Abstract
In this study, we addressed the extent of diversification of phages associated with nitrogen-fixing symbiotic Rhizobium species. Despite the ecological and economic importance of the Rhizobium genus, little is known about the diversity of the associated phages. A thorough assessment of viral diversity requires investigating both lytic phages and prophages harboured in diverse Rhizobium genomes. Protein-sharing networks identified 56 viral clusters (VCs) among a set of 425 isolated phages and predicted prophages. The VCs formed by phages had more proteins in common and a higher degree of synteny, and they group together in clades in the associated phylogenetic tree. By contrast, the VCs of prophages showed significant genetic variation and gene loss, with selective pressure on the remaining genes. Some VCs were found in various Rhizobium species and geographical locations, suggesting that they have wide host ranges. Our results indicate that the VCs represent distinct taxonomic units, probably representing taxa equivalent to genera or even species. The finding of previously undescribed phage taxa indicates the need for further exploration of the diversity of phages associated with Rhizobium species.
This article is part of the theme issue ‘The secret lives of microbial mobile genetic elements’.
Keywords: phages, prophages, Rhizobium, evolution, taxonomy, protein networks
1. Introduction
Bacteriophages, or phages, are abundant in every terrestrial and marine microbiome [1]. They play a significant role in the ecology and evolution of bacterial communities by enabling horizontal gene transfer (HGT) and influencing the global biochemical cycles [2]. The interaction of phages with a bacterium usually leads to cell lysis and death, reshaping bacterial communities [3,4]. Alternatively, phages can become integrated into the chromosome, remaining as prophages. Under particular environmental stressors, prophages are induced to replicate and kill the cell.
There is compelling evidence of the widespread occurrence of phage footprints left in bacterial genomes [5]. Complete prophages in bacterial genomes can be induced, but a significant proportion of large and small phage genome fragments can no longer be activated [6]. This latter category is thought to represent vestiges of ancient phage–bacteria interactions [7]. Alternatively, prophages may be part of functional phage-related mobile elements like phage inducible chromosomal islands (PICIs), and gene transfer agents (GTAs) widespread in bacterial species [8,9]. In both classes of mobile elements, phage structural proteins (capsid and tail) and DNA processing (terminases) are recruited to carry out chromosomal genes encoding virulence genes and other genes [10] instead of the phage genome [9]. Recently, it has been suggested that selection favours prophage retention, although the advantages to the bacteria are unclear [11–13]. One direct benefit of harbouring prophages is immunity to reinfection, but prophages may also increase bacterial fitness by modifying metabolism via auxiliary metabolic genes (AMGs) and by providing targets for recombination and HGT, allowing the bacteria to adapt to their ecological niche [12–15].
The wide diversity of bacterial viruses requires a taxonomy that reflects, ideally, their evolutionary nature. However, this cannot proceed without identifying the fundamental units of evolution, which could be designated species, viral populations or structural clusters [16]. Over the years, viral taxonomy has evolved from morphotypes and nucleic acid types to genome-based classification [17]. Approaches based purely on comparative genomics such as average nucleotide identity (ANI) have proved insufficient given their dependence on defined thresholds, genome mosaicism and variable mutation rates among viruses [18]. Recently, gene- and protein-sharing networks have been shown to deal better with the mosaic genome structure of viruses while preserving the advantages related to ANI and protein-level similarity [19,20].
In GenBank, there are 18 884 phage genomes, about 70% of which come from bacterial species of γ-proteobacteria, Actinobacteria and Firmicutes. By contrast, α-proteobacteria, β-proteobacteria, bacteroidetes and cyanobacteria phages are poorly represented (electronic supplementary material, figure S1). The bias in phage sampling may affect the phage discovery results in metagenomes (based on bioinformatics algorithms) and it may affect our understanding of phage diversification in the major classes of bacteria. Moreover, better characterization of prophages will improve the algorithms to distinguish them from other related phage mobile elements (PICIS and GTAs) [21].
In this study, we investigated Rhizobium phages to expand the knowledge of their diversification among closely related species [22]. Rhizobium is a bacterial genus involving species of great economic importance owing to their capacity to form symbiotic relationships with the roots of legume plants and to fix nitrogen. However, few studies focus specifically on Rhizobium phages and their roles in Rhizobium ecology and evolution [22–24]. Therefore, we aimed to place Rhizobium phages into a phylogenetic context, identify novel and unclassified phages and determine how they are distributed throughout the Rhizobium genus. Our approach relied on protein-sharing networks [20] and phylogenetic inferences, and on searching for prophages in complete and draft Rhizobium genomes. As a result, we identified viral clusters (VCs), which were relatively consistent in terms of genome structure and phylogeny. We unveiled the high level of mobility and large trans-species host range of certain phages, the host specialization of other phages and the hidden diversity of prophages in Rhizobium species.
