Abstract
Naturally occurring plasmids come in different sizes. The smallest are less than a kilobase of DNA, while the largest can be over three orders of magnitude larger. Historically, research has tended to focus on smaller plasmids that are usually easier to isolate, manipulate and sequence, but with improved genome assemblies made possible by long-read sequencing, there is increased appreciation that very large plasmids—known as megaplasmids—are widespread, diverse, complex, and often encode key traits in the biology of their host microorganisms. Why are megaplasmids so big? What other features come with large plasmid size that could affect bacterial ecology and evolution? Are megaplasmids 'just' big plasmids, or do they have distinct characteristics? In this perspective, we reflect on the distribution, diversity, biology, and gene content of megaplasmids, providing an overview to these large, yet often overlooked, mobile genetic elements.
This article is part of the theme issue ‘The secret lives of microbial mobile genetic elements’.
Keywords: plasmid, megaplasmid, mobile genetic element, genome evolution, horizontal gene transfer, pangenome
1. Megaplasmids: very large plasmids
The word plasmid was coined in 1952 to refer to any extra-chromosomal ‘hereditary determinant’ [1]. Though this broad term originally included endosymbionts, in the years since it has been narrowed to refer principally to DNA extrachromosomal genetic elements that, unlike phage, do not encode capsids for transmission. Nevertheless, plasmids can be extraordinarily diverse—in terms of composition, with G + C content ranging from 20% to almost 90% [2]; in structure, with linear and circular forms, and the ability to integrate into the chromosome or excise and replicate independently [3–5]; in host range [6]; in mechanism of replication [7]; in gene content [8]; and (like the microbes that host them) in the environments in which they are found [9–12]. Natural plasmids can be found in bacteria, archaea and eukaryotes, such as yeast and slime moulds [13–15]. Perhaps the most striking manifestation of plasmid diversity is their size. Plasmid genomes span three orders of magnitude, with the largest coming in at around 2.5 Mb—3500× bigger than the smallest plasmids (which can be less than 800 bp), and approximately 4.5× the size of the chromosome of some culturable bacteria [2,16]. Plasmids can clearly be very small, and they can also be very big.
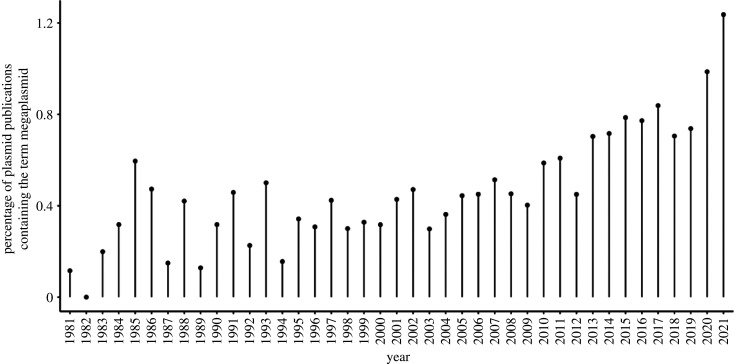
Very large plasmids were first called ‘megaplasmids’ by Rosenberg and colleagues in the early 1980s. By employing a gentle DNA extraction process to investigate the genetics of Sinorhizobium meliloti, they were able to identify extrachromosomal elements that were too large and fragile to be retrieved by their previously-used alkaline lysis approach [17,18]. With an apparent molecular weight ‘clearly larger’ than 300 × 106 Da—which is approximately 450 kb—they termed these elements megaplasmids. The term has persisted, and increased in popularity (figure 1) as a handle referring to very large plasmids, bounded on one side by size (i.e. a small megaplasmid is ‘just’ a plasmid), and on the other by the extent to which they are integrated into cellular physiology, with very large additional replicons that are essential for viability termed secondary chromosomes or ‘chromids’ [19–21].
Figure 1.
Percentage of plasmid publications containing the term ‘megaplasmid’. Metadata on publications containing the term ‘plasmid’ or ‘megaplasmid’ from 1981 to date were retrieved separately from Pubmed (https://pubmed.ncbi.nlm.nih.gov/advanced/) using the following search queries: for ‘plasmid’ - (plasmid[Title/Abstract]) AND ((‘1981’ [Date - Publication] : ‘3000’ [Date - Publication])); for ‘megaplasmid’ - (megaplasmid[Title/Abstract]) OR (mega-plasmid[Title/Abstract]) AND ((‘1981’[Date - Publication] : ‘3000’[Date - Publication])). The number of publications recorded for each term per year was used to calculate the percentages displayed in the plot.
It is apt that megaplasmids were initially defined operationally in the context of an extraction protocol, as megaplasmid identification in the years since has largely followed the prevailing technical norms. Many megaplasmids were first characterized during the period when microbial genetics was pursued principally through use of gel electrophoresis and hybridization [22,23]. The advent of accessible short-read sequencing technology precipitated a genomics revolution in microbiology, but while this flood of data vastly improved our understanding of microbial genome content, unambiguously resolving megaplasmids remained difficult, because megaplasmids are usually maintained at low copy number, and carry sequences which are repetitive, or similar to those in the chromosome. Assembly often failed to yield complete megaplasmid contigs, and it is likely that short-read databases hold many megaplasmids that are not annotated as such, as recently demonstrated for a family of megaplasmids identified in Pseudomonas aeruginosa [24]. The recent availability of long-read single-molecule sequencing technologies, such as the PacBio SMRT protocol and Nanopore devices, enables megaplasmids to be unambiguously resolved [25–27], and with this increased detection rate, recent years have seen a steep increase in the proportion of plasmid publications containing the term ‘megaplasmid’ (figure 1).
The dynamic, mosaic, modularity of microbial mobile genetic elements means that the borders of any category—plasmid, phage, transposon, integrative and conjugative element (ICE)—are somewhat fuzzy [28–30], and this issue poses a particular problem for megaplasmids. Are megaplasmids ‘just’ big plasmids, or is there something beyond size per se that sets megaplasmids apart? Is there any biological meaning in a term that was first used in relation to a DNA extraction procedure? In this perspective, we explain the problems with defining megaplasmids, and assess whether there is a distinct, coherent category of mobile genetic elements we can refer to as megaplasmids. We discuss what features of very big plasmids might distinguish them from other genetic compartments, how and why they became so large, and what they reveal about microbial evolutionary ecology. Throughout, we give some examples of how megaplasmids contribute to the biology of diverse microbes.
2. How big is a megaplasmid?
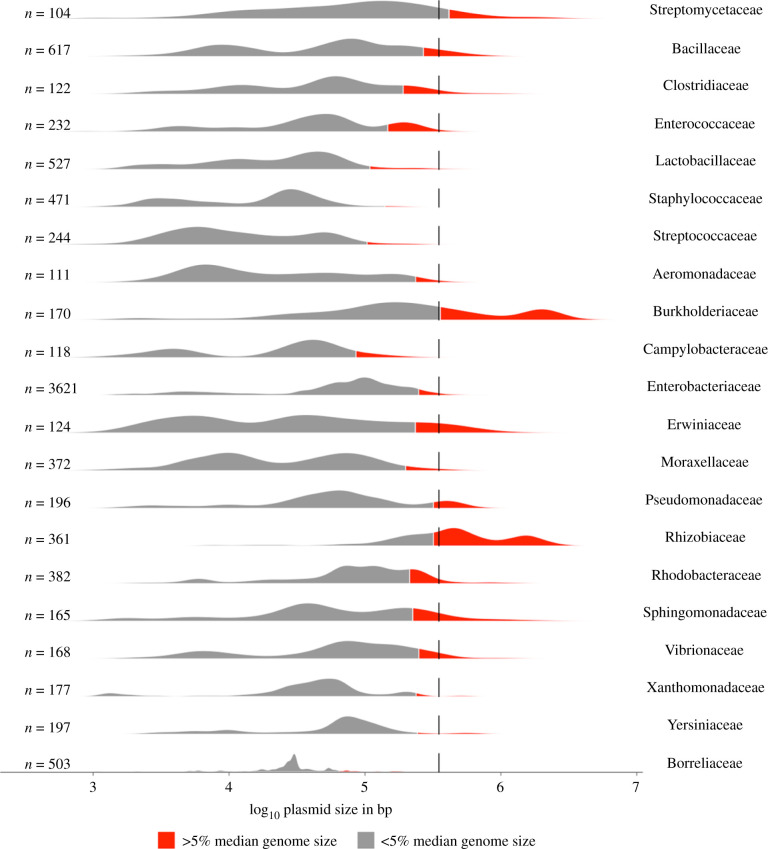
If plasmid size is plotted as a histogram, most species give a bimodal distribution (figure 2). The smaller peak, at around 103–104 bp, corresponds to (usually) multi-copy, non-conjugative plasmids, like the ColE1 plasmids, whereas the larger peak, at around 104–105 bp, corresponds to low-copy, often conjugative plasmids, like RP4 [32]. This bimodality reflects a natural division, roughly separating two classes of plasmid which have distinct biological features and behaviours [33]. Rather than a distinct third peak, megaplasmids tend to appear as part of a continuous spectrum, or shoulder, on the right-hand of the larger plasmid peak. There are some exceptions: distinct ‘megaplasmid’ peaks can be seen in the distributions of plasmids from Pseudomonadaceae, Rhizobiaceae, Burkholderiaceae and Enterococcaceae, for example. These mainly appear to represent related plasmids within each family, which could reflect underlying biological differences or fitness benefits associated with large plasmid size. Overall, megaplasmids appear to share many of the general features of plasmids comprising the large plasmid peak, including replication mechanism, copy number and transmissibility, and thus do not appear distinct from large plasmids the way large plasmids do from small ones [33,34]. More sensitive assignment may be possible as more sequences are collected, and methods developed to control for phylogenetic correlation in a mosaic and recombining group of elements that lacks core conserved genes, but currently, defining megaplasmids is usually achieved through a rather unsatisfying process of slicing the pool of plasmids at an arbitrary size threshold.
