Biological Chemistry, 1955. 215(1): p. 15–26. [PubMed] [Google Scholar]
- 4.Young SG, et al. , GPIHBP1 and Lipoprotein Lipase, Partners in Plasma Triglyceride Metabolism. Cell metabolism, 2019. 30(1): p. 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young SG and Zechner R, Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes & development, 2013. 27(5): p. 459–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstock PH, et al. , Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. The Journal of clinical investigation, 1995. 96(6): p. 2555–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersten S, Physiological regulation of lipoprotein lipase. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2014. 1841(7): p. 919–933. [DOI] [PubMed] [Google Scholar]
- 8.Catoire M, et al. , Fatty acid-inducible ANGPTL4 governs lipid metabolic response to exercise. Proceedings of the National Academy of Sciences, 2014. 111(11): p. E1043–E1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh AK, et al. , Brown adipose tissue derived ANGPTL4 controls glucose and lipid metabolism and regulates thermogenesis. Molecular metabolism, 2018. 11: p. 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birrane G, et al. , Structure of the lipoprotein lipase–GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proceedings of the National Academy of Sciences, 2019. 116(5): p. 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arora R, et al. , Structure of lipoprotein lipase in complex with GPIHBP1. Proceedings of the National Academy of Sciences, 2019. 116(21): p. 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Zeev O, et al. , Maturation of lipoprotein lipase. Expression of full catalytic activity requires glucose trimming but not translocation to the cis-Golgi compartment. Journal of Biological Chemistry, 1992. 267(9): p. 6219–6227. [PubMed] [Google Scholar]
- 13.Lo J-Y, Smith LC, and Chan L, Lipoprotein lipase: role of intramolecular disulfide bonds in enzyme catalysis. Biochemical and biophysical research communications, 1995. 206(1): p. 266–271. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Zeev O, et al. , Lipoprotein lipase and hepatic lipase: the role of asparagine-linked glycosylation in the expression of a functional enzyme. Journal of lipid research, 1994. 35(9): p. 1511–1523. [PubMed] [Google Scholar]
- 15.Paterniti JR, et al. , Combined lipase deficiency (cld): a lethal mutation on chromosome 17 of the mouse. Science, 1983. 221(4606): p. 167–169. [DOI] [PubMed] [Google Scholar]
- 16.Péterfy M, et al. , Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nature genetics, 2007. 39(12): p. 1483. [DOI] [PubMed] [Google Scholar]
- 17.Ehrhardt N, Bedoya C, and Péterfy M, Embryonic viability, lipase deficiency, hypertriglyceridemia and neonatal lethality in a novel LMF1-deficient mouse model. Nutrition & metabolism, 2014. 11(1): p. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Péterfy M, Lipase maturation factor 1: a lipase chaperone involved in lipid metabolism. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2012. 1821(5): p. 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts BS, et al. , Lipase maturation factor 1 affects redox homeostasis in the endoplasmic reticulum. The EMBO journal, 2018. 37(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi L, Tsai B, and Arvan P, New insights into the physiological role of endoplasmic reticulum-associated degradation. Trends in cell biology, 2017. 27(6): p. 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang J. and Qi L, Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends in biochemical sciences, 2018. 43(8): p. 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sha H, et al. , The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab, 2014. 20(3): p. 458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi G, et al. , ER-associated degradation is required for vasopressin prohormone processing and systemic water homeostasis. The Journal of clinical investigation, 2017. 127(10): p. 3897–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim GH, et al. , Hypothalamic ER–associated degradation regulates POMC maturation, feeding, and age-associated obesity. The Journal of clinical investigation, 2018. 128(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharya A, et al. , Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. The EMBO journal, 2018. 37(22): p. e99277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei J, et al. , HRD1-ERAD controls production of the hepatokine FGF21 through CREBH polyubiquitination. The EMBO journal, 2018. 37(22): p. e98942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankaitis VA and Wang Y, Lipoprotein Lipase Sorting: Sphingomyelin and a Proteoglycan Show the Way. Trends in Cell Biology, 2020. [DOI] [PubMed] [Google Scholar]