2. Material and methods
(a) . Phage isolation and genome sequencing
In this study, we isolated 25 phages from agriculture soils in localities of Mexico and Argentina, via the enrichment method using Rhizobium and Sinorhizobium hosts (electronic supplementary material, table S1) [23]. The DNA from the phages was purified according to the DNA Isolation Kit for Cells and Tissues (Roche Life Sciences, CA, USA) protocol, with modifications [22]. Libraries for sequencing 22 phage genomes were constructed using the Nextera Kit and processed using an Illumina NextSeq 500 system (Unidad Universitaria de Secuenciación Masiva de DNA (UUSMD)-Universidad Nacional Autónoma de México (UNAM)). Two phage genomes (RHEph15 and RHEph24) were sequenced by the Sanger method as previously described [23], and one phage genome (RHEph12) with PacBio technology (Macrogen, Korea). Details on assembly, coverage and GenBank accessions identifiers are provided in the electronic supplementary material, table S1. Open reading frames (ORFs) prediction and annotation were performed automatically with PROKKA [25]. The results were manually curated by inspection of BLASTx hits based on the non-redundant GenBank database, and BLASTp hits based on the virus orthologous groups [26] and InterPro databases.
(b) . Rhizobium genomes and prophage prediction
Rhizobium genomes used for prophage prediction and phylogenetic analysis were downloaded from the GenBank RefSeq database (release date 14 April 2021). We selected 612 genomes based on genome length greater than or equal to 6 Mb, N50 value greater than 20 kb and the presence of the nifH gene (electronic supplementary material, table S2). Prophages were predicted with VIBRANT (Virus Identification By iteRative ANnoTation) v. 1.2.1 [27], using default parameters (prophage sequence length greater than or equal to 1000 bp; number of ORFs greater than or equal to four).
The phage life cycle was predicted using the machine learning PhageAI program accessed through the phageai.lifecycle.classifier [28].
(c) . Network construction and analysis
We clustered the phages using vConTACT v. 2 [20]. It clusters similar proteins into protein clusters (PCs) and then calculates the VCs according to the maximum probabilities of sharing PCs (edges) between the genomes (nodes) in order to produce a bipartite network. The PCs were determined using the Markov cluster algorithm (MCL) and the VCs were determined using ClusterONE, with default parameters (MCL inflation: 2; penalty value: 2) and with an edge weight of 10. The networks generated by vConTACT were visualized using Cytoscape v. 3.8.2 (https://cytoscape.org/) [29]. The VCs involving Rhizobium phages and/or prophages were manually checked and adjusted as following: first, it was verified that within the VC all the individual genomes were related by a minimal edge value greater than 60. The genomes with edges less than 60 were removed. Second, the phage genomes included in VCs were examined for inconsistencies with the previously reported average nucleotide identity by Mummer (ANIm) groups already reported [22], and average amino acid identity groups.
To construct synteny maps of the genomes of the phages in the VCs, we used EasyFig v. 2.2.3 [30]. Pairwise comparisons between phage genomes were conducted using the BLASTn algorithm with an e-value cut-off of 0.001. The number and percentage of PCs shared between pairs of phage genomes were computed using a homemade perl script.
(d) . Phylogenetic analyses
To construct a phylogenetic tree of Rhizobium genomes, we aligned 1181 bp of the concatenated dnaA and recA gene segments from 612 Rhizobium genomes, and 42 dnaA and recA sequenced polymerase chain reaction fragments of Rhizobium genomes [31]. Multiple alignments were carried out by MUSCLE [32]. A phylogenetic tree was constructed in MEGA v. 7 using the maximum-likelihood method [33] with bootstrap of 1000 replicates.
We used the terminase large subunit (TerL) protein as a marker to construct the phage phylogenetic tree. TerL proteins of previously reported Rhizobium phages [22] were used to search for homologous TerL proteins in the 642 genomes of phages and prophages incorporated in the vConTACT network (electronic supplementary material, table S3). The searches were conducted using BLASTp (e-value cut-off: 1 × 10−6; coverage: 70%; identity: 30%). Using these criteria, 485 TerL proteins were obtained to make a multiple gapped alignment of TerL with ClustalW (-align option) [32]. The maximum-likelihood phylogenetic tree was constructed with IQ-Tree [34] based on the best substitution model found by ModelFinder (VT + F + R6 (Variable Time + Empirical Codon Frequencies + FreeRate)) with 1000 ultrafast bootstrap replicates. The same procedure was followed for performing a phylogeny of the major capside proteins (MCP). To further support the VCs, we used the VIPTree program, which is based on the similarity in proteins by tBlastx (e-value 10−2; identity 30%; amino acid length 30) converted to distance in a neighbor-joining tree [35].
The phylogenetic trees were visualized using iTOL v. 6 (https://itol.embl.de/).
3. Results
(a) . High genomic similarity of Rhizobium phages
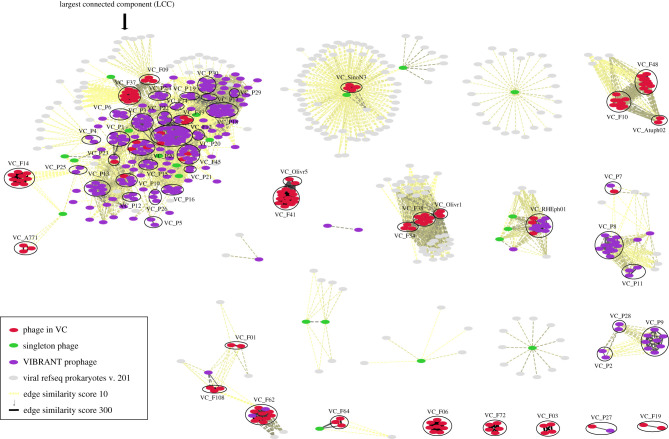
As the taxonomic relationships of Rhizobium phages were unknown, we constructed a vConTACT network [20], involving 425 Rhizobium isolated phages and predicted prophages. The phages comprised 155 phages already reported [22], of which 25 phages were sequenced in this study, and 270 prophages predicted using VIBRANT [27] (see next subsection). After an initial test with the default vConTACT settings (MCL inflation: 2; edge weight greater than 1), we decided to construct a stringent network using an edge weight greater than 10. The network consisted of 3751 nodes joined by 66 332 edges. There were 478 VCs, including 56 VCs involving Rhizobium phages and/or prophages, and there were 102 Rhizobium phage and prophage singletons (electronic supplementary material, figure S2). The VCs that involved Rhizobium phages and/or prophages were shown to be weakly related to other α as well as γ- and β-proteobacteria phages by edges that indicated lower probabilities of sharing PCs.
