SUMMARY
Viral RNA-dependent RNA polymerases (RdRps) are a target for broad-spectrum antiviral therapeutics. Recently, we demonstrated that incorporation of the T-1106 triphosphate, a pyrazine-carboxamide ribonucleotide, into nascent RNA increases pausing and backtracking by the poliovirus RdRp. Here, by monitoring Enterovirus A-71 RdRp dynamics during RNA synthesis using magnetic tweezers, we identify the “backtracked” state as an intermediate used by the RdRp for both copy-back RNA synthesis and homologous recombination. Cell-based assays and RNAseq experiments further demonstrate that the pyrazine-carboxamide ribonucleotide stimulates these processes during infection. These results suggest that pyrazine-carboxamide ribonucleotides do not induce lethal mutagenesis or chain termination, but function by promoting template switching and formation of defective viral genomes. We conclude that RdRp-catalyzed intra- and intermolecular template switching can be induced by pyrazine-carboxamide ribonucleotides, defining an additional mechanistic class of antiviral ribonucleotides with potential for broad-spectrum activity.
eTOC Blurb
Using a multidisciplinary approach, Janissen, Woodman et al. demonstrate that intra- and intermolecular recombination events in enteroviruses originate from a “backtracked” polymerase intermediate. Pyrazine-carboxamide ribonucleotides promote formation of this intermediate, revealing the antiviral nature of chemically induced recombination and an unique mechanistic class of antiviral ribonucleotides.
Graphical Abstract
INTRODUCTION
It is still not possible to predict the emergence of viruses capable of founding an epidemic, a fact that has been reconfirmed many times over the past few decades. Since the outbreak of West Nile virus in 1999, one RNA virus after another has emerged, causing significant morbidity and mortality. In 2019, yet another outbreak occurred and rapidly spread globally. While the world is under constant threat of an influenza pandemic (Carrasco-Hernandez et al., 2017), the ongoing pandemic was caused by a coronavirus, specifically severe acute respiratory syndrome coronavirus-2 (SARS CoV-2) (Ghanbari et al., 2020; Zhu et al., 2020). An increase of enterovirus D-68 (EV-D68) infections in the US, associated with acute flaccid myelitis in young children, has been registered in the past years, and future outbreaks are anticipated (Park et al., 2021). With recurrent outbreaks of enterovirus A71 in the Asia-Pacific region come hand-foot-and-mouth disease and severe acute flaccid paralysis (Puenpa et al., 2019). This state of affairs demands the availability and rapid development of broad-spectrum antiviral therapeutics to address the next unanticipated and/or unknown viral pathogen.
Viral polymerases have emerged as tractable and highly efficacious antiviral targets (Tsai et al., 2006). Past and present protocols to treat virus infections have included compounds targeting the viral RNA-dependent RNA polymerase (RdRp). To date, clinically approved antiviral nucleosides have functioned either by terminating nucleic acid synthesis (chain terminators) or by increasing mutational load on the viral genome (lethal mutagenesis) (Seley-Radtke and Yates, 2018; Yates and Seley-Radtke, 2019). Resistance to antiviral nucleotides usually comes with a fitness cost, thus increasing the long-term utility of these classes of antiviral agents (Irwin et al., 2016). The major obstacle to the development of antiviral nucleotides is off-target effects, caused primarily by cellular polymerase utilization of the compounds in question (Arnold et al., 2012). However, the specificity of antiviral nucleotides continues to improve (Coats et al., 2014).
Recently, pyrazine carboxamide nucleoside analogs were approved in Japan to treat influenza virus infection (Mifsud et al., 2019). The drug, commercially known as favipiravir, is also known as T-705. It is employed in several countries as a potential antiviral drug against SARS CoV-2 infection based on evidence for its effectiveness in the treatment thereof (Joshi et al., 2021; Shannon et al., 2020). Favipiravir is a fluorinated pyrazine carboxamide base analog and requires the cellular nucleotide salvage pathway to convert the base into a nucleoside triphosphate. A version of favipiravir lacking fluorine, known as T-1105, is also active, but its conversion to the triphosphate is less efficient (Furuta et al., 2009). The nucleoside version of T-1105 is referred to as T-1106, and the metabolism of this compound to the triphosphate yields a drug with efficacy superior to favipiravir (Furuta et al., 2009). The results of studies probing the mechanism of action of these compounds have been ambiguous. Some studies were consistent with chain termination (Sangawa et al., 2013), while recent results were consistent with lethal mutagenesis (de Avila et al., 2017). Of course, such uncertainty in the mechanism of action complicates worldwide approval beyond Japan.
To gain insights into the underlying mechanism of action of T-1106, we previously used a single-molecule magnetic-tweezers platform to monitor individual poliovirus (PV) RdRp elongation complexes over thousands of nucleotide-addition cycles and observed the ability of T-1106-TP (superior to that of ribavirin-TP) to cause the RdRp to pause and then backtrack (Dulin et al., 2017). The elongation complex was able to recover from the backtracked state, but recovery required tens to thousands of seconds. As a result, traditional polymerase elongation assays would view these backtracked states as prematurely terminated products (Dulin et al., 2017). This backtracking phenomenon has to date not been observed with either prototypical chain terminators or mutagens. These studies therefore provided very compelling evidence for the existence of a third mechanistic class of antiviral ribonucleoside analogs.
While the structure of the backtracked state of PV is not known, the nascent RNA was likely displaced from template to yield a single-stranded 3’ end with lengths greater than tens of nucleotides (Dulin et al., 2017). Such a structure is now known for the SARS CoV-2 RdRp (Malone et al., 2021). The ability of an RdRp to produce free single-stranded 3’ ends was intriguing, because such an end could undergo an intermolecular template switch by annealing to a second template, with resumed synthesis producing a recombinant RNA product.
The goals of the present study were to determine the extent to which the backtracked state represented an intermediate on path for recombination, and to identify the mechanism of action of the pyrazine carboxamide nucleoside analog T-1106. To do so, we introduced the RdRp from EV-A71, a virus prone to recombination in nature (Woodman et al., 2018), into our pipeline with the idea that perhaps such backtracked states might also be prevalent with this enzyme. By comparing the EV-A71 and PV RdRps, we provide evidence that the backtracked state is indeed a common intermediate for template switching. Unexpectedly, for EV-A71 RdRp we observed a high frequency of intramolecular template switching, which has been coined “copy-back RNA synthesis” in the literature (Vignuzzi and López, 2019). Importantly, the capacity for T-1106 to promote recombination was also observed in cells based on RNAseq data from cells infected with EV-A71 in the presence or absence of the drug. Our study therefore makes a clear mechanistic connection between copy-back RNA synthesis and homologous recombination, and provides compelling evidence that an enhancement of the probability of template switching results in an antiviral effect. The experimental paradigm reported here should prove useful in dissecting the contributions and determinants of the RdRp and the template to formation of defective viral genomes, a newly emerging strategy for interfering with virus multiplication and attenuating viral pathogenesis (Vignuzzi and López, 2019).
RESULTS
Pausing of EV-A71 RdRp promotes copy-back RNA synthesis
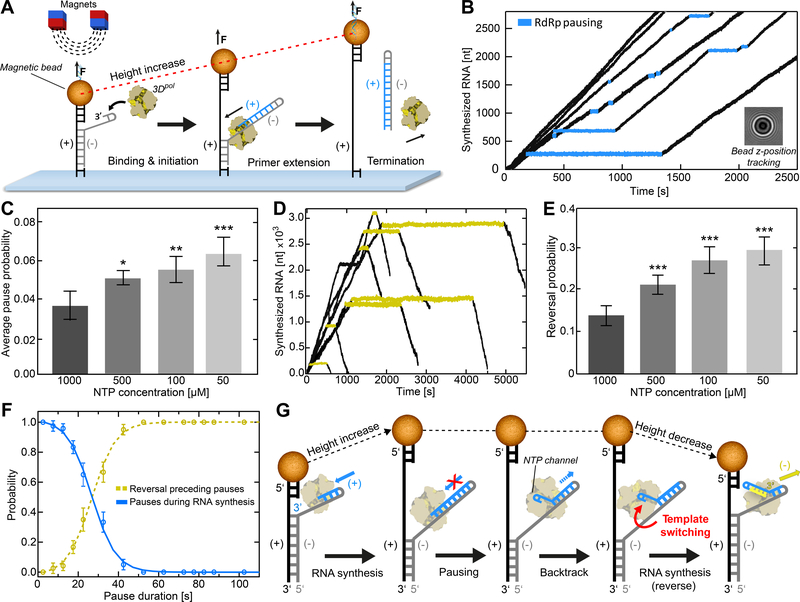
The development of a magnetic-tweezers platform to monitor nucleotide addition by nucleic acid polymerases is unmasking the stochastic behavior of viral polymerases and illuminating states of the polymerase induced by pausing and drugs (Dulin et al., 2015, 2017; Seifert et al., 2021). In this assay (Figure 1A), single-stranded RNA is tethered to the surface and a magnetic bead. A template ssRNA, including a 24 nt hairpin structure at its 3’ end, is annealed to the tethered ssRNA strand, creating a predominantly double-stranded RNA. Primer-extension from the 3’-end of template RNA will lead to displacement of the template RNA from the tethered RNA. At forces >8 pN, the conversion of dsRNA to ssRNA causes a corresponding increase in the distance of the bead from the surface, thus permitting measurement of nucleotide incorporation with few nucleotide resolution over thousands of cycles of nucleotide addition (Dulin et al., 2015, 2017; Seifert et al., 2021).
Figure 1. Magnetic tweezers assay of EV-A71 RdRp reveals pause-dependent copy-back RNA synthesis.
(A) Schematic of the single-molecule (+)-strand RNA synthesis assay, showing the binding of an RdRp to a hairpin at the 3’-end of the (−)-strand (grey) of the surface-attached RNA construct. A magnetic bead attached to the RNA construct is subject to a constant force of 25 pN during RNA synthesis (blue). (B) Sample EV-A71 RdRp trajectories showing stochastic pausing behavior over time, inferred via the change of the diffraction pattern of the attached magnetic bead (inset) at a rate of 50 Hz. Within individual trajectories, the data in blue highlight single pauses of different duration and template position during processive RNA synthesis. (C) Comparison of the average pause frequency (±SD) at different rNTP concentrations extracted from dwell-time distributions (Figure S1A). Dataset statistics and experimental parameters for each condition are provided in Table S1. (D) Sample individual RdRp elongation trajectories showing the occurrence of reversal events following extended pausing (yellow). (E) The average occurrence probability (±SD) of reversals per RdRp for all measured conditions. (F) The probability of observing pauses of specified pause durations during RNA synthesis (blue) co-plotted with the probability of reversals (yellow); the crossover point is found at ~26 s. (G) Proposed model underlying the reversal events, in which pause-induced elongation complex is followed by copy-back RNA synthesis of the newly synthesized (+)-strand RNA (blue). Synthesis of a new (−)-strand (yellow) results in reannealing of the original (+)-strand template (grey) to the complementary ssRNA tether (black), leading to an overall decrease in RNA tether extension. Statistical analyses were performed using ANOVA with comparative Tukey post-hoc test (significance levels p: *** = 0.001; ** = 0.01; * = 0.05). See also Figures S1 and S2.
In a previous study with PV RdRp, we showed that one consequence of prolonged pausing by the enzyme is backtracking, where the enzyme unwinds the 3’-terminus of nascent RNA (Dulin et al., 2017). It was possible that this state was an intermediate for homologous recombination by an intermolecular template-switching mechanism. Such a template switch would occur by annealing the 3’-terminus of the backtracked, product RNA to a second (acceptor) template. In the magnetic tweezers, there is no acceptor template; therefore, intermolecular template switching cannot occur. In most instances, the nascent RNA reannealed to the original template and RNA synthesis resumed (Dulin et al., 2017).
Here, we evaluate EV-A71 RdRp as it might provide additional insight into the backtracked state. As illustrated in Figure 1B, pauses of differing duration interrupted processive nucleotide addition by EV-A71 RdRp (quantified in Figure S1C). For PV RdRp, we previously observed an inverse correlation between nucleotide concentration and pause probability (Dulin et al., 2017). In this context, EV-A71 behaved similarly (Figures 1C, S1A–B). Pausing is thought to occur in response to misincorporation and happens more often at low nucleotide concentration (Dulin et al., 2017). Such a response might facilitate correction by pyrophosphorolysis (Dulin et al., 2017; Jin et al., 2013a).
At some frequency for PV RdRp, however, the paused state undergoes backtracking (Dulin et al., 2017). Surprisingly, under the same conditions, EV-A71 RdRp undergoes reverse translocation (referred to as “reversals”), based on the decrease in position of the bead relative to the surface (Figure 1D). Pauses induced by low nucleotide concentrations, found to be drivers of backtracking by PV RdRp (Dulin et al., 2017), are also drivers of reversals by EV-A71 RdRp (Figure 1E–F). While it was not possible to identify sequence motifs at locations where reversals occurred, GC-rich regions favored reversals (Figure S2). Given the assay design, the only ways for the bead to approach the surface would be for the EV-A71 RdRp to dissociate and initiate primer extension via the accessible 3’ ssRNA end of the bead handle, or to reanneal the displaced template ssRNA to the tethered ssRNA. Our control experiments excluded the former possibility, as in absence of the template ssRNA strand the RdRp was not able to perform primer-extension (Figure S1E–F). For the latter scenario to occur, nascent ssRNA would have to be displaced from the template ssRNA. We hypothesized that this could occur as a result of pausing by EV-A71 RdRp followed by backtracking, which produces a single-stranded 3’-end that can be used by EV-A71 RdRp as a primer for copy-back RNA synthesis (Figure 1G). This hypothesis was tested in the following experiments. That a single polymerase could carry out both the initial round of RNA synthesis and copy-back RNA synthesis is supported by the good agreement of the kinetic behavior of the polymerases during both processes (Figure S1D).
