Abstract
Leiomyosarcoma (LMS) is a mesenchymal malignancy with a complex karyotype. Despite accumulated evidence, the factors contributing to the development of LMS are unclear. Here, we investigated the role of tight-junction protein 1 (TJP1), a membrane-associated intercellular barrier protein during the development of LMS and the tumor microenvironment. We orthotopically transplanted SK-LMS-1 cells and their derivatives in terms of TJP1 expression by intramuscular injection, such as SK-LMS-1 Sh-Control cells and SK-LMS-1 Sh-TJP1. We observed robust tumor growth in mice transplanted with LMS cell lines expressing TJP1 while no tumor mass was found in mice transplanted with SK-LMS-1 Sh-TJP1 cells with silenced TJP1 expression. Tissues from mice were stained and further analyzed to clarify the effects of TJP1 expression on tumor development and the tumor microenvironment. To identify the TJP1-dependent factors important in the development of LMS, genes with altered expression were selected in SK-LMS-1 cells such as cyclinD1, CSF1 and so on. The top 10% of highly expressed genes in LMS tissues were obtained from public databases. Further analysis revealed two clusters related to cell proliferation and the tumor microenvironment. Furthermore, integrated analyses of the gene expression networks revealed correlations among TJP1, CSF1 and CTLA4 at the mRNA level, suggesting a possible role for TJP1 in the immune environment. Taken together, these results imply that TJP1 contributes to the development of sarcoma by proliferation through modulating cell-cell aggregation and communication through cytokines in the tumor microenvironment and might be a beneficial therapeutic target.
Keywords: cytokines, leiomyosarcoma, tjp1, tumor microenvironment
INTRODUCTION
According to the World Health Organization classification, sarcomas originating from soft tissues result in more than 50 histological subtypes, accounting for less than 1% of all cancers (Fletcher, 2014). Despite the heterogeneity and rarity, several subtypes are understood and diagnosed by molecular biomarkers and are considered simple karyotype soft-tissue sarcomas (Barretina et al., 2010; Helman and Meltzer, 2003). Others with few distinct and clear genomic changes, such as leiomyosarcoma (LMS) and undifferentiated pleomorphic sarcoma (UPS) (Barretina et al., 2010; Helman and Meltzer, 2003) are categorized as complex karyotypes. LMS originating from smooth muscle is a relatively common soft-tissue sarcoma (~10% of soft-tissue sarcomas) (Cloutier and Charville, 2019), therapeutic strategies for advanced stages are still being investigated (Barretina et al., 2010). The complexity index in sarcomas (CINSARC) has been studied as a prognostic aid in sarcomas, particularly for LMS (Chibon et al., 2010; Guo et al., 2015). The Cancer Genome Atlas (TCGA) network and others have enlarged our understanding of soft-tissue sarcomas including LMS at the molecular level (Cancer Genome Atlas Research Network, 2017; Kim et al., 2018). Furthermore, Chudasama et al. (2018) performed an integrated analysis of LMS cases, which revealed that tumor evolution to metastasis and advanced stages occurs via an abrogated tumor suppressor network followed by whole genome duplication and severe genomic instability, and that ‘BRCAness’ is potentially an actionable genetic trait (Chudasama et al., 2018).
It has been suggested that innate and adaptive immune cells modulate tumor progression (Grivennikov et al., 2010; Vesely et al., 2011). Natural defenses can be boosted by targeting the immune system; this strategy has recently been revolutionized (Hegde and Chen, 2020; Zhang and Zhang, 2020). In sarcomas, immunotherapy has been studied and tried continuously for better clinical outcomes especially in patients with recurrent or metastatic disease (Mata and Gottschalk, 2015). Several studies have shown an association between tumor-infiltrating lymphocytes and the prognosis (Rusakiewicz et al., 2013; Sorbye et al., 2011). Integrated analyses of multi-omics-based TCGA data have shown the possibility for sarcoma immunotherapy (Thorsson et al., 2018), in which six immune subtypes (C1-C6) based on immunogenomic analyses of 10,000 tumors, were classified regardless of cancer type (Thorsson et al., 2018). Accordingly, soft-tissue sarcomas represent five of the subtypes, except C5, which is “immunologically quiet” with the highest ratio of macrophages to lymphocytes (Thorsson et al., 2018), suggesting the possibility of an immune therapeutic strategy against soft-tissue sarcoma. George et al. (2017) presented the effect of anti-programmed death (PD)-1 checkpoint blockade therapy on metastatic uterine LMS and suggested a correlation between PTEN and resistance to pembrolizumab (George et al., 2017). B cells have been suggested to be associated with survival and the immunotherapeutic response in sarcoma, particularly LMS and UPS (Petitprez et al., 2020), implying that modulating the immune response is a good therapeutic strategy for soft-tissue sarcomas.
Tight-junction protein 1 (TJP1) has been implicated as a scaffolding protein in a variety of cellular processes. TJP1 has been identified as a major component of tight junctions by providing a link between occludin, a transmembrane tight-junction protein, and the actin cytoskeleton (Fanning et al., 1998), which are essential for barrier function (Martin and Jiang, 2009). TJP1 controls lamellae-formation-mediated motility in cancer cells by binding to integrin (Tuomi et al., 2009). TJP1 increases and contributes to cell motility in transforming growth factor β-stimulated lung cancer cells, implying that TJP1 may be more than a tight-junction member (Lee et al., 2015). TJP1 has been implicated in modulating proteasome capacity and its inhibitor sensitivity in multiple myeloma (Zhang et al., 2016). In previous study, we compared transcriptomic data from TCGA sarcoma as well as National Cancer Center (NCC) sarcoma, which revealed that TJP1 expression levels were higher in LMS than normal tissues (Lee et al., 2020). The role of TJP1 in sarcoma development was further demonstrated by comparing the colonies of SK-LMS-1 parental cells, SK-LMS-1 cells stably expressing short hairpin (Sh) RNA against TJP1 (Sh-TJP1) and SK-LMS-1 cells expressing untargeted ShRNA (Sh-Control) in anchorage-independent growth assays, and by examining cell-cell aggregation in non-adherent culture systems (Lee et al., 2020). We hypothesized that increased TJP1 expression contributes to the progression of LMS and is applicable as a therapeutic target. However, the in vivo contribution to cancers of TJP1 should be further validated.
