Abstract
Biomaterials is an interdisciplinary field of research to achieve desired biological responses from new materials, regardless of material type. There have been many exciting innovations in this discipline, but commercialization suffers from a lengthy discovery to product pipeline, with many failures along the way. Success can be greatly accelerated by harnessing machine learning techniques to comb through large amounts of data. There are many potential benefits of moving from an unstructured empirical approach to a development strategy that is entrenched in data. Here, we discuss the recent work on the use of machine learning in the discovery and design of biomaterials, including new polymeric, metallic, ceramics, and nanomaterials, and how machine learning can interface with emerging use cases of 3D printing. We discuss the steps for closer integration of machine learning to make this exciting possibility a reality.
Graphical abstract
1. Introduction
Many of the recent clinical challenges involve the need for biomaterials. One of the major stumbling blocks preventing the widespread use of biomaterials as implants are the poor interaction between the materials and the biological system. In addition, the complex interplay between composition, structures, and properties renders the materials discovery process tedious. For instance, for polyurethane alone, there are already a vast number of different possible compositions [1]. There are many aspects of input parameters that can be used to optimize the output properties. For instance, there is a complex relationship between surface properties and biological response, where much of the data-driven research focuses on solving. To tackle these issues, high-throughput platforms have been developed to generate an adequate number of inputs that consider physical and compositional cues [[2], [3], [4], [5], [6], [7]].
Despite the advent of high-throughput platforms, biomaterials research is still in its infancy in terms of multi-dimensional data generation and utilization. In molecular biology, a large amount of data helps data scientists to draw a meaningful conclusion. For instance, hundreds of thousands and even millions of gene expression profiles of calls are either derived genetically or pharmacologically, helping to form a connectivity map between inputs and outputs [8]. On the other hand, the structure-property relationship has long been quantitatively studied in materials science beyond composition properties. Likewise, the complexity of multi-dimensional data is the biggest challenge. Many compound compositions can be formed out of the 94 naturally occurring elements, with wide-ranging physical properties. For example, changing the alloy composition can lead to changes in metal properties such as hardness and corrosion resistance. Therefore, large volume screening and machine learning (ML) are actively pursued, motivated by their potential to shorten the time taken for scientific breakthroughs to commercial products. Experimentally, there is a fundamental shift of paradigm from a traditional empirical approach to a more systematic and guided approach via machine learning augmentation. Materials databases can be leveraged to accelerate both experimental and theoretical studies [[9], [10], [11], [12], [13]]. This is made easier by user-friendly data mining tools such as Matminer [14]. For instance, machine learning can drastically reduce the computational cost and time compared to traditional DFT (Density functional theory). Experimentally, owing to its ability to handle multi-dimensional data, rapid advancements in functional materials such as photovoltaics [15], piezoelectric [16], and thermoelectric [[17], [18], [19], [20], [21], [22]] have been demonstrated. However, most applications of machine learning in materials science to date have mainly been focusing on a narrow class of materials. There is still the lack of a comprehensive approach to accelerating materials discovery, development, characterization, all the way to the prototyping stage. A particular research area that will greatly benefit from the use of machine learning is biomaterials. Traditionally, it takes years of research and development for a material to be deemed good and safe enough for biological applications.
The current Biomaterials market size is estimated at more than 100 billion USD in 2019, with a CAGR of 15.9% [23], indicating that biomaterial development has significant therapeutic and commercial potential. Biomaterials is a multidisciplinary discipline that studies how biological systems interact with sophisticated materials. Biomaterials research aims to create and manipulate material qualities to produce a certain biological response [24]. We decided to concentrate our discussion on biomaterials because the wealth of scientific knowledge that has already been accumulated in this sector over many decades of research provides a fruitful foundation for making excellent use of machine learning. This field also has a multidisciplinary impact on how machine learning may be implemented across various materials, such as metals and polymers, with direct repercussions for improving people's quality of existence. It is important and relevant to transform the paradigm of thinking in advancing biomaterials, from conventional empirical methods to data-driven machine learning.
The training data may be supplied into the algorithms in both circumstances (whether theoretical or experimental). Supervised learning aims to develop a function that can predict output values from a collection of input data reliably. This may be done using training datasets that have several inputs and output sets. Several input variables (e.g., material composition) are included in training data (i.e., properties). The principal purpose of both techniques is for data to be modeled to learn about the underlying structure or distribution. In the next sections, we will look at circumstances in which supervised and unsupervised learning is important in the evolution of biomaterials.
In this paper, we outline the biomaterial realm of machine learning. A discussion will be placed on the applicability and promise of the data-driven approach in various biomaterials. The input and output qualities of generic biomaterial design are discussed briefly in Section 2. Polymer-based biomaterials will be explored in Section 3, Section 4 metals, Section 5 ceramics, and Section 6 nanomaterials will be discussed. Section 7 discusses the part on additive manufacturing and 3D printed biomaterials. Finally, section 8 outlines the prospects for expediting the creation and processing through the application of machine learning of these diverse kinds of biomaterials.
2. Biomaterials design - parameters and primer to machine learning
Biomaterials such as drug delivery depots, tissue engineering scaffolds, imaging/diagnostic systems, and device coatings have been employed extensively. There are various ways biomaterials can be used in the body, including implantation, blood vessel injection, skin application, or extracting bodily components for existing analysis [25]. In terms of material types, metallic and ceramic materials have been traditionally used as biomaterials, predominantly found in implants. Because of the impressive mechanical strength and relative inertness, metals have been employed [26]. Due to its capacity to encourage bone development and regeneration, ceramics are intriguing and are widely utilized as dental implants [27]. However, in recent times, polymeric biomaterials have found rapid growth due to their shape moldability. Furthermore, the advancement of additive manufacturing and high-resolution spatial patterning has given an extra dimension to biomaterials research in recent years [28,29].
Input factors for chemical and physical configurations exist to create a biomaterial to fulfil its desired purpose (Table 1). The input parameters for biomaterials may be modified to vary the output qualities. Inputs for chemical components often involves the purposeful inclusion of chemical moieties that impact material characteristics. This comprises pH switchable groups to respond to the biologcal environment [[30], [31], [32], [33], [34], [35]], adding charges for polymer-gene interactions in gene delivery agents [[36], [37], [38], [39], [40], [41]], Adding targeting moieties like folic acid for cancer targetting nanoparticles [30,31], bisphosphonate as a binding moiety to improve bone binding [27], and the proliferation and directed differentiation of stem cells induced by cell-matrix interacting ligands [42,43]. Physical inputs can also impact cell interactions [44]. These include the observation of enhanced permeability and retention (EPR) effects through the use of smaller nanoparticles [45,46], larger sized microparticles for the reduced extent of fibrosis [47], surface topology, and patterning to selectively enrich cells and extracellular vesicles, identified by microfluidic devices [28,48]. Materials that simulate extracellular matrices can potentially provide chemical and mechanical data on modifying cell behavior [49,50].
Table 1.
Biomaterials input and output parameters.
| Input Parameter | Output Property | Demonstrated application | Ref |
|---|---|---|---|
| Microstructure | Crack propagation | Materials improvement | [51] |
| Composition design | Steel corrosion crack resistance | [52] | |
| Molecular properties | Drug nanoparticle formation | [53] | |
| Chemical structure | Printability of drug-loaded formulations | [54] | |
| Processing parameters | Surface roughness | [55] | |
| Surface chemistry Positive charges |
Protein adsorption Antimicrobial |
In vitro | [56] [57] |
| Chemical structure | Cell adhesion | [58] | |
| Mechanical properties | cell morphology/differentiation | [39] | |
| Bulk shape | spatial organization of cells | [43] | |
| Adsorbed protein | Clearance pattern | In vivo | [59] |
| Chemical structure | Anti-fibrosis | [60] | |
| Chemical structure | organ targeting | [61] |
It is worth noting that the design of biomaterials needs to take account of extra complications in the body environment. Nanoparticles are traditionally tested with in-vitro cell tests for cell internalization. However, numerous additional physiological obstacles, such as renal filtration, need to be overcome. Therefore, tests that can replicate physiological function are crucial. Normally, an implant in the body forms a fibrotic layer over time causing implant failure. In biomaterial design, precise design can ensure that this does not happen. It is feasible to screen for compounds with anti-fouling and anti-fibrotic capabilities by combining a library of various chemical moieties [60]. The bodily environment may also change the material's functioning via degradation processes, which might have detrimental effects on performance over time. The different input parameters have also been tested to different extents for demonstrated applications, from improving material properties, to in vitro cell-based assays and in vivo studies in animal models.
It takes years for novel biomaterials to be made and used. The creation of biomaterials typically takes several phases from the optimization of material characteristics, biocompatibility studies, clinical tests, etc., due to the difficulties of the design parameter optimization process. These methods are time-consuming and laborious. For example, it took 20 years to produce the first product based on the PLGA counterpart for prostate cancer therapy from the initial PLA biocompatibility and biodegradation report [62]. Machine learning might address or expedite many of the challenges inherent in the creation of biomaterials in this data-driven age. For example, by using convoluted neural networks, it is possible to accelerate the process to obtain predictions for optimized composite materials to just 10h, as opposed to 5 days when extensive finite element method modelling is conducted for all the possible material compositions [63]. Searching through incredibly large parameter space to find the optimal biomaterial for in vivo usage can also become a less daunting task by using a large dataset with suitable data analysis [60]. It is necessary to understand the kind and amount of data available and the associated types of algorithms to make optimal use of machine training to speed biomaterial development.
In machine learning, supervised learning is commonly used, and the goal for supervised learning is to build a model that can map the input parameters to the corresponding output properties and derive a relationship between the inputs and outputs. It is especially important that this model is predictive and can accurately provide estimated output properties when new input data is provided. The other major class is unsupervised learning, where the goal is to assign the input parameters into groups based on their properties. For both cases, the training dataset is used to develop the ML model, while a separate test dataset is used to assess the performance of the model.
ML algorithms vary in terms of their use case and complexity (see Table 2). If the prediction can be accurately performed with few variables, then simpler models such as regression can be used. If accurate prediction necessitates accounting for subtle changes in many variables, then more complex models such as neural networks would be required. While there are clear benefits to a model that can navigate complex datasets and come up with accurate predictions, these models could be more difficult to explain and interpret. Furthermore, the amount of data required scales dramatically with the model complexity and can be a limiting factor. While a regression could be modeled with a dozen data points, a convolutional neural network can easily require more than several thousand data points for the model to be trained. Therefore, in addition to the complexity of the prediction task, the availability of data can often hamper the capability of ML models to provide accurate predictions.
Table 2.
Common machine learning algorithms.
| Type | Methods | When to use | Considerations |
|---|---|---|---|
| Simpler models |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Complex models |
|
|
|
|
|
|
|
|
|
|
|
| Smaller dataset models |
|
|
|
|
|
|
|
| Data sampling |
|
|
|
|
|
|
|
|
|
|
Datasets often need to be processed to become amenable to ML algorithms. For example, there could be class imbalance issues where there is too much data for a certain input parameter and too little data for a second input parameter, and therefore the model cannot satisfactorily predict for the second input. Another consequence of class imbalance is the underestimation of feature importance (how much certain input contributes to outputs) of the undersampled input parameter.
To avoid class imbalance, it is crucial to adopt proper data sampling, depending on the type of data. Data sampling methods including random oversampling and Latin hypercube sampling are some of the commonly used methods. The challenges with smaller datasets that biomaterials research frequently face can be partly overcome by specialized techniques such as transfer learning, and by techniques that incorporate prior knowledge such as Bayesian inference.
For more detailed information on machine learning and its application to other related fields, the readers are advised to consult extensive reviews available [[64], [65], [66], [67], [68]].
