Abstract
Members of the agrobacteria–rhizobia complex (ARC) have multiple and diverse plasmids. The extent to which these plasmids are shared and the consequences of their interactions are not well understood. We extracted over 4000 plasmid sequences from 1251 genome sequences and constructed a network to reveal interactions that have shaped the evolutionary histories of oncogenic virulence plasmids. One newly discovered type of oncogenic plasmid is a mosaic with three incomplete, but complementary and partially redundant virulence loci. Some types of oncogenic plasmids recombined with accessory plasmids or acquired large regions not known to be associated with pathogenicity. We also identified two classes of partial virulence plasmids. One class is potentially capable of transforming plants, but not inciting disease symptoms. Another class is inferred to be incomplete and non-functional but can be found as coresidents of the same strain and together are predicted to confer pathogenicity. The modularity and capacity for some plasmids to be transmitted broadly allow them to diversify, convergently evolve adaptive plasmids and shape the evolution of genomes across much of the ARC.
This article is part of the theme issue ‘The secret lives of microbial mobile genetic elements’.
Keywords: evolution, plasmids, symbiosis, virulence, agrobacteria, rhizobia
1. Introduction
Plasmids mediate horizontal gene transfer (HGT) and have major roles in the evolution of bacteria, such as causing transitions in lifestyles [1]. Plasmids are highly recombinogenic and can generate new and diverse combinations by shuffling genes with other replicons [2–5]. However, the degree to which plasmids can diversify bacteria probably has boundaries, as there are barriers to HGT and features that constrain interactions among plasmids [6–10]. Understanding the processes that promote and limit plasmid diversity is crucial for understanding the evolution of traits they encode.
The agrobacteria–rhizobia complex (ARC) is a genus-level group of bacteria [11]. Members of agrobacteria are recognized as plant pathogens [12]. The three major lineages, called biovars (BV1–3), are inferred to have emerged independently and at different times in the history of the ARC [13]. BV1 is diverse, subdivided into genomospecies that approximate species-level groups, and sister to narrow-host range species of agrobacteria [14]. BV2 is monophyletic and sister to the BV2-like clade while BV3 represents multiple species-level groups [11,15]. Members of rhizobia are known as nitrogen-fixing symbionts of legumes. Those in the ARC form several polyphyletic clades, which are interspersed with clades of agrobacteria [11]. The separation of pathogens and mutualists between agrobacteria and rhizobia in the ARC is not absolute as exceptions have been identified [11,16,17].
ARC members have multipartite genomes that include a primary and secondary chromosome and often multiple plasmids [18,19]. With the exception of the primary chromosome, most other replicons have a repABC replication locus and may have trb and tra loci necessary for mediating inter-bacterial conjugation [19–21]. Even secondary chromosomes, called ‘chromids’, have repABC genes. Based on analysis of chromid sequences from one strain representative of each of the three distantly related biovars, it was hypothesized that chromids originated from a common ancestral plasmid [18,19,22]. Among plasmids of ARC members, the best characterized are oncogenic and symbiosis plasmids necessary for virulence and symbiotic nitrogen fixation, respectively [12,23]. Oncogenic plasmids are subdivided into tumourigenic (Ti) and rhizogenic (Ri) plasmids, associated with crown gall and hairy root disease, respectively [12]. Symbiosis plasmids have genes necessary for triggering nodulation and fixing nitrogen and often genes that can influence the host range [24]. Most other accessory plasmids in members of the ARC are less understood but are broadly implicated in catabolism [25].
Oncogenic plasmids have a number of conserved genes necessary for virulence [12]. The virA and virG genes are necessary for detecting plant-derived signals and regulating vir gene expression [26,27]. The virB genes encode an apparatus for inter-kingdom gene transfer while virD1, 2 and 4 genes are important or necessary for processing T-DNAs, regions of oncogenic plasmids transferred into host cells [28]. Genes in the virC operon assist with T-DNA processing while virE1 and virE2 have functions in protecting the single-stranded T-DNA in plant cells and importing it to the host nucleus [29,30]. Some Ri plasmids lack virE and instead have GALLS, which despite lacking sequence similarity, can complement a virE2 mutant [31,32]. The T-DNA contains oncogenes, such as tms1, that cause dysregulation of plant hormone levels and disease symptoms. T-DNAs also include genes necessary for producing opines, which are nutrients for the pathogen and signals for plasmid replication and conjugation [33].
Modularity is a major driver of the evolution of plasmids [11]. Our goal was to determine evolutionary relationships among ARC replicons, with a focus on oncogenic plasmids. We sequenced strains of agrobacteria previously identified as having unusual characteristics or as carrying unusual oncogenic plasmids [34–37]. We grouped these plasmid sequences with over 4000 other plasmid-derived sequences in a network and used relationships revealed as components and cliques to infer the evolution of oncogenic plasmids.
2. Results
(a) . Agrobacteria-rhizobia complex plasmids are extremely diverse and complex
We sequenced genomes of agrobacteria predicted to have novel oncogenic plasmids and extracted the plasmid sequences (electronic supplementary material, table S1). We also mined genome sequences from 1251 members of the ARC and identified 4081 complete plasmid sequences and contigs predicted to correspond to plasmids (electronic supplementary material, tables S2 and S3). A network, constructed on the basis of similarities in k-mer signatures of sequences, yielded 939 distinct components, which are maximal connected subgraphs (figure 1). Within this dataset, most plasmid and contig sequences are grouped into few components while most components are poorly represented, as more than two-thirds are represented by only one plasmid sequence. The diversity is consistent with findings from phylogenetic analysis of translated sequences of repC and indicated that these plasmids, along with chromids in the ARC, are derived from a large and ancient family (electronic supplementary material, figure S1). Plasmids with finished sequences range from 0.009 Mb–2.58 Mb (mean 0.342 Mb; median 0.204 Mb), though some of the largest may be chromids that were not identified on the basis of relationships to previously characterized chromids and were not filtered out of the dataset.
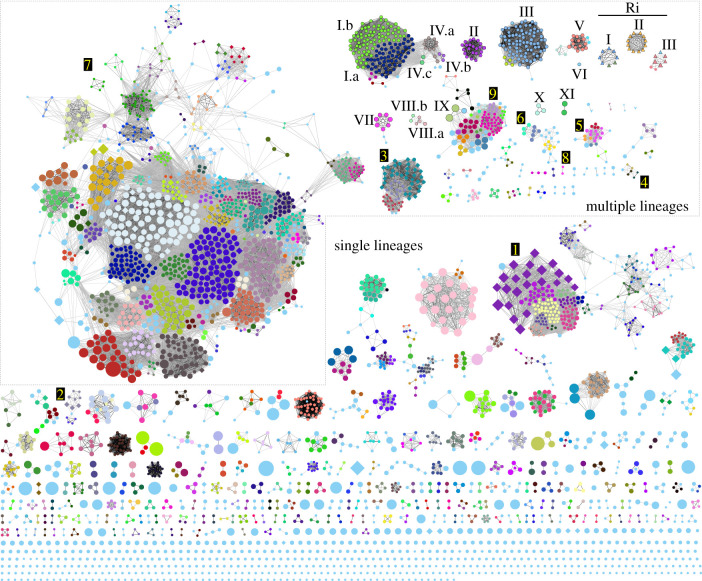
Figure 1.
