Abstract
Three novel Avian avulavirus species were discovered and isolated during 2017 from Gentoo penguins (Pygoscelis papua) at Kopaitic island in the Northwestern region of the Antarctic Peninsula. The viruses were officially named as Avian avulavirus 17 (AAV17), Avian avulavirus 18 (AAV18) and Avian avulavirus 19 (AAV19), collectively referred to as penguin avulaviruses (PAVs). To determine whether these viruses are capable of infecting the three species of Pygoscelis spp. penguins (Gentoo, Adelie and Chinstrap) and assess its geographical distribution, serum samples were collected from seven locations across the Antarctic Peninsula and Southern Shetland Islands. The samples were tested by Hemagglutination inhibition assay using reference viruses for AAV17, AAV18 and AAV19. A total of 498 sera were tested, and 40 were positive for antibodies against AAV17, 20 for AAV18 and 45 for AAV19. Positive sera were obtained for the penguin’s species for each virus; however, antibodies against AAV18 were not identified in Adelie penguins. Positive penguins were identified in all regions studied. Positive locations include Ardley Island and Cape Shirreff at Livingston Island (Southern Shetland Region); Anvers Island, Doumer Island and Paradise Bay in the Central Western region; and Avian Island at Southwestern region of the Antarctic Peninsula. The lowest occurrence was observed at the Southwestern region at Lagotellerie Island, where all samples were negative. On the other hand, Cape Shirreff and Paradise Bay showed the highest antibody titres. Field samples did not evidence cross-reactivity between viruses, and detection was significantly higher for AAV19 and lower for AAV18. This is the first serologic study on the prevalence of the novel Avian avulaviruses including different locations in the white continent. The results indicate that these novel viruses can infect the three Pygoscelis spp. penguins, which extend across large distances of the Antarctic Peninsula.
Keywords: Antarctica, Avian avulavirus, distribution, penguins, wildlife
1 |. INTRODUCTION
Three novel Avian avulaviruses (AAV) species were identified in Gentoo penguins (Pygoscelis papua) from Kopaitic Island, Northwestern Region of the Antarctic Peninsula, in 2017 (Neira et al., 2017). These viruses were officially named as Avian avulavirus 17 (AAV17), Avian avulavirus 18 (AAV18) and Avian avulavirus 19 (AAV19; Amarasinghe et al., 2018). There was no evidence of serum antibody cross-reactivity among the three PAVs in experimental studies (Neira et al., 2017; Wille et al., 2019). A limited number of Gentoo penguins have been tested for antibodies against these viruses, with few positive sera (Neira et al., 2017). In addition, other members of the genus Pygoscelis, Adélie penguin (Pygoscelis adeliae) and Chinstrap penguin (Pygoscelis antarcticus), cohabitate with Gentoo penguins in several locations of the Antarctic, including Kopaitic Island (Naveen & Lynch, 2011). To date, there is limited information about the susceptibility of these penguins to PAVs and the distribution of these viruses is also unknown. The goal of this study was to determine the distribution and prevalence of these novel AAVs in the three species of the genus Pygoscelis from seven locations of the Antarctic Peninsula. These locations include three large regions of the Antarctic Peninsula: South Shetland Islands, Central western and South-west regions. The results of this study will contribute to increasing the knowledge about potential emerging pathogens in the Antarctic continent.
2 |. MATERIALS AND METHODS
The sampling was carried out during the 53rd Chilean Antarctic Expedition supported by the Instituto Antártico Chileno (Chilean Antarctic Institute/INACH), between January and February 2017. Blood samples were collected from three species of penguins: Pygoscelis papua, Pygoscelis adeliae and Pygoscelis antarcticus. Sampling included mainly adult penguins, but at Ardley Island most of the samples were obtained from juvenile penguin just before fledging. Briefly, the penguins were trapped using a mesh landing net and were then freed carefully from the net and placed on the lap of the handler facing forward. The eyes were covered with a black cloth, and the beak and wings were held gently to restrain the animal, while exposing the feet. The blood was collected by venipuncture on the common plantar digital vein using a 23G needle with a 3-ml syringe. Blood was placed in 2-ml microtubes and centrifuged for 10 min at 800 g using a portable microcentrifuge. The sera were transferred to 1.5-ml microtubes and stored at −20°C until processing.