Then, we focused only on the 56 VCs of Rhizobium phages and/or prophages by selecting them with Cytoscape, as shown in figure 1. In this subset, there were 642 nodes (genomes) joined by 6378 edges. There was a largest connected component (LCC) that contained most of the Rhizobium phage and prophage VCs (figure 1). Although some of the VCs were exclusively formed by phages (encircled red ellipses) the majority of the VCs were formed by prophages (encircled purple ellipses). The edges within VCs had the highest scores (greater than 60), while the edges between VCs had scores greater than 10 but less than 60. This means that the phage genomes in a VC had a median percentage of shared PCs greater than 50% of the total (figure 2d).
Figure 1.
Viral clusters (VCs) of isolated phages and predicted prophages, and their network relationships. The network was obtained using vConTACT v2 with an edge weight greater than 10 (electronic supplementary material, figure S2) and was visualized using Cytoscape. The largest connected component (LCC) is shown on the left; besides the LCC, the other VCs have weak or no relationships to each other. Virulent phages are indicated by circles containing red ellipses exclusively; temperate phages in circles with red and purple ellipses; prophages are shown by encircled purple ellipses only. In the inset, the colour key indicates the network components.
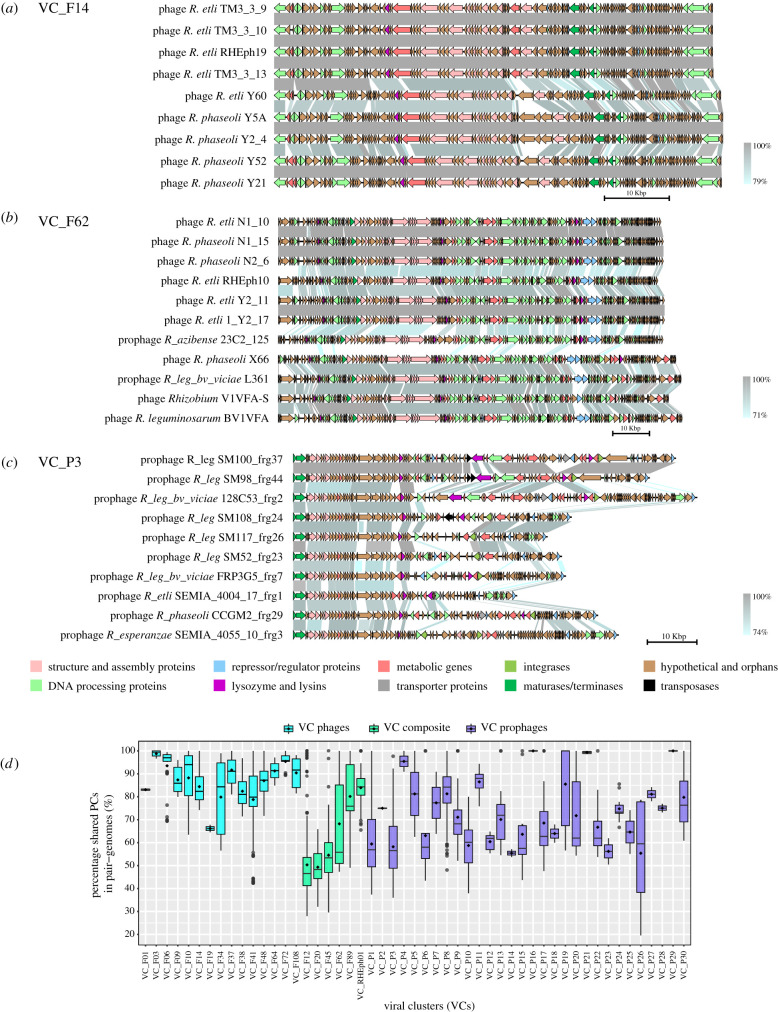
Figure 2.
Synteny variation between phages and prophages in VCs. (a) VC_F14 (lytic phages only); (b) VC_F62 (nine predicted temperate phages and two prophages); (c) VC_P3 (prophages only); and (d) percentage of shared PCs among pairs of phage genomes in the 51 VCs, with VCs comprising lytic phages only, composite, and prophages only, shown in different colours. Functional annotation is shown in the colour key, and the percentage of similarity between ORFs is shown in greyscale.
Of the 56 VCs, 20 VCs contained 106 of the 155 phages (68.4%) included in the network. These VCs did not include any prophage, and their phages were predicted to be virulent by the program PhageAI [28]. Additionally, six VCs were composite VCs that included at least one predicted prophage, and contained 22 phages isolated experimentally (14.4%). The composite VCs are formed by temperate phages as predicted by the PhageAI program. A further 30 VCs were composed mostly by predicted prophages and four isolated phages. The remaining 23 phages were singletons without relationships with the other phages.
