Abstract
DNA origami is a powerful nanomaterial for biomedical applications due in part to its capacity for programmable, site-specific functionalization. To realize these applications, scalable and efficient conjugation protocols are needed for diverse moieties ranging from small molecules to biomacromolecules. Currently, there are no facile and general methods for in situ covalent modification and label-free quantification of reaction conversion. Here, we investigate the post-assembly functionalization of DNA origami and the subsequent high-performance liquid chromatography-based characterization of these nanomaterials. Following this approach, we developed a versatile DNA origami functionalization and characterization platform. We observed quantitative in situ conversion using widely accessible click chemistry for carbohydrates, small molecules, peptides, polymers, and proteins. This platform should provide broader access to covalently functionalized DNA origami, as illustrated here by PEGylation for passivation and HIV antigen decoration to construct virus-like particle vaccines.
Keywords: DNA origami, DNA nanotechnology, nanoparticles, click chemistry, bioconjugation, reaction characterization
Introduction
Structural DNA nanotechnology1–3 has been extensively applied to biological applications,4, 5 including therapeutic delivery,6–10 due to its geometric and chemical programmability at the nanoscale. The DNA origami method uses a long single-stranded DNA (ssDNA) ‘scaffold’ and short oligonucleotides ‘staples’ to fold complex DNA nanostructures with quantitative yields.11–14 Polyhedral wireframe nanostructures have been developed to provide precise control over 2D and 3D structure.15, 16 Overcoming bottlenecks with recent efforts in top-down sequence design17–20 and large-scale, custom scaffold production,21–23 DNA origami is imminently positioned for translational research.3, 24 As recently reviewed by Madsen and Gothelf, this will require the efficient functionalization of DNA nanostructures, a field that remains underdeveloped.25
Precise functionalization of DNA origami is commonly achieved through post-assembly hybridizations, where ssDNA overhangs on the DNA nanostructure hybridize to a complementary nucleic acid strand attached to the desired conjugate. This method enables the facile and site-specific attachment of nucleic acid-modified proteins,26–32 peptides,7, 33, 34 lipids,35 and dyes31, 36 in an orthogonal and sequence-programmable manner. Disadvantages of this non-covalent strategy include potential dissociation and loss of resolution in 3D organization. Similarly, streptavidin-biotin systems are also viable for DNA origami functionalization, yet these systems are limited in nanoscale resolution and conjugate scope.37
Alternatively, covalent conjugation strategies can be implemented prior to self-assembling DNA nanostructures. Staples are modified using solution- or solid-phase chemistry, and are then self-assembled with the scaffold to yield functionalized DNA origami.38, 39 This approach presents its own challenges, however. Generating highly decorated nanostructures requires the individual synthesis, purification, and characterization of a high number of staples, which substantially impedes the fabrication workflow. Additionally, most folding protocols require high temperatures and salt concentrations that can denature proteins and bleach dyes, limiting the conjugate scope of this approach.
To address these challenges, in situ (post-assembly) covalent conjugation strategies have been reported. Covalent functionalization strategies reduce the potential for conjugate disassociation and increase spatial resolution, both of which are advantageous for biomedical applications. However, these strategies often require the synthesis of functional staple strands, suffer from slow reaction kinetics, and cannot easily incorporate heterovalent displays of conjugates. Gothelf and coworkers demonstrated single molecule orthogonal chemical reactions on origami nanostructures,40 and others have functionalized DNA origami using protein tags.41–43 Andersen and coworkers used click chemistry to attach an enzyme to DNA nanostructures.44 Recently, strain-promoted azide-alkyne cycloaddition (SPAAC) chemistry was employed to generate superstructures from monomeric DNA origami45 and dense arrays of fluorophores on DNA nanostructures.46 These reports rely on atomic force microscopy or transmission electron microscopy (TEM) to evaluate reaction conversion. Both methods are semi-quantitative for ensemble measurements and are not amenable to analyzing small molecule conjugates, dense arrays, and conjugations on 3D structures. Alternatively, Funke and Dietz used gel electrophoresis to characterize thiol-Michael additions on DNA nanostructures.47 While this characterization method measures reaction conversions quantitatively, it requires secondary labels, as do other quantitative methods.27, 48
Here, we report a facile workflow to fabricate and characterize covalently functionalized DNA origami virus-like particles (DNA-VLPs). We leverage in situ click chemistry49, 50 to functionalize the nanostructures, we quantify reaction conversions using liquid chromatography, and we structurally characterize the DNA-VLPs. We demonstrate the efficient functionalization and application of DNA-VLPs with therapeutically relevant conjugates, and develop a general workflow that can integrate different conjugation chemistries and conjugate classes for diverse therapeutic, vaccine, theranostic, and materials science applications.
Results and Discussion
Given the current limitations in characterizing covalent conjugations on DNA nanostructures, we hypothesized that using a hydrophobic SPAAC functional group could enable characterization via high-performance liquid chromatography (HPLC). DNA origami scaffold routing and staple sequences were designed by the top-down algorithm DAEDALUS17 with dibenzocyclooctyne-amine (DBCO) moieties at specific staple 5’ termini (Figure 1). I52-30xDBCO, an icosahedron with two DNA duplexes per edge, a 52 base pair edge length, and one DBCO group per edge, was chosen as a model system to investigate in situ covalent conjugations. Staples containing DBCO groups were synthesized using standard phosphoramidite chemistry and purified using HPLC (Figure S1). DNA-VLPs were fabricated and characterized (Figure S2) as previously reported.17 Reaction conditions were optimized for the efficient high-density functionalization of DNA-VLPs (Figure S3), which were subsequently purified using centrifugal filtration. Reversed-phase HPLC using BEH-C18 columns under denaturing conditions was then employed to separate functionalized staples from non-functionalized staples and the scaffold strand, enabling quantitative determination of reaction conversion. This characterization, in combination with structural characterization by agarose gel electrophoresis (AGE), dynamic light scattering (DLS), and TEM affords an improved analysis of covalently-functionalized DNA-VLPs.
