Abstract
The emergence of antibiotic resistant bacteria is a major threat to modern medicine. Rapid adaptation to antibiotics is often mediated by the acquisition of plasmids carrying antibiotic resistance (ABR) genes. Nonetheless, the determinants of plasmid-mediated ABR gene transfer remain debated. Here, we show that the propensity of ABR gene transfer via plasmids is higher for accessory chromosomal ABR genes in comparison with core chromosomal ABR genes, regardless of the resistance mechanism. Analysing the pattern of ABR gene occurrence in the genomes of 2635 Enterobacteriaceae isolates, we find that 33% of the 416 ABR genes are shared between chromosomes and plasmids. Phylogenetic reconstruction of ABR genes occurring on both plasmids and chromosomes supports their evolution by lateral gene transfer. Furthermore, accessory ABR genes (encoded in less than 10% of the chromosomes) occur more abundantly in plasmids in comparison with core ABR genes (encoded in greater than or equal to 90% of the chromosomes). The pattern of ABR gene occurrence in plasmids and chromosomes is similar to that in the total Escherichia genome. Our results thus indicate that the previously recognized barriers for gene acquisition by lateral gene transfer apply also to ABR genes. We propose that the functional complexity of the underlying ABR mechanism is an important determinant of ABR gene transferability.
This article is part of the theme issue ‘The secret lives of microbial mobile genetic elements’.
Keywords: horizontal gene transfer, antibiotic resistance, Escherichia, Klebsiella, Salmonella
1. Introduction
Bacteria harbour vast potential to rapidly adapt to different selective environments, including environments defined by antibiotics. Notably, antibiotic resistance (ABR) predates the clinical use of antibiotics: ABR genes have been reported from permafrost [1] and ancient human remains [2]. Today, the ability of bacteria to adapt to antibiotics forms the core of the ABR crisis [3]. Drug therapy is aimed at killing bacteria by targeting key components and processes that are crucial for their reproduction; the repertoire of genetic variants giving rise to ABR therefore includes many core genes [4]. For example, bacterial efflux pumps, which function in resistance to diverse antibiotics, have multiple roles in bacterial physiology in addition to removal of antibiotics from the cell, including the extrusion of heavy metals and cell signalling molecules [5]. Indeed, genes encoding efflux pumps and their regulation are typically part of bacterial core genomes, i.e. they are nearly universally present in bacterial genomes [6]. By contrast, other resistance mechanisms are encoded by genes in the accessory genome and can be found also on mobile genetic elements (e.g. genes involved in target replacement or target protection) [7]. While the emergence of ABR under long-term drug therapy is common via de novo mutations, lateral transfer of ABR genes by mobile genetic elements has the potential to disseminate resistance to various antibiotics within microbial communities in clinical settings, agriculture and also the environment [8,9].
The acquisition of ABR genes can be mediated by three main transfer mechanisms: transformation, transduction and conjugation. Bacterial organisms that are naturally competent may acquire ABR genes via natural transformation of mobile genetic elements carrying ABR genes, including transposons, insertion sequence (IS) elements and integrons (e.g. [10]). Transduction can cause the transfer of ABR genes encoded in the genome of lysogenic phages (e.g. [11,12]) or the transfer of ABR gene-containing plasmids by phages (e.g. [13]). Notwithstanding, plasmid-mediated ABR gene transfer occurs most commonly via conjugation, which facilitates the transfer of conjugative and mobilizable plasmids (e.g. [14]). The dissemination of ABR genes by plasmids is often associated with the evolution of plasmid-encoded integrons that may comprise multiple ABR genes [15]. The type of vehicle that meditates the ABR gene transfer may have implications for the phylogenetic range of the transfer event. Phage-mediated gene transfer is often restricted to closely related donors and recipients owing to phage host specificity [12]. Similarly, the apparent taxonomic pattern in networks of plasmid sequence similarity [16] suggests that plasmid host range constrains the range of plasmid-mediated gene transfer to closely related donor and recipients (with the exception of broad host range plasmids, e.g. [17]).
The prevalence of gene transfer may furthermore depend on genetic and functional characteristics of the transferred gene. Genes that encode proteins whose function depends on interaction with multiple proteins, e.g. within the same complex or cellular process, are rarely transferred, likely because their acquisition leads to interference with the component sociometry [18,19]. An increase in gene copy number following lateral gene transfer may often lead to disadvantageous dose effect, which has been shown to pose a barrier to lateral gene transfer [20,21]. The acquisition of novel laterally transferred genes may have deleterious effects on the recipient owing to diverse reasons ranging from the consequences of foreign DNA integration into the genome to proper expression and function of the protein product in the cell (e.g. [22]; reviewed in [23]). A potential disruption of chromosomal architecture may be minimized if genes are transferred by and expressed from extra-chromosomal genetic elements like plasmids (i.e. assuming non-integrative plasmids). Nonetheless, ABR gene expression from a plasmid locus may be accompanied by fitness cost to the host owing to various other reasons. For example, ABR mechanisms that rely on drug inactivation may require a lower energetic investment (i.e. in ATP) of the cell in comparison with efflux pumps [24]. Such trade-offs in the adaptation of ABR plasmids in newly colonized hosts are expected to have consequences for the evolution of plasmid ABR gene content.
Plasmid gene content is furthermore expected to be subject to purifying selection owing to genetic conflicts between plasmid and chromosomally encoded loci that affect the same phenotype [25]. Indeed, a recent study suggests that the evolution of plasmid ABR gene content depends on the level of genetic conflicts with chromosomally encoded genes, which is strongest for ABR genes encoding efflux pumps and weakest for ABR genes encoding antibiotic target inactivation mechanisms [7]. Considering the association between ABR gene classification into core or accessory genome and the ABR mechanism, we hypothesize that the prevalence of ABR genes on plasmids depends on the ABR gene occurrence in bacterial chromosomes. Here, we examine ABR gene occurrence in bacterial plasmids and chromosomes using a large-scale analysis of three Enterobacteriaceae genera: Escherichia, Salmonella and Klebsiella, for which a large number of genomes are publicly available. We further assess whether ABR gene occurrence on plasmids and chromosomes is associated with the resistance mechanism and inferred ABR gene transfer events using phylogenetics and the temporal pattern of gene occurrence in plasmids and chromosomes.
2. Data and methods
To study the pattern of ABR gene occurrence in bacterial replicons (chromosomes and plasmids), we examined all complete genomes of plasmid-carrying isolates in the genera Escherichia, Salmonella and Klebsiella from the NCBI genome database (electronic supplementary material, table S1). Assembly and annotation files of 2580 genomes were downloaded from the NCBI RefSeq database [26] (v. 01/2021). An additional 78 complete genomes lacking RefSeq annotation were downloaded from the GenBank database. Protein-coding genes of the 78 genomes were annotated using Prokka [27] (v. 1.14.6, with parameter --kingdom Bacteria --gcode 11 --genus (Escherichia/Salmonella/Klebsiella) --usegenus --evalue 1 × 10−9). Isolate metadata were downloaded from the BioSample database. Samples lacking host information were examined in detail. A total of 23 strains sequenced within the framework of laboratory experiments were excluded. The surveyed genomes include: 1169 chromosomes and 3286 plasmids from Escherichia; 846 chromosomes and 3020 plasmids from Klebsiella; 620 chromosomes and 1090 plasmids from Salmonella. Homologues to genes that may provide ABR were identified based on the Comprehensive Antibiotic Resistance Database (CARD; v. 3.1.0) using the Resistance Gene Identifier tool (RGI; v. 5.1.1, with parameter --clean) [28]. The application of RGI uncovered 126 910 protein-coding genes; of these, we excluded 4774 protein-coding genes whose sequence had less than 70% identity or was less than 90% shorter or greater than 30% longer in comparison with the CARD ABR genes. Of the total 3044 genes in CARD, 416 (14%) ABR genes had 122 136 homologues in the examined isolates. Of those, we further excluded 9 genes belonging to multiple ABR mechanism classes.