3. Polymeric biomaterials applications
Polymers are very useful as biomaterials due to their adaptability which is unparalleled by metals and ceramics [69,70]. The great range of physical and chemical characteristics of polymers have made numerous applications possible such as tissue engineering and delivery vehicles for drugs/genes [[71], [72], [73], [74], [75]]. It is predicted that medicinal polymers alone are around 1 billion dollars in market size. Polymers were originally designed for non-biomaterial applications using plastics, elastomers, and fibers. Nevertheless, enormous design, synthesis, and evaluation efforts have been done on polymers as biomaterials for certain purposes and various criteria have been used in the research of polymers as biomaterials. Due to the complexity and multi-dimensional factors that determine the characteristics of polymeric biomaterials the development of polymers as qualified biomaterials always present a tremendous challenge to clearly understand the relationships between performance and underlying chemistry and structures of polymers. Machine learning (ML) has been studied for the self-assembly of dipeptide hydrogels and the adherence of cells and protein profiles on polymeric surfaces.
3.1. Polymeric biomaterials examples
3.1.1. Self-assembled dipeptide hydrogels
Hydrogels are 3D cross-linked networks [[76], [77], [78], [79], [80], [81]]. Without dissolving its backbone, it can hold a large amount of water. In combination with its good biocompatibility, these unique features turn hydrogels into adaptable biomaterials for broad uses, such as tissue engineering, sensing, cell encapsulation, and drug administration. However, within recent years there has been a highly unclear link between the chemical structures of peptides and hydrogel properties. In Li Fei and others' work, a structural diverse hydrogel library was developed with a combinatorial strategy of more than 2000 peptides. To predict the gel formation potential, ML was employed to establish a relationship between the chemical structure of peptides and the self-assembly behavior. Furthermore, the stiffness of hydrogels has been discovered to berelated both to the crosslinking degree and to the nanofiber diameter [82].
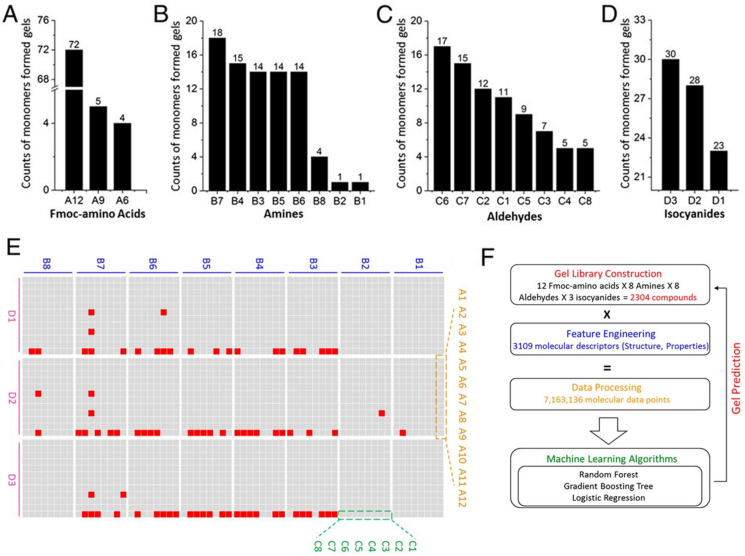
Fig. 1 shows the process of generating a library of the comprehensive chemical library as the testing pool. 31 Monomers were used, which includes 8 amines, 8 aldehydes/ketones, 12 Fmoc-amino acids, and 3 isocyanides to provide a combination of 2304 compounds through the Ugi reaction, shown in Fig. 1a–e. The overall process flow is shown in Fig. 1f. From these compounds, 7,163,136 structural parameters (3109 per molecule) were obtained based on the PaDEL-descriptor calculation. Due to class imbalance arising from the fact that less than 4% of hydrogels were successfully formed, data resampling was performed, such as random oversampling (RO), adaptive synthetic sampling (ADASYN), and synthetic minority oversampling technique (SMOTE). RO is the random addition of data points to the minority class. ADASYN and SMOTE are both more sophisticated methods to add these data points by the process of interpolation, with further changes in the number of data points corresponding to the complexity and difficulty for ADASYN.
Fig. 1.
A-D) 8 amines, 8 aldehydes/ketones, 12 Fmoc-amino acids, and 3 isocyanides used for generating the library of hydrogels. E) Screening results of hydrogels (red, a gel formed; gray, solution state. F) Preparation of peptides library. Adapted from the literature [82].
After resampling the data, multiple classification models were applied to the data. An extensive list of algorithms ranging from linear classifiers such as logistic regression to non-linear ones such as neural networks was used. Subsequently, after hyperparameter tuning for each of the algorithms, three best performing ones were selected, namely gradient boosting, random forest, and logistic regression. Gradient boosting was found to perform slightly better than the other two. Due to the highly imbalanced data, precision and recall were used to evaluate the quality of the algorithm performance. Precision is defined as the ratio of correct results to predicted results, while recall is the fraction of correct results in predicted positive samples. The precision of 54%, 57%, and 62% was achieved for the random forest, logistic regression, and gradient boosting, respectively.
In addition, feature importance was calculated with the top 20 descriptors obtained from each of the three machine learning algorithms. It was found that the descriptors Fmoc-amino acids, largest absolute of Burden modified eigenvalue, and smallest absolute of Burden modified eigenvalue contributed significantly to the successful formation of molecular hydrogels.
3.1.2. Response of cell and adsorption of proteins on polymeric surfaces
Polymers can be used as tissue engineering scaffolds and to develop substitutes for tissues and organs [40,71,83]. To develop scaffold polymer, understanding the cellular response and the relationship with polymeric properties is of utmost importance [[84], [85], [86]]. Factors such as surface wettability, surface chemistry, topography, and mechanical properties need to be accounted for when developing these polymers for scaffolds. De Boer and colleagues showed that surface topography can be screened to modulate cytokine secretion from stromal cells and support the phenotype of tendon tenocytes [87,88]. Machine learning may be done using a partial least squares regression approach to understand the association between adherence to human embryonic stem cells (hESC) and the different polyacrylate polymers [58]. An array with nearly 500 polyacrylates with a wide range of cross-linking density and hydrophobicity/hydrophilicity was synthesized from 22 acrylate monomers via combinatorial approaches [89]. A long-wavelength UV source was used to polymerize different combinations of monomers. Human embryonic stem cells were seeded onto arrays of polyacrylate after coating with fetal bovine serum. Cellular responses were quantified by laser scanning cytometry. Colony formation frequency was used to quantify the results, the ratio of polymer which hES cell colonies formed to the total number of replicate spots of the same kind of polymers on each array. The frequency of colony formation of hES was not correlated to the roughness of the surface, elastic modulus, or wettability. Instead, good agreement was found between the colony formation frequency and the predicted values from ToF SIMS of the polyacrylate surface for both the training and validation model. A regression coefficient may be produced for each ToF-SIMS ion for the quantitative understanding of the relative contribution to the frequency of colony formation. For cyclic ions, ions comprising oxygen and hydrocarbon ions, a positive regression coefficient was observed. Tertiary amine moiety and the tertiary butyl movement, however, exhibit a coefficient of negative regression. The use of computer descriptors for investigating hEB adherence to pre-treated polyacrylate micro arrays was designed exclusively using computer models [90,91]. The Bayesian Nonlinear Model (BRANNLP) was built utilizing the sparse Laplacian neural network fed with 23 molecular descriptors and 2 hidden layer nodes. For the hEB adhesive prediction, the r2 value of 0.80 and 0.82 was derived from the training set and the test set, whjich is higher than the predictive power of PLS models utilizing experimental parameters [90].
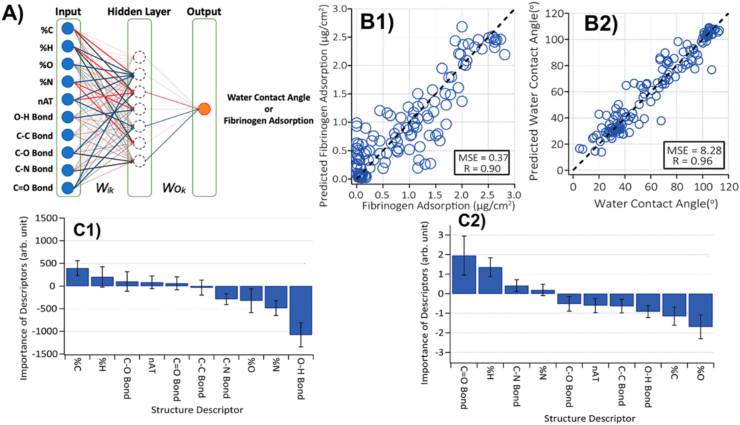
In other studies, Rudolf, et al. leveraged a data-driven approach to predict protein adsorption on self-assembled monolayers (SAMs) [92]. The study aimed at predicting the contact angle of the water (WCA) on SAMs and protein adsorption. Chemical structures of the constituent molecules of the SAMs were fed into machine learning with an artificial neural network (ANN) model. In addition, the importance of parameters describing chemical structure was also investigated. Protein surface adsorption has a direct impact on biological surface reactions that involve various biological activities, such as a cell adhesion profile [90,91]. As can be observed from Fig. 2A, the adsorption behavior of proteins was analyzed and predicted on the surfaces of the monolayers with a three-stage artificial neural network (ANN) [92]. Fig. 2B shows the relationship between predicted and actual fibrinogen and water contact angle, respectively. The high R-value of 0.98 indicates high accuracy and consistency between predicted and actual values. Fig. 2C shows the feature importance of structural descriptors. Each descriptor has either positive or negative importance. Positive importance contributes to the increase in the WCA while negative ones lower the WCA. Structural descriptors such as O–H bond and %N play important roles in lowering WCA. This is consistent with the general understanding that surfaces with high polarity show small WCAs. In contrast, %H and %C, which represent the nonpolar methyl terminal group and alkyl chain show positive importance. The lower part of Fig. 2c shows the quantitative importance of Whiteside's rules, which represents an empirical rule of protein resistance. In essence, by training ANN with 145 SAMs data, the algorithm can predict both WCA and protein adsorption accurately. In addition, the analysis provided by the trained ANN can reveal the quantitative importance of each structural parameter, providing an important guide for material design. The degree of importance also agrees well with a general perception of the physicochemical properties of SAMs.
Fig. 2.
(A) ANN model was used to evaluate and calculate the water contact angle and fibrinogen adsorption on the surfaces of self-assembled monolayers (SAM). The red and blue arrows illustrate positive and negative weights after the training, respectively. (B1-2) Amounts of adsorbed fibrinogen onto SAMs predicted by the trained ANN plotted from single-lab data against corresponding experimental values. (C1-2) Results of analytical importance following the one-laboratory data set training. Standard deviation (N = 2000) is the standard error bar. (d) Hypothetical SAMs adsorbed predicted quantity of fibrinogen. The amount of methylene units in their alkyl chain has been changed, preserving the same terminal groups, in (c) OH-, (d) CH3-, (e) NH2- and (f) COOH-terminated SAMs. Adapted from the literature [92].
4. Metallic biomaterials applications
4.1. Metallic biomaterials
Metallic biomaterials show typical features such as better thermal, electrical, and mechanical capabilities, inertness in the body and ability to perform structural functions. Stainless steel is generally the most frequently used implant as it is easy to handle and sterilize by conventional sterilization processes for an excellent finish [93]. Cobalt chromium alloy is used as a wear resilient implant, for example in prosthesis and implantable [26]. Metallic biodegradables have also demonstrated the possibility of orthopaedic implantables and cardiovascular stents [94]. One of the main issues is to develop metals with comparable mechanical properties to hard tissue [95]. Alloy doping is often used to change the characteristics of metals, but the wide range of potential alloy compositions make designing metals with desired attributes a challenging problem.