Samples were obtained from seven locations in the Antarctic Peninsula. The locations correspond to Antarctic Specially Protected Areas (ASPA), which were selected rationally according to accessibility and to obtain samples from the three penguin species. Sampling sites included three large areas: (a) the South Shetland Islands region, including Ardley Island (62°13′S, 58°56′W) and Cape Shirreff at Livingston Island (62°34′25″S, 60°48′36″W); (b) the Central western region, including Doumer Island near Yelcho Station (Chile; 64°52′33″S, 63°35′00″W), Paradise Bay near the Gabriel Gonzalez Videla (GGV) Station (Chile) (64°49′43″S, 62°49′13″W) and Biscoe Point at Anvers Island (64°49′42″S, 63°42′50″W); and (c) the South-west region, including Avian Island (67°46′S, 68°54′W) and Lagotellerie Island (67°51′29″S, 67°10′57″W; Figure 1).
FIGURE 1.
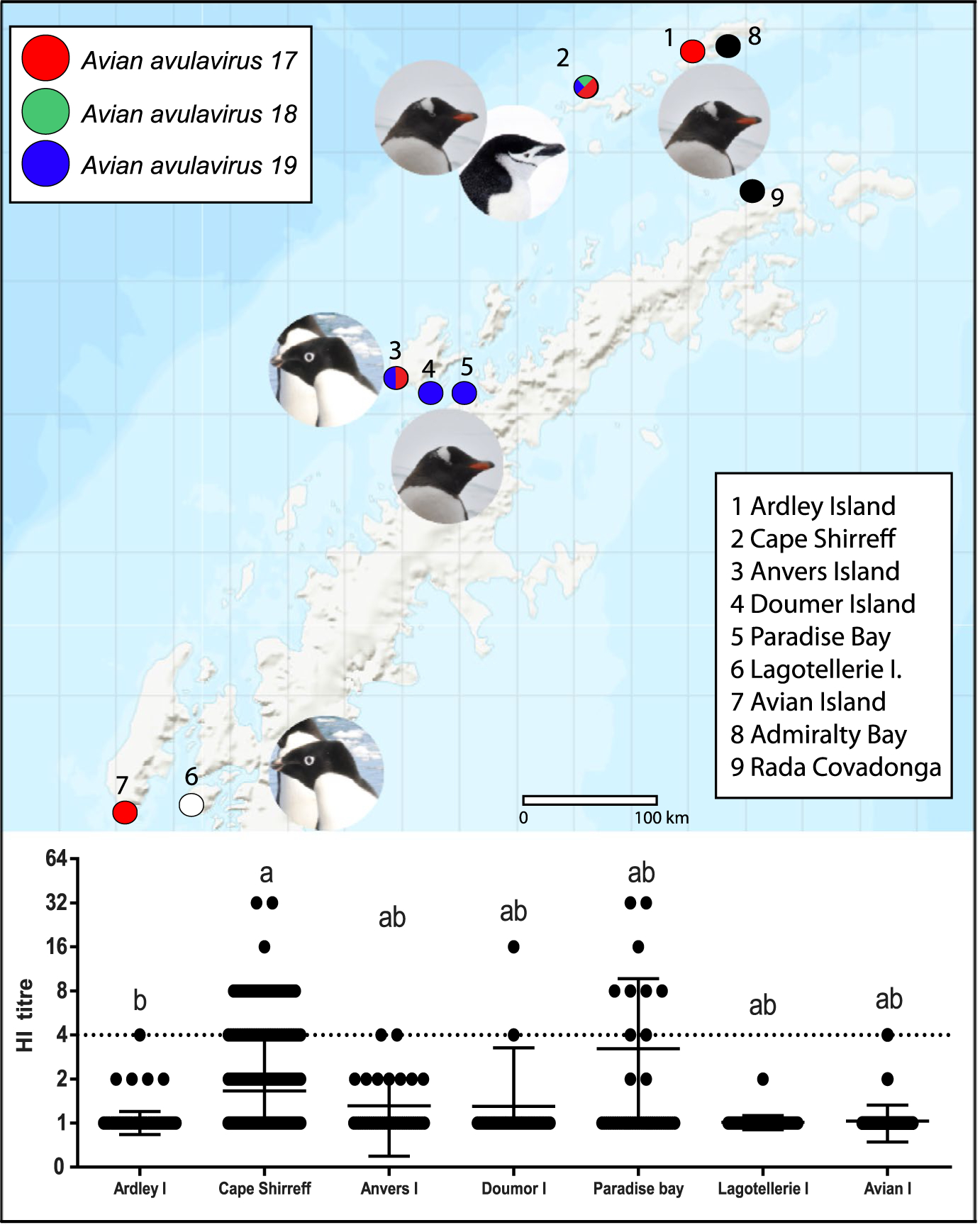
Antibody detection of novel penguin Avian avulaviruses in the Antarctic Peninsula. (a) Geographic representation of sampling locations; avulavirus and penguin species are showed in each location. (b) HI titres against Avian avulaviruses (17, 18 and 19) sorted by location
The sera obtained were tested by hemagglutination inhibition (HI) assay, using reference strains of each AAV species. Reference strains were as follows: AAV-17/Gentoo/Kopaitic Island/117/2016, AAV-18/Gentoo/Kopaitic Island/84/2016 and AAV-19/Gentoo/Kopaitic Island/132/2016, for AAVs 17, 18 and 19, respectively. Virus strains were propagated, and sera were inactivated before the HI assay. The strains were inoculated into the allantoic cavity of 10-day-old specific-pathogen-free embryonated chicken eggs. The eggs were then incubated for 72 hr at 37°C. Subsequently, the eggs were euthanized in a CO2 chamber and refrigerated at 4°C overnight. Allantoic fluid was collected and titrated by hemagglutination (HA) assay, using a standard protocol (Kitikoon, Gauger, & Vincent, 2014). Viruses were diluted to eight hemagglutination units (HAU)/50 μl for further HI assay.
