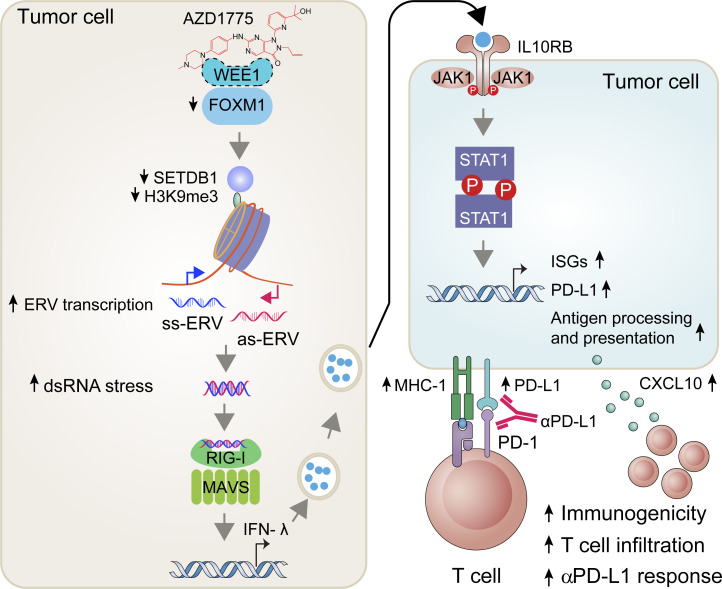
WEE1 inhibition modulates the efficacy of cancer immunotherapy by regulating dsRNA and interferon responses, which increases recruitment of anti-tumor T cells with concurrent PD-L1 elevation. This study provides a rationale for combination strategies between WEE1 inhibitors and anti–PD-L1 therapies.
Abstract
Targeted therapies represent attractive combination partners with immune checkpoint blockade (ICB) to increase the population of patients who benefit or to interdict the emergence of resistance. We demonstrate that targeting WEE1 up-regulates immune signaling through the double-stranded RNA (dsRNA) viral defense pathway with subsequent responsiveness to immune checkpoint blockade even in cGAS/STING-deficient tumors, which is a typical phenotype across multiple cancer types. WEE1 inhibition increases endogenous retroviral elements (ERVs) expression by relieving SETDB1/H3K9me3 repression through down-regulating FOXM1. ERVs trigger dsRNA stress and interferon response, increasing recruitment of anti-tumor T cells with concurrent PD-L1 elevation in multiple tumor models. Furthermore, combining WEE1 inhibition and PD-L1 blockade induced striking tumor regression in a CD8+ T cell–dependent manner. A WEE1 inhibition–induced viral defense signature provides a potentially informative biomarker for patient selection for combination therapy with WEE1 and ICB. WEE1 inhibition stimulates anti-tumor immunity and enhances sensitivity to ICB, providing a rationale for the combination of WEE1 inhibitors and ICB in clinical trials.
Graphical Abstract
Introduction
Immune checkpoint blockade (ICB) has improved outcomes in multiple malignancies (Carbognin et al., 2015). However, only a subset of cancer patients benefits from ICB (Zou et al., 2016). To extend the clinical benefits of ICB to a more significant number of patients, many trials combining ICB with small molecule inhibitors are currently underway. In preclinical models, DNA methyl-transferase inhibitors, histone deacetylase inhibitors, and bromodomain inhibitors restore cytotoxic T cell functions and reverse the immune-suppressive effects of the tumor microenvironment and demonstrate synergistic activity with ICB (Dunn and Rao, 2017), supporting clinical trials.
The binding of double-stranded DNA (dsDNA) to cyclic GMP-AMP synthase (cGAS) in tumor cells activates the stimulator of the IFN genes (STING) pathway. Activation cGAS/STING pathway induces IFN production and boosts host antitumor immunity (Kwon and Bakhoum, 2020). However, the cGAS/STING pathway is commonly aberrant in a number of cancer types, including colorectal carcinoma, melanoma, and ovarian cancer (de Queiroz et al., 2019). Therefore, it is essential to identify alternative approaches to induce IFN activation, especially in cGAS/STING-deficient tumors.
Wee1-like protein kinase (WEE1) ensures that cells with DNA damage arrest and allow time for DNA repair by inhibiting CDK1 (also known as CDC2) and CDK2 (Heald et al., 1993). WEE1 inhibition abrogates S and G2 arrest, causing cells with unrepaired DNA damage to enter mitosis and undergo mitotic catastrophe (Matheson et al., 2016). Recently, increasing clinical trials reported that WEE1 inhibitor (Adavosertib/AZD1775) monotherapy or combined with gemcitabine demonstrated promising clinical activity and synergistic anti-tumor effects in multiple cancer types with limited treatment options, such as recurrent and platinum-resistant serous ovarian carcinoma (Leijen et al., 2016; Lheureux et al., 2021; Liu et al., 2020), and also unresectable pancreatic cancer (Cuneo et al., 2019). However, the potential role of WEE1 inhibition in modulation of the efficacy of cancer immunotherapy remains poorly understood.
Expression of tumor endogenous retroviral elements (ERVs) leads to IFN pathway activation and improved responses to immunotherapy by altering both innate and adaptive immune responses (Cañadas et al., 2018). For instance, decreasing DNA methylation by DNA methyltransferase inhibitors (Chiappinelli et al., 2015) or diminishing histone methylation by LSD1 inhibition (Sheng et al., 2018) up-regulates ERV expression and activates IFN signaling, thus promoting antigen presentation, immunogenicity, and cytokine production, which subsequently synergizes with ICB. Herein, we demonstrate that WEE1 inhibition increases expression of a suite of IFN-responsive genes through up-regulating ERV expression and subsequent double-stranded RNA (dsRNA)–dependent pathways in cancer cells. Our findings provide a novel insight into mechanisms underlying the actions of WEE1 inhibitors, thereby providing a rationale for combination strategies between WEE1 inhibitors and ICB in cancers.
Results
AZD1775 activates tumor immune responses by activating the IFN pathway and increasing T cell infiltration
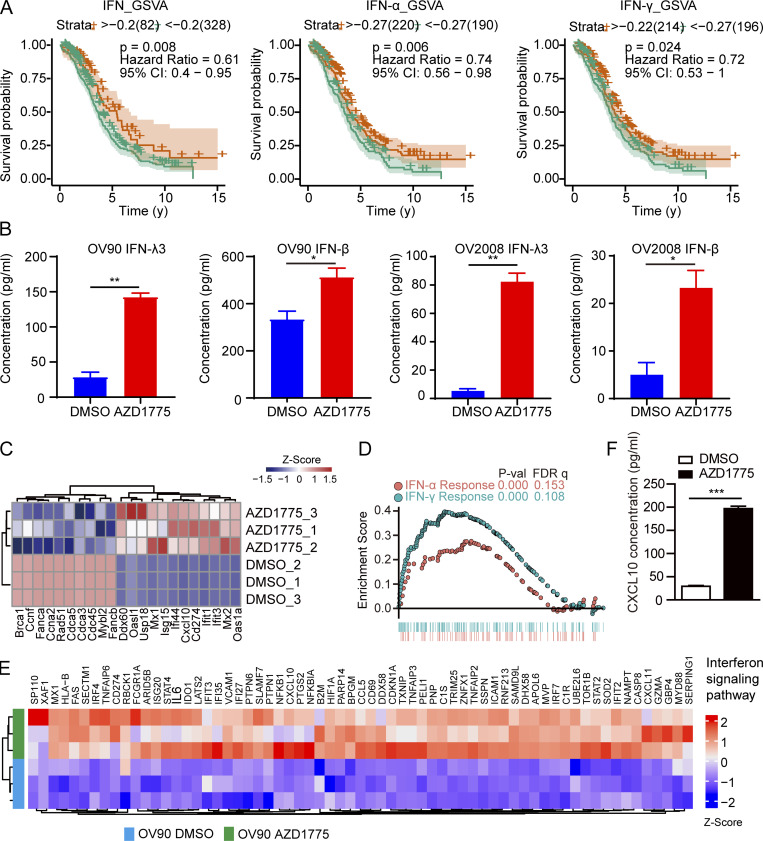
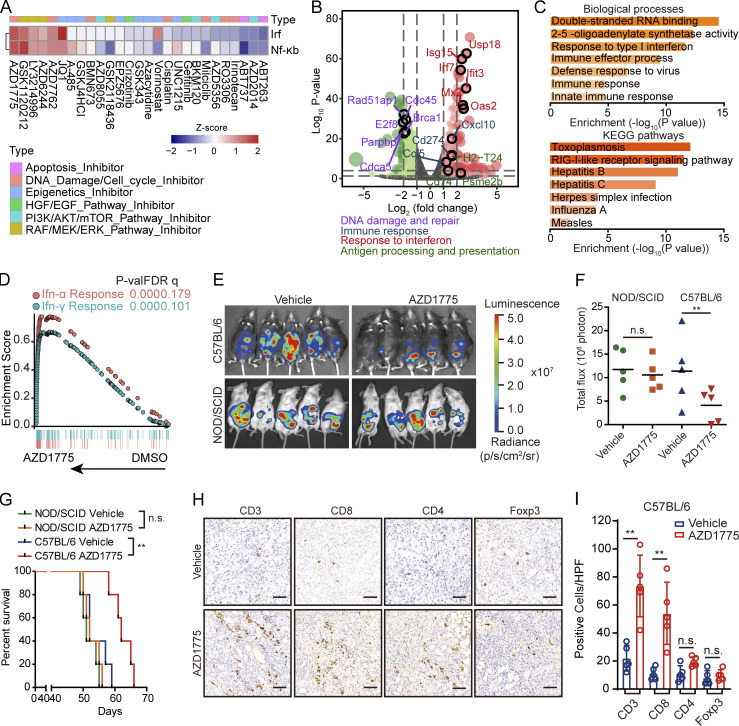
The IFNs, including type I IFN (IFN-α and IFN-β), type II IFN (IFN-γ), and type III IFN (IFN-λ), play a critical role in tumor immune system interactions (Garris et al., 2018; Lasfar et al., 2016; Zitvogel et al., 2015). Gene set variation analysis (GSVA) in The Cancer Genome Atlas (TCGA) database showed that higher IFN signaling in ovarian cancer is associated with improved prognosis (Fig. S1 A). IFN signaling activation has the potential to enhance ICB in various cancers (Chiappinelli et al., 2015). We thus used HCT116-Dual cells, a reporter cell that has been widely used for high throughput screening of IFN activation, to determine whether a series of targeted inhibitors induced IFN signaling (Fig. 1 A). Surprisingly, cell cycling checkpoint inhibitors, including the WEE1 inhibitor (WEE1i, AZD1775) and the CHK1/2 inhibitor (AZD7762), increased IFN signaling, with the WEE1i having the best effect (Fig. 1 A). Despite previous reports of poly (ADP-ribose) polymerase inhibitors (PARPi) altering IFN signaling, the PARPi, BMN673, did not up-regulate IFN signaling in HCT116 cells (Fig. 1 A), likely due to a KRAS mutation mediating resistance to PARPi monotherapy, as we previously reported (Sun et al., 2017). Interestingly, multiple MEK/ERK inhibitors induced profound IFN signaling potentially due to the abrogation of MAPK pathway in KRAS mutant HCT116 cells resulting in DNA damage and STING pathway activation (Sun et al., 2017). We verified up-regulation of IFN-β and IFN-λ (especially IFN-λ3) after WEE1 inhibition, with IFN-α and IFN-γ being undetectable in OV90 and OV2008 cells (Fig. S1 B). RNA sequencing (RNA-seq) in murine ovarian cancer cells (ID8) after AZD1775 treatment showed that DNA damage repair genes, especially the homologous recombination repair pathway, were down-regulated (Fig. 1 B). Consistent with the reporter assay, IFN-stimulated genes (ISGs), including Isg15, Ifit3, Mx2, and Oas2, were markedly up-regulated (Fig. 1 B), which we also validated by quantitative RT-PCR (RT-qPCR; Fig. S1 C). Moreover, AZD1775 also up-regulated a number of IFN-sensitive antigen processing and presentation genes (Tap1, Psme2, and H2-t24) in ID8 cells (Fig. 1 B). AZD1775-induced genes were consistently enriched in gene ontology (GO) terms related to dsRNA binding, IFN response signaling, and immune effector process (Fig. 1 C, top). Further, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis showed up-regulation of gene sets encompassing the RIG-I–like receptor signaling pathway and cellular defense response to the virus (Fig. 1 C, bottom). Gene set enrichment analysis (GSEA) also showed enrichment in “IFN response” (Fig. 1 D) after WEE1 inhibition. AZD1775 induced similar responses in OV90, supporting generalizability (Fig. S1, D and E). AZD1775 also significantly increased CXCL10 release from OV90 cells, an essential downstream IFN target involved in T cell chemotaxis (Fig. S1 F).
Figure S1.
AZD1775 activates immunogenic response and IFN pathway. (A) Kaplan–Meier survival curves according to different IFN pathway gene set expressions of ovarian cancer in TCGA. (B) ELISA detection of IFN-β and IFN-λ3 levels in the OV90 and OV2008 cell supernatant treated with AZD1775 or DMSO for 72 h (n = 2; three independent experiments). (C) Heatmap of qPCR quantification of selected genes differentially expressed in ID8 cells treated with DMSO or AZD1775 for 48 h (n = 3). (D) HALLMARK-IFN–enriched pathways were enriched with GSEA analysis in OV90 cells treated with AZD1775 (n = 3). (E) Heatmap of IFN pathway genes analyzed by RNA-seq in OV90 cells after treatment with 800 nM AZD1775 or DMSO for 48 h (n = 3). (F) ELISA detection of CXCL10 levels in OV90 cells treated with AZD1775 or DMSO for 72 h (n = 2; three independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by log-rank test (A) and unpaired t test (B and F). P-val, P value; FDR q, false discovery rate p-value.
Figure 1.
AZD1775 activates tumor immune response by IFN pathway and recruiting T cell infiltration. (A) Z-score of relative Irf and NF-κB activity in HCT116-Dual cells after treatment with various agents as indicated (treatment/DMSO) for 48 h. Data represent three independent experiments. (B–D) RNA-seq data analysis of ID8 cells treated with AZD1775 or DMSO (three repeats). (B) Volcano plot of differentially expressed genes. (C) GO and KEGG terms enriched after AZD1775 treatment. (D) GSEA of HALLMARK-IFN pathways induced by AZD1775. (E–I) In vivo experiment of ID8 treated with AZD1775 (n = 5; two independent experiments). Representative images (E) and relative total flux of luminescence (F) of ID8 tumors in C57BL/6 and NOD/SCID mice treated with vehicle or AZD1775. (G) Kaplan–Meier survival curves of C57BL/6 and NOD/SCID mice with ID8 tumors treated as indicated. (H and I) Representative images and quantification of immunohistochemistry analyses of CD3+, CD8+, CD4+, and Foxp3+ T reg cells in ID8 tumors after treatments in C57BL/6 mice. HPF, high power field; P-val, P value; FDR q, False Discovery Rate p-value. Scale bar, 50 µm. **, P < 0.01, n.s., not significant as determined by unpaired t test (F and I) and log-rank test (G). p, photons; sr, steradian.
