SUMMARY
Circulating memory CD8 T cell trafficking and protective capacity during liver-stage malaria infection remains undefined. We find that effector memory CD8 T cells (Tem) infiltrate the liver within 6 hours after malarial or bacterial infections and mediate pathogen clearance. Tem recruitment coincides with rapid transcriptional upregulation of inflammatory genes in Plasmodium-infected livers. Recruitment requires CD8 T cell-intrinsic LFA-1 expression and the presence of liver phagocytes. Rapid Tem liver infiltration is distinct from recruitment to other non-lymphoid tissues in that it occurs both in the absence of liver tissue resident memory “sensing-and-alarm” function and ~42 hours earlier than in lung infection by influenza virus. These data demonstrate relevance for Tem in protection against malaria and provide generalizable mechanistic insights germane to control of liver infections.
In brief
Lefebvre et al. describe the dynamics and mechanisms by which circulating memory CD8 T cells infiltrate the liver to control local malaria and bacterial infection. This work suggests that circulating memory CD8 T cells could be useful targets for developing vaccines and therapeutics for malaria and other liver-specific pathogens.
Graphical abstract
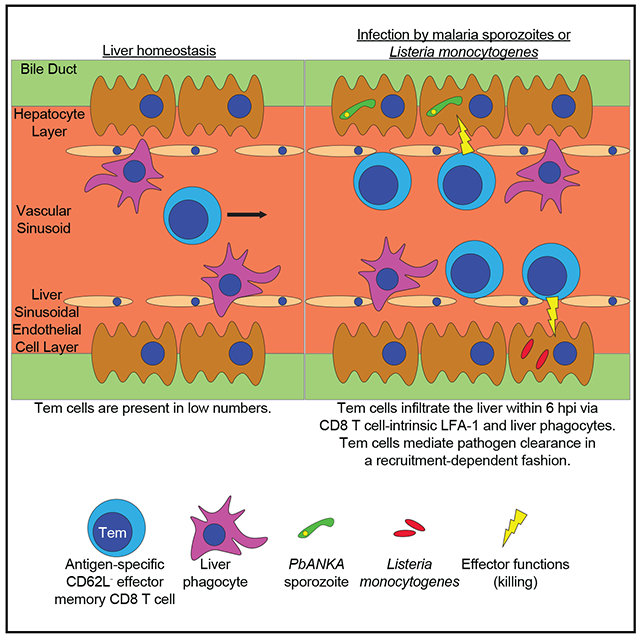
INTRODUCTION
There are no licensed highly efficacious vaccines for malaria, a devastating mosquito-borne disease responsible for millions of infections and hundreds of thousands of deaths annually (World Health Organization, 2020). Vaccine efforts targeting the liver- or blood-stages of the causative parasite Plasmodium suffer from either lack of long-term efficacy in endemic areas or field delivery constraints (Doll and Harty, 2014; Laurens, 2020; Agnandji et al., 2012), and anti-malarial drugs are frequently compromised by emerging parasite resistance (Crompton et al., 2014). Malaria vaccine development has been impeded by our lack of mechanistic understanding of induction of long-lived sterilizing anti-malarial immunity (Sauerwein, 2009).
Infected mosquitoes deliver Plasmodium sporozoites into host skin; from there, the parasites travel through the bloodstream to infect hepatocytes in the liver. Hepatocyte infection remains active for ~2 days in mice and ~7 days in humans, during which time parasites replicate and differentiate into merozoites in preparation for transition to blood-stage infection (Cowman et al., 2016; Kurup et al., 2019b). If all infected hepatocytes are not cleared by adaptive immune responses within this time frame, a process termed “sterilizing immunity,” parasites emerge from hepatocytes and infect erythrocytes to initiate potentially lethal blood-stage infection. Liver-stage malaria is susceptible to vaccine-induced memory CD8 T cell-mediated immunity (Olsen et al., 2018; Vaughan and Kappe, 2017).
Sterilizing anti-malarial immunity can been induced in humans living in malaria-free areas and in mouse models via intravenously (i.v.) or mosquito-delivered immunization with radiation attenuated sporozoites (RAS), which cause an abortive liver infection (Butler et al., 2010; Ishizuka et al., 2016; Nussenzweig et al., 1967). Memory CD8 T cells specific for sporozoite- and liver-stage-derived epitopes mediate RAS-induced sterilizing immunity (Overstreet et al., 2008; Schofield et al., 1987). Precise contributions to liver-stage immunity from distinct memory CD8 T cell subsets, however, remain incompletely understood (Lefebvre and Harty, 2020).
Memory CD8 T cells are composed of subpopulations that differ with respect to anatomical location, recirculation patterns, surface marker expression, and effector function (Jameson and Masopust, 2018). Circulating memory CD8 T cells include effector memory (Tem) and central memory (Tcm). Tem lack expression of secondary lymphoid organ (SLO)-homing receptors such as L-selectin (CD62L), patrol the blood and non-lymphoid tissues (NLT), and exert effector function during recall responses (Martin and Badovinac, 2018). Phenotypic heterogeneity exists within CD62L− Tem, and thus studies of Tem should delineate how these cells are defined (Gerlach et al., 2016; Jameson and Masopust, 2018; Olson et al., 2013). Tcm are CD62L+, are enriched in SLOs, and proliferate and differentiate into effector cells during recall responses (Martin and Badovinac, 2018). Non-recirculating resident memory (Trm) reside in NLTs and SLOs at homeostasis, express markers such as CD69 and CXCR3, and facilitate local and systemic immunity during recall responses (Masopust and Soerens, 2019; Skon et al., 2013).
RAS immunization generates antigen-specific liver Trm and CD62L− circulating memory CD8 T cells. The former patrol the liver sinusoids, whereas the latter exist in the liver, blood, and spleen at homeostasis (Fernandez-Ruiz et al., 2016; Ghilas et al., 2020). Liver Trm cells were indispensable for RAS-induced sterilizing immunity in mice (Fernandez-Ruiz et al., 2016). It remains unclear, however, if circulating memory CD8 T cells also mediate immunity. Mice with high frequencies of antigen-specific circulating memory CD8 T cells in the blood exhibit enhanced control of liver-stage malaria (Schmidt et al., 2008). Determining the capacity of circulating memory CD8 T cells to clear liver-stage malaria should inform vaccine development guided by a more complete understanding of memory CD8 T cell subset protective capacity.
Understanding of memory CD8 T cell dynamics during liver-stage malaria infection may inform preventative and therapeutic approaches. Much of what is known about memory CD8 T cell trafficking during homeostasis and inflammation has been studied in NLTs where tight endothelial barriers separate vasculature from parenchyma (Nolz, 2015). In the liver, blood flows comparatively slowly through sinusoids lined by a fenestrated endothelium (Lee and Kubes, 2008). Mechanistic understanding of circulating CD8 T cell dynamics and effector function during liver infection is limited primarily to studies of effector CD8 T cells (Cockburn et al., 2013, 2014; Guidotti et al., 2015). The liver is also a target of many clinically relevant non-malarial infections (Protzer et al., 2012), and it plays a role in controlling bacteremia and sepsis (Cousens and Wing, 2000; Hyde et al., 1990; Lee et al., 2010). Here, we demonstrate that antigen-specific Tem cells infiltrate the infected liver to mediate pathogen control by mechanisms that are temporally and mechanistically distinct from recruitment to other NLTs.
RESULTS
Accumulation of liver Tem after sporozoite challenge of RAS-vaccinated mice
We addressed the capacity of circulating memory CD8 T cells to mediate immunity to liver-stage malaria in a model of RAS vaccination using CB6F1 (BALB/c × C57BL/6 F1) mice, which respond to H-2d and H-2b-restricted liver-stage malaria protective CD8 T cell epitopes (Doll et al., 2016). This is advantageous because it permits study of a variety of protective and non-protective CD8 T cell epitopes that have been characterized on either mouse background. CB6F1 mice may therefore more closely model diverse human CD8 T cell responses than either parent strain does. Mice were twice vaccinated with Plasmodium berghei (Pb) RAS to generate memory CD8 T cells specific for liver-stage malaria (Figure 1A). These vaccines uniformly induced sterilizing liver-stage immunity against challenge with 103 virulent Pb sporozoites 4 weeks after the second RAS dose (data not shown).
Figure 1. Accumulation of liver Tem after sporozoite challenge of RAS-vaccinated mice.
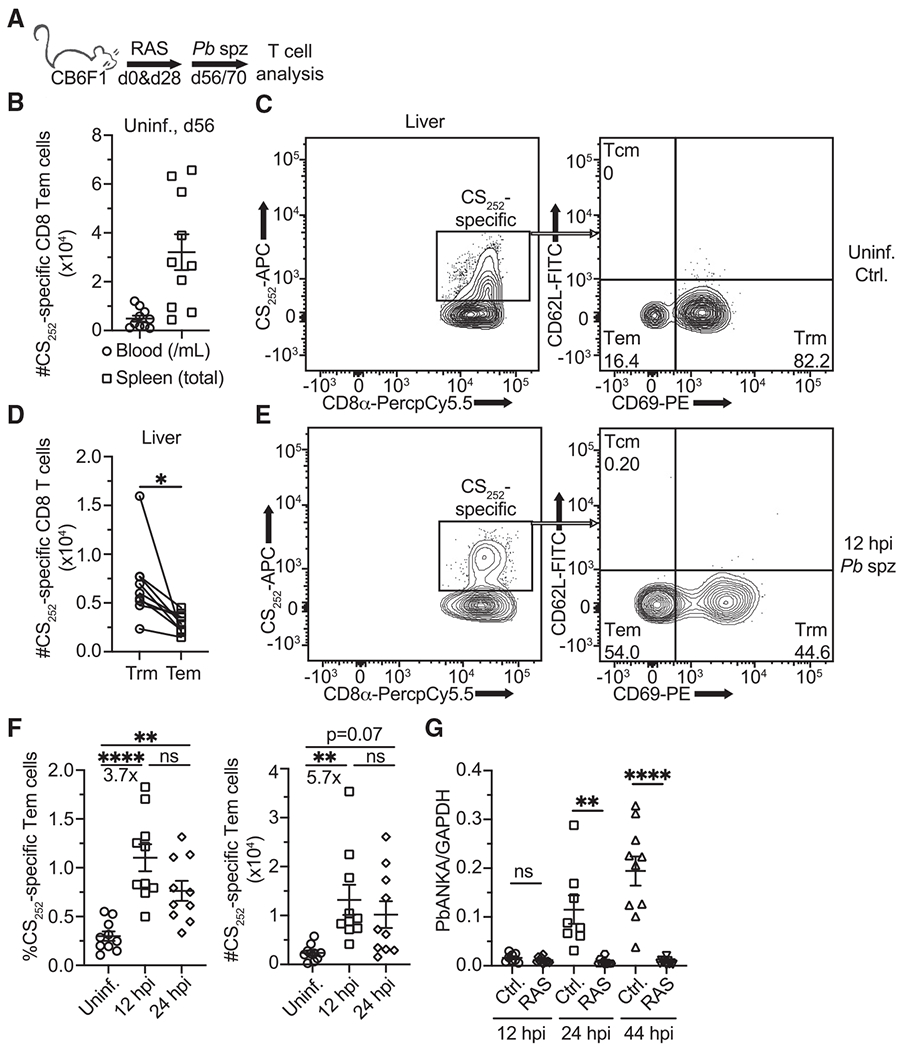
(A) Experimental schematic for (B)–(F). CB6F1 mice received intravenous (i.v.) vaccinations with 104 PbANKA (Pb) radiation attenuated sporozoites (RAS) 28 days apart. Mice were challenged with 2 × 104 virulent Pb sporozoites 56 or 70 days after the first RAS vaccination. Blood, spleens, and livers were collected for T cell analysis via flow cytometry prior to or 12- and 24-h post-infection (hpi).