Figure 2.
Plasmid size distributions for families with greater than 100 plasmids in the COMPASS database [13]. The number of plasmids analysed is displayed to the left of each graph. Red filled areas correspond to 5% of the median genome size for each family, according to EZBioCloud [31]. The dotted line indicates 350 kb [19]. Plots were generated in R using the ggplot and tidyverse packages. See the electronic supplementary material for further details.
Thresholds for minimum megaplasmid size have thus varied. diCenzo and Finan's seminal review on multipartite genomes set the threshold at 350 kb, based on 10% of median bacterial genome size [19], while 100 kb has been used for other studies [20], but as genome size itself varies widely across phyla, the very largest plasmids in some species might be only of middling size in others (figure 2). Fixed thresholds are likely to bias megaplasmids towards species with already large genomes, since plasmid size and genome size are correlated [33]. The ecological and evolutionary factors driving large plasmid size are likely to apply similarly to phylogenetically diverse species, so to understand the biology of very large plasmids we consider it more appropriate to regard plasmid size relative to other replicons in that species. For example, Campylobacter and Aliarchobacter plasmids of only approximately 100–150 kb can harbour greater than 7% of total genome content [35,36], which in Pseudomonas would be equivalent to greater than 450 kb. In figure 2, we highlight plasmids that are relatively large: greater than or equal to 5% of total genome size for that family. Nevertheless, as the figure shows, even proportional thresholds artificially divide what is in fact a continuous distribution, and so we emphasise that our use of 5% is intended to be illustrative rather than prescriptive.
Megaplasmids have also been distinguished from the other large replicons: secondary chromosomes and ‘chromids’ [21]. This difference is a qualitative one: chromids and secondary chromosomes carry essential housekeeping genes found on the chromosome in other species, often confirmed experimentally. Compared with plasmids, chromids and secondary chromosomes tend to have genomic signatures—such as GC content, codon usage and dinucleotide relative abundance—that more closely resemble those of the chromosome [19,21]. The distinction between secondary chromosomes and chromids comes down to their evolutionary history: secondary chromosomes formed by the split of an ancestral chromosome, while chromids most likely originated from megaplasmids [19]. Secondary chromosomes thus have replication machineries that are clearly distinct from those of chromids and megaplasmids [37], and on this basis it appears that secondary chromosomes are less common than other large replicons (though there are some possible natural and artificial examples [19,38]). Because carriage of essential genes is a discrete and measurable trait, distinguishing megaplasmids from chromids appears conceptually less arbitrary than distinguishing megaplasmids from large plasmids. However, determining essentiality is not always easy, and not always consistent across environments and strains [39,40]. Definitive classifications may also require the evaluation of replicons in a context that takes into account conservation through evolutionary time and over different levels of taxonomy. For instance, megaplasmids are considered to be more plastic whereas chromids and secondary chromosomes may be found more consistently in every member of a clade as a consequence of their essentiality [21]. Regardless of underlying definitions and classification schemes, it is currently apparent that ‘megaplasmid’, ‘chromid’ and ‘secondary chromosome’ are not used consistently across the literature.
Taxonomies of mobile genetic elements have been notoriously difficult owing to their mosaic structure, their diversity, their propensity to recombine, and horizontal transmission across phylogenetic boundaries [8,41–44]. For example, some ICEs are more closely related to plasmids than other ICEs, with plasmid-like features such as extrachromosomal replication and partitioning systems [45,46]. Recent network analyses have had some success in categorizing plasmids into clusters based on genome-level pairwise similarities, but such schemes ignore core functional identities of plasmids like replication and transmission, and can still produce edge cases and mosaics that defy unambiguous classification [8,47]. Overall, though there may be trends, exceptions are often the rule. Arguing over ‘what counts’ as a megaplasmid may not be helpful in understanding their biology, as replicons have the potential to cross size thresholds and transition between essentiality/non-essentiality with ease. With these concepts and ideas in mind, we would like to offer a somewhat provocative view that perhaps plasmids, megaplasmids, and chromids should be considered as a spectrum that varies in multiple characteristics including size and ‘essentiality’ rather than generic placement into defined orthogonal groups. There exist relatively small plasmids that are absolutely essential for physiological functioning of cells, such as the 9.4 kb ribosomal RNA-encoding plasmids of Aureimonas [48], and gigantic plasmids that are expendable and whose fitness effects are only apparent in specific niches and contexts, for example, the 1.35 Mb pSymA of Sinorhizobium meliloti, deletion of which has only minor effects on growth, transcription and proteome phenotypes [49–51]. Some small plasmids exist entirely as mosaics from multiple contributing replicons while some large plasmids have remained syntenic despite extensive divergence in sequence [52,53]. If one were to reconceptualize plasmid terminology, taken together, perhaps it would be best to simply discuss the characteristics of each replicon itself rather than worry about nomenclatural challenges. After all, just like a vacuum, nature often abhors cleanly delineated conceptual taxonomic schemes. We, therefore, caution against absolutes in determining what counts as a megaplasmid.
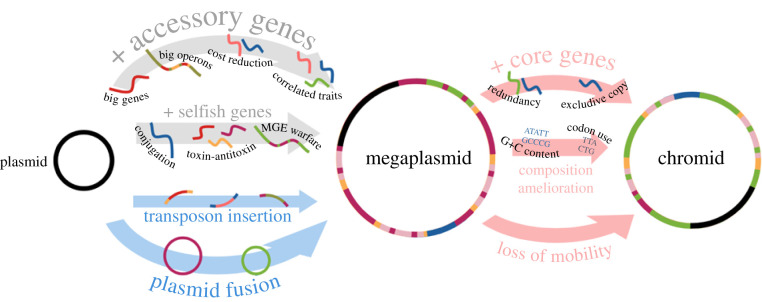
An analogy can perhaps be found in the use of the term ‘megafauna’ by conservation biologists to refer to animals larger than some threshold mass, a threshold that, as with megaplasmids, varies across studies and the type of organism in question [54]. Different megafauna species may be more closely related to smaller animals than they are to one another, for example, an elephant is more similar genetically to the pocket-sized golden mole than to the bison. However, similar evolutionary pressures are probably responsible for the large size of both bison and elephants, and genetically diverse megafaunal species can play similar ecological roles. We suggest that, just as ‘megafauna’ remains a useful term for conservation ecologists, ‘megaplasmid’ can remain a useful term to define plasmids that are big relative to the genomes they come from, even if megaplasmids cannot be considered a coherent group in terms of an unambiguous size threshold, evolutionary relatedness, or specific gene content. A loose term, like megaplasmid or megafauna, encourages us to generalize to understand the underlying biology. In the next sections, we discuss these features of megaplasmids—their size, genetics, and gene content—that explain their evolutionary and ecological role (figure 3).
Figure 3.
What makes a megaplasmid. Grey arrows; megaplasmids arise and persist when plasmids are selected to carry more accessory gene content: big genes or operons, multiple different environmentally correlated traits, and/or genes that reduce physiological burdens. At the same time, ‘selfish’ traits that promote plasmid vertical and horizontal transmission can also drive up plasmid size. Blue arrows; new genes can be acquired by the activities of other mobile genetic elements, including transposons, plasmids, ICEs and phage (not shown). Red arrows; chromids are thought to develop from megaplasmids by acquiring core genes, potentially through an intermediate step of carrying a redundant copy. Chromids also tend to lose the ability for horizontal transmission. Consequently, ‘locked in’ to a particular genome, the nascent chromid acquires the compositional signatures of the host chromosome.
3. Why are megaplasmids bigger?
Selection simultaneously acts on plasmids from multiple levels [55]. Selection acts on plasmids directly, favouring those that are more effective at persisting and transmitting. At the same time, selection acts at higher levels of biological organization, for example, at the level of the cell or the population, indirectly favouring plasmids that enhance the fitness of their microbial hosts. Selection also acts at lower organizational levels: that of transposons or insertion sequences, which can exploit plasmids to spread to new hosts. Plasmid gene content is shaped by these different selective pressures (alongside genetic drift, which is thought to be relatively weak in most microbial populations [56]). Selection at different organizational levels is not necessarily aligned—in fact, it is often assumed that there is some trade-off between host-level and plasmid-level selection, such that plasmids which are more effective at transmission are more of a burden on their hosts, while those that benefit their hosts become less proficient at horizontal transmission [57,58]. In the long run, plasmids might be expected to become either efficient parasites that have shed any genetic cargo that does not enhance their selfish reproduction, or domesticated mutualists that are incapable of autonomous replication or transmission [59]. Prokaryotic genomes are subject to a deletional bias, and thus plasmids exist under a constant threat of gene loss [60]. It may therefore seem surprising to find large megaplasmids that both confer many beneficial traits, and are capable (and even highly proficient) at spreading to new hosts: a case in point is the approximately 400 kb pBT2436-like family of Pseudomonas megaplasmids, which transmit multiple resistance determinants at a very high rate [24]. In fact, there are many ways in which megaplasmids might be successful because of, not despite, their large size.