Several phages from the RefSeq viral database were incorporated into the 56 Rhizobium phage VCs. Five VCs were formed exclusively by very similar lytic phages of Sinorhizobium and Agrobacterium (electronic supplementary material, table S4). Although, the phages P10VF, PRL2RES, RL38J1 from Rhizobium leguminosarum, phiM9 from Sinorhizobium and Atu_ph04 of Agrobacterium were connected with six other phages from Rhizobium etli included in VC_F41, they had the lowest edge weight in the cluster but greater than 60, indicating that phages in this VC have diversified to infect a broad range of species. Some phage genomes that were considered singletons in the previous analysis of ANI [22] were grouped into VCs. For instance, VC_F19 contained the phages X2-24 from Sinorhizobium americanum (Xoxocotla, México) and the phage P106B from Rhizobium gallicum (Saskatchewan, Canada). VC_F108 contained the phage i4 from Rhizobium phaseoli (from Argentina) and the phages L338C from R. leguminosarum (Saskatchewan, Canada) and P11VFA [24]. Other VCs contained the phage ort11 from Sinorhizobium (VC_F34) and the phage 16-3 from Rhizobium (VC_F20), which were from the RefSeq database.
(b) . Prophages are widespread in Rhizobium
To determine the extent of prophages in the Rhizobium genomes, we predicted prophages using VIBRANT for 612 Rhizobium genomes selected based on their completeness [27]. Of the 657 predicted prophages, 124 were high-quality, 146 were medium-quality and the rest (387) were low-quality. As we aimed to identify prophages with the highest probabilities of being complete, we only used the 270 high- and medium-quality prophage predictions in the subsequent analysis. The 270 prophages were located in genome segments of 20–100 kb, but more frequently around 40–50 kb (electronic supplementary material, figure S3). The length and gene content matched the estimated size distribution of the isolated Rhizobium phages. Therefore, we assumed that the predictions represented the most likely prophages in the Rhizobium genomes assessed (electronic supplementary material, table S2).
To assess the genome similarity relationships between prophages and phages, all 270 prophages were incorporated into the vConTACT network as described above (figure 1). There were 191 prophages in VCs that were either partly (six VCs) or exclusively (30 VCs) formed by prophages, while 79 prophages were singletons (figure 1; electronic supplementary material, table S4).
Prophages from diverse geographical origins had strong relationships with certain Rhizobium phages, expanding VC_F12, VC_F89, VC_F20, VC_F45, VC_F62 and VC_RHEph01. For instance, VC_F12 is composed of a phage isolated in México (TM3_3_3), a phage isolated in Argentina (N28) and 27 predicted prophages from diverse strains of R. leguminosarum and R. phaseoli isolated worldwide (electronic supplementary material, table S4). The addition of the 27 prophages to VC_F12 indicates successful expansion of its host range.
Isolated phages that were predicted as temperate were also found as prophages in six VCs (electronic supplementary material, table S4). For instance, VC_RHEph01 includes the previously characterised REHph01 phage, which infects a broad spectrum of R. etli and R. phaseoli strains [23]. Our study shows that this phage belongs to a VC of temperate phages that are highly conserved in Rhizobium species (such as R. leguminosarum and Rhizobium anhuenhense), confirming the wide host range and the diversification of the members of this VC across Rhizobium species (figure 1; electronic supplementary material, table S4).
To determine the range of distribution of species that harbour the prophages and isolated phages in VCs, all of them were mapped onto the dnaB-recA based maximum-likelihood bacterial phylogenetic tree (made with MEGA7). Although a few VCs appeared rather constrained in geography and host range (e.g. VC_F01, VC_F03 and VC_F06) (electronic supplementary material, figure S4a), the results generally suggest that some isolated phage and predicted prophage clusters are dispersed in a broad range of Rhizobium species (electronic supplementary material, figure S4b,c).
(c) . Higher synteny in VC_phage genomes than in VC_prophage genomes
To investigate the colinearity among the VC_phage genomes, we conducted pairwise comparisons using BLASTn. We discovered that the 15 VC phages (comprising experimentally isolated phages but no prophages) had highly conserved gene content and order, but less conserved genetic identity (72–100%) (figure 2). Geographical distance and local adaptation may explain the divergence exhibited by isolates from different parts of the world [22] (figure 2). However, VC_F14 contains highly similar phages, isolated in two nearby agriculture plots in México (Tepoztlán and Yautepec) [22] (figure 2a). The synteny analysis revealed only a single indel site in the middle of the map, which was occupied by an annotated estearase in the case of the phages from Tepoztlán (e.g. pTM3_3_9) and by hypothetical genes in the phages from Yautepec (e.g. pY5A).
The synteny maps showed absence of DNA rearrangements (e.g. inversions) or insertions within the virion structure- and replication-related genes. This is clear in VC_F62, which contains similar phages that were isolated in México and Argentina, two phages from Canada and two prophages in Rhizobium azabensis 23C2, a strain isolated in Tunisia [36], and in R. leguminosarum bv viciae L361. Although the prophage maintains the essential scaffold for phage structure and replication, its hypothetical gene content was different from the gene content of the phages. Additionally, the pX66 phage isolated from R. phaseoli INC2-5 in México shared fewer PCs with other phages while preserving its modular structure.