Plasmids of the ARC form diverse network components and cliques. Weighted undirected network of plasmids and contigs with a repABC locus. Plasmids and contigs are represented as nodes, scaled to represent their size and are linked into network components according to similarity (determined on the basis of k-mer signatures; minimum Jaccard similarity of 0.1 with darkest edges indicating greatest similarity). Nodes are coloured according to clique and shaped according to trait (circles, types I–XI Ti plasmids or accessory plasmids; triangles, types I–III Ri plasmids; inverted ‘V’ shape, only vir gene-containing plasmids; diamonds, symbiosis plasmid). Boxed region includes network components with plasmids distributed across multiple lineages, oncogenic and vir gene-containing plasmids. Numbered network components are referred to in the text.
Components can represent a single family in which all members are closely related with numerous homologous plasmid regions and/or clusters of families more loosely linked by fewer homologous plasmid regions. To distinguish between these, we analysed components for cliques, which are subgraphs in which all members are connected to each other [38]. To determine the degree to which cliques reflect evolutionary relationships of plasmids, we examined relationships of cliques with oncogenic plasmids that were previously grouped into types of evolutionarily related sequences (figure 1; electronic supplementary material, table S2; [11]). Outcomes of the two methods were largely congruent and only minor differences were identified potentially because of differences in thresholds used to group plasmids or inclusion of partial plasmid sequences.
We next determined relationships among cliques, components and lineages of bacteria. To this end, bacterial members of the ARC were assigned to one of 59 lineages, which approximate a symbiovar or biovar (electronic supplementary material, figure S2 and table S3). In general, and in this dataset, components, including several with accessory plasmids of agrobacteria and symbiosis plasmids of rhizobia, are limited mostly to single bacterial lineages (figure 1; electronic supplementary material, figure S2). Several components of accessory plasmids associated with BV1 are even more limited in being restricted to specific genomospecies. The pSymA symbiosis plasmids of component 1 are restricted to Ensifer (lineage 35) and are within a component with loose connections to several cliques of accessory plasmids. The relationships between cliques could reflect genetic exchange between plasmids of different cliques or connections to partially assembled pSymA plasmids. Component 2 is particularly notable because the plasmids, which include those conferring symbiotic nitrogen fixation, are limited to strains of BV2-like, a species-level group that includes members with oncogenic plasmids [11].
In this dataset, only 37 components have plasmids present in different bacterial lineages (figure 1). Components 3–5 have symbiosis plasmids that are found in more than one bacterial lineage and some accessory plasmids of agrobacteria are broadly distributed. For example, component 6 has plasmid members present in five bacterial lineages representing those of agrobacteria and rhizobia. Component 7 is the largest in the network and has many cliques, some of which are related by few connections and others with many connections. All but one plasmid of this component is present in strains of Rhizobium leguminosarum and Rhizobium phaseoli (lineage 51; figure 1; [39]). The features of component 7 suggest it consists of plasmids that are reshuffling genes, including those involved in symbiosis, across multiple families of plasmids that are largely limited to one bacterial lineage.
Oncogenic plasmids are some of the most broadly distributed plasmids in the dataset and are present across agrobacteria and rhizobia, different agrobacterial lineages and different species-level groups (figure 1; electronic supplementary material, figure S2). Type II Ri plasmids are present in lineage 53, a group with three newly sequenced strains of rhizobia isolated from hydroponically grown tomatoes (electronic supplementary material, figure S2). Ri plasmid-carrying rhizobia and strains in the Ochrobactrum genus have been previously shown to cause root mat disease to plants [40]. A type I.a Ti plasmid is present in a member of lineage 15 (Neorhizobium). As previously reported, some types of Ti and Ri plasmids are present in strains of both BV1 and BV2 [11]. However, the breadth of host range does not generalize to all oncogenic plasmids, as type IV and all type V Ti plasmids are mostly limited to BV3, and all type I Ri plasmids are limited to strains of BV1 [41]. It is unclear whether the broad distribution observed for types of oncogenic plasmid is unique, as most ARC plasmids are poorly sampled.
(b) . Relationships of important accessory plasmids of agrobacteria
We identified a plasmid related to pAgK84, which carries genes for agrocin 84, an inhibitor of leucyl–tRNA synthetase and an allele encoding a resistant variant of leucyl–tRNA synthetase (component 8; figure 1; [42,43]). The pAgK84 plasmid is in a BV2 strain used in biocontrol against agrobacteria that induce the synthesis and catabolism of agrocinopine opines [33,44]. The newly discovered plasmid is in strain CFBP5877 of genomospecies G6 of BV1. Alignment of the two plasmid sequences revealed few differences, none of which were predicted to affect the ability of strain CFBP5877 to produce agrocin 84. Strain CFBP5877 is also pathogenic and carries a type III Ti plasmid. A partially sequenced plasmid in pathogenic strain Bo542 also has homology to pAgK84 [43,45]. These two strains show that members of the pAgK84 family are present among agrobacterial populations, compatible with BV1 and BV2-like lineages and compatible with oncogenic plasmids. These revive concerns over the potential for continued applications of biocontrol agrobacteria to break down biocontrol.
Relationships among accessory plasmids may inform the origin(s) of chromids in the ARC. Members of BV2 have a replicon that is similar in composition to chromids, but it was predicted to lack essential genes and was thus not considered a true chromid [18]. Re-analysis revealed 10 and 30 homologues of genes, six of which are overlapping, identified as essential based on two separate TnSeq studies (electronic supplementary material, table S4; [46,47]). Regardless, the hypothesis that these chromid and chromid-like replicons emerged from a common ancestral plasmid predicts that those of sister lineages will have more homologous and colinear genes than those from more distantly related lineages, such as those previously analysed [18]. By contrast to predictions, chromids and chromid-like replicons of BV2, BV2-like and lineage 58, three closely related lineages, differ substantially in several regards (electronic supplementary material, figure S2; [11]). Most notable is that strains of BV2-like have two plasmids with approximately 72% and 38% of their regions homologous to those on the chromid-like molecule of a BV2 strain (electronic supplementary material, figure S3A–C). The two plasmids have 26 and four genes predicted to be essential and 19.4 and 51.2% of their regions are unique with no detectable homology to the genome of the reference BV2 strain. Members of lineage 58 have a chromid with 20 genes predicted to be essential, 37.5% of its regions homologous to the chromid-like replicon of the reference BV2 strain, and 47.9% with no homology to either chromid-like or chromosome of the reference strain. Of the essential genes, only seven are present on related molecules in all three strains representative of the closely related lineages (electronic supplementary material, table S4). In addition, genes homologous across replicons have little synteny and the degree to which they are shuffled is more similar to that observed in comparisons of more distantly related lineages (electronic supplementary material, figure S3D–F; [11]).