Figure 1. Covalent functionalization of DNA-VLPs.
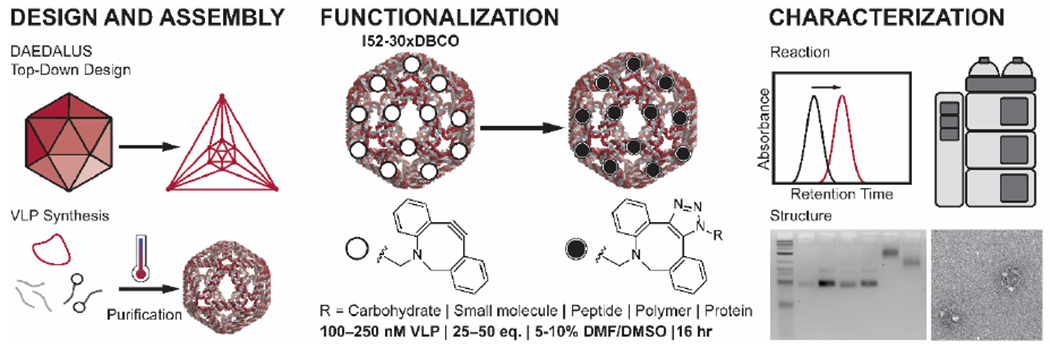
DNA-VLPs are designed using the top-down sequence design algorithm DAEDALUS,17 assembled with quantitative folding protocols, and covalently functionalized using SPAAC. A strained alkyne (DBCO) is installed onto staple strands, and is reacted with an azide-functionalized conjugate after assembly of nanostructures. The reaction conversion is analyzed via a liquid chromatography method, and the resulting DNA-VLPs are analyzed via standard structural characterization.
We directly injected the DNA-VLPs into the HPLC column (60 °C; 0.1 M triethylammonium acetate in water:acetonitrile gradient) and observed that hydrophobic staples were separated from the rest of the staples and scaffold of the nanostructure. The comparative HPLC analysis of the I52 scaffold, I52, and I52-30xDBCO demonstrated the ability of this denaturing technique to isolate origami staples with click-reactive moieties (Figure 2.A). To highlight the scope of this characterization method, we incubated I52-30xDBCO with diverse therapeutically relevant conjugates. HPLC traces of I52-30xDBCO and purified DNA-VLPs covalently functionalized with a carbohydrate, a small molecule, a peptide, a synthetic polymer, and a clinically relevant HIV protein antigen indicate quantitative coverage (>95%) of the DNA-VLP with only moderate stoichiometric excesses and reaction times (Figure 2.B and Table S1). The observed shifts in the HPLC were confirmed using a simpler oligonucleotide system (Figure S4). Note that for the Cy5 conjugate, an example of a small molecule, there was an additional spectroscopic signature, as it absorbs in the near-IR region. This offered further evidence that the DNA origami was sufficiently denatured and that all DBCO staples were separated quantitatively by HPLC (Figure 2.C). Additionally, reaction conversions were confirmed using an established spectroscopic ratiometric technique27 (Figure 2.D and Figure S5). We attribute the difference between methods when quantifying Cy5 coverage to DNA-induced fluorescence quenching of cyanine dyes.51 Note that while these techniques can be compared for conjugates with spectroscopic fingerprints (protein and fluorophore), only the HPLC method can monitor the reaction conversion for the other conjugates. AGE (Figure 2.E) and DLS (Figure S6) indicate the monodispersity and nanostructure integrity of the DNA-VLPs after functionalization and purification.
Figure 2. Characterization of covalently functionalized DNA-VLPs.