The inference of protein families in Escherichia was performed using a dataset of 599 completely sequenced Escherichia strains downloaded from the NCBI RefSeq database (v. 2018) (previously reported in [29]). Of the total strains, 416 plasmid-carrying isolates were retained for further analysis (electronic supplementary material, table S2). Genome-wise reciprocal best hits (RBHs) of protein sequences were identified using MMseqs2 [30] (v. 13.45111, with module easy-rbh, applying a threshold of E-value ≤ 1 × 10−10). RBHs were further compared by global alignment using Parasail-python [31] (v. 1.2.4, with the Needleman–Wunsch algorithm). Sequence pairs with greater than or equal to 30% identical amino acids were clustered into protein families using the Markov clustering algorithm (MCL) [32] (v. 12-135, with parameter --abc -I 2.0).
Data analysis, statistical inference and visualization were performed with R (v. 4.0) and MATLAB® (v. 2018b). Testing for a bias in ABR gene occurrence towards plasmids or chromosomes was performed by comparing the frequency of ABR gene occurrence on both replicons with the frequency of the remaining genes on both replicons (using Fisher's exact test with fisher.test function in R).
For the phylogenetic reconstruction, non-redundant protein sequences of selected ABR genes were aligned using MAFFT [33] (v. 7.475). Owing to the large number of identical protein sequences among the ABR gene homologues, the sequences were filtered to include non-redundant amino acid sequences at the resolution of replicon and genus. Phylogenetic trees were reconstructed using IQ-TREE [34] (v. 1.6.12) with restricted automatic model selection to the Le & Gascuel (LG) model [35] and the LG4X model [36] as additional alternative (with parameter -mset LG -madd LG4X). The LG model was chosen owing to its suitability for the inference of phylogenetic trees using protein sequences from bacterial organisms by means of maximum likelihood methods. The resulting trees were rooted using the midpoint criterion and visualized using iTOL [37] v. 6 (https://itol.embl.de/).
3. Results
(a) . Plasmid ABR gene content is associated with gene prevalence on chromosomes
The survey for genes that putatively confer ABR revealed 2810 (38%) plasmids that encode an ABR gene homologue (termed here ABR plasmids). The ABR plasmids typically encode multiple ABR genes, with the median number of ABR genes per plasmid ranging between four in Escherichia and six in Salmonella. The proportion of ABR plasmids was similar among the three genera: 34% (1131) in Escherichia, 42% (1256) in Klebsiella and 39% (423) in Salmonella. The genomes of all isolates in the three genera had at least one homologue to an ABR gene encoded in the chromosome.
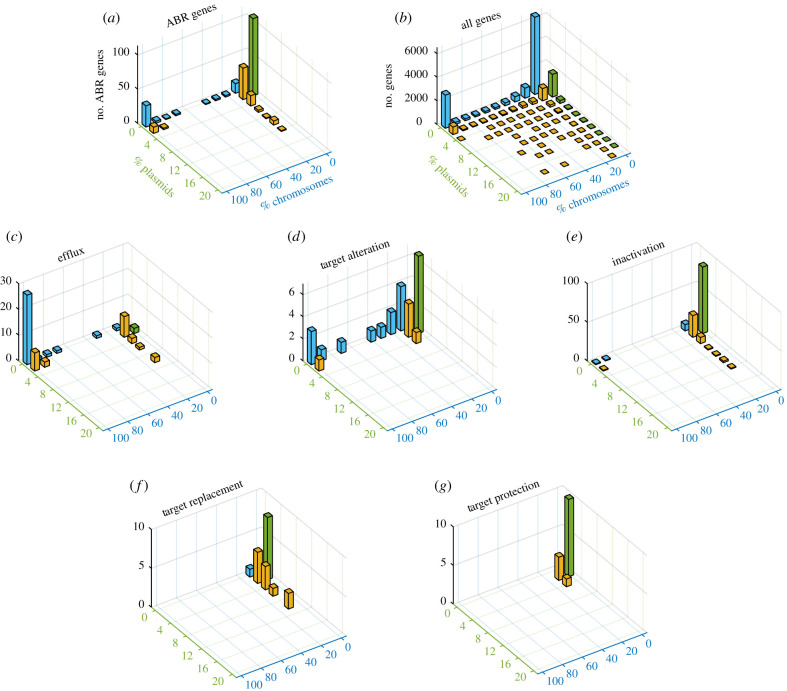
The ABR gene homologues in Escherichia plasmids and chromosomes fall within three classes: (1) ABR genes exclusively found on plasmids, (2) ABR genes found exclusively on chromosomes, and (3) ABR genes found on both plasmids and chromosomes (figure 1a). The pattern of gene occurrence on plasmids and chromosomes further reveals two main groups: ABR genes encoded on plasmids and shared with a small proportion of the chromosomes (less than or equal to 10%)—these genes are considered to be part of the accessory genome—and ABR genes that are encoded on plasmids and are shared with greater than or equal to 90% of the chromosomes—these genes are considered to be part of the core genome (see yellow bars in figure 1a). A similar pattern of ABR gene occurrence on plasmids and chromosomes is observed in the genomes of Klebsiella and Salmonella isolates (electronic supplementary material, figures S1 and S2). To compare the pattern of ABR gene occurrence with the total Escherichia genes, we repeated the analysis using a dataset of protein families in Escherichia isolates. The results show that the ABR gene occurrence in plasmids and chromosomes bears similarity to the distribution of the total gene families in Escherichia genomes (figure 1b): in both datasets homologues of core chromosomal genes found in greater than or equal to 90% of the chromosomes are less abundant in plasmids in comparison with homologues of accessory chromosomal genes found in fewer than or equal to 10% of the chromosomes. The similar pattern of shared genes among plasmids and chromosomes observed for the ABR genes and the total genome indicates that the association between ABR gene prevalence in plasmids and chromosomes is general within Escherichia genomes rather than a unique property of ABR genes.
Figure 1.
A comparison of gene prevalence in plasmids and chromosomes in Escherichia isolates. Plots in the figure present three-dimensional histograms of gene occurrence in plasmids (green axis) and chromosomes (blue axis). The height of bars in the histogram corresponds to the number of genes that are encoded in the same frequency group on chromosomes and plasmids. The bar coordinates correspond to the proportion of replicons (plasmids or chromosomes) where the genes occur. Note that no gene occurs in more than 20% of the plasmids; genes occurring in 100% of the chromosomes correspond to core gene families. Genes occurring only on chromosomes are shown with blue bars. Genes occurring only on plasmids are shown with green bars. Yellow bars correspond to genes that occur in both plasmids and chromosomes. (a) ABR genes in the examined isolates (data supplied in electronic supplementary material, table S3). (b) Gene occurrence of all Escherichia gene families. (c–g) ABR genes according to their classification into ABR mechanism (as in CARD [28]).
