Abstract
Horizontally transferred elements, such as plasmids, can burden host cells with various metabolic and fitness costs and may lead to other potentially detrimental phenotypic effects. Acquisition of the Pseudomonas syringae megaplasmid pMPPla107 by various Pseudomonads causes sensitivity to a growth-inhibiting substance that is produced in cultures by Pseudomonads during growth under standard laboratory conditions. After approximately 500 generations of laboratory passage of Pseudomonas stutzeri populations containing pMPPla107, strains from two out of six independent passage lines displayed resistance to this inhibitory agent. Resistance was transferable and is, therefore, associated with mutations occurring on pMPPla107. Resequencing experiments demonstrated that resistance is likely due to a large deletion on the megaplasmid in one line, and to a nonsynonymous change in an uncharacterized megaplasmid locus in the other strain. We further used allele exchange experiments to confirm that resistance is due to this single amino acid change in a previously uncharacterized megaplasmid protein, which we name SkaA. These results provide further evidence that costs and phenotypic changes associated with horizontal gene transfer can be compensated through single mutational events and emphasize the power of experimental evolution and resequencing to better understand the genetic basis of evolved phenotypes.
This article is part of the theme issue ‘The secret lives of microbial mobile genetic elements’.
Keywords: megaplasmid, experimental evolution, bacteriostatic
1. Introduction
Plasmids are extrachromosomal replicons that can often move between bacterial cells through conjugation, and that are a major contributor to horizontal gene transfer (HGT) events in bacteria. Thousands of genes can be exchanged via HGT in a single transfer and this potentially opens up new niches for each organism through the acquisition of genes encoding proteins involved in metabolism, antibiotic resistance, virulence and symbiosis [1–6]. Although plasmids can provide a variety of advantages to a bacterial cell in a given environment, HGT can also engender general fitness costs and can lead to additional phenotypic changes that could be costly under specific environmental contexts [7,8]. The existence of such costs has led some to generally question how plasmids are maintained within populations over evolutionary time given plasmid–chromosome conflicts, a discussion that is often referred to as the ‘plasmid paradox’ [9,10]. Identification of compensatory mutations selected to ameliorate plasmid–chromosome conflicts during laboratory passage can significantly inform our understanding of the mechanistic basis of these conflicts and may, therefore, ultimately provide deeper molecular insights into the costs of HGT as well as the ‘plasmid paradox’ [7,8].
We have previously shown that acquisition of the Pseudomonas syringae megaplasmid pMPPla107 by Pseudomonas stutzeri sensitizes this strain background to the presence of an inhibitory agent that has bacteriostatic properties [11–13]. Sensitivity is found in pMPPla107's original host strain P. syringae pv. lachrymans 107 and can be transferred to various Pseudomonas spp. upon their acquisition of pMPPla107, thus indicating that the phenotype is linked to the acquisition of pMPPla107 [12]. Production of this inhibitory agent is conserved across measured Pseudomonas spp., appears correlated with Pseudomonas physiology, and is likely linked to an essential process as all viable strains within a P. aeruginosa transposon library maintained activity [13].
Although the acquisition of plasmids by new host backgrounds often creates metabolic, physiological and fitness costs, previous research has shown that various types of compensatory mutations occur rapidly on either the chromosome or plasmid, with amelioration of costs enabling the persistence of plasmids [14–17]. For instance, experiments on P. fluorescens with the mercury-resistant pQBR103 demonstrate that compensatory mutations in gacA/gacS occur in strains with and without selection using mercury [14]. Moreover, mutations in two helicases and an RNA polymerase subunit resulted in host dependence on the plasmid RP4 while also increasing the uptake of additional plasmids [15]. Therefore, compensatory mutations may not only explain plasmid persistence mechanisms but also increased plasmid promiscuity.
For these reasons and to better understand the genetic basis of previously described phenotypic costs associated with pMPPla107 [11], we carried out the experimental passage of P. stutzeri populations under conditions that select for maintenance of pMPPla107. Our goal with these passage experiments was to identify strain backgrounds that evolve resistance to this inhibitory agent with the hope that identification of compensatory mutations would provide better understanding of the genetic basis of these costs.
2. Methods
(a) . Laboratory passage experiment
A frozen stock of strain DBL408 [11] was streaked to salt water lysogeny broth (SWLB) agar supplemented with rifampicin (50 ng µl−1) and tetracycline (10 ng µl−1). Hereafter, every instance mentioned for SWLB liquid and solid media will also include supplementation with these antibiotics at these concentrations unless otherwise noted. From this initial culture plate, six independent single colonies of P. stutzeri strain DBL408 were picked into 2 ml cultures of SWLB in 5 ml polypropylene tubes with caps. These cultures were grown under shaking (220 r.p.m.) conditions at 27°C for 2 days, at which point a subset of these cultures was frozen in 40% glycerol at −80°C and labelled as ‘passage 0’ while a 1 : 1000 (cells : media) dilution was carried out into fresh 2 ml of SWLB supplemented with the same antibiotics as above. Each passage, cells were plated to SWLB agar plates to observe colony morphology in case of contamination. Tetracycline in the media selects for maintenance of the megaplasmid in strain DBL408, while rifampicin protects against contamination from additional sources. Although specific experiments were not performed to evaluate this point directly, it is likely that megaplasmids would be lost from these strains and potentially whole populations during early passages given the cost of megaplasmid carriage in P. stutzeri [11]. Every 10 passages, a 750 µl sample of the culture was mixed to 40% final concentration with glycerol and stored at −80°C. This passage process was repeated for approximately 500 generations of growth (log2 of 1000 = 9.96 divisions per passage; 50 passages total).