Here, we transplanted three SK-LMS-1 cell lines, parental, Sh-Control and Sh-TJP1, into immunodeficient mice by intramuscular injection and monitored the mice until tumors were observed, which was up to 14 weeks. Tumors were observed only in mice with SK-LMS-1 cells expressing TJP1. The differentially expressed genes by TJP1 knockdown in the SK-LMS-1, parental, Sh-Control, and Sh-TJP1 cells were listed, and 40 genes were further analyzed in LMS from the TCGA dataset. Furthermore, the top 10% of genes with high expression in the TCGA LMS transcriptome were further analyzed for an association between TJP1 and immune characteristics of LMS.
MATERIALS AND METHODS
Materials
All cell lines were purchased from the American Type Culture Collection (ATCC, USA) and the culture methods and media followed ATCC recommendations. Eagle’s Minimum Essential Medium for the SK-LMS-1 cells was obtained from Corning (USA). Antibiotic-antimycotic and fetal bovine serum (FBS) were purchased from Gibco (USA). The polyclonal antibody against TJP1 (ZO-1, #617300) was obtained from Thermo Fisher Scientific (USA). The antibody against sequestosome-1 (SQSTM1, p62, #610832) was obtained from BD Biosciences (USA). The antibody against Ki67 (#ab15580) was purchased from Abcam (UK). Antibodies against EGF-like module-containing mucin-like hormone receptor-like 1 (EMR1, F4/80, #70076), phosphorylated JAK2Tyr1007/1008(#3776), and Bcl-extra-large (Bcl-xL, #2764) were purchased from Cell Signaling Technology (USA). Antibodies against cyclin D1 (#sc8369) and β-actin (#sc69879) were obtained from Santa Cruz Biotechnology (USA). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were purchased from Cell Signaling Technology. The Miracle-StarTM western blot detection system was obtained from iNtRON Biotechnology (Korea).
Cell culture
All cells were authenticated by short-tandem repeat polymerase chain reaction (PCR) in 2017 at the National Cancer Center Omics core facility (Perkin Elmer, USA). The cells were cultured every 2-3 days to maintain 40%-70% confluency in cell-specific media containing 10% (v/v) FBS and 1% (v/v) antimycotic-antibiotic solution. In some experiments, we compared cell lines, such as SK-LMS-1 Sh-Control and Sh-TJP1 cells (Lee et al., 2020) to investigate the role of TJP1. Briefly, to achieve stable lentivirus-mediated expression of shRNA targeting TJP1 in SK-LMS-1 cells, cells were cultured for 24 h, incubated with 5 μg/ml polybrene for 30 min, and infected as previously described (Lee et al., 2020). Then survived clones were selected and pooled to avoid any clonal variation for TJP1 expression and other experiments in our study.
Cell-cell aggregation assay
Cells (1 × 105) were seeded on a 35-mm dish without coating, cultured for 24 h, and digitized by inverted light microscopy (CKX53; Olympus, Japan) (Lee et al., 2020).
In vivo study
This study (NCC-18-439 to H.J.Y.) was reviewed and approved by the Institutional Animal Care and Use Committee of the National Cancer Center Research Institute. All mice were maintained under specific-pathogen-free conditions in the animal facility of the National Cancer Center in Korea. Animal experiments were conducted according to the guidelines on the care and use of laboratory animals from the Institute of Laboratory Animal Resources.
SK-LMS-1 cells were grown for 2-3 days to 70%-80% confluency. The cells were harvested with trypsin and counted using a hemocytometer and a microscope. Cells (5 × 106/100 μl/mouse) for the orthotopic xenograft mouse models (Babichev et al., 2016), were injected into the right hind limb of 9-week-old mice (Rag2-/- γc-/- immunodeficient mice; The Jackson Laboratory, USA) (Strowig et al., 2011), including a placebo. Eleven mice were weighed and monitored weekly for 14 weeks. The National Cancer Center animal molecular imaging team performed positron emission tomography/computed tomography (PET/CT) using 18F-fluorodeoxyglucose (FDG) after 9 and 12 weeks to identify any tumor masses in the animal models (Supplementary Table S1). The mice for molecular image scanning were fasted for 6 h and anesthetized with vaporized 2% isoflurane in oxygen on a PET/CT system (eXplore Vista-CT; GE Healthcare, USA). The images were normalized to determine the standardized uptake values (Kim et al., 2011). The widths of the left/right hind limbs of the mice were measured at 13 weeks after the injection (22-week-old mice) using calipers.
Tissue preparation and staining for the in vivo model
We sacrificed animals according to the ILAR guidelines. Tissues (tumors and adjacent normal tissues) were fixed in 10% neutral buffered formalin to prepare formalin-fixed paraffin-embedded tissue blocks. Immunohistochemistry and H&E staining were conducted by the National Cancer Center Animal Sciences Branch. Tumor tissues for immunohistochemistry were stained with antibodies against Ki67 or F4/80 and hematoxylin and colorized using 3,3′‐diaminobenzidine. The stained tissues were digitized at 20× magnification using an Aperio AT Turbo whole-slide scanner (Leica Biosystems, USA) equipped with a clinical-grade RGB camera (Verma et al., 2019). Regions of interest were prepared and annotated with scale bars. Images from slides not stained with a primary antibody were used as a negative control. Some tissues stained with H&E, Ki67 (a proliferation marker) or F4/80 (marker for macrophage) were quantified by using HistoQuest software (TissueGnostics) (Paek et al., 2017) and Vectra 3 (Akoya Biosciences, USA) with the help of a professional operator of NCC Omics Core Center and further analyzed statistically.
Reverse transcription (RT)-PCR
Total RNA was isolated using an RNeasy Mini Kit (Qiagen, USA). Total RNA (5 μg) was reverse transcribed using oligo-dT or random primers and SuperScriptTM III Reverse Transcriptase (Invitrogen, USA), according to the manufacturer’s instructions. The PCR was performed using gene-specific primers (Supplementary Table S2). The PCR products were subjected to 1.5% (w/v) agarose gel electrophoresis. The resulting bands were visualized with ethidium bromide and photographed using the GelDoc program (Bio-Rad Laboratories, USA).
Immunoblotting
The protein samples were heated at 95°C for 7 min and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 8%-15% acrylamide gels followed by transfer to polyvinylidene difluoride membranes. Immunoblotting was performed as described previously (Paek et al., 2019).
Bioinformatics analyses for TJP1 and related tumor microenvironments in sarcoma
The LMS transcriptome was obtained from the TCGA using cBioportal (Gao et al., 2013). Further analyses were performed using webtools Idep.92 (http://bioinformatics.sdstate.edu/idep92/) (Ge et al., 2018), Heatmapper (http://www.heatmapper.ca/) (Babicki et al., 2016), and Morpheus (https://software.broadinstitute.org/morpheus) to investigate the correlation between the genes and heatmaps. g:Profiler (http://biit.cs.ut.ee/gprofiler/gost) was used for pathway analyses (Raudvere et al., 2019). Finally, scatter plots were prepared using GraphPad Prism 5.0 (GraphPad Software, USA).