Next, we assessed the effect of AZD1775 on ID8 tumors in syngeneic immunocompetent (C57BL/6) and immunodeficient (NOD.Cg-Prkdcscid [NOD/SCID]) mice. Strikingly, while AZD1775 was highly active in C576BL/6 mice, these anti-tumor effects were markedly attenuated in immunodeficient NOD/SCID mice (Fig. 1, E–G). Consistently, AZD1775 markedly increased tumors infiltrating CD3+ T cells, mainly CD8+ T cells, without altering the CD4+ T cells and regulatory T (T reg) cells (Fig. 1, H and I).
cGAS/STING pathway is frequently defective in cancer cells
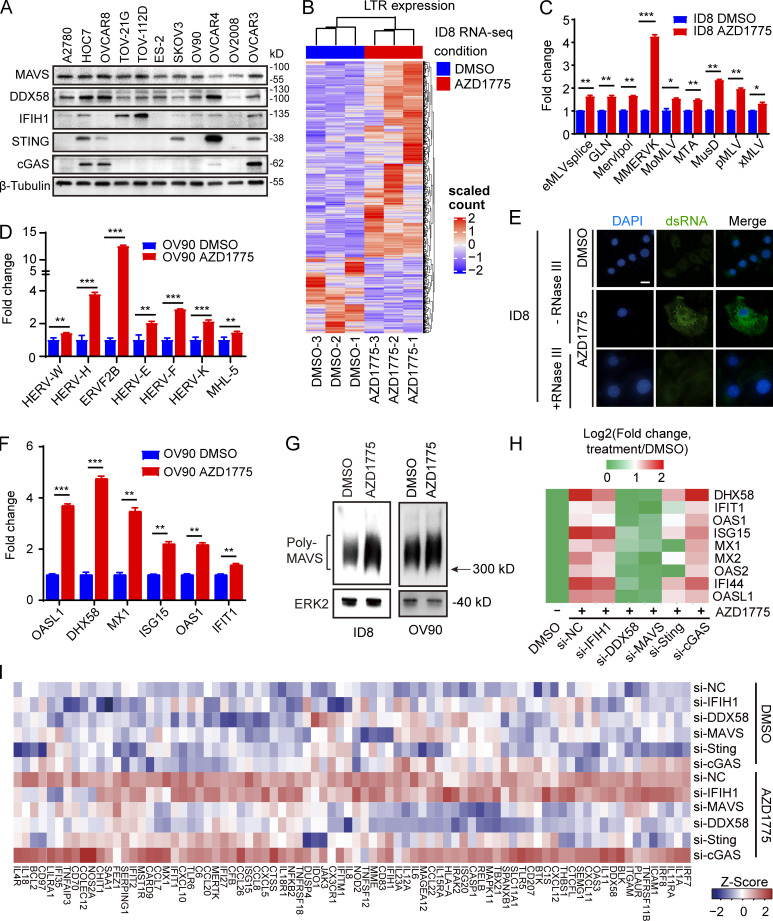
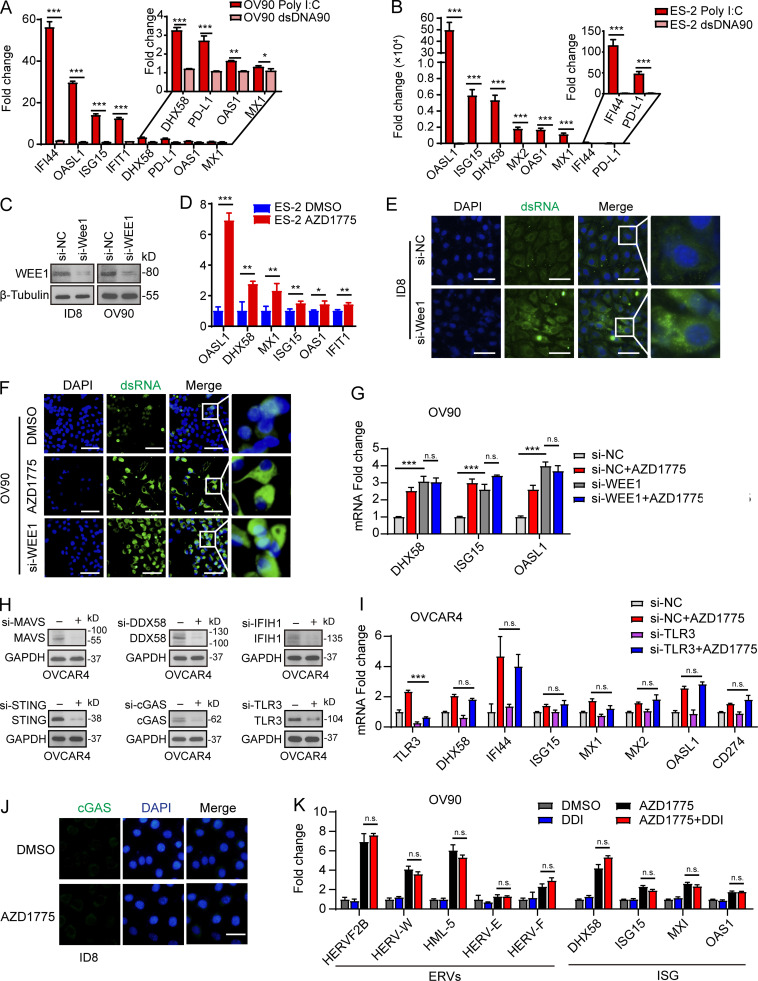
Drugs targeting DNA damage response pathways have been reported to promote anti-tumor immunity by activating the dsDNA-activated cGAS/STING pathway (Wayne et al., 2021). However, many studies reported the frequently defective cGAS/STING pathway in multiple cancer types (de Queiroz et al., 2019), wherein the ability of dsDNA to activate cGAS/STING would have limited clinical significance. GO terms associated with double-strand RNA binding and KEGG analysis with the RIG-I–like receptor signaling pathway suggested that WEE1 inhibition may activate IFN signaling through the alternative dsRNA. We thus evaluated the expression of cytosolic sensors for dsDNA and dsRNA in 11 human ovarian cancer cell lines. Consistent with previous reports, the expression of STING was shallow in 6 of the 11 cell lines examined (Fig. 2 A). Further, cGAS was also diminished in 7 of 11 ovarian cancer cell lines while the dsRNA sensors, mitochondrial antiviral-signaling protein (MAVS) and DDX58, but not IFIH1, were expressed at readily detectable levels in most cell lines (Fig. 2 A). To assess the ability of ovarian cancer cells to respond to dsRNA or dsDNA, we compared ISG expression after stimulation with poly I:C (a synthetic dsRNA analogue) or dsDNA90 (a synthetic dsDNA analogue). As expected, only poly I:C up-regulated ISG expression in cGAS/STING–deficient OV90 and ES-2 cells (Fig. S2, A and B).
Figure 2.
AZD1775 induced ERVs transcription are responsible for IFN responses in ovarian cancer cells. (A) Western blot of basic expression of dsDNA and dsRNA sensor proteins in 11 ovarian cancer cell lines. Data represent three independent experiments. (B) Heatmaps of differentially expressed ERVs after AZD1775 for 48 h in ID8 cells. (C and D) Quantification of nine randomly selected ERVs by qPCR in ID8 and OV90 cells treated with DMSO or AZD1775 for 48 h (three independent experiments). (E) Cellular dsRNA was evaluated with anti-dsRNA (J2) immunofluorescence in ID8 cells with DMSO or AZD1775 for 72 h. RNase III was used as a negative control for dsRNA signal. Scale bars, 20 µm (three independent experiments). (F) Quantification of ISGs by qPCR in OV90 cells after AZD1775 for 48 h (three independent experiments). (G) MAVS protein polymerization detected by immunoblot using SDD-AGE (Materials and methods) in ID8 and OV90 cells treated with DMSO or AZD1775 for 72 h. Data represent three independent experiments. (H) Heatmap of relative expression of ISGs are shown. OVCAR4 cells were transfected with or without siRNA as indicated and treated with DMSO or AZD1775 for 48 h. Data are representative of three experiments. (I) Heatmap of differentially expressed genes after AZD1775 detected by NanoString immune panel in OVCAR4 cells with siRNA inhibition (n = 1) of the indicated dsDNA/dsRNA sensors. The qPCR data were normalized to β-actin. Data across panels represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by unpaired t test (C, D, and F).
Figure S2.
WEE1 inhibition induces generation of dsRNA and activation of IFN pathway. (A and B) Quantification of the selected ISGs by qPCR in OV90 cells and ES-2 after stimulation with poly I:C or dsDNA90 (n = 2; three independent experiments). (C) Western blot validation of the effect of WEE1 siRNA knockdown in ID8 and OV90 cells. Data represent three independent experiments. (D) Quantification of ISGs by qPCR in ES-2 cells after AZD1775 treatment for 48 h (three independent experiments). (E and F) Cellular dsRNA was evaluated with anti-dsRNA (J2) immunofluorescence in ID8 cells transfected with either si-NC, si-Wee1, or AZD1775 for 72 h. Scale bar, 50 µm (three independent experiments). (G) Quantification of the selected ISGs by qPCR in OV90 cells transfected with either si-NC or si-WEE1 and treated with AZD1775 for 48 h (three independent experiments). (H) Western blot validation of the effect of dsRNA and dsDNA sensors siRNA knockdown in OVCAR4 cells. Data represent three independent experiments. (I) Quantification of the selected ISGs by qPCR in OVCAR4 cells. Cells were transfected with either si-NC or si-TLR3 and treated with AZD1775 for 48 h (three independent experiments). (J) Representative images of immunofluorescence staining of cGAS in ID8 cells treated with DMSO or AZD1775 for 48 h. Scale bar, 20 µm. Data represent three independent experiments. (K) Quantification of the selected ERV and ISGs by qPCR. OV90 cells were treated with DMSO or AZD1775 or didanosine (DDI) or combination for 48 h (three independent experiments). The real-time qPCR data were normalized to β-actin. Data across panels represent mean ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values were determined by unpaired t test (A, B, and D), and ANOVA with Bonferroni post hoc test (G, I, and K).
AZD1775 induces ERV expression in ovarian cancer cells
We next analyzed ERV expression using RepeatMasker in the RNA-seq data of ID8 cells. Remarkably, AZD1775 increased ERVs expression (Fig. 2 B), further verified by a subset of randomly selected ERVs using RT-qPCR in ID8 and OV90 cells (Fig. 2, C and D). Furthermore, staining with a dsRNA-specific antibody (J2) demonstrated WEE1i-induced accumulation of dsRNA in the cytoplasm (Fig. 2 E). To ascertain the on-target effects of WEE1 inhibition with AZD1775, we performed siRNA-mediated WEE1 knockdown in multiple cell lines (Fig. S2 C). Similar to pharmacological inhibition (Fig. 2 F and Fig. S2 D), WEE1 knockdown also induced dsRNA and ISG expression (Fig. S2, E–G). AZD1775 did not further enhance IFN signaling in WEE1 knockdown cells, further suggesting that the effects of AZD1775 are on-target (Fig. S2 G). Moreover, consistent with the report that dsRNA binding to cytosolic sensors induces MAVS protein polymerization (Wu et al., 2013), MAVS protein polymerization was observed after AZD1775 treatment (Fig. 2 G). Taken together, WEE1 is a key regulator of the expression of a group of ERVs in murine and human ovarian cancer models.
dsRNAs are mainly responsible for WEE1 inhibition mediated IFN responses
We next assessed the role of the dsDNA/dsRNA sensor pathway in WEE1 inhibition–induced IFN activation by knocking down different dsDNA and dsRNA sensors (Fig. S2 H). Interestingly, knockdown of cGAS did not blunt, while STING knockdown only modestly decreased, AZD1775-induced IFN signaling in OVCAR4 cells (Fig. 2 H), suggesting the cGAS/STING pathway does not appear to be a critical mediator of WEE1i-induced IFN signaling even in OVCAR4 cells with moderate cGAS and high STING protein levels (Fig. 2 A). In contrast, knockdown of MAVS and DDX58, but not IFIH1 and TLR3, completely reversed AZD1775-induced ISG expression (Fig. 2 H and Fig. S2 I), indicating that DDX58 is a key mediator. Previous studies have shown that DDX58 preferentially binds to short (<300 bp) dsRNAs that have blunt ends, whereas IFIH1 recognizes longer dsRNAs (>1,000 bp) with no end specificity and cooperatively assembles into a filament on the dsRNA (Reikine et al., 2014). Thus, the effects of WEE1i on IFN signaling may be mediated by short dsRNAs rather than long dsRNAs. Consistently, AZD1775 increased almost all immune mediators revealed by the NanoString platform, indicating a broad effect on immune signaling (Fig. 2 I). Unsurprisingly, knockdown of DDX58 or MAVS, but not cGAS or IFIH1, essentially reversed the induction of the suite of immune mediators, while STING knockdown modestly decreased the effects of WEE1 inhibition on immune mediator expression (Fig. 2 I). Consistently, in contrast with dsRNA accumulation, cGAS aggregation was not observed by immunofluorescence staining after AZD1775 treatment (Fig. S2 J). Inhibition of reverse transcription with didanosine did not significantly reduce AZD1775-induced IFN signaling (Fig. S2 K), suggesting that the effects of WEE1 inhibition on the IFN response were triggered by dsRNA from ERV transcription, but not by dsDNA after reverse transcription of ERVs.
Decreased H3K9me3 after WEE1 inhibition contributes to dsRNA stress and promotes IFN activation
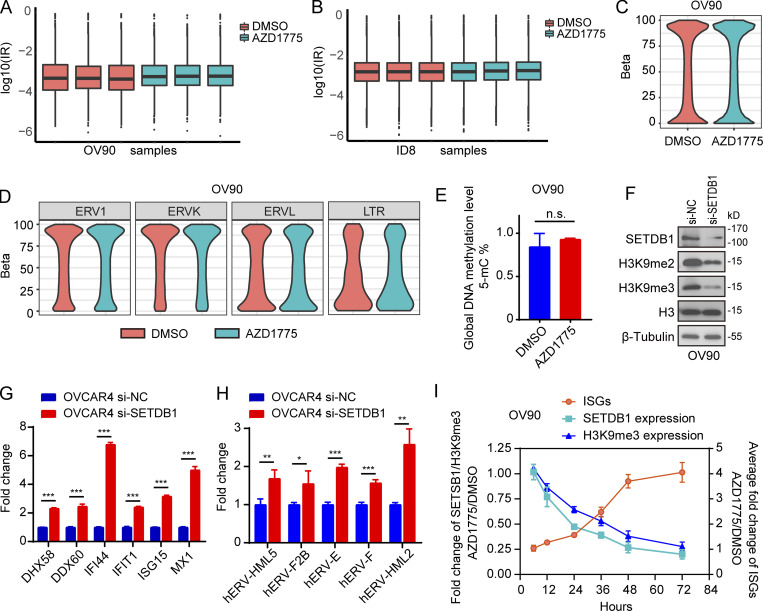
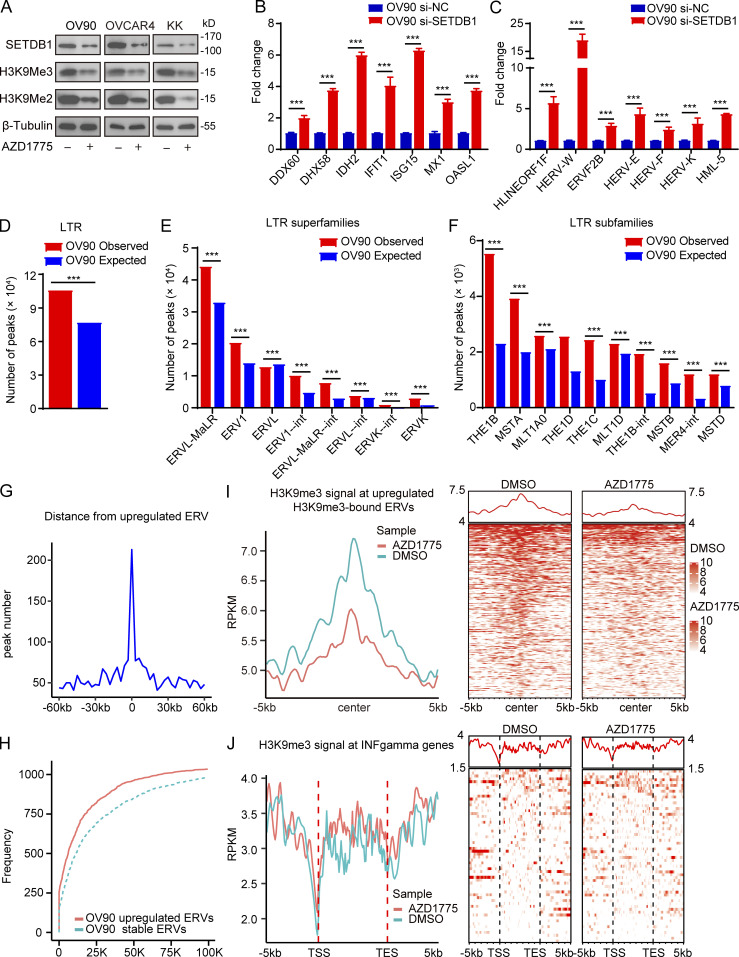
A recent study (Bowling et al., 2021) showed that the accumulation of mis-spliced transcripts results in dsRNA accumulation and IFN response in tumor cells. However, there was no enrichment of spliceosome-associated pathways (Fig. 1 C) and no significant accumulation of intron-retained transcripts (Fig. S3, A and B) after WEE1 inhibition, excluding dsRNA production owing to AZD1775-induced mis-splicing. DNA methylation and histone modifications represent fundamental epigenetic mechanisms regulating ERV expression (Mager and Lorincz, 2017). However, no significant differences in whole-genome DNA (Fig. S3 C) and LTR methylation levels (Fig. S3 D) were observed after WEE1i based on whole-genome bisulfite sequencing (WGBS). Furthermore, the MethylFlash Global DNA Methylation (5-mC) ELISA assay revealed that WEE1i did not detectably alter global methylation levels (Fig. S3 E). Histone H3 lysine 9 trimethylation (H3K9me3) plays a vital role in silencing active ERVs (Elsässer et al., 2015; Sharif et al., 2016). SETDB1, which catalyzes H3K9me3, maintains ERVs in a silent state (Sharif et al., 2016). Interestingly, histone H3 lysine 9 dimethylation (H3K9me2), H3K9me3, and SETDB1 were massively down-regulated by AZD1775 in OV90, OVCAR4, and KK cells (Fig. 3 A). Moreover, knockdown of SETDB1 increased ISG and ERV expression in OV9O (Fig. S3 F; and Fig. 3, B and C) and OVCAR4 cells (Fig. S3, G and H). Interestingly, SETDB1 and H3K9me3 had fallen by almost half after 24 h of AZD1775 treatment compared with control cells, then gradually declined over time with the ISGs massively increasing after 48 h of WEE1 inhibition (Fig. S3 I), indicating SETDB1 and H3K9me3 down-regulation may be the cause of IFN activation. Furthermore, chromatin immunoprecipitation sequencing (ChIP-seq) showed that H3K9me3 peaks were enriched at LTR fragments (Fig. 3, D–F). Integrated analysis of the RNA-seq and ChIP-seq data revealed that up-regulated ERVs were directly bound by H3K9me3 and enriched within 60 kb of an H3K9me3 peak (Fig. 3, G and H). Remarkably, AZD1775 decreased the H3K9me3 peaks at these up-regulated ERVs (Fig. 3 I). To exclude the possibility that H3K9me3 directly bound the up-regulated ISGs, we investigated H3K9me3 peak distributions near up-regulated ISGs. As expected, ChIP-seq analysis failed to identify H3K9me3 occupancy near up-regulated ISGs promoters (Fig. 3 J). Thus IFN/antiviral responses induced by AZD1775 appear to be, at least in part, due to up-regulation of ERVs by H3K9me3 depletion.