A recombination-deficient EV-A71 RdRp variant attenuates virus population
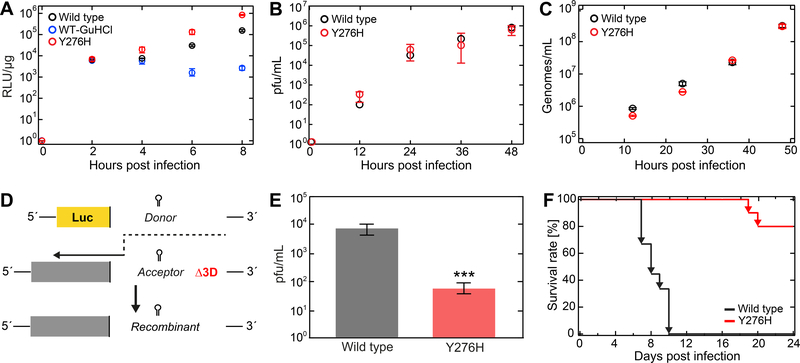
The availability of a recombination-defective EV-A71 RdRp derivative would prove very useful in making the strongest case for a relationship between the observation of backtracking or reversals and recombination. In PV RdRp, Y275H was identified as a substitution that impaired recombination substantially, although the molecular basis for the reduced recombination efficiency is yet not known (Acevedo et al., 2018; Kempf et al., 2020). We engineered the orthologous Y276H substitution into RdRp-coding sequence of EV-A71. In the context of a subgenomic replicon, Y276H EV-A71 had no growth defect when compared to WT (Figure 2A), in agreement with previous observations (Tee et al., 2019). By plaque assay and RT-qPCR, Y276H and WT EV-A71 were indistinguishable (Figure 2B–C). To assess the recombination efficiency, we transfected the subgenomic replicon RNA described above (donor template) and an EV-A71 genomic RNA deleted for 3Dpol-coding sequence (acceptor template) into cells; recombination between these RNAs produced viable virus (Figure 2D). The yield of recombinant virus was reduced by nearly 100-fold (Figure 2E).
Figure 2. EV-A71 Y276H RdRp variant exhibits significance decrease in recombination efficiency and virulence.
(A) Replication efficiency of the EV-A71 wild type (WT) and Y276H variant subgenomic replicons in RD cells as a function of time post transfection. GuHCl refers to the presence of guanidine hydrochloride, a potent inhibitor of replication. Luciferase activity is reported in relative light units (RLU; AVG±SD) per microgram of total protein (n = 3 replicates per time point). (B) EV-A71 WT (black) and Y276H variant (red) single-step growth curves (±SD; n = 3) confirm similar plaque formation. (C) EV-A71 WT (black) and Y276H variant (red) single-step growth curves (AVG ±SD; n = 3 for each time point). RD cells were infected with virus equivalent to 200 genomes/cell. Samples were taken at the indicated times and the genome amount of virus titer was quantified via RT-qPCR. (D) Cell-based recombination assay. EV-A71 C2-strain firefly luciferase-encoding sub-genomic replicon (donor) and full-length EV-A71 C2-MP4 strain genome (acceptor) carrying a lethal deletion of the 3Dpol region are co-transfected in RD cells. Only upon co-transfection can replication-competent virus be generated by RdRp-mediated template switch from donor to acceptor (indicated by dashed black arrow). (E) The Y276H mutation in the EV-A71 replicon inhibits recombinant yield. Resulting recombinant virus were quantified by pfu/ml (AVG±SD; n = 3). (F) EV-A71 C2-MP4 WT and Y276H virulence in hSCARB2 mice. 21-day old mice were orally inoculated with either WT or Y276H virus (n = 10 per virus) at a dose equivalent to 2×107 genomes and scored for survival post-infection. Effect of Y276H variant is severely attenuated relative to WT. Statistical analysis consisted of an unpaired, two-tailed t-test (significance level p: *** ≤ 0.001)
Previous studies have indicated that the ability of PV RdRp to catalyze recombination is required for PV to cause disease in a mouse model, even though growth of recombination-defective PV mutants in cell culture appears equivalent to or better than WT. We evaluated the virulence of Y276H EV-A71 in a mouse model that supports infection by oral inoculation. These mice express human SCARB2 protein, which is a receptor for EV-A71 (Yamayoshi et al., 2009). The population of viruses carrying the Y276H EV-A71 was highly attenuated in this model relative to WT (Figure 2F), in agreement with a recently published study (Tee et al., 2019).
Recombination deficiency of Y275(6)H RdRps originates from enhanced binding to nascent RNA and diminished backtracking
Based on the data gathered in the past for PV RdRp and those described above for EV-A71 RdRp, our mechanism for template switching begins with RdRp pausing, followed by backtracking, and ending with either resolution of the backtrack (PV) or reversals (EV-A71). The availability of a recombination-defective EV-A71 RdRp, Y276H RdRp, should facilitate establishment of a correlation between reversal events observed using the magnetic tweezers and the requirements of the template-switching process.
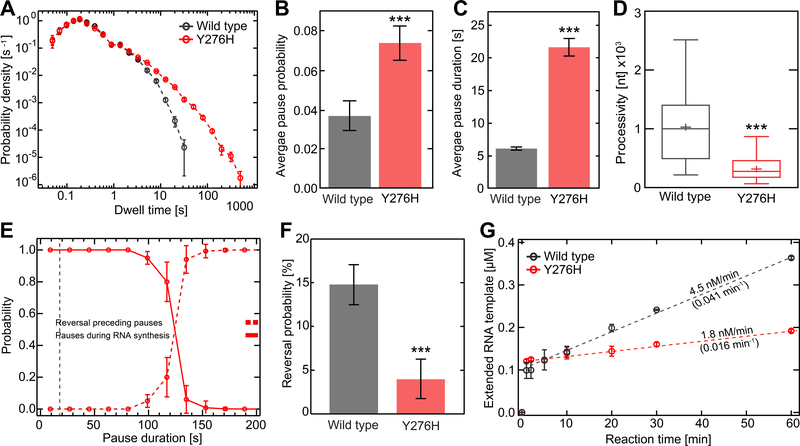
The kinetics of correct nucleotide addition were unchanged for Y276H RdRp relative to WT (note the overlap of probability density in the dwell-time distributions up to 3 s in Figure 3A). In contrast, the rate of processive nucleotide addition for the recombination-defective derivative appeared compromised by a 2-fold increase in pausing frequency (Figure 3B) and a 4-fold increase in the duration of the pause (Figure 3C). The overall processivity of this derivative was also reduced by 4-fold (Figure 3D). The reversals observed with Y276H RdRp derived from a pause-dependent mechanism as observed for WT (Figure 3E). Interestingly, because the typical duration of pauses beyond which reversals were observed increased by five-fold (~123 s vs. ~26 s, compare Figures 1F, 3E), more than the typical duration of pauses themselves, we predicted that the incidence of reversals would decrease for Y276H RdRp relative to WT. Such a decreased occurrence of reversals (by 4-fold, Figure 3F) was indeed observed experimentally. With pausing elevated and reversals diminished, the backtrack intermediate should accumulate. This was not observed, suggesting that an inability to backtrack is the functional defect associated with the Y276H RdRp derivative that interferes with recombination.
Figure 3. EV-A71 Y276H RdRp variant is impaired for copy-back RNA synthesis.
(A) Superimposed dwell-time distributions for EV-A71 Y276H RdRp (red) and WT (black) RdRp. The dwell times used for the construction of the distributions are the time needed for the RdRps to synthesize four consecutive nucleotides. The Y276H variant exhibits a broad increase in the probability and duration of long pauses compared to WT. The error bars (AVG±SD) result from bootstrapping with 1,000 iterations. (B-D) Quantification of the data in panel (A). In comparison to WT RdRp, the Y276H variant shows a significant increase in (B) average pausing probability (±SD) and (C) average apparent pause duration (±SEM). The processivity (D) is significantly decreased for the Y267H RdRp variant compared to WT. (E) The probability of observing pauses of specified durations during RNA synthesis (red line) co-plotted with the probability of observing reversals (red dashed line); the crossover point amounts to ~123 s. (F) The Y276H RdRp (red) variant causes significantly fewer reversal events (AVG±SD) than WT RdRp (grey). (G) In vitro bulk RNA synthesis assay results (AVG±SD; n = 3 repetitions) showing the amount of extended Sym/SubU template for EV-A71 WT and Y276H RdRp over time. Dashed lines represent linear regressions fitted to the data, revealing significantly lower approximated rates of RNA extension (values above the dashed lines) and turnover (values below the dashed lines; unit: RNA min−1 RdRp−1) for the Y276H RdRp variant compared to WT. Statistical analyses were performed using unpaired, two-tailed t-tests (significance level p: *** ≤ 0.001).
We next questioned the reason for the inability of Y276H RdRp to form a backtrack intermediate. For backtracking to occur, the paused polymerase needs to release the 3’-end from the active site. The stability of the RdRp at the 3’-end can be inferred from the steady-state rate constant for single-nucleotide incorporation (Arnold and Cameron, 2000). A reduction in the value of this rate constant implies increased affinity for the terminus. Indeed, the Y276H RdRp elongation complex was 3-fold more stable than that formed by WT RdRp on the synthetic template developed to monitor elongation by enteroviral RdRps (Figure 3G) (Arnold and Cameron, 2000; Shi et al., 2020). It is therefore likely that the increase in pausing frequency and pause duration originate from the enhanced affinity of the Y276H RdRp for the 3’-end of nascent RNA, which ultimately leads to the observed inability of the Y276H RdRp to backtrack.
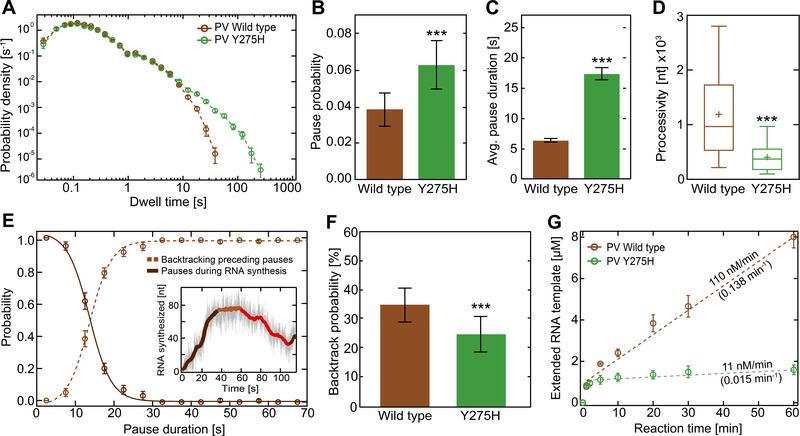
The availability of the orthologous derivative in PV, Y275H RdRp, provided the opportunity to determine if the mechanism causing the recombination defect is conserved for both viral polymerases. As shown in Figure 4A–D, Y275H RdRp and Y276H RdRp behaved similarly relative to their respective WT RdRp (based on features of the dwell-time distribution, including pause probability and average pause duration, as well as processivity). In contrast to EV-A71 RdRp, backtracking of PV RdRp is readily detectable (Figure 4E, inset) due to the fact that the backtracked intermediate is its end point and not followed by a reversal. Similar to the trigger of reversals in EV-A71 RdRp (Figure 1F), backtracking of PV RdRp derived from a pause-dependent mechanism (Figure 4E). Therefore, for Y275H RdRp, a decrease in the incidence of backtracking should be discernible, providing empirical validation of our hypothesis that recombination defects in both polymerases may originate from a conserved backtracked intermediate. Indeed, the probability of backtracking was reduced for Y275H RdRp relative to WT RdRp (Figure 4F). The significant reduction in backtracking probability for Y275H RdRp may reduce the incidence of template-switching-proficient backtracks. Similar to the recombination-defective EV-A71 polymerase, Y275H RdRp exhibits a substantially more stable elongation complex than WT (Figure 4G).
Figure 4. PV Y275H RdRp is impaired for backtracking.
(A) Superimposed dwell-time distributions of poliovirus Y275H RdRp variant (green) and WT (brown) RdRp. The 275H variant exhibits a broad increase in the probability and duration of long pauses compared to WT. The dwell-time window was set to 4 nt, and the error bars (AVG ±SD) result from bootstrapping with 1,000 iterations. (B-D) Quantification of the data in panel (A). In comparison to WT RdRp, the Y275H variant shows a significant increase in (B) average pausing probability (±SD) and (C) average apparent pause duration (±SEM) during RNA synthesis. The processivity (D) is significantly decreased for the Y265H RdRp variant compared to WT. (E) The probability of observing pauses of specified durations during PV WT RNA synthesis (dark brown) co-plotted with the probability of observing backtracking (light brown dashed line); the crossover point is found at ~14 s. (F) The Y275H variant (green) shows a significantly decreased backtracking probability (AVG±SD) than WT RdRp (brown). (G) In vitro bulk RNA synthesis assay results (AVG±SD; n = 3 repetitions) showing the amount of extended Sym/SubU template for PV WT and Y275H RdRp over time. Dashed lines represent linear regressions fitted to the data, revealing significantly lower approximated rates of RNA extension (values above the dashed lines) and turnover (values below the dashed lines; unit: RNA min−1 RdRp−1) for the Y275H RdRp variant compared to WT. Statistical analyses were performed using unpaired, two-tailed t-tests (significance level p: *** ≤ 0.001).