- 28.Vannier C. and Ailhaud G, Biosynthesis of lipoprotein lipase in cultured mouse adipocytes. II. Processing, subunit assembly, and intracellular transport. Journal of Biological Chemistry, 1989. 264(22): p. 13206–13216. [PubMed] [Google Scholar]
- 29.Sundberg EL, Deng Y, and Burd CG, Syndecan-1 Mediates Sorting of Soluble Lipoprotein Lipase with Sphingomyelin-Rich Membrane in the Golgi Apparatus. Developmental cell, 2019. 51(3): p. 387–398. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunn KH, et al. , The structure of helical lipoprotein lipase reveals an unexpected twist in lipase storage. Proceedings of the National Academy of Sciences, 2020. 117(19): p. 10254–10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An M, et al. , ULK1 prevents cardiac dysfunction in obesity through autophagy-meditated regulation of lipid metabolism. Cardiovascular research, 2017. 113(10): p. 1137–1147. [DOI] [PubMed] [Google Scholar]
- 32.Klinger SC, et al. , SorLA regulates the activity of lipoprotein lipase by intracellular trafficking. Journal of cell science, 2011. 124(7): p. 1095–1105. [DOI] [PubMed] [Google Scholar]
- 33.Ioka RX, et al. , Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. Journal of Biological Chemistry, 2003. 278(9): p. 7344–7349. [DOI] [PubMed] [Google Scholar]
- 34.Beigneux AP, et al. , Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell metabolism, 2007. 5(4): p. 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristensen KK, et al. , A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proceedings of the National Academy of Sciences, 2018. 115(26): p. E6020–E6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beigneux AP, et al. , Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. Journal of Biological Chemistry, 2009. 284(44): p. 30240–30247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mysling S, et al. , The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. elife, 2016. 5: p. e12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies BS, et al. , GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell metabolism, 2010. 12(1): p. 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allan CM, et al. , Mobility of “HSPG-bound” LPL explains how LPL is able to reach GPIHBP1 on capillaries. Journal of lipid research, 2017. 58(1): p. 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kristensen KK, et al. , Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism. Proceedings of the National Academy of Sciences, 2020. 117(8): p. 4337–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies BS, et al. , Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. Journal of lipid research, 2012. 53(12): p. 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulbourne CN, et al. , The GPIHBP1–LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell metabolism, 2014. 19(5): p. 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaRosa J, et al. , A specific apoprotein activator for lipoprotein lipase. Biochemical and biophysical research communications, 1970. 41(1): p. 57–62. [DOI] [PubMed] [Google Scholar]
- 44.Breckenridge WC, et al. , Hypertriglyceridemia associated with deficiency of apolipoprotein C-II. New England Journal of Medicine, 1978. 298(23): p. 1265–1273. [DOI] [PubMed] [Google Scholar]
- 45.Wolska A, et al. , Apolipoprotein C-II: new findings related to genetics, biochemistry, and role in triglyceride metabolism. Atherosclerosis, 2017. 267: p. 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma V, Ryan RO, and Forte TM, Apolipoprotein AV dependent modulation of plasma triacylglycerol: a puzzlement. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2012. 1821(5): p. 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaap FG, et al. , ApoAV reduces plasma triglycerides by inhibiting very low density lipoprotein-triglyceride (VLDL-TG) production and stimulating lipoprotein lipase-mediated VLDL-TG hydrolysis. Journal of biological chemistry, 2004. 279(27): p. 27941–27947. [DOI] [PubMed] [Google Scholar]