(c) . Virulence plasmid evolution
We next examined oncogenic plasmid types to understand how their evolutionary histories are shaped by others. Most of the oncogenic plasmid sequences obtained since Weisberg et al. [11] correspond to one of the previously defined types (figures 1 and 2a; electronic supplementary material, figure S4 and table S1; [11]). Twenty-one oncogenic plasmids sequenced after that study represent new types on the basis of network and phylogenetic analyses (figures 1 and 2a; electronic supplementary material, figures S5–S8 and table S1; [11,48]). Type VII and VIII Ti plasmids are present only in a specific lineage of agrobacteria (lineage 58; electronic supplementary material, figure S2; [34,49,50]). Type VIII Ti plasmids formed separate cliques but based on prior methods used to classify oncogenic plasmids, were defined as a and b subtypes (figure 1; electronic supplementary material, figure S4; [11]). Type IX Ti plasmids are large (0.590 Mb–1.0 Mb) and are present in one strain of BV2-like and two strains of genomospecies G1 in BV1 (electronic supplementary material, figure S2; [36]). Earlier studies of this type of Ti plasmid had identified a vir locus and only a small T-DNA lacking homologues of canonical oncogenes [37]. We identified a second larger T-DNA that has homologues of canonical oncogenes, but to more diverged homologues originally discovered in type VI Ti plasmids (figure 2d; electronic supplementary material, figure S7). These unusual oncogene homologues are unlike those of type I–V Ti plasmids and were previously predicted to have been acquired from non-agrobacterial strains [11]. These unusual oncogenes were not known at the time of earlier studies and could explain why previous efforts failed to identify the second T-DNA in type IX Ti plasmids [37]. Types X and XI Ti plasmids are present in narrow-host range species [51].
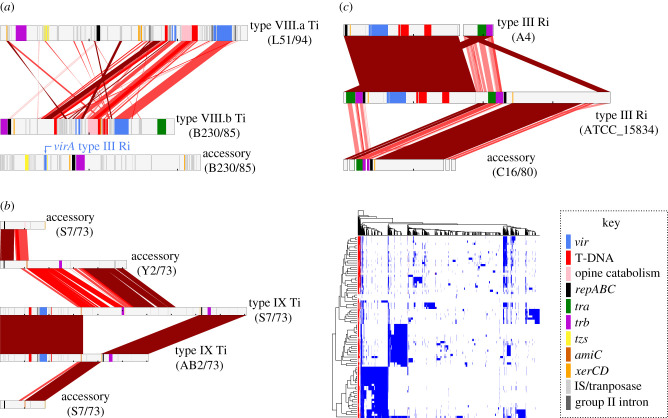
Figure 2.
The newly identified types of oncogenic Ti plasmids are mosaics of multiple plasmids. (a) Maximum-likelihood phylogeny for virD2. Clades with newly defined types of Ti plasmids are labelled. The alleles of type VII plasmids are numbered according to the locus they associate with. Strains that are described in the text are labelled in the tree. Black-coloured branches have UFBoot support greater than or equal to 95% and SH-aLRT support greater than or equal to 80%. Trees are midpoint rooted. Maps of the type (b) VII, (c) VIII and (d) IX Ti plasmids. The Ti plasmids from strains B21/90, L51/94 and S7/73, respectively, were used as the reference for each of the three types. Coloured bars indicate loci associated with virulence and plasmid replication. The three different vir loci of type VII Ti plasmids are distinguished. Unlabelled grey bars indicate genes predicted to encode an insertion sequence, transposase, type II intron or recombinase. Larger and more detailed representation of plasmid types are provided as supplemental figures (electronic supplementary material, figures S5–S7).
Type VII Ti plasmids are potentially recently formed types. They are large (0.384 Mb–0.455 Mb) mosaics probably derived from plasmids similar to type VI Ti, type III Ri and type IV.c Ti plasmids, which each contributed a partial vir locus (figure 2a,b; electronic supplementary material, figures S5 and S8). The vir 1 locus has most of the vir genes, but virD4, necessary for virulence, and the entire virE operon are absent. However, vir 2 and 3 loci have homologues of genes that are absent from vir 1 as well as genes redundant to those in vir 1. The vir 2 locus has virB, virC, virG and virD1–4 as well as the GALLS gene, which can fully substitute for the virE operon. The vir 3 locus has virD2–5 and the virE operon. Type VII Ti plasmids from strains isolated in the 1980s have three copies of virD2 while the plasmid from strain 1078, isolated in 2015, has two copies and lacks a region near the vir locus where a virD2 homologue would otherwise be present (figure 2a). T-DNAs and opine-encoding loci are also multiplexed and fragmented. One T-DNA is similar to the T-DNA that was previously identified as being the most common among oncogenic plasmids [11,52]. A second T-DNA contains only homologues of unusual tms2 and tms1 genes first discovered in type VI Ti plasmids. The third T-DNA contains only a predicted opine synthase-encoding gene. In addition, type VII Ti plasmids have unique arrangements of vir genes and T-DNAs and copies of mobile genetic elements often near and within these loci.
Type VIII Ti plasmids also have hallmarks of being mosaics of two or three oncogenic plasmids, one of which is an Ri plasmid (figure 2c; electronic supplementary material, figure S6). Type VIII plasmids have two partial vir loci in which one contains virA, virG and the virB, virC and virE operons and the other has virD1–4 genes. The virA homologues of most type VIII Ti plasmids are distantly related to those of other plasmid types (electronic supplementary material, figure S8A). By contrast, the virD2 homologues are most similar to those of type V and X Ti plasmids (figure 2a). Type VIII Ti plasmids have a T-DNA with rol as well as hypothetical protein- and opine synthase-encoding genes. A second T-DNA contains only homologues of unusual tms2 and tms1 genes first discovered in type VI Ti plasmids and tends to be associated with less-defined border sequences. The type VIII.a Ti plasmids also have a cluster of genes implicated in cytokinin biosynthesis and is present among some Ri plasmids, which could have been donors of the region [53–56].
Type VIII.b Ti plasmids are themselves predicted to be non-functional because they lack virA, a gene necessary for regulating the expression of vir genes, though we cannot exclude the possibility that a non-homologous regulatory system is present on type VIII.b Ti plasmids that can regulate virulence [27]. Nevertheless, strains with type VIII.b Ti plasmids are pathogenic [57]. We predict this is because they each have a virA homologue on a co-residing accessory plasmid. These accessory plasmids are related and like some Ri plasmids, have genes involved in cytokinin biosynthesis. Additionally, the virA genes are more similar to homologues of type III Ri plasmids than to those of type VIII.a Ti plasmids (electronic supplementary material, figure S8A).