(a) . Larger plasmids can be more effective parasites
Expanding gene content is one way in which larger plasmids can be more efficiently inherited (i.e. vertical transmission), and more efficiently spread (i.e. horizontal transmission). Vertical transmission requires viable offspring cells to receive a copy of the plasmid. Partitioning systems, which mechanically distribute copies of the plasmid to offspring [61], are encoded by low copy-number (approx. less than 5) plasmids (such as large plasmids tend to be [62]) out of necessity because the risks of loss by segregation are untenable [63]. However, such systems are imperfect, and if plasmid-free cells do not suffer the burdens of carriage, the plasmid can be driven out of the population by purifying selection. Therefore, larger plasmids tend to also carry toxin-antitoxin (TA) systems which ensure that plasmid-free cells do not succeed [64]. These linked genes, encoding a long-lived toxin and a short-lived antitoxin, result in an (often lethal) fitness cost if the antitoxin gene is lost [65], and are often found to be necessary for megaplasmid stability, as was seen for pTTS12 (583 kb) and pAtC58 (542 kb) [66,67]. In fact, evolution experiments selecting for increased stability of a 43 kb IncP-1beta plasmid found this plasmid reproducibly acquired a TA system from the chromosome, enhancing its persistence and increasing its size [68,69]. Some plasmids encode many different TA systems, with as many as seven detected on the 103 kb Synechocystis plasmid pSYSA [70]. TA systems are themselves relatively small, meaning that the genes themselves can make only a negligible contribution to plasmid size, but several TA systems distributed across a plasmid can stabilize larger regions that would otherwise be prone to deletion [71,72].
Horizontal transmission of plasmids is thought to occur mainly by conjugation [73,74]; this is particularly the case for larger plasmids which are likely to exceed the packaging limits of most phage capsids, thus preventing efficient transduction (though see [75]), and are more vulnerable to extracellular damage, thus inhibiting transformation [76]. Conjugation requires a conjugative pilus: a macromolecular structure comprising approximately 12 proteins or more [77,78], and thus the genetic size of the transfer region means that self-conjugative plasmids tend to be large [33]. Many megaplasmids are predicted to encode conjugative machinery, with experiments showing conjugation sometimes at very high rates [24,79–81]. Though rare [13,82], some plasmids carry multiple conjugative and replication machineries (e.g. [79]), potentially extending the range of conditions in which they can transfer and hosts in which they can be maintained, with bigger plasmids more likely to carry multiple replication modules (electronic supplementary material, figure S4). Horizontal transmission can also be enhanced by plasmid-encoded pili and fimbriae that promote biofilm formation and cell-cell adhesion [83,84]. However, though larger plasmids are more likely to be conjugative (electronic supplementary material, figure S3), some megaplasmids appear to lack conjugative machinery altogether, and either depend on the conjugative machinery of other plasmids to transmit (i.e. they are mobilizable) or are apparently not mobile at all [85]. Annotation algorithms that use the well-described conjugative machineries of Enterobacteriaceae as templates may underperform when detecting transfer genes in non-conventional organisms [86], and the thick seam of uncharacterized megaplasmid genes may yet show new mechanisms for horizontal transmission. Alternatively, non-mobilizable megaplasmids may be en route to becoming chromids, as discussed below.
Plasmids occupy a world inhabited by other plasmids. Interacting mobile genetic elements can exploit one another's transmission machineries, compete for cellular resources, and directly exclude one another (for example, plasmids of the same incompatibility group) [87,88]. Co-infection of a prokaryote with multiple different plasmids is common [89], and mechanisms have evolved to promote one plasmid over another. Some plasmids pack a punch by carrying their own CRISPR-Cas systems, to eliminate other plasmids [53,90,91]. CRISPR-encoding plasmids tend to be conjugative and relatively big (the CRISPR locus itself is approx. 6 kb [91]), and interestingly, the targets of these ‘plasmid-hunting plasmids' tend also to be relatively large, suggesting that CRISPR-mediated plasmid warfare tends to be a clash of titans [90,92].
(b) . Larger plasmids can be more effective mutualists
Many plasmids carry ‘accessory genes’: genes that do not confer a direct fitness benefit on plasmids, and instead benefit plasmids indirectly by improving the fitness of the microbes that carry them. Accessory genes confer functions as diverse as virulence, antibiotic resistance, metabolism, biofilm production, and gene regulation [83,93–95]. Plasmids that provide beneficial accessory genes have fitness interests aligned with those of their hosts—where hosts benefit, the plasmid benefits too. Under such circumstances, selection and drift would be expected to eventually rob plasmids of their autonomy, through the loss of horizontal transmission, and/or integration of beneficial accessory genes into the chromosome. However, beneficial plasmids persist [59,96]. A powerful explanation is the ‘local adaptation’ model of plasmid accessory gene content [55,97]. Briefly, the local adaptation model states that some traits are beneficial—or indeed, may be essential—only in certain specific situations or environments. New bacterial strains colonizing that environment are at a disadvantage compared with the incumbent if they do not have the trait; this will be the case even if the incoming strain has beneficial mutations at other positions in the genome. A plasmid that can transmit locally-adaptive traits into a competitive newcomer enables that newcomer to succeed, indirectly benefiting the transmissible, accessory-gene-encoding plasmids in the process [96,98].
The local adaptation model can explain why some plasmids are so large because in some cases, the genes involved are themselves very big. For example, the three megaplasmid-borne polyketide synthase genes MLSA1, MLSA2, MLSB, which enable Mycobacterium ulcerans to produce mycolactone toxin and thus cause infection, are 51 kb, 7 kb and 42 kb respectively, forming a locus that exceeds 100 kb in size [99]. While such giant genes are an exception, large gene clusters, encoding a series of enzymes all necessary for biosynthesis, catabolism, or virulence, are often found on megaplasmids: the nicotine catabolism pathway of the Arthrobacter nicotinovorans pAO1 [100] and the entry region of the Shigella virulence plasmid [93] provide just two examples.
Larger plasmids can carry many different locally adaptive genes. But in other cases, traits are encoded on different, coexisting replicons [101]. What might cause locally-adaptive traits to accumulate on a single large plasmid, rather than being distributed across multiple smaller plasmids? The ‘selfish operon’ hypothesis proposes that a group of accessory genes which contributes to a single beneficial function becomes linked or clustered in bacterial genomes, because each gene benefits from being co-transferred [102]. In other words, gaining the genes for this function separately provides no benefit until the whole set has been acquired. Genes that are clustered—and therefore probably transfer together—are more likely to provide recipients with the full set, and hence the corresponding fitness benefits. The argument can be extended in the light of the local adaptation model to explain multi-trait plasmids. Here, we can define the ‘function’ of the accessory genes as adaptation to a single specific environment, for example, an environment contaminated with several toxic substances. Lacking resistance to just one toxin might be lethal, and so locally adaptive plasmids will be most successful if they confer resistance to all. At a broader scale, this dynamic would play out to shape plasmid accessory gene content: where there is correlation between different local selective pressures, there is selection for the corresponding adaptive traits to become aggregated on a single mobile plasmid [98]. Where there are many of these locally correlated environmental pressures, we might expect to see larger plasmids.
There are several examples of multi-trait plasmids that may reflect these pressures. The use of antibiotics in clinical and agricultural settings means that microbes inhabiting those environments often face several specific chemical threats in contrast to closely-related strains in the wider environment. Large resistance plasmids often carry mechanisms to resist not only many different antibiotics (electronic supplementary material, figure S2), but also disinfectants, and metals, occasionally or historically used as antiseptics [103]. Toxic metals often co-occur in industrial waste from factories and mining, and similarly selects for plasmids with resistance to multiple metals/metalloids [104]. In soil, microbes can exploit diverse, chemically related, but low-abundant compounds produced by plants, thanks to multifunctional catabolic plasmids [94,105], which may explain why megaplasmids are more common in families such as Rhizobiaceae, Burkholderiaceae and Pseudomonadaceae, which often associated with soil (figure 2). Genes enabling nitrogen fixation and hydrogenotrophy are often conferred by the same plasmid because H2 is produced during the reduction of dinitrogen to ammonia, and hydrogenotrophy enables microbes to recover some of the energetic costs of nitrogen fixation [106]. Where adaptation to a new environment requires the concurrent expression of several different traits, larger plasmids can provide.
However, the functional traits encoded by megaplasmids are often only a small portion of the whole sequence. Many megaplasmid genes are of unknown function, and it is not clear whether these are selfish or mutualistic. Much work is required to characterize their biochemical and evolutionary roles, but recent work looking for genes that co-localize with known anti-phage mechanisms has enabled several new genome defence pathways to be identified [107]. Perhaps similar approaches with locally-adapted plasmids may shed light on the ‘known unknowns’ of the prokaryote pangenome.
Besides accessory gene functions, megaplasmids can become more effective mutualists by reducing the impact they have on the physiology of their hosts. Some plasmids come equipped with transfer RNAs (tRNAs), which appear to be more common on larger plasmids (electronic supplementary material, figure S5), and along with other translation- and transcription-related genes may alleviate the metabolic burdens associated with carriage [53], as has been shown for tRNA-encoding prophage [108]. Plasmid burden can also be ameliorated by nucleoid-associated proteins like histone-like nucleoid structuring, which are often found on larger plasmids and can regulate plasmid gene expression by bridging distant DNA sequences to shape nucleoid architecture [109]. Anti-SOS proteins likewise minimize plasmid-associated disruptions [110]. Furthermore, megaplasmids, and their prokaryotic hosts, can evolve and coevolve to minimize burden. Single mutations can largely ameliorate carriage costs of some megaplasmids, and laboratory studies show that at least under certain conditions, some megaplasmids have no detectable fitness cost, suggesting that size per se is not principally responsible for megaplasmid fitness costs [111]. Very big plasmids are not necessarily very burdensome plasmids.
