Abstract
The ETS protein ER81 is a DNA-binding factor capable of enhancing gene transcription and is implicated in cellular transformation, but presently the mechanisms of its actions are unclear. In this report, ER81 is shown to coimmunoprecipitate with the transcriptional coactivator CREB-binding protein (CBP) and the related p300 protein (together referred to as CBP/p300). Moreover, confocal laser microscopic studies demonstrated that ER81 and p300 colocalized to nuclear speckles. In vitro and in vivo interaction studies revealed that ER81 amino acids 249 to 429, which encompass the ETS DNA-binding domain, are responsible for binding to CBP/p300. However, mutation of a putative protein-protein interaction motif, LXXLL, in the ETS domain of ER81 did not affect interaction with CBP/p300, whereas DNA binding of ER81 was abolished. Furthermore, two regions within CBP, amino acids 451 to 721 and 1891 to 2175, are capable of binding to ER81. Consistent with the physical interaction between ER81 and the coactivators CBP and p300, ER81 transcriptional activity was potentiated by CBP/p300 overexpression. Moreover, an ER81-associated protein kinase activity was enhanced upon p300 overexpression. This protein kinase phosphorylates ER81 on serines 191 and 216, and mutation of these phosphorylation sites increased ER81 transcriptional activity in Mv1Lu cells but not in HeLa cells. Altogether, our data elucidate the mechanism of how ER81 regulates gene transcription, through interaction with the coactivators CBP and p300 and an associated kinase that may cell type specifically modulate the ability of ER81 to activate gene transcription.
The ETS family of transcription factors consists of a large number of proteins that perform diverse functions, such as serum stimulation of the c-fos promoter (52), activation of the herpes simplex virus immediate-early promoter (36), regulation of immunoglobulin light-chain enhancers (13), control of lymphoid and myeloid lineage commitment during hematopoiesis (15, 43), and Drosophila eye development (46). A characteristic feature of this class of proteins is a highly conserved, 85-amino-acid-long DNA-binding motif termed the ETS domain, which specifically binds to GGA(A/T)-containing DNA target sequences. Some ETS proteins also display homology outside the ETS domain, suggestive of a common ancestor as a founder of a subfamily of ETS proteins. For instance, one ETS subfamily consists of ER81 (7, 31, 41), PEA3 (23, 54), and ERM (40). These three proteins display ∼95% identity within the ETS domain, compared to only ∼60% identity with the prototypical ETS domain of c-Ets-1, and more than 85 and 50% in the N- and C-terminal transcription activation domains, respectively (35).
Northern blot analysis revealed that ER81 is not ubiquitously expressed: high expression is observed in brain, heart, and lung and moderate levels have been detected in spleen, pancreas, intestine, and colon, whereas little expression is observable in liver or skeletal muscle (7, 41). However, ER81 expression varies in some tissues within the Mammalia; for instance, the levels of ER81 mRNA in the kidney were high in mice (7) but very low in humans (41). Moreover, human ER81 is highly expressed in several tumor cell lines (41), and an analysis of breast cancer cell lines revealed that ER81 mRNA levels, but not those of PEA3 or ERM, were significantly elevated in some of these cell lines. Interestingly, ER81 expression was inversely correlated to the presence of mRNA for estrogen and progesterone receptor in these breast cancer cell lines (3), indicating that ER81 expression may be restricted to a subtype of breast cancer cells growing steroid hormone independently and thereby being refractory to tamoxifen treatment. These data suggest that ER81 may contribute to the transformation of certain cell lines.
Several members of the ETS family, such as c-Ets-1 and c-Ets-2 (10), c-fos-regulating ternary complex factors (19, 26, 39), the Drosophila Pointed-P2 protein (8, 46), and the transcriptional repressor ERF (49), are targeted by mitogen-activated protein kinases (MAPKs). Similarly, we have demonstrated that ER81 is phosphorylated by ERK1-MAPK in vitro and is a target of the Ras/Raf/MEK/ERK signaling cascade in vivo (24). However, it is still unknown whether ERK1-MAPK directly phosphorylates and activates ER81 in vivo.
A variety of transcription factors, including the AP-1 components Fos and Jun (2, 5), the cyclic AMP response element-binding protein (CREB) (9), as well as Ets-1 and Ets-2 (30, 55), interact with the homologous coactivators CREB-binding protein (CBP) and p300 (collectively referred to as CBP/p300) to mediate RNA polymerase II-dependent gene transcription. Although it is unclear how these protein-protein interactions lead to transactivation, one suggestion is that CBP/p300 acts as an adaptor between these transcription factors and components of the basal transcription machinery such as TFIID and TFIIB, or possibly RNA polymerase II itself (1, 32, 34). Since CBP/p300 possesses intrinsic histone acetyltransferase activity, CBP/p300 recruitment could also activate chromatin-repressed promoters and enhancers by acetylation of histones or other proteins involved in promoter regulation (4, 45).
Targeted gene disruption studies have demonstrated that p300 function is essential for normal embryonic cellular proliferation and morphogenesis and, similarly, CBP knock-out mice display an embryo-lethal phenotype (56). Interestingly, although heterozygous p300 and CBP knock-outs are viable, a p300+/− CBP+/− double heterozygote is embryo lethal, suggesting that a combined gene dosage of at least three active CBP and p300 alleles is required for viability and indicating that the CBP and p300 proteins perform similar functions in the cell. Furthermore, haploinsufficiency of CBP has been correlated to Rubinstein-Taybi syndrome, which is characterized by severe developmental abnormalities, including mental retardation, craniofacial and skeletal abnormalities, and increased cancer incidence (17, 48). Consistently, heterozygous CBP+/− knock-out mice display many but not all of the phenotypic changes associated with Rubinstein-Taybi syndrome (50).
In this report, we demonstrate that the nuclear protein ER81 physically associates with CBP/p300 in vitro as well as in vivo and that this physical interaction potentiates ER81-dependent transcription. In addition, an ER81-associated protein kinase has been detected, whose activity is elevated in the presence of CBP/p300. This ER81-associated kinase phosphorylates ER81 on serines 191 and 216, thereby affecting its transcriptional activity in a cell type-specific manner. Altogether, our data suggest mechanisms of gene regulation by a complex of ER81, an ER81-associated protein kinase, and the homologous coactivators CBP and p300.
MATERIALS AND METHODS
Plasmids.
Murine ER81 and truncations thereof were cloned into eukaryotic expression vectors providing an amino-terminal hemagglutinin (HA) tag or six copies of a Myc tag (6Myc) according to standard techniques. The TORU-luc (27), Fos-luc, and GAL4-luc (26) reporter plasmids have been described previously. Expression vectors for GAL4 and glutathione S-transferase (GST) fusion proteins have been reported (24, 28). Site-directed mutagenesis was accomplished by utilizing standard PCR procedures and subsequent verification by DNA sequencing.
Luciferase reporter gene assays.
Mink lung (Mv1Lu) or HeLa cells plated on 6-cm dishes were transiently transfected by the calcium phosphate coprecipitation method. Cells were harvested 36 h after transfection and lysed, and the cleared lysate was used to measure luciferase activity and also β-galactosidase activity from a cotransfected, constitutively active β-galactosidase expression vector (pEQ176). The latter was used to normalize luciferase activity for transfection efficiency (26).