Figure S3.
Down-regulation of SETDB1 increased ISG expression, along with ERVs up-regulation. (A and B) Intron-residing retrotransposons (IR) analysis in RNA-seq data of OV90 cells and ID8 cells (n = 3). (C and D) Whole-genome DNA (C) and LTR methylation levels (D) detected by WGBS in OV90 cells after AZD1775 treatment for 48 h (n = 3). (E) The quantification of global DNA methylation levels of OV90 cells after AZD1775 treatment for 48 h detected by MethylFlash Global DNA Methylation (5-mC) ELISA assay (n = 3; three independent experiments). (F) Western blot of indicated proteins in OV90 cells after SETDB1 silencing with siRNA for 72 h. Data represent three independent experiments. (G and H) Quantification of selected ISGs (G) and transcriptions of ERVs (H) by qPCR in OVCAR4 cells transfected with SETDB1 or scramble (si-NC) siRNA for 72 h (three independent experiments). (I) OV90 cells were treated with AZD1775 or DMSO and harvested at indicated hours. SETDB1 and H3K9me3 were detected by Western blot while ISGs (DHX58, DDX60, IFI44, IFIT1, ISG15, and MX1) were detected by qPCR. Fold changes of SETDB1 or H3K9me3 at each time point were calculated by comparing to DMSO after quantification using Image Lab software 6.0.1. The relative fold changes of ISGs (mean fold changes of DHX58, DDX60, IFI44, IFIT1, ISG15, and MX1) at each time point were also calculated by comparing to DMSO-treated cells. Data represent three independent experiments. The qPCR data were normalized to β-actin. Error bars represent SEM between three replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant, as determined by unpaired t test (E, G, and H).
Figure 3.
Decreased H3K9me3 after WEE1 inhibition contributes to dsRNA stress and promotes IFN activation. (A) Western blot of indicated proteins in OV90, OVCAR4, and KK cells treated with AZD1775 for 72 h. Data represent three independent experiments. (B and C) Quantification of ISGs (B) and ERVs (C) by qPCR in OV90 cells treated as indicated for 48 h (three independent experiments). (D–J) CHIP-seq of H3K9me3 analysis in OV90 cells treated with AZD1775 or DMSO (n = 3). (D–F) The observed and expected (10,000 bootstraps) H3K9me3 peaks located at LTR (D), LTR superfamilies (E), and LTR subfamilies (F) in OV90 cells. (G) Line plot showing the distance distribution from all up-regulated ERVs to H3K9me3 peaks in 60-kb flanking windows (3 kb per bin) in OV90 cells. (H) Cumulative frequencies of distance from up-regulated ERVs to the closest peaks. Dotted line represents the cumulative frequencies of distance from a random selection of stable ERVs to their closest peaks in OV90 cells. (I) Line plot of average H3K9me3 signal at up-regulated ERVs in the gene body plus 5 kb upstream of the transcription start site (TSS) and downstream of the transcription end site (TES) and heatmap showing the H3K9me3 signal for each up-regulated ERV in the gene body plus 5 kb upstream of the TSS and 5 kb downstream of TES in OV90 cells. (J) Density distribution (left) and heatmap (right) of H3K9me3 ChIP-seq signal across the up-regulated ISGs transcription start site (TSS, −5/ + 5 kb). ***, P < 0.001, as determined by unpaired t tests (B and C) and one-sided binomial test (D–F).
FOXM1 binds to the SETDB1 promoter regulating ERVs transcription
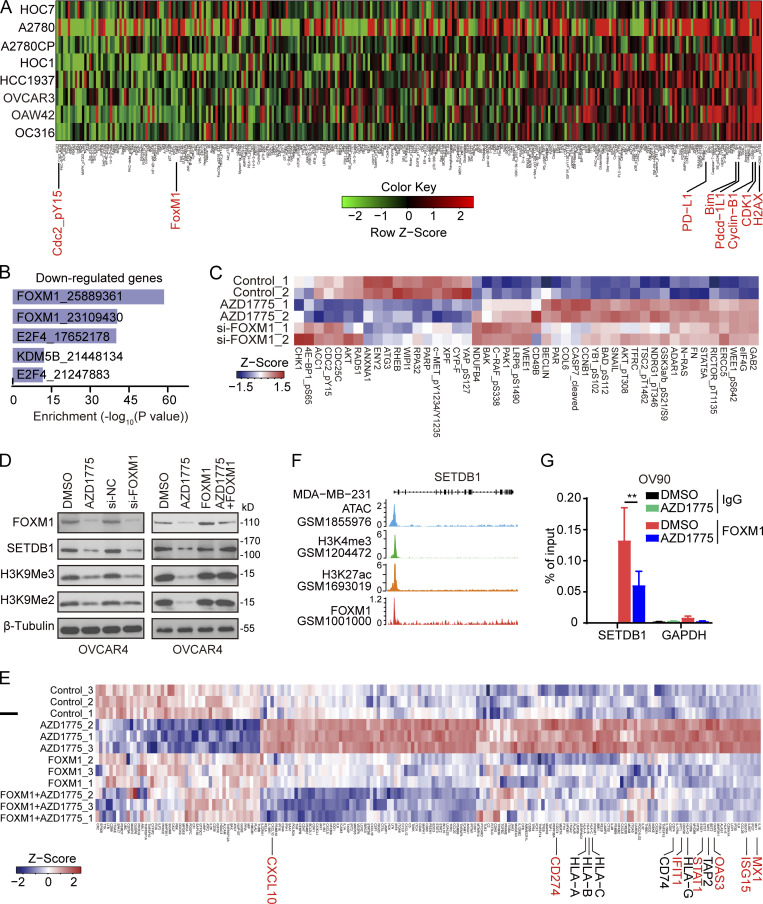
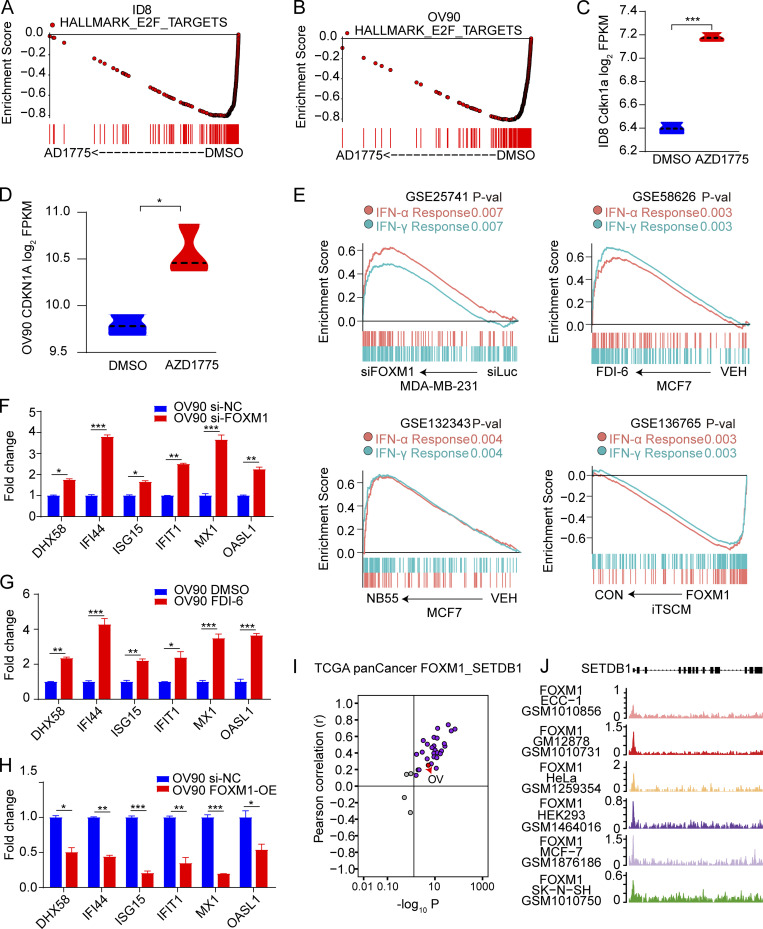
To identify mechanisms underlying the effect of AZD1775 on SETDB1 expression, we performed reverse phase protein array (RPPA) analysis to assess signaling pathway perturbations in response to AZD1775 in eight ovarian cancer cell lines. AZD1775 decreased phosphorylation of its direct target (CDC2_pY15) and increased DNA damage (H2AX-pS140; Fig. 4 A), suggesting these effects were on target and generalizable. Consistent with our previous studies (Fang et al., 2019), AZD1775 dramatically decreased FOXM1 (Fig. 4 A). Moreover, FOXM1 target genes were significantly down-regulated by WEE1i as indicated by Enrich analysis (Chen et al., 2013; Kuleshov et al., 2016; Fig. 4 B). The E2F pathway that regulates FOXM1 expression was also suppressed by WEE1i (Fig. S4, A and B). Furthermore, CDKN1A (encode p21), the major CDK inhibitor whose activation down-regulates FOXM1 activity, was up-regulated after WEE1i (Fig. S4, C and D). Overall, multiple orthogonal approaches indicate that WEE1i decreases FOXM1 activation.
Figure 4.
FOXM1 binds to the SETDB1 promoter regulating ERVs transcription. (A) Protein lysates from indicated cells treated with AZD1775 were analyzed by RPPA (n = 2). Heatmap of RPPA data representing “rank-ordered” changes induced by AZD1775. (B) Enrichr analysis of transcription factors associated with genes down-regulated in AZD1775-treated ID8. (C) RPPA profiling in OVCAR4 cells after AZD1775 or FOXM1 silencing with siRNA (n = 2). (D) Western blot of indicated proteins in OVCAR4 cells treated as indicated. The data represent three independent experiments. (E) Heatmap of differentially expressed genes detected by NanoString immune panel in parental or ectopic FOXM1 expressing OVCAR4 cells with or without AZD1775 treatment (n = 3). (F) Screenshot of assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq), H3K4me3, H3K27me3, and FOXM1 ChIP-seq tracks of SETDB1 in MDA-MB-231 cells. (G) ChIP-qPCR analysis of FOXM1 binding in the SETDB1 promoter region with or without AZD1775 treatment in OV90 cells (n = 3; three independent experiments). Data across panels were mean ± SEM. **, P < 0.01, as determined by unpaired t test.
Figure S4.
Association of FOXM1 with SETDB1 and IFN pathway. (A and B) GSEA analysis of enrichment of HALLMARK E2F PATHWAY in RNA-seq data of ID8 and OV90 cells after treatment with AZD1775 or DMSO for 48 h (n = 3). (C and D) The expression of CDKN1A after treatment with AZD1775 or DMSO for 48 h in RNA-seq data of ID8 and OV90 cells (n = 3). (E) GSEA analysis of IFN pathways after inhibition of FOXM1 by siRNA or inhibitors (FDI-6, NB55), or after FOXM1 overexpression (FOXM1; n = 3). (F–H) Quantification of selected ISGs in OV90 cells treated with FOXM1 inhibition (si-FOXM1 or FDI-6) or overexpression (FOXM1-OE; three independent experiments). (I) Pearson’s correlation of SETDB1 and FOXM1 expression in different cancer types from TCGA. Note that every dot represents one cancer type. (J) Screenshot of FOXM1 ChIP-seq tracks of SETDB1 in ECC-1, GM12878, HeLa, HEK293, MCF7, and SK-N-SH cells. Sample numbers are shown (n = 1 each). The qPCR data were normalized to β-actin. Data across panels represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, as determined by unpaired t test (C, D, and F–H). FPKM, fragments per kilobase of exon model per million mapped fragments; P-val, P value.
Interestingly, FOXM1 down-regulation recapitulated the effects of WEE1i on most target proteins in OVCAR4 cells, except for upstream cell cycle–related proteins (CHK1, CDC2_pY15, and CDC25C; Fig. 4 C), supporting that FOXM1 mediated many of the effects of WEE1i. Moreover, IFN response genes were positively enriched after knockdown of FOXM1 by siRNA or inhibition (FDI-6, NB55), while FOXM1 overexpression decreased these genes (Fig. S4 E). The consistent results were also observed in OV90 cells (Fig. S4, F–H). These results suggested that the activation of IFN pathway by AZD1775 is likely mediated by FOXM1.
We thus determined whether decreases in FOXM1 expression after AZD1775 decreased SETDB1 and its subsequent effect on ERV expression. In the TCGA pan-cancer database, there was a positive correlation between SETDB1 and FOXM1 in most cancer lineages, including ovarian cancer (Fig. S4 I). In addition, AZD1775 and knockdown of FOXM1 remarkably decreased FOXM1, SETDB1, H3K9me2, and H3K9me3 (Fig. 4 D, left) while ectopic FOXM1 reversed the down-regulation of SETDB1, H3K9me2, and H3K9me3 induced by AZD1775 (Fig. 4 D, right), further supporting FOXM1 as the critical player in WEE1i-mediated regulation of SETDB1 as well as H3K9me2 and H3K9me3. Furthermore, NanoString analysis showed that FOXM1 overexpression abrogated the effect of AZD1775 on ISGs and other immune-related genes (Fig. 4 E).
We next explored how FOXM1 regulates SETDB1 expression. The ENCODE database and ChIP-seq data in GSE40767 revealed FOXM1 enrichment at the SETDB1 promoter in MDA-MB-231 (Fig. 4 F). FOXM1 enrichment at the SETDB1 promoter was also observed in ECC-1, GM12878, HEK293, MCF-7, and SK-N-SH cells (Fig. S4 J). Consistent with genome-wide studies, we verified the binding of FOXM1 at the SETDB1 promoter using ChIP-PCR in OV90 cells, which was decreased by AZD1775 treatment (Fig. 4 G). Together, these data support the contention that the effects of AZD1775 on FOXM1 and SETDB1 are causal factors for the induction ERVs and IFN/antiviral responsive genes mediated by WEE1 inhibition.
AZD1775 induces PD-L1 expression through IFN signaling
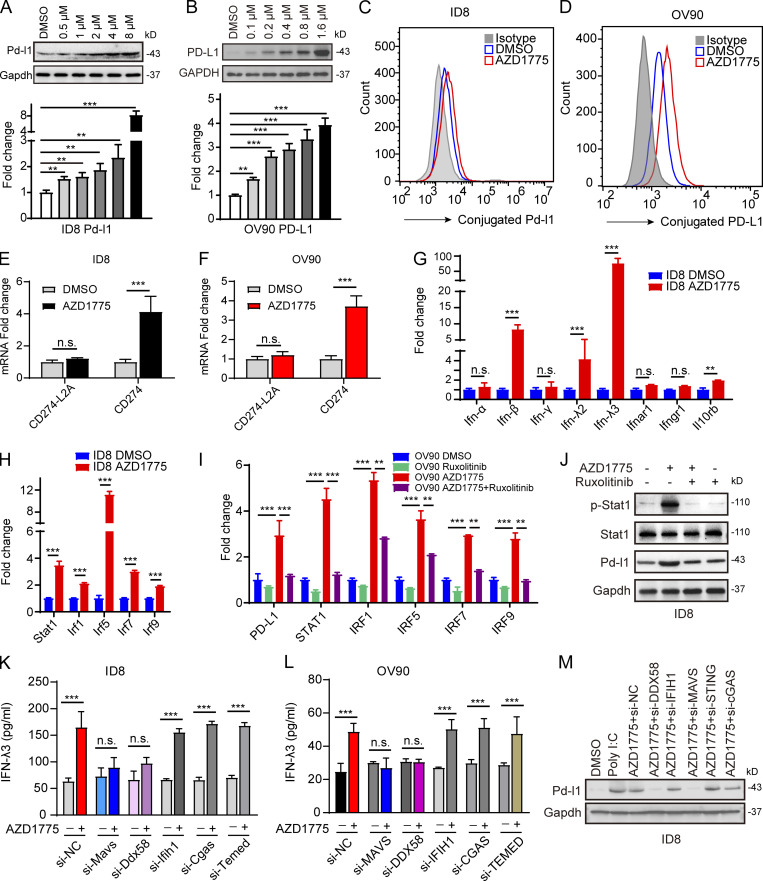
Both the RNA-seq and RPPA data showed that AZD1775 increased PD-L1 in ovarian cancer cells (Fig. 1 B and Fig. 4 A). We verified that AZD1775 induced PD-L1 mRNA and protein expression in a concentration-dependent manner (Fig. 5, A and B). Increased cell membrane expression of PD-L1 was confirmed after AZD1775 treatment by flow cytometry (Fig. 5, C and D). A recent study showed that the soluble PD-L1 isoform could be produced by exaptation of an intronic LINE-2A (L2A) endogenous retroelement in the CD274 gene (Ng et al., 2019). However, AZD1775 showed no effect on the expression of soluble PD-L1 (CD274-L2A) transcripts by qPCR primers specific to the soluble PD-L1 isoform (Fig. 5, E and F), while canonical full-length PD-L1 (CD274) was induced (Fig. 5, E and F). IFN can induce PD-L1 expression, which allows tumor cells to evade immune surveillance (Dorand et al., 2016). We found that IFN-β and IFN-λ, especially IFN-λ3 (Fig. S1 B and Fig. 5 G) and IFN-sensitive transcription factors (STAT1, IRF1, IRF5, IRF7, and IRF9), were elevated in response to AZD1775 (Fig. 5 H). JAKs mediate intracellular signaling after ligand-dependent activation of the IFN receptor (Shuai and Liu, 2003). Notably, ruxolitinib, a JAK inhibitor, mitigated AZD1775-induced increases in IFN-sensitive transcription factors (Fig. 5 I), as well as p-STAT1 and PD-L1 expression (Fig. 5 J), supporting an essential role for IFN activation in PD-L1 up-regulation. Significantly, knockdown of DDX58 and MAVS but not IFIH1 and cGAS blunted AZD1775-induced increases in IFN secretion (Fig. 5, K and L) and PD-L1 expression (Fig. 5 M), suggesting that WEE1 inhibition-induced production of dsRNA mediates IFN activation and PD-L1 up-regulation even in cGAS/STING–deficient cancer cells.