After 50 passages, a sample from each independent evolutionary population was streaked onto SWLB agar plates. Two single colonies (A-500 and B-500) were picked from each evolutionary passage line and frozen as single strain isolates from each passage population. We focused on isolating single colonies, rather than on whole population sequencing, with the goal of linking identifying distinct phenotypic changes to underlying genotypic changes to better understand the underlying molecular mechanisms rather than on profiling evolutionary dynamics across whole populations. In this way, our experiments described herein treat laboratory passage like a phenotypic screen or selection in a microbial genetics experiment [18].
Similar procedures to the above were used to isolate single colony strains from whole populations at generations 0 and 100 from population 4, except that frozen stock cultures were streaked to SWLB agar plates and approximately 10 colonies were picked in each instance as a rough way to sample population frequencies.
(b) . Inhibitory agent sensitivity test
We isolated single colonies from whole populations of six evolved lines as a means to facilitate linking observed phenotypic changes in these lines to underlying genetic events. We followed previously described protocols [11,13] to test the activity of the inhibitory agent against isolates from each of the six evolved populations. Briefly, overlays were prepared by diluting an overnight culture into fresh media and incubating this new culture on a shaker at 27°C for 4 h with one exception. In the case of skaA allelic exchange overlays, we report the results after 5 h of growth in this manuscript but also include data from 4 h incubation in electronic supplementary material, files S6–S10. Cells from this 4-h culture were then mixed with 0.4% molten agar and plated on King's media B (KB) agar media. This 0.4% agar overlay was allowed to solidify for approximately 15 min, at which point supernatant was added. The inhibitory agent was collected by growing P. stutzeri for 24–48 h, centrifuging cells at 10 000×g for 5 min and sterilizing supernatants through a 0.22 µm filter. After sterilization, 10 µl of supernatants were spotted onto the overlay plate and allowed to dry. Overlay plates were grown at 27°C for approximately 24 h, at which point zones of inhibition were observed. Raw photos from overlays for most experiments are included as electronic supplementary material, figures S1–S10, with each assay taking place on 30 mm petri dishes for scale.
(c) . Conjugation of evolved megaplasmids into ‘ancestral’ P. stutzeri
An ‘ancestral’ strain of P. stutzeri (DBL388), which has undergone fewer than 5 laboratory passages, was mixed with strains 3B-500, 4B-500 and 5B-500 for megaplasmid transfer experiments. DBL388 was created through Tn7 transposition of a transposon from plasmid AKN34 [19] into DBL332 [12] and is resistant to gentamicin. For conjugation experiments, overnight liquid cultures of each strain were mixed 1 : 1 and then centrifuged at 3000×g for 3 min. The supernatant was removed without disturbing the pellet, and pellets were washed and resuspended in 1 ml of 10 mM MgCl2. Strain mixtures were centrifuged and washed one additional time, after which 100 µl of resuspended cells were spread on KB plates with rifampicin (50 ng µl−1) and incubated for at least 48 h at 27°C. After 48 h, strain mixtures were resuspended in 10 mM MgCl2, and plated on KB agar plates supplemented with gentamicin (10 ng µl−1) and tetracycline (10 ng µl−1). Gentamicin- and tetracycline-resistant colonies arising on this plate were picked to liquid culture and underwent diagnostic PCR for the presence of pMPPla107 using primers from [20].
(d) . Genome sequencing and annotation
For sequencing experiments, a single colony of each strain was picked to 2 ml SWLB media and grown overnight, shaking at 27°C. DNA samples were extracted from these cultures using a Promega Wizard kit (Madison, WI) with RNAse treatments. P. stutzeri strain DBL408 (the progenitor strain of all experimental lines) as well as strains 1B-500, 2B-500, 4B-500 and 6B-500 were sequenced using 100 bp paired end reads on an Illumina HiSeq by the University of Arizona sequencing core (SRX11065275, SRX5391491, SRX5391490, SRX5391492, SRX5391486, respectively). Pseudomonas stutzeri lines 3B-500 and 5B-500 were sequenced using 250 bp paired end reads on an Illumina MiSeq by MicrobesNG (Birmingham, UK; SRX5391493, SRX5391487, respectively). For the identification of polymorphisms, we used gene annotations of pMPPla107 from a previous publication ([1], Accession: NZ_CP031226.1) and the annotations from the P. stutzeri 28a24 reference sequence ([21], Accession: CP007441.1). We additionally isolated a single colony from the passage 0 stock for experimental population 4, referred to herein as 4A-0, and the exconjugant strain DBL619 (arising from a mating between 4B-500 and DBL388). Genomic DNA for these isolates were isolated as above and sequenced using 150 bp paired end reads on an Illumina MiSeq by SNPsaurus (Eugene, OR; SRX11065277, SRX11065276, respectively). Lastly, we isolated single colonies from P. stutzeri strains DBL494 and DBL1802, and extracted genomic DNA as above. Genomic DNA for these two strains was sequenced using 150 bp paired end reads on an Illumina NovaSeq by MiGS (Pittsburgh, PA; SRX11559214 and SRX11559215, respectively).
We further assembled reads de novo from strains 4A-0, 4B-500 and 5B-500 using unicycler v. 0.8.4 and with default parameters. Contigs for these assemblies can be found in electronic supplementary material, file S2, 4A-0; file S3, 4B-500; file S4, 5B-500. We note that our best estimate based on read depth and coverage is that megaplasmid pMPPla107 is found in approximately the same copy number as the chromosome and there is no indication based on these new assemblies that the copy number changed over the course of the experimental passage.
(e) . Mapping reads and calling variants
Illumina reads from all six evolved lines were mapped to the pMPPla107 reference sequence (accession CP031226.1) using Breseq v. 0.35.5 [22] with default parameters. We only considered variants that were strongly supported for analyses within this manuscript.