Conformational dynamics of the RNA-binding channel as a determinant of the type and efficiency of RdRp-catalyzed template switching
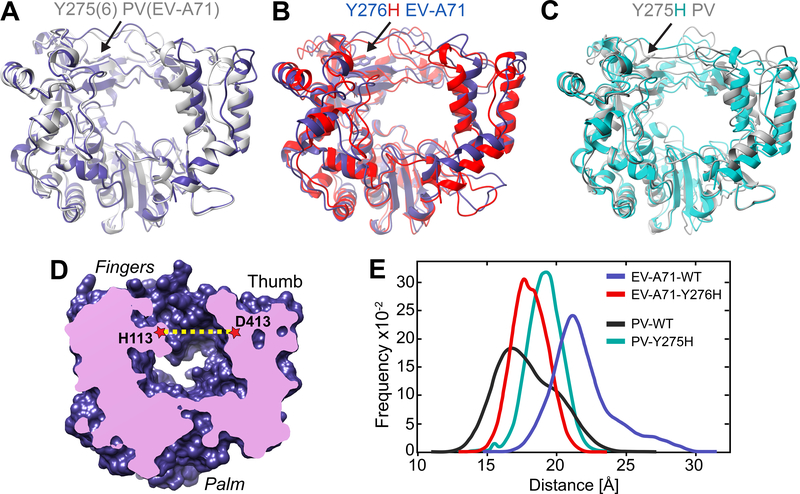
It has become increasingly clear that the biochemical properties of enzymes are governed as much by their conformational dynamics as by their three-dimensional structure (Cameron et al., 2009; Malone et al., 2021; Moustafa et al., 2011). The conformational dynamics of PV RdRp is an important contributor to the specificity and efficiency of its polymerase function (Cameron et al., 2016). The availability of structures of the RdRp from PV and EV-A71 (Gong and Peersen, 2010; Shu and Gong, 2016), and models of the PV Y275H and EV-A71 Y276H RdRps produced for this study, permitted us to determine whether structure and/or dynamics explain the differences observed in the propensity for a RdRp to catalyze copy-back RNA synthesis or the impact of the Y275(6)H substitution on backtracking or intramolecular template switching. Structural differences provided little, if any, insight (Figure 5A–C); therefore, we turned to dynamics.
Figure 5. RNA-duplex channel dimensions and conformational dynamics of EV-A71 and PV RdRps.
(A) Superimposed crystal structures of EV-A71 WT (blue) and PV WT (grey) shown as cartoons. (B) Superimposed crystal structures of EV-A71 WT (blue) and major conformation of its Y276H mutant (red). (C) Superimposed crystal structures of PV WT (grey) and major conformation of its Y275H mutant (cyan). The major conformation of the Y275(6)H mutants resulted from MD simulations. (D) Cut-through volume rendering of EV-A71 WT crystal structure (PDB 3N6L), where the RNA duplex channel can be observed in the center of the structure. The channel width was assessed by measuring the distance between His-113 at the fingers and Asp-413 at the thumb domains in EV-A71, or their equivalent residues Ser-112 and Asp-412 in the PV WT crystal structure (PDB 1RA6). (E) RNA duplex channel widths measured for EV-A71 WT (blue) and PV WT (black) from their crystal structures, and from MD simulations for the EV-A71 Y276H variant (red).
We have previously established a pipeline for evaluating the conformational dynamics of picornaviral polymerases by using molecular dynamics simulations (MD) (Moustafa et al., 2011, 2014). This pipeline evaluates correlated motions of the conserved structural motifs and in reports on the dynamics of the open and occluded states of the catalytic site that drive specificity of nucleotide selection (Moustafa et al., 2014). The pipeline also evaluates dynamics of the RNA-binding channel. Here, we have monitored the time-dependent changes in the distance of two residues lining opposite sides of the RNA-binding channel: H113 and D413 in case of EV-A71 (Figure 5D), and S112 and D412 in case of PV. The average size of the RNA-binding channel of PV WT RdRp was smaller (by ~4 Å) than that of EV-A71 WT RdRp (Figure 5E).
Interestingly, introduction of the EV-A71 Y7276H substitution decreased the average size of the RNA-binding channel by ~3 Å over the duration of time sampled (Figure 5E) to a similar size observed for PV WT RdRp. In contrast, PV Y275H exhibited a ~2 Å increase, rendering the channel size similar for both Y275(6)H RdRp variants.
The observed differences in the conformational dynamics of the RNA duplex channel strongly suggest that this parameter could influence the ability and degree of copy-back RNA synthesis. In particular, copy-back RNA synthesis of the nascent RNA strand by EV-A71 WT RdRp may be facilitated by increased spatial dynamics in the active center that allow the newly synthesized RNA strand to form a new primer by “snap-back” priming after backtracking.
RdRp backtracking in response to incorporation of the pyrazine carboxamide, T-1106, promotes intra- and intermolecular template switching in vitro and in cells.
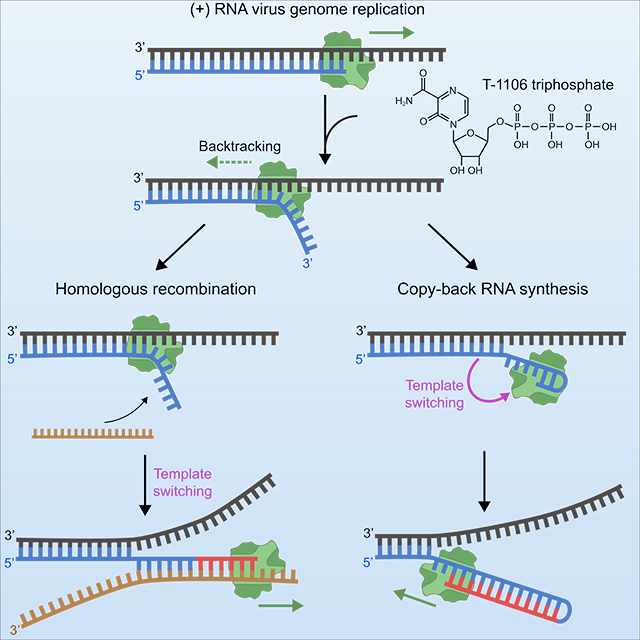
Our study thus far makes a very compelling case for the ssRNA 3’-end produced by RdRp backtracking serving as an intermediate for both intra- and intermolecular template switching. Our interest in backtracking, however, was motivated by our previous observation that incorporation of T-1106 ribonucleoside triphosphate (T-1106-TP) into nascent RNA induced backtracking in PV (Dulin et al., 2017). In the context of our current findings, we would expect T-1106 ribonucleotide to promote copy-back RNA synthesis and/or homologous recombination in EV-A71. If this is the case, then T-1106 may actually represent an antiviral ribonucleoside whose antiviral activity could be attributed to a post-incorporation event unrelated to termination or mispairing, in this case template switching.
We compared EV-A71 RdRp elongation dynamics in the magnetic tweezers in the absence and presence of T-1106-TP. Incorporation of T-1106 ribonucleotide induced elevated levels of pausing (Figure 6A–C), diminished processivity (Figure 6D), and led to an increased incidence of reversals (Figure 6E) whose dynamics (Figure 6A) were, as in the absence of drug, comparable to those of forward translocation. Together, these behaviors reflect increased intramolecular template switching (copy-back RNA synthesis) in the presence of T-1106-TP. We also verified that the outcome of T-1106-TP utilization by PV RdRp was as expected based on our previous study (Figure S3) (Dulin et al., 2017). Indeed, T-1106-TP induced detectable backtracking by PV RdRp (Figure S3D), consistent with a greater likelihood for intermolecular template switching (homologous RNA recombination).
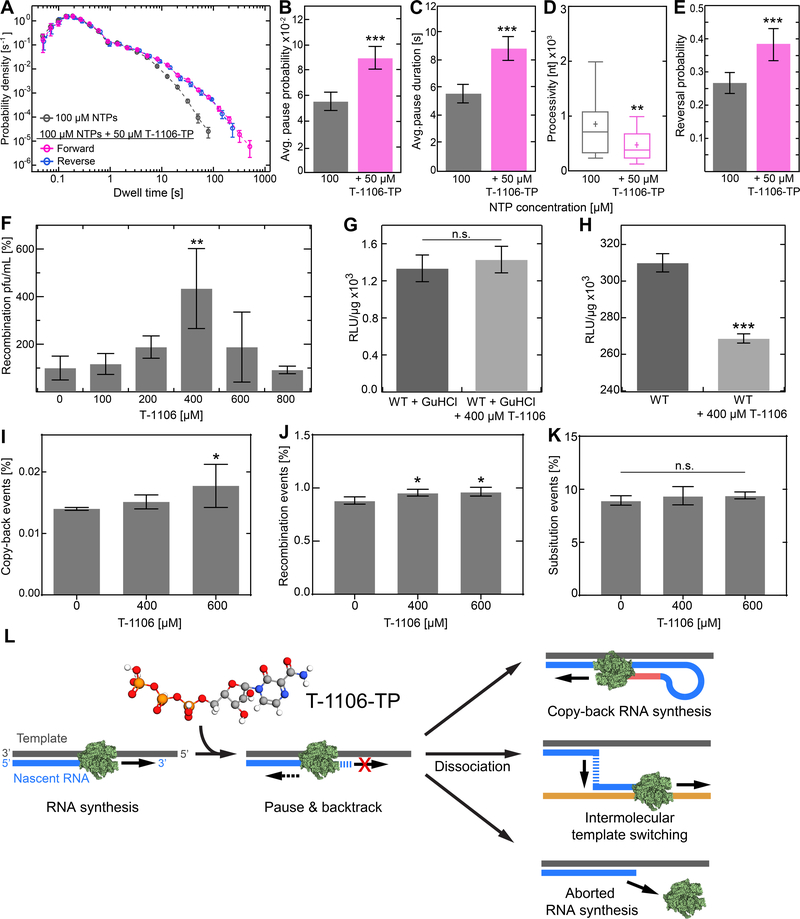
Figure 6. Pyrazine carboxymide T-1106 induces an increase in intra- and intermolecular template switching in vitro and in cell.
(A) Superimposed dwell-time distributions of EV-A71 RdRp forward RNA synthesis activity in the presence (magenta) or absence (grey) of the nucleotide analogue T-1106-TP. The reverse RNA synthesis dwell-time distribution in absence of T-1106-TP (blue) is superimposed. The dwell-time window was set to 4 nt, and the error bars (±SD) result from bootstrapping with 1,000 iterations. (B-E) The addition of T-1106-TP changed significantly the pausing behavior of WT RdRp, exhibiting significantly increased (B) pausing probability (±SD) and (C) average pause duration (±SEM), which led to a (D) significantly decreased in RNA synthesis processivity, and (E) vastly increased copy-back RNA synthesis probability. (F) Cell-based recombination assays conducted in presence of different concentrations of T-1106. Relative viable WT recombinant yield, normalized as a percentage of a carrier (DMSO) treated control (AVG ±SD; N = 3 replicates for each condition). Calculated IC50 amounts to 340±140 μM T-1106; the HeLa cell toxicity (CC50) of T-1106 was found to be >2 mM (Dulin et al., 2017). (G, H) EV-A71 donor translation (G) and replication (H) efficiency (AVG±SD) for WT RdRp. (I-K) The frequency of identified (I) copy-back RNA synthesis and (J) homologous recombination events, extracted from RNAseq, were significantly increased upon T-1106 addition, while (K) no change in mutation occurrence within the EV-A71 genome was observed. (L) Model mechanism of pyrazine carboxymide nucleotide analogue causing stall in RNA synthesis leading to copy-back RNA synthesis, intermolecular template switching, or abortive genome synthesis by RdRp dissociation. Statistical analyses were performed using ANOVA with comparative Tukey post-hoc test (significance levels α: *** = 0.001; ** = 0.01; * = 0.05; n.s. = non-significant), and unpaired, two-tailed t-tests (significance level p: *** ≤ 0.001). See also Figures S3 and S4.
T-1106 inhibits EV-A71 multiplication in cells (Figure S4A). However, in the presence of T-1106 concentrations between 200 μM and 600 μM, a clear increase in recombination was observed (Figure 6F). Notably, the resulting IC50 value of 340±140 μM for EV-A71 is comparable with those previously found (T-1106: 510±30 μM; clinically used ribavirin: 550±30 μM) for PV RdRp (Dulin et al., 2017). Using a subgenomic replicon expressing luciferase (Figure 6G), we showed that T-1106 did not affect reporter expression in the presence of the replication inhibitor guanidine hydrochloride (GuHCl), consistent with the drug having no effect on translation of the viral polyprotein. Replication-dependent, reporter expression was inhibited (Figure 6H). Antiviral ribonucleosides that function by lethal mutagenesis do not significantly reduce replication-dependent, reporter expression (Crotty et al., 2000). It is therefore possible that the observed inhibition reflects a reduction in the amount of replication-competent replicon RNA resulting from enhanced copy-back RNA synthesis in the presence of T-1106-TP, and concomitant production of truncated (defective) replicons.
Our results are so far consistent with the idea of ribonucleotide analogs promoting copy-back RNA synthesis and homologous RNA recombination. We performed the same experiments with the recombination-defective variant, Y276H EV-A71. With this variant, T-1106 failed to increase recombination (Figure S4C), consistent with the RdRp serving as the mediator of the effect of T-1106. Using the Y276H-encoding subgenomic replicon, we showed that translation was not impacted by the presence of T-1106, as observed for WT EV-A71 (Figure S4D). Replication-dependent reporter expression was not impaired by the presence of T-1106 as observed for WT EV-A71 (Figure S4E). This observation confirms that the reduction in replication-dependent reporter expression for WT EV-A71 (Figure 6H) was indeed caused by a backtracking-induced phenomenon like the production of defective viral genomes (Vignuzzi and López, 2019).
Given that the impact of the drug on template switching-dependent mechanisms underlying copy-back and homologous recombination is alleviated for the recombination-defective variant Y276H, does this mean that a facile route to the development of resistance to this mechanistic class of antiviral ribonucleotides exists? To address this possibility, we evaluated the sensitivity of Y276H EV-A71 to T-1106. There was no more than a 2-fold change in the observed sensitivity of this derivative to T-1106 relative to WT (compare panels A, B in Figure S4). Therefore, elimination of the template-switching dependent activity is insufficient for resistance to T-1106 and perhaps all members of the pyrazine-carboxamide class of antiviral ribonucleosides.