Western blotting.
ER81 proteins were separated in sodium dodecyl sulfate (SDS)–8% polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore) in 0.3 M Tris-HCl (pH 10.5)–20% methanol (anode buffer) and 25 mM Tris-HCl (pH 9.4)–40 mM ɛ-amino-n-hexanoic acid–20% methanol (cathode buffer) at 0.7 mA/cm2 for 80 min (semi-dry blotter, EBU-4000; C.B.S. Scientific). HA-tagged p300 and HA-tagged CBP were separated in SDS–5% polyacrylamide gels and then transferred to PVDF membrane in 20 mM Tris–150 mM glycine–0.1% SDS–10% methanol at 40 V for 16 h at 4°C (Trans-blot cell; Bio-Rad). Membranes were blocked with 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–0.05% Tween 20–3% bovine serum albumin for 1 h at room temperature and then incubated with anti-HA (12CA5; 1:5,000) or anti-Myc (9E10; 1:5,000) murine monoclonal antibodies for 2 h. The membranes were washed four times with 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–0.05% Tween 20 and then incubated with a 1:5,000 dilution of horseradish peroxidase-linked anti-mouse immunoglobulin G (IgG) secondary antibody. The immunoreactive proteins were finally detected with an enhanced chemiluminescence kit (ECL; Amersham-Pharmacia).
Production of GST fusion proteins.
GST fusion proteins were expressed in Escherichia coli BL21 cells. Enrichment of the fusion proteins was done on Ni2+-nitrilotriacetic acid-agarose columns (Qiagen) in 6 M guanidine HCl by virtue of a histidine tag present at the junction between the GST moiety and the fused proteins. Proteins were renatured by dialysis against 50 mM Tris-HCl (pH 7.5)–100 mM NaCl–0.2 mM dithiothreitol–0.5 mM phenylmethylsulfonyl fluoride.
GST pull-down assays.
293T cells were transiently transfected with HA-tagged or Myc-tagged proteins by the calcium phosphate coprecipitation method. Whole-cell extracts were prepared in 600 μl of lysis buffer (5 mM Tris-HCl [pH 7.1], 15 mM Na4P2O7, 25 mM NaCl, 25 mM NaF, 0.5% Triton X-100) supplemented with inhibitor mix (1 mM phenylmethylsulfonyl fluoride; 2 μg of aprotinin, 10 μg of leupeptin, and 1 μg of pepstatin A per ml; and 0.2 mM Na3VO4) and 0.2 mM dithiothreitol. Approximately 200 ng of GST fusion proteins was bound to glutathione-agarose (20-μl bed volume), washed in binding buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 0.2 mM EDTA, 0.2 mM Na3VO4, 0.05% Tween 20, 0.2 mM dithiothreitol) and incubated with 20 to 100 μl of 293T cell extracts in a final volume of 600 μl for 1 h at 4°C. Bound proteins were washed three times in binding buffer, then Laemmli sample buffer was added, and the beads were boiled for 2 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and Western blotting was subsequently performed.
Immunoprecipitations.
293T cells were transiently transfected with HA-tagged or Myc-tagged proteins. At 36 h after transfection, cells were washed once with phosphate-buffered saline (PBS) and then lysed as described above for 30 min at 4°C. In the case of anti-Myc coimmunoprecipitation of p300, cells were lysed in 5 mM Tris-HCl (pH 7.1)–15 mM Na4P2O7–50 mM NaCl–25 mM NaF–0.22% Triton X-100 supplemented with inhibitor mix, 0.2 mM dithiothreitol, and 2% bovine serum albumin. Lysates were cleared by centrifugation, 25 μl of protein A-agarose (Repligen) slurry was added, and the lysates were precleared for 45 min with rotation at 4°C. After centrifugation, the lysates were incubated with anti-HA or anti-Myc murine monoclonal antibodies (12CA5 and 9E10, respectively) for 2 h with rotation at 4°C. Precipitates were washed three times with lysis buffer and once with 20 mM HEPES (pH 7.4), and either resuspended in Laemmli sample buffer or used for in vitro phosphorylation assays. For the immunoprecipitation of endogenous ER81 proteins, HeLa cells were lysed as for the anti-Myc coimmunoprecipitations, and 1 μl of rabbit anti-ER81 antibodies was employed.
In vitro phosphorylation.
Immunoprecipitated Myc-tagged ER81 was phosphorylated on the protein A-agarose beads for 30 min at 30°C in a volume of 20 μl with 1 μCi of [γ-32P]ATP (3,000 Ci/mmol). The reaction buffer was composed of 20 mM HEPES (pH 7.4), 12 mM MgCl2, 15 mM sodium β-glycerophosphate, 0.5 mM EGTA, 0.2 mM dithiothreitol, 0.5 mM Na3VO4, and 10 μM ATP. The reaction was terminated by the addition of Laemmli sample buffer and boiling.
In vivo phosphorylation and phosphopeptide mapping.
293T cells were transiently transfected with Myc-tagged ER81. Thirty-six hours after transfection, cells were metabolically labeled with 32Pi for 4 h. Then, ER81 was immunoprecipitated with anti-Myc monoclonal antibody 9E10, and the immunoprecipitate was boiled in Laemmli sample buffer and subjected to SDS-PAGE. The phosphorylated protein was eluted from the gel, treated with performic acid, and digested with trypsin, and the resulting phosphopeptides were resolved by electrophoresis in the first dimension (pH 1.9 buffer, 1 kV, 30 min) and in the second dimension by ascending chromatography employing phosphochromatography buffer on cellulose thin-layer plates (53).
Gel retardation assay.
Gel retardation assays were performed with extracts from transiently transfected 293T cells and with the 32P-labeled E74 oligonucleotide. Complexes were resolved on a 5% native polyacrylamide gel at 4°C as previously described (25).
Immunofluorescence studies.
Mv1Lu cells were grown on coverslips and cotransfected with expression plasmids for Myc-tagged and HA-tagged proteins by the calcium phosphate coprecipitation method (26). The precipitate was left on the cells for 10 h and then washed away, and the cells were further incubated for 36 h. Then, the cells were washed twice in PBS and subsequently fixed for 10 min in PBS–2% sucrose–3.7% formaldehyde. After one wash with PBS–0.1 M glycine for 5 min, cells were blocked and permeabilized with PBS–2% normal donkey serum–0.4% Triton X-100 for 20 min. Coverslips were then incubated for 1 h with anti-Myc murine monoclonal antibody (9E10) and anti-HA rabbit polyclonal antibody (Y-11; Santa Cruz Biotechnology). After four washes with PBS–0.2% bovine serum albumin–0.1% Triton X-100, coverslips were incubated for 1 h with donkey anti-rabbit IgG antibody coupled to fluorescein isothiocyanate (FITC) and donkey anti-mouse IgG antibody coupled to Texas Red. Finally, coverslips were washed twice each with PBS–0.2% bovine serum albumin–0.1% Triton X-100 and PBS and then mounted on glass slides.