The sera were inactivated to eliminate non-specific inhibitors and hemagglutinating agents using heat treatment, red blood cell adsorption and kaolin treatment. Briefly, 100 μl of serum were incubated at 56°C for 30 min. Then, 600 μl of 25% kaolin (hydrated aluminium silicate), diluted in Borate buffered saline (BBS), was added. The solution was incubated at room temperature (RT) for 30 min and centrifuged at 800 g for 20 min. Finally, 600 μl of 20% turkey red blood cells, diluted in PBS, was added and the mixture was incubated for 2 hr at RT. Inactivated sera were considered to be at a 1:10 dilution. After a centrifugation at 800 g for 20 min, the supernatant was collected and stored at −20°C until used.
Hemagglutination inhibition assay was carried out following a standard protocol (Kitikoon et al., 2014). HI titre ≥40 was considered positive, and the limit of detection was 20. The HI results were analysed by Kruskal–Wallis and Dunn test to determine differences between groups sorted by location, species and viruses. True prevalence determinations were obtained for Cape Shirreff and Ardley Island. However, due to the scarce number of samples for the rest of the locations, disease freedom calculations were estimated. Based on the number of samples obtained for each place, a cut-off value was determined. One or more positive samples in those places indicate that the prevalence was higher that the cut-off value calculated. The epidemiological calculations were performed using AUSVET EpiTools (Sergeant, 2019)..
3 |. RESULTS AND DISCUSSION
In this study, 498 samples were tested: 211 samples were obtained from Gentoo penguin, 206 from Chinstrap penguin and 81 from Adélie penguin. Of the total, 75 (15.1%) samples were positive at least to one virus and the HI titres ranged between 20 and 320 (Figure 1). Forty (8%) samples were positive for AAV 17, 20 (4%) for AAV 18 and 45 (9%) for AAV 19. Twelve (2.4%) samples were positive for two AAVs, and 9 (1.8%) samples were positive for the three AAVs; however, results did not evidence cross-reactivity between viruses based on the field results. Results were summarized in Table 1.
