“Diffuse hemispheric glioma, H3 G34-mutant” (DHG) is being included as a new CNS grade 4 tumor type in the forthcoming 5th edition of the WHO Classification of Tumors of the Central Nervous System. DHG arises within the cerebral hemispheres of teenagers and young adults and can histologically resemble anaplastic astrocytoma, glioblastoma, or CNS embryonal tumor.1 Affected patients have a somewhat more favorable prognosis than both “Glioblastoma, IDH-wildtype” and “Diffuse midline glioma, H3 K27M-mutant,” but invariably suffer from disease recurrence and mortality using current treatment regimens.1–3 Exploration of genomically targeted therapeutic strategies is therefore needed.
DHG is molecularly defined by a recurrent glycine to arginine or valine substitution at codon 35 of the histone H3.3 gene H3F3A, corresponding to amino acid 34 of the mature H3.3 protein. This p.G34R/V mutation results in steric change which blocks di- and tri-methylation of lysine 36, thereby obstructing this posttranslational modification critical for glial differentiation.4,5 Additionally, DHG frequently have co-occurring mutational inactivation of TP53, and deleterious mutations in the ATRX chromatin remodeling gene associated with alternative lengthening of telomeres.1–3 However, the complete genomic landscape of this new tumor entity and potential targets for precision medicine therapy have yet to be fully defined.
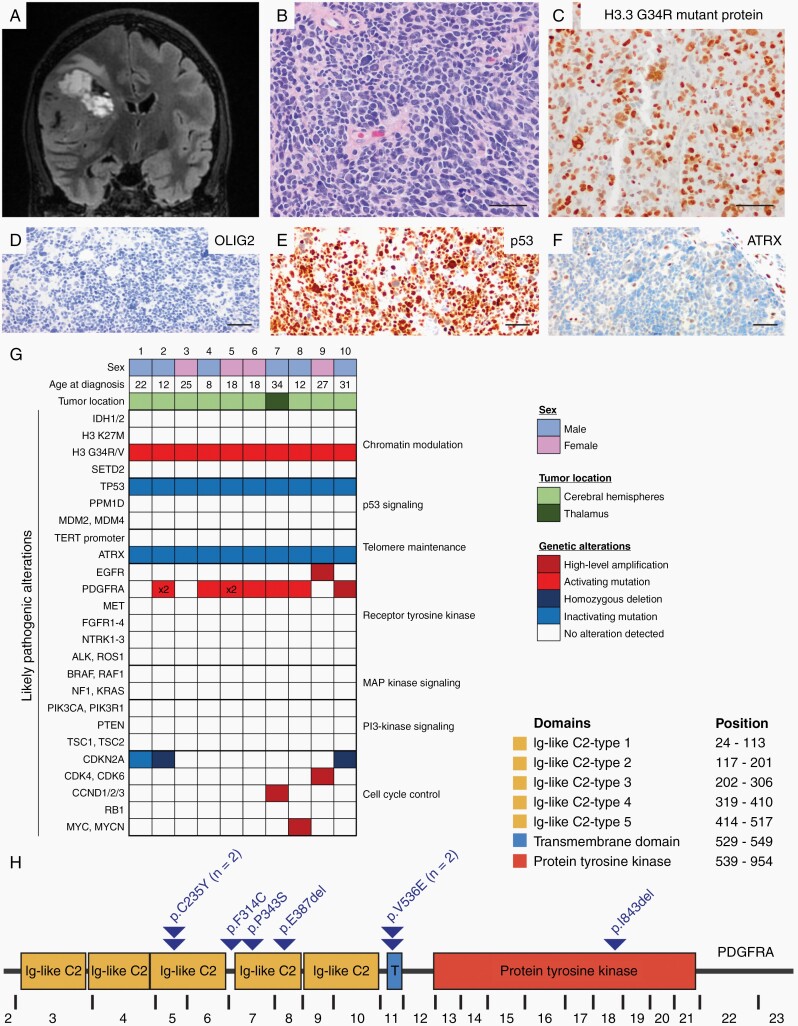
Here we report genomic characterization of 10 DHG tumors (Figure 1), which revealed two recurrent and potentially targetable genetic perturbations: activating mutations in PDGFRA and diverse alterations affecting the CDK4/6-cyclin D-p16INK4a-Rb cell cycle pathway. All tumors harbored H3F3A p.G34R mutation, along with inactivating TP53 and ATRX variants. Six tumors also harbored somatic missense mutations in PDGFRA, and one other tumor harbored focal high-level amplification of wildtype PDGFRA (70% PDGFRA alteration frequency). Additionally, three tumors harbored homozygous deletion or truncating mutation of CDKN2A, one had focal high-level amplification of CCND2, and one other had focal high-level amplification of CDK6 (50% cell cycle alteration frequency). This genomic analysis has revealed the possibility of targeting mutant PDGFRA and/or activated CDK4/6 using small molecule kinase inhibitors (eg, dasatinib and abemaciclib, respectively), and genomically guided clinical trials investigating these or similar agents in children and young adults with DHG may thus be warranted.