RESULTS
ER81 and CBP/p300 are in a complex in vivo.
CBP and p300 are versatile coactivators which have been shown to cooperate with the prototypical ETS proteins Ets-1 and Ets-2 (30, 55). Therefore, we wished to investigate whether CBP/p300 can interact with the ETS protein ER81 and thereby facilitate ER81-dependent gene transcription. To address this question, we first employed coimmunoprecipitation assays. 293T cells were transiently transfected with Myc-tagged, full-length ER81 and HA-tagged CBP, and the cells were lysed 36 h after transfection. The cell lysate was immunoprecipitated with anti-HA antibodies, the immunoprecipitated proteins were then separated by SDS-PAGE, and Western blotting with anti-Myc antibodies was performed. As shown in Fig. 1A (lane 3), ER81 coimmunoprecipitated with CBP. As negative controls, lysates of cells that were transfected with Myc-tagged ER81 or HA-tagged CBP alone were also immunoprecipitated (Fig. 1A, lanes 1 and 2, respectively). No signal for Myc-tagged ER81 could be detected, indicating that ER81 specifically coimmunoprecipitated with CBP.
FIG. 1.
ER81 coimmunoprecipitates with CBP and p300. (A) 293T cells were transiently transfected with HA-tagged CBP and 6Myc-ER81, and immunoprecipitations (IP) with anti-HA antibodies were performed. ER81 present in the immunoprecipitates was detected by immunoblotting with anti-Myc antibodies. The intracellular levels of ER81 (input) are also shown. (B) Similar coimmunoprecipitation assays with HA-tagged p300 and 6Myc-ER81. (C) Reverse-order, anti-Myc immunoprecipitation of 293T cell extracts transiently transfected with p300-HA and 6Myc-ER81. p300 present in the immunoprecipitates was detected by immunoblotting with anti-HA antibodies. The intracellular levels of p300 (input) are also shown. (D) HeLa cell extracts were immunoprecipitated with no antibody, anti-GAL4 antibodies, or anti-ER81 antibodies. Coimmunoprecipitated p300 was detected by Western blotting with anti-p300 antibodies (C-20; Santa Cruz).
Coimmunoprecipitation assays were also conducted with 293T cells that were transiently transfected with Myc-tagged ER81 and HA-tagged p300. As with CBP, a specific coimmunoprecipitation of ER81 with p300 was observed (Fig. 1B). We also performed a reverse-order coimmunoprecipitation experiment in which proteins immunoprecipitated with anti-Myc antibodies were separated by SDS-PAGE and ER81-associated proteins were detected by anti-HA Western blotting. In this experiment, p300 coimmunoprecipitated with ER81 (Fig. 1C, lane 3). Again, this coimmunoprecipitation was proven to be specific, since no signal for HA-tagged p300 was detectable with lysates from cells that had been transfected with HA-tagged p300 alone (Fig. 1C, lane 2).
Furthermore, we investigated whether endogenous ER81 and endogenous p300 would coimmunoprecipitate. To this end, HeLa cells were lysed and treated with anti-ER81 antibodies, the immunoprecipitates were resolved by SDS-PAGE, and the presence of p300 was tested by immunoblotting. As shown in Fig. 1D (lane 3), endogenous p300 indeed coimmunoprecipitated with endogenous ER81, whereas no p300 was detected when anti-GAL4 antibodies or no antibodies were utilized for the immunoprecipitation (lanes 1 and 2). Therefore, ER81 and CBP/p300 can interact in vivo.
To further substantiate that ER81 and p300 form a complex in vivo, we performed confocal laser scanning microscopy on Mv1Lu cells transiently transfected with Myc-tagged ER81 and HA-tagged p300. The cells were probed with anti-Myc murine monoclonal antibodies and anti-HA rabbit polyclonal antibodies, followed by detection with donkey anti-rabbit IgG antibody coupled to FITC and donkey anti-mouse IgG antibody coupled to Texas Red. Whereas nontransfected cells and cells that were not incubated with primary antibodies did not exhibit immunostaining (data not shown), both full-length ER81 (ER812–477) and p300 revealed a nuclear, punctate staining pattern (Fig. 2, top panel). Overlaying the two individual staining patterns demonstrated that they coincide to a large degree, as indicated by the appearance of yellow dots as a result of the merging of the red and green colors. As a control, we utilized a C-terminal truncation of ER81 (ER812–334) which has lost its ETS domain and its putative nuclear localization signal. ER812–334 displayed a predominantly cytoplasmic localization, whereas p300 still showed the punctate nuclear staining (Fig. 2, bottom panel), and consequently no colocalization could be observed in the merged picture. Similarly, ER81333–477, which is localized to the cell nucleus but does not interact with p300 (see Fig. 4), did not reveal colocalization with p300 (data not shown). Collectively, these data indicate that ER81 can physically associate and colocalize with the transcriptional coactivators CBP and p300 in vivo.
FIG. 2.
Colocalization of ER81 and p300 in the nucleus, assayed by confocal laser scanning microscopy. Mv1Lu cells were transiently transfected with full-length 6Myc-ER81 (ER812–477) and HA-tagged p300 (top panel). The cells were probed with anti-Myc mouse monoclonal antibodies and anti-HA rabbit polyclonal antibodies, followed by detection with donkey anti-rabbit IgG antibody coupled to FITC and donkey anti-mouse IgG antibody coupled to Texas Red. A C-terminal truncation of ER81 (ER812–334), which lacks the ETS domain and consequently the nuclear localization signal, was used as a negative control (bottom panel).
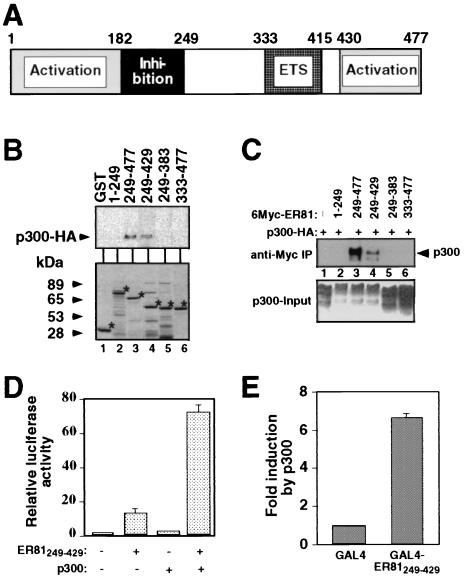
FIG. 4.
ER81 amino acids 249 to 429 are responsible for the interaction with p300. (A) Schematic representation of the murine ER81 molecule. Highlighted are the DNA-binding ETS domain, the N- and C-terminal transactivation domains, and amino acids 182 to 249, which exert an inhibitory effect on the two transactivation domains. (B) Pull-down of HA-tagged p300 by various GST-ER81 fusion proteins. p300 was detected by anti-HA immunoblotting (top panel). The bottom panel shows Coomassie-stained, purified GST-ER81 fusion proteins. Asterisks show the position of each protein. (C) 293T cells were transiently transfected with various truncated forms of Myc-tagged ER81 and HA-p300. Cell lysates were used for anti-Myc immunoprecipitations, and coimmunoprecipitated p300-HA was detected by anti-HA immunoblotting. (D) p300 potentiates transcriptional activity of ER81249–429 in Mv1Lu cells measured with the TORU-luc reporter construct. (E) Activation of GAL4-ER81249–429 by p300 in Mv1Lu cells. A GAL4 DNA-binding site-driven reporter construct was cotransfected with either the GAL4 DNA-binding domain or the GAL4-ER81249–429 fusion protein. Enhancement of luciferase activity upon overexpression of p300 is depicted.