4.2. Metallic biomaterials
One of the essential variables in understanding the service lives of implants in the human organism is their fatigue qualities for metallic biomaterials. For the prediction of the fatigue properties of nickel and aluminum alloys, Fujii et al. [96] and Kang et al. [97] reported the use of an artificial neural network (ANN). Iacoviello et al. described ANN application in the prediction of fatigue propagation in sintered duplex steel [98]. Mohanty et al. developed a mixed Gaussian method for enhanced prediction of fracture trends and development [99]. The ML approach for detecting fatigue cracks and forecasting fatigue creep rates has been developed by Wang et al. [100], Dinda et al. [101], and Rovinelli et al. [51] The approach for prediction of fatigue strength using the ANN method has also been published by Agrawal et al. [[102], [103], [104]] and Canyurt et al. [105] These studies offer an effective strategy to predict crack initation and growth rates of long-term metallic implants so that implant conditions in the human body are monitored in a timely manner.
In several metal biomaterials, stress corrosion crack (SCC) has been a problem that has reduced the long-term lifetime of metal implants. Cao et al. have reported an optimal SCC resistant alloy composition optimization technique with ML [52]. The authors employ the interactive feedback circuit as a representative of SCC resistance with electric conductivity. The properties of the new Al alloy (Al-6.05Zn-1.46Mg1·32Cu-0.13Zr-0.02Ti-0.50Y-0.23Ce) predicted using the efficient global (EGO) method showed better SCC resistance than the standard 7N01 alloy. The interactive feedback training loop indicates possible applicability for finding metallic biomaterials with enhanced resistance.
It is vital to have low modulus materials similar to human bone for the usage of metallic materials as bone implants to minimize stiffness mismatch and stress shielding. Wu et al. employed ANNs to search for novel low modulus titanium alloys. For Young's modulus and Martensitic temperature, the authors have employed two distinct data sets containing 164 and 112 compositions respectively. By training ANNs on these very small data sets. a new alloy Ti–12Nb–12Zn–12Sn (in wt. Percent) was discovered and validated with a low modulus of 41GPa and with low cost metals [106]. This work reveals that ML approaches can be used to design low-modulus metal biomaterials and to search successfully new compositions of titanium alloys.
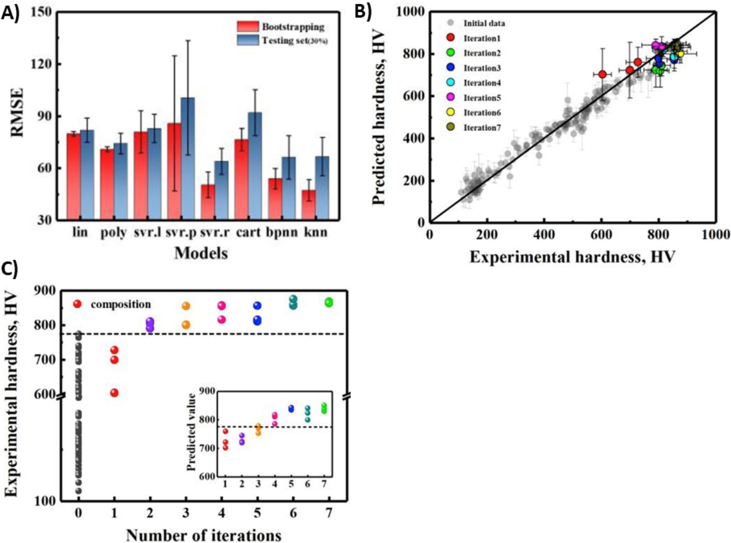
High hardness alloys often have a strong wear resistance to extended service life that is crucial for hip, joint and knee implants. High entropy alloys (HEA) are composed of several elements combined at close to equimolar ratios without any primary element, therefore a larger number of possible compositions exist. Cheng et al. [107] used an ML surrogate model to map between composition and hardness and used the design of the experiment and the EGO method to iteratively search for alloys with higher hardness. The authors were able to predict and validate a new HEA of Al–Co–Cr–Cu–Fe–Ni system with 10% higher hardness than the original training dataset (see Fig. 3). Chang et al. have demonstrated that ANNs are also used to study highly challenging HEA formulas, where the stimulated anneal algorithm is added [108]. Taken together, it shows that ML algorithms might improve the screening with the improved wear resistance of hard metallic biomaterials.
Fig. 3.
(A) Model evaluation and selection by estimating the test error for different models. A bootstrapping method was used, indicating that svr. r (radial basis function kernel) ML model has low test error and outperforms the others. (B) The results of the design of the experiment and EGO mediated iterative loops. The predicted hardness values were plotted versus the measured values for the alloys in the training data and experimental data. (C) The hardness of the newly synthesized alloys as a function of loop iteration number. The inset of (b) plots the predicted values as a function of iteration number, showing a similar tendency to the measured values. Adapted from the literature [107].
5. Ceramics biomaterials applications
5.1. Ceramic biomaterials
For so many years, ceramic materials were used to relieve pain and restore function of calcified tissue (bones and teeth) via the use of ceramic materials based on natural calcium phosphate [109]. Ceramic materials (HA and TCP) are frequently employed for artificial joints or tooth implants because of stable physicochemical features, good biocompatibility and being osteoconductive. Significant progress has been made in dental and medical applications of ceramics such as bioglass to rebuild the bone [110], using alumina for hip-endoprostheses femoral balls [111], and some pioneering efforts in the effective use of synthetic HA and β-TCP for dentistry and medical uses as bone substitute material [[112], [113], [114], [115], [116]]. Bioceramics, such as alumina and zirconia, can be bioinert, retaining their physical and mechanical qualities for long durations. The bone formation can also be supported actively by bioceramics such as bioglasses, orthophosphate calcium, and hydroxyapatite. Several synthetic bioceramic materials have been created in a variety of shapes and forms as alternate options to autogenous bone for restoration, replacement, and augmentation.
5.2. Ceramic biomaterials examples
Ceramics with high strength and hardness can reduce wear during its use. High entropy ceramics are composed of several metal cation species combined at close to equimolar ratios, and can potentially form high hardness materials and corrosion-resistant coatings [117]. However, the main hindrance of their discovery lies in predicting their formation. In work by Kauffman et al. machine learning methods leveraging on thermodynamics and compositional attributes are used to predict the synthesizability of disordered metal carbides. The feature importance of thermodynamics and compositional parameters are then explored and explained. As a result, up to 70 new compositions are predicted and validated by density functional theory calculations and experimental synthesis [118].
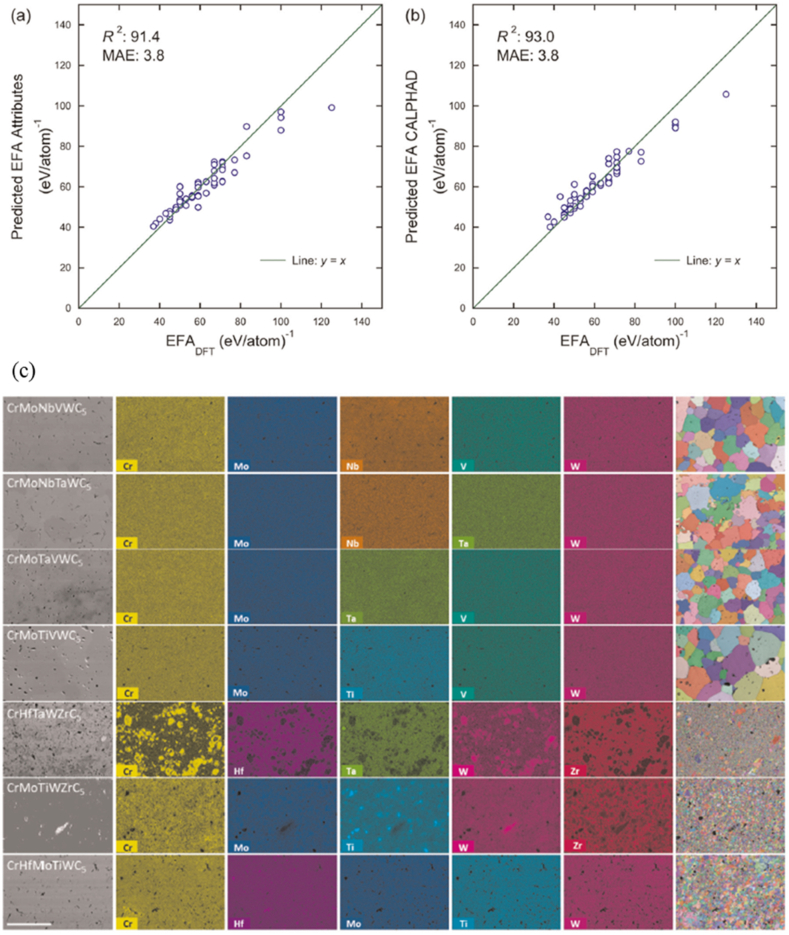
Random fittings were conducted for 56 previously known EFA (entropy forming capability) values to seek new high entropy ceramics. The datasets contain nine previously synthesized compositions, six of which are one phase. Only 8 carbide metal elements (Hf, Nb, Ta, Ti, Mo, V, W, and Zr) were used in earlier research [119]. The ML model performance was evaluated with five-fold cross-validation that may advise the optimal model hyperparameters. The performance of the ML model is compared in Fig. 4a with CALPHAD information and chemical characteristics only. While the MAEs are equal in all models (3.8 eV/atom), the determination coefficient (R2) predicts better results when using CALPHAD data. Fig. 4c provides a microstructural examination utilizing EDS to assess the homogeneity of the sintered pellets. The findings of both procedures revealed that the samples as treated were either single-phase, as well as chemically homogenous or chemically separated. This discovery demonstrates that the compositions are one phase. In contrast, because of the competing phases prohibiting further grain expansion during sintering, multi-phase samples have much lower grain sizes.
Fig. 4.
The ML models are assessed to match the data supplied. a) The ML-predicted EFA, using random forest fit b) ML forest random fit projected EFA values with 108 chemical characteristics and eight CALPHAD-evaluated features against the known DFT EFA. c) Synthesized material microstructural analysis. Bar scale 100 μm [119].
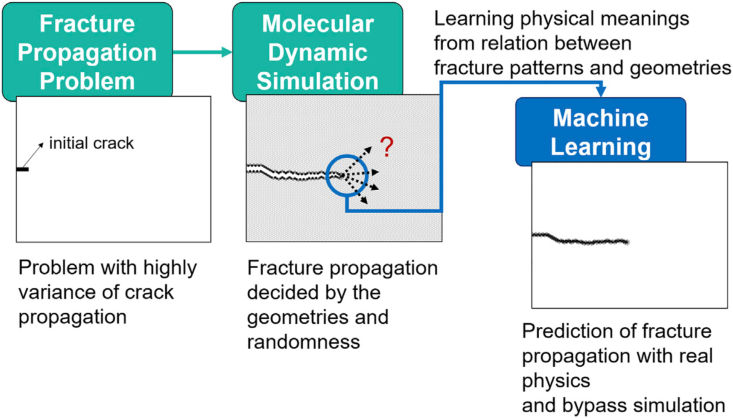
The complexity of fracture mechanics predictions in traditional molecular dynamics simulations with the high computational effort needed makes it difficult to provide results for materials design at the nanoscale. A team led by Buehler developed a quick screening system to measure fracture resistance of potential materials by examining the propagation of cracks through the molecular structure of the material to assess the processes in which various material systems fail, including composite, coated layers from crystalline structures (see Fig. 5) [118]. These huge quantities of data have now been included in the AI system, the fundamental physical principles of failure have been established and this can assist in the prediction of the performance of a new material/structure that is not in the training set.
Fig. 5.
De novo approach to study the fracture problem with the crack patterns and crack length, based on a training set derived from molecular dynamics (MD) simulations. It can be used to replace the stochastic problem of fracture propagation in MD simulations through a machine-learning model that can predict the overall crack propagation pathways [120]. This obviates the need for time-consuming MD simulation, opening novel avenues for materials design solutions with atomistic-level degrees of freedom.
6. Nanoengineered biomaterials applications
6.1. Biomaterials from nanoparticles
Most bulk biomaterials allow for a high mechanical strength when used in cardiovascular and orthopaedic applications. Nanoparticles have been used in diverse biomedical applications including drug release, regenerative medicine, and tissue engineering. Nanoparticles possess their own unique characteristics, and nanoparticle-bulk material composites can resolve issues of insufficient mechanical strength for biological applications with high mechanical loading.