Figure 5.
AZD1775 induces PD-L1 expression through IFN signaling. (A and B) Western blots and qPCR of PD-L1 levels in ID8 and OV90 cells treated with a concentration gradient of AZD1775 for 48 h. Data represent three independent experiments. (C and D) Cytofluorimetric histograms of PD-L1 obtained after AZD1775 for 48 h in ID8 and OV90 (three independent experiments). (E and F) Quantification of two CD274 variants by qPCR in ID8 cells and OV90 treated with AZD1775 or DMSO for 48 h. CD274 encoded the canonical full-length PD-L1 while CD274-L2A is the predominant solute PD-L1–encoding variant (three independent experiments). (G and H) Quantification of IFN/IFNR-related genes (G) and IFN-related transcription factors (H) by qPCR in ID8 cells treated with AZD1775 or DMSO for 48 h (three independent experiments). (I) Quantification of selected genes by qPCR in OV90 cells treated with AZD1775, JAK inhibitor (ruxolitinib, 5 µM), or combination for 48 h (three independent experiments). (J) Western blots for STAT1 activation and PD-L1 expression in ID8 cells treated with AZD1775, ruxolitinib, or combination. Data represent three independent experiments. (K and L) ELISA detection of IFN-λ3 levels treated as indicated in ID8 and OV90 cells (three independent experiments). (M) Western blots for PD-L1 treated as indicated. Data represent three independent experiments. The qPCR data were normalized to β-actin. Data across panels represent mean ± SEM. **, P < 0.01; ***, P < 0.001, n.s., not significant as determined by unpaired t test (E–H, K, and L), and ANOVA with Bonferroni post hoc test (A, B, and I).
AZD1775 combined with ICB represses tumor growth in a CD8+ T cell–dependent manner
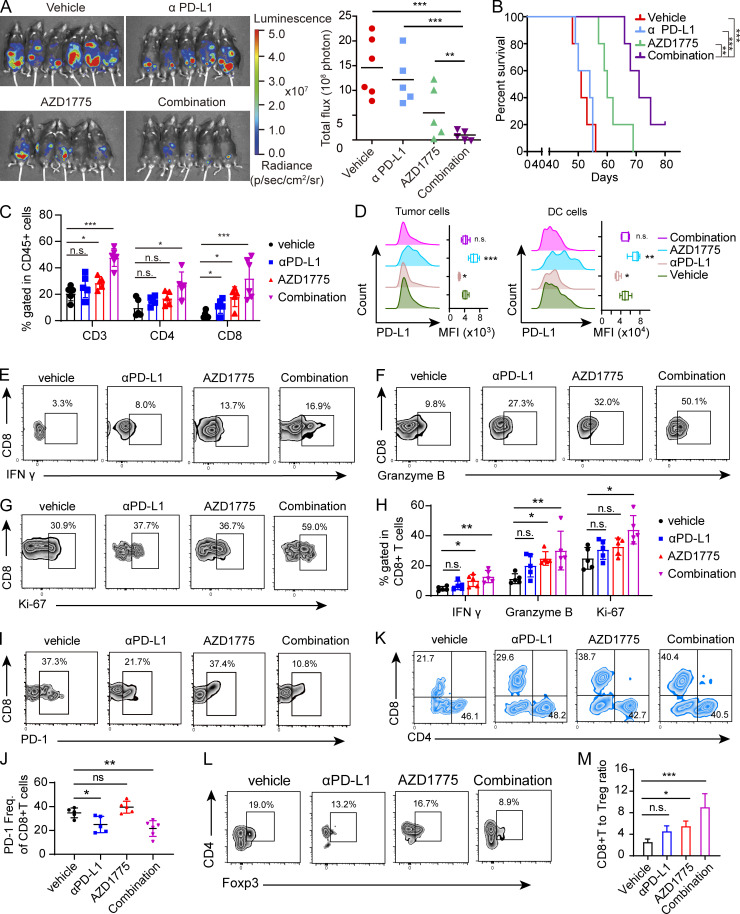
PD-L1 expression correlates with clinical response to anti–PD-L1 and anti–PD-1 therapy in ovarian cancer (Disis et al., 2019; Hamanishi et al., 2015). To determine the effects of combined AZD1775 and αPD-L1 on anti-tumor immunity in vivo, we used the ID8 (STING-deficient) syngeneic ovarian cancer mouse model. While anti–PD-L1 monotherapy did not significantly decrease or AZD1775 alone modestly decreased ID8 tumor growth, the combination of AZD1775 and anti–PD-L1 reduced tumor growth markedly (Fig. 6 A) and prolonged survival time (Fig. 6 B) without notable side effects (Fig. S5 A). Consistently, we observed increased immune cell infiltration characterized by positive CD45 cells (Fig. S5 B) and higher tumor-infiltrating CD3+ T cells in ID8 tumors composed predominantly of CD8+ T cells (Fig. 6 C). Of note, AZD1775 did not affect CD8+ T cell proliferation in vitro (Fig. S5 C), consistent with a lack of deleterious effects on CD8+ T cells. Furthermore, we observed increased PD-L1 on ID8 tumor cells by flow cytometry after AZD1775, while PD-L1 on DC cells was also induced (Fig. 6 D). Importantly, the proportion of cytotoxic CD8+ IFN-γ+ cells and other functional parameters, like granzyme B and Ki-67, significantly increased after AZD1775 monotherapy, further augmented by combination therapy (Fig. 6, E–H). Additionally, expression of the inhibitory PD-1 on CD8+ T cells was reduced after αPD-L1 treatment and combination treatment (Fig. 6, I and J). The T reg cells were moderately decreased after combination therapy, and the CD8/T reg cell ratio was increased by AZD1775 or combination treatments (Fig. 6, K–M).
Figure 6.
AZD1775 combined with ICB represses tumor growth in a CD8+ T cell–dependent manner. (A) Representative images and quantification of total flux of luminescence of ID8 tumors in C57BL/6 mice after treatment with vehicle, AZD1775, αPD-L1, or combination of AZD1775 and αPD-L1 (n = 5 or 6 each, two independent experiments). (B) Kaplan–Meier survival curves of C57BL/6 mice with ID8 tumors treated as described in A (n = 5 or 6 each, two independent experiments). (C) The percentage of CD3+, CD4+, and CD8+ T cells in CD45+ cells in tumor tissues from different treated groups were quantified via flow cytometry (n = 5 or 6 each). (D) PD-L1 expression determined by flow cytometry on tumor cells (left) and DC cells (right) in tumors treated as indicated (n = 5 or 6 each). (E–H) Representative flow cytometry plots and quantification of IFN-γ, granzyme B, and Ki-67–positive T cells (gated on CD8+ cells) in ID8 tumors treated as indicated (n = 5 or 6 each). (I and J) Representative flow cytometry plots (I) and quantification (J) of PD-1–positive T cells (gated on CD8+ cells) in ID8 tumors treated as indicated (n = 5 or 6 each). (K) Representative flow cytometry plots (n = 5 or 6 each) of CD4+ and CD8+ T cells (gated on CD3+ T cells) in ID8 tumors treated as indicated. (L) Representative flow cytometry plots of Foxp3+ T reg cells (gated on CD4+ cells) in ID8 tumors treated as indicated (n = 5 or 6 each). (M) The ratio of CD8+T cells to T reg cells is shown (n = 5 or 6 each). The data in C–M are from one representative experiment of two performed with similar results. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values were determined by log-rank test (B), and ANOVA with Bonferroni post hoc test (A, C, D, H, J, and M). MFI, mean fluorescent intensity.
Figure S5.
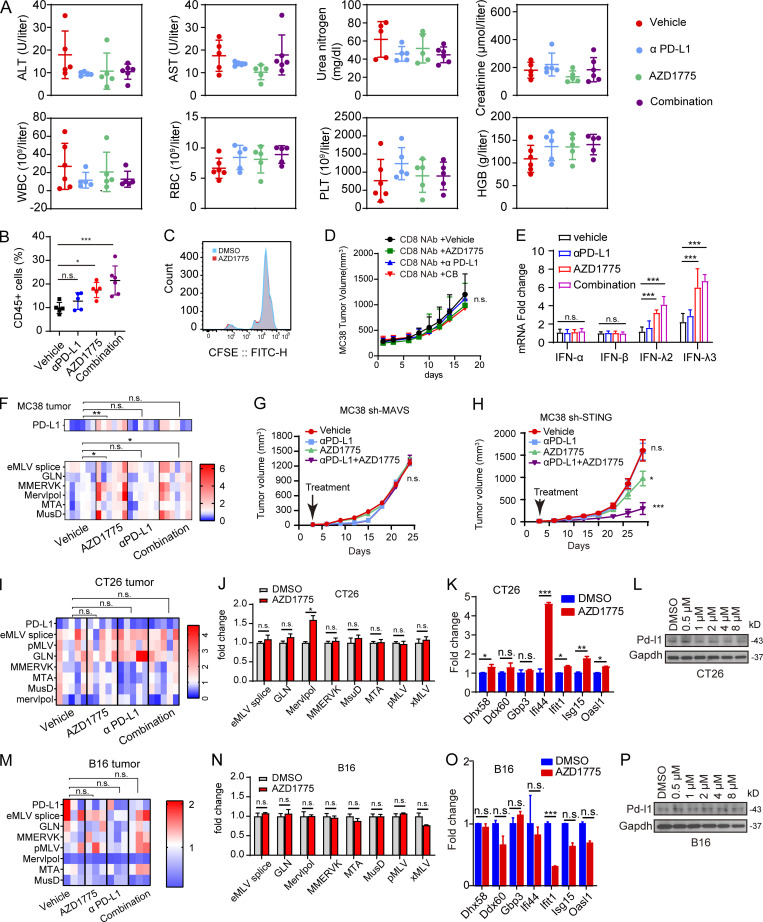
Efficacy of AZD1775, αPD-L1, and combination therapy in ID8, MC38, CT26, and B16 models. (A) Plot of alanine aminotransferase (ALT), aspartate aminotransferase (AST), urea nitrogen, creatinine, white blood cell (WBC), red blood cell (RBC), platelet (PLT) count, and hemoglobin (HGB) levels in mice with ID8 tumors treated with indicated treatments (n = 5 or 6). Data are representative of two independent experiments. (B) Percentage of CD45-positive cells in freshly isolated cells of tumor tissues from ID8 mice treated with indicated treatments analyzed by flow cytometry (n = 5 or 6; two independent experiments). (C) CD8+ T cells were obtained from human blood peripheral blood mononuclear cells using magnetic beads and labeled with CFSE. Then, CD8+ T cells were treated with DMSO or AZD1775 for 4 d. Proliferation activity was detected by flow cytometry with FITC. The data represent three independent experiments. (D) C57BL/6 mice received 200 μg anti-CD8α monoclonal antibody 3 d before MC38 challenge and consolidated on the 0, 3rd, 8th, 14th, 20th, and 26th day after MC38 challenge. These mice were treated from the third day after challenge with indicated treatments. The tumor growth was recorded every 3 d. (n = 5–7). (E) Quantification of IFNs in MC38 tumors treated with vehicle, AZD1775, αPD-L1, or combination (n = 5–7). (F) Heatmap of normalized expression of PD-L1 and selected ERVs versus β-actin in MC38 tumors treated with indicated treatments (n = 5 or 6). (G and H) Tumor growth curves of MAVS and STING-defective MC38 treated with indicated treatments (n = 5). (I and M) Heatmap of normalized expression of PD-L1 and selected ERVs versus β-actin in CT26 (n = 5) and B16 (n = 3) tumors treated with indicated treatments. (J and O) Quantification of selected ERVs (J and N) and ISGs (K and O) expression by qPCR in CT26 and B16 cells treated with AZD1775 or DMSO cells for 48 h in vitro (three independent experiments). (L and P) Western blot of PD-L1 expression in CT26 and B16 cells treated with a series of concentration of AZD1775 or DMSO. Three independent experiments. The qPCR data were normalized to β-actin. Data across panels represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values were determined by unpaired t test (J, K, N, and O) and ANOVA with Bonferroni post hoc test (B, D–I, and M).
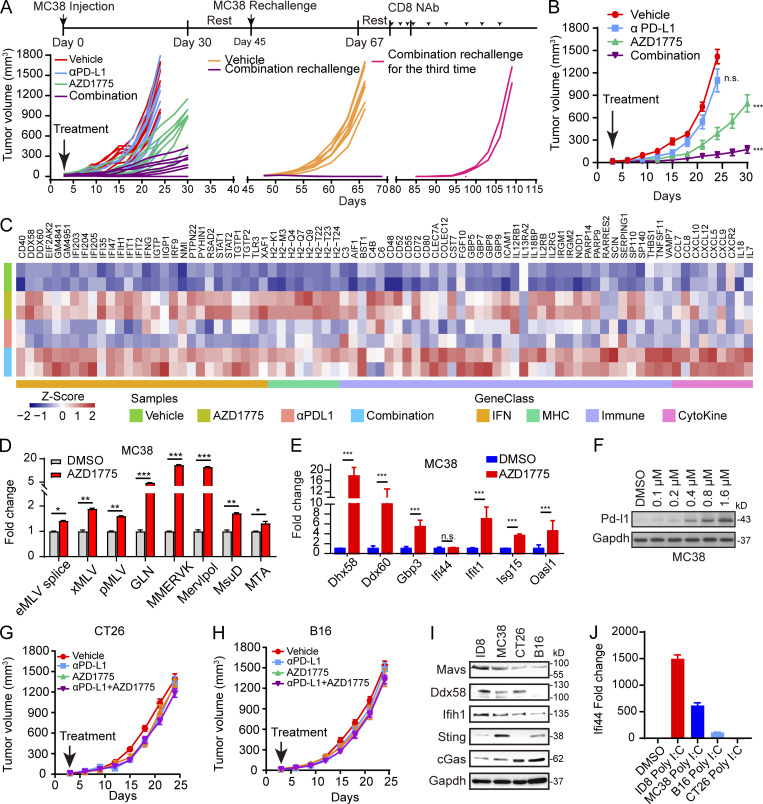
To assess the generalizability, we evaluated combinational therapy in MC38, CT26, and B16-bearing immunocompetent syngeneic mouse models. AZD1775 combined with αPD-L1 resulted in marked tumor growth delay in MC38-bearing mice (Fig. 7, A and B). Notably, the combination treatment resulted in complete tumor regression (CR) that persisted >50 d after treatment cessation in three of nine treated mice, while no CRs were achieved in the monotherapy group (Fig. 7 A). Moreover, combination therapy rendered mice resistant to a rechallenge with a subsequent injection of MC38 cells 2 wk after stopping therapy (Fig. 7 A). The observed tumor-suppressive effects were CD8+ T cell–dependent because the therapeutic benefit of the combination treatment was abrogated when MC38 cells were injected for the third time after antibody-mediated depletion of CD8+ T cells (Fig. 7 A). Furthermore, we depleted CD8+ T cells before and during the MC38 tumor challenge and treatment. Again, the anti-tumor effects of AZD1775 and combination therapy were eliminated by anti-CD8 treatment (Fig. S5 D). Notably, we verified that type III IFN, ERVs, and PD-L1 were up-regulated in MC38 tumors after AZD1775 and combination treatment in vivo (Fig. S5, E and F), consistent with the effects in vitro. Furthermore, RNA-seq analysis of MC38 tumors revealed that AZD1775 up-regulated the expression of genes related to IFN signaling, cytokine, and antigen processing and presentation (Fig. 7 C). At the same time, combination therapy showed more intense up-regulation of these genes, especially T cell chemokines, further supporting a role of T cells in the efficacy of the combination therapy (Fig. 7 C), which was also consistent with our in vitro results that AZD1775 increased ERVs, ISGs, and PD-L1 expression in MC38 cells (Fig. 7, D–F). Notably, the efficacy of WEE1i monotherapy and the synergistic anti-tumor effects of WEE1i and αPD-L1 were abrogated in MAVS-depleting MC38 tumors (Fig. S5 G) but not in STING-deficiency MC38 tumors (Fig. S5 H), further supporting the critical role of dsRNA sensor in response to WEE1i and the combination treatment in vivo. Compared with MC38-bearing mice, the overall anti-tumor effect was weaker in CT26- and B16-bearing mice (Fig. 7, G and H). In contrast with MC38, AZD1775 or combination treatment did not induce ERVs, ISGs, or PD-L1 in CT26 and B16 cells both in vivo and in vitro (Fig. S5, I–P). Interestingly, the basal expression levels of the dsRNA sensors DDX58 and MAVS were lower in CT26 and B16 cells than in MC38 cells (Fig. 7 I), which could at least partially explain the lack of IFN activation after WEE1 inhibition or poly I:C stimulation in CT26 and B16 cells (Fig. 7 J).