To directly assess the impact of T-1106 on the sequence of the viral genomes produced by WT EV-A71, we performed next-generation RNA sequencing on RD cells infected with EV-A71 WT in the absence and presence of T-1106. RNA sequencing revealed that T-1106 drug dosages between 400–600 μM increased the frequency of genomes with sequences consistent with copy-back RNA synthesis (Figure 6I) and homologous recombination (Figure 6J). We did not observe any evidence for triggers of template switching being related to sequence motifs in the absence or presence of T-1106 (Figure S5), in agreement with our magnetic tweezers results and our previous observations made in EV-A71 cell-based assays (Woodman et al., 2018). Interestingly, copy-back RNA synthesis occurred with higher probability in guanine- and cytosine-rich regions of the genome (Figure S5D), consistent with our in vitro single-molecule observations (Figure S2).
Collectively, these studies make an unambiguous mechanistic link between intra- and intermolecular template-switching processes by showing that they share the same backtracked intermediate produced by the same triggers. Notably, with respect to the potential mechanism of action of pyrazine carboxamide ribonucleoside analogs, the RNA sequencing results did not exhibit any change in the mutational load (Figure 6K) or termination of RNA synthesis. Instead, the mechanism of action of this class of antiviral ribonucleotides appears attributable to the induction of intra- und intermolecular template-switching leading to production of defective viral genomes.
DISCUSSION
Among the last frontiers in RdRp enzymology is a mechanistic description of RNA recombination. In most RNA viruses, recombination is primarily an RdRp-mediated process. In PV, which is one of the best studied RNA virus models, it has been shown that recombination involves an RdRp-mediated template-switching mechanism in cells (Kirkegaard and Baltimore, 1986) and that RdRp is sufficient to catalyze the template-switching reaction in vitro (Arnold and Cameron, 1999). Recently, a renaissance in the study of RNA recombination has begun, as evidenced by the development of cell-based assays to probe the incidence of recombination (Kempf et al., 2019; Lowry et al., 2014; Woodman et al., 2016, 2018).
In our earlier work that monitored PV RdRp elongation dynamics at the single-molecule level in vitro, we observed extensive RdRp pausing followed by backtracking in response to incorporation of diverse ribonucleotide analogs. This phenomenon was especially pronounced in the presence of the pyrazine carboxamide, T-1106 (Dulin et al., 2017), which is known now to also induce pauses and backtracking in SARS CoV-2 RdRp (Seifert et al., 2021). In a typical single-molecule experiment, we observed the extrusion of tens of nucleotides of ssRNA by PV RdRp in response to backtracking, the probability of which increased in response to T-1106-TP utilization (Dulin et al., 2017) (Figure S5E). Given sufficient time, the drug-induced ssRNA reannealed, the RdRp rebound to the primer-template junction, and elongation resumed.
Observation of such spatiotemporal dynamics of the nascent RNA-template-RdRp complex was unprecedented and motivated further study. Intriguingly, cell-based recombination studies in PV and other RNA viruses have shown that favipiravir and its derivate T-1106 increase recombination frequency (Figure 6E) (Abdelnabi et al., 2017; Arnold and Cameron, 2004; Eyer et al., 2018; Gowen et al., 2010; Julander et al., 2007). Since PV RdRp backtracking propensity represented the sole change in RNA synthesis dynamics in response to T-1106 treatment, it could represent a recombination intermediate. With that possibility in mind, this study added the EV-A71 RdRp, because EV-A71 has been suggested to undergo high rates of recombination in nature (Woodman et al., 2018). EV-A71 recombination is a major cause of the recurring outbreaks in Asia (Lee et al., 2018; Puenpa et al., 2019).
The first major conclusion of this study is that the viral RdRp has evolved to sense and to respond to incorporation of an incorrect nucleotide or nucleotide analogue by pausing and backtracking, thereby producing a recombinogenic 3’-end. Nucleotide misincorporation will be increased at lower nucleotide concentrations and in the presence of skewed nucleotide pools (Dulin et al., 2017). Under these conditions, both EV-A71 and PV RdRps exhibit an increase in the probability and duration of pausing (Figures 1C, S1B, respectively) (Dulin et al., 2017). Similarly, utilization of T-1106-TP increases the pause probability and duration for both polymerases (EV-A71: Figure 6B–C; PV: Figure S3B–C). The average pause duration is measured in the tens of seconds for both enzymes (EV-A71: Figure S2B; PV: Figure S3C), not milliseconds as typically observed for nucleotide addition (Dolan et al., 2018). We suggest that the consequence of a misincorporation- or ribonucleotide analogue-induced pause is backtracking in both systems. In the case of PV RdRp, there is no debate because the backtracked state was observed to accumulate during the course of an experiment (Figure 4F) and does so more frequently upon utilization of T-1106 triphosphate (Figure S5E) (Dulin et al., 2017). In contrast, several known chain terminators did not induce backtracking of PV RdRp (Dulin et al., 2017). With EV-A71 RdRp, the backtracked state does not accumulate. Instead, the probability of reversals increases (Figures 1D, 6D), which reflects the use of the 3’-end of nascent RNA as a new primer for copy-back RNA synthesis (Figure 1G). Backtracking would thus be the simplest mechanism for liberating the 3’-end for use as a primer.
While it has been speculated that RNA recombination and copy-back RNA synthesis are mechanistically related based on observations that circumstances which elevate RNA recombination also elevate copy-back RNA synthesis (Vignuzzi and López, 2019), how these processes were connected was unknown. Our study is consistent with two fates for the ssRNA produced by backtracking. The first is intermolecular template switching, which is homologous RNA recombination. The second is intramolecular template switching, which is copy-back RNA synthesis. This conclusion is supported by RNAseq analysis of EV-A71 genomes produced during replication in cells in the absence or presence of T-1106 (Figure 6I–J). Therefore, this study makes an unequivocal mechanistic link between these two template-switching processes by demonstrating that they share the same backtracked intermediate and triggers.
Why are there two different outcomes for the backtracked state? This state accumulates with the PV RdRp, but does not accumulate with the EV-A71 RdRp. For copy-back RNA synthesis to occur, the nucleic-acid-binding site needs to be sufficiently large to accommodate a three-stranded intermediate at the time of initiation (Figure 1G). Analysis of the dynamics of the nucleic-acid-binding site of EV-A71 RdRp demonstrated a clear ability of this enzyme to accommodate such an intermediate with an approximate diameter of ~24Å. (Figure 5D). The corresponding site in PV RdRp is on average 4 Å smaller (Figure 5D). Presumably, the dimensions of PV RdRp are too small to accommodate such an intermediate. If so, then the dimensions of the RNA-binding site sampled by an RdRp may predict the capacity of a virus polymerase to catalyze copy-back RNA synthesis.
The product of copy-back RNA synthesis will lead to a truncated dsRNA that is defective in its coding capacity and/or a potent activator of innate immune responses (Vignuzzi and López, 2019). Whether or not production of copy-back RNA is deliberate is unclear but may be for some viruses because the copy-back RNA can dampen the intensity of the infection (Vignuzzi and López, 2019). During replication in the cell when the backtracked state arises, templates complementary to the single-stranded 3’-end would be present. Under these conditions, perhaps intermolecular template switching would perhaps be favored for both enzymes.
Over the past few years, a connection has been made between RdRp fidelity and recombination (Korboukh et al., 2014; Lee et al., 2018; Li et al., 2019; Lowry et al., 2014; Poirier et al., 2016; Woodman et al., 2018). With more incorporation errors and/or consumption of nucleotide analogs by the RdRp comes an increase in the frequency of recombination both in cells and in test tubes (Lee et al., 2018). The reason for this has not been clear. Our studies provide a mechanism. Recombination is thought to provide a means to suppress the impact of deleterious mutations (Dolan et al., 2018; Kempf et al., 2019; Xiao et al., 2016). Our data would suggest that a balance between mutation and recombination must exist, because increasing the frequency of recombination exhibits antiviral activity in cells (Figures 6F,I, S4C) attributable to production of defective genomes (Figure 6G). Consistent with too much recombination being inhibitory to virus genome replication, not every event that could trigger recombination (Figures 1C, 6B, and S3B) actually does so; only a fraction of paused polymerases actually undergo copy-back synthesis (Figures 1E, 6D) or accumulate in the backtracked state (Figure S3E). Our previous study of PV RdRp suggested that only a subset of elongation complexes were competent for misincorporation or incorporation of certain nucleotide analogs (Dulin et al., 2017). If this is the case, then these misincorporation-competent elongation complexes may be the only complexes competent for backtracking and template switching.
For an event to trigger template switching, that event must first trigger backtracking. Our studies have focused on the perception of nucleotide addition as correct or incorrect as a mechanism to induce backtracking and consequentially template switching. However, other mechanisms to induce backtracking may exist. One intriguing possibility is that certain template sequences may direct the RdRp to pause and/or backtrack. Sequence- and factor-dependent pausing is well represented in the DNA-dependent RNA polymerase literature (Kang et al., 2019). In the case of the respiratory syncytial virus (RSV), it is known that guanine- and cytosine-rich regions promote copy-back RNA synthesis (Sun et al., 2019). In this system, these sequences alter polymerase elongation capacity. It is intriguing to speculate that these sequences promote backtracking. While our single-molecule and RNAseq experiments did not correlate backtracking to specific sequences, the incidence of copy-back RNA synthesis increased in guanine- and cytosine-rich regions of template (Figures S3, S5).
Our interpretation that the backtracked state and copy-back synthesis contributing to recombination are of biological relevance is supported by mechanistic evaluation of RdRp derivatives known to be defective for recombination in cells (Figure 2) (Acevedo et al., 2018). PV Y275H RdRp exhibits one of the strongest recombination-defective phenotypes described (Acevedo et al., 2018), but the molecular basis for the defect is not known for this mutant or the others reported in the PV system or other viral systems (Kempf et al., 2019, 2020; Xiao et al., 2016). Here, we show that EV-A71 Y276H RdRp exhibits an equally strong recombination defect in cells (Figure 2). Both the PV and EV-A71 recombination-defective derivatives retain the ability to sense and respond to errors based on their ability to pause even more frequently (and with longer duration) than WT (Figures 3B–C, 4B–C). The increased pausing did not translate to an equal increase in backtracks or copy-back synthesis (Figures 3F, 4F). The inability to produce the 3’ ssRNA intermediate following backtracking appeared to be caused by an increase in the stability of these enzymes with the 3’-end of nascent RNA-template duplex (Figures 3G, 4G). We conclude that release of the nascent RNA-template duplex by the RdRp is an obligatory step in converting the paused RdRp elongation complex into a backtracked state with a recombinogenic, single-stranded 3’-end. This level of detail for the mechanism of RdRp-mediated RNA recombination is unprecedented. Several recombination-defective/impaired RdRp variants are known (Kempf et al., 2019; Kim et al., 2019; Xiao et al., 2016), and it is possible that the mechanistic basis for the defects will be different (Kempf et al., 2020). Therefore, evaluation of these derivatives may enable a genetic dissection of the mechanism of template switching and illumination of steps and/or intermediates masked by analyzing WT RdRp.
Given the details on the mechanism of template switching elucidated by this study, it is now clear that the conformational dynamics of the RdRp required to support template switching are substantial. For intramolecular template switching (copy-back synthesis) to occur, the enzyme has to flip to move in the opposite direction and expand the RNA-binding channel to accommodate an additional strand of RNA. For intermolecular template switching (homologous recombination) to occur, the enzyme would have to relocate from the site of backtracking to the new primed-template junction without dissociating into solution. Such polymerase acrobatics are well documented for HIV RT (Abbondanzieri et al., 2008; Liu et al., 2008, 2010). Interestingly, nucleotide analogs also promote substantial changes in the conformational dynamics of HIV RT, as observed here for the EV-A71 RdRp (Liu et al., 2010).
The pyrazine carboxamide ribonucleoside analog T-1106 does not appear to induce chain termination (Figures 6E, S3D) or lethal mutagenesis (Figure 6K), in contrast to previous accounts (de Avila et al., 2017; Jin et al., 2013b; Sangawa et al., 2013). For structurally similar analogs like ribavirin, which contains a triazole carboxamide pseudo base, lethal mutagenesis represents the main underlying mechanism of action (Dulin et al., 2017; Vignuzzi et al., 2005). While both analogs induce backtracking intermediates (EV-A71: Figure 6D; PV: Figure S3E), the antiviral mechanism of action of T-1106 appears to be limited to induced template-switching (Dulin et al., 2017). Perhaps the ability of the T-1106 pseudo base to form a transient basepair within nascent RNA enables priming of copy-back synthesis, a point to further examine in future studies.
Both past observations and recent investigations suggest that the identified recombination mechanism and its susceptibility to drugs may be conserved across RNA viruses. Historical findings described defective virus genomes (DVG) that likely originated from copy-back RNA synthesis for vesicular stomatitis virus (Lazzarini et al., 1981), Sendai virus (Re et al., 1983), measles, and parainfluenza (Calain et al., 1992; WHISTLER et al., 1996; Yount et al., 2006). Recent findings have confirmed this hypothesis for several viruses (Vignuzzi and López, 2019), and searched for underlying mechanism and trigger(s): in RSV, G:C-rich genome regions (rather than genome motifs) were found to promote copy-back RNA synthesis (Sun et al., 2019), a similar trigger to the one we identify here for EV-A71, and in SARS CoV-2 a sequence motif-independent RdRp pause/backtracking intermediate susceptible to T-1106 was identified (Malone et al., 2021; Seifert et al., 2021).
In conclusion, our studies reveal the power of magnetic tweezers for the mechanistic characterization of RdRp-catalyzed RNA recombination. The ability to detect an important recombinogenic intermediate and follow the fate of that intermediate provides the first direct mechanistic connection between RNA recombination and copy-back RNA synthesis. We have further elaborated the mechanism of action of the pyrazine carboxamide class of compounds and discovered that T-1106 functions by promoting formation of the recombinogenic intermediate. This leads us to suggest that this class of compounds may be particularly efficacious against those viruses either known to produce defective viral genomes by copy-back RNA synthesis or associated with high rates of recombination. An important aspect of this class of antiviral compounds is that the barrier to resistance for the virus may be insurmountable. Mutants that were resistant to T-1106-induced template switching remain highly sensitive to the drug, presumably due to an inability to combat the deleterious effects of accumulated mutations (Dolan et al., 2018; Kempf et al., 2019; Xiao et al., 2016). Altogether, our results define inducible intra-and intermolecular template switching (Figure 6K) as a tractable mechanistic target with broad-spectrum appeal.