We identified nine partial virulence plasmids and contigs each with a vir locus, but no identifiable oncogenes or typical T-DNAs in the genome sequences of their corresponding strains (figure 1; electronic supplementary material, table S1). These vir genes form unique clades in phylogenies, suggesting that their corresponding plasmids originated from those unrelated to those within this dataset (figure 2a; electronic supplementary material, figure S8). The vir genes of plasmids present in rhizobial strains CFN42, 3841 (plasmid pRL7), BR 10423, FH14, N122 and agrobacteria BV3 strain F2/5 are predicted to be complete and sufficient for carrying out T-DNA transfer and delivery of protein effectors. Notably, p42a of strain CFN42 is demonstrably functional when paired with a T-DNA-bearing plasmid [58]. Prior studies failed to identify a T-DNA on p42a, suggesting it is ‘unarmed’. Though we also could not identify homologues of known oncogenes, we and others have identified a single T-DNA border sequence and genes encoding an opine synthase [59]. We also identified additional T-DNA border sequences and a gene encoding trehalose-6-synthase on p42a. Homologues of those encoding the putative opine synthase and trehalose-6-synthase were also identified in the draft genome sequences of strains FH14 and N122. Unexpectedly, a homologue with 91% identity in sequence to the trehalose synthase-encoding gene is also present in a T-DNA of type X Ti plasmids. Trehalose is a natural disaccharide with potential as a source of energy but also with implications in pathogenicity and in affecting plant development [60,61]. The three other partial virulence plasmids of strains MHM7a, STM61555 and WSM1369 were predicted to have non-functional vir loci and are probably not sufficient by themselves in conferring pathogenicity. In each case, virC and a complete or partial virD locus was found on a small contig bordered by sequences of insertion sequence elements and no complete homologues of virA, virG, virE operon or GALLS could be confidently identified in their corresponding genome sequences. In addition, strain MHM7a has an incomplete virB operon.
Interactions with accessory plasmids have diversified oncogenic plasmids. The component with type VIII Ti plasmids has two cliques connected by three incompletely assembled plasmid sequences (figure 1). These plasmids and contigs were related on the basis of homologous and syntenic regions surrounding and including vir genes, opine catabolism genes and T-DNAs. Conversely, regions that include repABC as well as trb and tra loci are diverse and correspond to different plasmid backbones (figure 3a; electronic supplementary material, figure S1). The two subtypes are predicted to have evolved by recombining homologous virulence regions into the backbones of different accessory plasmids.
Figure 3.
Recombination between oncogenic and non-oncogenic plasmids yield diverse molecules. (a) Alignment of type VIII.a and VIII.b Ti plasmids. The accessory plasmid of strain B230/85 with virA is represented at the bottom. (b) Left: alignment of Ti type IX plasmids with donor accessory plasmids (shown twice). Right: heatmap showing gene homologue presence/absence patterns in plasmid members of network component 9. Red/blue and white lines indicate presence and absence, respectively, of a gene (column) in a plasmid (row). Red lines indicate presence of genes homologous to those in the non-oncogenic plasmid of strain S7/73. Rows and column clustering were generated based on Ward's clustering of binary distances. (c) Alignment of the type III Ri plasmids with contigs predicted to correspond to an accessory plasmid. Breaks in the diagram indicate partially assembled sequences. In all panels, red lines and blocks link regions that are similar between plasmids; darker colours indicate greater similarity. Plasmids are displayed as linear molecules with coloured blocks representing functional modules, recombination breakpoints and mobile genetic elements (see key). Tick-marks indicate increments of 0.1 Mb.
Type IX Ti plasmids are connected to cliques of accessory plasmids predominantly restricted to strains of genomospecies 1 in BV1 (component 9; figure 1). Type IX Ti plasmids are related to each other because of homologous and colinear virulence regions, but these plasmids also have large variable regions that separate them into different cliques (figures 2d and 3b; electronic supplementary material, figure S7). Mirroring this relationship, the accessory plasmids of component 9 each have a small, conserved region that circumscribes the repABC locus, but the accessory plasmids separated into multiple cliques because of large polymorphic regions (figures 1 and 3b). Alignments of type IX Ti plasmids and related accessory plasmids revealed homologous regions flanked by amiC and xerC gene loci. Among the accessory plasmids of component 9, amiC and xerC flank variable regions and could be recombination hotspots that cause-related plasmids to diversify and for oncogenic and accessory plasmids to converge, thereby connecting cliques within components.
Last, a type III Ri plasmid has integrated with an accessory plasmid (figure 1). The Ri plasmid of strain ATCC 15834 is larger than others of the same type and is connected to an accessory plasmid (figure 3c). Alignments demonstrated that a region flanking repABC, tra and trb is homologous and colinear to the accessory plasmid of strain C16/80. Conversely, the virulence region of the type III Ri plasmid of ATCC 15834 is homologous and colinear to a region of a type III Ri plasmid in strain A4. The accessory plasmid of C16/80 is also similar to another accessory plasmid of strain A4, which is predicted to have cointegrated with the Ri plasmid of A4 [62]. Strain C16/80 is also pathogenic, but it carries a type II Ri plasmid so recombination between accessory and oncogenic plasmid probably occurred in other strains.
3. Discussion
We constructed a comprehensive plasmid network to gain a more holistic understanding of the evolution of ARC members (figure 1). Interactions among plasmids have had significant impacts on pathogenic members. Reshuffling has yielded plasmids with new combinations of virulence genes sufficient for conferring pathogenicity to new lineages of agrobacteria. For example, the type VII Ti plasmids acquired vir genes from multiple and distantly related oncogenic plasmids yet strains with these plasmids have a sufficient set of virulence genes and are predicted to be pathogenic. For type VIII.b Ti plasmids, we suggest a scenario in which an ancestral Ti plasmid-bearing strain acquired an Ri plasmid, allowing for the loss of the original virA homologue from the ancestral type VIII.b Ti plasmid and loss of most virulence functions from an ancestral Ri plasmid. A similar natural bipartite relationship with virA located on a co-residing accessory plasmid was reported for strain 1D1609 [63]. However, in this strain, we suggest virA was acquired directly from the type II Ti plasmid co-residing in strain 1D1609, as virA of the accessory plasmid is homologous to those of other type II Ti plasmids (electronic supplementary material, figure S8A). Discovery of these plasmids confirm that vir alleles are intercompatible and demonstrate that reshuffling has little cost to virulence (figure 2b; electronic supplementary material, figure S5). Plasmid evolution has also yielded molecules competent in only some virulence functions. These include p42a and others similar to it that are like a hypothesized proto-oncogenic plasmid predicted to have the capacity to transform plants and confer fitness benefits to bacteria without causing disease symptoms [33]. The ARC also has partial virulence plasmids in which some or most virulence genes are absent. Despite being functionally incomplete, these partial virulence plasmids represent an important genetic reservoir and can be paired together or recombined to generate new combinations sufficient for pathogenicity.