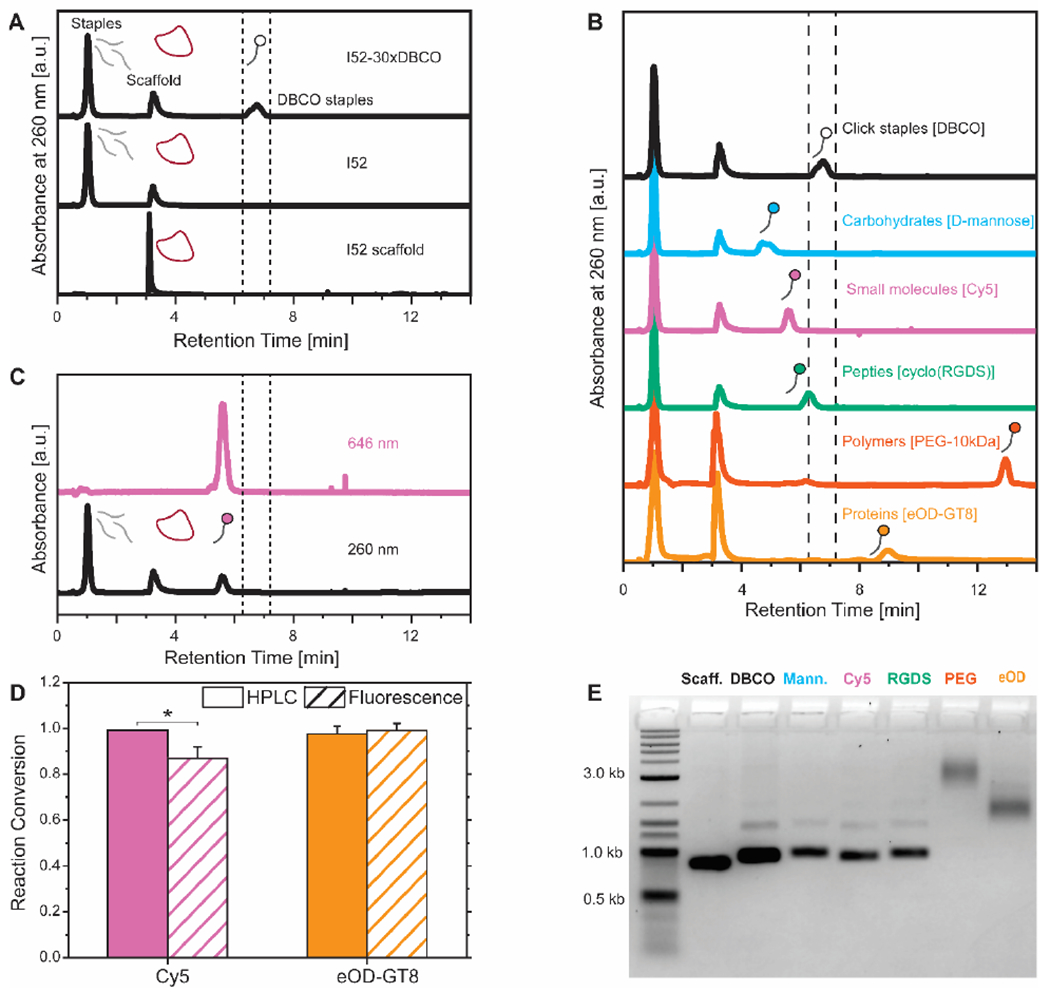
A) HPLC traces of the DNA-VLP scaffold (I52 Scaffold), DNA-VLP (I52), and DNA-VLP with DBCO groups at the 5’ terminus of staples (I52-30xDBCO). The hydrophobic click chemistry staples are separated as the DNA-VLP is denatured. B) HPLC traces separate functionalized staples allowing for the quantification of reaction conversions; quantitative conversions (>95%) were observed for all conjugate classes. C) Secondary spectroscopic signatures (646 nm for Cy5) indicate that all functionalized staples are separated from the rest of the DNA-VLP. D) Reaction conversion quantified by the HPLC method and a ratiometric fluorescent method. Error bars represent the standard error of the mean (n=3). P values are from a paired Student’s t-test (*P<0.05). E) AGE confirms structural integrity of nanostructures after functionalization and purification.
PEGylation is commonly used to passivate therapeutic materials, increase circulation time, and inhibit nuclease activity. To inhibit exonuclease activity, researchers have incorporated hexaethylene glycol onto 3’/5’-termini in DNA nanostructures.52, 53 PEGylation was also implemented non-covalently into DNA origami through the electrostatic complexation of poly(lysine)-co-poly(ethyelene glycol) block copolymers to the DNA origami surface,54 resulting in protection against endonucleases. To demonstrate the utility of this workflow, we functionalized I52-30xDBCO with PEG-10kDa-azide to generate a 5’-termini covalently PEGylated DNA-VLP (I52-PEG) (Figure 3.A). We then assessed the potential for this PEGylation strategy to protect against nuclease degradation in serum. We compared this strategy to a bare DNA-VLP (I52) and the poly(lysine)-PEG strategy.54 Using AGE to analyze the stability of covalently PEGylated DNA-VLPs, we observed enhanced stability compared to bare DNA-VLPs (Figure 3.B and Figure S7). We also observed a downshift in the gel overtime, presumably corresponding to the loss of PEG polymers as the nanostructure was degraded. The relative performance of the degradation protection strategies was assessed. Covalent PEGylation of the 5’-termini of staples offered enhanced nuclease protection on the order of several hours, yet was inferior to the protection provided by the poly(lysine)-PEG approach (Figure 3.C), likely because the 3’-termini and internal staple regions were unmodified, providing only partial coverage of the DNA nanostructure.
Figure 3. Stabilization of wireframe DNA origami with covalent PEGylation.