Previous studies suggested that the occurrence of ABR genes in plasmids may be related to the underlying mechanism of ABR [7,24]. Indeed, further examination of the ABR gene occurrence on plasmids and chromosomes reveals differences across the ABR mechanism classes. ABR genes that are involved in the expression of efflux pumps are typically core chromosomal genes that are rarely found on plasmids (figure 1c). By contrast, ABR genes classified as antibiotic target alteration include both chromosome- and plasmid-exclusive genes, with rare gene sharing between plasmids and chromosomes (figure 1d). ABR genes classified as antibiotic inactivation comprise mostly plasmid-specific genes, with some genes occurring in chromosomes as accessory genes and rarely as core genes (figure 1e). Genes classified as target protection and target replacement are encoded on a small number of plasmids and chromosomes (figure 1f,g).
To further examine the mode of ABR gene evolution, that is, if it is mainly by vertical inheritance or lateral gene transfer, we explored biases in the prevalence of ABR gene homologues towards plasmids and chromosomes (termed hereafter ‘replicon type’) for each of the ABR genes in our dataset. The results of the statistical analysis confirm that most of the ABR genes coding for efflux pumps are significantly more frequent on chromosomes (table 1; electronic supplementary material, table S3). Nonetheless, some exceptional efflux-coding genes are enriched on plasmids, including tetA/B, floR and qacEΔ1, whose homologues are mainly encoded in plasmids in all three genera (electronic supplementary material, table S3). The distribution of ABR genes that encode antibiotic target alteration proteins is balanced between chromosomes and plasmids in Escherichia, while in Klebsiella and Salmonella it is biased towards plasmids. Examples of chromosomal antibiotic target alteration genes include the gene encoding the translation elongation factor EF-Tu, which is found as an exclusive core gene in all genera, and bacA, an undecaprenyl pyrophosphate phosphatase [39], which is a core chromosomal gene in Escherichia and Salmonella. Examples of genes encoding antibiotic target alteration proteins that are overrepresented on plasmids include ermB and mcr-1 (electronic supplementary material, table S3).
Table 1.
Comparison of ABR frequency in plasmids and chromosomes. The number of ABR genes having a bias towards plasmids or chromosomes according to the resistance mechanism classification (using Fisher's exact test in contingency analysis, α = 0.05 and correction for multiple comparisons with false discovery rate (FDR) [38]). No bias to the chromosome was found in ABR genes for antibiotic target protection and replacement (marked with ‘—’).
| genus | antibiotic efflux |
antibiotic inactivation |
antibiotic target alteration |
antibiotic target protection |
antibiotic target replacement |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| chromosome | plasmid | chromosome | plasmid | chromosome | plasmid | chromosome | plasmid | chromosome | plasmid | |
| Escherichia | 40 (74%) | 14 (26%) | 3 (2%) | 126 (98%) | 10 (50%) | 10 (50%) | — | 13 (100%) | — | 17 (100%) |
| Klebsiella | 14 (50%) | 14 (50%) | 3 (3%) | 100 (97%) | 8 (40%) | 12 (60%) | — | 11 (100%) | — | 13 (100%) |
| Salmonella | 20 (61%) | 13 (39%) | 4 (5%) | 74 (95%) | 5 (38%) | 8 (62%) | — | 9 (100%) | — | 13 (100%) |
ABR genes in the inactivation class are typically encoded on plasmids, with rare genes showing a bias towards the chromosome (table 1). Those exceptional genes include ampH, which is a core chromosomal gene in all three genera. This gene encodes a penicillin binding protein that has a function in the determination of cell shape in Escherichia coli and can confer resistance to β-lactam antibiotics [40]. Another example is ampC1, which codes for a β-lactamase and is nearly universally encoded in Escherichia and Salmonella but absent in Klebsiella genomes. Additional examples are aac (6’)-Iy and aac (6’)-Iaa, both encoding aminoglycoside acetyltransferases, which are exclusively found in Salmonella chromosomes. The genes fosA5 and fosA6, mediating resistance against fosfomycin, have been reported in Escherichia [41], yet in our extensive dataset they are observed exclusively as accessory chromosomal genes in Klebsiella (electronic supplementary material, table S3).
Purifying selection on plasmid gene content due to genetic conflicts is expected to differ among taxa having different compositions of their core genome. Nonetheless, the bias of ABR genes towards specific replicon type (i.e. plasmids or chromosomes) is generally similar among the three tested genera in our dataset, likely since they are closely related and their core ABR genome largely overlaps. Nonetheless, 23 ABR genes stand out as exceptions (table 2). Those are mostly plasmid genes with variable presence on the chromosomes, i.e. they can be considered accessory genes. One extreme case is the antibiotic efflux gene oqxA, which is a core chromosomal gene in Klebsiella and enriched on plasmids in the other two genera. Among the exceptional genes encoding antibiotic inactivation proteins, the gene blaCARB-1 is found as an accessory chromosomal gene in Salmonella, with no bias towards specific replicon type, but only on plasmids in Escherichia and Klebsiella (electronic supplementary material, table S2).
Table 2.
ABR genes whose relative frequency in plasmids (Pls) and chromosomes (Chr) differs among the three tested genera. The table shows ABR gene occurrence on plasmids and chromosomes in the three genera. The ABR gene frequency on both replicons was compared using Fisher's exact test (the p-value is reported in addition to adjusted p-value for multiple comparisons using false discovery rate (FDR)). Chromosome- and plasmid-exclusive genes and genes absent from any of the genera were excluded from this analysis (a total of 98 ABR genes were tested). Genes discussed in the Results §§3(c)–3(e) are highlighted in italics.