(f) . Allelic replacement of skaA
To definitively demonstrate that the skaA allele from P. stutzeri 5B-500 is causative of the observed change in sensitivity to supernatants, we moved the P. stutzeri 5B-500 allele into a naive strain background. Briefly, a dsDNA block containing the P. stutzeri 5B-500 allele as well as a silent change in skaA that introduces an Eco53kI restriction site was obtained from IDT (Coralville, IA) and is found as file S1 in the electronic supplementary material. BP clonase (Invitrogen; Waltham, MA) was used to recombine this dsDNA block into the pDONR207 entry vector to create plasmid pDBL100. Then, LR clonase (Invitrogen; Waltham, MA) was used to recombine this dsDNA block from plasmid pDBL100 into destination vector pMTN1907 to create plasmid pDBL101. Plasmid pDBL101 was transformed into Escherichia coli strain S17 (to create strain DBL1804), and conjugated into P. stutzeri strain DBL494. Strain DBL494 is a nalidixic acid- and gentamicin-resistant derivative of P. stutzeri strain DBL492, which is a nalidixic acid (100 ng ul−1)-resistant derivative of strain 28a24. To create strain DBL494, a mariner transposon providing gentamicin resistance was transposed into the pMPPla107 megaplasmid in strain DBL453 and then conjugated into strain DBL492. The transposon in this strain has been localized to the megaplasmid by sequencing [23].
A tetracycline-resistant strain was isolated from the conjugation between E. coli DBL1804 and DBL494, in which plasmid pDBL101 was integrated into the megaplasmid in DBL494. A single tetracycline-resistant colony was picked to 2 ml plain KB media and grown overnight, at which point dilutions were plated onto KB media agar supplemented with 10% sucrose. Sucrose-resistant colonies were screened for tetracycline resistance and remaining tetracycline sensitive colonies were screened by PCR using forward primer 5′0-GCCTGGGTGACACCTATCAG-3′ and reverse primer 5′-AATGCTCAGCTGCAGTCGAT-3′ using an annealing temperature of 55°C and an extension time of 1 : 30. Colonies were considered to contain the allelic replacement if the PCR product from this reaction was not cut by Eco53kI.
(g) . Synteny plots
We used SynMap2 with the LAST algorithm and default parameters to compare the sequences of ancestral pMPPla107 and pMPPla107-4B500 [24]. DAGChainer Options were: nucleotide distance, −D = 20, and −A = 5. The Tandem duplication distance was set to 10 and the C-score was set to 0.
(h) . Gene function and structure predictions
To predict functional characteristics, we input the amino acid sequence of SkaA as input into a PsiBlast search of the NCBI non-redundant protein sequence database (date of search last search: 19 July 2021) [25]. PsiBlast was run for four iterations, at which point no new protein matches were added as results. The amino acid sequence of SkaA was also used as an input to the Phyre2 web server [26], with selection of the ‘intensive’ setting. Lastly, we used a Google collaboration implemented version of AlphaFold 2 to predict three-dimensional structure of the protein [27,28]. Briefly, sequences are picked through BLAST and aligned using MMseqs2. The resulting .pdb file can be found as electronic supplementary material, file S5, and was visualized through the Protein Data Bank and Mol* [29,30].
(i) . Supplemental data
All supplemental data for this manuscript can be found on Figshare at doi:10.6084/m9.figshare.15072537. Electronic supplementary material, table S1 lists all strains and plasmids used in this manuscript and includes citations for [31,32]. Electronic supplementary material, figures S1–S10 are raw pictures used for overlay experiments. Electronic supplementary material, figure S11 is a Mauve alignment of the 4A-0, 4B-500 and pMPPla107 megaplasmid sequences. Electronic supplementary material, file S1 is a .fasta file for the sequence that was ordered from IDT and used to create vectors for allelic exchange experiments with skaA. Electronic supplementary material, files S2–S4 are .fasta files for genome assemblies of strains 4A-0, 4B-500 and 5B-500. Electronic supplementary material, file S5 is a .pdb file for the SkaA structure prediction.
3. Results
(a) . Strains from 2 of 6 laboratory passage populations gain resistance to a previously described inhibitory agent
We previously identified that acquisition of plasmid pMPPla107 by P. stutzeri sensitizes this strain to an unknown inhibitory agent produced by numerous Pseudomonas strains [11]. To identify strain backgrounds that were resistant to this agent, we screened for the presence of inhibition in single colony isolates from these evolved populations. Single colony isolates from two out of six lines (referred to from here on as 4B-500 and 5B-500) revert to the non-pMPPla107 phenotype and demonstrate resistance to this inhibitory agent (figure 1).
Figure 1.
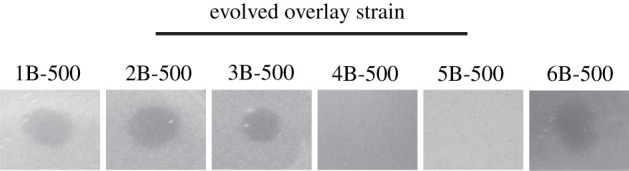
Testing inhibitory agent on all six evolved lines reveals two resistant lines. Six populations carrying pMPPla107 were passaged under laboratory conditions for 500 generations. Single colonies isolated from these populations were tested for sensitivity against inhibitory agent found in Pseudomonas spp. supernatants. Isolates from lines 4B-500 and 5B-500 revert to a non-pMPPla107 phenotype where a zone of inhibition is not present, indicating resistance to the inhibitory agent. All overlays were plated after 4 h of growth in KB and spotted with 10 µl of P. stutzeri filter-sterilized supernatants. The number (1, 2, 3, …) indicates the individual lines and B indicates the second of two isolates taken at Generation 500. All images are representative of three biological replicates.