(B) Numbers of CS252-specific Tem cells in the blood (per mL) and spleen of RAS-vaccinated mice 1 month after 2nd RAS dose. Tem cells are defined as live singlets that bind to the CS252 tetramer and are CD8α+CD11ahiCD62L−CD69−.
(C) Representative flow cytometry (gated on live CD8α+CD11ahi singlet events) depicting CS252-specific (left) Tem and Trm phenotype populations (right) in the liver of RAS-vaccinated mice 56 days after the first RAS vaccination. Tetramer staining identifies CS252-specific cells. Numbers represent frequencies.
(D) Pairwise comparison of CS252-specific Trm and Tem phenotype cells recovered from the livers of vaccinated mice in B). Liver Trm cells are defined as CD8α+CD11ahiCD62L−CD69+.
(E) Representative flow cytometry (gated on live CD8α+CD11ahi singlet events) depicting CS252-specific (left) Tem and Trm phenotype populations (right) in the liver of mice 12 hpi with virulent Pb sporozoites. Numbers represent frequencies.
(F) Frequencies and numbers of CS252-specific Tem phenotype cells recovered from livers prior to or 12 and 24 hpi with Pb sporozoites.
(G) Unvaccinated (infected control) and RAS-vaccinated (as in 1A) mice were challenged with 104 virulent Pb spz. RNA was extracted from the liver hepatocyte fractions at 12, 24, and 44 hpi. Pb 18S ribosomal RNA transcripts were quantified via real-time qRT-PCR and normalized to mouse GAPDH transcripts.
All data are combined from two independent experiments, each with at least 3 mice per group. Symbols represent individual mice and mean ± SEM are depicted. (D) Paired two-tailed t test was performed assuming similar standard deviation (SD). (F) One-way ANOVA (Tukey post hoc test) comparing the mean of each column to the mean of every other column was performed. (G) Unpaired two-tailed t tests assuming similar SD were performed between RAS- and un-vaccinated groups at each time point. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns, p > 0.05.
A prominent protective memory CD8 T cell population induced by RAS immunization in mice targets the major histocompatibility complex (MHC)-I restricted immunodominant protective circumsporozoite antigen (CS) (Kumar et al., 2006; Romero et al., 1989), which sporozoites shed during hepatocyte infection. This T cell population can be tracked in CB6F1 mice using tetramers specific for Kd-CS252–260 (SYIPSAEKI). Tissues were harvested from RAS-vaccinated mice, and CS252-specific memory CD8 T cells were quantified using the surrogate markers of CD8 T cell activation CD8α and CD11a (Rai et al., 2009) and Kd-CS252-tetramer via flow cytometry (Figures 1B, 1C, and S1A). Liver-localized CS252-specific T cells were primarily CD62L− 4–6 weeks after the second RAS vaccination, with most cells co-expressing CD69 (Figures 1D and S1A). Thus, both putative effector memory phenotype (Tem, CD69−CD62L−) and tissue resident memory (Trm, CD69+CD62L−) were represented. Few Tcm (CD69−CD62L+) were generated. Here, Tem will be defined as circulating memory CD8 T cells that are CD69−CD62L−.
Tem and Trm cells exhibited differential expression of tissue homing receptors in accordance with published findings. Liver Trm were CXCR3hi and CX3CR1lo, for example, whereas most Tem were CXCR3low and CX3CR1hi (Figure S1B) (Fernandez-Ruiz et al., 2016; Gerlach et al., 2016). Trm and Tem shared high expression of CXCR6 (Figure S1B), a chemokine receptor previously described as part of the core gene signature of tissue residency after viral infection (Mackay et al., 2016). This suggests that chemokine receptor expression by memory CD8 T cells may be regulated in an infection-specific manner.
RAS-vaccinated mice were challenged with virulent Pb sporozoites to study Tem dynamics early in liver-stage malaria infection (Figure 1A). Within 12 hours post-infection (hpi), the frequency and numbers of CS252-specific Tem cells rapidly increased in the liver whereas the number of liver Trm remained stable (Figures 1E, 1F, and S1C). These results suggested, but did not conclusively prove, rapid recruitment of circulating Tem to the liver.
The consequence of numerical increases in liver-localized Tem was assessed by determining liver parasite burden after infection of unvaccinated controls or RAS-vaccinated mice. Liver parasite burden in RAS-vaccinated mice did not increase between 12–44 hpi as compared to unvaccinated control mice, suggesting that increased Tem numbers correlated with parasite control, and that memory CD8 T cell-mediated control of liver-stage malaria occurs as early as 24 hpi (Figure 1G). These data demonstrate that malaria-specific Tem cell numbers rapidly increase in the sporozoite-infected liver during a window critical to parasite clearance.
Liver infection drives Tem infiltration of the liver
We sought to observe Tem cell dynamics during liver-stage malaria in a controlled, trackable fashion. Such a system was desirable because inflammation and/or infection can rapidly alter the expression of phenotypic markers used to identify Trm, including CD69 and CXCR3 (Cibrián and Sánchez-Madrid, 2017; Meiser et al., 2008).
CD45.1/.2 mice reconstituted with either CS252–260 (CS-R)-specific or OVA257–264 (OT-I-R)-specific TCR retrogenic (Rtg) T cells were generated (Figure S2A). Naive CD45.1/.2+ Rtg cells expressing either enhanced green fluorescent protein (eGFP, CS-R cells) or mCherry (mCh, OT-I-R cells) (Figure S2B) were transferred into naive CD45.2/.2 recipient mice. These mice subsequently received dendritic cells pulsed with either CS252 or OVA257 peptide followed by vaccination with recombinant Listeria monocytogenes (Lm) expressing a secreted homologous peptide, a technique that generates large epitope-specific circulating memory CD8 T cell responses (Badovinac et al., 2005; Schmidt et al., 2008). Rtg memory cells isolated from spleens of DC-Lm-vaccinated mice expressed surface markers associated with Tem (CD62L−CX3CR1hi) (Figure S2C), similar to those induced by RAS vaccination, and were CD69− (data not shown).
Memory CS-R (liver-stage Pb antigen specific) and OT-I-R (Pb antigen non-specific) cells were co-adoptively transferred into RAS-vaccinated mice, which were subsequently challenged with Pb sporozoites (Figure 2A). Circulating CD62L− CS-R and OT-I-R both increased in number and frequency in the liver at 12 hpi (Figures 2A, 2B, and S2D). Rtg Tem and endogenous CS252-specific Tem cell numbers were also increased in livers at 6 hpi, which unequivocally demonstrated that Tem cells are recruited in an expeditious manner during liver infection (Figures 2C and 2D).
Figure 2. Liver infection drives Tem infiltration of the liver.
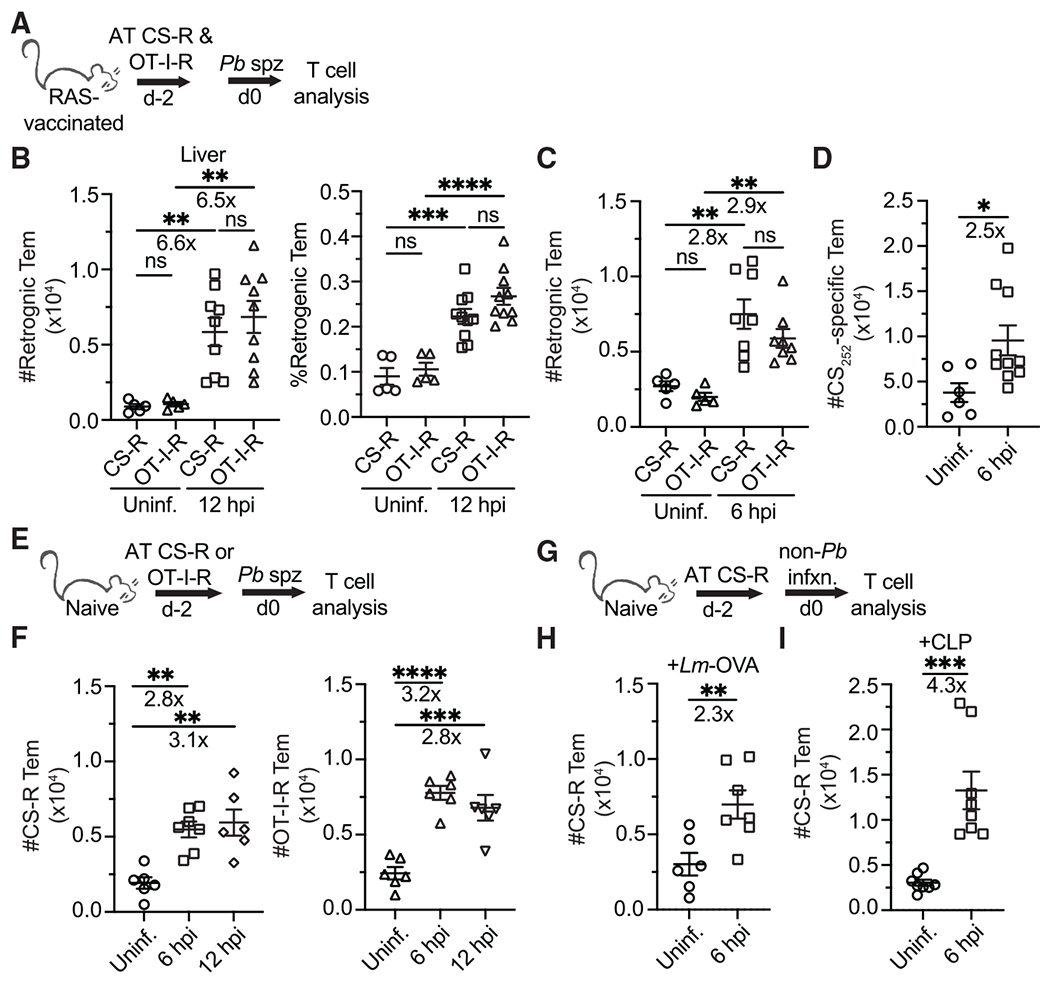
(A, E, and G) Experimental schematics. CS and/or OT-I TCR-retrogenic (Rtg) memory cells were enriched from the spleens of DC-Lm-vaccinated donor mice (see Figure S2) via a CD8α+ T cell enrichment and adoptively transferred (AT) i.v. (5 × 105 cells of one or both types) into either RAS-vaccinated mice that received 1 dose of 104 RAS 1–2 months prior to rechallenge or unvaccinated mice. Mice receiving Rtg cells remained uninfected or were challenged with Pb sporozoites. Livers were harvested at either 6 or 12 hpi.