Limitation of the study
A limitation of the study is the inability to interrogate directly the RNA products of RdRp activity that are produced in the magnetic tweezers. Therefore, we have relied on the intersection of the biophysical and biochemical experiments to associate reverse RNA synthesis signatures with copy-back RNA synthesis, and to demonstrate that copy-back synthesis and homologous recombination respond to similar triggers that result in kinetic intermediates of the RdRp.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Prof. Nynke H. Dekker (n.h.dekker@tudelft.nl).
Materials availability
This study did not generate new unique reagents.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table. The single-molecule data reported in this study have been deposited at a publicly accessible repository. The DOI is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Digoxigenin antibodies | Roche | RRID: AB_514496 |
| Bacterial and virus strains | ||
| SURE Competent Cells | Agilent technologies | Cat#200238 |
| Chemicals, peptides, and recombinant proteins | ||
| ApU dinucleotide | IBA Lifesciences GmbH | Cat#0-31004 |
| Biotin-16-dUTP | Roche | Cat#11093711103 |
| Digoxigenin-11-dUTP | Roche | Cat#11093681103 |
| rNTPs | GE Healthcare | Cat#27-2025-01 |
| Streptavidin-coated superparamagnetic beads | Thermo Fischer | Cat#65001 |
| T-1106 triphosphate | Blake Petersen Lab | |
| Ribavirin triphosphate | Jena Bioscience | Cat#NU-1105L |
| Dulbecco’s Modified Eagle’s Medium | Thermo Fischer | Cat#31966021 |
| Streptomycin | Thermo Fischer | Cat#15140122 |
| Superase RNase inhibitor | ThermoFischer | Cat#AM2694 |
| Critical commercial assays | ||
| Ribomax large scale RNA production kit | Promega | Cat#P1300 |
| T7 mMESSAGE mMACHINE kit | Thermo Fischer | Cat#AM1344 |
| Qubit RNA Broad Range assay kit | Thermo Fischer | Cat#Q10211 |
| TransIT-mRNA transfection kit | MirusBio | Cat#MIR2250 |
| Direct-zol-96 RNA purification kit | Zymo Research | Cat#R2056 |
| Quant-iT RNA assay kit | Thermo Fischer | Cat#Q33140 |
| Next rRNA Depletion Kit | New England Biolabs | Cat#E6310L |
| NEBNext Ultra II RNA Library preparation kit | New England Biolabs | Cat#E7775 |
| NextSeq500/550 Mid Output kit v2.5 | Illumina | Cat#20024907 |
| Mycoplasma Detection Kit | Lonza | Cat#LT07-318 |
| RNeasy MinElute cleanup kit | Quiagen | Cat#74204 |
| Deposited data | ||
| Single-Molecule Magnetic Tweezers data | This study | DOI: 10.4121/c.5608934 |
| Raw and processed RNAseq data | This study | NIH GEO: GSE183959 |
| Experimental models: Cell lines | ||
| RD cell line | ATCC | RRID: CVCL_1649 |
| Experimental models: Organisms/strains | ||
| Transgenic human hSCARB2–expressing mice | Fujii et al., 2013 | |
| Oligonucleotides | ||
| 5’-aacuguugguguacgcgaaagcgu | GE Healthcare Dharmacon | |
| 5’-taatacgactcactataggatcgccaagattagcggatcctacctgac | Biolegio | AB-For |
| 5’-ggttaacctcaacttccatttcc | Biolegio | AB-Rev |
| 5’-cccctcgaggggaaaaaaaaaaaaccgtatgacgctggaag | Biolegio | CD-For |
| 5’-taatacgactcactataggccggacgtttcggatcttccgacatgcgc | Biolegio | CD-Rev |
| 5’-aagattagcggatcctacctgac | Biolegio | Bio-For |
| 5’-bio-taatacgactcactataggaacggcttgatatccactttacg | Biolegio | Bio-Rev |
| 5’-agcgtaaaattcagttcttcgtggcg | Biolegio | Dig-For |
| 5’-dig-aatacgactcactatagggctaccggttaacctcaacttccatttcc | Biolegio | Dig-Rev |
| 5’-tgccattcagggactgccgatgtcggtgcagccg | Biolegio | SP-For |
| 5’-taatacgactcactataggagcgccgcttccatgtcctggaacgct | Biolegio | SP-Rev |
| 5’-acgttctcagtgcggactgtag | IDT | MP4-3156-Fwd |
| 5’-ccttggaaaagagcttc | IDT | C2/4-5057-Rev |
| 5’-gaagctcttttccaagg | IDT | C2/4-5057-Fwd |
| 5’-ctggttataacaaatttaccc | IDT | C2/4-7384-Rev |
| 5’-atcaatcacacccatcatgtgcatcgcaataaaacttattg | IDT | Y276H-Fwd |
| 5’-caataagttttattgcgatgcacatgatgggtgtgattgat | IDT | Y276H-Rev |
| 5’-acgtagcccagcgcgtcggccg | IDT | Eag-I-Rev |
| 5’-agcagtgctgactagtaaagacc | IDT | SpeI-Fwd |
| 5’-cggcagcccagaagaact | IDT | qPCR-Fwd |
| 5’-gccaccctatctccctgga | IDT | qPCR-Rev |
| 5’-FAM-tcaccatgaagttgtgtaaggatgcta-BHQ | Biosearch Technologies | qPCR-Probe |
| Recombinant DNA | ||
| Plasmid pBB10 | Petrushenko et al., 2006 | pBB10 |
| pSumoEV-A71-3D | This study | |
| pSumoEV-A71-3D-Y276H | This study | |
| pSumoPV-3D | Arnold J.J. & Cameron C.E., 2000 | |
| pSumoPV-3D-Y275H | Acevedo et al., 2018 | |
| pEV-A71-MP4 | Wang et al., 2004 | |
| pEV-A71-Y276H | This study | |
| pEV-A71-C2-replicon | Woodman et al., 2018 | |
| pEV-A71-MP4-Δ3D | Woodman et al., 2018 | |
| Software and algorithms | ||
| MatLab R13 | MathWorks Inc. | RRID: SCR_001622 |
| Igor Pro 6.37 | Wavemetrics | RRID: SCR_000325 |
| LabView 2011 | National Instruments | RRID: SCR_014325 |
| ImageQuant | GE Healthcare Life Sciences | RRID: SCR_014246 |
| ImageLab | BioRad | www.bio-rad.com/en-ch/product/image-lab-software |
| VODKA | (Sun et al., 2019) | |
| Bowtie2 | (Langmead and Salzberg, 2012) | RRID: SCR_016368 |
| BBTools | Joint Genome Institute | RRID: SCR_016968 |
| bcl2fastq v2.2 | Illumina | RRID: SCR_015058 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human embryonic rhabdomyosarcoma cell culture
Human embryonic rhabdomyosarcoma RD cells (ATCC CAT#CCL-136, RRID: CVCL_1649) were grown in Dulbecco’s Modified Eagle Medium (DMEM; ThermoFischer). Media was supplemented with 100 U/ml penicillin (ThermoFischer), 100 μg/ml streptomycin (ThermoFischer), and 10% Heat Inactivated (HI)-FBS (ThermoFischer). All cells were passaged in the presence of trypsin-EDTA. Cell were maintained at 37°C/5% CO2. Prior to the experiments, RD cells were tested for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza) and were negative: B/A ratio < 1; no mycoplasma reads were detected.
Generation of transgenic human hSCARB2–expressing mice
Transgenic mice expressing hSCARB2 were generated as described previously (Fujii et al., 2013), and kindly provided by Dr. Satoshi Koiki (Tokyo Metropolitan Institute of Medical Science, Japan). The mice were housed and fed at the National Laboratory Animal Center (NLAC), NARLabs, Taiwan. Experiments were carried out in accordance with the ‘Guide for the care and use of laboratory animals’, the recommendations of the Institute for Laboratory Animal Research and Association for Assessment and Accreditation of Laboratory Animal Care International standards. The animal experiment protocol was approved by the Institutional Animal Care and Use Committee in the Chang Gung University (CGU 106–117), Taiwan. The hSCARB2-expressing mice were housed at room temperature ranging between 20 and 23 °C with a relative humidity between 55 and 60%, and kept under a 12/12 h light/dark cycle. For the experiments, three-week-old female and male pathogen-free hSCARB2 mice were randomly distributed in the experimental groups of animals.
METHOD DETAILS
Plasmids
The mouse adapted EV-A71 C2-MP4 infectious clone was kindly provided by Dr. Jen-Reng Wang (Cheng Kung University, Taiwan) and modified by insertion of a ribozyme sequence between the T7 promoter and viral genome sequence in a pBR-derived plasmid (Kung et al., 2010; Petrushenko et al., 2006).The EV-A71 C2 replicon was modified from a previously described EV-A71 C2–2231 replicon by addition of a T7-ribozyme and polyA sequence inserted at the 5’ and 3’ end of the replicon sequence in a pBR-derived plasmid (Tang et al., 2016). The EV71Δ3D template was constructed from the full-length EV-A71 C2-MP4 infectious clone by removal ~800 nt between the blunt cutting restriction sites (ScaI and NruI) within the 3Dpol coding region. The Y276H mutant replicon and infectious clone were constructed by using site-directed mutagenesis. The sequences of the primers (Integrated DNA Technologies, Inc.; IDT) used for the plasmid construction are described in the Key Resources Table.
Purification, 5′−32P Labeling, and Annealing of sym/sub
RNA oligonucleotides were purified, labeled, and annealed as described previously (Cameron et al., 2009). Poliovirus RNA-dependent RNA polymerase (3Dpol): Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub).
Expression and purification of Enterovirus A-71 RdRp
Mutation of the Y276 codon was performed by standard PCR mutagenesis. Expression and purification of WT and mutant 3Dpol enzymes followed previous procedures with some minor modifications (Arnold and Cameron, 1999; Arnold et al., 2006; Gohara et al., 1999). 3Dpol is expressed as a fusion protein to SUMO and an N-terminal polyhistidine tag that increases protein production, eases purification and allows for production of 3Dpol with the naturally occurring Gly1 (Arnold et al., 2006; Gohara et al., 1999).
Protein purification: buffer B (100 mM potassium phosphate, 500 mM NaCl, 5 mM imidazole, 5 mM β-mercaptoethanol, 60 μM ZnCl2, 20% w/v glycerol, pH 8.0) and buffer C (100 mM potassium phosphate, 500 mM NaCl, 60 μM ZnCl2, 5 mM β-mercaptoethanol, 20% w/v glycerol, pH 8.0) were prepared. Cell pellets were resuspended in 50 ml lysis buffer (50 ml buffer B, 1.4 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 mM PMSF, 0.1% N-P40) and subjected to sonication. Cell lysates were centrifuged at 30,000 g and 4°C for 30 min. Supernatant was applied to Ni-NTA (Invitrogen) columns pre-equilibrated with buffer C1 (buffer C, 5 mM imidazole and 0.1% N-P40). The resin was washed with three bed volumes each of buffer C1 and buffer C2 (buffer C, 5 mM imidazole), and protein was eluted using high imidazole buffers C3 (buffer C, 50 mM imidazole) and C4 (buffer C, 500 mM imidazole). The polyhistidine tag and SUMO protein domain were cleaved from 3Dpol using the protease Ulp1. Protein solutions were dialyzed against 80 mM Tris-HCl, 500 mM NaCl, 20% w/v glycerol, 10 mM β-mercaptoethanol, 60 μM ZnCl2, pH 8.0 overnight (optimal buffer for protease cleavage), and then dialyzed against 100 mM potassium phosphate, 20% w/v glycerol, 10 mM β-mercaptoethanol, 60 μM ZnCl2, pH 8.0 for 2–3 h. A second Ni-NTA column was used to separate the purified 3Dpol from the cleaved polyhistidine tag/SUMO domain, using procedures identical to the first Ni-NTA column. For WT and Tyr276His 3Dpol, additional phospocellulose and Q-sepahrose columns were used to ensure protein solutions were free of trace contaminants of nuclease and phosphatase activities (Arnold et al., 2006; Gohara et al., 1999). Proteins were >95% homogeneous as estimated by coomassie-blue staining of SDS PAGE gels. Protein is stable at 4°C for 3–4 months under high salt conditions (80 mM Tris-HCl, 500 mM NaCl, 20% w/v glycerol, 10 mM β-mercaptoethanol, 60 μM ZnCl2, pH 8.0).
In vitro RNA synthesis, cell transfection, and recombinant virus quantification
The EV-A71 C2 replicon and C2-ΔIRES-replicon were linearized with SalI. The EV-A71-MP4, EV71Δ3D cDNA were linearized with EagI. All linearized cDNA was transcribed in vitro using T7 RNA Polymerase treated with 2U DNAse Turbo (ThermoFisher) to remove residual DNA template. The RNA transcripts were purified using RNeasy Mini Kit (Qiagen) before spectrophotometric quantification. Purified RNA (amounts as specified elsewhere) in RNase-free H2O were transfected into cell lines using TransMessenger (Qiagen). The mixture was incubated according to the manufacturer’s instructions and added to RD cell monolayers in 12-well tissue culture plates. Virus amount was quantified by plaque assay. Briefly, media supernatant and cells were harvested at time-points post transfection (specified in the main text), subjected to three freeze-thaw cycles and clarified. Supernatant was then used on fresh RD cells in 12-well plates, virus infection was allowed to continue for 30 min. Media was then removed, and cells were subjected to 2x PBS (pH 7.4) washes before a 1% (w/v) agarose-media overlay was added. Cells were incubated for 3–4 days and then fixed and stained with crystal violet for virus quantification.