(b) . Mutations providing resistance to the inhibitory agent are transferable and localized to the megaplasmid
In previous evolutionary studies focusing on plasmids, the burden of plasmid acquisition selected for compensatory mutations to arise on both the host chromosome [9,14,15,33] and on the plasmids themselves [17]. To identify whether resistance mutations to this inhibitory agent occurred on the chromosome or megaplasmid, we carried out conjugation experiments to move megaplasmids from 3B-500 (sensitive), 4B-500 (resistant) and 5B-500 (resistant) to an ancestral P. stutzeri background. Exconjugants with the ‘ancestral’ chromosome and evolved megplasmids from 4B-500 and 5B-500 (but not 3B-500) demonstrate resistance to this inhibitory agent (figure 2). Therefore, mutations providing resistance to inhibition from lines 4B-500 and 5B-500 can be positionally localized to the evolved megaplasmids.
Figure 2.
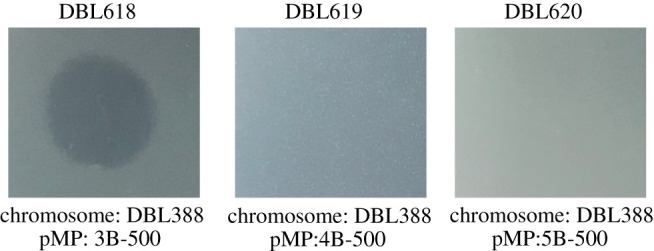
Exconjugants of pMPPla107 from populations 4 and 5 into an ancestral chromosomal background transfers resistance to the inhibitory agent. Given that strains 4B-500 and 5B-500 were known to display resistance against the inhibitory agent, we conjugated megaplasmids from these strains into an unevolved P. stutzeri background DBL388. We additionally used conjugations with the megaplasmid from 3B-500 as a sensitive (positive) control as we knew this evolved strain was still sensitive. While a megaplasmid from 3B-500 maintains sensitivity to the inhibitory agent in the DBL388 background, megaplasmids from 4B-400 and 5B-500 transfer resistance. All overlays were plated after 4 h of growth in KB and spotted with 10 µl of P. stutzeri filter-sterilized supernatants. All images are representative of three biological replicates. Raw image files can be found in electronic supplemental material at Figshare (doi:10.6084/m9.figshare.15072537.v3): DBL618 is electronic supplementary material, figure S1, DBL619 is electronic supplementary material, figure S2, DBL620 is electronic supplementary material, figure S3. (Online version in colour.)
(c) . Independent mutations likely provide resistance to the unknown inhibitory agent
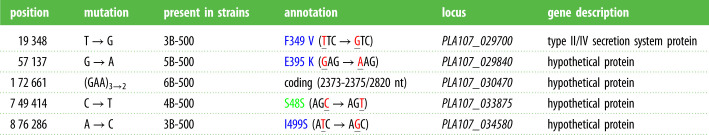
To identify the resistance mutations in lines 4B-500 and 5B-500, we resequenced and analysed genomes from single colony isolates arising from each of the six laboratory passage strains of P. stutzeri after 500 generations. We further resequenced the generation 0 isolate from population 4, 4A-0. Although mutations appear to have arisen on megaplasmids in all lines except 1B-500 and 2B-500, lines 4B-500 and 5B-500 carry single mutations that are not present in any of the other strains (table 1). 4B-500 contains a silent mutation in an uncharacterized gene (PLA107_033875), while 5B-500 contains a nonsynonymous mutation in an uncharacterized gene (PLA107_029840). Furthermore, there is a large deletion in 4B-500 that is not present in any other strain (see below).
Table 1.
Mutations occurring on megaplasmid pMPPla107 in experimental passage strains. Amino acid annotation in green is silent and those in blue are missense mutations. Nucleotides that are changed compared to wild type are highlighted in red in the codon.
 |
(d) . A 368 kb deletion in pMPPla107 is strongly linked to supernatant resistance in strain 4B-500
Conjugation of the megplasmid from 4B-500 into a naive background strongly suggests that mutations providing resistance to the inhibitory agent are associated with and colocalized to the megaplasmid (figure 2).
Analysis of the genome from isolate 4B-500 shows that it contains a large deletion of approximately 368 kb, which occurred within pMPPla107 between approximate positions 131–499 kb (figure 3a). This deletion region includes 451 predicted genes, 53 of which have some functional annotation and two tRNA loci (table 2). Interestingly, there appear to be no clearly repetitive or overlapping sites at the ends of the deletion site as visualized by CoGe [24] (https://genomevolution.org/r/uboj).
Figure 3.
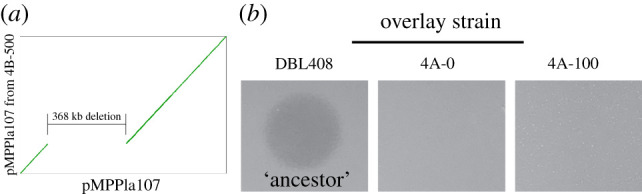
A large deletion occurs early during the passage of population 4 and provides resistance to the inhibitory agent. (a) SynMap dotplot visualizes the large deletion occurring from 131 to 499 kb in the evolved 4B pMPPla107 as a large shift across the x-axis. The remaining portions of the sequences maintain perfect synteny, indicating that a clean deletion occurred. The x-axis is ancestral pMPPla107 gene order where x1, …, N = gene1, …, N and the y-axis is the line 4B evolved pMPPla107 gene order where y1, …, N = gene1…N.]. (b) We tested single colony isolates from frozen stocks of the passage line 4 populations at generations 0 and 100 for sensitivity to the inhibitory agent. We found that resistance to the inhibitory agent is present in the generation 0 isolate from population 4. This resistance is maintained in generation 100 (shown) through generation 500 and is the only unique mutation in pMPPla107 other than a synonymous SNP. All overlays were plated after 4 h of growth n KB and spotted with 10 µl of P. stutzeri filter-sterilized supernatants. All images are representative of three biological replicates. Raw image files can be found in electronic supplementary material at Figshare (doi:10.6084/m9.figshare.15072537.v3): DBL408 is electronic supplementary material, figure S4, 4A-0 is electronic supplementary material, figure S5, 4A-100 is electronic supplementary material, figure S6. (Online version in colour.)