(B) Quantification of Rtg Tem cell numbers and frequencies in livers of RAS-vaccinated mice that were uninfected or 12 hpi with 3 × 3 104 virulent Pb sporozoites. Rtg Tem cells are defined by an expression profile of CD8α+CD11ahiCD45.1+ CD62L−. CS252-Rtg (CS-R) cells expressing eGFP are detected in the FITC channel, whereas OT-I-Rtg (OT-I-R) cells expressing mCherry are detected in the PE-TxRed channel.
(C and D) Quantification of liver-localized CS-R and OT-I Tem (C) and endogenous (D) CS252-specific Tem cells at 6 hpi with Pb sporozoites in RAS-vaccinated mice.
(E–I) Quantification of liver-localized CS-R and/or OT-I-R Tem cells in unvaccinated mice 6 h postinoculation with either Pb sporozoites (E and F), 106 recombinant virulent Lm expressing secreted OVA peptide (G and H), or cecal ligation and puncture surgery (G and I).
All data are combined from 2 independent experiments, each with 2–5 mice per group. Symbols represent individual mice and mean ± SEM are depicted. (B, C, and F) One-way ANOVA (Tukey post hoc test) comparing the mean of each column to the mean of every other column was performed. (D, H, and I) Unpaired two-tailed t tests assuming similar SD were performed. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns p > 0.05.
The circulation seemed a likely source of Tem cells. Indeed, the number of Rtg Tem cells in the blood simultaneously decreased in infected mice (Figure S2E). Ki67 expression by Tem cells, a marker of cellular proliferation (Scholzen and Gerdes, 2000), was not increased on liver-localized Rtg cells 12 hpi, suggesting that the increased number of Tem cells did not require reactivation and proliferation prior to liver localization (Figure S2F). CD62L+ Tcm cells exhibited similar liver recruitment kinetics during liver-stage malaria (data not shown), however, we focused on Tem cells as those were the predominant circulating memory population generated by RAS vaccination (Figure 1C). Liver-localized CS-R and OT-I-R cells exhibited increased CD69 expression at 12 hpi (Figure S2G). CD69 upregulation, which occurs via TCR- and/or cytokine-dependent mechanisms (Cibrián and Sánchez-Madrid, 2017), suggested that some recruited Tem cells underwent activation after liver entry. This highlighted the importance of using a system where circulating memory CD8 T cells can be distinguished from Trm after seeding a tissue.
Antigen-specific Trm cells produce interferon gamma (IFNγ) to exert “sensing and alarm” functions that mediate lymphocyte recruitment to reinfected tissues (Ariotti et al., 2014; Schenkel et al., 2013). To address a role for malaria-specific liver Trm in recruitment, we challenged unvaccinated Tem cell recipients with sporozoites. Rtg Tem cells were recruited to the liver after infection regardless of vaccination status (Figures 2E, 2F, and S2H). Endogenous activated CD8 T cells were also elevated in unvaccinated mice 6 hpi, suggesting that recruitment includes non-specific memory CD8 T cell populations (Figures S2I and S2J). These analyses demonstrated that rapid Tem recruitment to the liver occurs independently of malaria-specific liver Trm.
It remained unclear as to whether rapid Tem liver recruitment was malaria-specific. We tested two additional liver infection models: (1) Lm, a bacterial pathogen that rapidly localizes to the liver after i.v. injection (Cousens and Wing, 2000) and can be controlled by antigen-specific effector-like circulating memory CD8 T cells (Olson et al., 2013), (2) and cecal ligation and puncture (CLP), a sepsis model that causes rapid polymicrobial infiltration of the liver (Hyde et al., 1990). CS-R Tem cells were recruited to livers within 6 hpi of virulent Lm infection or CLP surgery in an antigen- and liver Trm-independent fashion (Figures 2G–I). These data indicated that liver infection by distinct pathogens induces rapid recruitment of Tem cells from the circulation.
Rapid recruitment of Tem is unique to the liver
Tem recruitment to the liver appeared to be unusually rapid compared to reports of memory CD8 T cell recruitment to other NLTs, which generally is not detected until 2–3 days post infection (Beura et al., 2018; Danahy et al., 2017; Kohlmeier et al., 2008; Osborn et al., 2019). Tem recruitment to the lungs after influenza virus infection was examined to ensure that rapid recruitment to other NLTs was not overlooked. CS-R and OT-I-R cells were co-adoptively transferred into influenza virus (X31, H3N2)-experienced mice to replicate a scenario where antigen-specific Trm cells might be able to aid in Tem recruitment (Figure 3A). Intravenous antibody exclusion was used to distinguish intravascular (iv+) and extravascular (iv−) cells because CD8 T cells need to extravasate into the lung parenchyma to access infected cells (Middleton et al., 2002).
Figure 3. Rapid recruitment of Tem is unique to the liver.
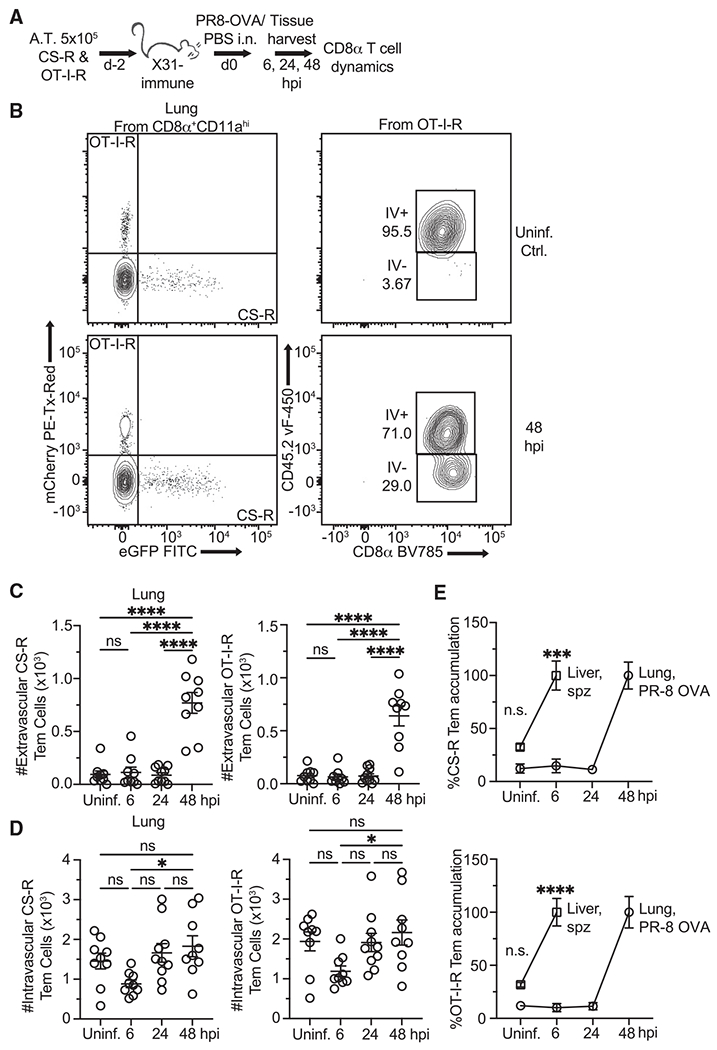
(A) Experimental schematic. Mice 5 weeks post-recovery from influenza A (IAV) X31 (H3N2) infection were injected with memory CS-R and OT-I-R Tem cells and treated by intranasal (in) infusion of PBS or IAV PR8-OVA (H1N1). Mice were injected i.v. with labeled anti-CD45.2 antibody prior to tissue harvest to distinguish vascular from parenchymal T cells. Lungs were harvested for T cell analysis 6–48 hpi.
(B) Representative flow cytometry showing how TCR-Rtg cells were categorized as IV+ (intravascular) or IV− (extravascular).
(C) Quantification of lung-localized extravascular (IV−) TCR-Rtg Tem cells from mice at various time points after PR8-OVA infection.
(D) Quantification of lung-localized intravascular (IV+) TCR-Rtg Tem cells from mice at various time points after PR8-OVA infection.
(E) Normalized comparison of Rtg Tem accumulation over time in the liver and lungs after infection with their respective pathogens.
All data are combined from 2 independent experiments, each with 3–4 mice per group. Symbols represent individual mice and mean ± SEM is depicted. (C and D) One-way ANOVA (Tukey post hoc test) comparing the mean of each column to the mean of every other column was performed. (E) Unpaired two-tailed t tests assuming similar SD comparing uninfected and 6 hpi groups were performed. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns, p > 0.05.
Upon challenge with heterologous influenza virus (PR8-OVA, H1N1), the number of Rtg Tem cells in the lung extravascular compartment did not increase until 48 hpi and did so regardless of antigen specificity (Figures 3B and 3C). The number of intravascular Tem cells remained unchanged between uninfected and infected mice at 48 hpi (Figure 3D). Comparison of peak Tem accumulation in the respective tissues revealed that Tem cells infiltrated the liver much more rapidly than the lung (Figure 3E). These findings highlight how the unique biology of the liver may impact circulating memory CD8 T cell recruitment during infection.