Continued positive selection for plasmid cargo genes means that plasmids can remain associated with a chromosome for a long time. Under these circumstances, adaptive and neutral processes favour an increasingly refined relationship between plasmid and chromosome, a process of plasmid 'domestication', the results of which are most clearly seen as chromids.
4. Are megaplasmids just chromids-in-training?
Most chromids characterized to date are relatively big, with replication machinery like that of plasmids, and it is likely that megaplasmids are the evolutionary progenitors of chromids [19,21]. Indeed, chromids often appear in plasmid databases and are sometimes considered as a form of large plasmid [13]. Two key features distinguish megaplasmids from chromids. First, chromids tend to more closely resemble their associated chromosomes in terms of nucleotide composition, suggestive of a prolonged association [19,21]. Consistent with this, chromids appear not to be horizontally transmitted, at least under natural conditions, and are instead inherited vertically [19,21]. Loss of horizontal transmission has been reported in evolution experiments with large conjugative plasmids, indicating the ease with which megaplasmids can become exclusively associated with a lineage, beginning the domestication process [112,113]. The other key distinction is that chromids encode core cellular functions, that in other species are on the chromosome, the implication being that cells cured of their chromids are not viable under any condition [21]. On a global level, core genes are less likely to undergo successful horizontal gene transfer, owing to the difficulties associated with integrating a foreign gene variant into the tight-knit protein interaction networks characteristic of core cellular activity [114]. However, like chromids, mobile megaplasmids also occasionally carry homologues of core genes. These genes may have distinct functions, but in some cases, such as the metabolic genes present on the resident extrachromosomal replicons of Sinorhizobium meliloti, or plasmid-borne chaparonin genes found in Escherichia, have been shown to complement mutations in their cognate chromosomal copies [115,116], indicative of genetic functional redundancy [117].
Redundant plasmid-borne core genes can play an important role in evolution. The loss of one copy of a redundant function, either through drift, or by selection (also known as Black Queen dynamics), renders the remaining copy essential, increasing interdependency between partners [118]. In a genome with both plasmid and chromosomal copies of a core gene, disruption of the chromosomal copy may have no phenotypic effect, but immediately renders the plasmid essential, dividing (albeit asymmetrically) core cellular functions across two replicons [40]. This increased genomic complexity may not have any immediate functional or mechanistic adaptive benefit [119], but it establishes the plasmid in the genome, exposing it to local mutational biases, and enabling subsequent evolutionary processes to exploit the gene regulatory or replication opportunities offered by a multipartite genome. Redundancy also enables core cellular functions to accumulate deleterious mutations, shielded from the purifying selection such genes would experience were they single-copy [120]. Plasmids thus act as ‘scribbling pads’ [121], with redundant genes and non-adaptive features providing a source of standing genetic variation that can enable evolution to traverse valleys in the fitness landscape [32].
The presence of redundant core genes on megaplasmids has inspired a model whereby megaplasmids are destined to either evolve into a chromid, or be lost [19]. However, distantly-related conjugative megaplasmids show clear gene synteny despite nucleotide sequence divergence, suggesting that these families have persisted and diversified as large, horizontally transmissible mobile genetic elements [24,25,53,80]. Some megaplasmids have clearly been around for a long time, suggesting that they have found success in a particular niche of the sub-cellular ecosystem, rather than representing a transitional state on the way to domestication.
5. Plasmid plasticity promotes megaplasmid emergence
While some megaplasmids seem to be ancient, the plasticity of plasmids provides many opportunities for new megaplasmids to arise [46]. There are many cases where large plasmids are clearly fusions, or mosaics, of two or more smaller replicons [43]. For example, in Bacillus cereus, a study found 29 out of 31 plasmids greater than 100 kb carried multiple different replication machineries, with phylogenetic analyses showing different evolutionary histories within each megaplasmid [122]. A megaplasmid in Klebsiella pneumoniae provides a concerning case where virulence and antimicrobial resistance (AMR) functions, which usually have distinct patterns of horizontal transmission, became conjoined, with unclear implications for public health [123,124]. Such fused megaplasmids may not be long-lived, but nevertheless provide powerful new evolutionary opportunities.
Plasmids can usually accumulate transposons more easily than chromosomal loci, because the genes they carry are not essential. As the transposon itself is redundant, it then provides a site for successive transposons to insert with negligible disruption. This generates 'hotspot' or 'junkyard' regions, which can accumulate new insertions, and thus standing genetic variation [125]. Such hotspot regions could also accelerate plasmid expansion, pushing large plasmids over the threshold into becoming megaplasmids. Transposons can also interact with one another or with their target sites, causing further rearrangements [126]. For example, homologous recombination between different copies of the same transposon can cause the capture of plasmid genes to the chromosome [127], and genes from other replicons to be acquired by plasmids [128], potentially mobilizing those genes to transmit onwards into new recipients. Conversely, interactions between transposons can also cause gene deletion from megaplasmids [113,129]. In many ways, the large size of megaplasmids reflects the plasticity of large plasmids more broadly, exemplifying the ease with which they can acquire and shuttle constituents of the pangenome.
6. Boxes: megaplasmids to watch
(a) . Salmonella enterica infantis, an emerging pathogen with an emerging megaplasmid: pESI (280 kb)
Salmonella enterica serovar Infantis (S. Infantis) is a globally emerging serovar of Salmonella enterica and a common cause of human salmonellosis [130]. The dominant lineage, which has become established in poultry and now poses most risk to humans owes its success to a virulence-resistance megaplasmid (approx. 280 kb) known as pESI [130–133]. pESI-like megaplasmids have now been found in S. Infantis isolates from around the world [134]. pESI confers several functions associated with pathogen success in environments of high antimicrobial usage: resistance to diverse antimicrobials, including mercury and oxidative stress, a siderophore cluster for iron acquisition during infection, and fimbriae genes, which might enable biofilm formation and epithelial adhesion [132]. Overall, the content of the megaplasmid varies between isolates, consistent with a mosaic structure that is derived from both IncI1 and IncP-1alpha plasmids, as well as different transposons [132,135]. pESI appears to have been principally inherited vertically by the emerging S. Infantis lineages, but there is evidence that it transmits readily between gut commensals and other pathogens [135].
(b) . Pseudomonas antimicrobial resistance megaplasmids, bridging the environment and the clinic: pBT2436 (423 kb) and relatives
Incompatibility typing and gel electrophoresis in the 1970s and 1980s revealed a large group of Pseudomonas megaplasmids (approx. 500 kb) known as IncP-2, which, in nosocomial isolates, carried antimicrobial resistance, and in soil isolates carried pathways for degradation of complex organic compounds such as camphor and octane [136]. Spurred on by the availability of long-read sequencing—often necessary to resolve their complex or repetitive structures [24,26]—recent years have seen an increase in the number of related Pseudomonas megaplasmid sequences [24,25,79,137], challenging the prevailing viewpoint that plasmids make only a minor contribution to antimicrobial resistance in Pseudomonas [138]. Found in clinical samples, soil isolates, and industrial processes, this family exemplifies the mosaic nature of megaplasmids and their ability to confer locally-adaptive traits. The conserved, syntenic region putatively encodes a metabolic gene cluster, chemotaxis apparatus, and a type IV pilus, as well as genes involved in plasmid replication, segregation, and transmission, but members of this family also possess a variable region with sample-specific gene content [24,26,139]. The apparently high stability, low fitness cost, and efficient transmission of these megaplasmids suggests that they could be effective vehicles of gene exchange in diverse habitats.
(c) . Acinetobacter megaplasmids, global distribution, local adaptation: pR4WN-1BD1 (285 kb) and relatives
Interestingly, a pattern similar to that of Pseudomonas AMR megaplasmids is emerging in another environmentally-widespread bacterium and occasional opportunistic pathogen: Acinetobacter spp. Acinetobacter plasmids are often big: 10% in NCBI GenBank were found to be greater than 100 kb [140]. A combination of PacBio and HiSeq sequencing recently revealed a family of megaplasmids (250–400 kb in size) transmitting multi-drug resistance across 11 different Acinetobacter species. A role for local adaptation in shaping plasmid accessory gene content was discernible in these data, as plasmids from similar geographical locations had similar accessory genes, even if they were not closely related [27]. Increased sampling of non-clinical sites may be necessary to reveal the full megaplasmid picture in Acinetobacter, as an analysis of Acinetobacter plasmids from hospitals and soil/water suggested that larger plasmids were obtained from environmental samples [141].
(d) . Haloarchaeal megaplasmids, gas vesicle giants
While plasmids have been identified across Archaea, large plasmids and megaplasmids seem more restricted, having been reported only in Haloarchaea and in some species of Methanosarcinales [14]. However, amongst the Haloarchaea, megaplasmids (greater than 130 kb) are common [142]. Recombination between plasmids and chromosomes occurs frequently, probably facilitated by the transposable elements often present at very high density on haloarchaeal megaplasmids [143]. As a consequence of this dynamic genome architecture, chromosomal genes are often found on haloarchaeal megaplasmids, sometimes resulting in the megaplasmid becoming an essential chromid [144]. Haloarchaeal megaplasmids are distinct amongst the mainly parasitic Archaeal extrachromosomal elements because they confer known accessory traits: in particular, gas vesicles that assist with buoyancy [14,144]. Some haloarchaeal megaplasmids do encode genes resembling bacterial conjugative apparatus [144], but horizontal gene transfer between Haloarchaea readily occurs via cell fusions which can transmit chromosomal as well as plasmid DNA [145]. The origin of the haloarchaeal megaplasmids has attracted speculation because megaplasmids are uncommon in Archaea. This has lead some to suggest that modern haloarchaeal megaplasmids emerged from an ancient inter-kingdom transmission event [14], which, considering the ease with which Haloarchaea form cytoplasmic bridges with their neighbours, along with phylogenetic analyses which indicate that the common ancestor of all Haloarchaea acquired approximately 1000 genes from Eubacteria [146], is a compelling hypothesis.