Single-step growth curve for EV-A71 WT and Y276H full-length virus
RD cells in 12-well plates were infected by each virus at a MOI of 0.1 in triplicate in serum-free media. After 1 h, cells were extensively washed by PBS and refreshed in 10% serum-containing media. Virus was harvested at different time-points post infection and the virus yield was quantified by plaque assay.
Real time qPCR analysis
Viral RNA was isolated with QiaAmp viral RNA purification kit (Qiagen), as recommended by the manufacturer. The real time qPCR analysis was performed by the Genomics Core Facility of The Pennsylvania State University. DNase-treated RNA was reverse-transcribed using the High Capacity cDNA reverse transcription kit (Applied Biosystems) and the protocol provided with the kit. Quantification by real time qPCR was done by adding 10 or 20 ng of cDNA in a reaction with 2x TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City CA) in a volume of 20 μl, with primers 5′-CGGCAGCCCAGAAG AACT-3′ (forward) and 5′-GCCACCCTATCTCCCTGGAT-3′ (reverse) and probe 5′-[6-Fam]-TCACCATGAAG TTGTGTAAGGATGCTA-3′ in a 7300 real time qPCR machine (Foster City). A standard curve was generated using in vitro transcribed RNA.
Luciferase assays
Supernatant was removed from transfected cell monolayers, and cells were briefly washed with PBS and lysed using 100 μl 1x Glo Lysis Buffer (Promega®) per well in a 12-well plate. The oxidation reaction was catalyzed by the addition of 10 μl cell lysate to 10 μl room temperature Bright-Glo Luciferase Assay System (Promega®) substrate. Luciferase activity was measured using a luminometer with values normalized to protein content of the extract using a protocol as previously described (Arnold et al., 2006).
Infection of transgenic human hSCARB2–expressing mice
For infection of mice expressing hSCARB2, 2×107 genome copies of WT and Y276H mutant EV-A71 C2-MP4 viruses were intragastrically inoculated into 21-day old mice using metallic gastric tubes. The inoculated mice were monitored daily for clinical disease symptoms, reflected by the disease scores defined as: 1. jerky movement; 2. paralysis of one hind leg; 3. paralysis of both hind legs; 4. death.
In vitro transcription of EV-A71 genomes for RNAseq
Mouse-adapted EV-A71 C2-MP4 cDNA was linearized with EagI. The linearized cDNA was in vitro transcribed using the T7 mMESSAGE mMACHINE kit (ThermoFisher) following manufacturer’s instructions. Reactions were treated with TURBO DNase (2U/reaction) to remove the DNA template. The RNA transcript was purified using the RNeasy MinElute Cleanup Kit (Qiagen). Purified RNA was quantified via Qubit 4 fluorometer using the Qubit RNA Broad Range assay kit (ThermoFisher). The quality of the transcripts was confirmed by agarose gel electrophoresis.
Virus infection with EV-A71 and rescue
In vitro transcribed RNA was transfected into RD cells via TransIT-mRNA transfection kit (Mirus Bio) following the manufacturer’s instructions. Briefly, RD cells were seeded in 6-well tissue culture plates at 8.0 × 105 cells/well (DMEM-GlutaMAX, 10% FBS, 1% P/S) and incubated overnight at 37°C/5% CO2. The day of the transfection, media was replaced with 2% FBS-containing media and then the transfection mixture containing 2.5 μg of EV-A71 C2-MP4 RNA was added dropwise. 48 h post-transfection, at nearly full CPE, the cells were freeze-thawed three times and then spun at 20,000 g for 10 min at 4°C to remove cellular debris. Viral titers were determined by plaque assay. This viral supernatant (P0) was passaged once to scale up and generate P1 virus, which was used for the subsequent RNAseq experiments.
Drug treatment of EV-A71-infected RD cells
RD cells were seeded in a 96-well tissue culture plate at 3.0 × 104 cells/well and incubated overnight at 37°C/5% CO2. Afterwards, cells were treated with T-1106 for 3 h before infection (doses are specified in Figure 6K). T-1106-treated cells were then infected at an MOI of 10 with EV-A71-C2-MP4 in triplicate. After infection, the cells were washed extensively with PBS, and the medium was replaced with T-1106. Infection proceeded until cytopathic effect was observed (48 h.p.i.).
Next-generation RNAseq
Total RNA was isolated with Direct-zol-96 RNA purification kit (Zymo Research) following manufacturer’s instructions. The recommended in-column DNase I treatment was also conducted to remove genomic DNA. Purified RNA was quantified with a microplate reader (Tecan M200 Infinite Pro) using the Quant-iT RNA assay kit (ThermoFisher). Ribosomal RNA was removed using the NEBNext rRNA Depletion Kit (Human/Mouse/Rat) (New England Biolabs). Sequencing libraries were prepared using the NEBNext Ultra II RNA Library preparation kit for Illumina (New England Biolabs) and the pooled library was loaded in a NextSeq500/550 Mid Output kit v2.5 (Illumina) for sequencing in a NextSeq500 (151 cycles, 8 nucleotides of index).
Bulk RdRp turnover experiments
1 μM WT and YH variants of poliovirus or EV-A71 C2-MP4 RdRp were incubated with 32P-labelled 20 μM RNA primer-template duplex (sym/subU) and 500 μM rNTPs. At various time points (described in text) the reaction was quenched by the addition of 500 mM EDTA and 35% formamide. Quenched reactions were mixed with an equal volume of loading buffer (75% formamide, 0.025% bromophenol blue and 0.25% xylene cyanol) and heated to 70°C prior to loading on a denaturing PAGE gel. Products were resolved from substrates by denaturing polyacrylamide gel electrophoresis (18.5% acrylamide, 1.5% bisacrylamide, 1x TBE buffer, 7 M Urea). Electrophoresis was performed in 1x TBE at 90 W. Gels were visualized using a PhosphorImager and quantified using ImageQuant software (GE Healthcare, RRID: SCR_014246).
RNA constructs for single-molecule RNA synthesis experiments
The RNA template used in our single-molecule assay is analogous to the sequences used in previous studies of PV and Φ6 RdRp RNA synthesis kinetics (Dulin et al., 2015, 2017). The RNA tether construct consists predominantly of a dsRNA, assembled by hybridization of a 2.8 kb template ssRNA to a 4.1 kb complementary strand, and two ~500 bases ssRNA strands containing either biotin or digoxigenin for the tethering between magnetic beads and the surface, as previously described in detail (Dulin et al., 2017). In contrast to the previously used hairpin for Poliovirus RdRp RNA synthesis initiation, terminating the 3’ end of the template, we used for the hairpin structure 24 bases with the following sequence: 5’-AACUGUUGGUGUACGCGAAAGCGU-3’. Essentially, the hairpin mimics a primer and promotes primer-dependent RNA synthesis initiation (Arnold and Cameron, 2000).
The RNA constructs were assembled by first subjecting Plasmid pBB10 to PCR amplification using primers AB-For, AB-Rev, CD-For, CD-Rev, Bio-For, Bio-Rev, Dig-For, Dig-Rev, SP-For, and SP-Rev, listed in the Key Resource Listing. Single-stranded RNA sequences were produced from these amplicons via in vitro run-off transcription using T7 RNA polymerase from the Ribomax large-scale RNA production system (Promega). Transcription reactions for the AB, CD and SP ssRNA fragments contained 500 ng DNA amplicon, 10 μl T7 buffer, 5 μl T7 polymerase, 1 μl 100 mM CTP, and 1 μl of 100 mM for all other NTPs (ATP, UTP, GTP) in a reaction volume of 50 μl. For the synthesis of BIO ssRNA, the UTP amount was reduced to half and the reaction mix was supplemented with 4.7 μl of 10 mM biotin-16-UTP (Roche), while for DIG ssRNA the UTP amount was reduced only to 0.63 μl with supplement of 3.7 μl or 10 mM digoxigenin-11-UTP (Roche, RRID: AB_514496). The synthesized ssRNA fragments were purified using a RNeasy MinElute cleanup kit (Qiagen) and eluted in 1 mM sodium citrate buffer (pH 6.4).
These different ssRNA fragments were then assembled into a dsRNA construct for the single-molecule studies conducted on Poliovirus and Enterovirus A-71 RdRp via hybridization. The ssRNA fragments were first mixed in equimolar ratio in 200 μl 0.5x SSC buffer, with the exception of the biotin (BIO) and digoxigenin (DIG)-enriched handles, which were added in four times molar excess in respect of the AB strand (1 μg AB, 1.4 μg CD, 360 ng SP, 450 ng BIO, and 516 ng DIG). Afterwards, the RNA mix was heated for 1 h to 65°C, cooled with a rate of 0.24°C/min down to 25°C. The final dsRNA construct was purified with RNeasy MinElute cleanup kit and eluted in 1 mM sodium citrate. All RNA concentrations were photometrically determined (Nanodrop).
Magnetic tweezers experimental configuration
The magnetic tweezers implementation used in this study has been described previously (Dulin et al., 2017; Janissen et al., 2018). Briefly, light transmitted through the sample was collected by a 50x oil-immersion objective (CFI 50XH, Plan Achromat, 50x, NA = 0.9, Nikon) and projected onto a 12-megapixel CMOS camera (#FA-80–12M1H, Falcon2, Teledyne Dalsa) with a sampling frequency of 50 Hz. The applied magnetic field was generated by a pair of vertically aligned permanent neodymium-iron-boron magnets (Webcraft) separated by a distance of 1 mm, suspended on a motorized stage (#M-126.PD2, Physik Instrumente) above the flow cell. Image processing of the collected light allows tracking the real-time position of both surface attached reference beads and superparamagnetic beads coupled to the dsRNA constructs in three dimensions over time. The bead x, y, z position tracking was achieved using a cross-correlation algorithm realized with custom-written software in LabView (2011, National Instruments Corporation, RRID: SCR_014325). In this software, bead positions were determined with spectral corrections to correct for camera blur and aliasing (Cnossen et al., 2014).
Single-molecule RdRp RNA synthesis assay
The flow cell preparation used in this study has been described in detail elsewhere (Dulin et al., 2017; Janissen et al., 2018). In brief, polystyrene reference beads (#17133, Polysciences GmbH) of 1.5 μm in diameter were diluted 1:1500 in PBS buffer (pH 7.4; Sigma Aldrich) and then adhered to the nitrocellulose-coated (Invitrogen) surface of the flow cell. Afterwards, digoxigenin antibodies (Roche, RRID: AB_514496) at a concentration of 0.1 mg/ml were incubated for 1 h within the flow cell, following a 2 h incubation of 10 mg/ml BSA (New England Biolabs) diluted in PBS (pH 7.4) buffer. After washing with PBS buffer, 100 μl of streptavidin-coated superparamagnetic beads (DynaBeads, LifeTechnologies; prior diluted 1:400 from stock) with a diameter of 1.5 μm were added resulting in the attachment of the beads to the surface-tethered dsRNA constructs. Afterwards, unbound beads were washed out with PBS buffer.
The preparation of RdRp:RNA ternary complexes was also performed as described previously (Dulin et al., 2015, 2017). Briefly, 1 μM RdRp in 100 μl EV buffer (50 mM HEPES, 5 mM MgCl2, 125 μg/ml BSA, 1 mM DTT, 1 U Superase RNase inhibitor, pH 6.6), supplemented with 600 μM ATP, 600 μM CTP, and 1.2 mM ApC, was flushed into the flow cells containing the dsRNA constructs. Stalled RdRp complexes were formed during 20 min of incubation at room temperature. Afterwards, the flow cell was washed with EV buffer and the RNA chain elongation was re-initiated by adding all four rNTP in an equimolar concentration of 1 mM, unless stated otherwise, and in presence or absence of nucleotide analogs. The single-molecule measurements were conducted for 2 h at constant pulling forces of 25 pN at 24°C with a camera acquisition rate of 50 Hz.
Molecular Dynamics simulations (MD)
The starting coordinates for the all-atom MD simulations systems prepared by the accessory program “tleap” of AMBER18 suite (Case et al., 2018). The crystal structure 3N6L of EV71 RdRp was used to prepare the starting coordinates for MD simulations of the WT system (Wu et al., 2010). The Y276H mutant system prepared by in silico replacement of Tyr at position-276 by a His; the steric clashes generated in the mutant system removed by subsequent energy minimization and equilibration. The WT PV RdRp was investigated previously by all-atom MD simulations (150 ns), using the 1RA6 structure as starting coordinates for the MD simulations (Arnold and Cameron, 1999; Moustafa et al., 2014). The Y275H mutant of PV RdRp was prepared by replacing Tyr-275 of 1RA6 structure by histidine; energy minimization and equilibration of the system removed any steric clashes in the mutant system.
For the performed MD simulations, the protein parameters of Amber14SB force field was used during calculations (Maier et al., 2015). All MD simulations were performed in explicit water (TIP3P model), imposing a minimal distance of 12 Å between the edge of the solvent box and any protein atom (Jorgensen et al., 1983). Calculations of the non-bonded interactions used a cutoff radius of 9 Å with periodic boundary conditions applied; particle mesh Ewald method was used to treat electrostatic interactions (Darden et al., 1993). The SHAKE algorithm was employed to constrain hydrogens bonded to heavy atoms (Ryckaert et al., 1977). The simulations were performed by first relaxing the systems in two cycles of energy minimization using SANDER program. Subsequently, the systems were slowly heated to 300 K using the parallel version PMEMD under NVT conditions (constant volume and temperature); Langevin dynamics with collision frequency (γ = 2) was used to regulate temperatures (Feller et al., 1995). The heated systems were then subjected to equilibration by running 100 ps of MD simulations under NPT conditions (constant pressure and temperature) with 1 fs integration time steps. MD trajectories were collected over 200 ns at 1 ps interval and 2 fs integration time steps. Analyses of the trajectories from MD simulations were performed using CPPTRAJ program (Roe and Cheatham, 2013). MD simulations were carried out on a multi-GPU workstation with 32-core processor (AMD Ryzen Threadripper 2990WX) and four Nvidia GTX 1080 Ti graphics cards.