TABLE 1.
Overall results of hemagglutination inhibition (HI) assay against Avian avulaviruses 17, 18 and 19
| Samples No. | AAV 17 (%) | AAV 18 (%) | AAV 19 (%) | Total (%) | |
|---|---|---|---|---|---|
| Penguin species | |||||
| P. papua | 211 | 7 (3) | 4 (2) | 14 (7) | 25 (12) |
| P. adeliae | 81 | 2 (2.5) | 0 (0) | 1 (1) | 3 (4) |
| P. antarcticus | 206 | 31 (15) | 16 (8) | 30 (15) | 77 (37) |
| Locations | |||||
| Ardley I. (P. papua) | 128 | 1 (1) | 0 (0) | 0 (0) | 1 (1) |
| Cape Shirreff (P. antarcticus/P. Papua) | 252 | 37 (15) | 20 (8) | 33 (13) | 90 (36) |
| Paradise Bay (P. papua) | 17 | 0 (0) | 0 (0) | 9 (53) | 9 (53) |
| Anvers I. (P. adeliae) | 14 | 1 (7) | 0 (0) | 1 (7) | 2 (14) |
| Doumer I. (P. papua) | 20 | 0 (0) | 0 (0) | 2 (10) | 2 (10) |
| Avian I. (P. adeliae) | 40 | 1 (3) | 0 (0) | 0 (0) | 1 (3) |
| Lagotellerie I. (P. adeliae) | 27 | 0 (0) | 0 (0 | 0 (0) | 0 (0) |
Note: Number and percentage of positive samples are shown per penguin species and locations.
All penguin species in this study (Gentoo, Chinstrap and Adélie) reported at least one positive sample to PAV (Figure S1). In the case of Adélie penguins, from the 81 samples collected, 2 (2.5%) were positive for AAV17, none for AAV18 and 1 (1.2%) for AAV19. For Chinstrap penguins, from a total of 206 sera tested, 31 (15%) were positive for AAV17, 16 (7.8%) for AAV18 and 30 (15%) for AAV19. From the 211 Gentoo’s samples, 7 (3.3%) were positive for AAV17, 4 (1.9%) for AAV18 and 14 (6.6%) for AAV19 (Table 1). All penguin species presented the lowest antibody titres to AAV18, which was statistically significant and only detected at Cape Shirreff (Figures S1,S2).
The results by location showed occurrence ranged between 0% and 53%. From the seven locations studied, only Lagotellerie Island was negative for all three viruses (Figure 1). The lower occurrence was observed at Ardley (0.8%) and Avian Island (2.5%), and each presented only one positive sample to AAV17. In the other hand, nine out of 17 (52.9%) samples obtained in Paradise Bay were positive to AAV19. Finally, Cape Shirreff was the only place on which two species (Chinstrap and Gentoo) were sampled in high numbers. Two hundred fifty-two samples were obtained, 37 (14.7%) were positive for AAV17, 20 (7.9%) for AAV18 and 33 (13.1%) for AAV19. Ten out of 46 (21.7%) samples obtained in Gentoo penguins were positive to at least one PAV, while 51 out of 206 (24.8%) samples obtained in Chinstrap penguins were positive to at least one PAV. Statistically, differences were recognized among locations per virus. The most remarkable finding was observed for AAV19, which showed three levels of seroreactivity. The higher reactivity was observed in Paradise Bay and Biscoe Point at Anvers Island, secondly in Cape Shirreff and the lower levels in Ardley and Avian Island (Table 1, Figure S2).
In Ardley Island, the prevalence was estimated in 0.8% for AAV17, with a confidence level of 99% and an absolute accepted error of 10% (with an adjustment of the sample size), while the prevalence for AAV18 and AAV19 was 0%. In Cape Shirreff, the true prevalence was calculated for Chinstrap penguins resulting 15% for AAV17 and AAV19, and 8% for AAV18, with a confidence level of 99.5% and an absolute error accepted of 10%.