- 48.Merkel M, et al. , Apolipoprotein AV accelerates plasma hydrolysis of triglyceriderich lipoproteins by interaction with proteoglycan-bound lipoprotein lipase. Journal of biological chemistry, 2005. 280(22): p. 21553–21560. [DOI] [PubMed] [Google Scholar]
- 49.Pennacchio LA, et al. , An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science, 2001. 294(5540): p. 169–173. [DOI] [PubMed] [Google Scholar]
- 50.Camporez JPG, et al. , ApoA5 knockdown improves whole-body insulin sensitivity in high-fat-fed mice by reducing ectopic lipid content. Journal of lipid research, 2015. 56(3): p. 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsson SK, et al. , Apolipoprotein AV; a potent triglyceride reducer. Atherosclerosis, 2011. 219(1): p. 15–21. [DOI] [PubMed] [Google Scholar]
- 52.Jong MC, et al. , Apolipoprotein C-III deficiency accelerates triglyceride hydrolysis by lipoprotein lipase in wild-type and apoE knockout mice. Journal of lipid research, 2001. 42(10): p. 1578–1585. [PubMed] [Google Scholar]
- 53.Westerterp M, et al. , Endogenous apoC-I increases hyperlipidemia in apoE-knockout mice by stimulating VLDL production and inhibiting LPL. Journal of lipid research, 2006. 47(6): p. 1203–1211. [DOI] [PubMed] [Google Scholar]
- 54.Ebara T, et al. , Chylomicronemia due to apolipoprotein CIII overexpression in apolipoprotein E-null mice. Apolipoprotein CIII-induced hypertriglyceridemia is not mediated by effects on apolipoprotein E. The Journal of clinical investigation, 1997. 99(11): p. 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berbée JF, et al. , Severe hypertriglyceridemia in human APOC1 transgenic mice is caused by apoC-I-induced inhibition of LPL. Journal of lipid research, 2005. 46(2): p. 297–306. [DOI] [PubMed] [Google Scholar]
- 56.Aalto-Setälä K, et al. , Mechanism of hypertriglyceridemia in human apolipoprotein (apo) CIII transgenic mice. Diminished very low density lipoprotein fractional catabolic rate associated with increased apo CIII and reduced apo E on the particles. The Journal of clinical investigation, 1992. 90(5): p. 1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu K. and Cooper AD, Postprandial lipoproteins and atherosclerosis. Front Biosci, 2001. 6: p. D332–D354. [DOI] [PubMed] [Google Scholar]
- 58.Rensen PC and van Berkel TJ, Apolipoprotein E effectively inhibits lipoprotein lipase-mediated lipolysis of chylomicron-like triglyceride-rich lipid emulsions in vitro and in vivo. Journal of Biological Chemistry, 1996. 271(25): p. 14791–14799. [DOI] [PubMed] [Google Scholar]
- 59.Kersten S, New insights into angiopoietin-like proteins in lipid metabolism and cardiovascular disease risk. Current opinion in lipidology, 2019. 30(3): p. 205–211. [DOI] [PubMed] [Google Scholar]
- 60.Dewey FE, et al. , Inactivating variants in ANGPTL4 and risk of coronary artery disease. New England Journal of Medicine, 2016. 374(12): p. 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Musunuru K, et al. , Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. New England Journal of Medicine, 2010. 363(23): p. 2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Helkkula P, et al. , ANGPTL8 protein-truncating variant and the risk of coronary disease, type 2 diabetes and adverse effects. medRxiv, 2020. [Google Scholar]
- 63.Aryal B, et al. , ANGPTL4 in Metabolic and Cardiovascular Disease. Trends in Molecular Medicine, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei X, et al. , Proteolytic processing of angiopoietin-like protein 4 by proprotein convertases modulates its inhibitory effects on lipoprotein lipase activity. Journal of Biological Chemistry, 2011. 286(18): p. 15747–15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biterova E, et al. , Structures of Angptl3 and Angptl4, modulators of triglyceride levels and coronary artery disease. Scientific reports, 2018. 8(1): p. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X. and Musunuru K, Angiopoietin-Like 3: From Discovery to Therapeutic Gene Editing. JACC: Basic to Translational Science, 2019. 4(6): p. 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yau M. h., et al. , A highly conserved motif within the NH2-terminal coiled-coil domain of angiopoietin-like protein 4 confers its inhibitory effects on lipoprotein lipase by disrupting the enzyme dimerization. Journal of Biological Chemistry, 2009. 