| resistance mechanism |
Escherichia |
Salmonella |
Klebsiella |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ARO | gene | Chr | Pls | Chr | Pls | Chr | Pls | p | padj | |
| 3003922 | oqxA | antibiotic efflux | 2 | 45 | 2 | 36 | 728 | 1 | 1.02 × 10−105 | 1.00 × 10−103 |
| 3003923 | oqxB | antibiotic efflux | 2 | 31 | 2 | 19 | 162 | 0 | 5.51 × 10−45 | 2.70 × 10−43 |
| 3000165 | tetA | antibiotic efflux | 62 | 283 | 20 | 165 | 13 | 232 | 1.02 × 10−5 | 7.71 × 10−5 |
| 3000166 | tetB | antibiotic efflux | 69 | 100 | 39 | 58 | 3 | 6 | 9.47 × 10−1 | 1.00 × 10−1 |
| 3005010 | qacEΔ1 | antibiotic efflux | 55 | 291 | 68 | 137 | 116 | 371 | 1.73 × 10−5 | 1.21 × 10−4 |
| 3000873 | blaTEM-1 | antibiotic inactivation | 65 | 333 | 53 | 133 | 9 | 404 | 6.65 × 10−22 | 2.17 × 10−20 |
| 3004290 | ampC | antibiotic inactivation | 1119 | 1 | 2 | 0 | 0 | 0 | — | — |
| 3003689 | mcr-1 | antibiotic target alteration | 13 | 95 | 0 | 26 | 0 | 8 | 1.10 × 10−1 | 2.83 × 10−1 |
| 3002790 | qnrS1 | antibiotic target protection | 8 | 105 | 1 | 41 | 2 | 132 | 6.61 × 10−2 | 2.06 × 10−1 |
| 3000412 | sul2 | antibiotic target replacement | 90 | 287 | 55 | 187 | 21 | 272 | 2.00 × 10−9 | 2.79 × 10−8 |
| 3002705 | floR | antibiotic efflux | 33 | 143 | 33 | 121 | 2 | 63 | 9.17 × 10−4 | 4.99 × 10−3 |
| 3000168 | tetD | antibiotic efflux | 6 | 1 | 1 | 4 | 2 | 50 | 3.69 × 10−6 | 3.28 × 10−5 |
| 3002660 | aph(6)-Id | antibiotic inactivation | 82 | 270 | 60 | 173 | 17 | 249 | 1.70 × 10−10 | 4.16 × 10−9 |
| 3002639 | aph(3″)-Ib | antibiotic inactivation | 82 | 263 | 59 | 167 | 18 | 245 | 4.26 × 10−10 | 8.34 × 10−9 |
| 3001059 | blaSHV-1 | antibiotic inactivation | 0 | 2 | 0 | 1 | 144 | 7 | 2.01 × 10−4 | 1.23 × 10−3 |
| 3002601 | aadA | antibiotic inactivation | 43 | 130 | 10 | 76 | 3 | 133 | 8.62 × 10−9 | 1.06 × 10−7 |
| 3001877 | blaCTX-M-14 | antibiotic inactivation | 23 | 43 | 2 | 13 | 7 | 51 | 7.64 × 10−3 | 3.57 × 10−2 |
| 3002013 | blaCMY-2 | antibiotic inactivation | 22 | 42 | 6 | 70 | 0 | 7 | 1.81 × 10−4 | 1.18 × 10−3 |
| 3002539 | aac(3)-IV | antibiotic inactivation | 5 | 37 | 4 | 45 | 12 | 21 | 4.48 × 10−3 | 2.20 × 10−2 |
| 3002603 | aadA3 | antibiotic inactivation | 5 | 29 | 27 | 29 | 97 | 44 | 1.70 × 10−8 | 1.85 × 10−7 |
| 3002683 | catI | antibiotic inactivation | 35 | 27 | 26 | 35 | 9 | 81 | 5.23 × 10−1 | 8.54 × 10−9 |
| 3001926 | blaCTX-M-65 | antibiotic inactivation | 4 | 13 | 1 | 20 | 0 | 76 | 5.21 × 10−4 | 3.00 × 10−3 |
| 3004656 | catII | antibiotic inactivation | 0 | 7 | 4 | 19 | 1 | 88 | 8.84 × 10−3 | 3.94 × 10−2 |
| 3002240 | blaCARB-1 | antibiotic inactivation | 0 | 4 | 25 | 0 | 0 | 2 | 1.36 × 10−6 | 1.33 × 10−5 |
| 3001397 | blaOXA-2 | antibiotic inactivation | 3 | 0 | 0 | 1 | 0 | 0 | 2.75 × 10−3 | 1.42 × 10−2 |
| 3000410 | sul1 | antibiotic target replacement | 53 | 286 | 66 | 140 | 76 | 371 | 8.05 × 10−6 | 6.58 × 10−5 |
| 3002862 | dfrA7 | antibiotic target replacement | 13 | 2 | 21 | 23 | 0 | 1 | 9.83 × 10−3 | 4.19 × 10−2 |
(b) . ABR gene transfer between plasmids and chromosomes
Our results reveal 133 (33%) ABR genes that are found on both replicons in at least one genus, thus indicating that they have been laterally transferred. Of those, 75 ABR genes were observed on both plasmid and chromosome within the same isolate genome in at least one strain. The majority of genes that we identified at least once as duplicated are enriched on plasmids (65 out of 75 genes); of those, 45 genes encode proteins that function in antibiotic inactivation (electronic supplementary material, table S4). Only five of the 75 duplicated genes are enriched for a chromosome location; these correspond to four genes encoding efflux pumps and one encoding translation elongation factor (target alteration). The presence of duplicated ABR genes on both replicons in the same isolate is relatively rare. We note here that deleterious effects in the context of gene gain following lateral gene transfer [23] are expected not only for the recipient, but rather also for the donor, following gene transfer from chromosomes to plasmids. Taken together, our results suggest that genes that are duplicated on the plasmid and chromosome in the same isolate often correspond to genes introduced into the genus as plasmid genes.
To further study the evolutionary history of genes shared between plasmids and chromosomes we reconstructed the phylogenetic trees of the ABR genes and examined their topology. In what follows, we present the results of 39 selected ABR genes exemplifying main trends in the data. The topology of ABR genes across replicon types and genera in the inferred phylogenetic trees confirms the occurrence of lateral gene transfer among genera and also replicon types in the history of all examined genes (electronic supplementary material, table S5). In the following, we present several prime examples.
(c) . Lateral transfer of genes encoding efflux pumps
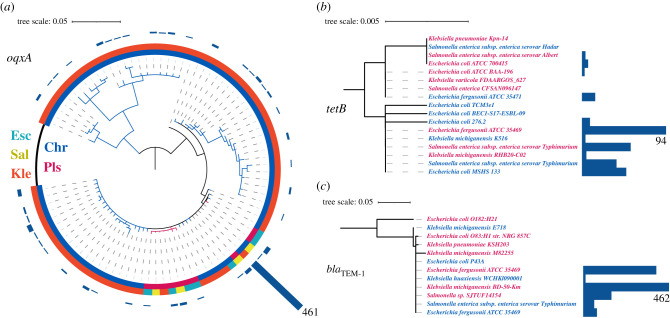
The genes oqxA and oqxB belong to the resistance–nodulation–cell division (RND) family of bacterial efflux pumps [42] and were first isolated from bacteria in swine manure from a farm where olaquinodox was used as a feed additive [43]. The proteins OqxA and OqxB, along with TolC, form an efflux pump that provides resistance against olaquinodox, chloramphenicol, quinolones, trimethoprim, tigecycline, nitrofurantoin, and also several detergents and disinfectants by reducing their concentrations in the cell [42,44]. Both oqxA and oqxB are almost strictly chromosomal genes in Klebsiella, with oqxA occurring in 86% and oqxB occurring in 19% of Klebsiella isolates. We identified a single Klebsiella isolate where oqxA is present on both chromosome and plasmid (electronic supplementary material, table S4). The two genes have homologues in Escherichia and Salmonella plasmids. The phylogenetic trees of both genes indicate lateral transfer between Klebsiella chromosomes and plasmids of the other two genera (figure 2a; electronic supplementary material, figure S3a). We note that the oqxA protein sequences are nearly identical, and hence the phylogenetic reconstruction does not allow a reliable inference of donors and recipients in the transfer event. To further study the history of the ABR genes, we collected information on the isolation date of strains included in our analysis. The gene oqxA was documented early in the 1950s as a chromosomal gene in Klebsiella (figure 3). The first documentation of oqxA on a plasmid in our dataset is from 2004 in Escherichia; in recent years, this gene has been found on both plasmids and chromosomes in all three genera. The gene oqxB was first documented in 2004 in Klebsiella chromosomes and Escherichia plasmids, while in recent years, it has been described for both replicon types in all three genera (figure 3). The example of oqxA suggests that barriers to gene transfer of core chromosomal genes may differ between genera. The first report on plasmid-encoded oqxAB in E. coli in the literature dates back to the late 1990s [45]; today, after more than 20 years, that gene is still very rare in Escherichia and Salmonella. It is tenable to speculate that barriers to plasmid-mediated oqxA acquisition are present also in Escherichia and Salmonella.
Figure 2.