By contrast to the isolated phages, the prophages in Rhizobium seemed to have lost more genes. The VC_prophages had lower percentages of shared PCs than the VC phages (median less than 60%) and showed more variability (figure 2d). Despite the conservation among prophages of the proteins required for virion synthesis and assembly and DNA processing, several alterations in synteny (loss of hypothetical genes and insertions related to transposases and integrases) had occurred. The loss of synteny in the VC_P3 prophages in R. leguminosarum strains suggests that these prophages were subjected to significant recombination events in their evolutionary history. Furthermore, the remaining genes in prophages had ratios of non-synonymous to synonymous mutations (Ka/Ks) < 1 that suggest that they are subject to purifying selection (electronic supplementary material, figure S5).
(d) . Phage phylogeny
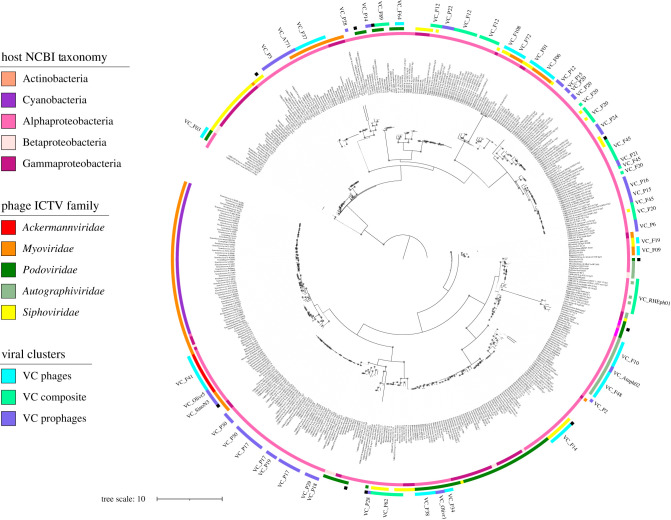
To understand the evolutionary relationships and taxonomy of Rhizobium phages, we constructed a phylogenetic tree using the maximum-likelihood method (in IQ-Tree software), based on the TerL protein encoded by the genomes in the vConTACT network (figure 1). The phylogenetic tree had viral clades with very close evolutionary relationships but separated from each other by long branches. It is likely that the tree topology reflects the insufficient sampling of phages of the α-proteobacteria class.
The placement of most of the phages in the TerL-based phylogenetic tree concurred with their assignments to International Committee on Taxonomy of Viruses (ICTV) families, which are based on tail morphology. However, some phages were placed in the same clade but predicted to correspond to different ICTV families (figure 3). Members of the, recently, incorporated ICTV families Ackermannviridae and Autographiviridae often appeared, according to the phylogenetic tree, to be related to Myoviridae and Podoviridae, respectively (figure 3, middle circle).
Figure 3.
Phage phylogeny and VCs distribution in clades. At the centre, the phage TerL—phylogenetic tree of 485 proteins, constructed using the IQ-Tree by maximum likelihood (see Material and Methods). The concentric circles indicate: inner, host NCBI taxonomy; middle, phages ICTV taxonomy; outer, VCs. The left inset indicates with colour code the distinct taxonomic ranks and VC types. Black dots in the branches of the tree indicate bootstrap greater than 75%. A lineal version of the TerL phylogeny is available to consult the details of the phylogeny (electronic supplementary material, figure S6).
To assess the association between clades and VCs, the members of VCs obtained using vConTACT were mapped onto the TerL-based phylogenetic tree. To this end, we defined a monophyletic group as that with boostrap greater than 75%, and looked for the distribution of members of VC clusters in single or separated clades. We were able to associate only 41 out of 56 VCs with the TerL clades. Most of the VCs were located in discrete viral clades (figure 3, third outermost circle; electronic supplementary material, figure S6). All the VCs formed by isolated phages belong to single TerL clades; five of six VCs composed of both isolated phages and predicted prophages showed congruency with clades; and only 15 out of 30 VCs, exclusively formed by predicted prophages, were in congruent clades (electronic supplementary material, table S7). Fifteen VCs with prophages were not analysed because the corresponding TerL proteins were excluded from the phylogeny owing to the BlastX cut-off used to select them.
Only three VCs (VC_F20, VC_P15 and VC_P28) were located in different clades of the phylogenetic tree, indicating that either phage recombination may have distorted the clustering results, or their association depend on the kind of phylogenetic marker used. To further evaluate this aspect, we did a phylogeny with the MCP protein and a VIP-Tree based on the presence and absence of proteins. Although the MCP phylogeny showed five separated VCs in distinct clades, the VIP-Tree agreed in most of the groups with the VCs (electronic supplementary material, table S7, figures S7 and S8). Clades either from TerL or MCP phylogenies, agreed well with the shared genomic structure indicated by the VCs (electronic supplementary material, table S7). Therefore, we argue that VCs have phylogenetic signals that indicate its process of diversification from a single ancestor.
In the recent ICTV release, only three genera of Rhizobium phages, two for Sinorhizobium and three for Agrobacterium have an approved status [37] (electronic supplementary material, tables S3 and S6). Two genera of lytic phages of Rhizobium named Cuernavacavirus and Rigallicvirus, agree with the VC_F48 and VC_F19 detected in the vConTACT network (electronic supplementary material, table S6). The other genus is Paadamvirus, which is a family of temperate phages formed with nine prophages and three phages from R. etli and R. leguminosarum.