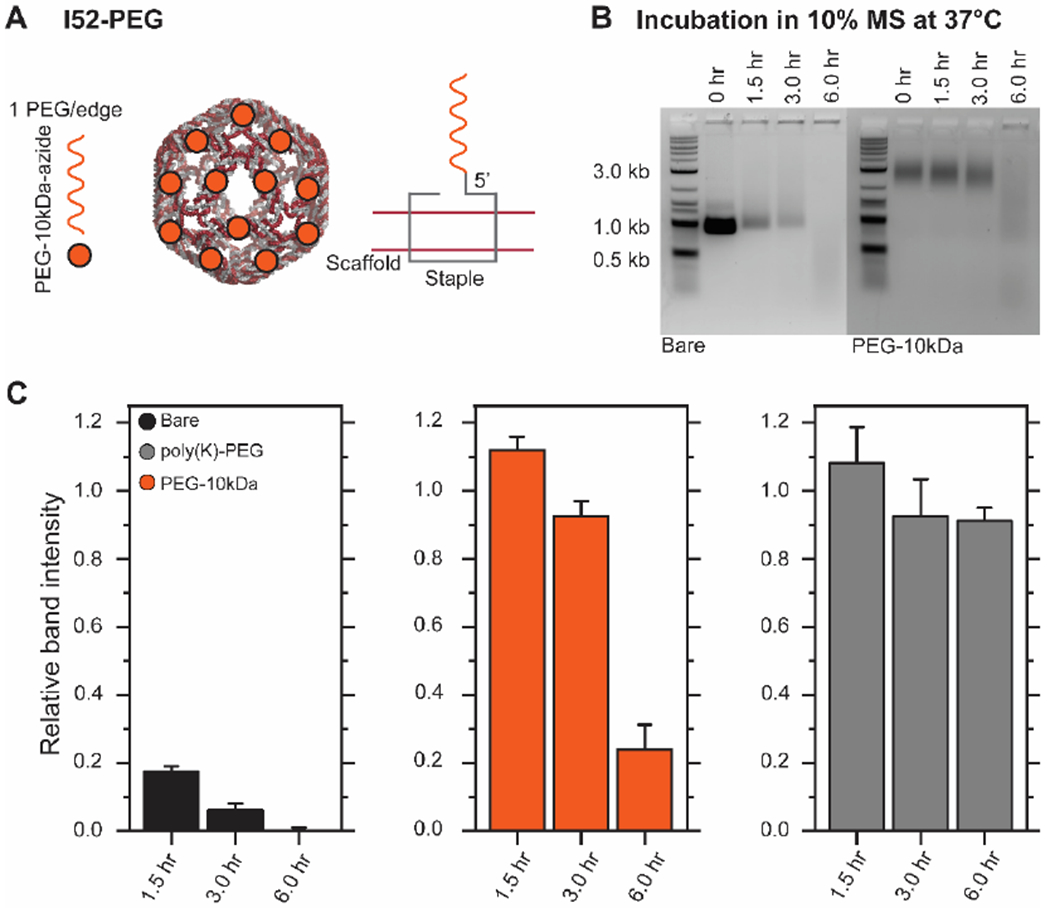
A) I52-PEG was fabricated using covalent functionalization resulting in PEGylation on each edge of the DNA-VLP. B) Representative AGE time series for bare and 5’-termini covalently PEGylated DNA-VLPs. DNA-VLPs were incubated in DMEM with 10% MS at 37 °C. C) Relative band intensities compared to the 0 hr data point for non-stabilized DNA-VLPs and stabilized DNA-VLPs (both 5’-termini covalent and non-covalent54 PEGylation strategies) are shown. Error bars represent the standard error of the mean (n=3).
To further demonstrate the application of this workflow to translational research, we explored fabricating a DNA-VLP vaccine. Our groups recently reported on DNA-VLPs that organize the HIV antigen eOD-GT8 (PDB: 5IDL) at the nanoscale to probe B-cell receptor (BCR) activation.27 The antigen was conjugated to the nanostructures through hybridization of the antigen onto ssDNA handles. Here, we fabricated I52-eOD (Figure 4.A) using the covalent functionalization methodology and observed quantitative coverage (Figure 2.B) while maintaining a monodisperse DNA-VLP (Figure 2.E and Figure S6). TEM characterization (Figure 4.B and Figure S8) validated a regular array of antigens scaffolded on the DNA-VLPs at the nanoscale. We then incubated this construct with Ramos B-cells that recombinantly express an IgM-BCR specific for eOD-GT8 and B-cell activation was quantified using a Ca2+ flux assay. The activation was comparable to our previously reported construct27 (I52-eOD-H) as well as a recombinant protein nanoparticle55 (P-VLP) (Figure 4.C and Figure S9). We conclude that the developed protocol for covalent functionalization is suitable to explore the role of nanoscale antigen organization on B-cell activation in future studies.
Figure 4. Antigen-functionalized DNA-VLPs activate B-cell receptors in vitro.