6.2. Nano-biomaterials
Nanomaterials have a high degree of complexity, which in addition to bulk properties such as composition and defects, can include other input parameters such as nanoparticle shape, size and surface properties which will have a more significant impact at the nanoscale. This large range of nanomaterial properties can be generated by varying the reaction conditions such as reaction temperature and time, and experimental parameters such as type of ligands and ratio of organic: aqueous solvents. Highly efficient flow chemistry has been combined with machine learning for combinatorial synthesis of quantum dots, and allowed the development of precise sizes and dispersity of quantum dots with suitable composition and bandgap for optoelectronic applications [121]. With a diverse selection of nanoparticles created, it is then possible to probe for biological functionalities with the assistance of machine learning. Metal nanoparticles were investigated to determine and predict the antibacterial properties, with nanoparticle core size being the key physicochemical feature [122]. Gold nanoparticles were also monitored for their fate in vivo by the combination of the tools of mass spectroscopy and supervised machine learning [59].
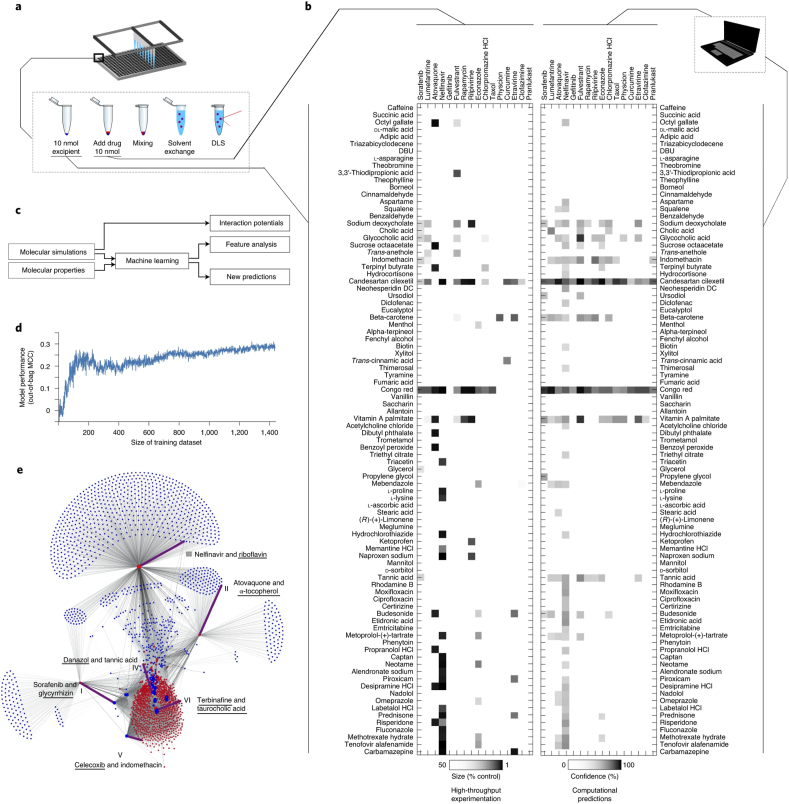
Machine learning can identify key features for improving performance, and subsequently allow for highly predictive synthesis of nanoparticles. In comparison to regular nanoparticle drug delivery systems which typically show low drug loading, self-assembling drug nanoparticles can have up to 95% high drug loading. Reker et al. rapidly screened through 1440 pairings of drugs and excipients for co-aggregation into nanoparticles by using a combination of liquid handling and high throughput dynamic light scattering characterization (Fig. 6) [53]. A random forest machine learning model was used to gain insight into the molecular features for co-aggregation, and subsequently used to predict additional co-aggregators (1.8%) from 2.1 million possible pairings of FDA approved drugs and excipients.
Fig. 6.
(a) Schematic of high throughput screening to create self-assembling drug nanoparticles. (b) High-throughput testing of all 1440 combinations of 16 drugs and 90 excipients (inactive ingredients, generally recognized-as-safe food and drug additives and other FDA-approved compounds). (c) Molecular properties of drug-excipient pairs are encoded, and molecular simulations (molecular dynamics) determines interaction potentials. These are fed into a random forest machine learning model to indicate important features and predict potential to co-aggregate and form nanoparticles. (d) Out-of-bag performance analysis on different training dataset sizes with Matthews correlation coefficient showing convergence. (e) The machine learning model was used to model 2.1 million pairs of drugs and excipients for ability to co-aggregate and form nanoparticles, and the six named pairs were validated experimentally. The novel component that was not part of the initial high throughput screen in part A are underscored [53].
The pharmacokinetics and eventual fate in the body are greatly affected by the biological components that interact with the nanoparticles. For anti-fouling PEGylated systems, the impetus is to prevent non-specific protein binding. Nanoparticle surfaces have been functionalized with a large library of ligands such as zwitterionic, hydroxyl acrylate and peptides to study the influence on preventing protein adsorption. Yarovsky et al. addressed this knowledge gap by using machine learning approaches to extract the quantitative relationship between material surface chemistry and the protein adsorption characteristics [56].
Using gold-labelled antibody nanoparticles, Hu et al. examined the surface mapping of protein bindings on particulate corona [23]. Functional composition, cellular recognition (e.g., cellular absorption by macrophages, releases of cytokines and immunological response), and nanotoxicity through the integration of machine learning and meta-analysis were forecast. Groups of cell membrane-coated nanoparticles have been employed as a platform for protein detection at the nanomaterial interface [123]. González-Díaz and colleagues looked to predictive design of nanoparticle drug delivery system by exploring changes in the nanoparticle, coating agent and drug [124]. They utilized a perturbation theory and machine learning model to develop a model for predicting the best components. The authors used pre-clinical drug test data sets and coated metal oxide nanoparticles from ChEMBL and public sources. Both data sets were fused to gain a final dataset of about 500,000 nanoparticles, which would be extremely valuable for further design of coated nanoparticles.
7. 3D printed biomaterials applications
7.1. 3D printed biomaterials
Additive manufacturing is an exciting emerging field due to the ease of creating complicated 3D geometric structures using computer aided design (CAD).
Different materials and techniques possess their own unique challenges to overcome (see Table 3). For example, laser powder bed fusion is commonly used for metals, but this will typically generate 3D prints with surface roughness, and metal leaching can also be a cause of inflammatory response and cell toxicity. Using extrusion or ink jet printing techniques will require additional post-sintering [125,126]. For ceramics, the process of 3D printing has an added challenge of intrinsic shrinkage issues during the sintering process, and the need to accurately generate complex hierarchical structures with porosity to allow for osteointegration [127]. High quality printing of polymers is mainly limited by material choice. Fused filament deposition can only be used for polymers that can melt without degrading, while stereolithography requires photopolymers [29]. Machine learning can affect 3D printing by facilitating design and materials selection and by driving and monitoring for process optimization.
Table 3.
Common additive manufacturing techniques for various materials.
| Type of material | Typical 3D printing techniques | Advantages | Challenges |
|---|---|---|---|
| Metal |
|
|
|
| Ceramic |
|
|
|
| Polymer |
|
|
|
7.2. Machine learning in 3D printed biomaterials
7.2.1. Materials and design screening
3D convoluted neural networks (CNN) has been performed which can effectively model the elasticity tensor and its gradients to predictions of microscale structure, and hence the resultant material properties [128]. The relationship between 3D fibrous substrates with cell confinement states have also been used with machine learning methods to classify cell shapes and map to the original substrate architectures [129].
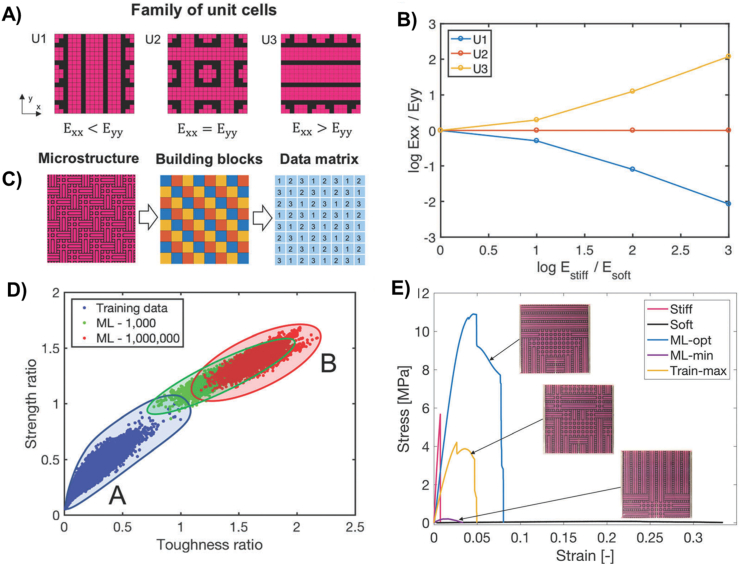
Gu et al. explored 3D printing of composite materials with microstructures composed of stiff and soft building blocks [63]. The authors defined 3 basic unit cells using patterns of stiff and soft building blocks, and generated 100,000 microstructures by combinations of unit cells which could be encoded into a data matrix (Fig. 7). By using CNNs where mechanical properties were calculated using a finite element method (FEM) to form the base truth, the machine learning model could impressively generate much stronger and tougher materials as compared to initial training set. The predicted composite materials were 3D printed and underwent stress-strain testing and were shown to be roughly 25 times tougher than the original stiff building block. While directly using FEM simulations for all the microstructures would take 5 days, the CNN required only 10 hours for training and under 1 min to output the machine learning optimized composite. This provides a promising new paradigm of smart additive manufacturing of better materials, driven by ML optimization of hierarchical building blocks.
Fig. 7.
(A) Stiff and soft building blocks are combined in patterns to form unit cells U1, U2 and U3. Pink refers to stiff building blocks, while black refers to soft building blocks. (B) The ratio between the modulus of stiff and soft blocks affects unit cell isotropy. (C) The microstructure of stiff and soft blocks is converted into unit cells U1 (blue), U2 (red) and U3 (yellow), which can be further converted into a data matrix with inputs of 1, 2, 3 corresponding to the type of unit cell. (D) Strength ratio is the strength of machine learning generated samples after 1000 loops (green), and after 1,000,000 loops (red), as compared to the highest training set strength. Toughness ratio is the toughness as compared to the highest training set toughness. (E) Stress-strain curves of machine learning generated 3D printed sample (ML-optimal and ML-minimum) [63].
Khadilkar et al. used machine learning for optimization as compared to FEM simulations [130]. 16,700 stereolithography 3D printed models underwent FEM simulations to estimate stress distributions, and this acted as the training set for the machine learning model. It was found that the two-stream CNN ML model performed better than single-stream CNN and artificial neural networks (ANN). ANN was trained on a lattice cell mechanical model, and was used to model the elastoplastic properties and predict the maximum Von Mises and stresses on the lattice cell [131]. While FEM simulations were conducted over 5–10 hours, the ANN approach took under a minute.
Marrone and colleagues investigated the Tg of polyhydroxyalkanoate homopolymers and copolymers [132]. The authors converted key polymer properties such as charge/polarity into a numerical matrix, and used a statistical machine learning model trained on experimentally determined polymer Tg, molecular weights and polydispersity. Their model identifies chemical features affecting Tg, and accurately predicts Tg of new polymers. The pharmaceutical software M3DISEEN was used to design 3D printable drug loaded formulations from a comprehensive list of 145 excipients [54]. The software could predict the printability of the drug-excipient formulation with an accuracy of 76% and the filament properties with an accuracy of 67%, and furthermore could suggest approximate processing temperatures for hot melt extrusion and the printing process. This can greatly facilitate the customizable 3D printing of pharmaceutical drugs.