Lateral gene transfer in the evolution of ABR genes. Bars next to the operational taxonomic units show the frequency of redundant amino acid sequences (with highest frequency noted). (a) The outer ring shows the genus of ABR gene origin; the inner ring shows whether the gene is encoded on a plasmid (Pls) or chromosome (Chr). (b,c) Isolate label is coloured according to replicon type (plasmid genes in pink and chromosomal genes in blue). Most phylogenetic trees we reconstructed here for the ABR genes included polytomies, hence donor and recipients in the gene transfer events cannot be reliably inferred. Nonetheless, considering the presence of the examined ABR genes on both plasmids and chromosomes supports the hypothesis that they have been transferred at least once in their evolutionary history.
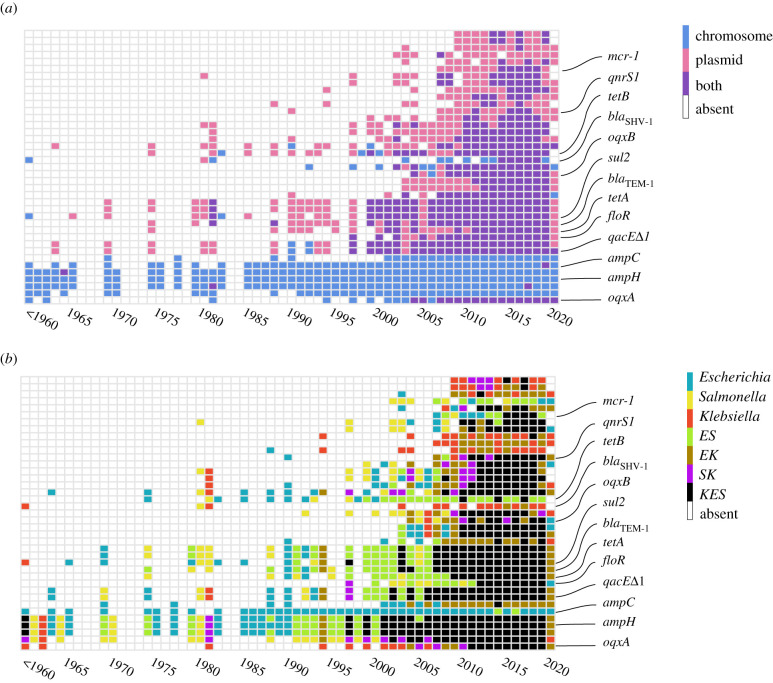
Figure 3.
Temporal pattern of ABR gene documentation in replicons and genera. Gene presence according to the isolation date of the strains is summarized. The information on the strains isolated between 1900 and 1960 is condensed in ‘<1960’. The figure shows data for 39 genes whose phylogenetic trees were closely examined (ordered as in electronic supplementary material, table S4). The names of 13 genes highlighted in the manuscript are marked. Different colours correspond to gene locations. (a) Temporal ABR gene pattern according to replicon type. (b) Temporal ABR gene pattern according to genus. ABR genes found in multiple genera are annotated by capitalized abbreviations of the three genera, i.e. E for Escherichia, K for Klebsiella, S for Salmonella.
The Tet efflux pumps were the first efflux systems discovered in bacteria [46,47]. These major facilitator (MF) family exporters are membrane–associated proteins that pump out tetracycline from the cell in an energy-dependent manner [48]. Unlike the OqxAB–TolC system, the Tet efflux pumps provide resistance only against tetracyclines. Since the original report of tetA, many more tet genes have been discovered, the vast majority in Gram-negative bacteria [49]. For our analysis, we focused on the tetA and tetB genes, since both genes are enriched on plasmids and rarely found on chromosomes (fewer than 6% in all genera). The phylogenetic reconstruction shows that the protein sequence of tetA is more diverged in comparison with that of tetB, and that for both genes, multiple transfer events occurred between chromosomes and plasmids, and also across bacterial genera (figure 2b; electronic supplementary material, figure S3d). Furthermore, we identified 13 isolates (from all genera) where tetA is encoded on both replicon types and four Escherichia isolates that encode tetB on both replicon types. The gene tetA in our data was documented firstly on a plasmid in Salmonella in 1974, while tetB was identified on a plasmid in Escherichia at the same time (figure 3). In recent years, both genes have been found on both replicon types and in all three genera, yet tetB is seldom documented in Klebsiella.
Another efflux gene that is mainly found on plasmids is qacEΔ1, a truncated variant of the qacE gene [50]. The qac genes are antiseptic efflux systems primarily exporting quaternary ammonium compounds. Their substrate range, however, is quite large and includes intercalating dyes, biguanidines, diamidines and xanthenes. The gene qacEΔ1 is frequently found in ABR gene cassettes isolated from bacteria exposed to these compounds compared with those that are not [51]. In our dataset, we found that qacEΔ1 is very often encoded on both plasmids and chromosomes in the same isolate (71 Klebsiella, 11 Escherichia and 7 Salmonella isolates). The protein sequence is highly conserved between replicons and across all three genera. In our dataset, qacEΔ1 was documented first on a plasmid in Escherichia in 1969, and then a Klebsiella plasmid in 1981; the first chromosomal qacEΔ1 was identified in Escherichia in 1990 (figure 3).
(d) . Lateral transfer of genes encoding antibiotic inactivation proteins
A prime example of antibiotic inactivation enzymes are the β-lactamases. These enzymes confer resistance to β-lactam antibiotics by cleaving their central β-lactam ring [52]. The gene blaTEM-1 was the first TEM β-lactamase gene identified;TEM β-lactamases are among the best-studied ABR enzymes [53,54]. Although the original blaTEM-1 gene confers resistance only to penicillins and early cephalosporins, its later variants also provide resistance against second-, third- and fourth-generation cephalosporins, monobactams and β-lactamase inhibitors [54]. The blaTEM-1 gene is mainly found on Klebsiella and Escherichia plasmids. The phylogenetic reconstruction indicates multiple transfer events and high protein sequence conservation of blaTEM-1 between replicons and across genera (figure 2c). It was first documented on plasmids in both Escherichia and Salmonella in 1974, and first reported on chromosomes in Salmonella in 1981 (figure 3).
The AmpC β-lactamase of Escherichia coli was the first reported bacterial enzyme capable of degrading penicillin [55]. Although active against penicillins, this inducible enzyme has higher activity against cephalosporins and is an important determinant of resistance against cephalosporins in clinical settings [56]. The ampC gene in Escherichia is nearly exclusively a chromosomal gene. Additionally, homologues of ampC were found on a plasmid in one Escherichia isolate and on the chromosome of two Salmonella isolates. The phylogenetic reconstruction indicates that the Salmonella homologues were acquired in two independent transfer events (electronic supplementary material, figure S3h).
(e) . Lateral transfer of ABR genes involved in target alteration, replacement and protection
The mcr-1 gene was the first plasmid-mediated colistin resistance determinant reported, signalling the breach of the last resort polymyxin antibiotics [57]. MCR-1 is a phosphoethanolamine transferase that alters lipid A in the outer membrane of Gram-negative bacteria, resulting in reduced affinity to colistin [58]. Homologues of MCR-1 are mainly found on plasmids in all three genera, with some rare homologues on chromosomes in Escherichia (1%). Three Escherichia coli isolates were identified with mcr-1 on both replicons. The nearly identical protein sequence of the homologues suggests that mcr-1 is laterally transferred among the genera by plasmids (electronic supplementary material, figure S3e). The gene was first documented on a plasmid in Escherichia in 2007 and on the chromosome in 2014; in recent years it has been found in all three genera (figure 3).