284(18): p. 11942–11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee E-C, et al. , Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL). Journal of Biological Chemistry, 2009. 284(20): p. 13735–13745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo M. and Peng D, ANGPTL8: an important regulator in metabolic disorders. Frontiers in endocrinology, 2018. 9: p. 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kersten S, et al. , Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. Journal of Biological Chemistry, 2000. 275(37): p. 28488–28493. [DOI] [PubMed] [Google Scholar]
- 71.Yoon JC, et al. , Peroxisome proliferator-activated receptor γ target gene encoding a novel angiopoietin-related protein associated with adipose differentiation. Molecular and cellular biology, 2000. 20(14): p. 5343–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim I, et al. , Hepatic expression, synthesis and secretion of a novel fibrinogen/angiopoietin-related protein that prevents endothelial-cell apoptosis. Biochemical Journal, 2000. 346(3): p. 603–610. [PMC free article] [PubMed] [Google Scholar]
- 73.Ge H, et al. , Oligomerization and regulated proteolytic processing of angiopoietin-like protein 4. Journal of Biological Chemistry, 2004. 279(3): p. 2038–2045. [DOI] [PubMed] [Google Scholar]
- 74.Dijk W, et al. , Angiopoietin-like 4 promotes intracellular degradation of lipoprotein lipase in adipocytes. Journal of lipid research, 2016. 57(9): p. 1670–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dijk W, et al. , Angiopoietin-like 4 promotes the intracellular cleavage of lipoprotein lipase by PCSK3/furin in adipocytes. Journal of Biological Chemistry, 2018. 293(36): p. 14134–14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lafferty MJ, et al. , Angiopoietin-like protein 4 inhibition of lipoprotein lipase evidence for reversible complex formation. Journal of Biological Chemistry, 2013. 288(40): p. 28524–28534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gutgsell AR, et al. , Mapping the sites of the lipoprotein lipase (LPL)–angiopoietin-like protein 4 (ANGPTL4) interaction provides mechanistic insight into LPL inhibition. Journal of Biological Chemistry, 2019. 294(8): p. 2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lookene A, et al. , Rapid subunit exchange in dimeric lipoprotein lipase and properties of the inactive monomer. Journal of Biological Chemistry, 2004. 279(48): p. 49964–49972. [DOI] [PubMed] [Google Scholar]
- 79.Ge H, et al. , Oligomerization state-dependent hyperlipidemic effect of angiopoietin-like protein 4. Journal of lipid research, 2004. 45(11): p. 2071–2079. [DOI] [PubMed] [Google Scholar]
- 80.Mysling S, et al. , The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. Elife, 2016. 5: p. e20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cushing EM, et al. , Angiopoietin-like 4 directs uptake of dietary fat away from adipose during fasting. Molecular metabolism, 2017. 6(8): p. 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ruppert PM, et al. , Fasting induces ANGPTL4 and reduces LPL activity in human adipose tissue. Molecular Metabolism, 2020: p. 101033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lichtenstein L, et al. , Angptl4 upregulates cholesterol synthesis in liver via inhibition of LPL-and HL-dependent hepatic cholesterol uptake. Arteriosclerosis, thrombosis, and vascular biology, 2007. 27(11): p. 2420–2427. [DOI] [PubMed] [Google Scholar]
- 84.Mandard S, et al. , The fasting-induced adipose factor/angiopoietin-like protein 4 is physically associated with lipoproteins and governs plasma lipid levels and adiposity. Journal of Biological Chemistry, 2006. 281(2): p. 934–944. [DOI] [PubMed] [Google Scholar]
- 85.Köster A, et al. , Transgenic angiopoietin-like (angptl) 4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology, 2005. 146(11): p. 4943–4950. [DOI] [PubMed] [Google Scholar]