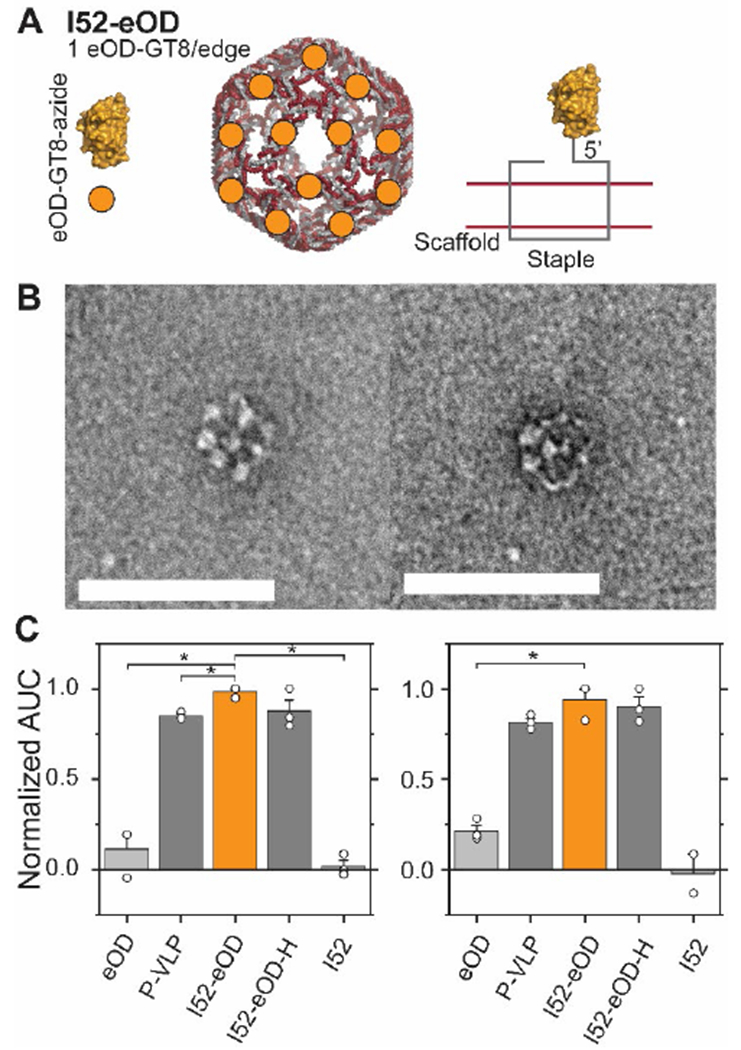
A) I52-eOD was fabricated using a covalent strategy by installing a reactive azide onto the eOD-GT8 antigen. B) Representative TEM micrographs of I52-eOD shows structural array of antigens at the nanoscale. Scale bars represent 100 nm. C) Calcium flux assays comparing covalent ligation to previously published constructs. P-VLP is a published protein nanoparticle presenting 60 copies of eOD-GT8.55 I52-eOD-H is a published DNA-VLP that hybridizes eOD-GT8 onto the nanostructure.27 The Ca2+ flux assay was conducted at a total eOD-GT8 concentration of 5 nM (left) and 1 nM (right). Error bars represent the standard error of the mean (n=3, n=2 for I52, 1nM). P values are from a paired Student’s t-test (*P<0.05).
Conclusions
We report a scalable and efficient fabrication protocol for covalently functionalized DNA origami. This work overcomes several key technical challenges, compared with previous work,44–46 towards fabricating and characterizing covalently functionalized DNA origami nanostructures through a post-assembly functionalization workflow. First, we identified and optimized reaction protocols to achieve quantitative reaction conversions with efficient reaction stoichiometries. Second, we report the post-assembly, covalent conjugations of carbohydrates, peptides, proteins, polymers, and small molecules that play central roles in the development of this technology as vaccines and therapeutic delivery vehicles. Third, we introduce an analytical HPLC technique that can quantitatively monitor reaction conversions for diverse conjugates of interest. In contrast with other characterization techniques, this advance allows for the characterization of functionalized DNA origami that was previously difficult to characterize, such as 1) nanostructures functionalized with small molecule conjugates; 2) nanostructures functionalized with conjugates that have no spectroscopic fingerprint; and 3) nanostructures functionalized with dense arrays of conjugates. Additionally, this analytical technique is agnostic to sample purity, and is applicable to different types of DNA nanostructures, including 1D, 2D, and 3D origami objects. Finally, we present an overall workflow that is agnostic to nanostructure type and conjugate type, allowing for other researchers to adopt the present methodology to their individual applications.
We demonstrated the utility of our protocol in two applications. PEGylation of the 5’-termini of staples offered several hours of protection against nucleases, which may be further inhibited by extending this approach to reactive nucleotides internal to staples. PEGylation also offers other desirable properties such as decreasing opsonization and increasing circulation time, which may be evaluated in future work.56 Conjugation of DNA-VLPs with a clinically relevant HIV antigen preserved its antigenic properties as evaluated in vitro using a B-cell reporter cell line. Evaluating whether this covalent approach affords increased stability when compared with the previously published hybridization27 approach will help direct future in vivo evaluations of these DNA-VLP vaccines. Because the present methodology is compatible with liquid handling conditions, we envision its integration into automated fabrication pipelines to generate libraries of covalently functionalized DNA-VLPs with distinct moieties for screening delivery vehicles’ and vaccine candidates’ performances.
Methods
DNA-VLP Design
DNA-VLP constructs were designed using DAEDALUS.17 For I52-30xDBCO, nick positions were modified for one staple on each edge of the nanostructure to position the 5’ end facing outwards from the nanostructure. These staples were then extended on the 5’ end with TT and TEG-DBCO.
DNA-VLP Fabrication
I52 and I52-30xDBCO were assembled as previously described.17 Briefly, 30 nM of scaffold and 150 nM of each oligonucleotide staple were dissolved in TAE buffer with 12 mM MgCl2 and thermally annealed. DNA-VLPs were purified into PBS using Amicon Ultra centrifugal filters (100 kDa) and stored at 4°C.
DNA-VLP Functionalization
I52-30xDBCO, at 250 nM (100 nM for eOD-GT8 reactions), was incubated with excess of azido-functionalized conjugates (25 eq. D-mannose, cyclo(RGDS), Cy5; 40 eq. eOD-GT8; 50 eq. PEG-10kDa) and allowed to react at room temperature for 16 hours. 5% DMF or 10% DMSO (for protein reactions) were added as co-solvents. Functionalized DNA-VLPs were then purified using Amicon Ultra centrifugal filters (100 kDa) into PBS. Pure functionalized DNA-VLPs were stored at 4°C.