Formerly, the ICTV proposed three other genera that included Rhizobium phages but they are no longer supported. However, we found 15 VCs that can be considered as new genera of Rhizobium phages. Among them, the VC_F37 that include the RHEph04 and -06 (formely Rheph4virus, Kleczkowskavirus), and the VC_F62 that forms a family of temperate phages similar to RHEph10 (formerly in the Nickievirus genus). The phage Nickie of Pseudomonas is unrelated to the VC_F62. It had a low number of shared PCs and edge values of less than 60, with genomes of the cluster.
4. Discussion
This study unveils novel phage taxa in symbiotic Rhizobium bacteria, their genomic relationships and their occurrence across Rhizobium species and strains. Our data reveal a complex network of structural and phylogenetic relationships, probably owing to coevolution and everchanging bacteria–phage interactions. Most of the 425 phages and prophages analysed here formed coherent clusters of relatively closely related phage genomes in terms of both structure and phylogeny. Furthermore, several phages and their prophage counterparts were found in various Rhizobium species and strains. Our results indicate that the identified VCs represent discrete taxonomic units and, most probably, units of evolution. Therefore, the VCs might be deemed taxa that are equivalent to genera or even species.
Evidence at the molecular level regarding viral species has been elusive owing to the lack of universal genes for phylogenetic reconstruction and pervasive genome mosaicism [18]. We showed that a given VC, defined based on PCs related to the genomes in the VC, represents a structural unit. The conserved patterns of synteny between isolated phage and prophage genomes clustered in VCs suggest that these isolated phages and predicted prophages might have diverged either recently or are under purifying selection. Still, in the TerL-based phylogenetic tree, most of the members of each VC clustered into a single clade, meaning that they have a single phylogenetic origin. However, the members of some VCs (VC_F20, VC_P15 and VC_P28) were divided into different clades. This suggests that the phages in these VCs had undergone recombination, which had rarely been studied until recently [18,38].
Discrete clusters of phages have been observed among phages associated with various bacterial species [39–41]. In cyanophage populations, clusters of closely related phages from a single Synechococcus strain were identified based on whole-genome ANI of about 99% [39]. This indicates that vertical evolution drove the diversification of these clusters of phages. By contrast, mycobacteriophages display a continuum of nucleotide diversity and phage clusters that exhibit shared genes between the clusters [40,42]. The mosaic genomes of mycobacteriophages originated as a result of HGT and recombination, thus reticulate evolution might explain the diversification of this group [43]. The vConTACT network reconciles both perspectives based on the statistical significance of phage genomic relationships within and between VCs. However, despite the robustness of the clustering methods used by vConTACT, the MCL inflation parameters may affect the size of the VCs and the identity of the included PCs. In the Rhizobium phage network, recombination appears to have only slightly distorted the VCs, except for in the abovementioned VCs. Therefore, the VCs represent genomic units of structure and evolution, which is relevant to understand the phage speciation process.
The spatial structure of Rhizobium-phage interactions indicate that they are locally adapted to their host bacteria. Sympatric isolation provides evolutionary opportunities for genetic disequilibrium between phage populations, as shown by the local adaptation at small and large geographical scales [22]. Phage host ranges vary from highly specialized to generalized. For instance, some Rhizobium phages infected only 2.2% of tested Rhizobium hosts (which were strains of the closely related species R. etli and R. phaseoli), while others infected up to 92.6% [22]. The host range of a phage is in part predictable based on host phylogeny, with closely related Rhizobium species being more likely to share phages than more distantly related Rhizobium species. However, we lack data on the extent to which individual phages in a VC share hosts. VCs can involve members with distinct host ranges. Members of VCs may be in the process of diverging from each other through specialization related to specific species and genera (e.g. VC_F62). Some of the VCs described here were found worldwide and in different species of Rhizobium and Sinorhizobium. It is likely that cosmopolitan phages will be recurrently found in diverse settings associated with symbiotic nitrogen-fixing bacteria. To improve our understanding, we need to sample phages from a broader range of hosts and conduct more comprehensive host range assessments.
Only a small proportion of the Rhizobium prophages were complete or almost complete, while the rest had preserved few of the essential genes required to enter a lytic cycle; the latter are known as prophage remnants, or ‘grounded’ phages, as described recently [12]. The scarcity of complete prophages in bacterial genomes may be explained by the cost involved in maintaining a harmful element, which at any moment can become activated and annihilate the cell. Thus, prophages become grounded by the selection of mutations that disrupt their lytic cycle. The lysogen has the advantage of being immune to reinfection and of harbouring ecologically relevant genes (e.g. antibiotic resistance genes, toxin genes and AMGs) [12,27,44]. The prophages may also serve as targets related to rearrangements and recombination with foreign mobile elements, providing variability for the bacterial population. The benefits to Rhizobium of harbouring prophages is not obvious from the predicted AMGs associated with them [27].