Figure 7.
AZD1775 combined with ICB assessment in mouse models. (A) Spider plot of MC38 tumor growth treated as indicated. Each line represents one mouse. (B) Tumor growth curves of MC38 tumors after treatment as indicated (n = 5–9). (C) Heatmap of differential expression of immune genes in MC38 tumor tissues from indicated groups (n = 2 each). (D and E) Quantification of selected ERV (D) and ISG (E) expression by qPCR in MC38 cells treated with AZD1775 or DMSO cells in vitro for 48 h (three independent experiments). (F) Western blot of PD-L1 expression in MC38 cells treated with a series of concentration of AZD1775 or DMSO for 72 h in vitro. Data represent three independent experiments. (G and H) Tumor growth curves of CT26 and B16 tumors after indicated treatment (n = 5 or 6). (I) Western blot of basic expression of dsDNA and dsRNA sensor protein in mouse cells. Data represent three independent experiments. (J) Quantification of IFI44 expression by qPCR in mouse cells treated with poly I:C (5 µg/ml) or DMSO cells for 48 h. The qPCR data were normalized to ACTIN. Data across panels represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. P values were determined by unpaired t test (D and E), and ANOVA with Bonferroni post hoc test (B, G, and H).
FOXM1, SETDB1, and IFN pathway activation may be informative biomarkers for response to AZD1775
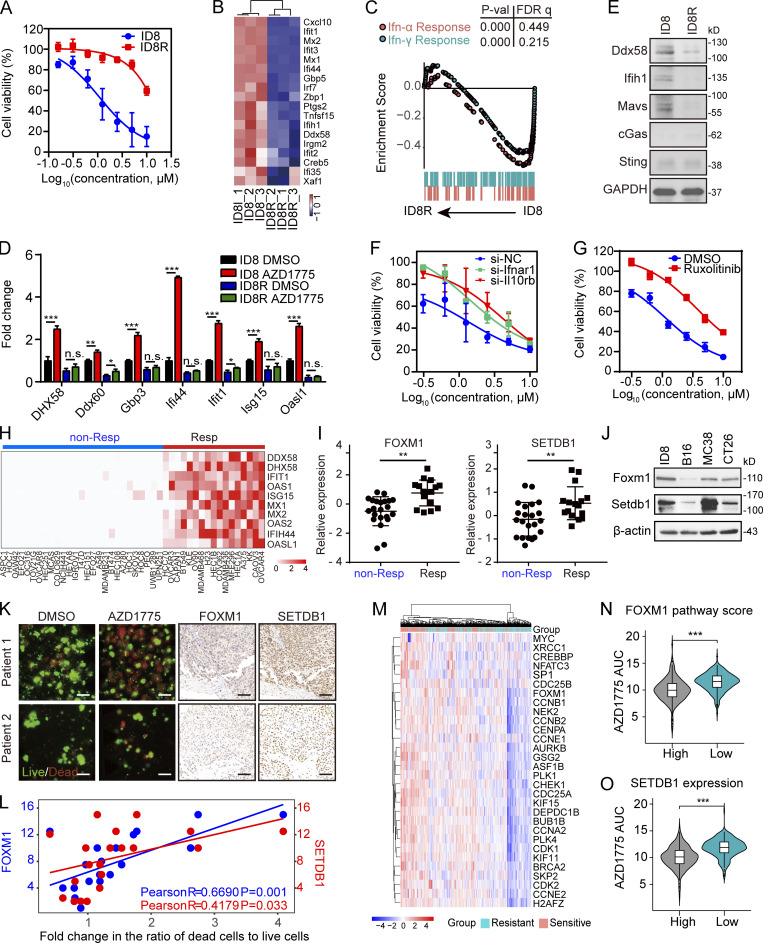
To further explore the molecular underpinning of responses to AZD1775, we developed AZD1775-resistant cells by culturing AZD1775-sensitive cells (ID8) in the continued presence of AZD1775 for 6 mo, at which time drug-resistant cells emerged (ID8R; Fig. 8 A). Interestingly, RNA-seq analysis demonstrated lower basal ISG expression in ID8R cells compared with ID8 (Fig. 8, B and C). Furthermore, AZD1775-induced ISG expression was also blunted in ID8R (Fig. 8 D). Interestingly, we found DDX58 and MAVS were massively down-regulated in ID8R cells (Fig. 8 E). Moreover, blocking the IFN pathway by IFN receptor (IFNR) knockdown or JAK inhibition increased the resistance of parental ID8 cells to AZD1775 (Fig. 8, F and G). In summary, the degree of IFN pathway activation induced by AZD1775 may reflect the ability of AZD1775 to induce cell death.
Figure 8.
FOXM1, SETDB1, and IFN pathway activity may be informative biomarkers for response to AZD1775. (A) Cell viability curves of parental or AZD1775-resistant ID8 (ID8R) cells treated with AZD1775 for 72 h (n = 3; three independent experiments). (B) Heatmap of differentially expressed ISGs in parental and ID8R cells. (C) GSEA of HALLMARK-IFN pathways in ID8R cells. (D) Quantification of selected ISGs by qPCR in ID8 and ID8R cells treated with AZD1775 or DMSO for 48 h (three independent experiments). The qPCR data were normalized to β-actin. Data across panels represent mean ± SEM. (E) Western blot of dsDNA and dsRNA sensors in ID8 and ID8R cells. Data represent three independent experiments. (F and G) Cell viability curves of ID8R cells to AZD1775 after transfection of IFNR siRNA or treated with 5 µM ruxolitinib for 48 h (n = 3; three independent experiments). (H) Heatmap of relative expression of selected ISGs (log2[AZD1775/DMSO]) by qPCR in 44 cancer cells (three independent experiments). Cell lines were classified as nonresponse (non-Resp) and response group (Resp) according to the mean fold-changes of ISGs (cutoff = 2). (I) The FOXM1 and SETDB1 levels in nonresponse and response cells are shown (n = 44). (J) Western blot of basic FOXM1 and SETDB1 expression in ID8, CT26, MC38, and B16 cells. Data represent three independent experiments. (K) Representative immunofluorescence images of AO/PI staining (left) in 3D microfluidic ex vivo culture of PDOS after treatment with AZD1775 or DMSO for 24 h, and IHC staining of FOXM1 and SETDB1 (right) in tumor tissues from the same patients (n = 20). (L) Scatter plot of the correlation between the ratio of live to dead cells measured by AO/PI staining and the IHC score of FOXM1 and SETDB1 in 20 ovarian cancer tissues. AO represents live nucleated cells (green). PI represents dead nucleated cells (red). Scale bar, 50 µm. (M) Unsupervised clustering of AZD1775-sensitive (n = 247) or -resistant (n = 247) cancer cells with FOXM1 pathway genes. (N and O) Comparison of AZD1775 sensitivity (AZD1775 AUC: higher AUC means cells were more resistant to AZD1775) according to FOXM1 pathway score (N) or SETDB1 expression (O). High (n = 247) and low (n = 247). *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant, as determined by unpaired t test. P-val, P value; FDR q, false discovery rate p-value.
To explore whether FOXM1 and SETDB1 could also predict IFN pathway activation in response to AZD1775, we assessed ISGs after AZD1775 on 44 cancer cell lines. As expected, higher FOXM1 and SETDB1 were associated with IFN pathway activation in response to AZD1775 (Fig. 8, H and I). ID8 and MC38, which have relatively high FOXM1 and SETDB1, were more responsive to AZD1775 than CT26 and B16 (Fig. 8 J). Then we used a three-dimensional (3D) microfluidic ex vivo culture of patient-derived organotypic tumor spheroids (PDOSs) to assess the therapeutic effect of AZD1775. As expected, patient samples with higher expression of FOXM1 and SETDB1 assessed by immunohistochemistry (IHC) showed a better response to AZD1775 (Fig. 8, K and L). Furthermore, analyses of the associations between gene expression signatures and AZD1775 sensitivity in the Cancer Therapeutics Response Portal (CTRP) showed that cells with a high FOXM1 pathway score (Fig. 8, M and N) or SETDB1 expression (Fig. 8 O) were more sensitive to AZD1775. These findings demonstrated that baseline levels of FOXM1/SETDB1 and induced IFN pathway activation could predict response to AZD1775.
Discussion
Taken together, our data demonstrate that targeting WEE1 up-regulates ERVs, IFN activation, and PD-L1 in tumors. This response may be critical in cGAS/STING-deficient cancer cells representing a common phenotype across multiple cancer lineages. The ability of WEE1i to induce ERV expression is due to interrupting H3K9Me3 repression of ERVs expression. Consequently, WEE1 inhibition increases anti-tumor T cell infiltration and sensitizes tumors to anti–PD-L1. Pre-treatment levels of FOXM1, SETDB1, and induced ISGs signature predicted response of tumor cells and patient PDOS to WEE1 inhibitor combinations, providing potential selection biomarkers for human trials.
WEE1 controls genomic stability by regulating replication origin firing or stabilization of replication forks (Domínguez-Kelly et al., 2011). WEE1 inhibition leads to DNA double-strand breaks and single-stranded lesions, stimulating immune responses through the dsDNA-dependent IFN pathway (Paludan and Bowie, 2013). However, cGAS/STING levels are deficient in most ovarian cancer cell lines, consistent with previous reports of cGAS/STING pathway defects in ovarian cancer (de Queiroz et al., 2019). Indeed, we showed that even though cGAS/STING is functional in a minority of cell lines (such as OVCAR4), STING inhibition only modestly decreased AZD1775-induced ISGs, supporting a critical role of the dsRNA pathway in WEE1i-induced IFN signaling and immune activation. We also demonstrated that WEE1i-induced ISGs require DDX58/MAVS, which was largely intact in most ovarian cancer cell lines. Together, these observations indicate that dsRNA signaling plays a critical role in AZD1775-mediated activation of the IFN signaling pathway, especially in cGAS/STING-deficient cancer cells. It is important that AZD1775 could still activate IFN signaling pathways through dsRNA and thus enhance the effect of αPD-L1 immunotherapy even in tumors such as ovarian cancers where STING signaling defects are common.
FOXM1 activity is generally up-regulated by CDKs, including CDK1 and CDK2 (CDK1/2), via phosphorylation (Liao et al., 2018). Since WEE1 is a negative regulator of CDK1/2, one would expect that WEE1 inhibition would up-regulate FOXM1 activity through relieving inhibition of CDK1/2. However, we found that AZD1775 remarkably decreased FOXM1 after 48–72 h of treatment in ovarian cancer cells, consistent with our previous publication (Fang et al., 2019). Our studies focused on the relatively long-term effects of the WEE1 inhibitor. Interestingly, a recent paper (Diab et al., 2020) showed that WEE1i caused an initial increase in CDK activity that lasted up to 8 h. However, this transient increase in CDK activity was followed by prolonged CDK inhibition throughout the remainder of WEE1i treatment and recovery in human papillomavirus (HPV)–negative head and neck squamous cell carcinoma (HNSCC) cells. Thus, rather than simply functioning as a CDK activator, WEE1i paradoxically suppressed CDK activity after prolonged treatment in HPV negative HNSCC cells, similar to what we observed in ovarian cancer cells. Mechanistically, the decreased CDK1/2 activity during and after prolonged WEE1i treatment (24 h) in HPV-negative HNSCC cells was associated with increased p21 (CDK inhibitor) binding to CDK1 and CDK2. Remarkably, we also found that WEE1 inhibition up-regulated p21 levels were up-regulated in our ovarian cancer models by RNA-seq. Moreover, the E2F pathway, a key regulator for FOXM1 expression, was suppressed after WEE1 inhibition by GSEA analysis. Therefore, relatively long-term effects and suppression of the E2F pathway may contribute to the down-regulation of FOXM1 by WEE1 inhibition in our studies.
ICB using single-agent antibodies has demonstrated modest activity in epithelial ovarian cancer, with a median response rate of 10–15% (Disis et al., 2016; Hamanishi et al., 2015; Infante et al., 2016; Matulonis et al., 2018). We have observed that targeting WEE1i elevated PD-L1 and enhanced CD8+ T cell recruitment in vivo, suggesting that these cancers could potentially benefit from the combination of AZD1775 and immunotherapy. Indeed, the combination of AZD1775 and PD-L1 blockade showed significant efficacy in controlling ID8 and MC38 tumors. High ISGs expression overlaps with the immune reactive subtype of TCGA ovarian cancer (Chiappinelli et al., 2015) and correlates with long-term benefit in patients treated with anti–CTLA-4 therapy in melanoma patients (Snyder et al., 2014). Similarly, ISGs were significantly increased after AZD1775 treatment in ID8 and MC38 cells. In contrast, in CT26 cells, only part of the ISG repertoire increased, and in B16 cells, there was almost no ISG increase, which is consistent with decreased responses to the combination of AZD1775 and anti–PD-L1 immunotherapy, suggesting that ISG induction may be an informative biomarker.
In summary, in addition to dsDNA/cGAS/STING responses, there is an alternative pathway for WEE1i-induced IFN production through the dsRNA/RIG-1/MAVS cascade. Given that STING responses are compromised in a significant number of tumors, and particularly ovarian cancer, this is of potential importance to the clinical implementation of WEE1 inhibitors. As several WEE1 inhibitors are now under clinical evaluation, there is an urgent need to understand better the potential mechanisms and combinations that could benefit patients. Further, our study has identified biomarkers that could potentially be used to enrich the population of patients likely to benefit from combination therapy with WEE1i and ICB. Together, these studies warrant exploration of the combination in clinical trials.
Materials and methods
Cell lines
All human cell lines were fingerprinted by short tandem repeat assays and verified to be free of mycoplasma contamination before use. HCT116-Dual cells were obtained from InvivoGen. A375, A549, ASPC1, B16, CAPAN1, COLO829, CT26, ES2, MC38, OV90, OVCAR3, SKOV3, TOV-112D, and TOV-21G cells were from the American Type Culture Collection. COV362 cells were from The European Collection of Authenticated Cell Cultures. A2780, BT474, BT549, CAOV3, EFO21, EFO27, H23, HCC70, HEC108, HEC151, HEC1A, HEC1B, HEC251, HEYA8, HOC1, HOC7, HOC8, IGROV1, KK, KLE, MC38, MCAS, MDA-MB-231, MDA-MB-436, MDA-MB-468, MFE296, NCIH441, OAW42, OC316, OVCAR4, OVCAR8, PEO, T47D, UPN251, and UWB1.289 cells were obtained from the M.D. Anderson Cancer Center characterized Cell line Core. The murine ovarian cancer cell line ID8 was a gift from K. Roby (University of Kansas, Lawrence, KS), derived from spontaneous malignant transformation of C57BL/6 mouse ovarian surface epithelium cells. Other cells were from the M.D. Anderson Cancer Center characterized Cell line Core. All cell lines were incubated at 37°C in an incubator with 5% CO2, and antibiotics (penicillin/streptomycin) were added to all cell culture media.
HCT116-Dual cells, ES2, HEC1A, HEC1B, and SKOV3 were cultured with McCoy’s 5a medium plus 10% FBS. A2780, BT549, HOC1, HOC7, HOC8, MDA-MB-231, OAW42, OC316, OV2008, OVCAR4, PEO, and T47D were cultured with RPMI-1640 medium plus 5% FBS. A375, B16, COV362, ID8, MC38, and MDA-MB-436 cells were cultured with DMEM medium plus 10% FBS. ASPC1, BT474, COLO829, CT26, EFO27, H23, HCC70, HEC108, HEC151, HEC251, HEYA8, IGROV1, KK, KLE, MCAS, NCIH441, and OVCAR8 cells were cultured with RPMI-1640 medium plus 10% FBS. UPN251 cells were cultured with RPMI-1640 plus 10% FBS and insulin. CAOV3 and EFO21 were cultured with RPMI-1640 plus 20% FBS. MDA-MB-468 cells were cultured with DMEM/F12 plus 10% FBS. A549 cells were cultured with F-12K plus 10% FBS. CAPAN1 cells were cultured with IMDM plus 20% FBS. OV90, TOV-112D, and TOV-21G cells were cultured with 199/MCDB105 plus 15% FBS. MFE296 and OVCAR3 cells were cultured with RPMI-1640 plus 20% FBS and insulin. UWB1.289 cells were cultured with 1:1 RPMI-1640 and 50% mammary epithelial growth medium plus 3% FBS. The cells were treated with inhibitors at 500 nM except where specifically indicated otherwise.