The gene sul2 confers resistance against the sulfonamide antibiotics [59,60]. Sulfonamides were among the first synthetic compounds for which specific antibacterial activities were discovered, and they were also among the first antibiotics to be used for medical treatment. They block folic acid synthesis in bacteria by competitively inhibiting dihydropteroate synthase (DHPS) [61]. Sul2 is an alternative drug-resistant variant of the DHPS enzyme that replaces the original drug-sensitive variant to provide resistance [62]. The gene is mainly found on plasmids and may occur also on chromosomes (6% of isolates). A total of 34 isolates encode a sul2 on both replicons, and the phylogenetic tree of sul2 indicates multiple transfer events between replicons and across genera (electronic supplementary material, figure S3f). Although typically a plasmid gene, sul2 was first documented on the chromosome in Klebsiella in the 1950s and only later on plasmids in Escherichia and Salmonella; in recent years, it has been reported in all three genera (figure 3).
The gene qnrS1 belongs to the Qnr family of proteins, which confer resistance to quinolones and flouroquinolones in Gram-negative bacteria by binding and protecting the target of these antibiotics, type II topoisomerase [63]. The binding of the target seems to destabilize the complex of topoisomerase with the drug, resulting in regeneration of the active enzyme [64]. Homologues of qnrS1 are mainly distributed on plasmids. We identified two isolates that carry this gene on both replicons, including E. coli strain LHM10-1 (isolated in 2017 in China from swine faeces) and Klebsiella pneumoniae strain C2421 (isolated in 2017 in China from human urine sample). The QnrS1 protein sequences are nearly identical in all plasmids and the phylogenetic tree topology indicates lateral gene transfer between replicons and in all three genera (electronic supplementary material, figure S3g). The gene was first documented on plasmids in Escherichia and Klebsiella in 2007 and in recent years it has been found on both replicons in all three genera (figure 3).
(f) . Temporal pattern of gene presence in replicons supports the phylogenetic inference
To further study the temporal pattern of ABR gene occurrence on specific replicon type—plasmids or chromosomes—we examined the collection date of isolate genomes encoding ABR genes. The available data reveal two general trends: core ABR genes were typically documented on chromosomes, with only rare findings on plasmids throughout the time range covered by the data. Notably, of the 75 core ABR genes that are enriched on chromosomes, 74 (99%) were documented first on chromosomes, with oqxB as the sole exception. Additionally, of the 226 genes found to be enriched on plasmids, 208 (92%) were first identified on plasmids. The temporal pattern of plasmid genes shows a trend of early documentation on plasmids and later presence on both replicons, which is in agreement with the phylogenetic trees reconstructed in this study (figure 2). The temporal pattern of ABR gene occurrence as observed in isolate genomes provides further support for the role of historical contingency in the bias of ABR gene occurrence on plasmids or chromosomes.
4. Discussion
In this study, we performed a comprehensive ABR gene analysis across 2635 complete genome sequences in the related bacterial genera Escherichia, Salmonella and Klebsiella, and identified an inverse association between ABR gene prevalence in plasmids and chromosomes. This inverse association strongly suggests that the presence of a homologous gene in the chromosome poses a barrier to plasmid-mediated ABR gene acquisition. Several non-exclusive mechanisms may account for the barrier to lateral gene transfer, as explained in detail below.
The bias of ABR genes towards specific replicon type—chromosomes or plasmids—is clearly associated with the ABR underlying mechanism [7]. ABR genes associated with efflux pumps are primarily encoded on chromosomes, those involved in target alteration can be encoded on both replicon types, and those for the remaining three mechanisms are almost exclusively encoded on plasmids. ABR genes encoding efflux pumps are frequently part of the core genome (e.g. in the genera we studied here), where they code for proteins having multiple functions in the cell outside the context of ABR. Examples are proteins involved in export of heavy metals [65], organic pollutants [66] and endogenous metabolites [67], and cell signalling molecules [68]. Furthermore, several efflux pumps are important for biofilm formation [69] and bacteria–plant interactions [70]. Core genes are expected to be highly integrated in the cellular transcriptional regulation network and their products are likely well integrated in protein–protein interaction networks. Proteins encoded by the core genome are therefore expected to have a high level of pleiotropic effects on bacterial physiology. Barriers for lateral transfer of core genes are expected to be high, e.g. owing to genetic conflicts [7] or the cost of their interference with well-coordinated cellular processes [23]. The same logic applies to core genes in the other classes of resistance mechanisms, including target alteration and antibiotic inactivation. Core ABR genes that code for antibiotic target alteration in our dataset include gyrA, parC and parE. These genes may provide resistance to antibiotics that target, e.g., type II topoisomerases, DNA gyrase and topoisomerase IV, which control topological transitions of DNA [71]; all of these are central components in the bacterial information processing mechanisms that are targets for drug therapy (e.g. with fluoroquinolones [4]). The frequency of ABR gene transfer in microbial evolution (i.e. transferability) is therefore expected to depend on the complexity of their function within the cell [18,19] rather than the exact mechanism of resistance, similarly to other genes that do not code for ABR mechanisms.
So far, we have discussed barriers to lateral transfer of ABR genes depending on the effect of gene acquisition on the bacterial organism. Notwithstanding, the acquisition of ABR genes may have consequences also for the evolution of stably inherited plasmids. Plasmids encoding ABR genes may also evolve a stable inheritance in a newly colonized host population in the absence of selective conditions for ABR [72]. By contrast, the presence of selective conditions for ABR can maintain in the population plasmids whose inheritance is unstable; such plasmids are at risk of extinction under non-selective conditions [73]. Indeed, plasmids equipped with mobility mechanisms have the capacity to persist via transfer to alternative host populations [74]. Nonetheless, the stability of plasmid vertical inheritance is paramount for plasmid survival and long-term evolution (reviewed in [75]). The potential deleterious effect of ABR gene acquisition in plasmid genomes on the plasmid fitness [76] may explain the rarity of ABR genes on plasmids (figure 1), specifically in non-mobile plasmids [73].
Plasmid-encoded ABR genes in our dataset are mostly classified as antibiotic target inactivation (figure 1). ABR genes coding for antibiotic inactivation frequently vary in their relative frequency on plasmids and chromosomes across the three compared genera. Many of those genes correspond to accessory chromosomal genes that are also prevalent on plasmids, indicating that they may be transferred between both replicon types (table 2). Indeed, accessory genes may prove essential depending on the isolate genetic background and environmental conditions [77]; nonetheless, their variable presence in a genus suggests that they are often dispensable. Genes whose evolution is characterized by rapid gain and loss dynamics are expected to encode proteins whose function is weakly dependent on specific genetic backgrounds, and are therefore more likely to evolve by lateral gene transfer [78]. The ABR gene content of plasmids reflects the propensity of specific ABR genes to be successfully transferred among donors and recipients within the plasmid host range.
Acknowledgements
We thank Giddy Landan for advice on data analysis. All calculations were performed on the HPC-system in Kiel University Computing Centre (Rechenzentrum).
Data accessibility
The data are provided in electronic supplementary material [79].
Authors' contributions
Y.W. and T.D. conceived the study. Y.W. performed data analysis and visualization. A.B. analysed distribution patterns in ABR gene families. All authors interpreted the results and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Funding
We are grateful for financial support from the German Science Foundation (projects 3 and 4 to T.D. and H.S. within the RTG 2501 TransEvo and grant no. SCHU 1415/12 to H.S.), the Leibniz Science Campus Evolutionary Medicine of the Lung (EvoLUNG to T.D. and H.S.), and Chinese Scholarship Council (CSC scholarship to Y.W.).