In this work, Rhizobium predicted prophages were less conserved than isolated phages experimentally isolated, judging by the synteny results and the gain and loss of genes. However, it is possible that the phage isolation process may create bias towards lower diversity, and under-represent the hidden virus diversity. Relatedly, prophage genes are under natural selection like any other housekeeping gene, and gene deletion allows prophages to remain in the bacterial genome [45]. Our analysis of the ratios of non-synonymous to synonymous substitutions (Ka/Ks) in three VCs comprising Rhizobium prophages suggested that the remnant phage genes are under selection like bacterial housekeeping genes (electronic supplementary material, figure S5). This is in agreement with previous results [11,45].
Rhizobium phages are only distantly related to other known groups of phages registered in the RefSeq viral database and the ICTV taxonomy. This is expected, owing to the poor representation in databases of the phages of α-proteobacteria and Rhizobiaceae. Although there are only three Rhizobium phage genera in the ICTV classification, our results indicate that 15 VCs (with high PC sharing probabilities; edge scores: 60–300) may represent new genera.
Although the ICTV has included genomic standards in its criteria [46], the wide diversity of phages and their low representation for some bacterial species prevent a consistent taxonomy. Additional efforts to obtain massive phage genomic information from the rhizosphere of various legumes may open a new insight into the diversity of Rhizobium-phage communities. We hope that modern approaches may recover the long history of Rhizobium phages given their importance for symbiotic association with legumes in sustainable agriculture [47].
Acknowledgements
We thank José Espíritu, Víctor del Moral, Gabriela Guerrero and Alfredo Hernández for support in perl scripts and software installation. Thanks to Gamaliel López-Leal for his help in the VirSorter program. We are very grateful to the international bioinformatics community that puts its codes and programs used in this work, freely available. Thanks are also extended to the reviewers for their valuable comments.
Data accessibility
The phage genome sequences were uploaded to GenBank. Accession numbers are provided in the electronic supplementary material, table S1. All data and perl programs are available if requested.
Authors' contributions
R.I.S.: data acquisition, network and phylogenetic analysis, synteny maps, manuscript review; P.B.: data acquisition, prediction and analysis of prophages, synteny maps, manuscript review; J.V.C.: data acquisition, literature review, drafting and editing of draft manuscripts; V.G.: data analysis, writing and editing of draft manuscripts, funding.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Support Program for Research and Technological Innovation Projects at Universidad Nacional Autónoma de México (PAPIIT-UNAM) (grant no. IN209817 to V.G.); Directorate General for Academic Personnel Affairs at UNAM (DGAPA-UNAM) (2016–2018 Postdoctoral Scholarship, to J.V.C.); J.V.C. was partially supported by National Science Foundation (NSF) (grant nos. DEB-1457508, IOS-1759048, to E.L. Simms).
References
- 1.Al-Shayeb B, et al. 2020. Clades of huge phages from across Earth's ecosystems. Nature 578, 425-431. ( 10.1038/s41586-020-2007-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux S, et al. 2016. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 537, 689-693. ( 10.1038/nature19366) [DOI] [PubMed] [Google Scholar]
- 3.Bouvier T, del Giorgio PA. 2007. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9, 287-297. ( 10.1111/j.1462-2920.2006.01137.x) [DOI] [PubMed] [Google Scholar]
- 4.Morella NM, Gomez AL, Wang G, Leung MS, Koskella B. 2018. The impact of bacteriophages on phyllosphere bacterial abundance and composition. Mol. Ecol. 27, 2025-2038. ( 10.1111/mec.14542) [DOI] [PubMed] [Google Scholar]
- 5.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67, 238-276. ( 10.1128/mmbr.67.2.238-276.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canchaya C, Fournous G, Brussow H. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53, 9-18. ( 10.1111/j.1365-2958.2004.04113.x) [DOI] [PubMed] [Google Scholar]
- 7.Lawrence JG, Ochman H. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl Acad. Sci. USA 95, 9413-9417. ( 10.1073/pnas.95.16.9413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penades JR, Christie GE. 2015. The phage-inducible chromosomal islands: a family of highly evolved molecular parasites. Annu. Rev. Virol. 2, 181-201. ( 10.1146/annurev-virology-031413-085446) [DOI] [PubMed] [Google Scholar]
- 9.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472-482. ( 10.1038/nrmicro2802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fillol-Salom A, Martinez-Rubio R, Abdulrahman RF, Chen J, Davies R, Penades JR. 2018. Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J. 12, 2114-2128. ( 10.1038/s41396-018-0156-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bobay LM, Touchon M, Rocha EP. 2014. Pervasive domestication of defective prophages by bacteria. Proc. Natl Acad. Sci. USA 111, 12 127-12 132. ( 10.1073/pnas.1405336111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramisetty BCM, Sudhakari PA. 2019. Bacterial ‘grounded’ prophages: hotspots for genetic renovation and innovation. Front. Genet. 10, 65. ( 10.3389/fgene.2019.00065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511-1520. ( 10.1038/ismej.2017.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obeng N, Pratama AA, Elsas JDV. 2016. The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol. 24, 440-449. ( 10.1016/j.tim.2015.12.009) [DOI] [PubMed] [Google Scholar]
- 15.Harrison E, Brockhurst MA. 2017. Ecological and evolutionary benefits of temperate phage: what does or doesn't kill you makes you stronger. Bioessays 39, 1700112. ( 10.1002/bies.201700112) [DOI] [PubMed] [Google Scholar]