Table 2.
Annotations of genes deleted from the megaplasmid in strain 4B-500.
| annotation | start | end | direction |
|---|---|---|---|
| HNH endonuclease | 131 589 | 132 017 | reverse |
| IcmE | 135 202 | 136 674 | reverse |
| IcmK | 136 674 | 137 732 | reverse |
| ribonucleotide-diphosphate reductase subunit beta | 142 392 | 143 558 | forward |
| nucleotidyl transferase AbiEii/AbiGii toxin family protein | 144 218 | 145 108 | reverse |
| endonuclease | 149 053 | 149 961 | forward |
| RtcB family protein | 149 987 | 151 210 | forward |
| transglycosylase SLT domain protein | 152 996 | 153 736 | forward |
| XRE family transcriptional regulator | 162 068 | 162 529 | forward |
| tRNA-Arg | 162 927 | 163 003 | forward |
| HDOD domain-containing protein | 166 715 | 168 022 | reverse |
| DUF805 domain-containing protein | 216 222 | 216 527 | reverse |
| RNA 2′-phosphotransferase | 220 194 | 220 772 | reverse |
| XRE family transcriptional regulator | 227 208 | 227 495 | reverse |
| SMI1/KNR4 family protein | 232 488 | 232 898 | reverse |
| DUF1127 domain-containing protein | 269 304 | 269 519 | forward |
| DUF21 domain-containing protein | 269 566 | 270 537 | forward |
| 3-oxoacyl-ACP synthase | 273 913 | 275 001 | forward |
| NAD-dependent epimerase/dehydratase family protein | 275 411 | 276 382 | forward |
| MBL fold metallo-hydrolase | 276 375 | 277 217 | forward |
| tRNA-Leu | 282 629 | 282 713 | forward |
| XRE family transcriptional regulator | 284 257 | 284 511 | forward |
| ASCH domain-containing protein | 284 869 | 285 198 | forward |
| adenylate-forming enzyme | 292 503 | 293 810 | forward |
| Arc family DNA-binding protein | 304 934 | 305 476 | forward |
| RND transporter | 312 173 | 313 522 | reverse |
| ParE | 328 197 | 330 101 | forward |
| ParC | 330 900 | 333 152 | forward |
| DUF721 domain-containing protein | 335 955 | 336 419 | reverse |
| SH3 domain-containing protein | 338 158 | 339 000 | forward |
| DUF4343 domain-containing protein | 358 704 | 359 468 | forward |
| thioredoxin TrxA | 368 277 | 368 594 | reverse |
| DUF541 domain-containing protein | 375 046 | 375 759 | forward |
| HNH endonuclease | 378 522 | 379 406 | forward |
| XRE family transcriptional regulator | 403 681 | 405 477 | forward |
| DUF262 domain-containing protein | 415 714 | 416 247 | forward |
| transposase | 433 226 | 434 356 | reverse |
| regulatory protein RecX | 441 216 | 441 695 | forward |
| FMN-binding glutamate synthase family protein | 444 888 | 446 483 | reverse |
| tryptophan–tRNA ligase | 450 046 | 451 407 | reverse |
| DUF4165 domain-containing protein | 452 760 | 456 818 | forward |
| SPFH domain-containing protein | 468 612 | 469 550 | reverse |
| Ig-like domain-containing protein | 473 955 | 474 710 | reverse |
| multidrug efflux RND transporter permease subunit | 475 218 | 478 319 | reverse |
| efflux RND transporter periplasmic adaptor subunit | 478 326 | 479 429 | reverse |
| DUF3828 domain-containing protein | 480 389 | 480 847 | forward |
| PH domain-containing protein | 481 802 | 482 416 | forward |
| conjugal transfer protein TraO | 484 704 | 485 081 | reverse |
| non-canonical purine NTP pyrophosphatase | 487 114 | 487 686 | forward |
| DUF4031 domain-containing protein | 488 336 | 488 689 | reverse |
| oxidoreductase | 48 9082 | 489 852 | forward |
| OfxX fusion product | 493 512 | 494 291 | reverse |
To further characterize this mutation, we sequenced a single colony isolate from the passage 0 stock of population line 4 (4A-0). Strain 4A-0 is resistant to this inhibitory agent (figure 3b), and sequencing demonstrates that this line also contains a large deletion on the megaplasmid (electronic supplementary material, file S2 and figure S11). Although these results indicate that the deletion in line 4B is responsible for resistance to the inhibitory agent, the large size of the deletion and the density of genes within this region make it difficult to discern which gene(s) are responsible for the resistance phenotype in this region. Resequencing comparisons of strain 4A-0 to DBL408 do not indicate that any additional mutations (besides the large deletion) have occurred in this strain in the megaplasmid (table 1). Depth and coverage assessments from assemblies of megaplasmids from strains 4A-0 and 4B-500 indicate that copy number remains approximately 1× and it has not dramatically increased or decreased from that in strain DBL408 (electronic supplementary material, files S2 and S3).
(e) . A single polymorphism present on the megaplasmid from strain 5B-500 is responsible for supernatant resistance
Conjugation of the megaplasmid from 5B-500 into a naive background strongly suggests that mutations providing resistance to the inhibitory agent are associated with and colocalized to the megaplasmid (figure 2).
Resequencing of strain 5B-500 demonstrated that a single polymorphism occurs on the megaplasmid from this strain (table 1). Depth and coverage assessments from assemblies of the megaplasmid from strain 5B-500 indicate that copy number remains approximately 1× and it has not dramatically increased or decreased from that in strain DBL408 (electronic supplementary material, file S4).