7. Conclusion and perspective
Megaplasmids are not the only maxi-size mobile genetic element to rise to prominence with new sequencing efforts. Large symbiosis ICEs (greater than 400 kb) are frequently spotted on Mesorhizobium spp. [147,148]. Huge bacteriophages, also known as jumbophage, have been identified and sequenced from isolates and from metagenomes, with the largest to date at 735 kb [149]. As with megaplasmids, many jumbophage genes are of unknown function, but some functions have been identified: CRISPR systems, nucleoid structuring proteins, and genes that support and manipulate host translation during the infection process [149,150]. ‘Borgs’—linear megaplasmids up to 1 Mbp in size—appear to confer locally-adaptive ecologically-significant functions to the methane-oxidising Archaeon, Methanoperedens [151]. The size and complexity of large mobile genetic elements demonstrates that sub-cellular life need not lack sophistication when undertaking large-scale reprogramming of cellular functions. As more—possibly even larger—elements are discovered through continued, broader, less-biased sequencing initiatives, we will develop an increasingly detailed picture of the intriguing ecologies and adaptations of huge mobile genetic elements.
From their original isolation and detection with modified DNA extraction techniques, to pulse-field gel analysis, to a renaissance facilitated by long-read sequencing, knowledge of megaplasmids in particular, and multipartite genome structure in general, has advanced with each technological development. What comes next? We anticipate that wider genomics and metagenomics surveys, informed by long-read sequencing, will expand our understanding of the habitat distribution of megaplasmids and chromids, as well as their accessory gene content. However, experiments, including laboratory evolution and detailed molecular and structural studies, will be crucial to test megaplasmid biochemical, ecological, and evolutionary functions. Contact sequencing approaches such as Hi-C and meta3C will provide powerful insight into the spatial structure of mobile genetic elements both within cells and within communities, enabling an understanding of how plasmids and chromids interact with chromosomal architecture, and population dynamics and drivers of transmission [152–154]. Finally, pangenomics analysis pipelines, combined with the ever-increasing abundance of sequencing data, will improve our knowledge of megaplasmid cargo, and how megaplasmids co-associate (or anti-associate) with other components of the accessory genome within cells [155]. Such work, alongside comparisons with other mobile genetic elements and routes of gene exchange, will help contextualize the role played by these high-capacity vehicles for horizontal gene transfer, giants of the sub-cellular world.
Acknowledgements
We thank Ellie Harrison for comments on an earlier draft. We apologize to all authors whose relevant work could not be cited for reasons of space.
Data accessibility
Data presented in this manuscript are already publicly available through accessible databases (NCBI, COMPASS, EZBioCloud). These databases are referenced in the text, and description is provided in the text for how they were accessed.
Authors' contributions
J.P.J.H.: conceptualization. All authors: writing; original draft. All authors: writing; review and editing.
Competing interests
We declare we have no competing interests.
Funding
A.C. has been supported by the EMBL-EBI/Wellcome Trust Sanger Institute Join Post-Doctoral Fellowship Program (ESPOD). J.B.J.H. is funded through the Max Planck Society (via a fellowship to Hinrich Schulenburg, Max-Planck Institute for Evolutionary Biology, Ploen, Germany).
References
- 1.Lederberg J. 1952. Cell genetics and hereditary symbiosis. Physiol. Rev. 32, 403-430. ( 10.1152/physrev.1952.32.4.403) [DOI] [PubMed] [Google Scholar]
- 2.Almpanis A, Swain M, Gatherer D, McEwan N. 2018. Correlation between bacterial G + C content, genome size and the G + C content of associated plasmids and bacteriophages. Microb. Genomics 4, 168. ( 10.1099/mgen.0.000168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinnebusch J, Tilly K. 1993. Linear plasmids and chromosomes in bacteria. Mol. Microbiol. 10, 917-922. ( 10.1111/j.1365-2958.1993.tb00963.x) [DOI] [PubMed] [Google Scholar]
- 4.Neale HC, Jackson RW, Preston GM, Arnold DL. 2018. Supercoiling of an excised genomic island represses effector gene expression to prevent activation of host resistance. Mol. Microbiol. 112, 444-454. ( 10.1111/mmi.14111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ices and conjugative plasmids is getting thinner. PLoS Genet. 11, e1005298. ( 10.1371/journal.pgen.1005298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shintani M, Takahashi Y, Yamane H, Nojiri H. 2010. The behavior and significance of degradative plasmids belonging to Inc groups in Pseudomonas within natural environments and microcosms. Microbes Environ. 25, 253-265. ( 10.1264/jsme2.me10155) [DOI] [PubMed] [Google Scholar]
- 7.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434-464. ( 10.1128/MMBR.62.2.434-464.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acman M, van Dorp L, Santini JM, Balloux F. 2020. Large-scale network analysis captures biological features of bacterial plasmids. Nat. Commun. 11, 1-11. ( 10.1038/s41467-020-16282-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Paulsen IT, Palenik B. 2012. Analysis of two marine metagenomes reveals the diversity of plasmids in oceanic environments. Environ. Microbiol. 14, 453-466. ( 10.1111/j.1462-2920.2011.02633.x) [DOI] [PubMed] [Google Scholar]
- 10.Heuer H, Smalla K. 2012. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36, 1083-1104. ( 10.1111/j.1574-6976.2012.00337.x) [DOI] [PubMed] [Google Scholar]
- 11.Suzuki Y, Nishijima S, Furuta Y, Yoshimura J, Suda W, Oshima K, Hattori M, Morishita S. 2019. Long-read metagenomic exploration of extrachromosomal mobile genetic elements in the human gut. Microbiome 7, 119. ( 10.1186/s40168-019-0737-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez MF, Kurth D, Farías ME, Soria MN, Castillo Villamizar GA, Poehlein A, Daniel R, Dib JR. 2020. First report on the plasmidome from a high-altitude Lake of the Andean Puna. Front. Microbiol. 11, 1343. ( 10.3389/fmicb.2020.01343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douarre P-E, Mallet L, Radomski N, Felten A, Mistou M-Y. 2020. Analysis of COMPASS, a new comprehensive plasmid database revealed prevalence of multireplicon and extensive diversity of IncF plasmids. Front. Microbiol. 11, 483. ( 10.3389/fmicb.2020.00483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forterre P, Krupovic M, Raymann K, Soler N. 2014. Plasmids from Euryarchaeota. Microbiol. Spectr 2, PLAS-0027-2014. ( 10.1128/microbiolspec.PLAS-0027-2014) [DOI] [PubMed] [Google Scholar]
- 15.Farrar NA, Williams KL. 1988. Nuclear plasmids in the simple eukaryotes Saccharomyces cerevisiae and Dictyostelium discoideum. Trends Genet. 4, 343-348. ( 10.1016/0168-9525(88)90054-6) [DOI] [PubMed] [Google Scholar]
- 16.Glass JI, et al. 2006. Essential genes of a minimal bacterium. Proc. Natl Acad. Sci. USA 103, 425-430. ( 10.1073/pnas.0510013103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg C, Casse-Delbart F, Dusha I, David M, Boucher C. 1982. Megaplasmids in the plant-associated bacteria Rhizobium meliloti and Pseudomonas solanacearum. J. Bacteriol. 150, 402-406. ( 10.1128/JB.150.1.402-406.1982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg C, Boistard P, Dénarié J, Casse-Delbart F. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184, 326-333. ( 10.1007/BF00272926) [DOI] [PubMed] [Google Scholar]
- 19.diCenzo GC, Finan TM. 2017. The divided bacterial genome: structure, function, and evolution. Microbiol. Mol. Biol. Rev. 81, e00019-17. ( 10.1128/MMBR.00019-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz E. 2009. Microbial megaplasmids. Berlin, Germany: Springer Science & Business Media. See https://market.android.com/details?id=book-25ZUrglbfhIC. [Google Scholar]
- 21.Harrison PW, Lower RPJ, Kim NKD, Young JPW. 2010. Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol. 18, 141-148. ( 10.1016/j.tim.2009.12.010) [DOI] [PubMed] [Google Scholar]
- 22.Maule J. 1998. Pulsed-field gel electrophoresis. Mol. Biotechnol. 9, 107-126. ( 10.1007/BF02760813) [DOI] [PubMed] [Google Scholar]
- 23.Lilley AK, Bailey MJ, Day MJ, Fry JC. 1996. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol. Ecol. 20, 211-227. ( 10.1111/j.1574-6941.1996.tb00320.x) [DOI] [Google Scholar]