Animal studies
All animal experiments had been approved by the Animal Experiment Ethics Committee of Tongji Hospital. 6–8-wk-old female C57BL/6 mice were purchased from Beijing Hua Fu Kang Biotechnology Co., Ltd. (HFK) and were used for ID8, MC38, and B16 xenografts. Female NOD/SCID mice (6–8 wk old) were purchased from HFK and were used for ID8 xenografts. 6–8-wk-old female BALB/c mice were purchased from HFK and were used for CT26 xenografts. All mice were housed in the Laboratory Animal Care Center of Tongji Hospital. All animal procedures and experiments were performed under the guidelines of the Laboratory Animal Care Center of Tongji Hospital.
Clinical specimens
The use of ovarian cancer samples was approved by the Ethics or Institutional Review Board of Tongji Hospital, following the Declaration of Helsinki. Informed consent was obtained from all subjects. 20 serous ovarian cancer samples were collected immediately after surgical resection and digested for 3D microfluidic ex vivo culture. Samples were also routinely fixed in 10% formalin, dehydrated, incubated in xylene, and embedded in paraffin for IHC staining.
IFN pathway response detection by HCT116-Dual assay
HCT116-Dual cells were treated with DMSO or inhibitors (500 nM). After a 48-h incubation, NF-κB activation was determined using QUANTI-Blue (InvivoGen; cat #rep-qbs), a secreted alkaline phosphatase detection reagent, by reading the OD at 655 nm. IRF activation was determined by measuring the relative light units in a luminometer using QUANTI-Luc (InvivoGen; cat #rep-qlc1).
The inhibitors of ABT-263/cat #S1001, AZD2014/cat #S2783, ABT737/cat #S1002, irinotecan HCl trihydrate/cat #S2217, RO-3306/cat #S7747, AZD5363/cat #S8019, BKM120/cat #S2247, azacitidine/cat #S1782, GSK343/cat #S7164, crizotinib/cat #S1068, EPZ5676/cat #S7062, GSK2118436/cat #S2807, AZD8055/cat #S1555, BMN673/cat #S7048, GSKJ4 HCl/cat #S7070, JQ1/cat #S7110, AZD7762/cat #S1532, AZD6244/cat #S1008, LY3214996/cat #S8534, GSK1120212/cat #S2673, AZD1775/cat #S1525, and milciclib/cat #S2751 were purchased from Selleck Chemicals. The milciclib/cat #HY-10424, gefitinib/cat #HY-50895, UNC1215/cat #HY-15649, cisplatin/cat #HY-17394, vorinostat/cat #HY-10221, and A-485/cat #HY-10745 were from MedChemExpress.
RNA-seq
Purified total RNA from ID8 and OV90 cells cultured in vitro or MC38 cells isolated from tumor-bearing mice was extracted by RNA extraction kit (TIANGEN, cat #DP419) and analyzed by an Agilent Bioanalyzer to assess RNA integrity. Total RNA from ID8 cells was used to generate poly(A)+ RNA for construction of an RNA-seq library by Annoroad Gene Technology Co., Ltd. RNA-seq was performed on the Illumina HiSeq 2000 platform. RNA from OV90 cells and MC38 tumor tissues were used to generate ribosomal RNA–depleted RNA or poly(A)+ RNA to construct RNA-seq libraries by the Beijing Genomics Institute. RNA-seq was performed on a BGISEQ-500 platform.
IHC staining
Tissues were fixed overnight in 4% paraformaldehyde and embedded in paraffin. All IHC staining was performed on 4-µm sections. After deparaffinization, rehydration, antigen unmasking, and endogenous peroxidase blocking, sections were blocked in 5% BSA with 0.1% Triton X-100 in PBS. Sections were incubated overnight at 4°C with primary antibodies. The details of antibodies are as follows: CD3 antibody (Abcam; cat #ab5690; 1:200); CD4 antibody (Abcam; cat #ab183685; 1:200); CD8 antibody (Cell Signaling Technology; cat #98941; 1:200); Foxp3 antibody (Cell Signaling Technology; cat #12653; 1:200); SETDB1 antibody (Abcam; cat #ab12317; 1:200); and FOXM1 antibody (Abcam; cat #ab232649; 1:200). Tissue sections were then stained using an Immunohistochemistry kit (Servicebio; cat #G1215; cat #G1215-200T; cat #G1216-200T). Tissues were observed under a microscope and photographed after being counterstained with hematoxylin, dried, and mounted. For assessment of CD3+, CD4+, CD8+, and Foxp3+ T cell infiltration, positively stained cells were counted from five high-power random fields of each sample, and the numbers averaged for each field. The IHC scores for SETDB1 and FOXM1 staining are assigned using a semi-quantitative five-category grading system as previously described (Sun et al., 2018).
RNA extraction and RT-qPCR
According to the manufacturer’s protocol, total RNA was isolated from cultured cells using a total RNA extraction kit (TIANGEN; cat #DP419). The RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific; cat #K1622) was used to synthesize cDNA on an RT-PCR System (Bio-Rad). RT-qPCR was performed on a CFX Connect quantitative real-time PCR System (Bio-Rad) using SYBR Green Master Mix (Vazyme; cat #R223-01). Relative mRNA expression was determined by ΔΔCt method and normalized by housekeeping gene β-actin. All primers were synthesized by TsingKe Co., Ltd. The sequences of primers used are listed in Table 1.
Table 1. Sequences of primers used.
| Gene | Forward primer, 5′-3′ | Reverse primer, 5′-3′ |
|---|---|---|
| Mouse | ||
| β-actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| eMLV spliced | CCAGGGACCACCGACCCACCG | TAGTCGGTCCCGGTAGGCCTCG |
| GLN | TGTGTAAGTCCAGACGCAG | CCAACCTACTCCAAAAACAG |
| MervlPol | ATCTCCTGGCACCTGGTATG | AGAAGAAGGCATTTGCCAGA |
| MMERVK | CAAATAGCCCTACCATATGTCAG | GTATACTTTCTTCTTCAGGTCCAC |
| MoMLV | CGAGACCTCATCACCCAGGTTAAG | CATCAGAGCAGCCGATTGTCTG |
| MTA | ATGTTTTGGGGAGGACTGTG | AGCCCCAGCTAACCAGAAC |
| MusD | GTGCTAACCCAACGCTGGTTC | CTCTGGCCTGAAACAACTCCTG |
| pMLV | CCGCCAGGTCCTCAATATAG | AGAAGGTGGGGCAGTCT |
| xMLV | TCTATGGTACCTGGGGCTC | GGCAGAGGTATGGTTGGAGTAG |
| Mx1 | GACCATAGGGGTCTTGACCAA | AGACTTGCTCTTTCTGAAAAGCC |
| Mx2 | GAGGCTCTTCAGAATGAGCAAA | CTCTGCGGTCAGTCTCTCT |
| Oas1b | GGCCTCTAAGGGGGTCAAG | CTGGCAGCACGTCAAACTTC |
| Isg15 | GGTGTCCGTGACTAACTCCAT | TGGAAAGGGTAAGACCGTCCT |
| Oasl1 | CAGGAGCTGTACGGCTTCC | CCTACCTTGAGTACCTTGAGCAC |
| Ddx58 | AAGAGCCAGAGTGTCAGAATCT | AGCTCCAGTTGGTAATTTCTTGG |
| Ifna1 | CTGTGCTTTCCTGATGGTCCT | TCCATGCAGCAGATGAGTCCT |
| Ifnb1 | AGCTCCAAGAAAGGACGAACA | GCCCTGTAGGTGAGGTTGAT |
| Ifng | ATAAGCGTCATTGAATCACAC | CTCAAACTTGGCAATACTCA |
| Ifnl2 | GCAGACCTGTACACAGCTTCA | CAGGTTGGAGGTGACAGAGG |
| Ifnl3 | GTTCAAGTCTCTGTCCCCAAAA | GTGGGAACTGCACCTCATGT |
| Ifnar1 | CTTGTAGCCTCACCGCCTA | CAACATCCACAAGCACGTCCA |
| Ifngr1 | GTGGAGCTTTGACGAGCACT | ATTCCCAGCATACGACAGGGT |
| Il10rb | ACCTGCTTTCCCCAAAACGAA | TGAGAGAAGTCGCACTGAGTC |
| Stat1 | AAATGCCCAAAGATTTAATCAGG | TTGGTCGCTCTTCGCCACA |
| Irf1 | ATGCCAATCACTCGAATGCG | TTGTATCGGCCTGTGTGAATG |
| Irf5 | GGTCAACGGGGAAAAGAAACT | CATCCACCCCTTCAGTGTACT |
| Irf7 | CACAGTCTTCCGCGTACCC | TCCCGGCTAAGTTCGTACACC |
| Irf9 | GAAATCACCATGCAAGCGAAG | TGAGTGGTTTACAACGCCAT |
| Gbp3 | GAGGCACCCATTTGTCTGGT | CCGTCCTGCAAGACGATTCA |
| CD274 (PD-L1) | GCTCCAAAGGACTTGTACGTG | TGATCTGAAGGGCAGCATTTC |
| CD274-L2A | TACAGCTGAATTGGTCATCCCA | GGATGGCCATCGTGCTAGGAA |
| Human | ||
| β-actin | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| HLINEORF1F | TTGGAAAACACTCTGCAGGATATTAT | TTGGCCTGCCTTGCTAGATT |
| HERV-W | TGAGTCAATTCTCATACCTG | AGTTAAGAGTTCTTGGGTGG |
| HERV-H | TGGTGCCGTGACTCG GAT | GCTGAGTCCGAAAAGAGAGTC |
| ERVF2B | AAAAAGGAAGAAGTTAACAGC | ATATAAAGACTTAGGTCCTGC |
| HERV-E | GGTGTCACTACTCAATACAC | GCAGCCTAGGTCTCTGG |
| HERV-F | CCTCCAGTCACAACAACTC | TATTGAAGAAGGCGGCTGG |
| HERV-K | AAAGAACCAGCCACCAGG | CAGTCTGAAAACTTTTCTCTC |
| HML-5 | TGAAAGGCCAGCTTGCTG | CAATTAGGAAATTCTTTTCTAC |
| OASL1 | CTGATGCAGGAACTGTATAGCAC | CACAGCGTCTAGCACCTCTT |
| DHX58 | GGGCCTCCAAACTCGATGG | TTCTGGGGTGACATGATGCAC |
| MX1 | GTTTCCGAAGTGGACATCGCA | CTGCACAGGTTGTTCTCAGC |
| ISG15 | CGCAGATCACCCAGAAGATCG | TTCGTCGCATTTGTCCACCA |
| OAS1 | GAAGGCAGCTCACGAAACC | AGGCCTCAGCCTCTTGTG |
| IFIT1 | TGTTGAAGCAGAAGCACACA | TCTACGCGATGTTTCCTACG |
| MX2 | CAGAGGCAGCGGAATCGTAA | TGAAGCTCTAGCTCGGTGTTC |
| IFI44 | GGTGGGCACTAATACAACTGG | CACACAGAATAAACGGCAGGTA |
| OAS1 | TGTCCAAGGTGGTAAAGGGTG | CCGGCGATTTAACTGATCCTG |
| OAS2 | ACGTGACATCCTCGATAAAACTG | GAACCCATCAAGGGACTTCTG |
| DDX60 | CAGCTCCAATGAAATGGTGCC | CTCAGGGGTTTATGAGAATGCC |
| IDH2 | CGCCACTATGCCGACAAAAG | ACTGCCAGATAATACGGGTCA |
| STAT1 | CAATGGAACTTGATGGCCCTA | TTCTACAGAGCCCACTATCCG |
| IRF1 | ATGCCCATCACTCGGATGC | CCCTGCTTTGTATCGGCCTG |
| IRF5 | GGGCTTCAATGGGTCAACG | GCCTTCGGTGTATTTCCCTG |
| IRF7 | GCTGGACGTGACCATCATGTA | GGGCCGTATAGGAACGTGC |
| IRF9 | GCCCTACAAGGTGTATCAGTTG | TGCTGTCGCTTTGATGGTACT |
| CD274 (PD-L1) | TGGCATTTGCTGAACGCATTT | TGCAGCCAGGTCTAATTGTTTT |
| CD274-L2A | TACAGCTGAATTGGTCATCCCA | AGGCAGACATCATGCTAGGTG |
ELISA
OV90 cells were stimulated with drugs for the indicated time, and media were replaced with fresh serum-free media 24 h before collecting supernatants. The cell debris in the supernatant was removed by centrifugation at 1,500 rpm. Human CXCL10 (Neobioscience; cat #EHC157.96), IFN-α (Neobioscience; cat #EHC144a.96), IFN-β (Neobioscience; cat #EHC026b.96), IFN-γ (Neobioscience; cat #EHC102g.96), and IFN-λ3/IL-29 (Neobioscience; cat #EHC015.48) were detected by ELISA according to the manufacturer’s protocol. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad). The concentrations of the samples were obtained according to the standard curve.
Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE)
ID8, OV90 cells were treated with DMSO or AZD1775 for 72 h and lysed with cold SDD-AGE lysis buffer (20 mM Tris-HCl, pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40, 2 mM EDTA, and 1× protease inhibitor) for 30 min. 5× SDD-AGE sample buffer (2.5× Tris-borate-EDTA, 2.5% SDS, 25% glycerol, and 0.25% bromophenol blue) was then added to SDD-AGE lysate to reach 1× and loaded onto a vertical 1.5% agarose gel. After electrophoresis for 30 min with a constant voltage of 100 V at 4°C, the SDD-AGE protein sample was transferred onto Immune-Blot PVDF Membrane for Western immunoblotting.
Western blot
Cells were lysed on the ice for 30 min with RIPA buffer (Servicebio; cat #G2002) supplemented with cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche; cat #05892970001) and phosphatase inhibitor (Servicebio; cat #G2007). Lysates were sonicated on ice before centrifugation at 12,000 rpm for 20 min at 4°C, and then supernatants were collected. Protein concentration was measured using Coomassie (Beyotime; cat #ST1119). 20 µg of total protein was used for all blots. Proteins were separated in SDS-PAGE gels (BioSci; cat #8012011), and electro-transferred onto polyvinylidene difluoride membranes (Cytiva Life Sciences; cat #10600023). Membranes were blocked using 5% BSA for 1 h. The membranes were incubated with following primary antibodies: WEE1 (Proteintech; cat #14375-1-AP; 1:1,000), β-tubulin (ABclonal; cat #AC021; 1:2,000), ERK2 (Abcam; cat #ab227134; 1:1,000), MAVS (Santa Cruz Biotechnology; cat #sc-166583; cat #sc-365334; 1:500), GAPDH (Abclonal; cat #AC001; 1:5,000), MDA5/IFIH1 (Cell Signaling Technology; cat #5321; 1:1,000), TMEM173/STING (Cell Signaling Technology; cat #13647; 1:1,000), cGAS (Santa Cruz Biotechnology; cat #sc-515777; 1:500), RIG-I/DDX58 (Cell Signaling Technology; cat #3743; 1:1,000), TLR3 (Abclonal; A11778; 1:1,000), SETDB1 (Abcam; cat #ab12317; 1:1,000), FOXM1 (Abcam; cat #ab180710; 1:1,000), TriMethyl-Histone H3K9 (Abcam; cat #ab176916; 1:1,000), DiMethyl-Histone H3K9 (Abcam; cat #ab32521; 1:1,000), Histone H3 (ABclonal; cat #A2348; 1:1,000), PD-L1 (Abcam; cat #ab205921; 1:1,000), STAT1 (Cell Signaling Technology; cat #9172; 1:1,000), and Phospho-Stat1 (Tyr701; D4A7; Cell Signaling Technology; cat #7649; 1:1,000) at 4°C overnight, followed by 1:5,000 secondary antibodies (Abclonal; cat #AS014 and cat #AS003) for 1 h at room temperature. Bands were visualized using a Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific; cat #32209).