Statistical analysis
All data are expressed as percentages of the control and shown as mean ± SE. Statistical comparisons between groups were made using Student’s t-tests. Values of P < 0.05 were considered significant.
RESULTS
TJP1 knockdown affects mesenchymal leiomyosarcoma tumor growth in vivo
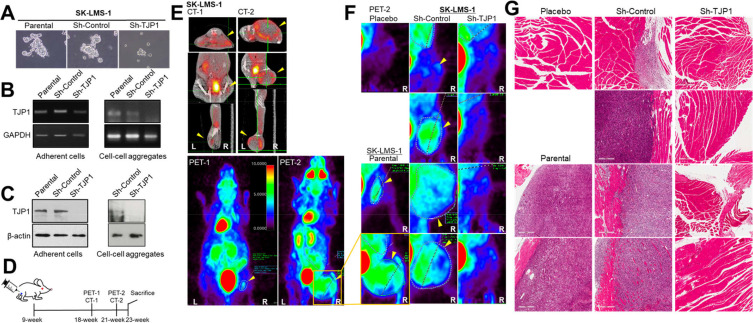
We previously suggested a role for TJP1 in LMS development via enhancing cancer cell growth in the three-dimensional environment. From cell-cell aggregation assay, SK-LMS-1 parental and Sh-Control cells formed more and larger cell-cell aggregates than Sh-TJP1 cells did (Fig. 1A), as we showed previously (Lee et al., 2020). Regardless of culture systems, TJP1 was stably less expressed in SK-LMS-1 Sh-TJP1 cells at RNA level and Protein levels (Figs. 1B and 1C). Thus, we investigated whether TJP1 contributed to tumor growth in vivo. We orthotopically transplanted the human LMS cell line, SK-LMS-1 and its derivatives in terms of TJP1 expression—SK-LMS-1 parental, Sh-Control, and SK-LMS-1 Sh-TJP1—into immunodeficient mice (9 weeks old) (Babichev et al., 2016; Ren et al., 2008). The placebo was the same volume of PBS. We measured the hind leg widths and body weights of the mice for 14 weeks and weighed the mice weekly after the intramuscular injection (Fig. 1D). Nine weeks after the injection, no significant tumor growth was observed on the hind legs of the mice; we examined the mice for possible tumor masses by 18F-FDG PET/CT. Of note, high uptake of 18F-FDG was detected in a variety of organs, including the kidneys, but a very weak but distinct signal was recognized in the hind leg only in one mouse in the SK-LMS-1 Sh-Control group (Supplementary Table S1). We performed an additional PET/CT scan 3 weeks later because of sudden tumor growth within 7 days (Figs. 1E and 1F, Supplementary Fig. S1, Supplementary Table S1). Tumor growth was observed in the SK-LMS-1 and SK-LMS-1 Sh-Control mice that expressed a moderate level of TJP1 (Fig. 1B). Surprisingly, we did not observe any tumor growth in SK-LMS-1 Sh-TJP1 mice (Table 1, Figs. 1E-1G).
Fig. 1. TJP1 contributes to LMS growth and development in vivo.
(A) SK-LMS-1 parental, Sh-Control and Sh-TJP1 cells were plated on coating-free culture plate for 24 h and images were obtained by an inverted light microscope. (B and C) Cells were plated on 35-mm dish with or without coating poly-L-lysine for 24 h and harvested for RT-PCR (B) or immunoblotting (C) to assess TJP1 expression. Data shown are representative of at least three independent experiments. Left panel data were obtained from adherent cells, and right panel from cell-cell aggregates from coating-free culture plates. (D) Timetable for the in vivo study and 18F-FDG PET/CT scanning (PET-1/CT-1 at 9 weeks and PET-2/CT-2 at 12 weeks after the injection, respectively. (E and F) Mice were scanned with a small animal 18F-FDG PET/CT imager. (E) Data shown are representative of animal with SK-LMS-1 parental cells at 9 weeks (CT-1 and PET-1) and 12 weeks (CT-2/PET-2) after the injection, respectively. The PET-2 images (the hind leg, injected area) of all mice were magnified (F) and tumors were indicated by yellow arrowheads. (G) Tissues from the injection site on the right hind leg were fixed for histological analyses followed by staining with H&E. The tissues were visualized on an Aperio AT Turbo whole-slide scanner (Leica Biosystems, USA). All images of (F) and (G) are arranged in same order.
Table 1.
Animal model statistics
| Injected | Tumorigenesis (rear leg) |
|---|---|
| Placebo | 0 (1) |
| SK-LMS-1 | 2 (2) |
| SK-LMS-1 Sh-Control | 4 (4) |
| SK-LMS-1 Sh-TJP1 | 0 (4) |
After the second-PET/CT scan, the mice were sacrificed within a few days as described in the Materials and methods. Tumor tissues from SK-LMS-1 and Sh-Control cell mice and hind-leg tissues, including where we injected the mice with Sh-TJP1 cells or placebo, were fixed and subjected to further investigation (Fig. 1G). We also examined lung tissues to identify any metastatic lesions or niches in the mice but did not find any specific tumor cell masses in lung tissues (data not shown).