Disease freedom was estimated for the rest of the locations. Thus, for Anvers Island, the prevalence was expected in ≥22% for both AAV17 and AAV19, but <22% for AAV18. In Doumer Island, the prevalence was calculated in ≥20% for AAV19 and <20% for AAV17 and AAV18. In Paradise Bay, the prevalence was estimated in ≥17% for AAV19 and <17% for AAV17 and AAV18. For Avian Island, it was ≥8.3 for AAV17 and <8.3% for AAV18 and AAV19. Finally, for Lagotellerie Island, it was <12% for the three viruses.
The aim of this study was to determine serologic evidence of PAVs in Pygoscelis spp. penguins in seven locations across the west side of the Antarctic Peninsula. HI assay was performed using reference strains of AAV17, AAV18 and AAV19, which were isolated and characterized in 2017 from Gentoo penguins in Kopaitic Island, Antarctic (Neira et al., 2017).
Our understanding of the epidemiology of Antarctic viruses requires investigation. The recent discovery of AAVs 17, 18 and 19 spotted new questions about avulavirus in Antarctica wildlife, for example if these viruses were or not infective for other penguin species or Antarctic birds. In this study, samples were collected from the three Pygoscelis penguins detecting antibodies in all of them against AAV17, AAV18 and AAV19 reference viruses, which strongly suggest that all PAVs are capable to infect Papua, Adélie and Chinstrap penguins. Antibodies against AAV19 presented the higher titres and were more frequently found and widespread compared with AAV17 and AAV18. On the other hand, Wille et al. (2019) detected higher proportion of AAV17 over AAV19, however that study was limited only to two locations at the Antarctic Peninsula, and the detection was performed by qPCR (Wille et al., 2019).
Other important question is the geographic distribution of these viruses. Previous to this study, only two locations had been sampled to determine the presence of PAV, Admiralty Bay at King George Island and Kopaitic Island, both located in the North of Antarctic Peninsula, with a distance of 130 km between them. We were able to conduct animal sampling at seven new locations, including three large regions in the Antarctic Peninsula, with a distance ranged between 15 to 770 km (Figure 1, Supplementary Table S1). Antibodies against AAV17 were found at Ardley and Avian Islands with a distance of 770 km approx., while AAV19 antibodies were detected in Cape Shirreff and the Central west locations (Anvers, Doumer and Paradise bay) located at a distance of 300 km approx. This shows the wide distribution of these viruses in the Antarctic Peninsula. If we consider the previous detections in Admiralty Bay and Kopaitic Island, there is evidence of PAVs at a distance >800 km across the Antarctic Peninsula. Pygoscelis penguins, especially Chinstrap and Adelie penguins, can migrate large distances (up to >1,000 km) in non-breeding season and may help in the virus dissemination (Dunn, Silk, & Trathan, 2011; Hinke et al., 2015; Levy et al., 2016). Up to date, PAVs have been not reported in other seabirds, but may have a role in the dissemination. In other hand, mi-grant birds have been considered disseminators of paramyxoviruses in the environment (Fornells et al., 2013). Miller, Watts, & Shellam, 2008, detected antibodies against NDV (a strain of AAV1) in south polar skuas (Catharacta maccormicki Saunders) in Vestfold Hills Region at the Princess Elizabeth Land, Antarctica (Miller et al., 2008). Skuas are migratory birds and penguin predators and may have an important role in AAVs dissemination, which need further studied.