The mutation identified on the megaplasmid in strain 5B-500 occurs at position 57 137 bp and leads to a nonsynonymous mutation, changing glutamate to a lysine (395 E > K) in a previously uncharacterized protein (PLA107_029840). Data from conjugation experiments and resequencing data strongly suggest that this polymorphism eliminates the sensitivity phenotype seen by strains that have acquired pMPPla107, thus we name this gene skaA for supernatant killing activity A. Given that the megaplasmid polymorphism identified in strain 5B-500 occurs outside the deletion region found in the 4B-500 megaplasmid, this also strongly implies that two separate compensatory strategies exist within pMPPla107 to provide resistance to the unknown inhibitory agent.
To definitively demonstrate that the single mutation present in skaA in P. stutzeri 5B-500 is causative of the change in supernatant sensitivity of this strain, we swapped alleles of skaA from 5B-500 into an otherwise ‘clean’ P. stutzeri genomic background containing the pMPPla107 megaplasmid (DBL494). As one can see in figure 4, the replacement of the wild-type allele in DBL494 with the skaA allele from 5B-500 leads to a loss of sensitivity to supernatant treatment under the measured conditions.
Figure 4.
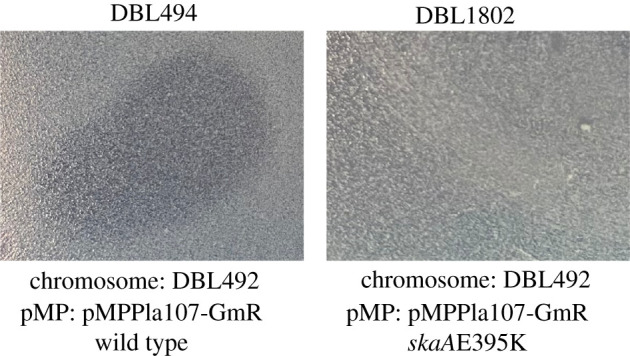
Allelic replacement of skaAE395 K provides resistance to inhibitory compounds in Pseudomonas supernatants. We recreated the skaAE395 K allele in a naive chromosomal and megaplasmid background (strain DBL494). A zone of inhibition can be seen for strain DBL494 but not for strain DBL1802. Strain DBL494 contains a wild-type allele of skaA on the megaplasmid while the skaAE395 K mutation has been recombined into the megaplasmid in strain DBL1802. Overlays were plated after 5 h of growth in KB liquid and spotted with 20 µl of P. stutzeri filter-sterilized supernatants. All images are representative of three biological replicates for these strains. Raw image files can be found in electronic supplementary material at Figshare (doi:10.6084/m9.figshare.15072537.v3): DBL494 is electronic supplementary material, figure S8, DBL1802 is electronic supplementary material, figure S10.
(f) . Protein predictions for SkaA
SkaA is characterized as a ‘hypothetical protein’ in the current annotation of the pMPPla107 megaplasmid, and does not appear to match any well-characterized proteins found in the current ( July 2021) version of the NR database at NCBI. The only PsiBlast hits that show relatively high sequence similarity to SkaA from plasmid pMPPla107 are found in other Pseudomonas genomes and are likely present on unconfirmed copies of other megaplasmids from the pMPPla107 family. However, SkaA also displays more distant (approx. 18% identity, approx. 37% positives) matches to ‘hypothetical’ proteins from various Vibrio and Staphylococcus species (e.g. WP_094133102.1).
Alphafold2 predicts that SkaA is composed of numerous alpha helices, potentially split up into roughly three domains (figure 5). Results from the Phyre2 server reinforce this prediction as there are no high quality (greater than 50% confidence) matches and all lower quality matches are proteins with domains composed of alpha helices. The aspartate to lysine change in SkaA from P. stutzeri 5B-500 is predicted to occur within one of the numerous predicted alpha helix domains of the protein (figure 5).
Figure 5.
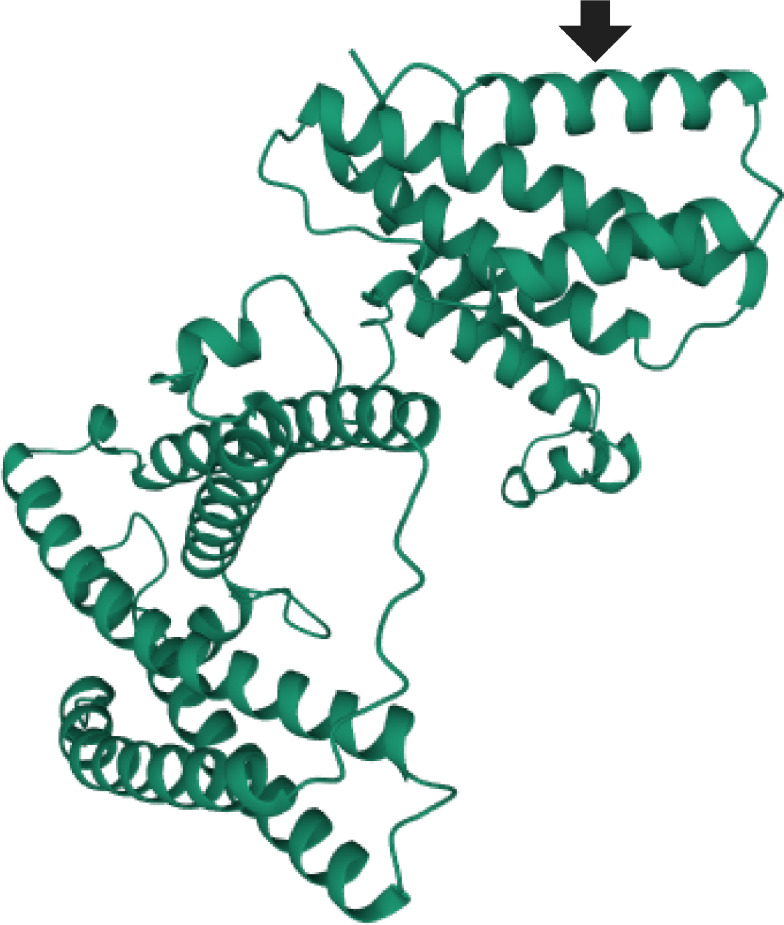
Predicted protein structure of SkaA. We used Alphafold2 to predict the protein structure of SkaA. The position of the E395 K change in the megaplasmid of 5B-500 is denoted with an arrow. (Online version in colour.)