Accessory plasmids impact the ecology of ARC members and evolutionary histories of other replicons, but their significance remains largely unknown [18,25,64–67]. The network provided a new framework for addressing their roles (figure 1). Recombination has yielded oncogenic plasmids with different backbones and novel genes with functions beyond pathogenicity, potentially impacting plasmid range and fitness of bacteria in diverse niches. Accessory plasmids can also capture virulence genes and diversify the genetic reservoir that contributes to the evolution of pathogenicity [63]. Recombination with symbiosis plasmids has been reported and the network suggests that processes influencing pathogenicity similarly shape nitrogen-fixing symbiosis [68,69]. Likewise, accessory plasmids have played an important role in the evolution of chromids [18]. Data cannot exclude the possibility that chromids evolved convergently, as allelic variation of chromid repABC loci, originally suggested to reflect reshuffling of the locus, also supports emergence of chromids from different plasmid backbones (electronic supplementary material, figures S1 and S3; [18]).
ARC members have critical roles in the health of plants and their ecosystems. These bacteria collectively have an enormously large and diverse set of replicons. Interactions among replicons drive convergent and divergent evolution, generating variation fundamental for robustness of traits. Interactions will continually result in the emergence, degradation and even re-emergence of oncogenic plasmids. Plasmids of the ARC are a genetic pool that diversify ARC members with unique combinations of functions to adapt them to various ecosystems and host species.
4. Methods
(a) . Genome sequencing, assembly and annotation
Strains of agrobacteria were obtained from the culture collections of Dr Larry Moore and the Oregon State University Plant Clinic. Previously described methods were used to extract genomic DNA as well as sequence, assemble and annotate genome assemblies [3]. Preparation of SeqWell libraries and sequencing on an Illumina HiSeq 3000 were done by the Center for Genome Research and Biocomputing (CGRB; Oregon State University, Corvallis, OR, USA). Some select strains (electronic supplementary material, table S1) were additionally sequenced on an Oxford Nanopore MinION, using the native genomic DNA library prep kit (LSK-109) with multiplexing on standard flowcells. For these, Illumina and Nanopore reads were used to generate hybrid assemblies.
Genome sequences of bacteria classified as members of the Rhizobiaceae and oncogenic plasmid sequences were downloaded from NCBI GenBank on 12 September 2019. Genome sequences of strains rho-1.1, rho-6.2 and rho-13.1 (Rhizobium tumorigenes) were also downloaded at this time [50].
(b) . Analyses and characterization of plasmids
Previously described methods were followed to identify and extract sequences of oncogenic plasmids from genome assemblies [11]. Complete or partial vir gene loci were also used to identify contigs corresponding to oncogenic plasmids. Oncogenic plasmids were classified following a previously described classification system [11]. Get_homologues v. 02062020 with the options ‘-M -t 0’ was used to cluster genes of oncogenic plasmids into orthologous groups [70]. The R package heatmap.plus was used to generate heatmaps of gene presence/absence [71]. The HMMER v. 3.3 program hmmsearch with the default options and a hidden Markov model (HMM) of T-DNA border sequences was used to identify T-DNA regions [11,72]. HMMs of translated virulence gene sequences from oncogenic plasmids of this dataset were built and used to verify the presence of vir genes on disarmed replicons. The program wgsim v. 0.3.1-r13 with the options ‘-N 50 000 -1 150 -2 150 -r 0 -R 0 –X 0 -e 0’ was used to generate simulated sequencing reads from genome assemblies for strains in which raw read data were not available in NCBI SRA [73]. Visuals of plasmid maps and associated information were generated for each novel plasmid type, using methods previously described [11].
For non-oncogenic plasmids, partial or complete sequences were identified on the basis of the presence of repABC genes. Translated sequences of alleles of repC from the type I.a Ti of strain C58, type VIII Ti of strain L51/94, and type X Ti of strain AF3.44 were used as queries in BLAST (v. 2.6.0 program blastp with the options ‘-max_target_seqs 5000’ and a minimum query coverage of 50%) searches. Increasing the maximum number of target sequences beyond the threshold of 5000 did not result in an increase in significant hits. Hits with less than 25% identity in amino acids were manually inspected to verify them as homologues of repC. Contigs corresponding to repC hits were retained as possible plasmids or plasmid fragments. Other non-repABC plasmids were extracted from completely assembled genome sequences and included in analyses. A tblastn search using translated alleles of nodC and fixC from strain R. leguminosarum bv. viciae 3841, as well as nodC from strain Sinorhizobum meliloti 2011 and a minimum query coverage and per cent identity of 50%, were used to identify sequences representing symbiosis plasmids.
Sourmash v. 3.5.0, with default parameters and a k-mer size of 31, was used to generate k-mer signatures, which were used to calculate a Jaccard index for all plasmid sequences [74]. A network was defined where edges connected plasmids in which one plasmid had a Jaccard index of greater than equal to 0.1 to the other. Cytoscape v. 3.8.1 was used to visualize the network [75]. The network was manually inspected to identify and eliminate contigs that have large blocks of homology to sequences of previously identified chromids [11]. The OSLOM v. 2.5 program oslom_undir with the options ‘-w -t 0.05 -r 50 -cp 0 -singlet -hr 0 -seed 1’ was used to define cliques and near cliques in a version of the plasmid network that only included nodes connected by edges with Jaccard index greater than or equal to 0.3 [38,76].
The LAST v. 1066 program lastal with the option ‘-f BlastTab+’ was used to compare plasmid and chromid sequences [77]. Homology of regions was visualized using the BioPython package GenomeDiagram v. 1.72 [78]. The program ProgressiveMauve v. 2.4.0 with the default options was used to align pAgK84 with a homologous plasmid [79].
(c) . Phylogenetic analyses
The programs automlsa2 v. 0.5.2 with the option ‘–allow_missing 4’ and IQ-Tree v. 2.1.2 with the options ‘-nt 8 -m MFP -B 1000 -alrt 1000 –msub nuclear –merge rclusterf’ were used to construct a multi-locus sequence analysis (MLSA) maximum-likelihood phylogeny [80–82]. Twenty-three translated reference gene sequences from agrobacteria strain C58 were used as queries to retrieve homologous sequences from all analysed genome sequences. Genes used were acnA, aroB, Atu0781, Atu1564, Atu2640, cgtA, coxC, dnaK, glyS, ham1, hemF, hemN, hom, leuS, lysC, murC, plsC, prfC, rplB, rpoB, rpoC, secA and truA [83].