HPLC Conversion Analysis
For all HPLC experiments, the following gradient was used, with a 0.1 M triethylammonium acetate in water: acetonitrile solvent system: t=0 min, 90:10; t=1 min, 90:10; t=11 min. 55:45 (linear ramp); t=16 min, 20:80 (linear ramp); t=20 min: 90:10 (step ramp). Injections of DNA-VLPs were at the following conditions: 60 nM [DNA-VLP], 50 μL. For calculating reaction conversion, full HPLC traces were baseline subtracted. Then, reagent and product peak areas were integrated. These peak areas were used to calculate the reaction conversion using the following formula: Reaction Conversion = Areaproduct/(Areaproduct+Areareagant)
Supplementary Material
Acknowledgements
We are grateful to Dr. Xiao Wang for synthesizing DBCO-modified oligonucleotide staples.
G.A.K., E-C.W., and M.B. were supported by NSF CCF-1564025, NSF DMREF CBET-1729397, NIH R21-EB026008, NIH R01-MH112694, ONR N00014-17-1-2609, ARO ISN W911NF-13-D-0001, and FastGrant AGMT EFF 4/15/20. E.-C. W. was additionally supported by the Feodor Lynen Fellowship of the Alexander von Humboldt Foundation. D.J.I. and B.R. were supported by the U. S. Army Research Office through the Institute for Soldier Nanotechnologies at MIT (Cooperative Agreement Number W911NF-18-2-0048), the Ragon Institute of MGH, MIT, and Harvard, the Marble Center for Nanomedicine, and the NIH (UM1 AI144462). DJI is an investigator of the Howard Hughes Medical Institute. This work made use of the MRSEC Shared Experimental Facilities at MIT, supported by NSF DMR-1419807, and a core center grant P30-ES002109, NIEHS.
Footnotes
Supporting Information
The supporting information is available free of charge online.
Extended experimental methods section; additional structural characterization of DNA-VLPs; reaction optimization analysis; additional information on HPLC trace analysis; additional information on the fluorescent-based reaction conversion assay; additional information on DNA-VLP stability; additional information on DNA-VLP B-cell signaling; table of additional information on reaction conversions; table of additional information on scaffold and staple sequences
References
- 1.Wamhoff EC; Banal JL; Bricker WP; Shepherd TR; Parsons MF; Veneziano R; Stone MB; Jun H; Wang X; Bathe M, Programming Structured DNA Assemblies to Probe Biophysical Processes. Annu Rev Biophys 2019, 48, 395–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bujold KE; Lacroix A; Sleiman HF, DNA Nanostructures at the Interface with Biology. Chem-Us 2018, 4 (3), 495–521. [Google Scholar]
- 3.Dobrovolskaia MA; Bathe M, Opportunities and Challenges for the Clinical Translation of Structured DNA Assemblies as Gene Therapeutic Delivery and Vaccine Vectors. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2021, 13 (1), e1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellmeier J; Platzer R; Eklund AS; Schlichthaerle T; Karner A; Motsch V; Schneider MC; Kurz E; Bamieh V; Brameshuber M; Preiner J; Jungmann R; Stockinger H; Schutz GJ; Huppa JB; Sevcsik E, DNA Origami Demonstrate the Unique Stimulatory Power of Single pMHCs as T Cell Antigens. Proc Natl Acad Sci U S A 2021, 118 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw A; Hoffecker IT; Smyrlaki I; Rosa J; Grevys A; Bratlie D; Sandlie I; Michaelsen TE; Andersen JT; Hogberg B, Binding to Nanopatterned Antigens is Dominated by the Spatial Tolerance of Antibodies. Nat Nanotechnol 2019, 14 (2), 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S; Jiang Q; Liu S; Zhang Y; Tian Y; Song C; Wang J; Zou Y; Anderson GJ; Han JY; Chang Y; Liu Y; Zhang C; Chen L; Zhou G; Nie G; Yan H; Ding B; Zhao Y, A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger in Vivo. Nat Biotechnol 2018, 36 (3), 258–264. [DOI] [PubMed] [Google Scholar]
- 7.Liu S; Jiang Q; Zhao X; Zhao R; Wang Y; Wang Y; Liu J; Shang Y; Zhao S; Wu T; Zhang Y; Nie G; Ding B, A DNA Nanodevice-Based Vaccine for Cancer Immunotherapy. Nat Mater 2021, 20 (3), 421–430. [DOI] [PubMed] [Google Scholar]
- 8.Bujold KE; Hsu JCC; Sleiman HF, Optimized DNA “Nanosuitcases” for Encapsulation and Conditional Release of siRNA. J Am Chem Soc 2016, 138 (42), 14030–14038. [DOI] [PubMed] [Google Scholar]
- 9.Ouyang C; Zhang S; Xue C; Yu X; Xu H; Wang Z; Lu Y; Wu ZS, Precision-Guided Missile-Like DNA Nanostructure Containing Warhead and Guidance Control for Aptamer-Based Targeted Drug Delivery into Cancer Cells in Vitro and in Vivo. J Am Chem Soc 2020, 142 (3), 1265–1277. [DOI] [PubMed] [Google Scholar]
- 10.Douglas SM; Bachelet I; Church GM, A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335 (6070), 831–4. [DOI] [PubMed] [Google Scholar]
- 11.Rothemund PW, Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440 (7082), 297–302. [DOI] [PubMed] [Google Scholar]