4. Discussion
Acquisition of megaplasmid pMPPla107 by many Pseudomonas strains sensitizes these strains to growth inhibition when treated with supernatants from other Pseudomonas cultures, and we used laboratory passage to isolate and identify mutations associated with resistance to this inhibitory agent. Although strains of P. stutzeri containing pMPPla107 are normally sensitized to the presence of a (currently) unidentified inhibitory agent produced by a variety of Pseudomonas strains under normal growth conditions, isolates from two of six experimental populations evolved resistance to this inhibition over 500 generations of passage. Numerous studies have found that compensatory mutations to plasmid carriage often occur on the chromosome (e.g. [14]), but we found that both mutations providing resistance (in lines 4B-500 and 5B-400/5B-500) likely occur on the megaplasmid.
Sequencing of line 4B-500 demonstrated that this line contains a 368 kb deletion within a region on the megaplasmid that we have previously identified as demonstrating extensive divergence between similar megaplasmids [1]. Since this deletion was present during the initial passage of this population (electronic supplementary material, figure S11), which was initiated from a single colony picked from the strain DBL408 stock, it is possible that this deletion was actually present and circulating as a polymorphism prior to the initiation of all experimental lines. This pattern suggests that this part of the megaplasmid is a potential cargo region where expendable genes may be more likely to provide benefits in certain environments rather than necessary genes for maintenance or transmission. Some of the genes found within this region include efflux pumps, antitoxins and multidrug resistance proteins, all of which could possibly affect resistance to the inhibitory agent (table 2). This region also includes genes annotated as parE and parC loci, which possibly contribute to contrasting sensitivity/resistance phenotypes to quinolone antibiotics by different Pseudomonad strain backgrounds [34]. Lastly, this region also appears to encode an Arc-like DNA-binding protein and two tRNA loci. It is unclear from our current data which of the hundreds of genes in this region is responsible for increased sensitivity to the inhibitory agent. We note that this is another example where large deletions on megaplasmids are shown to contribute to the amelioration of detrimental phenotypic consequences from these plasmids, and highlight that this trend suggests megaplasmid amelioration to new host strains may often occur through large scale deletion of unnecessary genes [35].
We have provided multiple lines of evidence suggesting that a single mutation on the megaplasmid from line 5B-500 imparts resistance to the inhibitory agent. This mutation generates a nonsynonymous change in an uncharacterized open reading frame, which we now formally name skaA. It is still unclear how skaA interacts with inhibitory agent or how the 395 E > K SNP changes these interactions, and protein structure predictions and amino acid alignments were largely uninformative. However, prior to creating the allelic swap of skaA as described above, we tried and failed numerous times to create a clean deletion of skaA in multiple strain backgrounds that were concurrently selected for megaplasmid maintenance. It is therefore possible that the SkaA protein is essential for proper replication and partitioning of megplasmid pMPPla107.
Combining comparative genomics, microbial genetics and evolutionary passage enabled us to identify two distinct genetic changes that affect sensitization of P. stutzeri to an inhibitory agent upon acquisition of genes [13]. We identify a region on pMPPla107 and a SNP in the gene we now call skaA that are responsible for resistance to the Pseudomonas inhibitory agent. Our data presented here are the framework on which to begin future work identifying the mechanism behind skaA and designing directed deletions within the 4B deletion that will be critical to identifying the other components regarding the inhibitory agent sensitivity phenotype associated with the acquisition of pMPPla107.
Data accessibility
We have included all raw data for the manuscript, either as accessioned sequencing files in the SRA or as electronic supplementary material files in Figshare. https://doi.org/10.6084/m9.figshare.15072537.v3.
Authors' contributions
B.A.S. conceived of the experiments, carried out molecular and bioinformatic work and wrote the manuscript. K.D. carried out experimental evolution passages. M.C. assisted with the preparation of materials and strain characterization. D.A.B. conceived of the experiments, helped to write the manuscript and carried out some bioinformatic analyses.
Competing interests
We declare we have no competing interests.
Funding
This work was supported through a grant from the United States National Science Foundation (NSF) IOS-1856556 to D.A.B.