For all other gene and protein phylogenies, MAFFT v. 7.471 with the default parameters was used to align gene or protein sequences [84]. Sequences of virD2 and virA from all strains were identified based on orthologue grouping from get_homologues analysis. Orthologue groups were combined prior to alignment. IQ-Tree v. 1.6.12 with the options ‘–bb 1000 –alrt 1000’ was used to generate phylogenies from gene or protein alignments [85]. The R package ggtree was used to visualize phylogenies [86]. Circos plots were generated using the R package circlize to visualize LAST alignments [87]. Base R and the R package ape were used to identify lineages of the MLSA phylogeny by hierarchical clustering (hclust function with method=‘single’) of pairwise tree distances (cophenetic.phylo function) cut at a height of 0.1 (cutree function) [88].
(d) . Analysis of essential genes
A bi-directional best BLAST hit approach was used to identify homologues of genes identified as essential [46,47]. The translated sequences from genes of strains Agrobacterium fabrum C58 or Sinorhizobium meliloti 1021 were used as queries in BLAST (v. 2.6.0 program blastp with the default options) searches against the complete annotated genomes of representative strains of BV2 (K84), BV2-like (AB2/73) and lineage 58 (B21/90). The best BLAST hit for each sequence was used as a query in reciprocal BLAST searches against the full genome sequence of strain C58 or strain 1021. Best BLAST hits to the original query sequence were identified as homologues.
Acknowledgements
We thank Melodie Putnam, Larry Hodges and Dr Joyce Loper for their assistance in selecting, providing and preparing agrobacterial strains for sequencing. We thank Dr Chih-Horng Kuo for providing the genome sequence for strain CFBP5877 and Dr Ed Davis for assistance in using autoMLSA2. Last, we thank Illumina for providing reagents, members of the Center for Genome Research and Biocomputing (CGRB) for sequencing services, and the Department of Botany and Plant Pathology (BPP) for supporting the computing infrastructure.
Data accessibility
Genome sequences for newly sequenced strains are deposited in the NCBI database under the BioProject ID PRJNA715259. Phylogenetic networks and trees in Newick format are made available at: https://github.com/osuchanglab/ARCPlasmidManuscript.
Authors' contributions
A.J.W. helped conceive and design the study, generated genome sequences for some strains, processed and analysed the data and drafted the manuscript; M.M. helped design the study, provided strains for sequencing and edited the manuscript; W.R. helped design the study, provided draft genome sequences for some strains and edited the manuscript; N.J.G. helped design the study and edited the manuscript; J.H.C. conceived and designed the study, coordinated the study and helped draft the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by the National Institute of Food and Agriculture, US Department of Agriculture awards grant nos. 2014-51181-22384 and 2020-51181-32154 to J.H.C. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Savory EA, et al. 2017. Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management. Elife 6, e30925. ( 10.7554/elife.30925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orlek A, et al. 2017. Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8, 182. ( 10.3389/fmicb.2017.00182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savory EA, Weisberg AJ, Stevens DM, Creason AL, Fuller SL, Pearce EM, Chang JH. 2020. Phytopathogenic Rhodococcus have diverse plasmids with few conserved virulence functions. Front. Microbiol. 11, 1022. ( 10.3389/fmicb.2020.01022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472-482. ( 10.1038/nrg3962) [DOI] [PubMed] [Google Scholar]
- 5.Che Y, Yang Y, Xu X, Břinda K, Polz MF, Hanage WP, Zhang T. 2021. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl Acad. Sci. USA 118, e2008731118. ( 10.1073/pnas.2008731118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711-721. ( 10.1038/nrmicro1234) [DOI] [PubMed] [Google Scholar]
- 7.Letek M, et al. 2010. The genome of a pathogenic Rhodococcus: cooptive virulence underpinned by key gene acquisitions. PLoS Genet. 6, e1001145. ( 10.1371/journal.pgen.1001145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489-495. ( 10.1016/j.tree.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 9.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25, 2034-2039. ( 10.1016/j.cub.2015.06.024) [DOI] [PubMed] [Google Scholar]
- 10.San Millan A, Toll-Riera M, Qi Q, MacLean RC. 2015. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 6, 6845. ( 10.1038/ncomms7845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisberg AJ, et al. 2020. Unexpected conservation and global transmission of agrobacterial virulence plasmids. Science 368, eaba5256. ( 10.1126/science.aba5256) [DOI] [PubMed] [Google Scholar]
- 12.Nester EW. 2015. Agrobacterium: nature's genetic engineer. Front. Plant Sci. 5, 730. ( 10.3389/fpls.2014.00730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keane PJ, Kerr A, New PB. 1970. Crown gall of stone fruit II. Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23, 585-596. ( 10.1071/BI9700585) [DOI] [Google Scholar]
- 14.Mougel C, Thioulouse J, Perrière G, Nesme X. 2002. A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. Evol. Microbiol. 52, 573-586. ( 10.1099/00207713-52-2-573) [DOI] [PubMed] [Google Scholar]
- 15.Kuzmanović N, Puławska J, Hao L, Burr TJ. 2018. The ecology of Agrobacterium vitis and management of crown gall disease in vineyards. In Agrobacterium biology (ed. Gelvin SB), pp. 15-53. Cham, Switzerland: Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- 16.Weller S, Stead D, Young JP. 2006. Recurrent outbreaks of root mat in cucumber and tomato are associated with a monomorphic, cucumopine, Ri-plasmid harboured by various Alphaproteobacteria. FEMS Microbiol. Lett. 258, 136-143. ( 10.1111/j.1574-6968.2006.00214.x) [DOI] [PubMed] [Google Scholar]
- 17.Haryono M, Tsai Y-M, Lin C-T, Huang F-C, Ye Y-C, Deng W-L, Hwang H-H, Kuo C-H. 2018. Presence of an Agrobacterium-type tumor-inducing plasmid in Neorhizobium sp. nchu2750 and the link to phytopathogenicity. Genome Biol. Evol. 10, 3188-3195. ( 10.1093/gbe/evy249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slater SC, et al. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 191, 2501-2511. ( 10.1128/JB.01779-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison PW, Lower RPJ, Kim NKD, Young JPW. 2010. Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol. 18, 141-148. ( 10.1016/j.tim.2009.12.010) [DOI] [PubMed] [Google Scholar]
- 20.Castillo-Ramírez S, Vázquez-Castellanos JF, González V, Cevallos MA. 2009. Horizontal gene transfer and diverse functional constrains within a common replication-partitioning system in Alphaproteobacteria: the repABC operon. BMC Genom. 10, 536. ( 10.1186/1471-2164-10-536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cevallos MA, Cervantes-Rivera R, Gutiérrez-Ríos RM. 2008. The repABC plasmid family. Plasmid 60, 19-37. ( 10.1016/j.plasmid.2008.03.001) [DOI] [PubMed] [Google Scholar]