CBP/p300 enhances ER81-mediated transactivation.
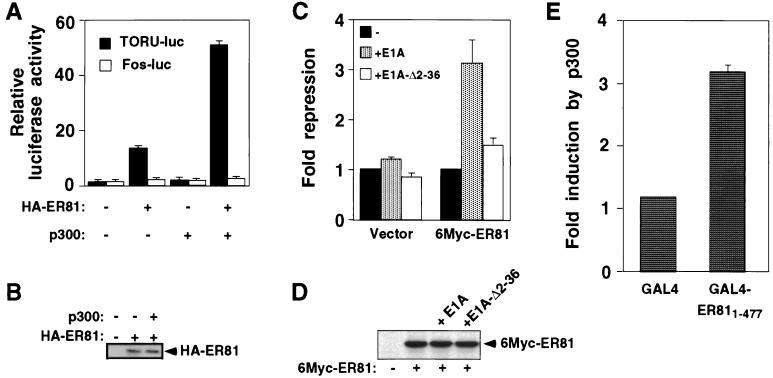
To test whether CBP/p300 has an impact on ER81-mediated transcription, a luciferase reporter construct (TORU-luc) was employed. This reporter plasmid contains an ETS binding site, to which ER81 can bind and thereby facilitate gene transcription (27). Mv1Lu cells were transiently transfected with full-length, HA-tagged ER81 and p300. At 36 h after transfection, cells were lysed and luciferase activity was measured. As shown in Fig. 3A and reported before (27), ER81 on its own already activates the TORU-luc reporter, whereas overexpression of p300 alone had no effect. However, p300 was able to potentiate ER81-mediated transcription by threefold, indicating that ER81 and p300 can synergistically upregulate gene transcription. The same effect was observed when CBP was used instead of p300 (data not shown). Control experiments revealed that the amount of ER81 was not affected by overexpression of p300 (Fig. 3B). Furthermore, we used a c-fos promoter-driven luciferase reporter construct (Fos-luc), which is not targeted by ER81. The Fos-luc reporter was unaffected by the presence of ER81 and p300 (Fig. 3A), indicating that the observed collaboration between ER81 and p300 is promoter specific.
FIG. 3.
Stimulation of ER81-mediated transcription by p300. (A) Mv1Lu cells were transiently transfected with the TORU-luc (black bars) or Fos-luc (white bars) reporter construct. Where indicated, 0.1 μg of HA-tagged ER81 and/or 0.5 μg of p300 was cotransfected. Luciferase activity derived from the reporter constructs is depicted. (B) Detection of HA-tagged ER81 levels by anti-HA immunoblotting of Mv1Lu cell extracts. Where indicated, p300 had been coexpressed with ER81. (C) Mv1Lu cells were transfected with 100 ng of 6Myc-ER81 or empty expression vector and 5 ng of E1A or E1A-Δ2-36, as indicated. Fold repression of the TORU-luc reporter was measured; arbitrarily, fold repression was set to 1 independently in case of the empty expression vector and 6Myc-ER81. (D) Detection of Myc-tagged ER81 levels by anti-Myc immunoblotting of Mv1Lu cell extracts. Where indicated, E1A or E1A-Δ2-36 was coexpressed. (E) p300 potentiates GAL4-ER811–477 activity in transiently transfected Mv1Lu cells. A GAL4-luc reporter construct was transfected with either 0.2 μg of the GAL4 DNA-binding domain or 0.2 μg of the GAL4-ER811–477 fusion. Enhancement of luciferase activity by coexpression of p300 (0.5 μg) was measured.
We wondered whether the transcriptional activation observed with ER81 alone involves an interaction with endogenous CBP/p300. One way of analyzing this is to sequester endogenous CBP/p300 by overexpression of the adenoviral E1A protein (18). Indeed, E1A repressed the transcriptional activity of ER81 by more than threefold, whereas transcription in the absence of overexpressed ER81 was unaffected by E1A (Fig. 3C); no changes in ER81 protein levels were detected upon E1A expression (Fig. 3D). Furthermore, an E1A mutant that is deficient in CBP/p300 binding (E1A-Δ2-36) barely repressed ER81-dependent transcription (Fig. 3C). Thus, ER81 relies on endogenous CBP/p300 in order to fully activate gene transcription.
Next, we tested whether DNA binding of ER81 is required for the functional synergism between ER81 and p300. To this end, we fused full-length ER81 to the GAL4 DNA-binding domain and tested the resulting fusion protein (GAL4-ER811–477) with a luciferase reporter construct driven by GAL4 DNA-binding sites. In this context, DNA binding is mediated by the GAL4 DNA-binding domain but not by the ETS domain of ER81. Whereas barely any activation mediated by p300 was observable with the GAL4 DNA-binding domain, the GAL4-ER811–477 fusion protein was stimulated threefold by p300 (Fig. 3E). Thus, DNA binding of ER81 via its ETS domain is not required for the functional collaboration with p300.
Mapping of the CBP/p300 interaction domain in ER81.
To delineate the regions of ER81 (see Fig. 4A for a diagram of ER81) required for the interaction with CBP/p300, GST pull-down assays were carried out. Various truncated forms of ER81 were overexpressed as GST fusion proteins in E. coli. Figure 4B (bottom panel) shows a Coomassie blue-stained gel of the GST-ER81 fusion proteins after purification. Approximately 200 ng of the purified GST-ER81 fusion proteins was bound to glutathione agarose beads and mixed with 100 μg of protein from lysates of 293T cells transiently overexpressing HA-tagged p300. Bound proteins were separated by SDS-PAGE, transferred to PVDF membrane, and probed with anti-HA antibodies. Figure 4B (upper panel) shows that the N-terminal amino acids of ER81 (GST-ER811–249, lane 2) did not interact with p300, whereas the C-terminal amino acids of ER81 (GST-ER81249–477, lane 3) bound p300. Additional deletion of 48 C-terminal amino acids in GST-ER81249–429 (lane 4) did not prevent binding of p300, whereas further C-terminal truncation into the ETS domain abolished binding of p300 (GST-ER81249–383, lane 5). Thus, ER81 amino acids 384 to 429 are required for the interaction with p300. In contrast to GST-ER81249–477, GST-ER81333–477 (lane 6) did not interact with p300, suggesting that ER81 amino acids 249 to 332 are also indispensable for the interaction with p300. Altogether, amino acids 249 to 429 of ER81 mediate the interaction with p300 in vitro.