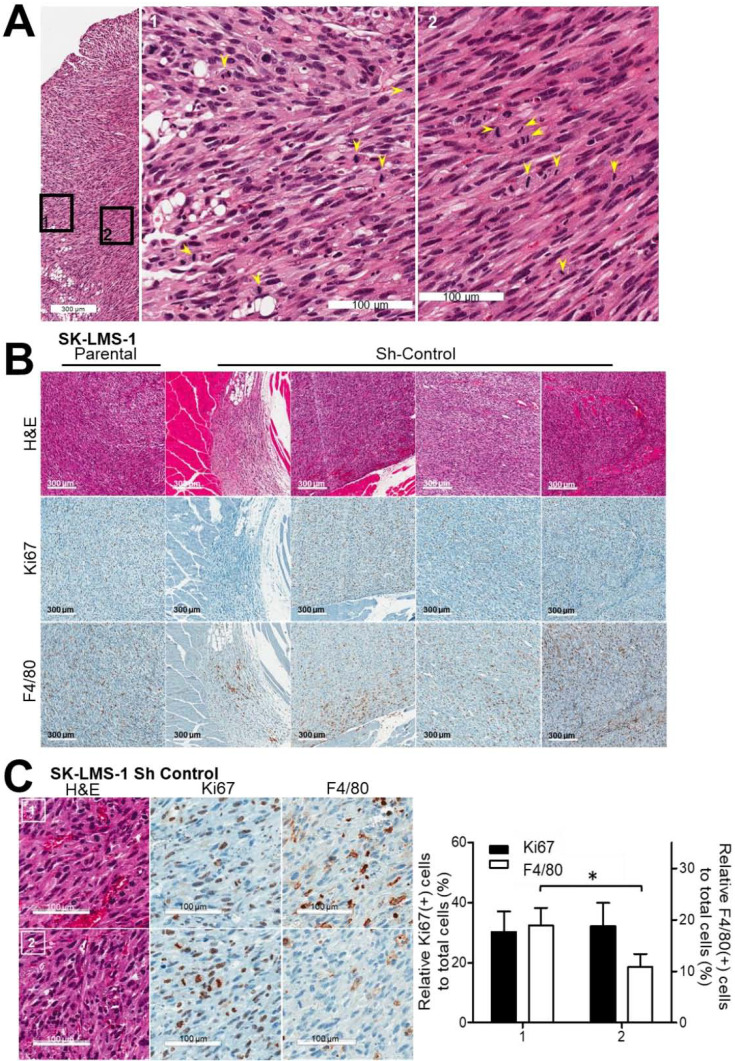
Tumor tissues from SK-LMS-1 and SK-LMS-1 Sh-Control mice were examined by a pathologist for further characterization. The tumor cells were mitotic and pleomorphic, implying high-grade tumor development (Fig. 2A). Mostly, many mitotic cells were detected in the sarcoma tumor tissues (Fig. 2A, arrowheads). Some regions of tumor tissues showed less mitotic (Figs. 2B and 2C) than the other tumor area, especially closed to normal tissues, suggesting an additional role of TJP1 beyond supporting cell-cell aggregates. We used immunohistochemistry to investigate the possibility of communication within the tumor microenvironment, as a factor in tumor development and to confirm the role of TJP1 on tumor growth by enhancing mitosis. Some regions with many Ki67(+) cells, which is also known as the Ki67 marker of proliferation (MKI67) (Hoos et al., 2001), had inflammatory cells, such as neutrophils (data not shown) and macrophages (Fig. 2B, F4/80), while others had either Ki67 or F4/80, a mouse macrophage-restricted protein (Lin et al., 2005) (Figs. 2B and 2C), supporting a role of TJP1 for cell-cell aggregation as well as for tumor microenvironments leading LMS development. In our study, tumor growth was apparent almost 3 months after the intramuscular injection in the right hind legs of all mice injected with parental and Sh-Control cells expressing TJP1, while none of the Sh-TJP1 cell mice expressed less TJP1, suggesting possible contribution of TJP1 on microenvironmental adjustment through cell-cell interaction as well as on communication for tumor growth and development of sarcoma, particularly LMS.
Fig. 2. Tumor tissues from mice injected with SK-LMS-1 parental and Sh-Control cells were characterized as aggressive and high-grade LMS.
(A) Tumors from SK-LMS-1 Sh-Control cells exhibited the mitotic (yellow arrowheads) and high-grade characteristics of LMS, as shown by H&E staining. Data are repre- sentative of all tumor tissues from the animal model. (B) Tumor tissues were further characterized by Ki67 (proliferation, mitotic cells) and F4/80 (macrophages) immunohistochemistry. Data shown are representative of all tumor tissues from the animal model. (C) Relative populations of Ki67(+), F4/80(+) cells in the regions of interest in tumor tissues were quantified with HistoQuest® and Vectra 3.0. Representative regions of interest are shown in the left panel; the right panel contains a bar graph showing the relative populations of Ki67(+) and F4/80(+) cells in five tumor tissues. 1, regions with large F4/ 80 populations; 2, regions with small F4/80 populations. The short and long scale bars are 300 and 100 µm, respectively. *P<0.05.
We further investigated several regions of interest in each tumor tissues available and quantified the relative number of cell with Ki67(+) or F4/80(+) to total nuclei (total cells) (Fig. 2C, Supplementary Fig. S2). Interestingly some regions showed significant population of F4/80(+) cells within tumors (Fig. 2C, “1”), especially, adjacent to normal tissues.
These results imply a contribution of aggressive proliferation and the microenvironment to tumor development of LMS with TJP1 expression. High-grade LMS tissues from our in vivo models, some of which showed high Ki67 expression, were too aggressive and pleomorphic to obtain support from the tumor microenvironment. Other cells, such as macrophages may have cooperated with the tumor cells and the microenvironment.
TJP1 knockdown affects gene expression related to cell death/survival and the tumor microenvironment
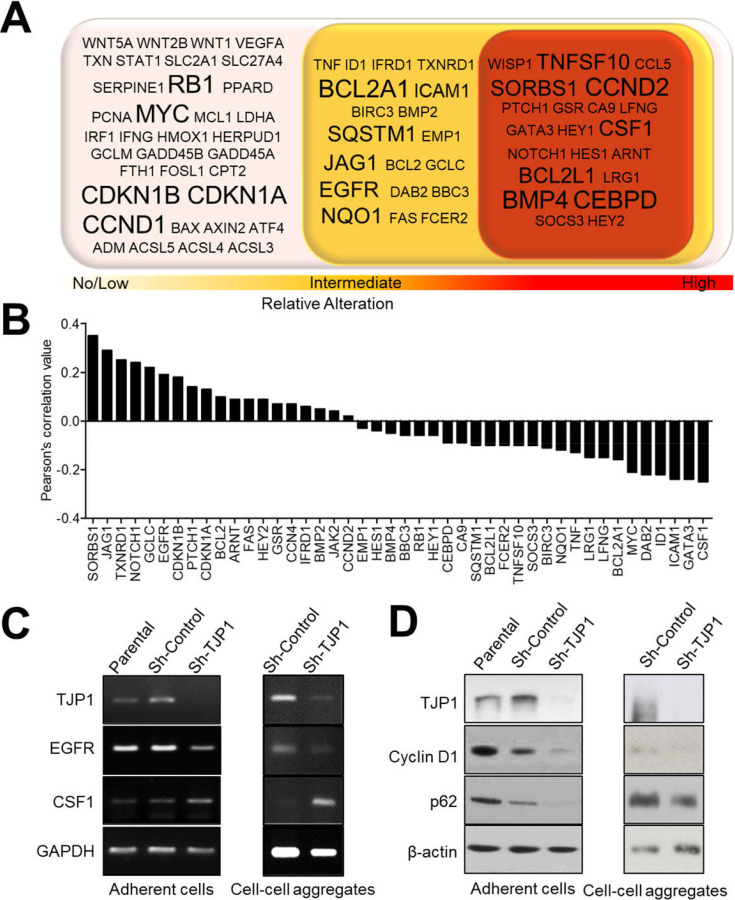
Because no in vivo tumors were formed in SK-LMS-1 Sh-TJP1 injected mice, we further examined SK-LMS-1 Sh-TJP1 cells grown in vitro to investigate the genes affected by TJP1 expression. Previously, we compared 84 genes involved in signal transduction pathway in SK-LMS-1 parental, Sh-Control and Sh-TJP1 cells by conducting PCR array (Lee et al., 2020). Here, we classified the 84 genes into three groups based on relative changes in mRNA levels (regardless whether up or downregulated) (Fig. 3A). We further investigated whether genes affected by TJP1 in SK-LMS-1 cells might be correlated to TJP1 in LMS tissues by searching the TCGA public database (Fig. 3B). Genes related to cell-cycle progression, such as G1/S-specific cyclin-D1 (CCND1, Cyclin D1 in protein) and CCND2, were classified into two groups of no change and intermediate, respectively and correlated to TJP1 positively