- 22.diCenzo GC, Finan TM. 2017. The divided bacterial genome: structure, function, and evolution. Microbiol. Mol. Biol. Rev. 81, e00019-17. ( 10.1128/MMBR.00019-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero D, Brom S. 2004. The symbiotic plasmids of the Rhizobiaceae. In Plasmid biology (eds Funnell BE, Phillips GJ), pp. 271-290. Chichester, UK: John Wiley & Sons, Ltd. [Google Scholar]
- 24.Masson-Boivin C, Sachs JL. 2018. Symbiotic nitrogen fixation by rhizobia-the roots of a success story. Curr. Opin. Plant Biol. 44, 7-15. ( 10.1016/j.pbi.2017.12.001) [DOI] [PubMed] [Google Scholar]
- 25.Platt TG, Morton ER, Barton IS, Bever JD, Fuqua C. 2014. Ecological dynamics and complex interactions of Agrobacterium megaplasmids. Front. Plant Sci. 5, 635. ( 10.3389/fpls.2014.00635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winans SC, Ebert PR, Stachel SE, Gordon MP, Nester EW. 1986. A gene essential for Agrobacterium virulence is homologous to a family of positive regulatory loci. Proc. Natl Acad. Sci. USA 83, 8278-8282. ( 10.1073/pnas.83.21.8278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachel SE, Nester EW. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 5, 1445-1454. ( 10.1002/j.1460-2075.1986.tb04381.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christie PJ, Whitaker N, González-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843, 1578-1591. ( 10.1016/j.bbamcr.2013.12.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelvin SB. 1998. Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J. Bacteriol. 180, 4300-4302. ( 10.1128/JB.180.16.4300-4302.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toro N, Datta A, Carmi OA, Young C, Prusti RK, Nester EW. 1989. The Agrobacterium tumefaciens virC1 gene product binds to overdrive, a T-DNA transfer enhancer. J. Bacteriol. 171, 6845-6849. ( 10.1128/jb.171.12.6845-6849.1989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodges LD, Cuperus J, Ream W. 2004. Agrobacterium rhizogenes GALLS protein substitutes for Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J. Bacteriol. 186, 3065-3077. ( 10.1128/JB.186.10.3065-3077.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ream W. 2009. Agrobacterium tumefaciens and A. rhizogenes use different proteins to transport bacterial DNA into the plant cell nucleus. Microb. Biotechnol. 2, 416-427. ( 10.1111/j.1751-7915.2009.00104.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessaux Y, Petit A, Farrand S, Murphy P. 1998. Opines and opine-like molecules involved in plant-Rhizobiaceae interactions. In The rhizobiaceae: molecular biology of model plant-associated bacteria (eds Spaink HP, Kondorosi A, Hooykaas P), pp. 173-197. Berlin, Germany: Springer. [Google Scholar]
- 34.Canfield M, Moore L. 1989. Unusual characteristics of Agrobacterium tumefaciens strains isolated from wild blackberry. Phytopathology 79, 1181. [Google Scholar]
- 35.Anderson AR, Moore LW. 1979. Host specificity in the genus Agrobacterium. Phytopathology 69, 320. ( 10.1094/Phyto-69-320) [DOI] [Google Scholar]
- 36.Unger L, Ziegler SF, Huffman GA, Knauf VC, Peet R, Moore LW, Gordon MP, Nester EW. 1985. New class of limited-host-range Agrobacterium mega-tumor-inducing plasmids lacking homology to the transferred DNA of a wide-host-range, tumor-inducing plasmid. J. Bacteriol. 164, 723-730. ( 10.1128/jb.164.2.723-730.1985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otten L, Schmidt J. 1998. A T-DNA from the Agrobacterium tumefaciens limited-host-range strain AB2/73 contains a single oncogene. Mol. Plant-Microbe Interact. 11, 335-342. ( 10.1094/MPMI.1998.11.5.335) [DOI] [PubMed] [Google Scholar]
- 38.Acman M, van Dorp L, Santini JM, Balloux F.. 2020. Large-scale network analysis captures biological features of bacterial plasmids. Nat. Commun. 11, 2452. ( 10.1038/s41467-020-16282-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young JPW, et al. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 7, R34. ( 10.1186/gb-2006-7-4-r34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weller SA, Stead DE, Young JPW. 2004. Acquisition of an Agrobacterium Ri plasmid and pathogenicity by other α-proteobacteria in cucumber and tomato crops affected by root mat. Appl. Environ. Microbiol. 70, 2779-2785. ( 10.1128/AEM.70.5.2779-2785.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burr TJ, Bazzi C, Süle S, Otten L.. 1998. Crown gall of grape: biology of Agrobacterium vitis and the development of disease control strategies. Plant Dis. 82, 1288-1297. ( 10.1094/PDIS.1998.82.12.1288) [DOI] [PubMed] [Google Scholar]
- 42.Farrand SK, Slota JE, Shim J-S, Kerr A. 1985. Tn5 insertions in the agrocin 84 plasmid: the conjugal nature of pAgK84 and the locations of determinants for transfer and agrocin 84 production. Plasmid 13, 106-117. ( 10.1016/0147-619X(85)90063-0) [DOI] [PubMed] [Google Scholar]
- 43.Kim J-G, Park BK, Kim S-U, Choi D, Nahm BH, Moon JS, Reader JS, Farrand SK, Hwang I. 2006. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc. Natl Acad. Sci. USA 103, 8846-8851. ( 10.1073/pnas.0602965103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis JG, Murphy PJ. 1981. Four new opines from crown gall tumours—their detection and properties. Mol. Gen. Genet. 181, 36-43. ( 10.1007/BF00339002) [DOI] [Google Scholar]
- 45.Slota JE, Farrand SK. 1982. Genetic isolation and physical characterization of pAgK84, the plasmid responsible for agrocin 84 production. Plasmid 8, 175-186. ( 10.1016/0147-619X(82)90055-5) [DOI] [PubMed] [Google Scholar]
- 46.Curtis PD, Brun YV. 2014. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol. Microbiol. 93, 713-735. ( 10.1111/mmi.12686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.diCenzo GC, Benedict AB, Fondi M, Walker GC, Finan TM, Mengoni A, Griffitts JS. 2018. Robustness encoded across essential and accessory replicons of the ecologically versatile bacterium Sinorhizobium meliloti. PLoS Genet. 14, e1007357. ( 10.1371/journal.pgen.1007357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuller SL, Savory EA, Weisberg AJ, Buser JZ, Gordon MI, Putnam ML, Chang JH. 2017. Isothermal amplification and lateral-flow assay for detecting crown-gall-causing Agrobacterium spp. Phytopathology 107, 1062-1068. ( 10.1094/phyto-04-17-0144-r) [DOI] [PubMed] [Google Scholar]
- 49.Kuzmanović N, Smalla K, Gronow S, Puławska J. 2018. Rhizobium tumorigenes sp. nov., a novel plant tumorigenic bacterium isolated from cane gall tumors on thornless blackberry. Sci. Rep. 8, 9051. ( 10.1038/s41598-018-27485-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuzmanović N, Behrens P, Idczak E, Wagner S, Götz M, Spröer C, Bunk B, Overmann J, Smalla K. 2019. A novel group of Rhizobium tumorigenes-like agrobacteria associated with crown gall disease of rhododendron and blueberry. Phytopathology 109, 1840-1848. ( 10.1094/PHYTO-05-19-0167-R) [DOI] [PubMed] [Google Scholar]