Liver-stage malaria infection rapidly triggers an inflammatory state in the liver
We examined gene expression during malaria infection to contextualize our Tem recruitment data in the broader scope of liver-localized responses to infection. Tissue infection drives rapid changes in gene expression that mediate downstream consequences in pathogen control and tissue repair, and antigen-specific Trm cells may facilitate these changes (Ariotti et al., 2014). Studies have determined that a type I IFN gene signature expression is highly upregulated late (>40 hpi) in liver-stage malaria infection (Liehl et al., 2014; Portugal et al., 2011), whereas others have characterized gene expression by hepatocytes and infiltrating CD11c+ monocytes during infection (Kurup et al., 2019a; Posfai et al., 2018). Much remains unknown, however, about the early liver transcriptional response to infection.
RNA-sequencing was performed on whole liver tissue 6 hpi with virulent Pb sporozoites to determine the liver transcriptional response to infection. mRNA expression patterns were examined in unvaccinated and RAS-vaccinated mice because Tem recruitment appeared independent of vaccination status (Figure 4A). Indeed, liver gene expression signatures clustered based on sporozoite challenge status but not vaccination status (Figure 4B).
Figure 4. Liver-stage malaria infection rapidly triggers an inflammatory state in the liver.
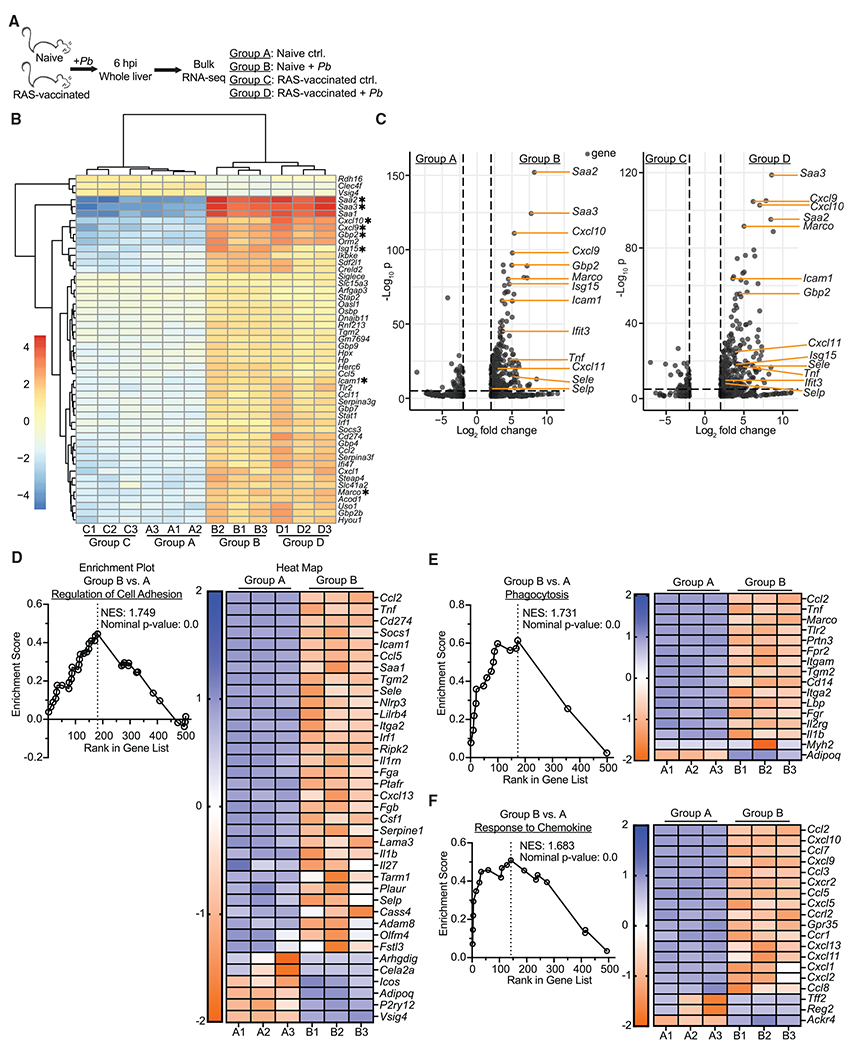
(A) Experimental schematic. Age matched female naive and RAS-vaccinated (one dose of 104 RAS i.v. 28 days before sporozoite challenge) CB6F1 mice were unchallenged or received 3 × 104 virulent Pb sporozoites. Livers were harvested at 6 hpi for RNA-seq.
(B) Heatmap displaying the top 60 differentially expressed genes (defined by adjusted p value) across the groups. Genes are sorted based on hierarchical clustering.
(C) Volcano plots comparing individual gene expression differences between various groups. Genes with a log2 fold change between −2 and 2 are excluded. Genes of particular interest are labeled. Dashed lines indicate thresholds of significance for log2 fold change (x axis, ±2) and adjusted p value (y axis, <0.05).
(D–F) Enrichment plots and associated heatmaps for gene pathways previously defined by the Gene Ontology database.
Data are from a single experiment with 3 biological replicates per group. Expression values were obtained via analysis with DESeq2, R, and edgeR. Thresholds for significance: log2 fold change >2 and <−2; normalized p value <0.05.
Inflammatory RNAs, including those for encoding proteins such as markers of integrin ligands (Icam1), scavenger receptors (Marco), proteins associated with a type I IFN response (Isg15, Ifit3, and Gbp2), CXCR3 ligands (Cxcl9, Cxcl10, and Cxcl11), and liver inflammation (Saa2 and Saa3) were similarly upregulated in infected mice (selected genes are highlighted by asterisks in Figure 4B, others are labeled in Figure 4C). No aforementioned genes were differentially expressed based on prior immunization history, although mRNAs for a few cell metabolism genes and a pattern recognition receptor were differentially regulated (Figure S3A). This result corroborated our findings that Tem recruitment dynamics were indistinguishable between unvaccinated and RAS-vaccinated hosts. Gene sequence enrichment analysis (GSEA) using previously defined transcriptional patterns (Gene Ontology database) revealed that networks associated with regulation of cell adhesion, phagocytosis, and chemokine signaling were upregulated in infected mice (Figures 4D–4F).
Several genes of interest were identified due to their potential as mechanisms for Tem recruitment to the liver: (Icam1) ICAM-1:LFA-1 interactions facilitated effector CD8 T cell localization to the liver and liver Trm surveillance and immune function in prior studies (John and Crispe, 2004; McNamara et al., 2017; Mehal et al., 1999); (Marco) macrophage receptor with collagenase structure (MARCO) is a scavenger receptor that is upregulated on macrophages that may facilitate inflammatory responses (Kraal et al., 2000; van der Laan et al., 1999); (Ifit3) type I IFN signaling has been shown to facilitate protective leukocyte responses during liver-stage malaria (Liehl et al., 2014); and (Cxcl9/10/11) CXCR3 ligands can be induced by type I IFN or IFNγ signaling to facilitate CXCR3-expressing T cell recruitment to infected tissues (Griffith et al., 2014). These data both indicated that the liver rapidly underwent substantial inflammatory changes in response to sporozoite infection regardless of vaccination status and suggested potential mechanisms by which Tem cells might rapidly localize to the liver.
Mechanisms underpinning rapid Tem recruitment to the liver
We sought to determine the mechanism(s) by which circulating Tem rapidly infiltrated the liver. Because Tem recruitment dynamics and gene expression patterns were indiscernible between unvaccinated and RAS-vaccinated mice, mechanistic inquiry was primarily performed using naive mice that received CS-R Tem cells prior to liver infection.
mRNA for ICAM-1 was upregulated at 6 hpi in the liver (Figure 4). Blockade of LFA-1:ICAM-1 interactions did not affect Tem liver-localization at homeostasis (Figure S4A), however, blockade reduced rapid Tem recruitment to the liver during liver-stage malaria infection (Figures 5A and 5B). LFA-1 was also required for Tem recruitment in the Lm infection model (Figure S4B). Flow cytometry studies of NLTs that rely on cellular extractions may underestimate numbers of tissue lymphocytes (Borges da Silva et al., 2019; Steinert et al., 2015). Microscopic analysis of liver sections demonstrated that circulating OT-I memory CD8 T cells were numerically enriched in liver tissue 6 hpi in C57BL/6 mice in an LFA-1-dependent fashion (Figures S4C–S4E). CD8 T cell-intrinsic LFA-1 was responsible for Tem recruitment (Figures S4F and S4G), which ruled out other LFA-1-expressing lymphocytes and granulocytes as causes for diminished Tem recruitment (Springer et al., 1987).
Figure 5. Mechanisms underpinning rapid Tem recruitment to the liver.
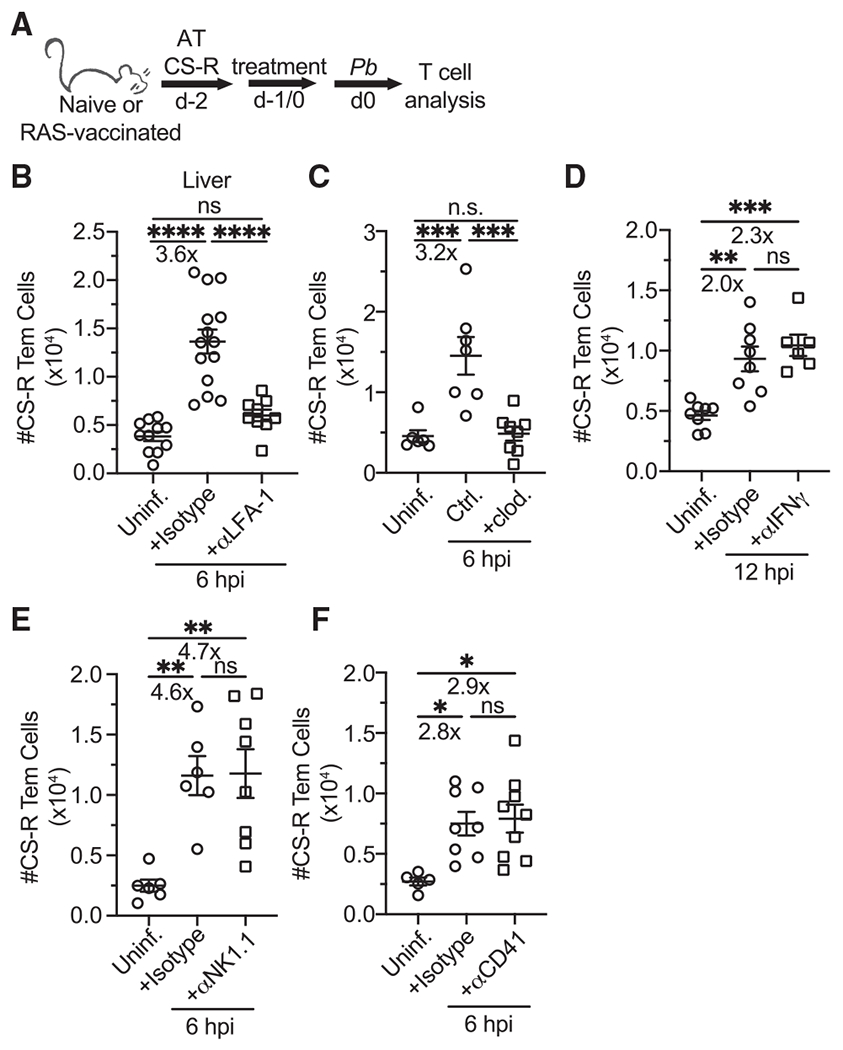
(A) Experimental schematic. Naive CD45.2 mice received 5 × 105 CD45.1+eGFP+ memory CS-R Tem cells via adoptive transfer, and some recipients were challenged with Pb sporozoites. Some mice also received various treatments including blocking antibodies/drugs (α-LFA-1 and α-IFNγ) and depleting antibodies/drugs (clodronate liposomes, α-NK1.1, α-CD41). Isotype control antibodies were delivered at the same time as the corresponding experimental antibody.