in LMS TCGA data. A few genes, such as BCL2L1, BCL2, BCL2A1, BIRC3, BBC3, TNFSF10 (related to apoptosis), SQSTM1 (also known as protein p62, related to autophagy), EGFR (related to anoikis-resistance and proliferation) are closely related to cell survival and death, whose correlation to TJP1 in LMS TCGA dataset were various. ICAM1, a well-known cell-cell adhesion gene, was also affected by TJP1 expression, negatively. LRG1, GATA3 (related to Janus kinase [JAK]-signal transducer and activator of transcription [STAT] pathway), and CSF1 (colony-stimulating factor 1, nuclear factor-kappa B pathway) are in a class with significant changes in response to TJP1 expression. Among them, genes related to survival and growth, EGFR and Cyclin D1, were shown to correlate to TJP1 expression positively in adherent cells as well as cell-cell aggregates (Figs. 3C and 3D). TNFSF10 expression was negatively correlated to TJP1 expression in SK-LMS-1 cells and TCGA LMS transcriptome (data not shown), whose role will be elucidated later. Another, CSF1, one of altered genes by TJP1 knockdown in SK-LMS-1 cells (Fig. 3), which has been implied in regulating tumor associated macrophages (Dwyer et al., 2017; Gyori et al., 2018), was shown to have negative correlation with TJP1 in adherent cells as well as cell-cell aggregates, suggesting communication between LMS tumor cells and the tumor microenvironment, particularly innate immunity.
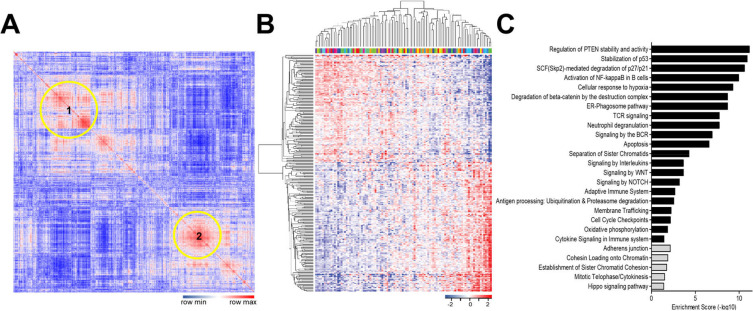
Fig. 3. TJP1 in LMS tissues was ranked within the top 10% of all transcriptomes, belonged to a cluster related to cell-cell communication and the cell cycle.
(A) The top 10% of all transcriptomes (1,816 of 18,160 genes) in LMS from the TCGA dataset were analyzed for gene-gene correlation using the MORPHEUS web tool and presented as Pearson’s Correlation Heatmap. Red represents a high correlation, and blue represents a low correlation in the heatmap. Yellow circles on the heatmap are clusters with a high correlation between genes. (B) Expression levels of genes from two clusters of Fig. 4A (Cluster 1 with 152 genes, Cluster 2 with 126 genes) are presented as heatmap. In the heatmap, rows are genes and lanes are the LMS patients’ IDs. (C) Genes from two clusters of Fig. 4B were categorized by signaling pathways and are listed based on gene-enrichment scores on a negative log scale.
TJP1 was enhanced in the TCGA LMS transcriptome
Our data from the animal models and TJP1 knockdown led us to hypothesize that TJP1 contribute to tumor progression in vivo by enhancing cell proliferation, survival, and modulating the tumor microenvironment in LMS. To investigate if TJP1 may contribute to tumorigenesis in LMS tissues, we obtained transcriptomic data of 106 LMS tissues from TCGA (Supplementary Fig. S3). In total, 18,166 genes were listed by expression level, and 1,816 genes (top 10% of expression level) were selected for further analyses. Using the MORPHEUS web tool (https://software.broadinstitute.org/morpheus/), we generated a similarity matrix analysis of 1,816 genes in terms of expression level. Pearson’s correlation analysis revealed two large clusters that exhibited a positive linear correlation (Fig. 4A). Genes related to two clusters were shown by hierarchical clustering by the MORPHEUS (Fig. 4B). Cluster 1 had 152 genes, which were related to cell-cycle regulation, the immune system, and cytokine-related pathways (Fig. 4C, black bars), while cluster 2, with 126 genes, was associated with adherens junctions, cell communication, and cell-cycle-related pathways (Fig. 4C, grey bars), suggesting enhanced cell cycle progression and cell-cell communication via cytokines within tumor microenvironment in LMS tissues. TJP1 was included in cluster 2, indicating its role in LMS development and progression.
Fig. 4. TJP1 knockdown alters gene expression related to cell survival/death and the tumor microenvironment.
(A) Eighty-four genes related to cancer cell signaling were classified into three groups based on changes caused by TJP1 knockdown in SK-LMS-1 cells. (B) Correlation of 40 genes from the intermediate and high groups of (A) with TJP1 were analyzed using LMS data from TCGA sarcoma. (C and D) Growing cells were harvested and subjected to RT-PCR (C) and immunoblotting (D). Data shown are representative of at least three independent experiments. Some cells were grown on 35-mm dish without coating poly-L-lysine for 24 h and harvested for RT-PCR and immunoblotting (cell-cell aggregates).