Next, we analyzed whether ER81 amino acids 249 to 429 interact with p300 in vivo. To this end, we performed coimmunoprecipitation assays. 293T cells were transiently transfected with the Myc-tagged truncation ER81249–429 in the absence and presence of HA-tagged p300. Precipitations were performed with anti-Myc antibodies, and the immunoprecipitated proteins were separated by SDS-PAGE, transferred to PVDF membrane, and probed with anti-HA antibodies (Fig. 4C). Indeed, p300 interacted with ER81 amino acids 249 to 429 in this coimmunoprecipitation assay. Expectedly, p300 also coimmunoprecipitated with ER81249–477 but not with ER811–249, ER81249–383, or ER81333–477 (Fig. 4C). We conclude that ER81 amino acids 249 to 429 represent the minimal region required for both in vitro and in vivo binding to p300.
Next, we analyzed whether the minimal p300 interaction domain in ER81 (ER81249–429) is activated by p300. Mv1Lu cells were cotransfected with the TORU-luc reporter and ER81249–429. This truncation on its own activates the TORU-luc reporter by 12-fold, which was further enhanced by p300 to ∼70-fold (Fig. 4D). Similarly, a GAL4-ER81249–429 fusion protein was potentiated in its activity by p300 (Fig. 4E). Thus, ER81 amino acids 249 to 429 are able to activate gene expression synergistically with p300.
An LXXLL motif is not involved in ER81-CBP/p300 interactions, but affects DNA binding.
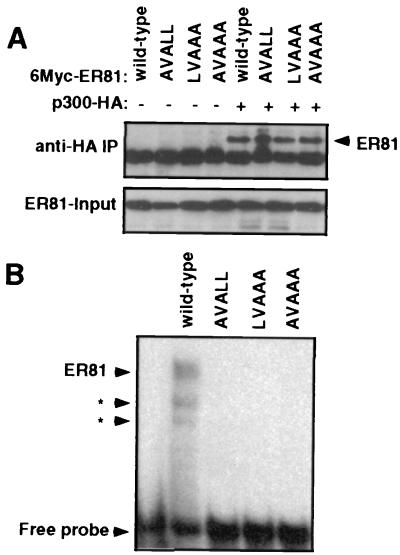
A recent study suggested that protein-protein interactions between nuclear hormone receptors and its coactivators are mediated by a short sequence motif, LXXLL, within the coactivators (22). Such an LXXLL motif is highly conserved in helix α1 of the ETS domain (37). Therefore, we investigated whether mutation of the LVALL motif (ER81 amino acids 341 to 345) affects ER81 binding to CBP/p300. To this end, three different ER81 mutants were generated, AVALL, LVAAA, and AVAAA, in which one or more of the leucine residues in the LVALL motif were replaced by alanine.
We first examined whether these mutants coimmunoprecipitated with p300. Myc-tagged, wild-type and mutant ER81 were coexpressed with HA-tagged p300 in 293T cells, and anti-HA immunoprecipitations were performed. As shown by anti-Myc Western blotting (Fig. 5A), both the wild-type and all of the mutant ER81 molecules coimmunoprecipitated with p300. Similarly, both wild-type and mutant ER81 interacted in vitro with CBP (data not shown). Thus, mutation of the LXXLL motif in ER81 does not interfere with binding to CBP/p300, suggesting that this motif does not contribute to CBP/p300 binding.
FIG. 5.
Impact of the LVALL motif in the ETS domain on ER81 function. (A) Anti-HA immunoprecipitations (IP) of 293T cells transiently transfected with HA-tagged p300 and Myc-tagged wild-type or mutant ER81. ER81 present in the immunoprecipitates was detected by anti-Myc immunoblotting. The intracellular levels of ER81 (input) are also shown. (B) Gel retardation assay with HA-tagged ER81249–477 and mutants thereof and the 32P-labeled E74 oligonucleotide. Arrowheads with asterisks point to two DNA complexes formed by proteolytic degradation products of ER81249–477.
However, all the mutants were inactive in transactivation assays, and none of them could collaborate with p300 (data not shown). These results led us to hypothesize that the LVALL motif might be important for DNA binding. Therefore, we performed gel retardation assays with wild-type and mutant ER81 and the 32P-labeled E74 oligonucleotide, to which ER81 does bind (24). The autoradiogram in Fig. 5B shows that only wild-type ER81 formed complexes with the E74 oligonucleotide, while none of the mutants bound the E74 oligonucleotide. Thus, the LXXLL motif in ER81 plays a profound role in DNA binding but none in CBP/p300 interaction.
In vitro binding studies identify two ER81 binding domains in CBP.
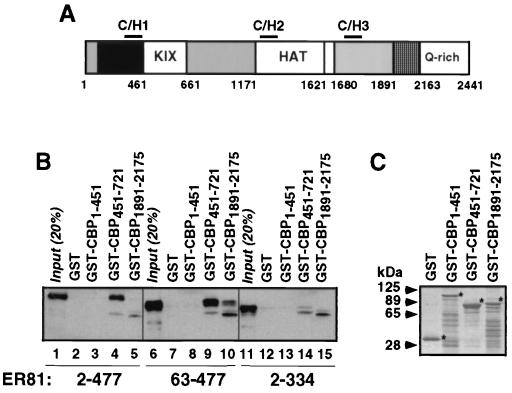
In order to define regions of CBP necessary for ER81 binding (see Fig. 6A for a diagram of CBP), we utilized various portions of CBP fused to GST in pull-down assays. Full-length ER812–477 bound to CBP amino acids 451 to 721 but not to GST, GST-CBP1–451, or to GST-CBP1891–2175 (Fig. 6B), although comparable amounts of GST fusion proteins were employed (Fig. 6C); in addition, other regions of CBP were also unable to interact with ER81 (data not shown). The control, ER812–334, did not interact with any of the GST-CBP fusion proteins (Fig. 6B). Identical results were obtained with full-length ER81 and with ER812–429 (data not shown), confirming that the C-terminal activation domain of ER81 is not required for binding to CBP. Surprisingly, deletion of just 62 N-terminal amino acids in ER8163–477 resulted not only in binding to CBP amino acids 451 to 721 but also in binding to CBP amino acids 1891 to 2175 (Fig. 6B, lane 10). Similarly, ER81 amino acids 249 to 429 bound to both GST-CBP451–721 and GST-CBP1891–2175 (data not shown). In conclusion, CBP amino acids 451 to 721 are capable of interacting with ER81 in vitro, whereas CBP amino acids 1891 to 2175 can only do so when N-terminal amino acids in ER81 are deleted.
FIG. 6.
CBP domains responsible for interaction with ER81. (A) Schematic representation of the CBP molecule. C/H, cysteine-histidine-rich domain; KIX, binding domain for the CREB transcription factor; HAT, histone acetyltransferase domain; Q-rich, glutamine-rich domain. (B) Pull-down of various truncated forms of Myc-tagged ER81 by GST-CBP fusion proteins. Bound ER81 was detected by anti-Myc immunoblotting. (C) Coomassie staining of GST-CBP fusion proteins. Asterisks indicate the respective full-length proteins.
Protein kinase associated with the ER81-CBP/p300 complex.