The 3D printing design process using CAD is iterative and time-consuming, driving the need for data-driven design. Maiden et al. showed that by employing machine learning, designers could be recommended to use existing CAD models most similar to what they are intending to design, thus speeding up the design process [133]. There is also errors in translating from CAD to 3D printed materials due to materials shrinkage or warping. The PrintFixer system was developed which uses data from previous 3D printing jobs to train its convolution based machine learning model to predict shape distortions, thus improving 3D printing accuracy by up to over 50% [134]. PrintFixer can work with a variety of materials and applications – from metals to thermal plastics and aerospace engineering. Machine learning can also assist in predicting the functional properties of 3D printed implants. CNN could be used to predict whether 3D printed CAD/CAM crowns would debond from stereolithography models [135]. CAD/CAM composite resin crowns were split into 2 categories: trouble-free and debonding (n = 12) for training the CNN model for predicting probability of debonding. The predicted result was impressively 98.5% accurate and 97.0% precise, showing that CNN could be used in practical dental applications.
7.2.2. 3D printing process optimization
Using machine learning, optimization of AM processes can be performed in a short time with best performance. Machine learning algorithms have been used to determine the process-structure-properties (PSP) relationship for many 3D printing processes, and ANN is the most commonly used technique for process optimization [136,137]. Various studies have compared machine learning algorithms with conventional optimization methods such as Taguchi method [138], polynomial regression model [139], ANOVA [140]. ANN was found to achieve a lower mean of errors registering 1.922% and 2.104% as compared to a second-order regression model with a mean of errors of 2.633% and 2.308% for bead width and bead height predictions. Machine learning has a high potential to successfully discover complex PSP relationships, overcoming many of the limitations associated with the techniques such as finite element modelling (FEM) methods. It focuses on understanding either process response or performance response, by either using data-driven approach, or combining both physics-based and data-driven approaches [141].
To optimize the process for 3D printing, there are several process parameters which will affect the printing quality. Zhang et al. used a Latin hypercube sampling method to fully explore the process parameter design space for aerosol jet printing [142]. The 2 most important parameters of sheath gas flow rate and carrier gas flow rate were studied in more detail by K-means clustering approach and a support vector machine. To allow determining of the operating window under different print speeds, transfer learning was adopted to speed up the process. The full 3D operating window is then created using incremental classification. By using a hybrid machine learning approach, it is possible to systematically improve the printing process as opposed to traditional experimental approaches. Rankouhi et al. studied how to predict part density and surface roughness when process parameters for selective laser melting were varied [55]. The authors used a multivariate Gaussian approach to transfer knowledge from copper and the 316L alloy individually to predicting properties of the composite material. High throughput measurement of part density and surface roughness were used to train the dataset, and process parameter maps were generated. These process maps allow for prediction of process-property in a compositional gradient zone of a multi-material printed part. As a model for 3D bioprinting, Bone et al. studied the process parameters for 3D printing of soft alginate hydrogels [143]. The authors experimentally established an initial dataset of 48 high and low fidelity alginate prints by varying print parameters, which were scored for similarity of the prints to the CAD designs. Hierarchical machine learning was used to identify dominant variables and predict process parameters that would generate high fidelity prints.
While additive manufacturing has many advantages, there is the issue of print quality of 3D printed objects. Wu and colleagues used an ensemble learning algorithm to predict surface roughness [144]. They incorporated 5 sensors to monitoring the printing conditions and used a profilometer to measure surface roughness. A random forest approach was used to determine feature importance and fed into the machine learning model. It was found that the build plate frequency amplitude, extruder vibration and build plate temperatures were the top 3 most important features, and the ensemble learning algorithm was found to outperform the individual learning algorithms with a high accuracy of prediction. The 3D printed product quality could also be monitored by laser-based process control system. A CNN machine learning model could classify the in-plane anomalies accurately, and can both reduce the height variation error and correct anomalies as they happen [145]. This is important for improving the additive manufacturing process, especially as it is upscaled for reproducible manufacturing in the future.
8. Conclusion and perspectives
Machine learning has advanced in the field of biomaterials, with more accessible high-throughput experimental equipment that generate training data, better data-mining from literature throughout the years and increasing understanding of how to apply suitable algorithms. Machine learning is used in several facets of biomaterial development: 1) as part of the initial screen to find new hydrogel forming materials if certain features are present in the material; 2) as part of optimizing the material's inner properties, such as enhancing the mechanical properties; (3) the determination and optimization of significant process parameters; and (4) the specialized investigation of material interactions with biological systems, such as anti-fouling and biologicalfate. Biomaterials covers a range of materials with starkly different properties, but there has been application of machine learning techniques to the field. An extensive training dataset with a large sample size is very useful to employ machine learning techniques with higher predictive power, and this is currently often obtained by the authors performing a set of high throughput experimentation. In cases of lower sample size, the parameter space can be explored by using techniques such as Latin hypercube samples, and by using Bayesian inference models which incorporate prior knowledge. Machine learning can greatly accelerate the innovation or optimization process by allowing the extrapolation of insights gained from thousands of data points to predict hits in much larger databases. There is the reduction in the number of experiments required to achieve optimal formulations, and a reduction in computational cost when simulations such as finite element analysis do not need to be extensively performed. In polymeric biomaterials, machine learning can screen for new materials for hydrogel formation and polymer moieties to mitigate the foreign body response. In metallic materials, high-entropy alloys (HEA) have benefited from machine learning applications for multi-parametric optimization. For ceramics materials, beyond prediction of material failure, there is also screening for antimicrobial properties in bioactive glass. In the space of nanomaterials development, modelling results from screening for co-aggregation can allow for extrapolation to a huge FDA approved drug and excipient database. Machine learning can also be used not only to improve 3D printing processes, but also in concert with 3D printing for prediction and validation of composite materials.
In various examples shown, machine learning was proven to facilitate and speed up the creation of biomaterials. Machine learning was nonetheless only carried out using a tiny subset of research on biomaterials and used as a technique to handle certain biomaterial issues. Biomaterials experiments and data collecting in different laboratories and nations are not standardised. In many situations the metadata or annotations for the data collection are not given by the original researchers, in addition to experimental details, making it nearly impossible to manipulate data for machine learning. As an appropriate training data set with proper annotations is often not available, experimental researchers either need to perform a set of high throughput experiments for the specific biomaterial problem or they are faced with a small sample size. Machine learning models are also encountering challenges as they are trained on an initial data set and expect to predict for improved performance in other settings. Recently, there has been the realization that under specification in machine learning models can lead to poor model performance when used for real life applications and needs to be accounted for [146]. While authors often mention the potential of their advances, it is often not clear whether the machine learning models presented is useful beyond the specific setting or can be extrapolated to a larger database.
We must make a couple of adjustments in terms of thinking and technological improvements to include machine learning into biomaterial research. First and foremost, data sharing is needed for larger datasets and for easier analysis by machine learning [147]. A uniform common standard must be developed for input characterization data, output testing assays, and metadata to guarantee that the obtained data are similar and generalizable across each institution. A more viable solution is to initiate multi-group collaborations that standardizes protocols and annotations to create generalisable data. The human genome project, where multiple groups worldwide have been able to efficiently combine capacities and exchange knowledge to fulfil the taxing task, is a viable previous reference point [148]. Next, the community should provide an open access repository for data, and reward authors if others use their submitted data. Computational groups can perform secondary analysis by bringing together multiple datasets, and credit the original authors accordingly. Finally, the collaboration between computer scientists and experimental biomaterials scientists needs to start early and a close working relationship fostered to generate datasets accessible to machine learning. In the case of an incomplete and poorly annotated dataset, there is not much useful insight that can be gained. With all these efforts, we believe that machine learning can be firmly integrated into the biomaterials development process for an incredibly fruitful era of biomaterials innovation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
K.X, F.W, A.S, M.H, P.T, P.W, W.S, contributed equally to the manuscript. Correspondence to E. Y, Z. L, and X.J.L. This work is financially supported by A∗STAR (Agency for Science, Technology and Research, Singapore).
Contributor Information
Enyi Ye, Email: yeey@imre.a-star.edu.sg.
Zibiao Li, Email: lizb@imre.a-star.edu.sg.
Xian Jun Loh, Email: lohxj@imre.a-star.edu.sg.
References