Fig. 1.
Clinicopathologic features and genomic landscape of diffuse hemispheric glioma, H3 G34-mutant. (A) MR imaging typically reveals an expansile, contrast-enhancing tumor within the cerebral hemispheres. (B) Histologically these tumors are diffuse high-grade gliomas, often with a primitive embryonal-like appearance. (C) Mutant-specific antibodies have been developed to detect the H3.3 G34R or G34V-mutant protein that molecularly defines these tumors (RevMAb clones RM240 and RM307). Additional immunohistochemical findings usually include the absence of OLIG2 expression (D), p53 overexpression (E), and loss of ATRX expression (F). (G) Oncoprint summary table of genomic findings in a cohort of 10 DHG patients. (H) The majority of PDGFRA mutations localize in the extracellular immunoglobulin-like domains but may also occur within the transmembrane domain or intracellular tyrosine kinase domain (annotation per RefSeq transcript NM_006206). Scale bars, 50 µm. Abbreviations: DHG, diffuse hemispheric glioma; MR, magnetic resonance.
A recent landmark study by Chen et al also found a high frequency of PDGFRA mutations in DHG, and demonstrated that mutant Pdgfra potently fuels gliomagenesis in vivo in a mouse model in concert with Atrx and Tp53 inactivation.6 Through elegant functional studies, they identified that DHG arises from GSX2-expressing interneuron progenitor cells and that H3.3 G34R mutation stalls differentiation by epigenetic silencing of mature neuronal genes and hijacking of the active cis-regulatory elements of the GSX2 gene to drive increased PDGFRA expression. In combination with the frequent PDGFRA mutations, these functional studies indicate a fundamental role for activation of PDGFRA signaling in DHG. Analysis of the specific PDGFRA variants from our tumor cohort (Figure 1H) and that of Chen et al reveals most mutations occur in the extracellular immunoglobulin-like domains, but also occasionally in the transmembrane domain or the autoinhibition site in the intracellular tyrosine kinase domain (p.D842 within exon 18) commonly mutated in gastrointestinal stromal tumors (GIST). While first-generation tyrosine kinase inhibitors such as imatinib effectively inhibit select mutant isoforms of PDGFRA, exon 18 mutations are associated with imatinib resistance. The agent avapritinib was recently approved by the FDA for the treatment of adults with advanced GIST with PDGFRA exon 18 mutations. Thus, selection of the tyrosine kinase inhibitor needs to be individually tailored to the specific PDGFRA mutation driving each patient’s tumor.
In light of these findings, we recommend prospective genomic interrogation for DHG patients to inform potential personalized therapeutic approaches and enrollment in precision medicine clinical trials investigating PDGFRA and CDK4/6 inhibitors.
Acknowledgments
We thank the staff of the UCSF Clinical Cancer Genomics Laboratory for assistance with genetic profiling.
Funding
C.G.L. is supported by the UCSF Training Program in Translational Brain Tumor Research, National Cancer Institute, NIH (T32 CA151022). D.A.S. is supported by the NIH Director’s Early Independence Award from the Office of the Director, National Institutes of Health (DP5 OD021403), the Morgan Adams Foundation, and the Yuvaan Tiwari Foundation. This study was supported in part by the philanthropically funded UCSF Glioblastoma Precision Medicine Program and the Biorepository Core (J.J.P.) of the UCSF Brain Tumor SPORE from the National Cancer Institute, NIH (P50 CA097257).
Conflict of interest statement. None of the authors have any relevant conflicts of interest to disclose.
Data availability statement. Sequencing data files are available from the authors upon request.
References
- 1. Korshunov A, Capper D, Reuss D, et al. Histologically distinct neuroepithelial tumors with histone 3 G34 mutation are molecularly similar and comprise a single nosologic entity. Acta Neuropathol. 2016;131(1):137–146. [DOI] [PubMed] [Google Scholar]
- 2. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 3. Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 4. Bjerke L, Mackay A, Nandhabalan M, et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013;3(5):512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jain SU, Khazaei S, Marchione DM, et al. Histone H3.3 G34 mutations promote aberrant PRC2 activity and drive tumor progression. Proc Natl Acad Sci U S A. 2020;117(44):27354–27364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen CCL, Deshmukh S, Jessa S, et al. Histone H3.3G34-mutant interneuron progenitors co-opt PDGFRA for gliomagenesis. Cell. 2020;183(6):1617–1633.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]