Previously, it has been shown that protein kinases can be associated with CBP/p300 (42, 47). Thus, we wished to test whether a protein kinase associated with the ER81-CBP/p300 complex could phosphorylate ER81. To this end, 293T cells were transiently transfected with full-length, Myc-tagged ER81 in the absence and presence of HA-tagged p300. ER81 was immunoprecipitated with anti-Myc antibodies and then incubated with [γ-32P]ATP. After boiling in Laemmli sample buffer, proteins were resolved by SDS-PAGE, and the dried gel was exposed to film. The autoradiograph in Fig. 7A (lane 3) shows that Myc-tagged ER81 was phosphorylated by a protein kinase present in the immunoprecipitate. Several scenarios can be envisaged, among others that this protein kinase is directly associated with ER81 or that this protein kinase is associated with endogenous CBP/p300, which in turn had coimmunoprecipitated with ER81. The fact that overexpression of p300 resulted in enhanced phosphorylation of ER81 (Fig. 7A, lane 4) may support the latter hypothesis.
FIG. 7.
A protein kinase coimmunoprecipitates with the ER81-p300 complex. (A) 6Myc-tagged ER81 was overexpressed in 293T cells. Where indicated, p300-HA was cotransfected. After anti-Myc immunoprecipitation, an in vitro phosphorylation assay was performed. The autoradiograph in the top panel shows the result of this assay. The bottom panel reveals ER81 levels by anti-Myc immunoblotting. (B) Two-dimensional analysis of tryptic phosphopeptides derived from 6Myc-tagged ER81 phosphorylated in vitro or in vivo. The right panel shows a mixture of in vitro- and in vivo-labeled phosphopeptides. Electrophoresis was performed in the first dimension (anode on the left), and ascending chromatography was performed in the second dimension.
To determine if the coimmunoprecipitated kinase activity is responsible for phosphorylation of ER81 in vivo, we performed metabolic 32Pi labeling of Myc-tagged ER81 in transiently transfected 293T cells. Radioactively labeled ER81 was then immunoprecipitated with anti-Myc antibodies, separated by SDS-PAGE, eluted from the gel, and subjected to trypsin digestion. Resulting phosphopeptides were resolved in two dimensions on cellulose thin-layer plates and visualized by autoradiography. As shown in Fig. 7B (middle panel), four major phosphopeptides designated a, b, c1, and c2 were detected (see Fig. 8A for a better resolution of the c1 and c2 phosphopeptides), which were also obtained after trypsin digestion of Myc-tagged ER81 that had been phosphorylated in vitro by the coimmunoprecipitated protein kinase (Fig. 7B, left and right panels). These data suggest that the protein kinase coimmunoprecipitating with ER81 can phosphorylate ER81 in vivo.
FIG. 8.
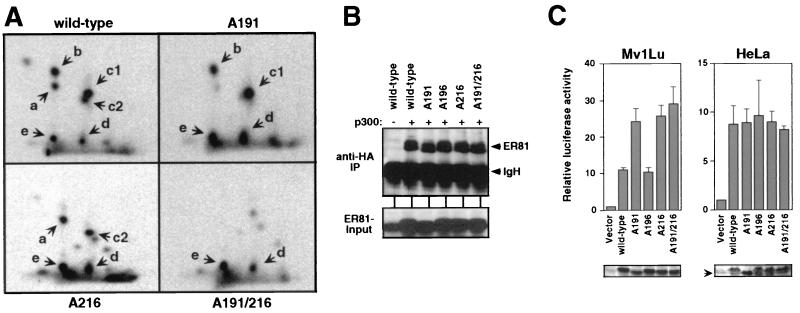
Serine residues 191 and 216 are phosphorylated by the ER81-associated kinase. (A) 6Myc-tagged ER81 (wild-type and indicated alanine mutants) was overexpressed in 293T cells and labeled with 32P in vivo. After anti-Myc immunoprecipitation (IP), the ER81 proteins were digested with trypsin and the resulting phosphopeptides were separated in two dimensions as in Fig. 7. (B) 6Myc-ER81 (wild-type and the indicated alanine mutants) was coexpressed with p300-HA in 293T cells. After anti-HA immunoprecipitation, ER81 was detected by anti-Myc immunoblotting. The bottom panel shows the intracellular levels of ER81. (C) Either 30 or 200 ng of 6Myc-tagged ER81 proteins was transiently transfected into Mv1Lu or HeLa cells, respectively, and luciferase activity from the cotransfected TORU-luc reporter was measured. The bottom panels show anti-Myc Western blots of protein extracts, demonstrating equal protein expression. Please note that the A191 mutant comigrates with an endogenous protein (arrow) that is also detected by the anti-Myc antibodies.
Phosphorylation of serines 191 and 216 by the ER81-associated protein kinase.
In order to determine how phosphorylation of ER81 by the associated protein kinase affects the function of ER81, we set out to map the respective phosphorylation sites with the aim of mutating them. In vivo metabolic 32Pi labeling of various ER81 truncations and subsequent analysis of tryptic phosphopeptides revealed that the phosphorylation sites of the ER81-associated protein kinase must reside within ER81 amino acids 182 to 249; furthermore, phosphoamino acid analysis revealed that the phosphorylated residues must be serines (data not shown). This limited the number of potential phosphorylation sites to five, serines 191, 196, 216, 247, and 249. All of these sites were mutated to alanine, and the respective in vivo 32P-labeled ER81 proteins were subjected to tryptic phosphopeptide analysis. Whereas the A196, A247, and A249 mutants did not reveal any change in tryptic phosphopeptide pattern compared to the wild-type ER81 molecule (data not shown), the phosphopeptide pattern of the A191 and A216 mutants was different (Fig. 8A). In particular, phosphopeptides a and c2 were missing in the A191 mutant, and phosphopeptides b and c1 were missing in the A216 mutant. It is not unusual to observe the disappearance of two phosphopeptides upon mutation of one phosphorylation site due to incomplete tryptic digestion, chymotrypsin contamination, or incomplete oxidation of cysteine residues (53). Finally, we also analyzed the double alanine mutant A191 A216: as expected, the major phosphopeptides a, b, c1, and c2 were no longer observable (Fig. 8A). In conclusion, we have identified serine residues 191 and 216 as the major phosphorylation sites of the ER81-associated protein kinase.
Next, we analyzed whether mutation of serine 191 or serine 216 in ER81 affects its ability to interact with CBP/p300. To this end, Myc-tagged ER81 and HA-tagged p300 were coexpressed in 293T cells, and anti-HA immunoprecipitations were performed. ER81 was subsequently detected in the immunoprecipitates by anti-Myc immunoblotting. As shown in Fig. 8B, no significant difference between wild-type and mutated ER81 in the ability to associate with p300 was observed. Thus, phosphorylation of ER81 at serine residues 191 and 216 appears not to affect the affinity towards CBP/p300.