- 86.Aryal B, et al. , Absence of ANGPTL4 in adipose tissue improves glucose tolerance and attenuates atherogenesis. JCI insight, 2018. 3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kroupa O, et al. , Linking nutritional regulation of Angptl4, Gpihbp1, and Lmf1 to lipoprotein lipase activity in rodent adipose tissue. BMC physiology, 2012. 12(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Georgiadi A, et al. , Induction of cardiac Angptl4 by dietary fatty acids is mediated by PPARβ/δ and protects against oxidative stress: P18. European Journal of Clinical Nutrition, 2009. 63. [Google Scholar]
- 89.Koishi R, et al. , Angptl3 regulates lipid metabolism in mice. Nature genetics, 2002. 30(2): p. 151–157. [DOI] [PubMed] [Google Scholar]
- 90.Liu J, et al. , Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. Journal of Biological Chemistry, 2010. 285(36): p. 27561–27570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katrine T-BS, et al. , O-Glycosylation Modulates Proprotein Convertase Activation of Angiopoietin-like Protein 3 POSSIBLE ROLE OF POLYPEPTIDE GalNAc-TRANSFERASE-2 IN REGULATION OF CONCENTRATIONS OF PLASMA LIPIDS. Journal of Biological Chemistry, 2010. 285(47): p. 36293–36303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haller JF, et al. , ANGPTL8 requires ANGPTL3 to inhibit lipoprotein lipase and plasma triglyceride clearance. Journal of lipid research, 2017. 58(6): p. 1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chi X, et al. , ANGPTL8 promotes the ability of ANGPTL3 to bind and inhibit lipoprotein lipase. Molecular metabolism, 2017. 6(10): p. 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen YQ, et al. , Angiopoietin-like protein 8 differentially regulates ANGPTL3 and ANGPTL4 during postprandial partitioning of fatty acids. Journal of Lipid Research, 2020: p. jlr. RA120000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Quagliarini F, et al. , Atypical angiopoietin-like protein that regulates ANGPTL3. Proceedings of the National Academy of Sciences, 2012. 109(48): p. 19751–19756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ren G, Kim JY, and Smas CM, Identification of RIFL, a novel adipocyte-enriched insulin target gene with a role in lipid metabolism. American Journal of Physiology-Endocrinology and Metabolism, 2012. 303(3): p. E334–E351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang R, Lipasin, a novel nutritionally-regulated liver-enriched factor that regulates serum triglyceride levels. Biochemical and biophysical research communications, 2012. 424(4): p. 786–792. [DOI] [PubMed] [Google Scholar]
- 98.Zhang R, The ANGPTL3–4-8 model, a molecular mechanism for triglyceride trafficking. Open biology, 2016. 6(4): p. 150272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kersten S, Angiopoietin-like 3 in lipoprotein metabolism. Nature Reviews Endocrinology, 2017. 13(12): p. 731. [DOI] [PubMed] [Google Scholar]
- 100.Fu Z, Abou-Samra AB, and Zhang R, A lipasin/Angptl8 monoclonal antibody lowers mouse serum triglycerides involving increased postprandial activity of the cardiac lipoprotein lipase. Scientific reports, 2015. 5: p. 18502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, et al. , Hepatic ANGPTL3 regulates adipose tissue energy homeostasis. Proceedings of the National Academy of Sciences, 2015. 112(37): p. 11630–11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, et al. , Mice lacking ANGPTL8 (Betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proceedings of the National Academy of Sciences, 2013. 110(40): p. 16109–16114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kovrov O, et al. , On the mechanism of angiopoietin-like protein 8 for control of lipoprotein lipase activity. Journal of lipid research, 2019. 60(4): p. 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oldoni F, et al. , ANGPTL8 has both endocrine and autocrine effects on substrate utilization. JCI insight, 2020. 5(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chi X, et al. , Angiopoietin-like 4 modifies the interactions between lipoprotein lipase and its endothelial cell transporter GPIHBP1. Journal of Biological Chemistry, 2015. 290(19): p. 11865–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shetty SK, Walzem RL, and Davies BS, A novel NanoBiT-based assay monitors the interaction between lipoprotein lipase and GPIHBP1 in real time. Journal of Lipid Research, 2020. 61(4): p. 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Benlian P, et al. , Premature atherosclerosis in patients with familial chylomicronemia caused by mutations in the lipoprotein lipase gene. New England Journal of Medicine, 1996. 335(12): p. 848–854. [DOI] [PubMed] [Google Scholar]