QUANTIFICATION AND STATISTICAL ANALYSIS
Next-generation RNAseq analysis
Reads were first demultiplexed using the bcl2fastq v2.2 conversion software (Illumina, RRID: SCR_015058). The reads were adapter- and quality-trimmed using BBDuk and the optical duplicates removed using Clumpify, all part of the BBTools suite (RRID: SCR_016968). Reads were aligned to EV-A71 WT reference genomes using bowtie2 (Langmead and Salzberg, 2012, RRID: SCR_016368). To identify deletions, the unaligned reads were processed via BBMap (a splice-aware aligner). To identify copybacks, reads were processed via VODKA, where the entire viral genome length was used as a template to generate the copyback index (Sun et al., 2019). In both cases, the output data was analyzed using an in-house R script.
Single-molecule data processing
RNA synthesis trajectories were processed using custom-written Igor v6.37 (RRID: SCR_000325) and MatLab R2013b (RRID: SCR_001622)-based custom-written scripts. The measured z-positions of magnetic beads during the RNA synthesis process were converted to synthesized RNA products as a function of time, using the empirically determined force-extension relationships for dsRNA and ssRNA molecules under the employed buffer conditions (Dulin et al., 2015, 2017). Briefly, for the conversion from bead z-position to synthesized nucleotides, we used the formula:
where for the applied force F, N represents the number of nucleotides of the template (2,820 nt), L(F) is the measured extension of the tether, Lds(F) is the full dsRNA length, and Lss(F) is the full length of a ssRNA tether. To reduce the effect of Brownian noise in the applied statistical analyses, all measured elongation trajectories were filtered to 1 Hz using a sliding mean average filter.
Statistical dwell time analysis of single RdRp elongation trajectories
The stochastic RNA synthesis dynamics of EV-A71 and PV RdRp were quantitatively assessed by a statistical analysis of elongation and pausing using a recently described bias-free dwell-time analysis (Dulin et al., 2015, 2017; Janissen et al., 2018). Using this approach, the times needed for RdRp to elongate through consecutive dwell time windows of four nucleotides - defined as dwell times - were determined for all measured RdRp trajectories under the same conditions to construct dwell time probability distributions (e.g. Figure S1A). The dwell times were bootstrapped 1,000 times to estimate the standard deviation and confidence intervals of the distributions (Dulin et al., 2017; Janissen et al., 2018). All dwell-time distributions qualitatively exhibit the same features, such as a peak at short time scales (~200 ms) and a tail of gradually decreasing probability in time scales ranging between 1 and 1,000 s. While the peak reflects fast kinetic processes consisting of nucleotide addition, NTP hydrolysis, PPi release, and translocation, the tail originates from off-pathway pauses, consistent with the previous observations for the PV RdRp (Dulin et al., 2017).
The quantitative description of the duration and probability of pauses during RNA synthesis derive from these dwell time distributions. To calculate the average pause probabilities (±SD), the dwell time distributions were integrated starting from a chosen threshold of 3 s, where single pauses are detectable within a dwell time window of 4 nt, similar to the value obtained in our previous single-molecule study of PV RdRp dynamics (Dulin et al., 2017). The apparent lifetime of these pauses is determined by averaging all dwell times (±SEM) exceeding this threshold of 3 s.
The probabilities of EV-A71 copy-back RNA synthesis (also referred as reversals) and PV backtracking were determined by dividing their occurrence by the total amount of RNA synthesis trajectories per pooled data set.
The enzyme processivities reflect the measured length of synthesized RNA chains (in nucleotides) for each measured RdRp defining their termination event, and the average velocity (± SD) resulted from dividing individual RdRp processivities by their active RNA synthesis duration.
Sequence analysis of copy-back locations from single-molecule and RNAseq assays
The sequence analysis was performed similar to the analysis used in the previously published study of EV-A71 recombinants yielding from transfected cell-based assays (Woodman et al., 2018).
From the in vitro single-molecule experiments, the sequences at which template switching occurred were extracted for analysis. The locations of template switching on the template RNA strand were determined by measuring the distance of the bead (in nucleotides) where reversals occur relative to the bead position before RNA synthesis was induced with NTPs. We used a sequence window of 31 nucleotides (−15 to +15 nucleotides around the detected locations) for extracting the sequences that were subjected to sequence analysis. This window size was determined based on two parameters. First, in a recently reported EV-A71 cell-based recombination assay, ‘copy-choice’ recombination occurred in regions with a sequence homology of between 5–11 nucleotides between the two parental template strands (Woodman et al., 2018). Second, the accuracy of the used assay is limited by the stiffness of the ssRNA construct and the corresponding degree of Brownian noise which accounts for a resolution of ±10 nucleotides. The window size of 31 nucleotides therefore guarantees to comprise any possible sequence motif that could be encountered in cells.
The analysis of the RNA sequencing data obtained from infected cells provided the exact copy-back RNA synthesis locations. We used a sequence window of 25 nucleotides (−12 to +12 nucleotides around the detected locations) to guarantee capturing potential sequence motifs with a length between 5 – 11 nucleotides (Woodman et al., 2018) up- and downstream the encountered copy-back locations.
From all extracted sequences of copy-back locations, we calculated the G:C and A:U densities, as well as the amount of successive G|C or A|U nucleotides. De novo sequence motif search for these location sequences were performed using the MEME suite (Version 5.0.1) (Bailey et al., 2009) with 1st-order background modelling and variable sequence motif width between 4–11 nt.
Supplementary Material
Highlights.
RdRp “backtracked” state is an intermediate for template switching
Intra- and intermolecular template switching use the same “backtracked” intermediate
Pyrazine-carboxamide nucleotides dysregulate recombination, increasing defective genomes
Alternative mechanistic class and target of antiviral nucleotides are revealed.
ACKNOWLEDGMENTS
We acknowledge funding to S.-R.S., C.E.C., and N.H.D. by the Human Frontiers Science Program (RPG0011/2015), to C.E.C. and J.J.A. by National Institutes of Health (R01AI45818), to A.W. by the American Heart Association (18POST33960071), and to S.-R.S. by the Ministry of Science and Technology, Taiwan (MOST 107-3017-F-182-001) and the Research Center for Emerging Viral Infections. We thank Friso Douma and Wessel Teunisse for experimental assistance, Theo van Laar for RdRp purification and RNA construct synthesis, and Martin Depken, David Dulin, and Behrouz Eslami-Mossallam for fruitful discussions.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SFJ, and Zhuang X (2008). Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature 453, 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelnabi R, Morais A.T.S. de, Leyssen P, Imbert I, Beaucourt S, Blanc H, Froeyen M, Vignuzzi M, Canard B, Neyts J, et al. (2017). Understanding the Mechanism of the Broad-Spectrum Antiviral Activity of Favipiravir (T-705): Key Role of the F1 Motif of the Viral Polymerase. J. Virol. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo A, Woodman A, Arnold JJ, Yeh M. Te, Evans D, Cameron CE, and Andino R (2018). Genetic recombination of poliovirus facilitates subversion of host barriers to infection. BioRxiv 273060. [Google Scholar]
- Arnold JJ, and Cameron CE (1999). Poliovirus RNA-dependent RNA Polymerase (3D pol) Is Sufficient for Template Switching in Vitro. J. Biol. Chem. 274, 2706–2716. [DOI] [PubMed] [Google Scholar]
- Arnold JJ, and Cameron CE (2000). Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275, 5329–5336. [DOI] [PubMed] [Google Scholar]
- Arnold JJ, and Cameron CE (2004). Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Bernal A, Uche U, Sterner DE, Butt TR, Cameron CE, and Mattern MR (2006). Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal. Biochem. 350, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Sharma SD, Feng JY, Ray AS, Smidansky ED, Kireeva ML, Cho A, Perry J, Vela JE, Park Y, et al. (2012). Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 8, e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila AI, Moreno E, Perales C, and Domingo E (2017). Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus. Virus Res. 233, 105–112. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calain P, Curran J, Kolakofsky D, and Roux L (1992). Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology 191, 62–71. [DOI] [PubMed] [Google Scholar]
- Cameron CE, Moustafa IM, and Arnold JJ (2009). Dynamics: the missing link between structure and function of the viral RNA-dependent RNA polymerase? Curr. Opin. Struct. Biol. 19, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Moustafa IM, and Arnold JJ (2016). Fidelity of Nucleotide Incorporation by the RNA-Dependent RNA Polymerase from Poliovirus. pp. 293–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Hernandez R, Jácome R, López Vidal Y, and Ponce de León S (2017). Are RNA Viruses Candidate Agents for the Next Global Pandemic? A Review. ILAR J 58, 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D, Ben-Shalom I, Brozell S, Cerutti D, Cheatham T III, Cruzeiro V, Darden T, Duke R, Ghoreishi D, et al. (2018). AMBER 2018 (University of California, San Francisco: ). [Google Scholar]
- Cnossen JP, Dulin D, and Dekker NH (2014). An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 85, 103712. [DOI] [PubMed] [Google Scholar]
- Coats SJ, Garnier-Amblard EC, Amblard F, Ehteshami M, Amiralaei S, Zhang H, Zhou L, Boucle SRL, Lu X, Bondada L, et al. (2014). Chutes and ladders in hepatitis C nucleoside drug development. Antiviral Res. 102, 119–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Maag D, Arnold JJ, Zhong W, Lau JYN, Hong Z, Andino R, and Cameron CE (2000). The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6, 1375–1379. [DOI] [PubMed] [Google Scholar]
- Darden T, York D, and Pedersen L (1993). Particle mesh Ewald: An N ·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. [Google Scholar]
- Dolan PT, Whitfield ZJ, and Andino R (2018). Mechanisms and Concepts in RNA Virus Population Dynamics and Evolution. Annu. Rev. Virol. 5, 69–92. [DOI] [PubMed] [Google Scholar]
- Dulin D, Vilfan ID, Berghuis BA, Hage S, Bamford DH, Poranen MM, Depken M, and Dekker NH (2015). Elongation-Competent Pauses Govern the Fidelity of a Viral RNA-Dependent RNA Polymerase. Cell Rep. 10, 983–992. [DOI] [PubMed] [Google Scholar]
- Dulin D, Arnold JJ, van Laar T, Oh H-S, Lee C, Perkins AL, Harki DA, Depken M, Cameron CE, and Dekker NH (2017). Signatures of Nucleotide Analog Incorporation by an RNA-Dependent RNA Polymerase Revealed Using High-Throughput Magnetic Tweezers. Cell Rep. 21, 1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyer L, Nencka R, de Clercq E, Seley-Radtke K, and Růžek D (2018). Nucleoside analogs as a rich source of antiviral agents active against arthropod-borne flaviviruses. Antivir. Chem. Chemother. 26, 2040206618761299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, and Brooks BR (1995). Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621. [Google Scholar]
- Fujii K, Nagata N, Sato Y, Ong KC, Wong KT, Yamayoshi S, Shimanuki M, Shitara H, Taya C, and Koike S (2013). Transgenic mouse model for the study of enterovirus 71 neuropathogenesis. Proc. Natl. Acad. Sci. 110, 14753–14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, and Morrey JD (2009). T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 82, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari R, Teimoori A, Sadeghi A, Mohamadkhani A, Rezasoltani S, Asadi E, Jouyban A, and Sumner SC (2020). Existing antiviral options against SARS-CoV-2 replication in COVID-19 patients. Future Microbiol. 15, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohara DW, Ha CS, Kumar S, Ghosh B, Arnold JJ, Wisniewski TJ, and Cameron CE (1999). Production of “Authentic” Poliovirus RNA-Dependent RNA Polymerase (3Dpol) by Ubiquitin–Protease-Mediated Cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138. [DOI] [PubMed] [Google Scholar]