Detection of dsRNA by immunofluorescence
Cells were seeded onto 20-mm glass slides in a 12-well plate at 50% confluence. The 20-mm glass slides with cells were collected after being treated for 72 h. The glass slides were washed with PBS, fixed with 4% polyoxymethylene, and permeabilized with 0.25% Triton X-100. When indicated, cells were incubated with 4U RNaselll (Biosystems; cat #A2290) in reaction buffer at 37°C for 4 h as instructed after permeabilization. Glass slides were blocked with 5% goat serum in PBS for 30 min. The glass slides were stained with J2 antibody (Biocompare; cat #10010500; 1:100) overnight at 4°C. The secondary antibody of Alexa Fluor 488 donkey anti-mouse immunoglobulin G (Antgene; cat #ANT023; 1:200) was performed for 1 h at room temperature. DAPI (Servicebio; cat #G1012) staining for 5–10 min was conducted to show the nucleus. Cells were observed and photographed on the machine: fluorescence, NIKON Eclipse Ti; software: Eclipse C2, Nikon. Image-pro Plus 6.0 was used for image analysis.
Gene interference and ectopic expression
All siRNAs employed in this study were purchased from Sigma-Aldrich and are listed in Table 2.
Table 2. siRNAs employed in this study.
| si-RNA | #1 | #2 | #3 |
|---|---|---|---|
| Mouse WEE1 | SASI_Mm01_00122232 | SASI_Mm01_00122233 | SASI_Mm02_00316314 |
| Human IFIH1 | SASI_Hs01_00171929 | SASI_Hs01_00171930 | SASI_Hs01_00171931 |
| Human DDX58 | SASI_Hs01_00047980 | SASI_Hs01_00047981 | SASI_Hs02_00345407 |
| Human MAVS | SASI_Hs01_00128708 | SASI_Hs01_00128711 | SASI_Hs01_00128712 |
| Human STING | SASI_Hs02_00371843 | SASI_Hs01_00031029 | SASI_Hs01_00031030 |
| Human CGAS | SASI_Hs01_00197466 | SASI_Hs01_00197467 | SASI_Hs01_00197468 |
| Human TLR3 | SASI_Hs01_00231802 | SASI_Hs01_00231803 | SASI_Hs01_00231804 |
| Human SETDB1 | SASI_Hs02_00344323 | SASI_Hs02_00344324 | SASI_Hs01_00150485 |
| Human FOXM1 | SASI_Hs01_00243977 | SASI_Hs01_00243978 | SASI_Hs01_00243979 |
| Mouse IL-10rb | SASI_Mm01_00056625 | SASI_Mm02_00313688 | SASI_Mm01_00056627 |
RNA interference transfections were performed according to the procedure for Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific; cat #L3000015). siRNAs were transfected at 100 µM final concentration. Poly I:C was obtained from Sigma-Aldrich (cat #P9582). Two single-strand DNA oligos of DNA90 (forward primer: 5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACATACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′; reverse primer: 5′-TGTAGATCATGTACAGATCAGTCATAGATCACTAGTAGATCTGTATGTAGATCATGTACAGATCAGTCATASGATCACTAGTAGATCTGTA-3′) were synthesized by Tsingke Biological Technology and separately dissolved at a final DNA duplex concentration of 20 mM. The solution was incubated for 1 min at 90–95°C and slowly cooled down to room temperature (for no less than 30 min) and then stored at 4°C or on ice before use. The transfection method of poly I:C and dsDNA90 were the same as that of siRNA transfection at a final concentration of 5 µg/ml.
cDNA of FOXM1 (NM_202003) was cloned into a GV493 vector (Genechem Inc.). To establish stable FOXM1 overexpression cell lines, OVCAR4 cells were infected with LV-FOXM1 according to the manufacturer’s instructions. After 48 h, OVCAR4 cells were treated with 2 µg/ml puromycin (Thermo Fisher Scientific; cat #L3000015) for 7 d. Puromycin-resistant clones were collected and expanded for further studies.
NanoString
For NanoString profiling, an nCounter Immunology Panel (Human V2; cat #XT-CSO-HIM2-12) was performed according to the standard protocol of NanoString with analysis and normalization of the raw NanoString data conducted using nSolver Analysis Software v3.0 (NanoString Technologies, Inc.).
RPPA
Protein lysates extracted from eight ovarian cancer cell lines (OC316, OAW42, OVCAR3, HCC1937, HOC1, A2780CP, A2780, and HOC7) treated with DMSO or AZD1775 (200 nM) for 48 h were analyzed using RPPA at the M.D. Anderson Functional Proteomics RPPA Core. Antibodies and approach details are described at the RPPA website (https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core/antibody-information-and-protocols.html). “Rank-ordered” changes induced by AZD1775 treatment are represented by heatmaps calculated by summing the median-centered protein amount normalized to DMSO.
ChIP-seq
Briefly, chromatin immunoprecipitation was performed in OV90 cells after treatment with DMSO or AZD1775 for 48 h. OV90 cells were fixed with 1% formaldehyde and quenched with glycine. Cells were disrupted by lysis buffer and glass beads. Lysates were sonicated to produce 200–500-bp DNA fragments. The cellular lysate was used for immunoprecipitation by incubating with magnetic beads bound to an anti-histone H3K9me3 antibody. DNA was purified, and DNA libraries were prepared using the TruSeq ChIP Library Prep Kit (Illumina) and sequenced on an Illumina HiSeq 2500 System as 2× 100-bp paired ends.
ChIP real-time qPCR (ChIP-qPCR)
ChIP assays were performed using SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads; Cell Signaling Technology; cat #9003) according to the instructions. OV90 cells were harvested and lysed after being cross-linked with 1% formaldehyde. The chromatin-protein supernatants were collected after centrifugation following chromatin digestion and sonication. Chromatin samples were incubated with FOXM1 primary antibody (Cell Signaling Technology; cat #20459) at 4°C overnight rotationally. Chromatin-antibody complexes were pulled down by magnetic A/G beads. Purified DNAs were obtained after immunoprecipitated chromatin was washed, eluted, de–cross-linked, and purified. Real-time PCR was used to quantify expression. Primers for ChIP-qPCR were as follows: GAPDH: forward primer 5′-CTGCAGTACTGTGGGGAGGT-3′, reverse primer 5′-CAAAGGCGGAGTTACCAGAG-3′; SETDB1: forward primer 5′-CTTCCCTTCCTGGGTGCATT-3′, reverse primer 5′-AGTTCCTCAACCACTGCCTG-3′.
WGBS
Genomic DNA was isolated from OV90 cells after treatment with DMSO or AZD1775 for 72 h using the DNeasy Blood and Tissue Kit (QIAGEN; cat #69504). WGBS library preparation was based on Illumina’s “WGBS for Methylation Analysis” protocol. The adaptor was ligated to the sonicated DNA, and then these fragments were processed for bisulfite conversion according to the manufacturer’s protocol. Libraries were prepared with 100 ng bisulfite-converted DNA using the TruSeq DNA Methylation kit (Illumina; cat #FC-151-1002) and sequenced on Illumina HiSeq 2000 platform, yielding 100-bp paired-end reads. Each sample was spiked with 5% Illumina PhiX genomic DNA control.
Global DNA methylation (5-mC) ELISA assay
The total DNA of cell samples was isolated using the Blood & Cell Culture DNA Midi Kit (QIAGEN; cat #13343) for the DNA methylation analysis. Global DNA hydroxymethylation (5hmC) was assessed using the MethylFlash Global DNA Methylation (5-mC) ELISA assay from EpiGentek (cat #P-1030-96) according to the manufacturer’s protocol.
Flow cytometry
For tissues of ID8 xenografts, tumor tissues were cut into small pieces and digested using a tumor dissociation kit (Miltenyi Biotec; cat #4035581) according to the instructions, on a shaking table for 1 h. A 200-mesh stainless steel filter was used to remove the remaining mass of tissue. After centrifugation (300 g, 10 min), single immune cells were collected and resuspended in stain buffer (BD Biosciences; cat #4032357). Samples were either directly incubated with the following antibodies from BD Biosciences: CD45/cat #557659, CD3/cat #555275, CD4/cat #563151, CD8a/cat #553030, CD11c/cat #558079, CD279/cat #744546, CD274/cat #741014, MHCII/cat #746197; or incubated with Foxp3/cat #560401, ki-67/cat #561283, granzyme B/Ebioscience/cat #46-8898-82, or IFN-γ/BD Biosciences/cat #563376 after permeabilization in the dark at room temperature for 40 min. As for IFN-γ detection, the single cells were stimulated with Leukocyte Activation Cocktail (BD Biosciences; cat #550583) for 6 h before being incubated with IFN-γ antibodies. Beckman Coulter flow cytometry (Cytoflex LX) was used to analyze the components of immune cells in the tumor, and FlowJo V10 software was used to quantify populations.
CFSE labeled CD8+ T cell proliferation assay
Peripheral blood mononuclear cells were obtained from healthy humans and CD8+ cells from the suspension were isolated by magnetic bead purification using CD8a microbeads (Miltenyi Biotec; cat #130-117-044) according to the manufacturer’s protocol. CD8+ cells were labeled with 1 µM CFSE (TOPSCIENCE; cat #150347-59-4) in PBS for 10 min at 37°C and stop staining using an equal volume of complete medium. CFSE-labeled CD8+ T cells were collected by centrifuge, and the supplement was removed. Cells were resuspended using a culture medium with 1 μg/ml anti-CD28 (BD PharMingen; cat #555725) and 50 IU/ml IL-2 (BD PharMingen; cat #554562). 1 × 105 CD8+ T cells per well were seed into a 96-well plate coated with anti-CD3e (BD PharMingen; cat #555329). Cells were treated with DMSO or 400 nM AZD1775 at 37°C in an incubator with 5% CO2 for 4 d and harvested, followed by flow cytometry. FlowJo V10 software was used to quantify populations. The level of proliferation was based on the FITC/FITC channel.
Generation of AZD1775-resistant cells
ID8 cells were subjected to AZD1775 in gradual increasing concentrations to generate AZD1775-resistant cells until cells adapted to 50 µM AZD1775. Monoclonal cell populations of the ID8-resistant cells were isolated by limiting dilution. Individual clones demonstrated different degrees of resistance to AZD1775. Cells were cultured in the absence of AZD1775 for a minimum of 1 mo before using them for experiments.
Cytotoxicity measurement
Cell viability measurements were performed by Cell Counting Kit-8 analysis (Dojindo Laboratories; cat #CK04). 3,000–5,000 (depending on cells) cells were seeded in 96-well plates for 24 h, followed by drug administration for 48 h. The media were carefully replaced with 100 μl fresh media supplemented with 10% CCK8 in the dark. Fresh media were used as a control. The plates were then incubated at 37°C for 2 h in the 5% CO2 humidified incubator. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad). The graphics were generated in GraphPad Prism 8.
3D microfluidic ex vivo culture
AIM Biotech 3D Cell Culture Chip (AIM Biotech; cat #DAX-1) was used for 3D culture. Human ovarian cancer tissues were digested by a Tissue Dissociation Kit (Miltenyi Biotec; cat #130-095-929). Following tumor dissociation, the samples were filtrated at a diameter between 40 and 100 µm and the filtered suspension was collected. A microsphere suspension consisting of 10 µl of above filtered suspension, 143 µl of rat tail collagen IV, 19 µl of PBS, 23 µl of H2O, and 5 µl of 0.5 M NaOH was mixed, and every 10 µl of suspension was pipetted into one gel channel. After culturing 3D culture chips in the 37°C thermostatic incubator for 30 min, 120 µl of complete culture medium was pipetted into each material channel. Microspheres were cultured for 24 h at 37°C in a thermostatic incubator and then treated with DMSO or AZD1775 for 24 h. Last, the living cells (green) and dead cells (red) were stained by acridine orange (AO) and propidium iodide (PI; Nexcelom Bioscience; cat #CS2-0106).
In vivo drug studies
The reagents used in vivo include AZD1775 (Selleck; cat #S1525), anti-mouse PD-L1 (B7-H1; CLONE: 10F.9G2; Bio X Cell; cat #BE0101), and anti-mouse CD8α (CLONE: 2.43; Bio X Cell; cat #BE0061).
ID8 xenografts
ID8 cells were resuspended in PBS at a final concentration of 1 × 107 cells/100 µl and then injected intraperitoneally into the 8–10-wk-old female C57BL/6 and NOD/SCID mice (HuaFu Kang Biotechnology). C57BL/6 or NOD/SCID mice were randomly divided into two groups (n = 5 or 6 per group) 2 wk later and treated with vehicle (0.5% hydroxypropylmethylcellulose and 0.2% Tween 80, oral gavage) or AZD1775 (60 mg/kg/d, oral gavage), 5 d on, 2 d off. For the ID8 xenografts model of combined AZD1775 and immune checkpoint suppression, the C57BL/6 mice were randomly divided into four groups (n = 5 or 6 per group) and treated with vehicle (oral gavage, 5 d on, 2 d off), AZD1775 (60 mg/kg/d, oral gavage, 5 d on, 2 d off), αPD-L1 (200 µg per mouse, intraperitoneal injection, every 3 d for six doses), or combination of AZD1775 and αPD-L1. Fluorescence images were conducted after 4 wk of drugs administration to show the residual tumor. Survival curves were generated by Kaplan–Meier. Tumor tissue and blood were obtained after the mice died or were sacrificed for further tissue analysis.
MC38, B16, and CT26 xenografts
106 MC38 or sh-MAVS or sh-STING MC38, 5 × 105 B16 and 5 × 105 CT26 cells were inoculated into the subcutaneous tissue flank of 8–10-wk-old female C57BL/6 mice (MC38 and B16) or BALB/c mice (CT26). Mice were randomly divided into four groups on the third day after cell injection, and drugs were administered by vehicle (5 d on, 2 d off, oral gavage), AZD1775 (60 mg/kg/d, 5 d on, 2 d off, oral gavage), and αPD-L1 (200 µg per mouse, intraperitoneal injection, every 3 d for six doses), or combinations of AZD1775 and αPD-L1. Tumor volumes were measured every 3 d and calculated using the formula V = (L × W2)/2 (L, length; W, width). Mice were treated for 25 d.
The mice of complete tumor regression in the MC38 model were re-challenged with MC38 cells injected subcutaneously into the contralateral flank. Tumor volumes were measured every 3 d until the 67th day to have a second stop. CD8 neutralizing antibody was used from day 80 (200 µg per mouse every other day, intraperitoneal injection, for three doses) for mice in the group of combination before injection of MC38 cells for the third time on day 85 (200 µg per mouse twice weekly) and tumor volumes measured and calculated as mentioned above. Mice were treated until day 25 and sacrificed for tissue harvest.
RNA-seq analysis
Raw reads were trimmed, and the collected clean data were aligned to hg38 or mm10 using STAR (Dobin et al., 2013). Reads per kilobase of exon model per million mapped reads (RPKM) normalized bigwigs were generated by deepTools (Ramírez et al., 2014). Differential expression genes analysis between groups of samples was performed in R with DESeq (Anders and Huber, 2010) from a bioconductor. GSEA was performed with R package ClusterProfiler (Yu et al., 2012). The prerank mode was used. The signed fold change × −log10 (P value) metric was used for preranking the genes. Heatmaps for RNA-seq were generated using R package pheatmap (https://cran.r-project.org/package=pheatmap). The RNA-seq, ChIP-seq, and WGBS datasets generated during this study are available at the National Genomics Data Center, part of the China National Center for Bioinformation (accession no. CRA003527, National Genomics Data Center–Genome Sequence Archive; http://cncb.ac.cn).
ERV elements identification
All transposable element (TE) analysis was performed using the Repeatmasker annotations downloaded from http://www.repeatmasker.org (Repeat Library 20140131). ERV elements identified by RepeatMasker (class “LTR”) were quantified. Differential expression for repeats was performed with edgeR the same way as for genes.
ChIP-seq analysis
Raw reads were mapped by Bowtie1 (Langmead et al., 2009), and duplicate reads were removed. Only uniquely mapped reads were retained. RPKM normalized bigwigs were generated by deepTools, and the tracks were visualized with the Integrative Genomics Viewer (IGV; Robinson et al., 2011). Peaks were called using epic2 (Stovner and Sætrom, 2019) for broad peaks with a false discovery rate of 0.05. Heatmaps for ChIP-seq data were generated using the EnrichedHeatmap (Gu et al., 2018) bioconductor package.
Genomic coordinates for TEs were downloaded from the University of California, Santa Cruz (UCSC) genome browser (RepeatMasker track) of the GRCh38 assembly. H3K9me3 overlapping ERVs were computed with the intersect module of the bedtools suite, defining overlap when at least 1 bp of the regions overlapped. To test for enriched H3K9me3 binding at TE classes and ERV families, we used the TEanalysis software shuffle_bed.pl (https://github.com/4ureliek/TEanalysis).
WGBS analysis
Raw reads were mapped by bwa-meth (https://arxiv.org/abs/1401.1129) after trimming adaptors with Trim Galore (Martin, 2011). Methylation levels were extracted using MethylDackel (https://github.com/dpryan79/MethylDackel), and signal track bedgraph files were generated using MethylDackel first and then converted to bigwig format using the UCSC bedGraphToBigwig (Kent et al., 2010) utility for visualizing in IGV.