- 12.Torring T; Voigt NV; Nangreave J; Yan H; Gothelf KV, DNA Origami: A Quantum Leap for Self-Assembly of Complex Structures. Chem Soc Rev 2011, 40 (12), 5636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong F; Zhang F; Liu Y; Yan H, DNA Origami: Scaffolds for Creating Higher Order Structures. Chem Rev 2017, 117 (20), 12584–12640. [DOI] [PubMed] [Google Scholar]
- 14.Dey S; Fan C; Gothelf KV; Li J; Lin C; Liu L; Liu N; Nijenhuis MAD; Saccà B; Simmel FC; Yan H; Zhan P, DNA Origami. Nat Rev Methods Primers 2021, 1 (1), 13. [Google Scholar]
- 15.Zhang F; Jiang S; Wu S; Li Y; Mao C; Liu Y; Yan H, Complex Wireframe DNA Origami Nanostructures with Multi-Arm Junction Vertices. Nat Nanotechnol 2015, 10 (9), 779–84. [DOI] [PubMed] [Google Scholar]
- 16.Benson E; Mohammed A; Gardell J; Masich S; Czeizler E; Orponen P; Hogberg B, DNA Rendering of Polyhedral Meshes at the Nanoscale. Nature 2015, 523 (7561), 441–4. [DOI] [PubMed] [Google Scholar]
- 17.Veneziano R; Ratanalert S; Zhang K; Zhang F; Yan H; Chiu W; Bathe M, Designer Nanoscale DNA Assemblies Programmed from the Top Down. Science 2016, 352 (6293), 1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun H; Zhang F; Shepherd T; Ratanalert S; Qi X; Yan H; Bathe M, Autonomously Designed Free-Form 2D DNA Origami. Sci Adv 2019, 5 (1), eaav0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun H; Shepherd TR; Zhang K; Bricker WP; Li S; Chiu W; Bathe M, Automated Sequence Design of 3D Polyhedral Wireframe DNA Origami with Honeycomb Edges. ACS Nano 2019, 13 (2), 2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jun H; Wang X; Bricker WP; Bathe M, Automated Sequence Design of 2D Wireframe DNA Origami with Honeycomb Edges. Nat Commun 2019, 10 (1), 5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veneziano R; Shepherd TR; Ratanalert S; Bellou L; Tao C; Bathe M, In Vitro Synthesis of Gene-Length Single-Stranded DNA. Sci Rep 2018, 8 (1), 6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shepherd TR; Du RR; Huang H; Wamhoff EC; Bathe M, Bioproduction of Pure, Kilobase-Scale Single-Stranded DNA. Sci Rep 2019, 9 (1), 6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelhardt FAS; Praetorius F; Wachauf CH; Bruggenthies G; Kohler F; Kick B; Kadletz KL; Pham PN; Behler KL; Gerling T; Dietz H, Custom-Size, Functional, and Durable DNA Origami with Design-Specific Scaffolds. ACS Nano 2019, 13 (5), 5015–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afonin KA; Dobrovolskaia MA; Church G; Bathe M, Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2020, 14 (8), 9221–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madsen M; Gothelf KV, Chemistries for DNA Nanotechnology. Chem Rev 2019, 119 (10), 6384–6458. [DOI] [PubMed] [Google Scholar]
- 26.Hahn J; Chou LYT; Sorensen RS; Guerra RM; Shih WM, Extrusion of RNA from a DNA-Origami-Based Nanofactory. ACS Nano 2020, 14 (2), 1550–1559. [DOI] [PubMed] [Google Scholar]
- 27.Veneziano R; Moyer TJ; Stone MB; Wamhoff EC; Read BJ; Mukherjee S; Shepherd TR; Das J; Schief WR; Irvine DJ; Bathe M, Role of Nanoscale Antigen Organization on B-Cell Activation Probed Using DNA Origami. Nat Nanotechnol 2020, 15 (8), 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marth G; Hartley AM; Reddington SC; Sargisson LL; Parcollet M; Dunn KE; Jones DD; Stulz E, Precision Templated Bottom-Up Multiprotein Nanoassembly through Defined Click Chemistry Linkage to DNA. ACS Nano 2017, 11 (5), 5003–5010. [DOI] [PubMed] [Google Scholar]
- 29.Fu J; Liu M; Liu Y; Woodbury NW; Yan H, Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures. J Am Chem Soc 2012, 134 (12), 5516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephanopoulos N; Liu M; Tong GJ; Li Z; Liu Y; Yan H; Francis MB, Immobilization and One-Dimensional Arrangement of Virus Capsids with Nanoscale Precision Using DNA Origami. Nano Lett 2010, 10 (7), 2714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw A; Benson E; Hogberg B, Purification of Functionalized DNA Origami Nanostructures. ACS Nano 2015, 9 (5), 4968–75. [DOI] [PubMed] [Google Scholar]
- 32.Fisher PDE; Shen Q; Akpinar B; Davis LK; Chung KKH; Baddeley D; Saric A; Melia TJ; Hoogenboom BW; Lin C; Lusk CP, A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement. ACS Nano 2018, 12 (2), 1508–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin J; Baker EG; Wood CW; Bath J; Woolfson DN; Turberfield AJ, Peptide Assembly Directed and Quantified Using Megadalton DNA Nanostructures. ACS Nano 2019, 13 (9), 9927–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchberger A; Simmons CR; Fahmi NE; Freeman R; Stephanopoulos N, Hierarchical Assembly of Nucleic Acid/Coiled-Coil Peptide Nanostructures. J Am Chem Soc 2020, 142 (3), 1406–1416. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y; Wang J; Shigematsu H; Xu W; Shih WM; Rothman JE; Lin C, Self-Assembly of Size-Controlled Liposomes on DNA Nanotemplates. Nat Chem 2016, 8 (5), 476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreiber R; Do J; Roller EM; Zhang T; Schuller VJ; Nickels PC; Feldmann J; Liedl T, Hierarchical Assembly of Metal Nanoparticles, Quantum Dots and Organic Dyes Using DNA Origami Scaffolds. Nat Nanotechnol 2014, 9 (1), 74–8. [DOI] [PubMed] [Google Scholar]