(B–F) Quantification of CS-R Tem cells 12 (D) or 6 (B, C, E, and F) hpi with 3 × 104 Pb sporozoites. Treatment doses, route of administration, and treatment administration time: α-LFA-1,300 μg i.v. 3 h prior; clodronate liposomes, 200 μL i.v. 1 day prior; α-IFNγ, 1 mg intraperitoneally (i.p.) 1 day prior; α-NK1.1, 300 μg and 100 μg i.v. 2 and 1 days prior, respectively; and α-CD41, 500 μg i.v. 3 h prior. Experiments were performed in previously naive CB6F1 mice, except for (D), in which CS-R cells were transferred into RAS-vaccinated mice.
All data are combined from 2–3 independent experiments, each with 2–5 mice per group. Symbols represent individual mice and mean ± SEM is depicted. (B–F) One-way ANOVA (Tukey post hoc test) comparing the mean of each column to the mean of every other column was performed. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns, p > 0.05.
Depletion of liver-resident and circulating phagocytic cells with clodronate liposomes inhibited Tem recruitment to the infected liver (Figure 5C), but it did not change Tem homeostatic localization (Figure S4H). This may relate to the ability of resident and circulating myeloid populations to mediate inflammatory responses and induce lymphocyte accumulation during liver infection and inflammation (Brempelis and Crispe, 2016; Ju and Tacke, 2016; Kurup et al., 2019a). Reduced Tem recruitment after clodronate depletion also occurred in RAS-vaccinated mice (data not shown), suggesting that any liver Trm “sensing-and-alarm” function was incapable of overcoming the absence of phagocytes. Neither depletion of Kupffer cells, liver-resident macrophages that coordinate local inflammatory responses and interact with circulating lymphocytes (Lee et al., 2010; Mehal et al., 1999; Sakai et al., 2019), nor depletion of CD11c+ cells, which are either tissue resident or recruited from the circulation (Kurup et al., 2019a), affected Tem recruitment (Figures S5A–S5D). This suggested that redundancy exists in phagocyte populations critical for recruitment of Tem to the liver.
Various putative mechanisms did not contribute to Tem liver recruitment. These included: IFNγ signaling (Figure 5D), which can be initiated by Trm cells and induce expression of CXCR3 ligands CXCL9/10/11 (Griffith et al., 2014; Schenkel et al., 2013); natural killer (NK) cells and natural killer T (NKT) cells (Figure 5E), numerically abundant liver lymphocytes that produce cytokines during infection (Lee et al., 2010; Peng et al., 2016); type I IFN signaling (data not shown), which mediates leukocyte trafficking (Liehl et al., 2014); and platelets (Figure 5F), which form intra-sinusoidal clusters that effector CD8 T cells utilize to localize to the infected liver (Guidotti et al., 2015). Tem recruitment was also not affected by CD44 blockade (data not shown), which facilitates effector CD8 T cell adhesion to platelet clusters in the sinusoids (Guidotti et al., 2015). Target cell depletion was confirmed (Figures S4I–S4K, S5B, and S5D). These results highlight that Tem recruitment to the liver involves various pathway(s) that exhibit key similarities and differences when compared to recruitment of effector and memory CD8 T cell subsets to the liver and other NLTs.
Tem cells mediate immunity to liver infection in a recruitment-dependent fashion
Evaluation of Tem protective capacity against liver-stage malaria was performed to determine the functional outcome of Tem infiltration of the liver. As demonstrated previously in the RAS vaccination model (Fernandez-Ruiz et al., 2016), CXCR3 antibody depletes antigen-specific liver Trm cells while not significantly affecting Tem numbers in DC-Lm-CS252 immunized mice (Figures 6A and 6B). Mice with an intact CS252-specific Tem cell compartment reduced liver parasite burden even in the absence of CS252-specific liver Trm cells (Figure 6C). To further substantiate the ability of Tem to mediate clearance of liver-stage malaria, parabiosis surgery was performed to join age-matched DC-Lm-CS252 (Pb liver-stage protective epitope)-vaccinated and DC-Lm-GAP5041 (a non-protective Pb epitope)-vaccinated mice (Doll et al., 2016) to allow circulating cells to equilibrate between partner mice (Figure 6D). CS252-specific Tem cells equilibrated between mice whereas liver Trm cells did not (Figures 6E and 6F). Similar results were observed for the GAP5041-specific memory CD8 T cell populations (data not shown). DC-Lm-GAP5041-vaccinated parabionts—that possess CS252-specific Tem cells but lack CS252-specific liver Trm cells—significantly reduced liver parasite burden upon challenge (Figure 6G).
Figure 6. Tem cells mediate immunity to liver infection in a recruitment-dependent fashion.
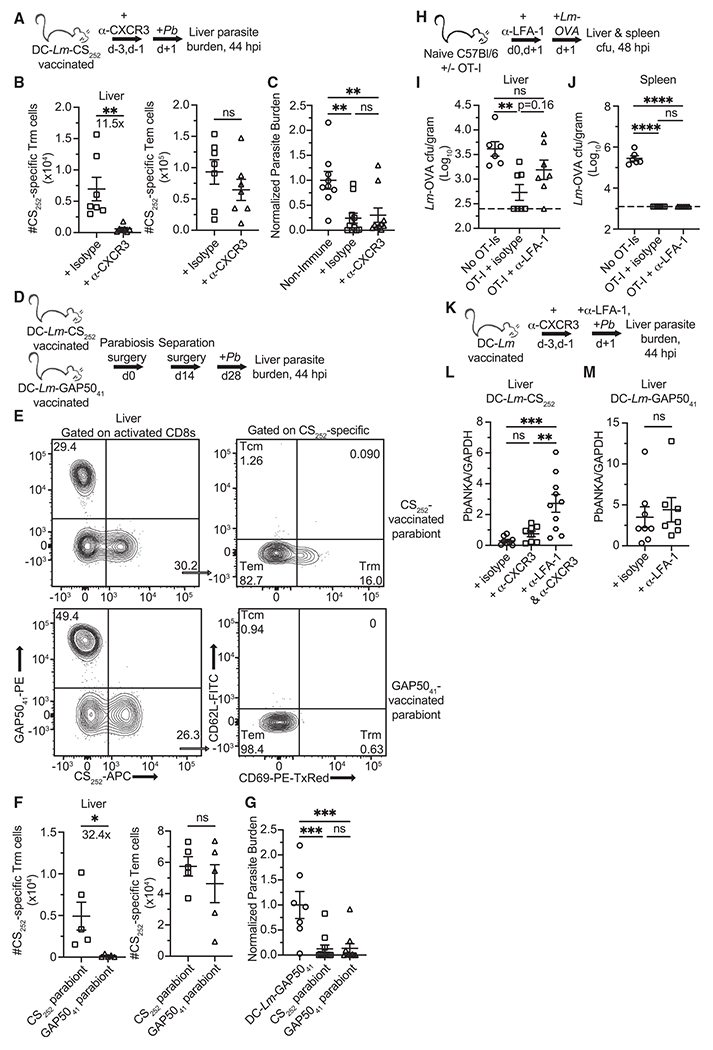
(A, D, and K) Experimental schematics for protection against virulent Pb sporozoites. 104 live sporozoites were injected i.v. into mice. Liver parasite burden was measured 44 hpi.
(B) Quantification of liver localized CS252-specific liver Trm and Tem cells in DC-Lm-CS252-vaccinated mice 2 days after the 2nd α-CXCR3 dose.
(C) Normalized parasite burden showing degree of liver sporozoite infection 44 hpi in naive or isotype control and α-CXCR3-treated DC-Lm-CS252-vaccinated mice.
(E) Representative flow from separated parabiont mice demonstrating that antigen-specific circulating memory CD8 T cell populations equilibrate in the liver, whereas liver Trm compartments do not equilibrate. Top row: DC-Lm-CS252-vaccinated parabiont; bottom row: DC-Lm-GAP5041-vaccinated parabiont.
(F) Mice that received either DC-Lm-CS252 or DC-Lm-GAP5041 vaccinations were joined via parabiosis. Mice were surgically separated 14 days after parabiosis surgery. 14 days post separation, tissues were harvested for quantification of liver localized CS252-specific liver Trm and Tem cells.
(G) Normalized parasite burden showing degree of liver sporozoite infection 44 hpi in mice that were DC-Lm-GAP5041 immunized or CS252− and GAP5041−-vaccinated parabionts.
(H) Experimental schematic for protection against virulent Lm-OVA. 5 × 105 OT-I Tem cells were isolated from the spleens of DC-Lm-OVA vaccinated C57BL/6 mice and adoptively transferred into naive recipient mice.
(I and J) Quantification of virulent Lm-OVA colony-forming unit (CFU) recovered from the livers and spleens of infected mice 48 hpi. Horizontal dashed lines indicate the limit of detection.
(L and M) Parasite burden showing degree of liver sporozoite infection 44 hpi in DC-Lm-CS252 and DC-Lm-GAP5041-vaccinated mice that were pre-treated with various antibodies.