- Gong P, and Peersen OB (2010). Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 107, 22505–22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Wong M-H, Jung K-H, Smee DF, Morrey JD, and Furuta Y (2010). Efficacy of favipiravir (T-705) and T-1106 pyrazine derivatives in phlebovirus disease models. Antiviral Res. 86, 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin KK, Renzette N, Kowalik TF, and Jensen JD (2016). Antiviral drug resistance as an adaptive process. Virus Evol 2, vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janissen R, Arens MMA, Vtyurina NN, Rivai Z, Sunday ND, Eslami-Mossallam B, Gritsenko AA, Laan L, de Ridder D, Artsimovitch I, et al. (2018). Global DNA Compaction in Stationary-Phase Bacteria Does Not Affect Transcription. Cell 174, 1188–1199.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Leveque V, Ma H, Johnson KA, and Klumpp K (2013a). NTP-mediated nucleotide excision activity of hepatitis C virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 110, E348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Smith LK, Rajwanshi VK, Kim B, and Deval J (2013b). The Ambiguous Base-Pairing and High Substrate Efficiency of T-705 (Favipiravir) Ribofuranosyl 5′-Triphosphate towards Influenza A Virus Polymerase. PLoS One 8, e68347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- Joshi S, Parkar J, Ansari A, Vora A, Talwar D, Tiwaskar M, Patil S, and Barkate H (2021). Role of favipiravir in the treatment of COVID-19. Int. J. Infect. Dis. 102, 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG, Furuta Y, Shafer K, and Sidwell RW (2007). Activity of T-1106 in a hamster model of yellow Fever virus infection. Antimicrob. Agents Chemother. 51, 1962–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mishanina TV, Landick R, and Darst SA (2019). Mechanisms of Transcriptional Pausing in Bacteria. J. Mol. Biol. 431, 4007–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BJ, Watkins CL, Peersen OB, and Barton DJ (2019). Picornavirus RNA Recombination Counteracts Error Catastrophe. J. Virol. 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BJ, Watkins CL, Peersen OB, and Barton DJ (2020). An Extended Primer Grip of Picornavirus Polymerase Facilitates Sexual RNA Replication Mechanisms. J. Virol. 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ellis VD, Woodman A, Zhao Y, Arnold JJ, and Cameron CE (2019). RNA-Dependent RNA Polymerase Speed and Fidelity are not the Only Determinants of the Mechanism or Efficiency of Recombination. Genes (Basel). 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K, and Baltimore D (1986). The mechanism of RNA recombination in poliovirus. Cell 47, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korboukh VK, Lee CA, Acevedo A, Vignuzzi M, Xiao Y, Arnold JJ, Hemperly S, Graci JD, August A, Andino R, et al. (2014). RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem. 289, 29531–29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung Y-H, Huang S-W, Kuo P-H, Kiang D, Ho M-S, Liu C-C, Yu C-K, Su I-J, and Wang J-R (2010). Introduction of a strong temperature-sensitive phenotype into enterovirus 71 by altering an amino acid of virus 3D polymerase. Virology 396, 1–9. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarini RA, Keene JD, and Schubert M (1981). The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26, 145–154. [DOI] [PubMed] [Google Scholar]
- Lee K-M, Gong Y-N, Hsieh T-H, Woodman A, Dekker NH, Cameron CE, and Shih S-R (2018). Discovery of Enterovirus A71-like nonstructural genomes in recent circulating viruses of the Enterovirus A species. Emerg. Microbes Infect. 7, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang H, Shi J, Yang D, Zhou G, Chang J, Cameron CE, Woodman A, and Yu L (2019). Senecavirus-Specific Recombination Assays Reveal the Intimate Link between Polymerase Fidelity and RNA Recombination. J. Virol. 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Abbondanzieri EA, Rausch JW, Grice SFJL, and Zhuang X (2008). Slide into Action: Dynamic Shuttling of HIV Reverse Transcriptase on Nucleic Acid Substrates. Science (80-.). 322, 1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Harada BT, Miller JT, Le Grice SFJ, and Zhuang X (2010). Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat. Struct. Mol. Biol. 17, 1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry K, Woodman A, Cook J, and Evans DJ (2014). Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length ‘Imprecise’ Intermediates. PLoS Pathog. 10, e1004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C (2015). ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone B, Chen J, Wang Q, Llewellyn E, Choi YJ, Olinares PDB, Cao X, Hernandez C, Eng ET, Chait BT, et al. (2021). Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. BioRxiv 2021.03.13.435256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud EJ, Hayden FG, and Hurt AC (2019). Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res. 169, 104545. [DOI] [PubMed] [Google Scholar]
- Moustafa IM, Shen H, Morton B, Colina CM, and Cameron CE (2011). Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 410, 159–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa IM, Korboukh VK, Arnold JJ, Smidansky ED, Marcotte LL, Gohara DW, Yang X, Sánchez-Farrán MA, Filman D, Maranas JK, et al. (2014). Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase. J. Biol. Chem. 289, 36229–36248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Pons-Salort M, Messacar K, Cook C, Meyers L, Farrar J, and Grenfell BT (2021). Epidemiological dynamics of enterovirus D68 in the United States and implications for acute flaccid myelitis. Sci. Transl. Med 13, eabd2400. [DOI] [PubMed] [Google Scholar]
- Petrushenko ZM, Lai C-H, Rai R, and Rybenkov VV (2006). DNA Reshaping by MukB Right-Handed Knotting, Left-Handed Supercoiling. J. Biol. Chem. 281, 4606–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier EZ, Mounce BC, Rozen-Gagnon K, Hooikaas PJ, Stapleford KA, Moratorio G, and Vignuzzi M (2016). Low-Fidelity Polymerases of Alphaviruses Recombine at Higher Rates To Overproduce Defective Interfering Particles. J. Virol. 90, 2446–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puenpa J, Wanlapakorn N, Vongpunsawad S, and Poovorawan Y (2019). The History of Enterovirus A71 Outbreaks and Molecular Epidemiology in the Asia-Pacific Region. J. Biomed. Sci. 26, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re GG, Gupta KC, and Kingsbury DW (1983). Genomic and copy-back 3’ termini in Sendai virus defective interfering RNA species. J. Virol. 45, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe DR, and Cheatham TE (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- Ryckaert J-P, Ciccotti G, and Berendsen HJ. (1977). Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341. [Google Scholar]
- Sangawa H, Komeno T, Nishikawa H, Yoshida A, Takahashi K, Nomura N, and Furuta Y (2013). Mechanism of Action of T-705 Ribosyl Triphosphate against Influenza Virus RNA Polymerase. Antimicrob. Agents Chemother. 57, 5202–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert M, Bera SC, van Nies P, Kirchdoerfer RN, Shannon A, Le T-T-N, Meng X, Xia H, Wood JM, Harris LD, et al. (2021). Inhibition of SARS-CoV-2 polymerase by nucleotide analogs: a single molecule perspective. BioRxiv 2020.08.06.240325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seley-Radtke KL, and Yates MK (2018). The evolution of nucleoside analogue antivirals: A review for chemists and non-chemists. Part 1: Early structural modifications to the nucleoside scaffold. Antiviral Res. 154, 66–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A, Selisko B, Le NTT, Huchting J, Touret F, Piorkowski G, Fattorini V, Ferron F, Decroly E, Meier C, et al. (2020). Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. BioRxiv 2020.05.15.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Ye H-Q, Deng C-L, Li R, Zhang B, and Gong P (2020). A nucleobase-binding pocket in a viral RNA-dependent RNA polymerase contributes to elongation complex stability. Nucleic Acids Res. 48, 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu B, and Gong P (2016). Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. 113, E4005–E4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Kim EJ, Felt SA, Taylor LJ, Agarwal D, Grant GR, and López CB (2019). A specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes. PLoS Pathog. 15, e1007707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W-F, Huang R-T, Chien K-Y, Huang J-Y, Lau K-S, Jheng J-R, Chiu C-H, Wu T-Y, Chen C-Y, and Horng J-T (2016). Host MicroRNA miR-197 Plays a Negative Regulatory Role in the Enterovirus 71 Infectious Cycle by Targeting the RAN Protein. J. Virol. 90, 1424–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee HK, Tan CW, Yogarajah T, Lee MHP, Chai HJ, Hanapi NA, Yusof SR, Ong KC, Lee VS, Sam I-C, et al. (2019). Electrostatic interactions at the five-fold axis alter heparin-binding phenotype and drive enterovirus A71 virulence in mice. PLOS Pathog. 15, e1007863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-H, Lee P-Y, Stollar V, and Li M-L (2006). Antiviral Therapy Targeting Viral Polymerase. Curr. Pharm. Des. 12, 1339–1355. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, and López CB (2019). Defective viral genomes are key drivers of the virus–host interaction. Nat. Microbiol. 4, 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, and Andino R (2005). Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 107, 173–181. [DOI] [PubMed] [Google Scholar]
- WHISTLER T, BELLINI WJ, and ROTA PA (1996). Generation of Defective Interfering Particles by Two Vaccine Strains of Measles Virus. Virology 220, 480–484. [DOI] [PubMed] [Google Scholar]
- Woodman A, Arnold JJ, Cameron CE, and Evans DJ (2016). Biochemical and genetic analysis of the role of the viral polymerase in enterovirus recombination. Nucleic Acids Res. 44, 6883–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman A, Lee K-M, Janissen R, Gong Y-N, Dekker N, Shih S-R, and Cameron CE (2018). Predicting Intraserotypic Recombination in Enterovirus 71. J. Virol. JVI.02057–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Lou Z, Miao Y, Yu Y, Dong H, Peng W, Bartlam M, Li X, and Rao Z (2010). Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China. Protein Cell 1, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, Brodsky L, and Andino R (2016). RNA Recombination Enhances Adaptability and Is Required for Virus Spread and Virulence. Cell Host Microbe 19, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, and Koike S (2009). Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801. [DOI] [PubMed] [Google Scholar]
- Yates MK, and Seley-Radtke KL (2019). The evolution of antiviral nucleoside analogues: A review for chemists and non-chemists. Part II: Complex modifications to the nucleoside scaffold. Antiviral Res. 162, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Kraus TA, Horvath CM, Moran TM, and López CB (2006). A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol. 177, 4503–4513. [DOI] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table. The single-molecule data reported in this study have been deposited at a publicly accessible repository. The DOI is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Digoxigenin antibodies | Roche | RRID: AB_514496 |
| Bacterial and virus strains | ||
| SURE Competent Cells | Agilent technologies | Cat#200238 |
| Chemicals, peptides, and recombinant proteins | ||
| ApU dinucleotide | IBA Lifesciences GmbH | Cat#0-31004 |
| Biotin-16-dUTP | Roche | Cat#11093711103 |
| Digoxigenin-11-dUTP | Roche | Cat#11093681103 |
| rNTPs | GE Healthcare | Cat#27-2025-01 |
| Streptavidin-coated superparamagnetic beads | Thermo Fischer | Cat#65001 |
| T-1106 triphosphate | Blake Petersen Lab | |
| Ribavirin triphosphate | Jena Bioscience | Cat#NU-1105L |
| Dulbecco’s Modified Eagle’s Medium | Thermo Fischer | Cat#31966021 |
| Streptomycin | Thermo Fischer | Cat#15140122 |
| Superase RNase inhibitor | ThermoFischer | Cat#AM2694 |
| Critical commercial assays | ||
| Ribomax large scale RNA production kit | Promega | Cat#P1300 |
| T7 mMESSAGE mMACHINE kit | Thermo Fischer | Cat#AM1344 |
| Qubit RNA Broad Range assay kit | Thermo Fischer | Cat#Q10211 |
| TransIT-mRNA transfection kit | MirusBio | Cat#MIR2250 |
| Direct-zol-96 RNA purification kit | Zymo Research | Cat#R2056 |
| Quant-iT RNA assay kit | Thermo Fischer | Cat#Q33140 |
| Next rRNA Depletion Kit | New England Biolabs | Cat#E6310L |
| NEBNext Ultra II RNA Library preparation kit | New England Biolabs | Cat#E7775 |
| NextSeq500/550 Mid Output kit v2.5 | Illumina | Cat#20024907 |
| Mycoplasma Detection Kit | Lonza | Cat#LT07-318 |
| RNeasy MinElute cleanup kit | Quiagen | Cat#74204 |
| Deposited data | ||
| Single-Molecule Magnetic Tweezers data | This study | DOI: 10.4121/c.5608934 |
| Raw and processed RNAseq data | This study | NIH GEO: GSE183959 |
| Experimental models: Cell lines | ||
| RD cell line | ATCC | RRID: CVCL_1649 |
| Experimental models: Organisms/strains | ||
| Transgenic human hSCARB2–expressing mice | Fujii et al., 2013 | |
| Oligonucleotides | ||
| 5’-aacuguugguguacgcgaaagcgu | GE Healthcare Dharmacon | |
| 5’-taatacgactcactataggatcgccaagattagcggatcctacctgac | Biolegio | AB-For |
| 5’-ggttaacctcaacttccatttcc | Biolegio | AB-Rev |
| 5’-cccctcgaggggaaaaaaaaaaaaccgtatgacgctggaag | Biolegio | CD-For |
| 5’-taatacgactcactataggccggacgtttcggatcttccgacatgcgc | Biolegio | CD-Rev |
| 5’-aagattagcggatcctacctgac | Biolegio | Bio-For |
| 5’-bio-taatacgactcactataggaacggcttgatatccactttacg | Biolegio | Bio-Rev |
| 5’-agcgtaaaattcagttcttcgtggcg | Biolegio | Dig-For |
| 5’-dig-aatacgactcactatagggctaccggttaacctcaacttccatttcc | Biolegio | Dig-Rev |
| 5’-tgccattcagggactgccgatgtcggtgcagccg | Biolegio | SP-For |
| 5’-taatacgactcactataggagcgccgcttccatgtcctggaacgct | Biolegio | SP-Rev |
| 5’-acgttctcagtgcggactgtag | IDT | MP4-3156-Fwd |
| 5’-ccttggaaaagagcttc | IDT | C2/4-5057-Rev |
| 5’-gaagctcttttccaagg | IDT | C2/4-5057-Fwd |
| 5’-ctggttataacaaatttaccc | IDT | C2/4-7384-Rev |
| 5’-atcaatcacacccatcatgtgcatcgcaataaaacttattg | IDT | Y276H-Fwd |
| 5’-caataagttttattgcgatgcacatgatgggtgtgattgat | IDT | Y276H-Rev |
| 5’-acgtagcccagcgcgtcggccg | IDT | Eag-I-Rev |
| 5’-agcagtgctgactagtaaagacc | IDT | SpeI-Fwd |
| 5’-cggcagcccagaagaact | IDT | qPCR-Fwd |
| 5’-gccaccctatctccctgga | IDT | qPCR-Rev |
| 5’-FAM-tcaccatgaagttgtgtaaggatgcta-BHQ | Biosearch Technologies | qPCR-Probe |
| Recombinant DNA | ||
| Plasmid pBB10 | Petrushenko et al., 2006 | pBB10 |
| pSumoEV-A71-3D | This study | |
| pSumoEV-A71-3D-Y276H | This study | |
| pSumoPV-3D | Arnold J.J. & Cameron C.E., 2000 | |
| pSumoPV-3D-Y275H | Acevedo et al., 2018 | |
| pEV-A71-MP4 | Wang et al., 2004 | |
| pEV-A71-Y276H | This study | |
| pEV-A71-C2-replicon | Woodman et al., 2018 | |
| pEV-A71-MP4-Δ3D | Woodman et al., 2018 | |
| Software and algorithms | ||
| MatLab R13 | MathWorks Inc. | RRID: SCR_001622 |
| Igor Pro 6.37 | Wavemetrics | RRID: SCR_000325 |
| LabView 2011 | National Instruments | RRID: SCR_014325 |
| ImageQuant | GE Healthcare Life Sciences | RRID: SCR_014246 |
| ImageLab | BioRad | www.bio-rad.com/en-ch/product/image-lab-software |
| VODKA | (Sun et al., 2019) | |
| Bowtie2 | (Langmead and Salzberg, 2012) | RRID: SCR_016368 |
| BBTools | Joint Genome Institute | RRID: SCR_016968 |
| bcl2fastq v2.2 | Illumina | RRID: SCR_015058 |