Finally, we investigated how abolishment of phosphorylation at residues 191 and 216 affects ER81-dependent transcriptional activation. As described for Fig. 3A, expression of the wild-type ER81 molecule stimulated the TORU-luc reporter by 11-fold in Mv1Lu cells (Fig. 8C). Mutation of either serine 191 or serine 216 to alanine raised the luciferase levels by ∼25-fold, while the double mutant A191 A216 was not significantly more active than the single alanine mutants. In addition, mutation of the nonphosphorylated serine 196 to alanine did not affect ER81 transcriptional activity, indicating that mutation of serine to alanine residues in ER81 does not grossly affect its structure. We conclude that the ER81-associated protein kinase can negatively regulate ER81 activity.
However, we also analyzed the impact of mutating serine residues 191 and 216 in HeLa cells (Fig. 8C). In contrast to the results shown for Mv1Lu cells, mutation of the phosphorylation sites did not alter the transactivation potential of ER81. Thus, the effect of serine 191 and serine 216 phosphorylation on ER81 activity may be cell type specific.
DISCUSSION
In this study, ER81 was demonstrated to interact physically with the transcriptional coactivators CBP and p300 both in vitro and in vivo. Importantly, ER81 and CBP/p300 synergized to activate gene transcription. Thus, we have unraveled the recruitment of CBP/p300 as a mechanism for how ER81 activates gene transcription. This collaboration between CBP/p300 and ER81 may also be involved in the transformation of cells caused by dysregulated ER81. In addition, ER81 may be one of the factors required to mediate the essential functions of CBP/p300 during embryonal development.
Our domain mapping data show that ER81 amino acids 249 to 429 are responsible for binding to CBP/p300. These amino acids encompass the ETS domain (amino acids 333 to 415), which may therefore essentially contribute to the interaction between ER81 and CBP/p300. However, since ER81 amino acids 249 to 332 are indispensable for interaction with CBP/p300, the ETS domain is not sufficient to mediate binding to CBP/p300. Interestingly, none of the known transactivation domains of ER81 are contained within ER81249–429, yet this fragment synergized with CBP/p300 to activate transcription. Thus, the transactivation domains of ER81 are not required for the cooperation between ER81 and CBP/p300. Furthermore, both full-length ER81 and ER81249–429 were able to activate the TORU-luc reporter in the absence of overexpressed p300. This activation of transcription appears to be dependent on endogenous CBP/p300, since sequestration of endogenous CBP/p300 by E1A inhibited the ability of ER81 to activate transcription. Altogether, these results indicate that the main gene-regulatory function of ER81 is to tether CBP/p300, which in turn stimulates gene transcription.
A common feature of the ETS domain conserved in almost all of the family members is an LXXLL sequence at its N terminus (37). An LXXLL motif was shown to be involved in protein-protein interactions between nuclear hormone receptors and their cofactors (22) and may represent a general protein-protein interaction motif. However, our data revealed that mutation of the leucine residues in the LVALL sequence of ER81 did not affect ER81-CBP/p300 interactions, excluding the possibility that this LXXLL motif participates in binding of CBP/p300. On the other hand, mutation of the LVALL sequence in ER81 abrogated DNA binding. Structural studies have shown that the LXXLL motif in the ETS domain is part of helix α1, which is not involved in the specific interaction with DNA target sequences, yet helix α1 appears to be important for the structural integrity of the ETS domain (20). Thus, although the ETS domain of ER81 is part of the minimal CBP/p300 interaction domain, altering structural features such as the LXXLL motif can be without effect on the interaction with CBP/p300. This suggests that only portions of the ETS domain participate in CBP/p300 binding.
In this report, we have demonstrated that CBP amino acids 451 to 721 bind to ER81. This region of CBP has been shown to interact with a variety of other transcription factors, including CREB (9), Jun (2), Myb (11, 44), Sap-1a (28), and the viral Tax protein (33). One unresolved question is whether CBP/p300 can accommodate simultaneous binding to ER81 and one or more of these proteins. If not, ER81 could compete with the aforementioned proteins for CBP/p300 recruitment and thereby negatively interfere with their function, and vice versa. On the other hand, CBP/p300 could still function as a “glue” to bring together ER81 and transcription factors binding outside CBP region 451 to 721, such as Fos or nuclear hormone receptors (18), and thereby facilitate cooperation between different classes of transcription factors at selected promoters containing respective DNA-binding sites.
Interestingly, ER81 was able to interact with CBP amino acids 1891 to 2175 in vitro when the N-terminal 62 amino acids were deleted. We hypothesized that possible interactions between the N and C termini of full-length ER81 might prevent binding to CBP amino acids 1891 to 2175. However, pull-down assays of various truncated ER81 proteins by different GST-ER81 fusion proteins did not reveal any intramolecular interactions in ER81 (data not shown). In that regard, binding of regulatory Smad proteins to CBP amino acids 1891 to 2175 depends on Smad phosphorylation by transforming growth factor beta receptor kinases (14, 29, 51). Whether a similar posttranslational modification regulates ER81 binding to CBP amino acids 1891 to 2175 remains to be studied.
We have also shown that CBP/p300 enhanced ER81 transcriptional activity measured with the TORU-luc reporter. The degree of activation of ER81 transcriptional activity by p300 was approximately threefold. Similar degrees of activation by CBP/p300 have been observed with other transcription factors, including Ets-1 (55), CREB (2), MyoD (12), E47 (12), p65 (16), and Sap-1a (28). The TORU-luc reporter construct is bound by ER81, and thus the ETS domain is complexed with DNA, which might be a prerequisite for functional interaction with CBP/p300. However, our experiments with the GAL4-ER81 fusion argue against this: upon binding to the GAL4 binding site, GAL4-ER81 was stimulated by p300 overexpression. Since the ETS domain is supposed to not bind DNA at the GAL4 binding site, ER81 can collaborate with CBP/p300 independently of DNA binding.
CBP and p300 possess intrinsic histone acetyltransferase activity that could potentially activate chromatin-repressed promoters and enhancers by acetylation of histone N-terminal lysine residues or other proteins involved in transcription (4, 45). Moreover, CBP/p300 directly acetylates transcription factors such as GATA-1 (6) and p53 (21). Acetylation of p53 has at least two consequences: increased sequence-specific DNA-binding activity (21) and stabilization of p53 in the cellular response to ionizing radiation (57). It will be interesting to elucidate whether ER81 serves as a substrate for acetylation by CBP/p300 and to study the impact of this hypothetical modification on its transcriptional activity in vivo.
ER81 is phosphorylated by the ERK1-MAPK (24), and ERK-MAPKs interact with CBP/p300 (38). However, the ER81-associated protein kinase characterized in this study does not represent an ERK-MAPK, since MAPK phosphorylation of ER81 results in a different tryptic phosphopeptide pattern than that obtained with this unidentified kinase (data not shown). Interestingly, overexpression of p300 enhanced the activity of the ER81-associated kinase. One model would be that p300 recruits another protein kinase into the ER81-p300 complex, which can activate the unidentified protein kinase. Such protein kinases could be pp90RSK (42) and cyclinE/cdk2 (47), which have been shown to interact with CBP/p300 outside the ER81 interaction domain(s). Alternatively, the unidentified protein kinase itself is bound to p300, and expression of exogenous p300 just enhances the total amount of p300 and thus of the unidentified protein kinase that coimmunoprecipitates with ER81.