All data are combined from 2 independent experiments, each with 2–5 mice per group. Symbols representing individual mice and mean ± SEM are depicted. (C, G, I, J, and L) One-way ANOVA (Tukey post hoc test) comparing the mean of each column to the mean of every other column was performed. (B, F, and M) Unpaired two-tailed t tests assuming similar SD were performed. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns, p > 0.05.
LFA-1 was blocked prior to infection with Lm or Pb sporozoites to determine if Tem recruitment mediated pathogen control. Adoptive transfer of circulating memory OT-I cells into naive C57BL/6 prior to infection facilitated clearance of virulent Lm expressing secreted OVA peptide from the livers and spleens of infected mice (Figures 6H and 6I). LFA-1 blockade prevented OT-I cells from significantly reducing liver bacterial burden versus the non-immune control, however, this treatment did not diminish Lm clearance in the spleen (Figure 6J). This suggested that liver Tem recruitment is inhibited in this model. LFA-1 blockade in Trm-depleted mice reduced CS252-specific Tem-mediated protection against liver-stage malaria, whereas it did not affect liver parasite burden in DC-Lm-GAP5041-vaccinated control mice (Figures 6K–M). These results demonstrate that rapid recruitment of antigen-specific Tem to the liver facilitates control of bacterial and malarial liver infection.
DISCUSSION
Here we demonstrate that antigen-specific circulating Tem cells rapidly infiltrate the liver during infection to mediate immunity against liver-stage malaria and liver bacterial infection. This draws a parallel to other NLT infections in which circulating memory lymphocytes can exert pathogen control (Labuda et al., 2021). Complete understanding of optimal liver-stage immunity should account for both circulating and resident memory CD8 T cell populations because translationally relevant vaccines such as RAS and “prime-and-trap” strategies induce both (Gola et al., 2018; Ishizuka et al., 2016; Valencia-Hernandez et al., 2020).
Engineering malaria vaccine platforms to induce heightened numbers of Tem and liver Trm could enhance the durable efficacy of vaccine-induced immunity and facilitate evaluation of protective immune responses in humans. Mathematical modeling predicted that a minimum numerical threshold of antigen-specific liver Trm is required to survey all hepatocytes during the time-critical window of liver-stage infection (Fernandez-Ruiz et al., 2016). Tem immunity might lower the number of liver Trm cells required to achieve sterilizing immunity and thus lower the requisite number of vaccine doses per patient, a limiting factor in human vaccine efforts (Jongo et al., 2018; Kurup et al., 2019b; Seder et al., 2013). Prior studies demonstrate that circulating effector and memory cells infiltrate NLTs to form Trm during inflammatory responses (Beura et al., 2018; Holz et al., 2018; Osborn et al., 2019). This suggests that subsequent recall responses, such as after booster vaccination or sporozoite inoculation via mosquito bite, might induce circulating memory CD8 T cells to replenish Trm populations in humans residing in malaria-endemic regions. This is important because liver Trm numbers wane over time, resulting in loss of sterilizing immunity (Valencia-Hernandez et al., 2020). Finally, circulating memory CD8 T cells can be tracked with relative ease via blood draw, while tracking human liver Trm requires invasive liver biopsy (Malik et al., 1991; Rockey et al., 2009; Seder et al., 2013). Determining if and how antigen-specific Tem and Trm cells cooperate during liver-stage malaria to mediate immunity should refine understanding of how to optimize vaccine-mediated protection.
Rapid Tem recruitment appears to be conserved across different liver infections because it was observed clinically relevant models of Lm infection and CLP-induced bacteremia/sepsis. Antigen-specific Tem recruitment mediated control of liver infections with Lm and Plasmodium. These findings suggest that rapid Tem recruitment may play a key role in immunity against a variety of liver infections.
Tem liver recruitment exhibited stark differences with regard to speed and mechanism when compared to our own experiments with influenza infection of the lung and other reports of circulating memory CD8 T cell behavior in non-hepatic NLTs. mRNA for genes involved in inflammatory pathways were upregulated by 6 hpi with sporozoites, and literature review identified proteins and cell types that might mediate Tem recruitment. CD8 T cell-expressed LFA-1 and liver phagocytes were critical mediators of rapid Tem liver recruitment. We hypothesize that Tem-expressed LFA-1 facilitated Tem binding to ICAM-1+ cells in infected livers, which include liver sinusoidal endothelial cells and Kupffer cells (Crispe, 2012; Panés et al., 1995). Suppositions concerning expeditious Tem liver recruitment can be clarified in future assessments of how the liver microenvironment dictates memory CD8 T cell dynamics and effector functions.
A role for other factors in Tem recruitment to the liver was not detected, including liver Trm presence and/or IFNγ signaling, type I IFN signaling, and platelets. This distinguishes Tem recruitment to the liver from previous rigorous studies of (1) effector CD8 T cell recruitment to the liver in a model of viral hepatitis, in which platelets mediated effector CD8 T cell accumulation (Guidotti et al., 2015), and (2) circulating memory CD8 T cell recruitment to infected non-hepatic NLTs. IFNγ, which may be released by Trm, and type I IFN signaling both may facilitate CXCR3 ligand production (Ariotti et al., 2014; McNab et al., 2015). The CXCR3low status of Tem (Figure S1B) may explain why type I IFN or IFNγ blockade did not affect Tem recruitment to the liver, and why antigen-specific liver Trm were not required for rapid Tem recruitment.
Our data provide evidence for a previously undescribed role of antigen-specific Tem in controlling liver-stage malaria while highlighting distinct mechanisms by which Tem rapidly traffic to the infected liver. Vaccine developers may want to consider antigen-specific Tem cell contributions to liver-stage immunity when designing efficacious anti-malarial vaccines. These findings indicate the need for further inquiry into the role of Tem cells in the pathogenesis of mammalian diseases.
Limitations of study
Utilizing large pathogen doses to elucidate immunological mechanisms in animal models is a common technique. It is not clear whether Tem recruitment occurs in the livers of mice infected with a low sporozoite dose (103) used to evaluate sterilizing immunity in mice (Doll et al., 2016). We hypothesize that recruitment to areas with infected hepatocytes occurs regardless of sporozoite dose, however, Tem recruitment might not be discernable in liver extracts after low dose sporozoites challenge because few hepatocytes would be infected. Intravital microscopy could determine T cell dynamics in areas proximal to infected hepatocytes.
Here, we provide evidence that Tem cells can control liver-stage malaria infection; however, the relative capacity of Tem cells and Trm cells to clear infected hepatocytes remains unclear. Such a comparison has not, to our knowledge, been made in the context of malaria and will likely be a complicated issue to address as it will require the generation of mice with antigen-specific Tem or liver Trm and not both, enumeration of these cells prior to challenge and precise detection of their ability to clear liver-stage malaria. Although information as to whether Tem or Trm are “better” at controlling liver-stage malaria has the potential to shape vaccine design, currently there are no vaccines that elicit just Trm without additionally generating circulating memory CD8 T cell populations. Thus, future studies may benefit most from assessing the complimentary and distinct contributions of Trm and Tem in providing immunity to liver-stage malaria.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John T. Harty (john-harty@uiowa.edu).
Materials availability
Many reagents and mice used in this study are available for purchase from the listed vendors. Reagents and organisms unique to this study may be available, with shipping fees paid by the requesting lab, upon request to the Lead Contact. Available materials include bacteria, viruses, and plasmids used to generate retrogenic mice. Due a lack of external centralized repository results in restrictions on availability of in house prepared tetramers and Clec4f-DTR mice (which can be generated by breeding parental mice available from Jackson Labs).
Data and code availability
The data that support the RNA sequencing results (Figure 4) have the NCBI identifier GSE178821 and are publicly available as of the date of publication.
No unique code was generated in this study.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mice were housed in specific pathogen free facilities at the University of Iowa. All experiments were performed with approval from the University of Iowa Institutional Animal Care and Use Committee (IACUC protocol #0051102), and within federal and institutional guidelines. CB6F1, C57BL/6 (B6), BALB/c, B6 Thy1.1 OT-I, B6 Thy1.1 P14, CD11c-DTR, LFA-1 KO, and Clec4f-DTR mice were used in this study. CD45.2/.2 CB6F1 mice were purchased from NCI, Charles Rivers. CD45.1/.2 CB6F1 mice (used as bone marrow donors) were generated by breeding CD45.1/.1 B6 mice and CD45.2/.2 BALB/c mice (purchased from NCI). Clec4f-DTR mice were generated by breeding Clec4f-Cre-tdTomato mice with ROSA26iDTR mice (purchased from Jackson labs) (Sakai et al., 2019). B6, CD11c-DTR, LFA-1 KO, B6 Thy1.1 OT-I, and B6 Thy1.1 P14 mice were purchased from Jackson laboratories. 6–10-week-old female mice were used for generation of trackable memory CD8 T cells; 6–12-week-old male and female mice were used as recipients for studies of CD8 T cell dynamics, with no differences observed in dynamics based on sex; 10–12-week-old female mice were used for parabiosis and liver-stage immunity studies.
Bacteria
Epitope expressing Listeria monocytogenes (Lm) strains were produced and maintained by the Harty lab as described (Doll et al., 2016). All Lm strains are streptomycin resistant, and thus Lm is grown in TSB broth containing streptomycin at 50 μg/mL. Lm strains are grown in an incubator kept at 37°C and shaking at 220 RPM until reaching an optical density (600 nm wavelength) of 0.06-0.1, at which point the solution is spun down for two minutes at 8,600 g to pellet the bacteria. The bacteria concentration is determined from multiplying the optical density by 1x109/mL. Lm is then resuspended in normal saline in preparation for mouse inoculation. This protocol has been used by our lab before (Badovinac et al., 2005; Schmidt et al., 2008). Acta−/−Inlb−/− Lm were used for immunizations, whereas virulent Lm were used for T cell recruitment and protection assays. Lm expresses secreted versions of the described peptides.
Viruses
PR8-OVA (H1N1) and X31 (H3N2) influenza virus strains were maintained by the Harty lab. LD50 calculations for each virus were made previously by the lab (data not shown). Viruses were diluted in normal saline and delivered intranasally (i.n.) with 30 μL of solution. Descriptions of influenza virus protocols can also be found in previous publications (Slütter et al., 2013; Van Braeckel-Budimir et al., 2018).