- 1.Thaburet J.F., Mizomoto H., Bradley M. High-throughput evaluation of the wettability of polymer libraries. Macromol. Rapid Commun. 2004;25(1):366–370. [Google Scholar]
- 2.Hook A.L., Chang C.Y., Yang J., Atkinson S., Langer R., Anderson D.G., Davies M.C., Williams P., Alexander M.R. Discovery of novel materials with broad resistance to bacterial attachment using combinatorial polymer microarrays. Adv. Mater. 2013;25(18):2542–2547. doi: 10.1002/adma.201204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hook A.L., Chang C.-Y., Yang J., Luckett J., Cockayne A., Atkinson S., Mei Y., Bayston R., Irvine D.J., Langer R. Combinatorial discovery of polymers resistant to bacterial attachment. Nat. Biotechnol. 2012;30(9):868–875. doi: 10.1038/nbt.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaim C.J., Chien S., Bhatia S.N. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 5.Unadkat H.V., Hulsman M., Cornelissen K., Papenburg B.J., Truckenmüller R.K., Carpenter A.E., Wessling M., Post G.F., Uetz M., Reinders M.J. An algorithm-based topographical biomaterials library to instruct cell fate. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108(40):16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Won Y.-W., Patel A.N., Bull D.A. Cell surface engineering to enhance mesenchymal stem cell migration toward an SDF-1 gradient. Biomaterials. 2014;35(21):5627–5635. doi: 10.1016/j.biomaterials.2014.03.070. [DOI] [PubMed] [Google Scholar]
- 7.Soen Y., Mori A., Palmer T.D., Brown P.O. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol. Syst. Biol. 2006;2(1):37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171(6):1437–1452. doi: 10.1016/j.cell.2017.10.049. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtarolo S., Setyawan W., Wang S., Xue J., Yang K., Taylor R.H., Nelson L.J., Hart G.L., Sanvito S., Buongiorno-Nardelli M., AFLOWLIB. ORG A distributed materials properties repository from high-throughput ab initio calculations. Comput. Mater. Sci. 2012;58:227–235. [Google Scholar]
- 10.Belsky A., Hellenbrandt M., Karen V.L., Luksch P. New developments in the Inorganic Crystal Structure Database (ICSD): accessibility in support of materials research and design. Acta Crystallogr. Sect. B Struct. Sci. 2002;58(3):364–369. doi: 10.1107/s0108768102006948. [DOI] [PubMed] [Google Scholar]
- 11.Saal J.E., Kirklin S., Aykol M., Meredig B., Wolverton C. Materials design and discovery with high-throughput density functional theory: the open quantum materials database (OQMD) JOM (J. Occup. Med.) 2013;65(11):1501–1509. [Google Scholar]
- 12.Jain A., Ong S.P., Hautier G., Chen W., Richards W.D., Dacek S., Cholia S., Gunter D., Skinner D., Ceder G. Commentary: the Materials Project: a materials genome approach to accelerating materials innovation. Apl. Mater. 2013;1(1) [Google Scholar]
- 13.Hutchinson M. Citrine Inf. Lolo. 2016 [Google Scholar]
- 14.Ward L., Dunn A., Faghaninia A., Zimmermann N.E., Bajaj S., Wang Q., Montoya J., Chen J., Bystrom K., Dylla M. Matminer: an open source toolkit for materials data mining. Comput. Mater. Sci. 2018;152:60–69. [Google Scholar]
- 15.Brandt R.E., Kurchin R.C., Steinmann V., Kitchaev D., Roat C., Levcenco S., Ceder G., Unold T., Buonassisi T. Rapid photovoltaic device characterization through Bayesian parameter estimation. Joule. 2017;1(4):843–856. [Google Scholar]
- 16.Somnath S., Law K.J., Morozovska A., Maksymovych P., Kim Y., Lu X., Alexe M., Archibald R., Kalinin S.V., Jesse S. Ultrafast current imaging by Bayesian inversion. Nat. Commun. 2018;9(1):513. doi: 10.1038/s41467-017-02455-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furmanchuk A.o., Saal J.E., Doak J.W., Olson G.B., Choudhary A., Agrawal A. Prediction of seebeck coefficient for compounds without restriction to fixed stoichiometry: a machine learning approach. J. Comput. Chem. 2018;39(4):191–202. doi: 10.1002/jcc.25067. [DOI] [PubMed] [Google Scholar]
- 18.Gorai P., Stevanović V., Toberer E.S. Computationally guided discovery of thermoelectric materials. Nat. Rev. Mater. 2017;2(9):17053. [Google Scholar]
- 19.Samsonidze G., Kozinsky B. Accelerated screening of thermoelectric materials by first-principles computations of electron–phonon scattering. Adv. Energy Mater. 2018:1800246. [Google Scholar]
- 20.Yamawaki M., Ohnishi M., Ju S., Shiomi J. Multifunctional structural design of graphene thermoelectrics by Bayesian optimization. Sci. Adv. 2018;4(6) doi: 10.1126/sciadv.aar4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Recatala-Gomez J., Suwardi A., Nandhakumar I., Abutaha A., Hippalgaonkar K. Toward accelerated thermoelectric materials and process discovery. ACS Appl. Energy Mater. 2020;3(3):2240–2257. [Google Scholar]
- 22.Suwardi A., Bash D., Ng H.K., Gomez J.R., Repaka D.M., Kumar P., Hippalgaonkar K. Inertial effective mass as an effective descriptor for thermoelectrics via data-driven evaluation. J. Mater. Chem. 2019;7(41):23762–23769. [Google Scholar]
- 23.Ban Z., Yuan P., Yu F., Peng T., Zhou Q., Hu X. Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(19):10492–10499. doi: 10.1073/pnas.1919755117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratner B.D. Biomaterials: been there, done that, and evolving into the future. Annu. Rev. Biomed. Eng. 2019;21(1):171–191. doi: 10.1146/annurev-bioeng-062117-120940. [DOI] [PubMed] [Google Scholar]
- 25.Li J., Mooney D.J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 2016;1:16071. doi: 10.1038/natrevmats.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermawan H., Ramdan D., Djuansjah J.R. Biomed. Eng. Theory Appl.; 2011. Metals for Biomedical Applications; pp. 411–430. [Google Scholar]
- 27.Koons G.L., Diba M., Mikos A.G. Materials design for bone-tissue engineering. Nat. Rev. Mater. 2020;5(8):584–603. [Google Scholar]
- 28.Zhang T., Hong Z.-Y., Tang S.-Y., Li W., Inglis D.W., Hosokawa Y., Yalikun Y., Li M. Focusing of sub-micrometer particles in microfluidic devices. Lab Chip. 2020;20(1):35–53. doi: 10.1039/c9lc00785g. [DOI] [PubMed] [Google Scholar]
- 29.González-Henríquez C.M., Sarabia-Vallejos M.A., Rodriguez-Hernandez J. Polymers for additive manufacturing and 4D-printing: materials, methodologies, and biomedical applications. Prog. Polym. Sci. 2019;94:57–116. [Google Scholar]
- 30.Ulbrich K., Holá K., Šubr V., Bakandritsos A., Tuček J., Zbořil R. Targeted drug delivery with polymers and magnetic nanoparticles: covalent and noncovalent approaches, release control, and clinical studies. Chem. Rev. 2016;116(9):5338–5431. doi: 10.1021/acs.chemrev.5b00589. [DOI] [PubMed] [Google Scholar]
- 31.Rosenblum D., Joshi N., Tao W., Karp J.M., Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018;9(1):1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loh X.J. Poly (DMAEMA-co-PPGMA): dual-responsive “reversible” micelles. J. Appl. Polym. Sci. 2013;127(2):992–1000. [Google Scholar]
- 33.Loh X.J., Ong S.J., Tung Y.T., Choo H.T. Dual responsive micelles based on poly [(R)-3-hydroxybutyrate] and poly (2-(di-methylamino) ethyl methacrylate) for effective doxorubicin delivery. Polym. Chem. 2013;4(8):2564–2574. [Google Scholar]
- 34.Loh X.J., Tsai M.-H., del Barrio J., Appel E.A., Lee T.-C., Scherman O.A. Triggered insulin release studies of triply responsive supramolecular micelles. Polym. Chem. 2012;3(11):3180–3188. [Google Scholar]
- 35.Loh X.J., Wu Y.-L. Cationic star copolymers based on β-cyclodextrins for efficient gene delivery to mouse embryonic stem cell colonies. Chem. Commun. 2015;51(54):10815–10818. doi: 10.1039/c5cc03686k. [DOI] [PubMed] [Google Scholar]
- 36.Li Z., Chee P.L., Owh C., Lakshminarayanan R., Loh X.J. Safe and efficient membrane permeabilizing polymers based on PLLA for antibacterial applications. RSC Adv. 2016;6(34):28947–28955. [Google Scholar]
- 37.Loh X.J., Wu Y.-L., Seow W.T.J., Norimzan M.N.I., Zhang Z.-X., Xu F.-J., Kang E.-T., Neoh K.-G., Li J. Micellization and phase transition behavior of thermosensitive poly (N-isopropylacrylamide)–poly (ϵ-caprolactone)–poly (N-isopropylacrylamide) triblock copolymers. Polymer. 2008;49(23):5084–5094. [Google Scholar]
- 38.Loh X.J., Zhang Z.-X., Mya K.Y., Wu Y.-l., He C.B., Li J. Efficient gene delivery with paclitaxel-loaded DNA-hybrid polyplexes based on cationic polyhedral oligomeric silsesquioxanes. J. Mater. Chem. 2010;20(47):10634–10642. [Google Scholar]
- 39.Rauwald U., del Barrio J., Loh X.J., Scherman O.A. “On-demand” control of thermoresponsive properties of poly (N-isopropylacrylamide) with cucurbit [8] uril host–guest complexes. Chem. Commun. 2011;47(21):6000–6002. doi: 10.1039/c1cc11214g. [DOI] [PubMed] [Google Scholar]
- 40.Su X., Tan M.J., Li Z., Wong M., Rajamani L., Lingam G., Loh X.J. Recent progress in using biomaterials as vitreous substitutes. Biomacromolecules. 2015;16(10):3093–3102. doi: 10.1021/acs.biomac.5b01091. [DOI] [PubMed] [Google Scholar]
- 41.Yang D.P., Oo M.N.N.L., Deen G.R., Li Z., Loh X.J. Nano-star-shaped polymers for drug delivery applications. Macromol. Rapid Commun. 2017;38(21):1700410. doi: 10.1002/marc.201700410. [DOI] [PubMed] [Google Scholar]
- 42.Kratochvil M.J., Seymour A.J., Li T.L., Paşca S.P., Kuo C.J., Heilshorn S.C. Engineered materials for organoid systems. Nat. Rev. Mater. 2019;4(9):606–622. doi: 10.1038/s41578-019-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., Zheng S., Ye K., He J., Shen Y., Cui S., Huang J., Gu Y., Ding J. Cell migration regulated by RGD nanospacing and enhanced under moderate cell adhesion on biomaterials. Biomaterials. 2020;263:120327. doi: 10.1016/j.biomaterials.2020.120327. [DOI] [PubMed] [Google Scholar]
- 44.Mitragotri S., Lahann J. Physical approaches to biomaterial design. Nat. Mater. 2009;8(1):15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai Y., Xu C., Sun X., Chen X. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chem. Soc. Rev. 2017;46(12):3830–3852. doi: 10.1039/c6cs00592f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arvanitis C.D., Ferraro G.B., Jain R.K. The blood–brain barrier and blood–tumour barrier in brain tumours and metastases. Nat. Rev. Cancer. 2020;20(1):26–41. doi: 10.1038/s41568-019-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veiseh O., Doloff J.C., Ma M., Vegas A.J., Tam H.H., Bader Andrew R., Li J., Langan E., Wyckoff J., Loo W.S., Jhunjhunwala S., Chiu A., Siebert S., Tang K., Hollister-Lock J., Aresta-Dasilva S., Bochenek M., Mendoza-Elias J., Wang Y., Qi M., Lavin D.M., Chen M., Dholakia N., Thakrar R., Lacík I., Weir Gordon C., Oberholzer J., Greiner D.L., Langer R., Anderson D.G. Size- and shape-dependent foreign body immune response to materials implanted in rodents and non-human primates. Nat. Mater. 2015;14(6):643–651. doi: 10.1038/nmat4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeo T., Tan S.J., Lim C.L., Lau D.P.X., Chua Y.W., Krisna S.S., Iyer G., Tan G.S., Lim T.K.H., Tan D.S.W., Lim W.-T., Lim C.T. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci. Rep. 2016;6(1):22076. doi: 10.1038/srep22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang G., Li F., Zhao X., Ma Y., Li Y., Lin M., Jin G., Lu T.J., Genin G.M., Xu F. Functional and biomimetic materials for engineering of the three-dimensional cell microenvironment. Chem. Rev. 2017;117(20):12764–12850. doi: 10.1021/acs.chemrev.7b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charrier E.E., Pogoda K., Wells R.G., Janmey P.A. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat. Commun. 2018;9(1):449. doi: 10.1038/s41467-018-02906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rovinelli A., Sangid M.D., Proudhon H., Ludwig W. Using machine learning and a data-driven approach to identify the small fatigue crack driving force in polycrystalline materials. njp Comput. Mater. 2018;4(1):35. [Google Scholar]