References
- 1.Smith BA, Leligdon C, Baltrus DA. 2019. Just the two of us? A family of Pseudomonas megaplasmids offers a rare glimpse into the evolution of large mobile elements. Genome Biol. Evol. 11, 1192-1206. ( 10.1093/gbe/evz066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hülter N, Ilhan J, Wein T, Kadibalban AS, Hammerschmidt K, Dagan T. 2017. An evolutionary perspective on plasmid lifestyle modes. Curr. Opin. Microbiol. 38, 74-80. ( 10.1016/j.mib.2017.05.001) [DOI] [PubMed] [Google Scholar]
- 3.Kado CI. 1998. Origin and evolution of plasmids. Antonie Leeuwenhoek 73, 117-126. ( 10.1023/A:1000652513822) [DOI] [PubMed] [Google Scholar]
- 4.Okubo T, Piromyou P, Tittabutr P, Teaumroong N, Minamisawa K. 2016. Origin and evolution of nitrogen fixation genes on symbiosis islands and plasmid in Bradyrhizobium. Microbes Environ. 31, 260-267. ( 10.1264/jsme2.ME15159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hynes MF, McGregor NF. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4, 567-574. ( 10.1111/j.1365-2958.1990.tb00625.x) [DOI] [PubMed] [Google Scholar]
- 6.Johnson TJ, Nolan LK. 2010. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 74, 477-478. ( 10.1128/MMBR.00002-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Millan A, Craig MacLean R. 2019. Fitness costs of plasmids: a limit to plasmid transmission. In Microbial transmission (eds F Baquero, E Bouza, J Gutiérrez-Fuentes, TM Coque), pp. 65-79. Washington, DC: ASM Press. ( 10.1128/9781555819743.ch4) [DOI] [Google Scholar]
- 8.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489-495. ( 10.1016/j.tree.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 9.Carroll AC, Wong A. 2018. Plasmid persistence: costs, benefits, and the plasmid paradox. Can. J. Microbiol. 64, 293-304. ( 10.1139/cjm-2017-0609) [DOI] [PubMed] [Google Scholar]
- 10.MacLean RC, San Millan A. 2015. Microbial evolution: towards resolving the plasmid paradox. Curr. Biol. 25, R764-R767. ( 10.1016/j.cub.2015.07.006) [DOI] [PubMed] [Google Scholar]
- 11.Dougherty K, Smith BA, Moore AF, Maitland S, Fanger C, Murillo R, Baltrus DA. 2014. Multiple phenotypic changes associated with large-scale horizontal gene transfer. PLoS ONE 9, e102170. ( 10.1371/journal.pone.0102170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romanchuk A, Jones CD, Karkare K, Moore A, Smith BA, Jones C, Dougherty K, Baltrus DA. 2014. Bigger is not always better: transmission and fitness burden of ∼1MB Pseudomonas syringae megaplasmid pMPPla107. Plasmid 73, 16-25. ( 10.1016/j.plasmid.2014.04.002) [DOI] [PubMed] [Google Scholar]
- 13.Smith B, Feinstein Y, Clark M, Baltrus D. 2019. A moving target: the megaplasmid pMPPla107 sensitizes cells to an inhibitory agent conserved across Pseudomonas spp. bioRxiv. ( 10.1101/537589) [DOI]
- 14.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism–mutualism continuum. Curr. Biol. 25, 2034-2039. ( 10.1016/j.cub.2015.06.024) [DOI] [PubMed] [Google Scholar]
- 15.Loftie-Eaton W, et al. 2017. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 1, 1354-1363. ( 10.1038/s41559-017-0243-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano H, Wegrzyn K, Loftie-Eaton W, Johnson J, Deckert GE, Rogers LM, Konieczny I, Top EM. 2016. Evolved plasmid–host interactions reduce plasmid interference cost. Mol. Microbiol. 101, 743-756. ( 10.1111/mmi.13407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton ER, Merritt PM, Bever JD, Fuqua C. 2013. Large deletions in the pAtC58 megaplasmid of Agrobacterium tumefaciens can confer reduced carriage cost and increased expression of virulence genes. Genome Biol. Evol. 5, 1353-1364. ( 10.1093/gbe/evt095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shuman HA, Silhavy TJ. 2003. The art and design of genetic screens: Escherichia coli. Nat. Rev. Genet. 4, 419-431. ( 10.1038/nrg1087) [DOI] [PubMed] [Google Scholar]
- 19.Lambertsen L, Sternberg C, Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6, 726-732. ( 10.1111/j.1462-2920.2004.00605.x) [DOI] [PubMed] [Google Scholar]
- 20.Baltrus DA, et al. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7, e1002132. ( 10.1371/journal.ppat.1002132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith BA, Dougherty KM, Baltrus DA. 2014. Complete genome sequence of the highly transformable Pseudomonas stutzeri Strain 28a24. Genome Announc. 2, e00543-14. ( 10.1128/genomeA.00543-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165-188. ( 10.1007/978-1-4939-0554-6_12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baltrus DA, Medlen J, Clark M. 2019. Identifying transposon insertions in bacterial genomes through nanopore sequencing. BioRxiv. ( 10.1101/765545) [DOI] [Google Scholar]
- 24.Haug-Baltzell A, Stephens SA, Davey S, Scheidegger CE, Lyons E. 2017. SynMap2 and SynMap3D: web-based whole-genome synteny browsers. Bioinformatics 33, 2197-2198. ( 10.1093/bioinformatics/btx144) [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 26.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845-858. ( 10.1038/nprot.2015.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumper J, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583-589. ( 10.1038/s41586-021-03819-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirdita M, Steinegger M, Söding J. 2019. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35, 2856-2858. ( 10.1093/bioinformatics/bty1057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res. 28, 235-242. ( 10.1093/nar/28.1.235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sehnal D, et al. 2021. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 49, W431-W437. ( 10.1093/nar/gkab314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sikorski J, Teschner N, Wackernagel W. 2002. Highly different levels of natural transformation are associated with genomic subgroups within a local population of Pseudomonas stutzeri from soil. Appl. Environ. Microbiol. 68, 865-873. ( 10.1128/AEM.68.2.865-873.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baltrus DA, Nishimura MT, Dougherty KM, Biswas S, Mukhtar MS, Vicente J, Holub EB, Dangl JL. 2012. The molecular basis of host specialization in bean pathovars of Pseudomonas syringae. Mol. Plant. Microb. Interact. 25, 877-888. ( 10.1094/MPMI-08-11-0218) [DOI] [PubMed] [Google Scholar]
- 33.San Millan A, Peña-Miller R, Toll-Riera M, Halbert ZV, McLean AR, Cooper BS, MacLean RC. 2014. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208. ( 10.1038/ncomms6208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baltrus DA, Smith C, Derrick M, Leligdon C, Rosenthal Z, Mollico M, Moore A, Clark M. 2021. Genomic background governs opposing responses to nalidixic acid upon megaplasmid acquisition in Pseudomonas. mSphere 6, e00008-21. ( 10.1128/mSphere.00008-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee M-C, Marx CJ. 2012. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 8, e1002651. ( 10.1371/journal.pgen.1002651) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have included all raw data for the manuscript, either as accessioned sequencing files in the SRA or as electronic supplementary material files in Figshare. https://doi.org/10.6084/m9.figshare.15072537.v3.