METHOD DETAILS
Retrogenic (Rtg) mice
Following an established protocol (Holst et al., 2006), bone marrow cells were harvested from female CD45.1/.2 CB6F1 donor mice and transduced with retrovirus expressing the TCRα/β construct specific for Kd-CS252-260 (prepared in house) and GFP or Kb-OVA257-264 and mCherry (Addgene). Transduced cells were transferred into irradiated CD45.2/.2 CB6F1 recipient mice via intravenous (i.v.) tail vein. Mice were screened for reconstitution 6 weeks post transfer. Naive Rtg cells were harvested from the spleens of donor retrogenic mice and adoptively transferred into naive CD45.2/.2 recipient mice. Recipients received a dendritic cell (DC) prime-Lm boost immunizations as described previously (Schmidt et al., 2008) and described below (Immunizations) to generate Rtg memory populations. Spleens were harvested 4-6 weeks post Lm-vaccination, and subsequent enrichment of CD8α T cells and adoptive transfer into recipients was performed for recruitment experiments.
Immunizations
Mice were injected i.v. with 104 radiation attenuated Pb sporozoites (RAS) for RAS immunizations. Dendritic cell (DC) prime-Lm boost immunizations were performed as previously described (Badovinac et al., 2005; Mach et al., 2000; Schmidt et al., 2008). Briefly, B16-Flt3 tumor cells were injected into CB6F1 or C57BL/6 mice. Two to three weeks later tumor recipient mice were injected i.p. with 2 μg purified LPS to cause DC maturation and migration to the spleen. Spleens were harvested from the mice twelve hours after LPS injection. Splenocyte suspensions were cultured with purified 1 μM CS252, GAP5041, or OVA257 peptide, after which CD11c+ dendritic cells were purified from suspensions using CD11c+ beads and LS columns (Miltenyi Biotec). 106 DCs were injected iv into recipient mice. Mice received iv 107 Acta−/− InlB−/− Lm expressing homologous secreted peptide seven days later. For influenza vaccinations, mice received in X31 (H3N2) diluted at 1:100,000 (0.1x LD50) in 30 μl of normal saline while under anesthesia as described previously (Van Braeckel-Budimir et al., 2018).
Infections
Mice were injected iv with 2x104 (Figure 1) or 3x104 (Figures 2–5) freshly isolated Plasmodium berghei ANKA (Pb) sporozoites for T cell recruitment and RNA-sequencing studies. 104 sporozoites were injected iv for qRT-PCR protection assays (Figures 1 and 6). 106 cfu of virulent Lm were injected iv for T cell recruitment assays (Figure 3). Cecal ligation and puncture was performed by opening the peritoneal cavity via the anterior abdomen, ligating the cecum, perforating the cecum twice and extruding cecal contents into the peritoneal cavity, and then suturing the peritoneal cavity and abdominal wall back in place, as described (Jensen et al., 2018). Mice received PR8-OVA(H1N1) diluted to 1:10,000 (10x LD50 for influenza challenge infections as described) (Van Braeckel-Budimir et al., 2018) (Figure 3).
Sporozoites
PbANKA sporozoites were dissected and purified from the salivary glands of female Anopheles stephensi mosquitos using centrifugation (Ozaki et al., 1984). Mosquito heads (with salivary glands attached) were removed from the thorax, added to DMEM with 1:1,000 mouse CD1 serum, and washed through nylon wool at 3,800 g for 90 s approximately five times in a microcentrifuge. Sporozoites were counted with a light microscope, and viability was assayed with 25 μg/mL propidium iodide (Invitrogen) under a fluorescent microscope. Radiation attenuated sporozoites (RAS) were created by irradiating virulent sporozoites with 200Gy using the University of Iowa Free Radical and Radiation Biology Program core facility. Mosquitos were sourced from the Harty lab insectary, the NYU Langone Health Insectary Core & Parasite Culture, and the SporoCore, University of Georgia, Athens, GA. No differences in sporozoite virulence, as measured by ability to cause subsequent blood-stage infection after iv injection into naive mice, were observed based on insectary source.
In-house prepared antibodies
Hybridoma cells expressing anti-mouse NK1.1 (PK136) or anti-mouse IFNγ (XMG1.2) were resuspended in serum free medium (CytivaHyClone SFM4MAB, #SH3051302) and incubated at 37C/5% CO2 in a bioreactor (Cole-Parmer Celline CL1000, # WCL1000-1) according to manufacturer protocol. Supernatants were harvested, protein concentrations determined and antibodies were diluted with sterile PBS to 2 mg/ml and stored at −80C until use. Purity of antibodies was confirmed by SDS-PAGE. Specificity was confirmed by in vivo cell depletion (NK1.1) or exacerbation of mouse Lm infection (XMG1.2).
Cell Blocking and depleting treatments
Blocking antibodies and matching isotype controls were administered i.v. 3 hours prior to Pb or Lm challenge. Depleting treatments were administered as following: α-CD31 iv 3 hours prior to infection, α-NK1.1 i.v. 48 and 24 hours before infection, diphtheria toxin (DT) ip 12 hours prior to challenge. Treatment doses: α-LFA-1 300 μg, α-IFNγ 1mg, α-CD41 500 μg, α-NK1.1 300 μg, DT 200 ng, clodronate liposomes 200 μL i.v. 12 hours prior to challenge.
Tetramers
Tetramers were conjugated from the following MHC class I monomers: Kd-CS252-260(SYIPSAEKI), Db-GAP5041-48(SQLLNAKYL), and Kb-OVA257-264 (SIINFEKL). These conjugates included either streptavidin allophycocyanin (APC) or phycoerythrin (PE). 1:100 24.2G Fc block was used to prevent non-specific tetramer binding to CD8 T cells.
Isolation and flow cytometry
Livers were finely chopped and digested in liver digest medium (ThermoFisher, 17703034) for 30 minutes in an incubator at 37°C and shaking at 110 RPM. Digested tissue was mashed through 70 μM cell strainers. Cell suspensions were spun at 5 min 500 g in a 35% Percoll(Cytiva)/65% HBSS(GIBCO) gradient and the supernatant, which contains debris and parenchymal cells, was discarded. Experiments extracting Kupffer cells and CD11c+ cells did not use a Percoll gradient and additionally used GentleMacs (Miltenyi Biotec, 130-093-235) processing to increase cell yield. Spleens were mashed through 70 μM cell strainers. All suspensions were subject to RBC lysis with 1x Vitalyse (Cmdg). All washes of lymphocyte preparations were performed for 5 minutes at 450 g. Cells were stained in PBS with 2-mercaptoethanol (ThermoFisher) to prevent fluorophore deconjugation (Jensen et al., 2020). Cell viability was assayed using the Fix Viability Dye 780. To analyze Ki67 expression, stained cells were fixed and permeabilized with the Biolegend Cell Staining Buffer according to the manufacturer’s instructions. To isolate CD8α+ T cells for adoptive transfer, the CD8α+ T cell isolation kit and LS columns (Miltenyi Biotec) were used according to manufacturer instructions.
RNA-sequencing
Whole liver tissue was isolated and processed using the RNEasy kit (QIAGEN, #74104) according to the manufacturer’s instructions (“Purification of Total RNA from Animal Tissues” with RNAprotect stabilization). RNA purity and quality were assessed using an Agilent 2100 Bioanalyzer (RNA Integrity Number range 8.8-9.7). Samples were subsequently sent to the University of Minnesota Genomics Core for preparation via NovaSeq SPrime 2x50. Raw normalized expression values were calculated using edgeR by the University of Minnesota Informatics Institute. Creation of PCA plots, heatmaps, volcano plots, and gene sequence enrichment analysis was performed using Trim Galore, Kallisto, DESeq2, and Gene Set Enrichment Analysis.
Liver parasite burden
For evaluation of liver parasite burden, hepatocytes were extracted with a 30-minute liver digestion, mashing through 100 αM cell strainers, and a hepatocyte spin (55 g, 3 minutes, low brake). Pellets were resuspended in TRIzol (Ambion) and flash frozen in liquid nitrogen. RNA was extracted and purified from the pellets using the RNA Clean & Concentrator kit (Zymo Research, #R1018) according to the manufacturer instruction. RNA samples were amplified and quantified using Taq polymerase (Applied Biosystems, #4444427) Pb 18S rRNA forward and reverse primers and probe (Integrated DNA Technologies, #142265234, 142265235, 115842913), mouse GAPDH forward and reverse primers and probe (Applied Biosystems, #4308313) the University of Iowa Genomics Core facility (7900 HT). Pb 18S rRNA transcript counts were normalized to mouse GAPDH counts.
Parabiosis
Female CB6F1 mice were surgically joined via suturing of the skin flap between the elbow and knee joints as previously described (Kamran et al., 2013; Van Braeckel-Budimir et al., 2018). Mice were separated 14 days later by removing existing sutures, debriding the surgical site, and suturing the skin flaps back into their original positions. Mice received anesthesia, pain management, and supportive care according to the IACUC protocol.
Microscopy
6 hpi with Pb sporozoites, livers were harvested and cut into sections using a scalpel and a PELCO easiSlicer vibratome (#11000) as previously described (Pearen et al., 2020). Liver sections were washed in PBS and stained for one hour in PBS containing antibodies and Fc block at 1:100 dilution. Sections were washed after staining completion and mounted onto glass slides with a ProLong gold anti-fade reagent (Invitrogen #P36934). All images were acquired between 1-5 days post mounting on a SP8 NLO Microscope (Leica) using a 10x motor collar-corrected objective (1.0 NA). High resolution stacks (512 × 512 733 format) of 1.01mm2 sections with a depth of 50-120 μm were captured with a z-interval of 5 μm and merged with Leica X software version 3.1.5.16308. All images were acquired sequentially using the following Excitation/Emission parameters: CD54-BV421 412-450, CD8α-AF488 479-505, Thy1.1-PE 565-605, and F4/80-AF647 638-665. To reduce noise a post-acquisition kernel-3 median filter was applied to images. Sequences of acquired image stacks were transformed into volume-rendered 3D images with Imaris x64 Version 9.7.1. The spots function was utilized to determine PE-Thy1.1+ OT-Is with consistent thresholds for XY diameter (greater than or equal to 4.5 μm) and Z diameter (greater than or equal to 20 μm).