- 52.Xinyu C., Yingbo Z., Jiaheng L., Hui C. Composition design of 7XXX aluminum alloys optimizing stress corrosion cracking resistance using machine learning. Mater. Res. Express. 2020;7(4) [Google Scholar]
- 53.Reker D., Rybakova Y., Kirtane A.R., Cao R., Yang J.W., Navamajiti N., Gardner A., Zhang R.M., Esfandiary T., L'Heureux J., von Erlach T., Smekalova E.M., Leboeuf D., Hess K., Lopes A., Rogner J., Collins J., Tamang S.M., Ishida K., Chamberlain P., Yun D., Lytton-Jean A., Soule C.K., Cheah J.H., Hayward A.M., Langer R., Traverso G. Computationally guided high-throughput design of self-assembling drug nanoparticles. Nat. Nanotechnol. 2021 doi: 10.1038/s41565-021-00870-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elbadawi M., Muñiz Castro B., Gavins F.K.H., Ong J.J., Gaisford S., Pérez G., Basit A.W., Cabalar P., Goyanes A. M3DISEEN: a novel machine learning approach for predicting the 3D printability of medicines. Int. J. Pharm. 2020;590:119837. doi: 10.1016/j.ijpharm.2020.119837. [DOI] [PubMed] [Google Scholar]
- 55.Rankouhi B., Jahani S., Pfefferkorn F.E., Thoma D.J. Compositional grading of a 316L-Cu multi-material part using machine learning for the determination of selective laser melting process parameters. Add. Manuf. 2021;38:101836. [Google Scholar]
- 56.Le T.C., Penna M., Winkler D.A., Yarovsky I. Quantitative design rules for protein-resistant surface coatings using machine learning. Sci. Rep. 2019;9(1):1–12. doi: 10.1038/s41598-018-36597-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalelkar P.P., Riddick M., García A.J. Biomaterial-based antimicrobial therapies for the treatment of bacterial infections. Nat. Rev. Mater. 2021 doi: 10.1038/s41578-021-00362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mei Y., Saha K., Bogatyrev S.R., Yang J., Hook A.L., Kalcioglu Z.I., Cho S.W., Mitalipova M., Pyzocha N., Rojas F., Van Vliet K.J., Davies M.C., Alexander M.R., Langer R., Jaenisch R., Anderson D.G. Combinatorial development of biomaterials for clonal growth of human pluripotent stem cells. Nat. Mater. 2010;9(9):768–778. doi: 10.1038/nmat2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazarovits J., Sindhwani S., Tavares A.J., Zhang Y., Song F., Audet J., Krieger J.R., Syed A.M., Stordy B., Chan W.C. Supervised learning and mass spectrometry predicts the in vivo fate of nanomaterials. ACS Nano. 2019;13(7):8023–8034. doi: 10.1021/acsnano.9b02774. [DOI] [PubMed] [Google Scholar]
- 60.Vegas A.J., Veiseh O., Doloff J.C., Ma M., Tam H.H., Bratlie K., Li J., Bader A.R., Langan E., Olejnik K., Fenton P., Kang J.W., Hollister-Locke J., Bochenek M.A., Chiu A., Siebert S., Tang K., Jhunjhunwala S., Aresta-Dasilva S., Dholakia N., Thakrar R., Vietti T., Chen M., Cohen J., Siniakowicz K., Qi M., McGarrigle J., Graham A.C., Lyle S., Harlan D.M., Greiner D.L., Oberholzer J., Weir G.C., Langer R., Anderson D.G. Combinatorial hydrogel library enables identification of materials that mitigate the foreign body response in primates. Nat. Biotechnol. 2016;34(3):345–352. doi: 10.1038/nbt.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sago C.D., Lokugamage M.P., Islam F.Z., Krupczak B.R., Sato M., Dahlman J.E. Nanoparticles that deliver RNA to bone marrow identified by in vivo directed evolution. J. Am. Chem. Soc. 2018;140(49):17095–17105. doi: 10.1021/jacs.8b08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blasi P. Poly (lactic acid)/poly (lactic-co-glycolic acid)-based microparticles: an overview. J, Pharmaceut. Invest. 2019:1–10. [Google Scholar]
- 63.Gu G.X., Chen C.-T., Richmond D.J., Buehler M.J. Bioinspired hierarchical composite design using machine learning: simulation, additive manufacturing, and experiment. Mater. Horizon. 2018;5(5):939–945. [Google Scholar]
- 64.Carleo G., Cirac I., Cranmer K., Daudet L., Schuld M., Tishby N., Vogt-Maranto L., Zdeborová L. Machine learning and the physical sciences. Rev. Mod. Phys. 2019;91(4) [Google Scholar]
- 65.Rajkomar A., Dean J., Kohane I. Machine learning in medicine. N. Engl. J. Med. 2019;380(14):1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 66.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G., Li B., Madabhushi A., Shah P., Spitzer M., Zhao S. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jablonka K.M., Ongari D., Moosavi S.M., Smit B. Big-data science in porous materials: materials genomics and machine learning. Chem. Rev. 2020;120(16):8066–8129. doi: 10.1021/acs.chemrev.0c00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmidt J., Marques M.R.G., Botti S., Marques M.A.L. Recent advances and applications of machine learning in solid-state materials science. njp Comput. Mater. 2019;5(1):83. [Google Scholar]
- 69.Nair L.S., Laurencin C.T. Biodegradable polymers as biomaterials. Prog. Polym. Sci. 2007;32(8–9):762–798. [Google Scholar]
- 70.Tian H.Y., Tang Z.H., Zhuang X.L., Chen X.S., Jing X.B. Biodegradable synthetic polymers: Preparation, functionalization and biomedical application. Prog. Polym. Sci. 2012;37(2):237–280. [Google Scholar]
- 71.Khademhosseini A., Langer R. A decade of progress in tissue engineering. Nat. Protoc. 2016;11(10):1775–1781. doi: 10.1038/nprot.2016.123. [DOI] [PubMed] [Google Scholar]
- 72.Uhrich K.E., Cannizzaro S.M., Langer R.S., Shakesheff K.M. Polymeric systems for controlled drug release. Chem. Rev. 1999;99(11):3181–3198. doi: 10.1021/cr940351u. [DOI] [PubMed] [Google Scholar]
- 73.Fenton O.S., Olafson K.N., Pillai P.S., Mitchell M.J., Langer R. Advances in biomaterials for drug delivery. Adv. Mater. 2018;30(29):29. doi: 10.1002/adma.201705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riley R.S., June C.H., Langer R., Mitchell M.J. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 2019;18(3):175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Xinxin, Xian Jun Loh Anti-Angiogenic Nanomicelles For the Topical Delivery of Aflibercept to Treat Retinal Neovascular Disease. Adv. Mater. 2021 doi: 10.1002/adma.202108360. In press. [DOI] [PubMed] [Google Scholar]
- 76.Barouti G., Liow S.S., Dou Q., Ye H., Orione C., Guillaume S.M., Loh X.J. New linear and star-shaped thermogelling poly ([R]-3-hydroxybutyrate) copolymers. Chem. Euro. J. 2016;22(30):10501–10512. doi: 10.1002/chem.201601404. [DOI] [PubMed] [Google Scholar]
- 77.Gan L., Deen G.R., Loh X., Gan Y. New stimuli-responsive copolymers of N-acryloyl-N′-alkyl piperazine and methyl methacrylate and their hydrogels. Polymer. 2001;42(1):65–69. [Google Scholar]
- 78.Loh X.J., Nguyen V.P.N., Kuo N., Li J. Encapsulation of basic fibroblast growth factor in thermogelling copolymers preserves its bioactivity. J. Mater. Chem. 2011;21(7):2246–2254. [Google Scholar]
- 79.Loh X.J., Yee B.J.H., Chia F.S. Sustained delivery of paclitaxel using thermogelling poly (PEG/PPG/PCL urethane) s for enhanced toxicity against cancer cells. J. Biomed. Mater. Res. 2012;100(10):2686–2694. doi: 10.1002/jbm.a.34198. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen V.P.N., Kuo N., Loh X.J. New biocompatible thermogelling copolymers containing ethylene-butylene segments exhibiting very low gelation concentrations. Soft Matter. 2011;7(5):2150–2159. [Google Scholar]
- 81.Xue K, Xian Jun Loh Advanced TherapeuticsProgress ReportHydrogels as Emerging Materials for Translational Biomedicine. Adv. Therapeut. 2019;2(1) doi: 10.1002/adtp.201800088. In this issue. [DOI] [Google Scholar]
- 82.Li F., Han J.S., Cao T., Lam W., Fan B.E., Tang W., Chen S.J., Fok K.L., Li L.X. Design of self-assembly dipeptide hydrogels and machine learning via their chemical features. Proc. Natl. Acad. Sci. U. S. A. 2019;116(23):11259–11264. doi: 10.1073/pnas.1903376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin Q., Lim J.Y.C., Xue K., Su X., Loh X.J. Polymeric hydrogels as a vitreous replacement strategy in the eye. Biomaterials. 2021;268:120547. doi: 10.1016/j.biomaterials.2020.120547. [DOI] [PubMed] [Google Scholar]
- 84.Cai P., Hu B., Leow W.R., Wang X., Loh X.J., Wu Y.L., Chen X. Biomechano-interactive materials and interfaces. Adv. Mater. 2018;30(31):1800572. doi: 10.1002/adma.201800572. [DOI] [PubMed] [Google Scholar]
- 85.Guo S., Zhu X., Loh X.J. Controlling cell adhesion using layer-by-layer approaches for biomedical applications. Mater. Sci. Eng. C. 2017;70:1163–1175. doi: 10.1016/j.msec.2016.03.074. [DOI] [PubMed] [Google Scholar]
- 86.Loh X.J., Gong J., Sakuragi M., Kitajima T., Liu M., Li J., Ito Y. Surface coating with a thermoresponsive copolymer for the culture and non-enzymatic recovery of mouse embryonic stem cells. Macromol. Biosci. 2009;9(11):1069–1079. doi: 10.1002/mabi.200900081. [DOI] [PubMed] [Google Scholar]
- 87.Leuning D.G., Beijer N.R.M., du Fossé N.A., Vermeulen S., Lievers E., van Kooten C., Rabelink T.J., Boer J.d. The cytokine secretion profile of mesenchymal stromal cells is determined by surface structure of the microenvironment. Sci. Rep. 2018;8(1):7716. doi: 10.1038/s41598-018-25700-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vermeulen S., Vasilevich A., Tsiapalis D., Roumans N., Vroemen P., Beijer N.R.M., Dede Eren A., Zeugolis D., de Boer J. Identification of topographical architectures supporting the phenotype of rat tenocytes. Acta Biomater. 2019;83:277–290. doi: 10.1016/j.actbio.2018.10.041. [DOI] [PubMed] [Google Scholar]
- 89.Hook A.L., Anderson D.G., Langer R., Williams P., Davies M.C., Alexander M.R. High throughput methods applied in biomaterial development and discovery. Biomaterials. 2010;31(2):187–198. doi: 10.1016/j.biomaterials.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 90.Yang J., Mei Y., Hook A.L., Taylor M., Urquhart A.J., Bogatyrev S.R., Langer R., Anderson D.G., Davies M.C., Alexander M.R. Polymer surface functionalities that control human embryoid body cell adhesion revealed by high throughput surface characterization of combinatorial material microarrays. Biomaterials. 2010;31(34):8827–8838. doi: 10.1016/j.biomaterials.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Epa V.C., Yang J., Mei Y., Hook A.L., Langer R., Anderson D.G., Davies M.C., Alexander M.R., Winkler D.A. Modelling human embryoid body cell adhesion to a combinatorial library of polymer surfaces. J. Mater. Chem. 2012;22(39):20902–20906. doi: 10.1039/C2JM34782B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwaria R.J., Mondarte E.A.Q., Tahara H., Chang R., Hayashi T. Data-driven prediction of protein adsorption on self-assembled monolayers toward material screening and design. ACS Biomater. Sci. Eng. 2020;6(9):4949–4956. doi: 10.1021/acsbiomaterials.0c01008. [DOI] [PubMed] [Google Scholar]
- 93.M. Niinomi, Metallic biomaterials, J. Artif. Organs, pp. 105-110. [DOI] [PubMed]
- 94.Han H.-S., Loffredo S., Jun I., Edwards J., Kim Y.-C., Seok H.-K., Witte F., Mantovani D., Glyn-Jones S. Current status and outlook on the clinical translation of biodegradable metals. Mater. Today. 2019;23:57–71. [Google Scholar]
- 95.Niinomi M. Recent progress in research and development of metallic structural biomaterials with mainly focusing on mechanical biocompatibility. Mater. Trans. 2018;59(1):1–13. [Google Scholar]