- 51.Chou L, et al. 2021. Species boundaries in the Agrobacterium tumefaciens complex and multi-level modular evolution of their antibacterial type VI secretion system and tumor-inducing plasmids. bioRxiv, 2021.05.20.444927. (doi:10/gk873g)
- 52.Paulus F, Otten L. 1993. Functional and mutated agrocinopine synthase genes on octopine T-DNAs. Mol. Plant-Microbe Interact. 6, 393-402. ( 10.1094/MPMI-6-393) [DOI] [PubMed] [Google Scholar]
- 53.Esberg B, Leung H-CE, Tsui H-CT, Björk GR, Winkler ME. 1999. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181, 7256-7265. ( 10.1128/JB.181.23.7256-7265.1999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P. 2011. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62, 2431-2452. ( 10.1093/jxb/err004) [DOI] [PubMed] [Google Scholar]
- 55.Hwang I, Sheen J, Müller B. 2012. Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353-380. ( 10.1146/annurev-arplant-042811-105503) [DOI] [PubMed] [Google Scholar]
- 56.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652-655. ( 10.1038/nature05504) [DOI] [PubMed] [Google Scholar]
- 57.Moore LW, Chilton WS, Canfield ML. 1997. Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl. Environ. Microbiol. 63, 201-207. ( 10.1128/aem.63.1.201-207.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lacroix B, Citovsky V. 2016. A functional bacterium-to-plant DNA transfer machinery of Rhizobium etli. PLoS Pathog. 12, e1005502. ( 10.1371/journal.ppat.1005502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otten L. 2021T-DNA regions from 350 Agrobacterium genomes: maps and phylogeny. Plant Mol. Biol. 106, 239-258. ( 10.1007/s11103-021-01140-0) [DOI] [PubMed] [Google Scholar]
- 60.Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. 2008. Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59, 417-441. ( 10.1146/annurev.arplant.59.032607.092945) [DOI] [PubMed] [Google Scholar]
- 61.Tournu H, Fiori A, Dijck PV. 2013. Relevance of trehalose in pathogenicity: some general rules, yet many exceptions. PLoS Pathog. 9, e1003447. ( 10.1371/journal.ppat.1003447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jouanin L, Tourneur J, Tourneur C, Casse-Delbart F. 1986. Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid 16, 124-134. ( 10.1016/0147-619X(86)90071-5) [DOI] [PubMed] [Google Scholar]
- 63.Haryono M, Cho S-T, Fang M-J, Chen A-P, Chou S-J, Lai E-M, Kuo C-H. 2019. Differentiations in gene content and expression response to virulence induction between two Agrobacterium strains. Front. Microbiol. 10, 1554. ( 10.3389/fmicb.2019.01554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaudequin-Dransart V, Petit A, Chilton WS, Dessaux Y. 1998. The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. MPMI 11, 583-591. ( 10.1094/MPMI.1998.11.7.583) [DOI] [Google Scholar]
- 65.Lang J, Planamente S, Mondy S, Dessaux Y, Moréra S, Faure D. 2013. Concerted transfer of the virulence Ti plasmid and companion At plasmid in the Agrobacterium tumefaciens-induced plant tumour. Mol. Microbiol. 90, 1178-1189. ( 10.1111/mmi.12423) [DOI] [PubMed] [Google Scholar]
- 66.Barton IS, Platt TG, Rusch DB, Fuqua C. 2019. Destabilization of the tumor-inducing plasmid from an octopine-type Agrobacterium tumefaciens lineage drives a large deletion in the co-resident at megaplasmid. G3 9, 3489-3500. ( 10.1534/g3.119.400554) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barton IS, Eagan JL, Nieves-Otero PA, Reynolds IP, Platt TG, Fuqua C. 2020. Co-dependent and interdigitated: dual quorum sensing systems regulate conjugative transfer of the Ti plasmid and the At megaplasmid in Agrobacterium tumefaciens 15955. Front. Microbiol. 11, 605896. ( 10.3389/fmicb.2020.605896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang XX, Kosier B, Priefer UB. 2001. Symbiotic plasmid rearrangement in Rhizobium leguminosarum bv. viciae VF39SM. J. Bacteriol. 183, 2141-2144. ( 10.1128/JB.183.6.2141-2144.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson M, Guhlin J, Epstein B, Tiffin P, Sadowsky MJ. 2018. The complete replicons of 16 Ensifer meliloti strains offer insights into intra- and inter-replicon gene transfer, transposon-associated loci, and repeat elements. Microb. Genom. 4, e000174. ( 10.1099/mgen.0.000174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696-7701. ( 10.1128/AEM.02411-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Day A. 2012. heatmap.plus. See https://cran.r-project.org/web/packages/heatmap.plus/index.html.
- 72.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. ( 10.1371/journal.pcbi.1002195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078-2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brown CT, Irber L. 2016. sourmash: a library for MinHash sketching of DNA. J. Open Source Softw. 1, 27. ( 10.21105/joss.00027) [DOI] [Google Scholar]
- 75.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498-2504. ( 10.1101/gr.1239303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lancichinetti A, Radicchi F, Ramasco JJ, Fortunato S. 2011. Finding statistically significant communities in networks. PLoS ONE 6, e18961. ( 10.1371/journal.pone.0018961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487-493. ( 10.1101/gr.113985.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pritchard L, White JA, Birch PRJ, Toth IK. 2006. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics 22, 616-617. ( 10.1093/bioinformatics/btk021) [DOI] [PubMed] [Google Scholar]
- 79.Darling AE, Mau B, Perna NT. 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147. ( 10.1371/journal.pone.0011147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davis EW II. 2021. automlsa2. See https://github.com/davised/automlsa2.
- 81.Davis EW II, Weisberg AJ, Tabima JF, Grunwald NJ, Chang JH. 2016. Gall-ID: tools for genotyping gall-causing phytopathogenic bacteria. PeerJ 4, e2222. ( 10.7717/peerj.2222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530-1534. ( 10.1093/molbev/msaa015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang YM, Tian CF, Sui XH, Chen WF, Chen WX. 2012. Robust markers reflecting phylogeny and taxonomy of rhizobia. PLoS ONE 7, e44936. ( 10.1371/journal.pone.0044936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268-274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28-36. ( 10.1111/2041-210X.12628) [DOI] [Google Scholar]
- 87.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811-2812. ( 10.1093/bioinformatics/btu393) [DOI] [PubMed] [Google Scholar]
- 88.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289-290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome sequences for newly sequenced strains are deposited in the NCBI database under the BioProject ID PRJNA715259. Phylogenetic networks and trees in Newick format are made available at: https://github.com/osuchanglab/ARCPlasmidManuscript.