AZD1775 sensitivity analysis in the Cancer Cell Line Encyclopedia and CTRP
Gene expression data were downloaded from Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle). The sensitivity data of AZD1775 were obtained from CTRP (https://portals.broadinstitute.org/ctrp/). The FOXM1_PATHWAY gene list was recruited from the literature, and the FOXM1 pathway score was calculated by the mean expression value of all genes in the pathway. Based on the drug area under the curve (AUC) values of AZD1775, cell lines were divided into three equal parts, with the high one third of the AUC values being defined as the AZD1775-resistant group and the low third of the AUC values being defined as the AZD1775-sensitive group. A heatmap for FOXM1 pathway genes in the AZD1775-resistant and -sensitive groups was generated using pheatmap. Cell lines were also divided into high or low groups based on FOXM1 pathway score or the expression of SETDB1 using the same method.
TCGA data analysis
For the association of IFN pathway genes (IFN_GSVA: REACTOME INTERFERON SIGNALING; IFN-α_GSVA: HALLMARK INTERFERON ALPHA RESPONSE; IFN-γ_GSVA: HALLMARK INTERFERON GAMMA RESPONSE) expression with survival, vital patient status (dead and alive) was used as a surrogate end point, and patients were dichotomized by GSVA score (Hänzelmann et al., 2013). The hazard ratios (HRs) and log-rank test were performed using the R Survival package (version 2.41.3).
TCGA pan-cancer gene expression data were downloaded from UCSC Xena (http://xena.ucsc.edu/). Pearson’s correlation coefficient for SETDB1 and FOXM1 expression was calculated in each cancer species.
GEO data analysis
We collected seven FOXM1 ChIP-seq datasets from GEO (https://www.ncbi.nlm.nih.gov/geo/) via the Cistrome Data Browser (Mei et al., 2017). Dataset information is listed in Table 3. The quality control and data analyses of these datasets were reported by ChiLin (Qin et al., 2016). RPKM-normalized bigwigs were generated by deepTools, and the tracks were visualized with IGV.
Table 3. Information of FOXM1 ChIP-seq datasets from GEO.
| GEO ID | Factor Name | Cell Line | Cell Type | Tissue |
|---|---|---|---|---|
| GSM1001000 | FOXM1 | MDA-MB-231 | Epithelium | Mammary gland |
| GSM1259354 | FOXM1 | HeLa | Epithelium | Cervix |
| GSM1010856 | FOXM1 | ECC-1 | Epithelium | Endometrium |
| GSM1010731 | FOXM1 | GM12878 | Lymphoblastoid | Blood |
| GSM1010750 | FOXM1 | SK-N-SH | Neuron | Brain |
| GSM1464016 | FOXM1 | HEK293 | Epithelium | Embryonic kidney |
| GSM1876186 | FOXM1 | MCF-7 | Epithelium | Mammary gland |
Statistical analysis
Significant differences between the two groups were analyzed using the unpaired two-tailed Student’s t test. P < 0.05 was considered statistically significant. Pearson’s correlation coefficient was used to compute correlations between variables, using a t test to assess the significance of the correlation. For comparisons of multiple groups, one-way ANOVA with Bonferroni post hoc test was used. All statistical analyses were done using SPSS 17.0 (SPSS Inc.). Data were analyzed and plotted using GraphPad Prism 8 software.
Online supplemental material
Fig. S1 shows that AZD1775 activates immunogenic response and the IFN pathway. Fig. S2 shows that WEE1 inhibition induces the generation of dsRNA and activation of the IFN pathway. Fig. S3 shows that down-regulation of SETDB1 increased ISG expression, along with ERV up-regulation. Fig. S4 shows the association of FOXM1 with SETDB1 and the IFN pathway. Fig. S5 shows the efficacy of AZD1775, αPD-L1, and combination therapy in ID8, MC38, CT26, and B16 models.
Acknowledgments
The current studies were supported by the Nature and Science Foundation of China (81874106 to G. Chen, 81974408 to C. Sun, 82002762 to E. Guo, and 82060472 to F. Li). G.B. Mills has support from the National Institutes of Health (P50CA217685 and U01 CA217842), Susan G. Komen Breast Cancer Foundation (SAC110052), the Breast Cancer Research Foundation, and a kind gift from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Author contributions: G. Chen, C. Sun, G.B. Mills, and E. Guo conceived the project. G. Chen and C. Sun performed the supervision and project administration. E. Guo, R. Xiao, Y. Wu, and F. Lu designed, performed, and validated the experiments with the help of C. Liu, B. Yang, X. Li, Y. Fu, Z. Wang, Y. Li, Y. Huang, F. Li, X. Wu, L. You., and T. Qin. C. Sun, E. Guo, and Y. Lu analyzed ChIP-seq, RNA-seq, WGBS, and public TCGA data. E. Guo, R. Xiao, and C. Sun wrote the manuscript, and the manuscript was revised by G.B. Mills, C. Sun, and G. Chen; D. Ma, and X. Huang provided resources. G. Chen, C. Sun, E. Guo, and F. Li acquired the funding.
References
- Anders, S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, E.A., Wang J.H., Gong F., Wu W., Neill N.J., Kim I.S., Tyagi S., Orellana M., Kurley S.J., Dominguez-Vidaña R., et al. 2021. Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell. 184:384–403.e21. 10.1016/j.cell.2020.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadas, I., Thummalapalli R., Kim J.W., Kitajima S., Jenkins R.W., Christensen C.L., Campisi M., Kuang Y., Zhang Y., Gjini E., et al. 2018. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat. Med. 24:1143–1150. 10.1038/s41591-018-0116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbognin, L., Pilotto S., Milella M., Vaccaro V., Brunelli M., Caliò A., Cuppone F., Sperduti I., Giannarelli D., Chilosi M., et al. 2015. Differential Activity of Nivolumab, Pembrolizumab and MPDL3280A according to the Tumor Expression of Programmed Death-Ligand-1 (PD-L1): Sensitivity Analysis of Trials in Melanoma, Lung and Genitourinary Cancers. PLoS One. 10:e0130142. 10.1371/journal.pone.0130142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli, K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. 2015. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 162:974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuneo, K.C., Morgan M.A., Sahai V., Schipper M.J., Parsels L.A., Parsels J.D., Devasia T., Al-Hawaray M., Cho C.S., Nathan H., et al. 2019. Dose Escalation Trial of the Wee1 Inhibitor Adavosertib (AZD1775) in Combination With Gemcitabine and Radiation for Patients With Locally Advanced Pancreatic Cancer. J. Clin. Oncol. 37:2643–2650. 10.1200/JCO.19.00730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz, N.M.G.P., Xia T., Konno H., and Barber G.N.. 2019. Ovarian Cancer Cells Commonly Exhibit Defective STING Signaling Which Affects Sensitivity to Viral Oncolysis. Mol. Cancer Res. 17:974–986. 10.1158/1541-7786.MCR-18-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diab, A., Gem H., Swanger J., Kim H.Y., Smith K., Zou G., Raju S., Kao M., Fitzgibbon M., Loeb K.R., et al. 2020. FOXM1 drives HPV+ HNSCC sensitivity to WEE1 inhibition. Proc. Natl. Acad. Sci. USA. 117:28287–28296. 10.1073/pnas.2013921117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disis, M.L., Patel M.R., Pant S., Hamilton E.P., Lockhart A.C., Kelly K., Beck J.T., Gordon M.S., Weiss G.J., Taylor M.H., et al. 2016. Avelumab (MSB0010718C; anti-PD-L1) in patients with recurrent/refractory ovarian cancer from the JAVELIN Solid Tumor phase Ib trial: Safety and clinical activity. J. Clin. Oncol. 34(15_suppl):5533. 10.1200/JCO.2016.34.15_suppl.5533 [DOI] [Google Scholar]
- Disis, M.L., Taylor M.H., Kelly K., Beck J.T., Gordon M., Moore K.M., Patel M.R., Chaves J., Park H., Mita A.C., et al. 2019. Efficacy and Safety of Avelumab for Patients With Recurrent or Refractory Ovarian Cancer: Phase 1b Results From the JAVELIN Solid Tumor Trial. JAMA Oncol. 5:393–401. 10.1001/jamaoncol.2018.6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Kelly, R., Martín Y., Koundrioukoff S., Tanenbaum M.E., Smits V.A., Medema R.H., Debatisse M., and Freire R.. 2011. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J. Cell Biol. 194:567–579. 10.1083/jcb.201101047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorand, R.D., Nthale J., Myers J.T., Barkauskas D.S., Avril S., Chirieleison S.M., Pareek T.K., Abbott D.W., Stearns D.S., Letterio J.J., et al. 2016. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 353:399–403. 10.1126/science.aae0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, J., and Rao S.. 2017. Epigenetics and immunotherapy: The current state of play. Mol. Immunol. 87:227–239. 10.1016/j.molimm.2017.04.012 [DOI] [PubMed] [Google Scholar]
- Elsässer, S.J., Noh K.M., Diaz N., Allis C.D., and Banaszynski L.A.. 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 522:240–244. 10.1038/nature14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., McGrail D.J., Sun C., Labrie M., Chen X., Zhang D., Ju Z., Vellano C.P., Lu Y., Li Y., et al. 2019. Sequential Therapy with PARP and WEE1 Inhibitors Minimizes Toxicity while Maintaining Efficacy. Cancer Cell. 35:851–867.e7. 10.1016/j.ccell.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, C.S., Arlauckas S.P., Kohler R.H., Trefny M.P., Garren S., Piot C., Engblom C., Pfirschke C., Siwicki M., Gungabeesoon J., et al. 2018. Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-γ and IL-12. Immunity. 49:1148–1161.e7. 10.1016/j.immuni.2018.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Eils R., Schlesner M., and Ishaque N.. 2018. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics. 19:234. 10.1186/s12864-018-4625-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanishi, J., Mandai M., Ikeda T., Minami M., Kawaguchi A., Murayama T., Kanai M., Mori Y., Matsumoto S., Chikuma S., et al. 2015. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J. Clin. Oncol. 33:4015–4022. 10.1200/JCO.2015.62.3397 [DOI] [PubMed] [Google Scholar]
- Hänzelmann, S., Castelo R., and Guinney J.. 2013. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., McLoughlin M., and McKeon F.. 1993. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 74:463–474. 10.1016/0092-8674(93)80048-J [DOI] [PubMed] [Google Scholar]
- Infante, J., Braiteh F., Emens L., Balmanoukian A., Oaknin A., Wang Y., Liu B., Molinero L., Fassò M., Hear C.O., and Gordon M.S.. 2016. Safety, clinical activity and biomarkers of atezolizumab (atezo) in advanced ovarian cancer (OC). Ann. Oncol. 27:vi300. 10.1093/annonc/mdw374.18 [DOI] [Google Scholar]
- Kent, W.J., Zweig A.S., Barber G., Hinrichs A.S., and Karolchik D.. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 26:2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov, M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90-7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, J., and Bakhoum S.F.. 2020. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 10:26–39. 10.1158/2159-8290.CD-19-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., Trapnell C., Pop M., and Salzberg S.L.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasfar, A., Gogas H., Zloza A., Kaufman H.L., and Kirkwood J.M.. 2016. IFN-λ cancer immunotherapy: new kid on the block. Immunotherapy. 8:877–888. 10.2217/imt-2015-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijen, S., van Geel R.M., Sonke G.S., de Jong D., Rosenberg E.H., Marchetti S., Pluim D., van Werkhoven E., Rose S., Lee M.A., et al. 2016. Phase II Study of WEE1 Inhibitor AZD1775 Plus Carboplatin in Patients With TP53-Mutated Ovarian Cancer Refractory or Resistant to First-Line Therapy Within 3 Months. J. Clin. Oncol. 34:4354–4361. 10.1200/JCO.2016.67.5942 [DOI] [PubMed] [Google Scholar]
- Lheureux, S., Cristea M.C., Bruce J.P., Garg S., Cabanero M., Mantia-Smaldone G., Olawaiye A.B., Ellard S.L., Weberpals J.I., Wahner Hendrickson A.E., et al. 2021. Adavosertib plus gemcitabine for platinum-resistant or platinum-refractory recurrent ovarian cancer: a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet. 397:281–292. 10.1016/S0140-6736(20)32554-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, G.B., Li X.Z., Zeng S., Liu C., Yang S.M., Yang L., Hu C.J., and Bai J.Y.. 2018. Regulation of the master regulator FOXM1 in cancer. Cell Commun. Signal. 16:57. 10.1186/s12964-018-0266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.F., Xiong N., Campos S.M., Wright A.A., Krasner C.N., Schumer S.T., Horowitz N.S., Veneris J.T., Tayob N., Morrissey S., et al. 2020. A phase II trial of the Wee1 inhibitor adavosertib (AZD1775) in recurrent uterine serous carcinoma. J. Clin. Oncol. 38(15_suppl):6009. 10.1200/JCO.2020.38.15_suppl.6009 [DOI] [PubMed] [Google Scholar]
- Mager, D.L., and Lorincz M.C.. 2017. Epigenetic modifier drugs trigger widespread transcription of endogenous retroviruses. Nat. Genet. 49:974–975. 10.1038/ng.3902 [DOI] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matheson, C.J., Backos D.S., and Reigan P.. 2016. Targeting WEE1 Kinase in Cancer. Trends Pharmacol. Sci. 37:872–881. 10.1016/j.tips.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Matulonis, U.A., Shapira-Frommer R., Santin A., Lisyanskaya A.S., Pignata S., Vergote I., Raspagliesi F., Sonke G.S., Birrer M., Provencher D.M., et al. 2018. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Interim results from the phase 2 KEYNOTE-100 study. J. Clin. Oncol. 36(15_suppl):5511. 10.1200/JCO.2018.36.15_suppl.5511 [DOI] [PubMed] [Google Scholar]
- Mei, S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L., et al. 2017. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45(D1):D658–D662. 10.1093/nar/gkw983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K.W., Attig J., Young G.R., Ottina E., Papamichos S.I., Kotsianidis I., and Kassiotis G.. 2019. Soluble PD-L1 generated by endogenous retroelement exaptation is a receptor antagonist. eLife. 8:e50256. 10.7554/eLife.50256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan, S.R., and Bowie A.G.. 2013. Immune sensing of DNA. Immunity. 38:870–880. 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Q., Mei S., Wu Q., Sun H., Li L., Taing L., Chen S., Li F., Liu T., Zang C., et al. 2016. ChiLin: a comprehensive ChIP-seq and DNase-seq quality control and analysis pipeline. BMC Bioinformatics. 17:404. 10.1186/s12859-016-1274-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Dündar F., Diehl S., Grüning B.A., and Manke T.. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42(Web Server issue, W1):W187-91. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikine, S., Nguyen J.B., and Modis Y.. 2014. Pattern Recognition and Signaling Mechanisms of RIG-I and MDA5. Front. Immunol. 5:342. 10.3389/fimmu.2014.00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, J., Endo T.A., Nakayama M., Karimi M.M., Shimada M., Katsuyama K., Goyal P., Brind’Amour J., Sun M.A., Sun Z., et al. 2016. Activation of Endogenous Retroviruses in Dnmt1(-/-) ESCs Involves Disruption of SETDB1-Mediated Repression by NP95 Binding to Hemimethylated DNA. Cell Stem Cell. 19:81–94. 10.1016/j.stem.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Sheng, W., LaFleur M.W., Nguyen T.H., Chen S., Chakravarthy A., Conway J.R., Li Y., Chen H., Yang H., Hsu P.H., et al. 2018. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell. 174:549–563.e19. 10.1016/j.cell.2018.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai, K., and Liu B.. 2003. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3:900–911. 10.1038/nri1226 [DOI] [PubMed] [Google Scholar]
- Snyder, A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S., et al. 2014. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371:2189–2199. 10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovner, E.B., and Sætrom P.. 2019. epic2 efficiently finds diffuse domains in ChIP-seq data. Bioinformatics. 35:4392–4393. 10.1093/bioinformatics/btz232 [DOI] [PubMed] [Google Scholar]
- Sun, C., Fang Y., Yin J., Chen J., Ju Z., Zhang D., Chen X., Vellano C.P., Jeong K.J., Ng P.K., et al. 2017. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci. Transl. Med. 9:eaal5148. 10.1126/scitranslmed.aal5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C., Yin J., Fang Y., Chen J., Jeong K.J., Chen X., Vellano C.P., Ju Z., Zhao W., Zhang D., et al. 2018. BRD4 Inhibition Is Synthetic Lethal with PARP Inhibitors through the Induction of Homologous Recombination Deficiency. Cancer Cell. 33:401–416.e8. 10.1016/j.ccell.2018.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, J., Brooks T., Landras A., and Massey A.J.. 2021. Targeting DNA damage response pathways to activate the STING innate immune signaling pathway in human cancer cells. FEBS J. 288:4507–4540. 10.1111/febs.15747 [DOI] [PubMed] [Google Scholar]
- Wu, B., Peisley A., Richards C., Yao H., Zeng X., Lin C., Chu F., Walz T., and Hur S.. 2013. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 152:276–289. 10.1016/j.cell.2012.11.048 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel, L., Galluzzi L., Kepp O., Smyth M.J., and Kroemer G.. 2015. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15:405–414. 10.1038/nri3845 [DOI] [PubMed] [Google Scholar]
- Zou, W., Wolchok J.D., and Chen L.. 2016. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8:328rv4. 10.1126/scitranslmed.aad7118 [DOI] [PMC free article] [PubMed] [Google Scholar]