- 108.Kobayashi J, et al. , A naturally occurring mutation at the second base of codon asparagine 43 in the proposed N-linked glycosylation site of human lipoprotein lipase: in vivo evidence that asparagine 43 is essential for catalysis and secretion. Biochemical and biophysical research communications, 1994. 205(1): p. 506–515. [DOI] [PubMed] [Google Scholar]
- 109.Hoffmann MM, et al. , Type I hyperlipoproteinemia due to a novel loss of function mutation of lipoprotein lipase, Cys239→ Trp, associated with recurrent severe pancreatitis. The Journal of Clinical Endocrinology & Metabolism, 2000. 85(12): p. 4795–4798. [DOI] [PubMed] [Google Scholar]
- 110.Abifadel M, et al. , Identification of the first Lebanese mutation in the LPL gene and description of a rapid detection method. Clinical genetics, 2004. 65(2): p. 158–161. [DOI] [PubMed] [Google Scholar]
- 111.Martínez M, et al. , The Mutation Gly Glu in Human Lipoprotein Lipase Produces a Missorted Protein That Is Diverted to Lysosomes. Journal of Biological Chemistry, 1996. 271(4): p. 2139–2146. [DOI] [PubMed] [Google Scholar]
- 112.Voss CV, et al. , Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proceedings of the National Academy of Sciences, 2011. 108(19): p. 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gin P, et al. , Chylomicronemia mutations yield new insights into interactions between lipoprotein lipase and GPIHBP1. Human molecular genetics, 2012. 21(13): p. 2961–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ariza MJ, et al. , Novel mutations in the GPIHBP1 gene identified in 2 patients with recurrent acute pancreatitis. Journal of clinical lipidology, 2016. 10(1): p. 92–100. e1. [DOI] [PubMed] [Google Scholar]
- 115.Ailhaud G, Cellular and secreted lipoprotein lipase revisited. Clinical biochemistry, 1990. 23(5): p. 343–347. [DOI] [PubMed] [Google Scholar]
- 116.Osborne JC Jr, et al. , Studies on inactivation of lipoprotein lipase: role of the dimer to monomer dissociation. Biochemistry, 1985. 24(20): p. 5606–5611. [DOI] [PubMed] [Google Scholar]
- 117.Garfinkel AS, et al. , Lipoprotein lipase: size of the functional unit determined by radiation inactivation. Journal of lipid research, 1983. 24(6): p. 775–780. [PubMed] [Google Scholar]
- 118.Iverius P-H and Ostlund-Lindqvist A, Lipoprotein lipase from bovine milk. Isolation procedure, chemical characterization, and molecular weight analysis. Journal of Biological Chemistry, 1976. 251(24): p. 7791–7795. [PubMed] [Google Scholar]
- 119.Beigneux AP, et al. , Lipoprotein lipase is active as a monomer. Proceedings of the National Academy of Sciences, 2019. 116(13): p. 6319–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang L, et al. , Calcium triggers folding of lipoprotein lipase into active dimers. Journal of Biological Chemistry, 2005. 280(52): p. 42580–42591. [DOI] [PubMed] [Google Scholar]
- 121.Wong H, et al. , A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proceedings of the National Academy of Sciences, 1997. 94(11): p. 5594–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ben-Zeev O. and Doolittle MH, Determining lipase subunit structure by sucrose gradient centrifugation, in Lipase and Phospholipase Protocols. 1999, Springer. p. 257–266. [DOI] [PubMed] [Google Scholar]
- 123.Lichtenstein L, et al. , Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell metabolism, 2010. 12(6): p. 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mattijssen F, et al. , Angptl4 serves as an endogenous inhibitor of intestinal lipid digestion. Molecular metabolism, 2014. 3(2): p. 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fujimoto K, et al. , Angptl3-null mice show low plasma lipid concentrations by enhanced lipoprotein lipase activity. Experimental animals, 2006. 55(1): p. 27–34. [DOI] [PubMed] [Google Scholar]
- 126.van der Vliet HN, et al. , Adenoviral overexpression of apolipoprotein AV reduces serum levels of triglycerides and cholesterol in mice. Biochemical and biophysical research communications, 2002. 295(5): p. 1156–1159. [DOI] [PubMed] [Google Scholar]
- 127.Fruchart-Najib J, et al. , Mechanism of triglyceride lowering in mice expressing human apolipoprotein A5. Biochemical and biophysical research communications, 2004. 319(2): p. 397–404. [DOI] [PubMed] [Google Scholar]