Next, we investigated whether TJP1 is associated with the tumor microenvironment and likely to be of interest for a therapeutic strategy with immune checkpoint inhibitors, such as antibodies against PD-1 or cytotoxic T lymphocyte-associated molecule 4 (CTLA4), which are a new class of monoclonal antibody immunotherapy (Zhang and Zhang, 2020). In the subpopulation of complex karyotype sarcomas that have been characterized as unbalanced and nonredundant genomic aberrations, immunotherapy is applicable in cases that the tumor is validated by a high density of B cells and the presence of tertiary lymphoid structures, as the specific sarcoma immune class (SIC) E (Petitprez et al., 2020). Thus, we first investigated whether the LMS data from TCGA showed a similar classification based on the gene signature (Fig. 5A). The TCGA LMS transcriptomes of 120 immune-system-related genes (Petitprez et al., 2020), were analyzed using hierarchical clustering in a heatmap. After clustering, 81 selected genes were repeatedly analyzed by excluding genes that did not form clusters in Fig. 5A. Based on the heatmap, we also observed some tissues with expression profiles similar to those of SIC E, such as strong expression of the B cell lineage and immune cell signatures. Two categories of genes were correlated with TJP1 and related genes (Fig. 3), such as TJP1, ICAM1, CSF1, EGFR, and BIRC3, and are listed in the same order as the tissues (Fig. 5B). The expression of CSF1, which regulates macrophages leading to modulation of the tumor microenvironment (Gyori et al., 2018; Pyonteck et al., 2013; Quail et al., 2016), led us to further investigate the relationship between TJP1 and immune-checkpoint inhibitors, such as PD-1, PDL-1, and CTLA4 (Fig. 5C); TJP1 expression was negatively correlated with PD-1, PDL-1, and CTLA4. Among these, CTLA4 was inversely correlated (Fig. 5D), suggesting that TJP1 could be applied as a therapeutic immune checkpoint inhibitor.
Fig. 5. TJP1 expression is significantly associated with CTLA4 in LMS.
(A) The LMS transcriptome was analyzed according to the SIC classification. (B) Selected genes in SK-LMS-1 cells related to TJP1 expression were clustered by expression level in the same tissue order as shown in Fig. 5A. Blue brackets contain EGFR and TJP1, and the red brackets contain the three genes CSF1, ICAM1, and BIRC3, respectively. (C) Similarly, TJP1 and PDL-1 (CD274) and PD-1 (PDCD1) and CTLA4 were visualized as a heatmap using the same tissue order as shown in Figs. 5A and 5B. (D) Pearson’s correlation coefficients between TJP1 and CTLA4 or CTLA4 and CSF1 were expressed in scatter plots using GraphPad Prism software. All data are from the LMS transcriptome in the TCGA dataset, obtained from cBioportal.
DISCUSSION
The major findings of this study are as follows: TJP1 knockdown affected the development of LMS in vivo (Fig. 1). Aggressive mitotic areas, mitotic/macrophage-infiltrated areas, and fewer mitotic/macrophage-infiltrated areas (Fig. 2) were observed in LMS tumor tissues from the animal model. TJP1 knockdown affected genes related to the cell cycle, apoptosis, cell-cell aggregation, and cytokines (Fig. 3). TJP1 was included in the top 10% of genes in the TCGA LMS transcriptome and belonged to a specific cluster with high positive gene-gene correlation (Fig. 4), and some LMS tissues with high immune function according to SIC classification (Petitprez et al., 2020), had low levels of TJP1 expression but high levels of CTLA4 expression (Fig. 5). Taken together, our results imply a role for TJP1 in the development of LMS.
Here, cells expressing TJP1 that were injected into immunodeficient mice developed into LMS in vivo, which was aggressively mitotic, mitotic with infiltrated macrophages, and less mitotic with aggressive infiltrated macrophages. However, injected cells with reduced TJP1 expression did not develop LMS, suggesting roles for TJP1 in tumor progression. For example, TJP1 may play a role in cell-cell aggregation (Fig. 1) and growth by avoiding anoikis and apoptosis through the EGFR and BCL2 signaling pathways, as suggested previously (Lee et al., 2020). In addition, TJP1 may modulate the tumor microenvironment during progression of LMS by regulating CSF1-linked innate immune pathways. Thus, we investigated the role of TJP1 and CSF1 in monocyte differentiation and macrophage polarization to determine whether targeting TJP1 might be beneficial for combined immunotherapy.
We observed dramatic tumor growth at 12 weeks after the intramuscular injection, which led us to hypothesize a potential role of TJP1 in aging and cell-cycle regulation. It is still unclear whether TJP1 is associated with aging in terms of cell-cycle regulation during tumor growth and development. Therefore, we are currently investigating the possible role of TJP1 in senescence. In this study, we injected SK-LMS-1 parental, Sh-Control and Sh-TJP1 cells into immunodeficient mice (Rag2-/- γc-/- immunodeficient mice; The Jackson Laboratory) and found mitotic cells within the resulting tumor tissues (Fig. 2); this suggested a role of TJP1 in LMS development. Some areas of the tumor tissues, particularly those that were close to normal issues, contained infiltrated macrophages, suggesting communication between SK-LMS-1 cells and macrophages within the tumor microenvironment (Fig. 2). The expression of CSF1, a well-established macrophage factor, was affected by TJP1 expression. Possible crosstalk between TJP1 and CSF1 in tumor cells and immune cells within the tumor microenvironment was explored via bioinformatic analyses of TCGA LMS transcriptomic data (Figs. 4 and 5).
Immunotherapy is quite attractive for treating complex karyotype sarcomas, as there are a few clear biomarkers for diagnosis and therapeutics. A few groups have recently performed integrated analyses based on immune signatures regardless of the classifying cancer type (Nirmal et al., 2018; Thorsson et al., 2018). Profiling immune-cell infiltration based on gene signatures or the expression of immune-checkpoint markers by immunohistochemistry is applicable in several cancers, including soft-tissue sarcoma (Dancsok et al., 2020; Mlecnik et al., 2016; Petitprez et al., 2020) suggesting that it is valuable for identifying genes that are important in immune-system modulation. One study showed that immune-checkpoint genes are negatively correlated with TJP1 but positively correlated with vimentin in lung cancer (Chae et al., 2018). Here, we identified a few genes with changed expression after TJP1 knockdown, such as EGFR, NOTCH1, BCL2, and CSF1 (Fig. 3), which have been implicated in a variety of cellular processes and cancer cell signaling (Sanchez-Vega et al., 2018). We have shown that TJP1 knocked-down cells respond more than control cells to gefitinib (Lee et al., 2020). Furthermore, CSF1, which was affected by TJP1 expression in our cell-based study (Fig. 3), was negatively correlated with CTLA4 in TCGA LMS transcriptome (Fig. 5), suggesting a possible role for TJP1 in CSF1 expression leading to modulation of the tumor microenvironment through the monocyte/macrophage axis. CSF1/CSF1R blockade reprogrammed tumor-infiltrating macrophages and improved the response to T-cell-checkpoint immunotherapy in a pancreatic cancer model (Zhu et al., 2014). In the current study, CXCL8 expression was negatively correlated with CSF1 levels in ovarian tumors, and increased CSF1 expression was related to low levels of the neutrophil signature, implying that that cotreatment with CSF1R and CXCR2 inhibitors decreases the population of tumor-associated macrophages, which contribute to the effect of PD1 immunotherapy (Kumar et al., 2017).