- 96.Fujii H., Djc M., Hkdh B. Bayesian neural network analysis of fatigue crack growth rate in nickel base superalloys. ISIJ Int. 1996;36(11):1373–1382. [Google Scholar]
- 97.Kang J.-Y., Song J.-H. Neural network applications in determining the fatigue crack opening load. Int. J. Fatig. 1998;20(1):57–69. [Google Scholar]
- 98.Iacoviello F., Iacoviello D., Cavallini M. Analysis of stress ratio effects on fatigue propagation in a sintered duplex steel by experimentation and artificial neural network approaches. Int. J. Fatig. 2004;26(8):819–828. [Google Scholar]
- 99.Mohanty S., Teale R., Chattopadhyay A., Peralta P., Willhauck C. International workshop on structural heath monitoring; Citeseer: 2007. Mixed Gaussian Process and State-Space Approach for Fatigue Crack Growth Prediction; pp. 1108–1115. [Google Scholar]
- 100.Wang H., Zhang W., Sun F., Zhang W. A comparison study of machine learning based algorithms for fatigue crack growth calculation. Materials. 2017;10(5):543. doi: 10.3390/ma10050543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dinda S., Kujawski D. Correlation and prediction of fatigue crack growth for different R-ratios using Kmax and ΔK+ parameters. Eng. Fract. Mech. 2004;71(12):1779–1790. [Google Scholar]
- 102.Agrawal A., Deshpande P.D., Cecen A., Basavarsu G.P., Choudhary A.N., Kalidindi S.R. Exploration of data science techniques to predict fatigue strength of steel from composition and processing parameters. Integr. Mater. Manuf. Inn. 2014;3(1):90–108. [Google Scholar]
- 103.Agrawal A., Choudhary A. Proceedings of the 25th ACM International on Conference on Information and Knowledge Management. 2016. A fatigue strength predictor for steels using ensemble data mining: steel fatigue strength predictor; pp. 2497–2500. [Google Scholar]
- 104.Agrawal A., Choudhary A. An online tool for predicting fatigue strength of steel alloys based on ensemble data mining. Int. J. Fatig. 2018;113:389–400. [Google Scholar]
- 105.Canyurt O.E. Estimation of welded joint strength using genetic algorithm approach. Int. J. Mech. Sci. 2005;47(8):1249–1261. [Google Scholar]
- 106.Wu C.-T., Chang H.-T., Wu C.-Y., Chen S.-W., Huang S.-Y., Huang M., Pan Y.-T., Bradbury P., Chou J., Yen H.-W. Machine learning recommends affordable new Ti alloy with bone-like modulus. Mater. Today. 2020;34:41. [Google Scholar]
- 107.Wen C., Zhang Y., Wang C., Xue D., Bai Y., Antonov S., Dai L., Lookman T., Su Y. Machine learning assisted design of high entropy alloys with desired property. Acta Mater. 2019;170:109–117. [Google Scholar]
- 108.Chang Y.-J., Jui C.-Y., Lee W.-J., Yeh A.-C. Prediction of the composition and hardness of high-entropy alloys by machine learning. JOM (J. Occup. Med.) 2019;71(10):3433–3442. [Google Scholar]
- 109.Bobbio A. The first endosseous alloplastic implant in the history of man. Bull. Hist. Dent. 1972;20(1):1–6. [PubMed] [Google Scholar]
- 110.Hench L.L., Splinter R.J., Allen W., Greenlee T. Bonding mechanisms at the interface of ceramic prosthetic materials. J. Biomed. Mater. Res. 1971;5(6):117–141. [Google Scholar]
- 111.Boutin P. Total arthroplasty of the hip by fritted aluminum prosthesis. Experimental study and 1st clinical applications. Revue de chirurgie orthopedique et reparatrice de l'appareil moteur. 1972;58(3):229. [PubMed] [Google Scholar]
- 112.Jarcho M. Calcium phosphate ceramics as hard tissue prosthetics. Clin. Orthop. Relat. Res. 1981;157:259–278. [PubMed] [Google Scholar]
- 113.de Groot K. CRC Press; Boca Raton, FL: 1983. Ceramics of Calcium Phosphate: Preparation and Properties. [Google Scholar]
- 114.Metsger D.S., Driskell T., Paulsrud J. Tricalcium phosphate ceramic--a resorbable bone implant: review and current status. JADA (J. Am. Dent. Assoc.) 1982;105(6):1035. doi: 10.14219/jada.archive.1982.0408. 1939. [DOI] [PubMed] [Google Scholar]
- 115.Akao M., Aoki H., Kato K. Mechanical properties of sintered hydroxyapatite for prosthetic applications. J. Mater. Sci. 1981;16(3):809–812. [Google Scholar]
- 116.LeGeros R. Calcium phosphate materials in restorative dentistry: a review. Adv. Dent. Res. 1988;2(1):164–180. doi: 10.1177/08959374880020011101. [DOI] [PubMed] [Google Scholar]
- 117.Oses C., Toher C., Curtarolo S. High-entropy ceramics. Nat. Rev. Mater. 2020;5(4):295–309. [Google Scholar]
- 118.Kaufmann K., Maryanovsky D., Mellor W.M., Zhu C., Rosengarten A.S., Harrington T.J., Oses C., Toher C., Curtarolo S., Vecchio K.S. Discovery of high-entropy ceramics via machine learning. Npj Comput. Mater. 2020;6(1):1–9. [Google Scholar]
- 119.Sarker P., Harrington T., Toher C., Oses C., Samiee M., Maria J.-P., Brenner D.W., Vecchio K.S., Curtarolo S. High-entropy high-hardness metal carbides discovered by entropy descriptors. Nat. Commun. 2018;9(1):1–10. doi: 10.1038/s41467-018-07160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsu Y.-C., Yu C.-H., Buehler M.J. Using deep learning to predict fracture patterns in crystalline solids. Matter. 2020;3(1):197–211. [Google Scholar]
- 121.Epps R.W., Bowen M.S., Volk A.A., Abdel-Latif K., Han S., Reyes K.G., Amassian A., Abolhasani M. Artificial chemist: an autonomous quantum dot synthesis bot. Adv. Mater. 2020:2001626. doi: 10.1002/adma.202001626. [DOI] [PubMed] [Google Scholar]
- 122.Mirzaei M., Furxhi I., Murphy F., Mullins M. A machine learning tool to predict the antibacterial capacity of nanoparticles. Nanomaterials. 2021;11(7):1774. doi: 10.3390/nano11071774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xuan M., Shao J., Li J. Cell membrane-covered nanoparticles as biomaterials. Nat. Sci. Rev. 2019;6(3):551–561. doi: 10.1093/nsr/nwz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Santana R., Zuluaga R., Gañan P., Arrasate S., Onieva E., Gonzalez-Diaz H. Predicting coated-nanoparticle drugs release systems with perturbation-theory machine learning (PTML) models. Nanoscale. 2020 doi: 10.1039/d0nr01849j. [DOI] [PubMed] [Google Scholar]
- 125.Zhao D., Liang H., Han C., Li J., Liu J., Zhou K., Yang C., Wei Q. 3D printing of a titanium-tantalum Gyroid scaffold with superb elastic admissible strain, bioactivity and in-situ bone regeneration capability. Add. Manuf. 2021;47 [Google Scholar]
- 126.Taylor S.L., Ibeh A.J., Jakus A.E., Shah R.N., Dunand D.C. NiTi-Nb micro-trusses fabricated via extrusion-based 3D-printing of powders and transient-liquid-phase sintering. Acta Biomater. 2018;76:359–370. doi: 10.1016/j.actbio.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 127.Chen Z., Li Z., Li J., Liu C., Lao C., Fu Y., Liu C., Li Y., Wang P., He Y. 3D printing of ceramics: a review. J. Eur. Ceram. Soc. 2019;39(4):661–687. [Google Scholar]
- 128.Yilin G., Fuh Ying Hsi J., Wen Feng L. Virtual and Physical Prototyping; 2021. Multiscale Topology Optimisation with Nonparametric Microstructures Using Three-Dimensional Convolutional Neural Network (3D-CNN) Models; pp. 1–12. [Google Scholar]
- 129.Tourlomousis F., Jia C., Karydis T., Mershin A., Wang H., Kalyon D.M., Chang R.C. Machine learning metrology of cell confinement in melt electrowritten three-dimensional biomaterial substrates. Microsyst. Nanoeng. 2019;5(1):15. doi: 10.1038/s41378-019-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khadilkar A., Wang J., Rai R. Deep learning–based stress prediction for bottom-up SLA 3D printing process. Int. J. Adv. Manuf. Technol. 2019;102(5–8):2555–2569. [Google Scholar]
- 131.Koeppe A., Padilla C.A.H., Voshage M., Schleifenbaum J.H., Markert B. Efficient numerical modeling of 3D-printed lattice-cell structures using neural networks. Manuf. Lett. 2018;15:147–150. [Google Scholar]
- 132.Pilania G., Iverson C.N., Lookman T., Marrone B.L. Machine-learning-based predictive modeling of glass transition temperatures: a case of polyhydroxyalkanoate homopolymers and copolymers. J. Chem. Inf. Model. 2019;59(12):5013–5025. doi: 10.1021/acs.jcim.9b00807. [DOI] [PubMed] [Google Scholar]
- 133.Maidin S.B., Campbell I., Pei E. Assembly Automation; 2012. Development of a Design Feature Database to Support Design for Additive Manufacturing. [Google Scholar]
- 134.Huang Q., Wang Y., Lyu M., Lin W. Shape deviation generator—a convolution framework for learning and predicting 3-D printing shape accuracy. IEEE Trans. Autom. Sci. Eng. 2020;17(3):1486–1500. [Google Scholar]
- 135.Yamaguchi S., Lee C., Karaer O., Ban S., Mine A., Imazato S. Predicting the debonding of CAD/CAM composite resin crowns with AI. J. Dent. Res. 2019;98(11):1234–1238. doi: 10.1177/0022034519867641. [DOI] [PubMed] [Google Scholar]
- 136.Deng L., Feng B., Zhang Y. An optimization method for multi-objective and multi-factor designing of a ceramic slurry: combining orthogonal experimental design with artificial neural networks. Ceram. Int. 2018;44(13):15918–15923. [Google Scholar]
- 137.Zhang W., Mehta A., Desai P.S., Higgs C. Int. Solid Free Form Fabr. Symp. Austin, TX. 2017. Machine learning enabled powder spreading process map for metal additive manufacturing (AM) pp. 1235–1249. [Google Scholar]
- 138.Ding D., Pan Z., Cuiuri D., Li H., van Duin S., Larkin N. Bead modelling and implementation of adaptive MAT path in wire and arc additive manufacturing. Robot. Comput. Integrated Manuf. 2016;39:32–42. [Google Scholar]
- 139.Mohamed O.A., Masood S.H., Bhowmik J.L. Investigation of dynamic elastic deformation of parts processed by fused deposition modeling additive manufacturing. Adv. Prod. Eng. Manag. 2016;11(3) [Google Scholar]
- 140.Bayraktar Ö., Uzun G., Çakiroğlu R., Guldas A. Experimental study on the 3D-printed plastic parts and predicting the mechanical properties using artificial neural networks. Polym. Adv. Technol. 2017;28(8):1044–1051. [Google Scholar]
- 141.Smith J., Xiong W., Yan W., Lin S., Cheng P., Kafka O.L., Wagner G.J., Cao J., Liu W.K. Linking process, structure, property, and performance for metal-based additive manufacturing: computational approaches with experimental support. Comput. Mech. 2016;57(4):583–610. [Google Scholar]
- 142.Zhang H., Moon S.K., Ngo T.H. Hybrid machine learning method to determine the optimal operating process window in aerosol jet 3D printing. ACS Appl. Mater. Interfaces. 2019;11(19):17994–18003. doi: 10.1021/acsami.9b02898. [DOI] [PubMed] [Google Scholar]
- 143.Bone J.M., Childs C.M., Menon A., Póczos B., Feinberg A.W., LeDuc P.R., Washburn N.R. Hierarchical machine learning for high-fidelity 3D printed biopolymers. ACS Biomater. Sci. Eng. 2020;6(12):7021–7031. doi: 10.1021/acsbiomaterials.0c00755. [DOI] [PubMed] [Google Scholar]
- 144.Loh X.J., Li Z.B. Soft materials research at IMRE. Macromol. Rapid Commun. 2019;40(5) doi: 10.1002/marc.201800914. [DOI] [PubMed] [Google Scholar]
- 145.Lyu J., Manoochehri S. Online convolutional neural network-based anomaly detection and quality control for fused filament fabrication process. Virtual Phys. Prototyp. 2021;16(2):160–177. [Google Scholar]
- 146.D'Amour A., Heller K., Moldovan D., Adlam B., Alipanahi B., Beutel A., Chen C., Deaton J., Eisenstein J., Hoffman M.D., Hormozdiari F. 2020. Underspecification presents challenges for credibility in modern machine learning. arXiv preprint arXiv:2011.03395. [Google Scholar]
- 147.Goh G.D., Sing S.L., Yeong W.Y. A review on machine learning in 3D printing: applications, potential, and challenges. Artif. Intell. Rev. 2021;54(1):63–94. [Google Scholar]
- 148.Collins F.S., Morgan M., Patrinos A. The human genome project: lessons from large-scale biology. Science. 2003;300(5617):286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]