Penguin seroreactors were identified in all sampling sites, excepting Lagotellerie Island. Geographically, the seroreactors were concentrated in Cape Shirreff at Livingston Island (Southern Shetlands Island Region) and in the Central west part of the Antarctic Peninsula. On the other hand, the South-west region, including Avian Island and Lagotellerie Island, evidence the lower occurrence, which could indicate the low prevalence or absence of these viruses in this region. In Ardley Island, most of the penguins were juvenile; this could be a plausible explanation to the scarce serology detection of PAVs in this location (1/123). Young penguins could be not good specimens for serology studies because they have short time to be exposed and to develop detecting antibodies; on the contrary, they born naïve to pathogens and could be good for direct viral detection. Adélie chicks previously showed a significantly higher overall prevalence than adults using direct detection (Wille et al., 2019). The low detection in Ardley Island and in other regions may be associated with the absence of viruses or the presence of other avian avulaviruses in the region. Other avian avulaviruses species have been reported previously in the Antarctic, such as AAV1 and AAV10 using both, serology and sequencing (Austin & Webster, 1993; Morgan & Westbury, 1981; Thomazelli et al., 2010; Wille et al., 2019). We were not able to perform serology using these reference viruses, which could be considered a limitation of this study. In other hand, avian avulaviruses generally present cross-reactivity between species but demonstrated low cross-reactivity of PAVs to other avulaviruses (Neira et al., 2017; Wille et al., 2019), and then, false-positive results are unlikely.
Another question is whether these novel viruses are pathogenic. Previous results and recent studies identified and isolated the viruses from clinically healthy penguins suggesting low pathogenicity of these viruses (Neira et al., 2017; Wille et al., 2019). In general, avian avulaviruses are considered non-pathogenic, except AAV1 (NDV), AAV2 and AAV3, which have consistently shown infect and cause disease in poultry (Alexander, 2000). Recently, Wille et al. (2019) performed experimental inoculations of AAV17, AAV18 and AAV19 in chickens, but no clinical signs were observed. Also, shedding and seroconversion were limited (Wille et al., 2019).
One important limitation of this study was the low number of samples collected because of the extreme environmental conditions or the limited time in most of the locations, which difficult the epidemiological estimations. This is the first study that evidences the widespread distribution of the novel PAVs across the Antarctic Peninsula. Further investigations are needed to understand the pathogenic role of these viruses in penguins as well as other seabirds in Antarctica.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the staff of Instituto Antártico Chileno for all their support during the Antarctic expedition 2017, especially to Pablo Espinoza, Patricio Barraza and Wendy Rubio. We thank to the Chilean Navy, especially to the AP-46 Almirante Oscar Viel 2017 crew and support personal from Marine Corps and Naval Aviation, for its assistance in field sampling. We also thank the staff of the Instituto de Salud Pública de Chile for biological supplies. This study was partly funded by the Programa Fondecyt de Iniciación N° 11170877 to V.N; the Programa Beca Doctorado Nacional de CONICYT N° 3344/2016 to J.M; CONICYT PIA/BASAL FB0002 to R.B.C; Programa de Investigación Asociativa from the Comisión Nacional de Investigación Científica y Tecnológica, project CONICYT-PIA Anillo ACT 1408 to R.A.M. and V.N. and by the INACH RT-4616 and the Center for Research in Influenza Pathogenesis (CRIP), a National Institute of Allergy and Infectious Diseases-funded Center of Excellence in Influenza Research and Surveillance (CEIRS), contract number HHSN272201400008C to R.A.M
Funding information