In this study, we found that TJP1 expression in LMS cell lines was critical for cell-cell aggregation in coating-free three-dimensional culture systems, and that TJP1 expression promoted tumor formation in vivo. Bioinformatic analyses were performed by profiling the genes affected by TJP1 knockdown in SK-LMS-1 cells, and by analyzing the top 10% of genes within all TCGA LMS transcriptomic data. These analyses provided further evidence for a role of TJP1 in LMS development and progression. The correlations among TJP1, CSF1, and CTLA4 support the possibility that targeting TJP1 may have therapeutic applications.
In conclusion, the data from this study strongly support a role for TJP1 in the progression of LMS and the tumor microenvironment. Although further confirmation should be performed in the future, we suggest that targeting TJP1 in LMS with high immune cell infiltration might be beneficial for anticancer therapeutics with immune-checkpoint inhibitors, particularly CTLA4.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
We thank Mi Sun Park (V.M.D.) and Bo Ra Kim (V.M.D.) of Animal Laboratory (National Cancer Center) and Dr. Se Hun Kang and colleagues of National Cancer Center Animal Molecular Imaging Team, Dr. Eun Kyung Hong, professional pathologist, Department of Pathology, National Cancer Center Hospital, Dr. Jong Kwang Kim of NCC Omics Core Center for their expert assistance and helpful suggestions. We also thank the NCC sarcoma research group (National Cancer Center) for their advice.
This research was funded by National Cancer Center grant NCC-1710252 (to H.J.Y.), NCC-1810865 (to H.J.Y.), NCC-2110521 (to H.J.Y.) and by the Korean Medical Device Development Fund Grant funded by the Korean Government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) NTIS-202012E12-02 (to D.H.K.).
Footnotes
AUTHOR CONTRIBUTIONS
E.Y.L. performed the experiments. M.K. and B.K.C. gave technical support and analyzed the data. H.J.Y. conceived and supervised the study. I.C. and D.H.K. provided expertise and feedback. H.J.Y. and E.Y.L. wrote and edited the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Babichev Y., Kabaroff L., Datti A., Uehling D., Isaac M., Al-Awar R., Prakesch M., Sun R.X., Boutros P.C., Venier R., et al. PI3K/AKT/mTOR inhibition in combination with doxorubicin is an effective therapy for leiomyosarcoma. J. Transl. Med. 2016;14:67. doi: 10.1186/s12967-016-0814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J., Taylor B.S., Banerji S., Ramos A.H., Lagos-Quintana M., Decarolis P.L., Shah K., Socci N.D., Weir B.A., Ho A., et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, author. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171:950–965.e28. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae Y.K., Chang S., Ko T., Anker J., Agte S., Iams W., Choi W.M., Lee K., Cruz M. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC) Sci. Rep. 2018;8:2918. doi: 10.1038/s41598-018-21061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibon F., Lagarde P., Salas S., Perot G., Brouste V., Tirode F., Lucchesi C., de Reynies A., Kauffmann A., Bui B., et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat. Med. 2010;16:781–787. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- Chudasama P., Mughal S.S., Sanders M.A., Hubschmann D., Chung I., Deeg K.I., Wong S.H., Rabe S., Hlevnjak M., Zapatka M., et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat. Commun. 2018;9:144. doi: 10.1038/s41467-017-02602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier J.M., Charville G.W. Diagnostic classification of soft tissue malignancies: a review and update from a surgical pathology perspective. Curr. Probl. Cancer. 2019;43:250–272. doi: 10.1016/j.currproblcancer.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Dancsok A.R., Gao D., Lee A.F., Steigen S.E., Blay J.Y., Thomas D.M., Maki R.G., Nielsen T.O., Demicco E.G. Tumor-associated macrophages and macrophage-related immune checkpoint expression in sarcomas. Oncoimmunology. 2020;9:1747340. doi: 10.1080/2162402X.2020.1747340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer A.R., Greenland E.L., Pixley F.J. Promotion of tumor invasion by tumor-associated macrophages: the role of CSF-1-activated phosphatidylinositol 3 kinase and Src family kinase motility signaling. Cancers (Basel) 2017;9:68. doi: 10.3390/cancers9060068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning A.S., Jameson B.J., Jesaitis L.A., Anderson J.M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 1998;273:29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- Fletcher C.D. The evolving classification of soft tissue tumours - an update based on the new 2013 WHO classification. Histopathology. 2014;64:2–11. doi: 10.1111/his.12267. [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S., Miao D., Demetri G.D., Adeegbe D., Rodig S.J., Shukla S., Lipschitz M., Amin-Mansour A., Raut C.P., Carter S.L., et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.3390/ijms21228470,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Jo V.Y., Mills A.M., Zhu S.X., Lee C.H., Espinosa I., Nucci M.R., Varma S., Forgo E., Hastie T., et al. Clinically relevant molecular subtypes in leiomyosarcoma. Clin. Cancer Res. 2015;21:3501–3511. doi: 10.1158/1078-0432.CCR-14-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyori D., Lim E.L., Grant F.M., Spensberger D., Roychoudhuri R., Shuttleworth S.J., Okkenhaug K., Stephens L.R., Hawkins P.T. Compensation between CSF1R+ macrophages and Foxp3+ Treg cells drives resistance to tumor immunotherapy. JCI Insight. 2018;3:e120631. doi: 10.1172/jci.insight.120631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde P.S., Chen D.S. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52:17–35. doi: 10.1016/j.immuni.2019.12.011. [DOI] [PubMed] [Google Scholar]