References
- 1.D'Costa VM, et al. 2011. Antibiotic resistance is ancient. Nature 477, 457-461. ( 10.1038/nature10388) [DOI] [PubMed] [Google Scholar]
- 2.Warinner C, et al. 2014. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 46, 336-344. ( 10.1038/ng.2906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laxminarayan R, et al. 2013. Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 13, 1057-1098. ( 10.1016/S1473-3099(13)70318-9) [DOI] [PubMed] [Google Scholar]
- 4.Baquero F, Levin BR. 2021. Proximate and ultimate causes of the bactericidal action of antibiotics. Nat. Rev. Microbiol. 19, 123-132. ( 10.1038/s41579-020-00443-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez MB, Martinez JL. 2016. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4, 14. ( 10.3390/microorganisms4010014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher LA, Lee SA, Manoil C. 2017. Importance of core genome functions for an extreme antibiotic resistance trait. mBio 8, e01655-17. ( 10.1128/mBio.01655-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez-Beltrán J, Sørum V, Toll-Riera M, de la Vega C, Peña-Miller R, San Millan A. 2020. Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria. Proc. Natl Acad. Sci. USA 117, 15 755-15 762. ( 10.1073/pnas.2001240117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson DI, Balaban NQ, Baquero F, Courvalin P, Glaser P, Gophna U, Kishony R, Molin S, Tønjum T. 2020. Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 44, 171-188. ( 10.1093/femsre/fuaa001) [DOI] [PubMed] [Google Scholar]
- 9.Heuer H, Schmitt H, Smalla K. 2011. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236-243. ( 10.1016/j.mib.2011.04.009) [DOI] [PubMed] [Google Scholar]
- 10.Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. 2012. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 8, e1002837. ( 10.1371/journal.ppat.1002837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fard RMN, Barton MD, Heuzenroeder MW. 2011. Bacteriophage-mediated transduction of antibiotic resistance in enterococci. Lett. Appl. Microbiol. 52, 559-564. ( 10.1111/j.1472-765X.2011.03043.x) [DOI] [PubMed] [Google Scholar]
- 12.Popa O, Landan G, Dagan T. 2016. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 11, 543-554. ( 10.1038/ismej.2016.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varga M, Kuntová L, Pantůček R, Mašlaňová I, Růžičková V, Doškař J. 2012. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol. Lett. 332, 146-152. ( 10.1111/j.1574-6968.2012.02589.x) [DOI] [PubMed] [Google Scholar]
- 14.Blau K, Bettermann A, Jechalke S, Fornefeld E, Vanrobaeys Y, Stalder T, Top EM, Smalla K. 2018. The transferable resistome of produce. mBio 9, 464-515. ( 10.1128/mBio.01300-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Che Y, Yang Y, Xu X, Břinda K, Polz MF, Hanage WP, Zhang T. 2021. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl Acad. Sci. USA 118, e2008731118. ( 10.1073/pnas.2008731118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redondo-Salvo S, Fernandez-Lopez R, Ruiz R, Vielva L, de Toro M, Rocha EPC, Garcillan-Barcia MP, de la Cruz F. 2020. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11, 3602. ( 10.1038/s41467-020-17278-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jørgensen TS, Xu Z, Hansen MA, Sørensen SJ, Hansen LH. 2014. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS ONE 9, e87924. ( 10.1371/journal.pone.0087924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen O, Gophna U, Pupko T. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481-1489. ( 10.1093/molbev/msq333) [DOI] [PubMed] [Google Scholar]
- 19.Jain R, Rivera MC, Lake JA. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA 96, 3801-3806. ( 10.1073/pnas.96.7.3801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449-1452. ( 10.1126/science.1147112) [DOI] [PubMed] [Google Scholar]
- 21.Kirit HA, Lagator M, Bollback JP. 2020. Experimental determination of evolutionary barriers to horizontal gene transfer. BMC Microbiol. 20, 326. ( 10.1186/s12866-020-01983-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Socha RD, Chen J, Tokuriki N. 2019. The molecular mechanisms underlying hidden phenotypic variation among metallo-β-lactamases. J. Mol. Biol. 431, 1172-1185. ( 10.1016/j.jmb.2019.01.041) [DOI] [PubMed] [Google Scholar]
- 23.Baltrus DA. 2013. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489-495. ( 10.1016/j.tree.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 24.Turner PE, Williams ESCP, Okeke C, Cooper VS, Duffy S, Wertz JE. 2014. Antibiotic resistance correlates with transmission in plasmid evolution. Evolution 68, 3368-3380. ( 10.1111/evo.12537) [DOI] [PubMed] [Google Scholar]
- 25.Werren JH. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl Acad. Sci. USA 108, 10 863-10 870. ( 10.1073/pnas.1102343108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Leary NA, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733-D745. ( 10.1093/nar/gkv1189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068-2069. ( 10.1093/bioinformatics/btu153) [DOI] [PubMed] [Google Scholar]
- 28.Alcock BP, et al. 2019. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 10, 226-229. ( 10.1093/nar/gkz935) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wein T, Wang Y, Barz M, Stücker FT, Hammerschmidt K, Dagan T. 2021. Essential gene acquisition destabilizes plasmid inheritance. PLoS Genet. 17, e1009656. ( 10.1371/journal.pgen.1009656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinegger M, Söding J. 2017. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026-1028. ( 10.1038/nbt.3988) [DOI] [PubMed] [Google Scholar]
- 31.Daily J. 2016. Parasail: SIMD C library for global, semi-global, and local pairwise sequence alignments. BMC Bioinf. 17, 81. ( 10.1186/s12859-016-0930-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enright AJ, Dongen SV, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575-1584. ( 10.1093/nar/30.7.1575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530-1534. ( 10.1093/molbev/msaa015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307-1320. ( 10.1093/molbev/msn067) [DOI] [PubMed] [Google Scholar]
- 36.Le SQ, Dang CC, Gascuel O. 2012. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol. Biol. Evol. 29, 2921-2936. ( 10.1093/molbev/mss112) [DOI] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. 2019. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256-W259. ( 10.1093/nar/gkz239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289-300. ( 10.1111/j.2517-6161.1995.tb02031.x) [DOI] [Google Scholar]
- 39.Ghachi ME, Bouhss A, Blanot D, Mengin-Lecreulx D. 2004. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J. Biol. Chem. 279, 30 106-30 113. ( 10.1074/jbc.M401701200) [DOI] [PubMed] [Google Scholar]
- 40.González-Leiza SM, Pedro MA, de Ayala JA. 2011. AmpH, a bifunctional dd-endopeptidase and dd-carboxypeptidase of Escherichia coli. J. Bacteriol. 193, 6887-6894. ( 10.1128/JB.05764-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo Q, Tomich AD, McElheny CL, Cooper VS, Stoesser N, Wang M, Sluis-Cremer N, Doi Y. 2016. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J. Antimicrob. Chemother. 71, 2460-2465. ( 10.1093/jac/dkw177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X-Z, Nikaido H. 2016. Antimicrobial drug efflux pumps in Escherichia coli. In Efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation and clinical implications (eds Li X-Z, Elkins CA, Zgurskaya HI), pp. 219-259. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 43.Hansen LH, Johannesen E, Burmølle M, Sørensen AH, Sørensen SJ. 2004. Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 48, 3332-3337. ( 10.1128/AAC.48.9.3332-3337.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Zhang H, Ning J, Sajid A, Cheng G, Yuan Z, Hao H. 2019. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control 8, 44. ( 10.1186/s13756-019-0489-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen LH, Sørensen SJ, Jørgensen HS, Jensen LB. 2005. The prevalence of the OqxAB amongst olaquindox-resistant multidrug efflux pump Escherichia coli in pigs. Microb. Drug Resist. 11, 378-382. ( 10.1089/mdr.2005.11.378) [DOI] [PubMed] [Google Scholar]