- 24.Cazares A, et al. 2020. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 11, 1370. ( 10.1038/s41467-020-15081-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Botelho J, Lood C, Partridge SR, van Noort V, Lavigne R, Grosso F, Peixe L. 2019. Combining sequencing approaches to fully resolve a carbapenemase-encoding megaplasmid in a Pseudomonas shirazica clinical strain. Emerg. Microbes Infect. 8, 1186-1194. ( 10.1080/22221751.2019.1648182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid M, Frei D, Patrignani A, Schlapbach R, Frey JE, Remus-Emsermann MNP, Ahrens CH. 2018. Pushing the limits of de novo genome assembly for complex prokaryotic genomes harboring very long, near identical repeats. Nucleic Acids Res. 46, 8953-8965. ( 10.1093/nar/gky726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaly TM, Paulsen IT, Sajjad A, Tetu SG, Gillings MR. 2020. A novel family of acinetobacter mega-plasmids are disseminating multi-drug resistance across the globe while acquiring location-specific accessory genes. Front. Microbiol. 11, 605952. ( 10.3389/fmicb.2020.605952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeifer E, de Sousa JAM, Touchon M, Rocha EPC. 2021. Bacteria have numerous phage-plasmid families with conserved phage and variable plasmid gene repertoires. Nuc. Acids Res. 49, 2655-2673. ( 10.1093/nar/gkab064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erdmann S, Tschitschko B, Zhong L, Raftery MJ, Cavicchioli R. 2017. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat. Microbiol. 2, 1446-1455. ( 10.1038/s41564-017-0009-2) [DOI] [PubMed] [Google Scholar]
- 30.Fillol-Salom A, Martínez-Rubio R, Abdulrahman RF, Chen J, Davies R, Penadés JR. 2018. Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J. 12, 2114-2128. ( 10.1038/s41396-018-0156-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613-1617. ( 10.1099/ijsem.0.001755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Beltrán J, DelaFuente J, León-Sampedro R, MacLean RC, San Millán Á. 2021. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 19, 347-359. ( 10.1038/s41579-020-00497-1) [DOI] [PubMed] [Google Scholar]
- 33.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434-452. ( 10.1128/MMBR.00020-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinto UM, Pappas KM, Winans SC. 2012. The ABCs of plasmid replication and segregation. Nat. Rev. Microbiol. 10, 755-765. ( 10.1038/nrmicro2882) [DOI] [PubMed] [Google Scholar]
- 35.Marasini D, Karki AB, Bryant JM, Sheaff RJ, Fakhr MK. 2020. Molecular characterization of megaplasmids encoding the type VI secretion system in Campylobacter jejuni isolated from chicken livers and gizzards. Sci. Rep. 10, 12514. ( 10.1038/s41598-020-69155-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller E, Hotzel H, Ahlers C, Hänel I, Tomaso H, Abdel-Glil MY. 2020. Genomic analysis and antimicrobial resistance of Aliarcobacter cryaerophilus strains from German water poultry. Front. Microbiol. 11, 1549. ( 10.3389/fmicb.2020.01549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fournes F, Val M-E, Skovgaard O, Mazel D. 2018. Replicate once per cell cycle: replication control of secondary chromosomes. Front. Microbiol. 9, 1833. ( 10.3389/fmicb.2018.01833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausiannikava D, Mitchell L, Marriott H, Smith V, Hawkins M, Makarova KS, Koonin EV, Nieduszynski CA, Allers T. 2018. Evolution of genome architecture in archaea: spontaneous generation of a new chromosome in Haloferax volcanii. Mol. Biol. Evol. 35, 1855-1868. ( 10.1093/molbev/msy075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dziewit L, Czarnecki J, Wibberg D, Radlinska M, Mrozek P, Szymczak M, Schlüter A, Pühler A, Bartosik D. 2014. Architecture and functions of a multipartite genome of the methylotrophic bacterium Paracoccus aminophilus JCM 7686, containing primary and secondary chromids. BMC Genomics 15, 124. ( 10.1186/1471-2164-15-124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rousset F, Cabezas-Caballero J, Piastra-Facon F, Fernández-Rodríguez J, Clermont O, Denamur E, Rocha EPC, Bikard D. 2021. The impact of genetic diversity on gene essentiality within the Escherichia coli species. Nat. Microbiol. 6, 301-312. ( 10.1038/s41564-020-00839-y) [DOI] [PubMed] [Google Scholar]
- 41.Garcillán-Barcia MP, de la Cruz F. 2013. Ordering the bestiary of genetic elements transmissible by conjugation. Mob. Genet. Elements 3, e24263. ( 10.4161/mge.24263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman's guide to bacterial insertion sequences. Microbiol. Spectr. 3, MDNA3-0030-2014. ( 10.1128/microbiolspec.MDNA3-0030-2014) [DOI] [PubMed] [Google Scholar]
- 43.Pesesky MW, Tilley R, Beck DAC. 2019. Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid 102, 10-18. ( 10.1016/j.plasmid.2019.02.003) [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Lopez R, Redondo S, Garcillan-Barcia MP, de la Cruz F. 2017. Towards a taxonomy of conjugative plasmids. Curr. Opin. Microbiol. 38, 106-113. ( 10.1016/j.mib.2017.05.005) [DOI] [PubMed] [Google Scholar]
- 45.Carraro N, Burrus V. 2015. The dualistic nature of integrative and conjugative elements. Mob. Genet. Elements 5, 98-102. ( 10.1080/2159256X.2015.1102796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cury J, Oliveira PH, de la Cruz F, Rocha EPC. 2018. Host range and genetic plasticity explain the coexistence of integrative and extrachromosomal mobile genetic elements. Mol. Biol. Evol. 35, 2230-2239. ( 10.1093/molbev/msy123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, Rocha EPC, Garcillán-Barcia MP, de la Cruz F. 2020. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11, 3602. ( 10.1038/s41467-020-17278-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anda M, Ohtsubo Y, Okubo T, Sugawara M, Nagata Y, Tsuda M, Minamisawa K, Mitsui H. 2015. Bacterial clade with the ribosomal RNA operon on a small plasmid rather than the chromosome. Proc. Natl Acad. Sci. USA 112, 14343-14 347. ( 10.1073/pnas.1514326112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.diCenzo GC, MacLean AM, Milunovic B, Golding GB, Finan TM. 2014. Examination of prokaryotic multipartite genome evolution through experimental genome reduction. PLoS Genet. 10, e1004742. ( 10.1371/journal.pgen.1004742) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fei F, diCenzo GC, Bowdish DME, McCarry BE, Finan TM. 2015. Effects of synthetic large-scale genome reduction on metabolism and metabolic preferences in a nutritionally complex environment. Metabolomics 12, 23. ( 10.1007/s11306-015-0928-y) [DOI] [Google Scholar]
- 51.diCenzo GC, Wellappili D, Golding GB, Finan TM. 2018. Inter-replicon gene flow contributes to transcriptional integration in the Sinorhizobium meliloti multipartite genome. G3 Genes|Genomes|Genetics 8, 1711-1720. ( 10.1534/g3.117.300405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fecskeová L, Kovařík M, Javorský P, Pristaš P. 2012. Mosaic structure of the small cryptic plasmid pKST23 from Escherichia coli. Folia Microbiol. 57, 277-279. ( 10.1007/s12223-012-0124-7) [DOI] [PubMed] [Google Scholar]
- 53.Smith BA, Leligdon C, Baltrus DA. 2019. Just the two of us? A family of pseudomonas megaplasmids offers a rare glimpse into the evolution of large mobile elements. Genome Biol. Evol. 11, 1192-1206. ( 10.1093/gbe/evz066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malhi Y, Doughty CE, Galetti M, Smith FA, Svenning J-C, Terborgh JW. 2016. Megafauna and ecosystem function from the Pleistocene to the Anthropocene. Proc. Natl Acad. Sci. USA 113, 838-846. ( 10.1073/pnas.1502540113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eberhard WG. 1990. Evolution in bacterial plasmids and levels of selection. Q. Rev. Biol. 65, 3-22. ( 10.1086/416582) [DOI] [PubMed] [Google Scholar]
- 56.Rocha EPC. 2018. Neutral theory, microbial practice: challenges in bacterial population genetics. Mol. Biol. Evol. 35, 1338-1347. ( 10.1093/molbev/msy078) [DOI] [PubMed] [Google Scholar]
- 57.Turner PE, Cooper VS, Lenski RE. 1998. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52, 315-329. ( 10.2307/2411070) [DOI] [PubMed] [Google Scholar]
- 58.Dimitriu T, Matthews A, Buckling A. 2020. Increased copy number couples the evolution of plasmid horizontal transmission and antibiotic resistance. bioRxiv ( 10.1101/2020.08.12.248336) [DOI] [PMC free article] [PubMed]
- 59.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262-267. ( 10.1016/j.tim.2012.04.003) [DOI] [PubMed] [Google Scholar]
- 60.Kuo C-H, Ochman H. 2009. Deletional bias across the three domains of life. Genome Biol. Evol. 1, 145-152. ( 10.1093/gbe/evp016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ebersbach G, Gerdes K. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39, 453-479. ( 10.1146/annurev.genet.38.072902.091252) [DOI] [PubMed] [Google Scholar]
- 62.Zhong C, Peng D, Ye W, Chai L, Qi J, Yu Z, Ruan L, Sun M. 2011. Determination of plasmid copy number reveals the total plasmid DNA amount is greater than the chromosomal DNA amount in Bacillus thuringiensis YBT-1520. PLoS ONE 6, e16025. ( 10.1371/journal.pone.0016025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Summers DK. 1991. The kinetics of plasmid loss. Trends Biotechnol. 9, 273-278. ( 10.1016/0167-7799(91)90089-Z) [DOI] [PubMed] [Google Scholar]