- Helman L.J., Meltzer P. Mechanisms of sarcoma development. Nat. Rev. Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- Hoos A., Stojadinovic A., Mastorides S., Urist M.J., Polsky D., Di Como C.J., Brennan M.F., Cordon-Cardo C. High Ki-67 proliferative index predicts disease specific survival in patients with high-risk soft tissue sarcomas. Cancer. 2001;92:869–874. doi: 10.1002/1097-0142(20010815)92:4<869::aid-cncr1395>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Kim C., Kim I.H., Kim S.I., Kim Y.S., Kang S.H., Moon S.H., Kim T.S., Kim S.K. Comparison of the intraperitoneal, retroorbital and per oral routes for F-18 FDG administration as effective alternatives to intravenous administration in mouse tumor models using small animal PET/CT studies. Nucl. Med. Mol. Imaging. 2011;45:169–176. doi: 10.1007/s13139-011-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim J.H., Kang H.G., Park S.Y., Yu J.Y., Lee E.Y., Oh S.E., Kim Y.H., Yun T., Park C., et al. Integrated molecular characterization of adult soft tissue sarcoma for therapeutic targets. BMC Med. Genet. 2018;19(Suppl 1):216. doi: 10.1186/s12881-018-0722-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Donthireddy L., Marvel D., Condamine T., Wang F., Lavilla-Alonso S., Hashimoto A., Vonteddu P., Behera R., Goins M.A., et al. Cancer-associated fibroblasts neutralize the anti-tumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell. 2017;32:654–668.e5. doi: 10.1016/j.ccell.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.Y., Yu J.Y., Paek A.R., Lee S.H., Jang H., Cho S.Y., Kim J.H., Kang H.G., Yun T., Oh S.E., et al. Targeting TJP1 attenuates cell-cell aggregation and modulates chemosensitivity against doxorubicin in leiomyosarcoma. J. Mol. Med. (Berl.) 2020;98:761–773. doi: 10.1007/s00109-020-01909-8. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Paek A.R., Yoon K., Kim S.H., Lee S.Y., You H.J. Tight junction protein 1 is regulated by transforming growth factor-beta and contributes to cell motility in NSCLC cells. BMB Rep. 2015;48:115–120. doi: 10.5483/BMBRep.2015.48.2.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.H., Faunce D.E., Stacey M., Terajewicz A., Nakamura T., Zhang-Hoover J., Kerley M., Mucenski M.L., Gordon S., Stein-Streilein J. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J. Exp. Med. 2005;201:1615–1625. doi: 10.1084/jem.20042307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.A., Jiang W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta. 2009;1788:872–891. doi: 10.1016/j.bbamem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Mata M., Gottschalk S. Adoptive cell therapy for sarcoma. Immunotherapy. 2015;7:21–35. doi: 10.2217/imt.14.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B., Bindea G., Angell H.K., Maby P., Angelova M., Tougeron D., Church S.E., Lafontaine L., Fischer M., Fredriksen T., et al. Integrative analyses of colorectal cancer show Immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Nirmal A.J., Regan T., Shih B.B., Hume D.A., Sims A.H., Freeman T.C. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol. Res. 2018;6:1388–1400. doi: 10.1158/2326-6066.CIR-18-0342. [DOI] [PubMed] [Google Scholar]
- Paek A.R., Mun J.Y., Hong K.M., Lee J., Hong D.W., You H.J. Zinc finger protein 143 expression is closely related to tumor malignancy via regulating cell motility in breast cancer. BMB Rep. 2017;50:621–627. doi: 10.5483/bmbrep.2017.50.12.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek A.R., Mun J.Y., Jo M.J., Choi H., Lee Y.J., Cheong H., Myung J.K., Hong D.W., Park J., Kim K.H., et al. The role of ZNF143 in breast cancer cell survival through the NAD(P)H quinone dehydrogenase 1(-)p53(-)Beclin1 axis under metabolic stress. Cells. 2019;8:296. doi: 10.3390/cells8040296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitprez F., de Reynies A., Keung E.Z., Chen T.W., Sun C.M., Calderaro J., Jeng Y.M., Hsiao L.P., Lacroix L., Bougouin A., et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F., Olson O.C., Quick M.L., Huse J.T., Teijeiro V., et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Bowman R.L., Akkari L., Quick M.L., Schuhmacher A.J., Huse J.T., Holland E.C., Sutton J.C., Joyce J.A. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352:aad3018. doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Korchin B., Lahat G., Wei C., Bolshakov S., Nguyen T., Merritt W., Dicker A., Lazar A., Sood A., et al. Combined vascular endothelial growth factor receptor/epidermal growth factor receptor blockade with chemotherapy for treatment of local, uterine, and metastatic soft tissue sarcoma. Clin. Cancer Res. 2008;14:5466–5475. doi: 10.1158/1078-0432.CCR-08-0562. [DOI] [PubMed] [Google Scholar]
- Rusakiewicz S., Semeraro M., Sarabi M., Desbois M., Locher C., Mendez R., Vimond N., Concha A., Garrido F., Isambert N., et al. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res. 2013;73:3499–3510. doi: 10.1158/0008-5472.CAN-13-0371. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbye S.W., Kilvaer T., Valkov A., Donnem T., Smeland E., Al-Shibli K., Bremnes R.M., Busund L.T. Prognostic impact of lymphocytes in soft tissue sarcomas. PLoS One. 2011;6:e14611. doi: 10.1371/journal.pone.0014611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T., Rongvaux A., Rathinam C., Takizawa H., Borsotti C., Philbrick W., Eynon E.E., Manz M.G., Flavell R.A. Transgenic expression of human signal regulatory protein alpha in Rag2-/-gamma(c)-/- mice improves engraftment of human hematopoietic cells in humanized mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13218–13223. doi: 10.1073/pnas.1109769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomi S., Mai A., Nevo J., Laine J.O., Vilkki V., Ohman T.J., Gahmberg C.G., Parker P.J., Ivaska J. PKCepsilon regulation of an alpha5 integrin-ZO-1 complex controls lamellae formation in migrating cancer cells. Sci. Signal. 2009;2:ra32. doi: 10.1126/scisignal.2000135. [DOI] [PubMed] [Google Scholar]
- Verma V., Paek A.R., Choi B.K., Hong E.K., You H.J. Loss of zinc-finger protein 143 contributes to tumour progression by interleukin-8-CXCR axis in colon cancer. J. Cell. Mol. Med. 2019;23:4043–4053. doi: 10.1111/jcmm.14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely M.D., Kershaw M.H., Schreiber R.D., Smyth M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- Zhang X.D., Baladandayuthapani V., Lin H., Mulligan G., Li B., Esseltine D.W., Qi L., Xu J., Hunziker W., Barlogie B., et al. Tight junction protein 1 modulates proteasome capacity and proteasome inhibitor sensitivity in multiple myeloma via EGFR/JAK1/STAT3 signaling. Cancer Cell. 2016;29:639–652. doi: 10.1016/j.ccell.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020;17:807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Knolhoff B.L., Meyer M.A., Nywening T.M., West B.L., Luo J., Wang-Gillam A., Goedegebuure S.P., Linehan D.C., DeNardo D.G. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.