- 16.Peterson AT. 2014. Defining viral species: making taxonomy useful. Virol. J. 11, 131. ( 10.1186/1743-422X-11-131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson D. 2004. Phage taxonomy: we agree to disagree. J. Bacteriol. 186, 7029-7031. ( 10.1128/JB.186.21.7029-7031.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bobay LM, Ochman H. 2018. Biological species in the viral world. Proc. Natl Acad. Sci. USA 115, 6040-6045. ( 10.1073/pnas.1717593115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iranzo J, Krupovic M, Koonin EV. 2017. A network perspective on the virus world. Commun. Integr. Biol. 10, e1296614. ( 10.1080/19420889.2017.1296614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang B, et al. 2019. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 37, 632-639. ( 10.1038/s41587-019-0100-8) [DOI] [PubMed] [Google Scholar]
- 21.Kogay R, Neely TB, Birnbaum DP, Hankel CR, Shakya M, Zhaxybayeva O. 2019. Machine-learning classification suggests that many alphaproteobacterial prophages may instead be gene transfer agents. Genome Biol. Evol. 11, 2941-2953. ( 10.1093/gbe/evz206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Cauwenberghe J, Santamaría RI, Bustos P, Juárez S, Ducci MA, Figueroa Fleming T, Etcheverry AV, González V. 2021. Spatial patterns in phage-Rhizobium coevolutionary interactions across regions of common bean domestication. ISME J. 15, 2092-2106 ( 10.1038/s41396-021-00907-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santamaría RI, et al. 2014. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl. Environ. Microbiol. 80, 446-454. ( 10.1128/AEM.02256-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halmillawewa AP, Restrepo-Cordoba M, Yost CK, Hynes MF. 2015. Genomic and phenotypic characterization of Rhizobium gallicum phage vB_RglS_P106B. Microbiology 161, 611-620. ( 10.1099/mic.0.000022) [DOI] [PubMed] [Google Scholar]
- 25.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068-2069. ( 10.1093/bioinformatics/btu153) [DOI] [PubMed] [Google Scholar]
- 26.Grazziotin AL, Koonin EV, Kristensen DM. 2017. Prokaryotic virus orthologous groups (pVOGs): a resource for comparative genomics and protein family annotation. Nucleic Acids Res. 45, D491-D498. ( 10.1093/nar/gkw975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieft K, Zhou Z, Anantharaman K. 2020. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90. ( 10.1186/s40168-020-00867-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piotr Tynecki AG, Kazimierczak J, Jadczuk M, Dastych J, Onisko A. 2020. PhageAI - bacteriophage life cycle recognition with machine learning and natural language processing. BioRxiv ( 10.1101/2020.07.11.198606) [DOI] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498-2504. ( 10.1101/gr.1239303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009-1010. ( 10.1093/bioinformatics/btr039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González V, et al. 2019. Phylogenomic Rhizobium species are structured by a continuum of diversity and genomic clusters. Front. Microbiol. 10, 910. ( 10.3389/fmicb.2019.00910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113. ( 10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher G, Tamura K, Kumar S. 2020. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237-1239. ( 10.1093/molbev/msz312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268-274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S. 2017. ViPTree: the viral proteomic tree server. Bioinformatics 33, 2379-2380. ( 10.1093/bioinformatics/btx157) [DOI] [PubMed] [Google Scholar]
- 36.Mnasri B, Liu TY, Saidi S, Chen WF, Chen WX, Zhang XX, Mhamdi R. 2014. Rhizobium azibense sp. nov., a nitrogen fixing bacterium isolated from root-nodules of Phaseolus vulgaris. Int. J. Syst. Evol. Microbiol. 64, 1501-1506. ( 10.1099/ijs.0.058651-0) [DOI] [PubMed] [Google Scholar]
- 37.International Committee on Taxonomy of Viruses (ICTV). See https://ictv.global/taxonomy/.
- 38.Meier-Kolthoff JP, Uchiyama J, Yahara H, Paez-Espino D, Yahara K. 2018. Investigation of recombination-intense viral groups and their genes in the Earth's virome. Sci. Rep. 8, 11496. ( 10.1038/s41598-018-29272-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. 2014. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature 513, 242-245. ( 10.1038/nature13459) [DOI] [PubMed] [Google Scholar]
- 40.Pope WH, et al. 2015. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 4, e06416. ( 10.7554/eLife.06416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159, 406-414. ( 10.1016/j.resmic.2008.03.005) [DOI] [PubMed] [Google Scholar]
- 42.Hatfull GF. 2010. Mycobacteriophages: genes and genomes. Annu. Rev. Microbiol. 64, 331-356. ( 10.1146/annurev.micro.112408.134233) [DOI] [PubMed] [Google Scholar]
- 43.Hatfull GF, et al. 2010. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397, 119-143. ( 10.1016/j.jmb.2010.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 147. ( 10.1038/ncomms1146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bobay LM, Rocha EP, Touchon M. 2013. The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 30, 737-751. ( 10.1093/molbev/mss279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmonds P, et al. 2017. Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 15, 161-168. ( 10.1038/nrmicro.2016.177) [DOI] [PubMed] [Google Scholar]
- 47.Vandecaveye SC, Katznelson H. 1936. Bacteriophage as related to the root nodule bacteria of alfalfa. J. Bacteriol. 31, 465-477. ( 10.1128/jb.31.5.465-477.1936) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The phage genome sequences were uploaded to GenBank. Accession numbers are provided in the electronic supplementary material, table S1. All data and perl programs are available if requested.