- 64.Sengupta M, Austin S. 2011. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79, 2502-2509. ( 10.1128/IAI.00127-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Melderen L, De Bast M. Saavedra. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5, e1000437. ( 10.1371/journal.pgen.1000437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kusumawardhani H, van Dijk D, Hosseini R, de Winde JH. 2020. A novel toxin-antitoxin module SlvT-SlvA regulates megaplasmid stability and incites solvent tolerance in Pseudomonas putida S12. Appl. Environ. Microbiol. 86, e00686-20. ( 10.1128/AEM.00686-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morton ER, Platt TG, Fuqua C, Bever JD. 2014. Non-additive costs and interactions alter the competitive dynamics of co-occurring ecologically distinct plasmids. Proc. R. Soc. B 281, 20132173. ( 10.1098/rspb.2013.2173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loftie-Eaton W, et al. 2016. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol. Biol. Evol. 33, 885-897. ( 10.1093/molbev/msv339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stalder T, Rogers LM, Renfrow C, Yano H, Smith Z, Top EM. 2017. Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance. Sci. Rep. 7, 4853. ( 10.1038/s41598-017-04662-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kopfmann S, Hess WR. 2013. Toxin-antitoxin systems on the large defense plasmid pSYSA of synechocystis sp. PCC 6803*. J. Biol. Chem. 288, 7399-7409. ( 10.1074/jbc.M112.434100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bardaji L, Añorga M, Echeverría M, Ramos C, Murillo J. 2019. The toxic guardians: multiple toxin-antitoxin systems provide stability, avoid deletions and maintain virulence genes of Pseudomonas syringae virulence plasmids. Mob. DNA 10, 7. ( 10.1186/s13100-019-0149-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pilla G, McVicker G, Tang CM. 2017. Genetic plasticity of the Shigella virulence plasmid is mediated by intra- and inter-molecular events between insertion sequences. PLoS Genet. 13, e1007014. ( 10.1371/journal.pgen.1007014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Popa O, Landan G, Dagan T. 2017. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 11, 543-554. ( 10.1038/ismej.2016.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. 2010. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl Acad. Sci. USA 107, 127-132. ( 10.1073/pnas.0908978107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hua J, Huet A, Lopez CA, Toropova K, Pope WH, Duda RL, Hendrix RW, Conway JF. 2017. Capsids and genomes of jumbo-sized bacteriophages reveal the evolutionary reach of the HK97 Fold. MBio 8, e01579-17. ( 10.1128/mBio.01579-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ. Biosafety Res. 6, 37-53. ( 10.1051/ebr:2007031) [DOI] [PubMed] [Google Scholar]
- 77.Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7, 703-714. ( 10.1038/nrmicro2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guglielmini J, Néron B, Abby SS, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. 2014. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42, 5715-5727. ( 10.1093/nar/gku194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Urbanowicz P, Bitar I, Izdebski R, Baraniak A, Literacka E, Hrabák J, Gniadkowski M. 2021. Epidemic territorial spread of IncP-2-Type VIM-2 carbapenemase-encoding megaplasmids in nosocomial Pseudomonas aeruginosa populations. Antimicrob. Agents Chemother. 65, e02122-20. ( 10.1128/AAC.02122-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hall JPJ, Harrison E, Lilley AK, Paterson S, Spiers AJ, Brockhurst MA. 2015. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 17, 5008-5022. ( 10.1111/1462-2920.12901) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romanchuk A, Jones CD, Karkare K, Moore A, Smith BA, Jones C, Dougherty K, Baltrus DA. 2014. Bigger is not always better: transmission and fitness burden of 1 MB Pseudomonas syringae megaplasmid pMPPla107. Plasmid 73, 16-25. ( 10.1016/j.plasmid.2014.04.002) [DOI] [PubMed] [Google Scholar]
- 82.Orlek A, et al. 2017. Ordering the mob: Insights into replicon and MOB typing schemes from analysis of a curated dataset of publicly available plasmids. Plasmid 91, 42-52. ( 10.1016/j.plasmid.2017.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burmølle M, Bahl MI, Jensen LB, Sørensen SJ, Hansen LH. 2008. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154, 187-195. ( 10.1099/mic.0.2007/010454-0) [DOI] [PubMed] [Google Scholar]
- 84.Ishiwa A, Komano T. 2004. PilV adhesins of plasmid R64 thin pili specifically bind to the lipopolysaccharides of recipient cells. J. Mol. Biol. 343, 615-625. ( 10.1016/j.jmb.2004.08.059) [DOI] [PubMed] [Google Scholar]
- 85.Li Z, et al. 2020. Molecular genetic analysis of an XDR Pseudomonas aeruginosa ST664 clone carrying multiple conjugal plasmids. J. Antimicrob. Chemother. 75, 1443-1452. ( 10.1093/jac/dkaa063) [DOI] [PubMed] [Google Scholar]
- 86.Carattoli A, Hasman H. 2020. PlasmidFinder and in silico pmlst: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol. Biol. 2075, 285-294. ( 10.1007/978-1-4939-9877-7_20) [DOI] [PubMed] [Google Scholar]
- 87.Dionisio F, Zilhão R, Gama JA. 2019. Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid 102, 29-36. ( 10.1016/j.plasmid.2019.01.003) [DOI] [PubMed] [Google Scholar]
- 88.Cooper TF, Heinemann JA. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl Acad. Sci. USA 97, 12 643-12 648. ( 10.1073/pnas.220077897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.San MA, Heilbron K, MacLean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J. 8, 601-612. ( 10.1038/ismej.2013.182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pinilla-Redondo R, Mayo-Muñoz D, Russel J, Garrett RA, Randau L, Sørensen SJ, Shah SA. 2019. Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 48, 2000-2012. ( 10.1093/nar/gkz1197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Newire E, Aydin A, Juma S, Enne VI, Roberts AP. 2020. Identification of a Type IV-A CRISPR-Cas system located exclusively on IncHI1B/IncFIB plasmids in Enterobacteriaceae. Front. Microbiol. 11, 1937. ( 10.3389/fmicb.2020.01937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pinilla-Redondo R, Russel J, Mayo-Muñoz D, Shah SA, Garrett RA, Nesme J, Madsen JS, Fineran PC, Sørensen SJ. In press. CRISPR-Cas systems are widespread accessory elements across bacterial and archaeal plasmids. Nuc. Acids Res. 2021, gkab859. ( 10.1093/nar/gkab859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pilla G, Tang CM. 2018. Going around in circles: virulence plasmids in enteric pathogens. Nat. Rev. Microbiol. 16, 484-495. ( 10.1038/s41579-018-0031-2) [DOI] [PubMed] [Google Scholar]
- 94.Dennis JJ. 2005. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16, 291-298. ( 10.1016/j.copbio.2005.04.002) [DOI] [PubMed] [Google Scholar]
- 95.Peter S, et al. 2020. Tracking of antibiotic resistance transfer and rapid plasmid evolution in a hospital setting by nanopore sequencing. mSphere 5, e00525-20. ( 10.1128/mSphere.00525-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bergstrom CT, Lipsitch M, Levin BR. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505-1519. ( 10.1093/genetics/155.4.1505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eberhard WG. 1989. Why do bacterial plasmids carry some genes and not others? Plasmid 21, 167-174. ( 10.1016/0147-619X(89)90040-1) [DOI] [PubMed] [Google Scholar]
- 98.Niehus R, Mitri S, Fletcher AG, Foster KR. 2015. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat. Commun. 6, 8924. ( 10.1038/ncomms9924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stinear TP, Pryor MJ, Porter JL, Cole ST. 2005. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology 151, 683-692. ( 10.1099/mic.0.27674-0) [DOI] [PubMed] [Google Scholar]
- 100.Mihasan M, Brandsch R. 2013. pAO1 of Arthrobacter nicotinovorans and the spread of catabolic traits by horizontal gene transfer in gram-positive soil bacteria. J. Mol. Evol. 77, 22-30. ( 10.1007/s00239-013-9576-x) [DOI] [PubMed] [Google Scholar]
- 101.Frank O, Göker M, Pradella S, Petersen J. 2015. Ocean's twelve: flagellar and biofilm chromids in the multipartite genome of Marinovum algicola DG898 exemplify functional compartmentalization. Environ. Microbiol. 17, 4019-4034. ( 10.1111/1462-2920.12947) [DOI] [PubMed] [Google Scholar]
- 102.Lawrence JG, Roth JR. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143, 1843-1860. ( 10.1093/genetics/143.4.1843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. 2015. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16, 964. ( 10.1186/s12864-015-2153-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, van der Lelie D, Mergeay M. 2007. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 189, 7417-7425. ( 10.1128/JB.00375-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vedler E. 2009. Megaplasmids and the degradation of aromatic compounds by soil bacteria. In Microbial megaplasmids (ed. Schwartz E), pp. 33-53. Berlin, Germany: Springer. [Google Scholar]
- 106.Schwartz E, Henne A, Cramm R, Eitinger T, Friedrich B, Gottschalk G. 2003. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H(2)-based ithoautotrophy and anaerobiosis. J. Mol. Biol. 332, 369-383. ( 10.1016/s0022-2836(03)00894-5) [DOI] [PubMed] [Google Scholar]
- 107.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120. ( 10.1126/science.aar4120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bailly-Bechet M, Vergassola M, Rocha E. 2007. Causes for the intriguing presence of tRNAs in phages. Genome Res. 17, 1486-1495. ( 10.1101/gr.6649807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takeda T, Yun C-S, Shintani M, Yamane H, Nojiri H. 2011. Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int. J. Evol. Biol. 2011, 685015. ( 10.4061/2011/685015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. 2009. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol. Cell 36, 121-130. ( 10.1016/j.molcel.2009.07.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hall JPJ, Wright RCT, Harrison E, Muddiman KJ, Jamie Wood A, Paterson S, Brockhurst MA. 2021. Plasmid fitness costs are caused by specific genetic conflicts. bioRxiv ( 10.1101/2021.04.10.439128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Turner PE, Williams ESCP, Okeke C, Cooper VS, Duffy S, Wertz JE. 2014. Antibiotic resistance correlates with transmission in plasmid evolution. Evolution 68, 3368-3380. ( 10.1111/evo.12537) [DOI] [PubMed] [Google Scholar]