- 37.Ora A; Jarvihaavisto E; Zhang H; Auvinen H; Santos HA; Kostiainen MA; Linko V, Cellular Delivery of Enzyme-Loaded DNA origami. Chem Commun (Camb) 2016, 52 (98), 14161–14164. [DOI] [PubMed] [Google Scholar]
- 38.Andersen ES; Dong M; Nielsen MM; Jahn K; Subramani R; Mamdouh W; Golas MM; Sander B; Stark H; Oliveira CL; Pedersen JS; Birkedal V; Besenbacher F; Gothelf KV; Kjems J, Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature 2009, 459 (7243), 73–6. [DOI] [PubMed] [Google Scholar]
- 39.Jahn K; Torring T; Voigt NV; Sorensen RS; Bank Kodal AL; Andersen ES; Gothelf KV; Kjems J, Functional Patterning of DNA Origami by Parallel Enzymatic Modification. Bioconjug Chem 2011, 22 (4), 819–23. [DOI] [PubMed] [Google Scholar]
- 40.Voigt NV; Torring T; Rotaru A; Jacobsen MF; Ravnsbaek JB; Subramani R; Mamdouh W; Kjems J; Mokhir A; Besenbacher F; Gothelf KV, Single-Molecule Chemical Reactions on DNA Origami. Nat Nanotechnol 2010, 5 (3), 200–3. [DOI] [PubMed] [Google Scholar]
- 41.Sacca B; Meyer R; Erkelenz M; Kiko K; Arndt A; Schroeder H; Rabe KS; Niemeyer CM, Orthogonal Protein Decoration of DNA Origami. Angew Chem Int Ed Engl 2010, 49 (49), 9378–83. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen TM; Nakata E; Saimura M; Dinh H; Morii T, Design of Modular Protein Tags for Orthogonal Covalent Bond Formation at Specific DNA Sequences. J Am Chem Soc 2017, 139 (25), 8487–8496. [DOI] [PubMed] [Google Scholar]
- 43.Timm C; Niemeyer CM, Assembly and Purification of Enzyme-Functionalized DNA Origami Structures. Angew Chem Int Ed Engl 2015, 54 (23), 6745–50. [DOI] [PubMed] [Google Scholar]
- 44.Grossi G; Dalgaard Ebbesen Jepsen M; Kjems J; Andersen ES, Control of Enzyme Reactions by a Reconfigurable DNA Nanovault. Nat Commun 2017, 8 (1), 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z; Xiong Y; Xiang S; Gang O, Controllable Covalent-Bound Nanoarchitectures from DNA Frames. J Am Chem Soc 2019, 141 (17), 6797–6801. [DOI] [PubMed] [Google Scholar]
- 46.Heck C; Torchinsky D; Nifker G; Gularek F; Michaeli Y; Weinhold E; Ebenstein Y, Label as You Fold: Methyltransferase-Assisted Functionalization of DNA Nanostructures. Nanoscale 2020, 12 (39), 20287–20291. [DOI] [PubMed] [Google Scholar]
- 47.Funke JJ; Dietz H, Placing Molecules with Bohr Radius Resolution Using DNA Origami. Nat Nanotechnol 2016, 11 (1), 47–52. [DOI] [PubMed] [Google Scholar]
- 48.Mathur D; Medintz IL, Analyzing DNA Nanotechnology: A Call to Arms for the Analytical Chemistry Community. Anal Chem 2017, 89 (5), 2646–2663. [DOI] [PubMed] [Google Scholar]
- 49.Kolb HC; Finn MG; Sharpless KB, Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl 2001, 40 (11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- 50.El-Sagheer AH; Brown T, Click Chemistry with DNA. Chem Soc Rev 2010, 39 (4), 1388–405. [DOI] [PubMed] [Google Scholar]
- 51.Kretschy N; Sack M; Somoza MM, Sequence-Dependent Fluorescence of Cy3- and Cy5-Labeled Double-Stranded DNA. Bioconjug Chem 2016, 27 (3), 840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conway JW; McLaughlin CK; Castor KJ; Sleiman H, DNA Nanostructure Serum Stability: Greater than the Sum of Its Parts. Chem Commun (Camb) 2013, 49 (12), 1172–4. [DOI] [PubMed] [Google Scholar]
- 53.Lacroix A; Vengut-Climent E; de Rochambeau D; Sleiman HF, Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale. ACS Cent Sci 2019, 5 (5), 882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ponnuswamy N; Bastings MMC; Nathwani B; Ryu JH; Chou LYT; Vinther M; Li WA; Anastassacos FM; Mooney DJ; Shih WM, Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation. Nat Commun 2017, 8, 15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jardine J; Julien JP; Menis S; Ota T; Kalyuzhniy O; McGuire A; Sok D; Huang PS; MacPherson S; Jones M; Nieusma T; Mathison J; Baker D; Ward AB; Burton DR; Stamatatos L; Nemazee D; Wilson IA; Schief WR, Rational HIV Immunogen Design to Target Specific Germline B Cell Receptors. Science 2013, 340 (6133), 711–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suk JS; Xu Q; Kim N; Hanes J; Ensign LM, PEGylation as a Strategy for Improving Nanoparticle-Based Drug and Gene Delivery. Adv Drug Deliv Rev 2016, 99 (Pt A), 28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