- 46.McMurry L, Petrucci RE, Levy SB. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl Acad. Sci. USA 77, 3974-3977. ( 10.1073/pnas.77.7.3974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ball PR, Shales SW, Chopra I. 1980. Plasmid-mediated tetracycline resistance in Escherichia coli involves increased efflux of the antibiotic. Biochem. Biophys. Res. Commun. 93, 74-81. ( 10.1016/S0006-291X(80)80247-6) [DOI] [PubMed] [Google Scholar]
- 48.Roberts MC. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19, 1-24. ( 10.1111/j.1574-6976.1996.tb00251.x) [DOI] [PubMed] [Google Scholar]
- 49.Poole K. 2007. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162-176. ( 10.1080/07853890701195262) [DOI] [PubMed] [Google Scholar]
- 50.Kazama H, Hamashima H, Sasatsu M, Arai T. 1999. Characterization of the antiseptic-resistance gene qacEΔ1 isolated from clinical and environmental isolates of Vibrio parahaemolyticus and Vibrio cholerae non-O1. FEMS Microbiol. Lett. 174, 379-384. ( 10.1111/j.1574-6968.1999.tb13593.x) [DOI] [PubMed] [Google Scholar]
- 51.Oliveira DMPD, Forde BM, Kidd TJ, Harris PNA, Schembri MA, Beatson SA, Paterson DL, Walker MJ. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181-19. ( 10.1128/CMR.00181-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livermore DM. 1995. beta-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8, 557-584. ( 10.1128/CMR.8.4.557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Datta N, Kontomichalou P. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208, 239-241. ( 10.1038/208239a0) [DOI] [PubMed] [Google Scholar]
- 54.Salverda MLM, De Visser JAGM, Barlow M. 2010. Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol. Rev. 34, 1015-1036. ( 10.1111/j.1574-6976.2010.00222.x) [DOI] [PubMed] [Google Scholar]
- 55.Abraham EP, Chain E. 1940. An enzyme from bacteria able to destroy penicillin. Nature 146, 837. ( 10.1038/146837a0) [DOI] [PubMed] [Google Scholar]
- 56.Jacoby GA. 2009. AmpC β-Lactamases. Clin. Microbiol. Rev. 22, 161-182. ( 10.1128/CMR.00036-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y-Y, et al. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161-168. ( 10.1016/S1473-3099(15)00424-7) [DOI] [PubMed] [Google Scholar]
- 58.Hinchliffe P, et al. 2017. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Scient. Rep. 7, 39392. ( 10.1038/srep39392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swedberg G, Sköld O. 1983. Plasmid-borne sulfonamide resistance determinants studied by restriction enzyme analysis. J. Bacteriol. 153, 1228-1237. ( 10.1128/JB.153.3.1228-1237.1983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rådström P, Swedberg G. 1988. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob. Agents Chemother. 32, 1684-1692. ( 10.1128/AAC.32.11.1684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sköld O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3, 155-160. ( 10.1054/drup.2000.0146) [DOI] [PubMed] [Google Scholar]
- 62.Sköld O. 1976. R-factor-mediated resistance to sulfonamides by a plasmid-borne, drug-resistant dihydropteroate synthase. Antimicrob. Agents Chemother. 9, 49-54. ( 10.1128/AAC.9.1.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson DN, Hauryliuk V, Atkinson GC, O'Neill AJ. 2020. Target protection as a key antibiotic resistance mechanism. Nat. Rev. Microbiol. 18, 637-648. ( 10.1038/s41579-020-0386-z) [DOI] [PubMed] [Google Scholar]
- 64.Jacoby GA, Strahilevitz J, Hooper DC. 2014. Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2, 2.5.33. ( 10.1128/microbiolspec.PLAS-0006-2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313-339. ( 10.1016/S0168-6445(03)00048-2) [DOI] [PubMed] [Google Scholar]
- 66.Rojas A, Duque E, Mosqueda G, Golden G, Hurtado A, Ramos JL, Segura A. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183, 3967-3973. ( 10.1128/JB.183.13.3967-3973.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruiz C, Levy SB. 2014. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J. Antimicrob. Chemother. 69, 390-399. ( 10.1093/jac/dkt352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180, 5443-5447. ( 10.1128/JB.180.20.5443-5447.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fahmy A, Srinivasan A, Webber MA. 2016. The relationship between bacterial multidrug efflux pumps and biofilm formation. In Efflux-mediated antimicrobial resistance in bacteria: mechanisms, regulation and clinical implications (eds Li X-Z, Elkins CA, Zgurskaya HI), pp. 651-663. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 70.García-León G, Hernández A, Hernando-Amado S, Alavi P, Berg G, Martínez JL. 2014. A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots. Appl. Environ. Microbiol. 80, 4559-4565. ( 10.1128/AEM.01058-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J. Antimicrob. Chemother. 51, 1109-1117. ( 10.1093/jac/dkg222) [DOI] [PubMed] [Google Scholar]
- 72.Wein T, Hülter NF, Mizrahi I, Dagan T. 2019. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 10, 2595. ( 10.1038/s41467-019-10600-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wein T, Wang Y, Hülter NF, Hammerschmidt K, Dagan T. 2020. Antibiotics interfere with the evolution of plasmid stability. Curr. Biol. 30, 3841-3847. ( 10.1016/j.cub.2020.07.019) [DOI] [PubMed] [Google Scholar]
- 74.Hall JPJ, Wood AJ, Harrison E, Brockhurst MA. 2016. Source–sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl Acad. Sci. USA 113, 8260-8265. ( 10.1073/pnas.1600974113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wein T, Dagan T. 2020. Plasmid evolution. Curr. Biol. 30, R1158-R1163. ( 10.1016/j.cub.2020.07.003) [DOI] [PubMed] [Google Scholar]
- 76.Hülter NF, Wein T, Effe J, Garoña A, Dagan T. 2020. Intracellular competitions reveal determinants of plasmid evolutionary success. Front. Microbiol. 11, 2062. ( 10.3389/fmicb.2020.02062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rousset F, Cabezas-Caballero J, Piastra-Facon F, Fernández-Rodríguez J, Clermont O, Denamur E, Rocha EPC, Bikard D. 2021. The impact of genetic diversity on gene essentiality within the Escherichia coli species. Nat. Microbiol. 6, 301-312. ( 10.1038/s41564-020-00839-y) [DOI] [PubMed] [Google Scholar]
- 78.Novick A, Doolittle WF. 2020. Horizontal persistence and the complexity hypothesis. Biol. Phil. 35, 2. ( 10.1007/s10539-019-9727-6) [DOI] [Google Scholar]
- 79.Wang Y, Batra A, Schulenburg H, Dagan T. 2021. Gene sharing among plasmids and chromosomes reveals barriers for antibiotic resistance gene transfer. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Wang Y, Batra A, Schulenburg H, Dagan T. 2021. Gene sharing among plasmids and chromosomes reveals barriers for antibiotic resistance gene transfer. Figshare. [DOI] [PMC free article] [PubMed]
Data Availability Statement
The data are provided in electronic supplementary material [79].