The major sites of phosphorylation by the ER81-associated protein kinase are serine 191 and serine 216. Both serine residues are located within the inhibitory domain of ER81 (see Fig. 4 for a sketch of ER81) and may modulate the activity of this domain. Mutation of serine 191 and/or serine 216 to alanine enhanced the ER81-dependent activation of the TORU-luc reporter in Mv1Lu cells, suggesting that phosphorylation at these sites negatively regulates ER81-dependent transcription. This suggests that phosphorylation of serines 191 and 216 may switch on the inhibitory domain of ER81.
What is the biological significance of this phosphorylation? On the one hand, recruitment of CBP/p300 enhances ER81-dependent transcription, while on the other hand, CBP/p300 overexpression stimulates the ER81-associated protein kinase, resulting in a decrease in ER81-dependent transcription. Thus, this protein kinase may be utilized to fine tune the collaboration between ER81 and CBP/p300. But how is this ER81-associated protein kinase regulated? Is it suppressed in HeLa cells, in which mutation of serines 191 and 216 has no effect on ER81 transcriptional activity? Could this protein kinase be downregulated in breast cancer cells, thereby promoting ER81's suspected contribution to breast tumorigenesis (3)? To answer these and other questions, it will be necessary to identify the protein kinase associated with the ER81-CBP/p300 complex, towards which our efforts are now directed.
ACKNOWLEDGMENTS
This work was supported by the Mayo Foundation and by the NCI-funded Mayo Clinic Cancer Center.
We thank Laura Cassiday and Kari Rossow for help in the construction of 6Myc-ER81 truncations and assistance in pull-down experiments with GST-CBP fusion proteins. We are grateful to Denis Bosc for critical comments on the manuscript as well as for providing the anti-ER81 antibodies and to Mike Getz for providing E1A-12S expression plasmids.
REFERENCES
- 1.Abraham S E, Lobo S, Yaciuk P, Wang H G, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 2.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Baert J L, Monte D, Musgrove E A, Albagli O, Sutherland R L, de Launoit Y. Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int J Cancer. 1997;70:590–597. doi: 10.1002/(sici)1097-0215(19970304)70:5<590::aid-ijc17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 7.Brown T A, McKnight S L. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 8.Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- 9.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 10.Coffer P, de Jonge M, Mettouchi A, Binetruy B, Ghysdael J, Kruijer W. junB promoter regulation: Ras mediated transactivation by c-Ets-1 and c-Ets-2. Oncogene. 1994;9:911–921. [PubMed] [Google Scholar]
- 11.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myc. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 12.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 13.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 14.Feng X H, Zhang Y, Wu R Y, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher R C, Scott E W. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 16.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giles R H. Update CBP/p300 transgenic mice. Trends Genet. 1998;14:214. doi: 10.1016/s0168-9525(98)01500-5. [DOI] [PubMed] [Google Scholar]
- 18.Giles R H, Peters D J, Breuning M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 19.Gille H, Sharrocks A D, Shaw P E. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 20.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 21.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 22.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 23.Higashino F, Yoshida K, Fujinaga Y, Kamio K, Fujinaga K. Isolation of a cDNA encoding the adenovirus E1A enhancer binding protein: a new human member of the ets oncogene family. Nucleic Acids Res. 1993;21:547–553. doi: 10.1093/nar/21.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol Cell Biol. 1996;16:1550–1556. doi: 10.1128/mcb.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janknecht R, Ernst W H, Nordheim A. SAP1a is a nuclear target of signaling cascades involving ERKs. Oncogene. 1995;10:1209–1216. [PubMed] [Google Scholar]
- 26.Janknecht R, Ernst W H, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janknecht R, Monte D, Baert J L, de Launoit Y. The ETS-related transcription factor ERM is a nuclear target of signaling cascades involving MAPK and PKA. Oncogene. 1996;13:1745–1754. [PubMed] [Google Scholar]
- 28.Janknecht R, Nordheim A. Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene. 1996;12:1961–1969. [PubMed] [Google Scholar]
- 29.Janknecht R, Wells N J, Hunter T. TGF-beta-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayaraman G, Srinivas R, Duggan C, Ferreira E, Swaminathan S, Somasundaram K, Williams J, Hauser C, Kurkinen M, Dhar R, Weitzman S, Buttice G, Thimmapaya B. p300/cAMP-responsive element-binding protein interactions with ets-1 and ets-2 in the transcriptional activation of the human stromelysin promoter. J Biol Chem. 1999;274:17342–17352. doi: 10.1074/jbc.274.24.17342. [DOI] [PubMed] [Google Scholar]
- 31.Jeon I S, Davis J N, Braun B S, Sublett J E, Roussel M F, Denny C T, Shapiro D N. A variant Ewing's sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–1234. [PubMed] [Google Scholar]
- 32.Kee B L, Arias J, Montminy M R. Adaptor-mediated recruitment of RNA polymerase II to a signal-dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 33.Kwok R P, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 34.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 35.Laget M P, Defossez P A, Albagli O, Baert J L, Dewitte F, Stehelin D, de Launoit Y. Two functionally distinct domains responsible for transactivation by the Ets family member ERM. Oncogene. 1996;12:1325–1336. [PubMed] [Google Scholar]
- 36.LaMarco K, Thompson C C, Byers B P, Walton E M, McKnight S L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- 37.Laudet V, Hanni C, Stehelin D, Duterque-Coquillaud M. Molecular phylogeny of the ETS gene family. Oncogene. 1999;18:1351–1359. doi: 10.1038/sj.onc.1202444. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y Z, Thomas N S, Latchman D S. CBP associates with the p42/p44 MAPK enzymes and is phosphorylated following NGF treatment. Neuroreport. 1999;10:1239–1243. doi: 10.1097/00001756-199904260-00016. [DOI] [PubMed] [Google Scholar]
- 39.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 40.Monte D, Baert J L, Defossez P A, de Launoit Y, Stehelin D. Molecular cloning and characterization of human ERM, a new member of the Ets family closely related to mouse PEA3 and ER81 transcription factors. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- 41.Monte D, Coutte L, Baert J L, Angeli I, Stehelin D, de Launoit Y. Molecular characterization of the ets-related human transcription factor ER81. Oncogene. 1995;11:771–779. [PubMed] [Google Scholar]
- 42.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 43.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 45.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 46.O'Neill E M, Rebay I, Tjian R, Rubin G M. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 47.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 48.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 49.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Topper J N, DiChiara M R, Brown J D, Williams A J, Falb D, Collins T, Gimbrone M A., Jr CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor beta transcriptional responses in endothelial cells. Proc Natl Acad Sci USA. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 53.van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis. 1994;15:544–554. doi: 10.1002/elps.1150150173. [DOI] [PubMed] [Google Scholar]
- 54.Xin J H, Cowie A, Lachance P, Hassell J A. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 1992;6:481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]
- 55.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. A role for CREB-binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 57.Yuan Z M, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Shioya H, Utsugisawa Y, Yokoyama K, Weichselbaum R, Shi Y, Kufe D. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]