Instituto Antártico Chileno, Grant/Award Number: RT-4616; National Institute of Allergy and Infectious Diseases, Grant/Award Number: HHSN272201400008C; Fondo Nacional de Desarrollo Científico y Tecnológico, Grant/Award Number: 11170877; Comisión Nacional de Investigación Científica y Tecnológica, Grant/Award Number: CONICYT-PIA Anillo ACT 1408 National Doctorate Scholarship Program CONICYT N°; CONICYT; National Institute of Allergy and Infectious Diseases, Grant/Award Number: HHSN272201400008C
Footnotes
ETHICS STATEMENT
The protocols for animal sampling were approved by the Ethical Scientific Committee for Animals and Environment Care from Pontificia Universidad Católica de Chile, certificate N°150430002. Also, virus propagation was approved by the Biosecurity Committee of the Universidad de Chile, certificate N°113.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Alexander DJ (2000). Newcastle disease and other avian paramyxoviruses Aetiology, Revue Scientifique Et Technique, 19, 443–462. 10.20506/rst.19.2.1231 [DOI] [PubMed] [Google Scholar]
- Amarasinghe GK, Aréchiga Ceballos NG, Banyard AC, Basler CF, Bavari S, Bennett AJ, … Kuhn JH (2018). Taxonomy of the order mononegavirales: Update 2018. Archives of Virology, 163(8), 2283–2294. 10.1007/s00705-018-3814-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin FJ, & Webster RG (1993). Evidence of ortho- and paramyxoviruses in fauna from Antarctica. Journal of Wildlife Diseases, 29, 568–571. 10.7589/0090-3558-29.4.568 [DOI] [PubMed] [Google Scholar]
- Dunn MJ, Silk JRD, & Trathan PN (2011). Post-breeding dispersal of Adélie penguins (Pygoscelis adeliae) nesting at Signy Island, South Orkney Islands. Polar Biology, 34(2), 205–214. 10.1007/s00300-010-0870-4 [DOI] [Google Scholar]
- Fornells LAMG, Travassos CEPF, Costa CM, Novelli R, Petrucci MP, Soffiati FL, … Couceiro JNSS (2013). Detection of avian paramyxoviruses in migratory and resident birds in the State of Rio de Janeiro, Brazil. Avian Diseases, 57, 780–784. 10.1637/10548-040813-ResNote.1 [DOI] [PubMed] [Google Scholar]
- Hinke JT, Polito MJ, Goebel ME, Jarvis S, Reiss CS, Thorrold SR, … Watters GM (2015). Spatial and isotopic niche partitioning during winter in chinstrap and Adélie penguins from the South Shetland Islands. Ecosphere, 6(7), art125. 10.1890/ES14-00287.1 [DOI] [Google Scholar]
- Kitikoon P, Gauger PC, & Vincent AL (2014). Hemagglutinin Inhibition assay with swine sera. In Spackman E (Ed.), Animal influenza virus. Methods in molecular biology (methods and protocols) (Vol. 1161). New York, NY: Humana Press. [DOI] [PubMed] [Google Scholar]
- Levy H, Clucas GV, Rogers AD, Leaché AD, Ciborowski KL, Polito MJ, … Hart T (2016). Population structure and phylogeography of the gentoo penguin (Pygoscelis papua) across the Scotia Arc. Ecology and Evolution, 6, 1834–1853. 10.1002/ece3.1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GD, Watts JM, & Shellam GR (2008). Viral antibodies in south polar skuas around Davis Station, Antarctica. Antarctic Science, 20(5), 455–461. 10.1017/S0954102008001259 [DOI] [Google Scholar]
- Morgan IR, & Westbury HA (1981). Virological studies of adelie penguins (Pygoscelis adeliae) in Antarctica. Avian Diseases, 25, 1019–1026. 10.2307/1590077 [DOI] [PubMed] [Google Scholar]
- Naveen R, & Lynch H (2011). Antarctic Peninsula Compendium3rd EditionThird Edit. In Naveen R, & Lynch H (Eds.). Chevy Cahse, MD, USA: Oceanites Inc. [Google Scholar]
- Neira V, Tapia R, Verdugo C, Barriga G, Mor S, Ng TFF, … González-Acuña D (2017). Novel avulaviruses in penguins, Antarctica. Emerging Infectious Diseases, 23(7), 1212–1214. 10.3201/eid2307.170054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant E (2019). EpiTools epidemiological calculators [Online] Retrieved from http://epitools.ausvet.com.au/content.php?page=home (accessed May 6, 2019).
- Thomazelli LM, Araujo J, Oliveira DB, Sanfilippo L, Ferreira CS, Brentano L, … Durigon EL (2010). Newcastle disease virus in penguins from King George Island on the Antarctic region. Veterinary Microbiology, 146, 155–160. 10.1016/j.vetmic.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Wille M, Aban M, Wang J, Moore N, Shan S, Marshall J, … Hurt AC (2019). Antarctic penguins as reservoirs of diversity for avian avulaviruses. Journal of Virology, 93(11), e00271–e319. 10.1128/JVI.00271-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