- 113.Porse A, Schønning K, Munck C, Sommer MOA. 2016. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860-2873. ( 10.1093/molbev/msw163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cohen O, Gophna U, Pupko T. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481-1489. ( 10.1093/molbev/msq333) [DOI] [PubMed] [Google Scholar]
- 115.diCenzo GC, Finan TM. 2015. Genetic redundancy is prevalent within the 6.7 Mb Sinorhizobium meliloti genome. Mol. Genet. Genomics 290, 1345-1356. ( 10.1007/s00438-015-0998-6) [DOI] [PubMed] [Google Scholar]
- 116.Wein T, Wang Y, Barz M, Stücker FT, Hammerschmidt K, Dagan T. 2021. Essential gene acquisition destabilizes plasmid inheritance. PLoS Genet. 17, e1009656. ( 10.1371/journal.pgen.1009656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Láruson ÁJ, Yeaman S, Lotterhos KE. 2020. The importance of genetic redundancy in evolution. Trends Ecol. Evol. 35, 809-822. ( 10.1016/j.tree.2020.04.009) [DOI] [PubMed] [Google Scholar]
- 118.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3, e00036-12. ( 10.1128/mBio.00036-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McShea DW, Brandon RN. 2010. Biology‘s first Law: the tendency for diversity and complexity to increase in evolutionary systems. Chicago, IL: University of Chicago Press. [Google Scholar]
- 120.Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. 2010. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput. Biol. 6, e1000732. ( 10.1371/journal.pcbi.1000732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Norris V, Merieau A. 2013. Plasmids as scribbling pads for operon formation and propagation. Res. Microbiol. 164, 779-787. ( 10.1016/j.resmic.2013.04.003) [DOI] [PubMed] [Google Scholar]
- 122.Zheng J, Peng D, Ruan L, Sun M. 2013. Evolution and dynamics of megaplasmids with genome sizes larger than 100 kb in the Bacillus cereus group. BMC Evol. Biol. 13, 262. ( 10.1186/1471-2148-13-262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lam MMC, Wyres KL, Wick RR, Judd LM, Fostervold A, Holt KE, Löhr IH. 2019. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J. Antimicrob. Chemother. 74, 1218-1222. ( 10.1093/jac/dkz028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wyres KL, et al. 2019. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 15, e1008114. ( 10.1371/journal.pgen.1008114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Oliveira PH, Touchon M, Cury J, Rocha EPC. 2017. The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun. 8, 841. ( 10.1038/s41467-017-00808-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBio 6, e00762. ( 10.1128/mBio.00762-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang X, Deatherage DE, Zheng H, Georgoulis SJ, Barrick JE. 2019. Evolution of satellite plasmids can prolong the maintenance of newly acquired accessory genes in bacteria. Nat. Commun. 10, 5809. ( 10.1038/s41467-019-13709-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. 2016. Mechanisms of evolution in high-consequence drug resistance plasmids. MBio 7, e01987-16. ( 10.1128/mBio.01987-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee M-C, Marx CJ. 2012. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 8, e1002651. ( 10.1371/journal.pgen.1002651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gymoese P, Kiil K, Torpdahl M, Østerlund MT, Sørensen G, Olsen JE, Nielsen EM, Litrup E. 2019. WGS based study of the population structure of Salmonella enterica Serovar Infantis. BMC Genomics 20, 870. ( 10.1186/s12864-019-6260-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gal-Mor O, Valinsky L, Weinberger M, Guy S, Jaffe J, Schorr YI, Raisfeld A, Agmon V, Nissan I. 2010. Multidrug-resistant Salmonella enterica Serovar Infantis, Israel. Emerg. Infect. Dis. 16, 1754-1757. ( 10.3201/eid1611.100100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, Rahav G, Grassl GA, Gal-Mor O. 2014. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica Serovar Infantis strain. Environ. Microbiol. 16, 977-994. ( 10.1111/1462-2920.12351) [DOI] [PubMed] [Google Scholar]
- 133.Cohen E, Rahav G, Gal-Mor O. 2020. Genome sequence of an emerging Salmonella enterica Serovar Infantis and genomic comparison with other S. Infantis strains. Genome Biol. Evol. 12, 151-159. ( 10.1093/gbe/evaa048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alba P, et al. 2020. Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genomics 6, 365. ( 10.1099/mgen.0.000365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aviv G, Rahav G, Gal-Mor O. 2016. Horizontal transfer of the Salmonella enterica Serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. MBio 7, e01395-16. ( 10.1128/mBio.01395-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jacoby GA, Sutton L, Knobel L, Mammen P. 1983. Properties of IncP-2 plasmids of Pseudomonas spp. Antimicrob. Agents Chemother. 24, 168-175. ( 10.1128/AAC.24.2.168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pincus NB, et al. 2020. Long-term persistence of an extensively drug-resistant subclade of globally distributed Pseudomonas aeruginosa clonal complex 446 in an academic medical center. Clin. Infect. Dis. 71, 1524-1531. ( 10.1093/cid/ciz973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088-17. ( 10.1128/CMR.00088-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weiser R, et al. 2019. Not all Pseudomonas aeruginosa are equal: strains from industrial sources possess uniquely large multireplicon genomes. Microb. Genomics 5, 0.000276. ( 10.1099/mgen.0.000276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Walter T, Klim J, Jurkowski M, Gawor J, Köhling I, Słodownik M, Zielenkiewicz U. 2020. Plasmidome of an environmental Acinetobacter lwoffii strain originating from a former gold and arsenic mine. Plasmid 110, 102505. ( 10.1016/j.plasmid.2020.102505) [DOI] [PubMed] [Google Scholar]
- 141.Salto IP, Tejerizo GT, Wibberg D, Pühler A, Schlüter A, Pistorio M. 2018. Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci. Rep. 8, 1-12. ( 10.1038/s41598-018-26180-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang H, Peng N, Shah SA, Huang L, She Q. 2015. Archaeal extrachromosomal genetic elements. Microbiol. Mol. Biol. Rev. 79, 117-152. ( 10.1128/MMBR.00042-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Filée J, Siguier P, Chandler M. 2007. Insertion sequence diversity in archaea. Microbiol. Mol. Biol. Rev. 71, 121-157. ( 10.1128/MMBR.00031-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.DasSarma S, Capes M, DasSarma P. 2009. Haloarchaeal megaplasmids. In Microbial Megaplasmids (ed. Schwartz E), pp. 3-30. Berlin, Germany: Springer. [Google Scholar]
- 145.Wagner A, Whitaker RJ, Krause DJ, Heilers J-H, van Wolferen M, van der Does C, Albers S-V. 2017. Mechanisms of gene flow in archaea. Nat. Rev. Microbiol. 15, 492-501. ( 10.1038/nrmicro.2017.41) [DOI] [PubMed] [Google Scholar]
- 146.Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl Acad. Sci. USA 109, 20 537-20 542. ( 10.1073/pnas.1209119109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Haskett TL, Ramsay JP, Bekuma AA, Sullivan JT, O'Hara GW, Terpolilli JJ. 2017. Evolutionary persistence of tripartite integrative and conjugative elements. Plasmid 92, 30-36. ( 10.1016/j.plasmid.2017.06.001) [DOI] [PubMed] [Google Scholar]
- 148.Botelho J, Schulenburg H. 2021. The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 29, 8-18. ( 10.1016/j.tim.2020.05.011) [DOI] [PubMed] [Google Scholar]
- 149.Al-Shayeb B, et al. 2020. Clades of huge phages from across Earth's ecosystems. Nature 578, 425-431. ( 10.1038/s41586-020-2007-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Guan J, Bondy-Denomy J. 2020. Intracellular organization by jumbo bacteriophages. J. Bacteriol. 203, e00362-20. ( 10.1128/JB.00362-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Al-Shayeb B, et al. 2021. Borgs are giant extrachromosomal elements with the potential to augment methane oxidation. bioRxiv ( 10.1101/2021.07.10.451761) [DOI] [Google Scholar]
- 152.Marbouty M, Cournac A, Flot J-F, Marie-Nelly H, Mozziconacci J, Koszul R. 2014. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. Elife 3, e03318. ( 10.7554/eLife.03318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Stalder T, Press MO, Sullivan S, Liachko I, Top EM. 2019. Linking the resistome and plasmidome to the microbiome. ISME J. 13, 2437-2446. ( 10.1038/s41396-019-0446-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kent AG, Vill AC, Shi Q, Satlin MJ, Brito IL. 2020. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379. ( 10.1038/s41467-020-18164-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Whelan FJ, Rusilowicz M, McInerney JO. 2020. Coinfinder: detecting significant associations and dissociations in pangenomes. Microb. Genomics 6, 338. ( 10.1099/mgen.0.000338) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data presented in this manuscript are already publicly available through accessible databases (NCBI, COMPASS, EZBioCloud). These databases are referenced in the text, and description is provided in the text for how they were accessed.