QUANTIFICATION AND STATISTICAL ANALYSIS
Details on statistical tests are in the figure legends. Statistical tests for non-RNA sequencing experiments were performed using GraphPad Prism software. Analyses for RNA sequencing experiments were conducted on trimmed data and all alignments were mapped to the mouse genome GRCm38. Transcript quantifications were performed with Kallisto (Bray et al., 2016) using default parameters. The Bioconductor packages edgeR (version 3.4.0) and DESeq2 (version 1.26.0) were used to normalize counts and output count-per-million values. Only protein-coding genes were used for downstream analyses. GSEA desktop application (version 4.1.0 [build 27]) was used for gene set enrichment analysis (GSEA).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3-APC (clone 17A2) | eBioscience | Cat# 12-0032-82 |
| CD69-PE (clone H1.2F3) | eBioscience | Cat# 12-0691-83 |
| CD69-PE-CF594 (clone H1.2F3) | BD Horizon | Cat# 562455 |
| CXCR3-BV421 (clone 173) | Biolegend | Cat# 126522 |
| Ly-6G-FITC(Clone 1A8) | Biolegend | Cat# 127605 |
| TER-119-APC/Cy7 (Clone TER-119) | Biolegend | Cat# 116223 |
| CD62L-PE/Dazzle (clone MEL-14) | Biolegend | Cat# 104448 |
| CD62L-PerCP-Cy5.5 (clone MEL-14) | Biolegend | Cat #104432 |
| CD62L-APC (clone MEL-14) | BD | Cat# 553152 |
| CD62L-FITC (clone MEL-14) | eBioscience | Cat #11-0621-85 |
| MHC-II (I-A/I-E)-BV510 (clone M5/114.15.2) | Biolegend | Cat# 107635 |
| CX3CR1-PE/Cy7 (clone SA011F11) | Biolegend | Cat# 149016 |
| Ly6C-FITC (clone AL-21) | BD Biosciences | Cat# 553104 |
| Ly-6C-PE/Cy7 (Clone AL-21) | BD | 560593 |
| F4/80-PE (clone BM8) | Biolegend | Cat# 123110 |
| F4/80-AF647 (clone BM8) | Biolegend | Cat# 123122 |
| F4/80-BV421 (clone BM8) | Biolegend | Cat# 123137 |
| CD45.2-vF450 (clone 104) | Tonbo | Cat# 75-0454-U100 |
| CXCR6-PE (Clone SA051D1) | Biolegend | Cat# 151104 |
| CD64-BV786 (Clone X54-5/7.1) | BD | Cat# 741024 |
| CD11b-PE (Clone M1/70) | Tonbo | Cat# 50-0112-U100 |
| CD8α-BV785 (Clone 53-6.7) | Biolegend | Cat# 100750 |
| CD8α-PercpCy5.5 (clone 53-6.7) | BioLegend | Cat# 100734 |
| CD11a-BV510 (clone M17/4) | BD Horizon | Cat# 624144 |
| CD90.1-PE (clone OX-7) | BioLegend | Cat# 202524 |
| CD90.1-APC (clone OX-7) | eBioscience | Cat# 17-0900-82 |
| CD45.1-FITC (clone A20) | BD | Cat# 553775 |
| CD45.1-ef450 (clone A20) | Invitrogen | Cat# 48-0453-82 |
| CD49b-PE-Cy7 (clone DX5) | eBioscience | Cat# 25-5971-81 |
| CD42d-APC (clone 1C2) | Biolegend | Cat# 148505 |
| CD11c-APC (clone N418) | Biolegend | Cat# 117310 |
| B220-APC (clone RA3-6B2) | Tonbo | Cat# 20-0452-U100 |
| CD49a-BB700 (clone Ha31/8) | BD Horizon | Cat# 742164 |
| CD54-BV421 (clone YN1/1.7.4) | Biolegend | Cat# 116141 |
| CD43-PE/Dazzle 594 (clone 1B11) | Biolegend | Cat# 121226 |
| Ki67-BV421 | BD | Cat# 562899 |
| Anti-mouse LFA-1a (clone M17/4) | BioXcell | Cat# BE0006 |
| Anti-mouse CXCR3 (clone 173) | BioXcell | Cat# BE0249 |
| Anti-mouse/human CD44 (clone IM7) | BioXcell | Cat# BE0039 |
| Anti-mouse CD41 (clone MWReg30) | Biolegend | Cat# 133902 |
| Anti-mouse IFNγ (clone XMG1.2) | Prepared in house | N/A |
| Anti-mouse NK1.1 (clone PK136) | Prepared in house | N/A |
| Mouse IgG2a isotype control (clone C1.18.4) | BioXcell | Cat# BE0085 |
| Anti-horseradish peroxidase (clone HRPN) | BioXcell | Cat# BE0088 |
| Armenian hamster IgG isotype control (polyclonal) | BioXcell | Cat# BE0091 |
| H-2Db-GAP5040-48 | Prepared in house | N/A |
| H-2Kb-OVA257-264 | Prepared in house | N/A |
| H-2Db-CS252-260 | Prepared in house | N/A |
| 24.2G Fc block | Prepared in house | N/A |
| Bacterial and virus strains | ||
| P. berghei ANKA parasitized-Anopheles stephensi | Harty lab insectary | N/A |
| P. berghei ANKA-sOva parasitized-Anopheles stephensi | Harty lab insectary | N/A |
| P. berghei ANKA GFP-parasitized Anopheles stephensi | NYU Langone Health Insectary Core & Parasite Culture | N/A |
| P. berghei ANKA luciferase-parasitized Anopheles stephensi | SporoCore, University of Georgia, Athens, GA | N/A |
| Listeria monocytogenes expressing Pb-CS252, attenuated (ΔactA, ΔInlB-deficient) | Harty lab | N/A |
| Listeria monocytogenes expressing Pb-GAP5041, attenuated (ΔactA, ΔInlB-deficient) | Harty lab | N/A |
| Listeria monocytogenes expressing OVA257-264, attenuated (ΔactA, ΔInlB-deficient) | Harty lab | N/A |
| Listeria monocytogenes expressing LCMV-GP33, attenuated (ΔactA, ΔInlB-deficient) | Harty lab | N/A |
| Listeria monocytogenes expressing OVA257-264, virulent | Harty lab | N/A |
| Recombinant influenza PR8-OVA virus | Harty lab | N/A |
| X31 influenza virus | Harty lab | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RNAprotect® Tissue Reagent | QIAGEN | Cat# 76106 |
| Liver Digest Medium | Thermo Fisher Scientific | Cat# 17703034 |
| LPS | Sigma | Cat# L-8274 |
| Brefeldin A (1,000x) | BioLegend | Cat# 420601 |
| Collagenase Type II | GIBCO | Cat# 17101-015 |
| DNase | Sigma-Aldrich | Cat# D4513-1VL |
| DPBS | GIBCO | Cat# 14190-144 |
| DMEM | GIBCO | Cat# 11965-092 |
| HBSS | GIBCO | Cat# 14025-092 |
| RPMI | GIBCO | Cat# 11875-093 |
| Percoll | GE Healthcare | Cat# 17-0891-01 |
| Vitalyse | CMDG | Cat# WBL0100 |
| Fixable Viability Stain | BD Horizon | Cat# 565388 |
| Cytofix Fixation Buffer | BD Bioscience | Cat# 554655 |
| Cell Staining Buffer | Biolegend | Cat# 420201 |
| TRIzol | Ambion | Cat# 1559601 |
| Propidium iodide | Invitrogen | Cat# V35118 |
| Mouse CD1 serum | Innovative Research | Cat# IGMSCD1SER10ML-36003 |
| Critical commercial assays | ||
| Anti-CD11c MicroBeads UltraPure mouse | Miltenyi Biotec | Cat# 130-108-338 |
| CD8α+ T Cell Isolation Kit mouse | Miltenyi Biotec | Cat# 130-104-075 |
| LS Columns | Miltenyi Biotec | Cat# 130-042-401 |
| RNeasy Plus Mini Kit (RNA sequencing) | QIAGEN | Cat# 74134 |
| QIAshredder (RNA sequencing) | QIAGEN | Cat# 79654 |
| RNA Clean & Concentrator −25 (RT-qPCR) | Zymo Research | Cat# R1018 |
| Deposited data | ||
| RNA Sequencing (Figure 4) | https://www.ncbi.nlm.nih.gov/geo/ | GSE178821 |
| Experimental models: cell lines | ||
| B16-Flt-3L cell line | Mach et al., 2000 | B16-FLT3L (RRID:CVCL_IJ12) |
| Experimental models: organisms/strains | ||
| Mouse: CB6F1/Crl | Harty Laboratory and National Cancer Institute | #176 |
| Mouse: C57BL/6J | Harty Laboratory and National Cancer Institute | #556 |
| Mouse: C57BL/6J | Harty Laboratory and The Jackson Laboratory | #000664 |
| Mouse: CB6F1 with CS252-260- specific TCRαβ chain retrogenic | This manuscript | N/A |
| Mouse: CB6F1 with OVA257-264- specific TCRαβ chain retrogenic | This manuscript | N/A |
| Mouse: B6.129S7-Itgaltm1Bll/J (LFA-1 KO) | The Jackson Laboratory | #005257 |
| Mouse: C57BU6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (ROSA26iDTR) | The Jackson Laboratory | #007900 |
| Mouse: C57BL/6J-Clec4fem1(cre)Glass/J (CLec4f-Cre-tdTomato) | The Jackson Laboratory | #033296 |
| Mouse: B6.FVB-1700016L21RikTg(Itgax-DTR/EGFP)57Lan/J (CD11c-DTR) | Harty Laboratory and The Jackson Laboratory | #004509 |
| Mouse: C57BL/6-Tg(Tcracrb)1100Mjb/J (OT-1) | Harty Laboratory and The Jackson Laboratory | #003831 |
| Mouse: B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz/TacMmjax (P14 TCRVα2Vβ8) | Harty Laboratory and The Jackson Laboratory | #37394-JAX |
| Software and algorithms | ||
| FlowJo 10.7.1 | FlowJo (BD) | https://www.fiowjo.com/solutions/flowjo/downloads |
| GraphPad Prism 9.1.1 (223) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Adobe Illustrator 25.2.3 | Adobe | https://www.adobe.com/products/illustrator.html |
| IMGT | IMGT®, the international ImMunoGeneTics information system | http://www.imgt.org |
| RStudio 3.6.1 | RStudio | https://www.rstudio.com/ |
| edgeR 3.4.0 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| DSeq2 1.26.0 | Bioconductor | https://bioconductor.org/packages/3.13/bioc/html/DESeq2.html |
| GSEA desktop application 4.1.0 [build 27] | Gene Set Enrichment Analysis (UC San Diego) | https://www.gsea-msigdb.org/gsea/index.jsp |
| Trim Galore 0.6.5 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Kallisto | Patcher lab | https://pachterlab.github.io/kallisto/ |
| Leica X 3.1.5.16308 | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Imaris x64 9.7.1 | Imaris (Oxford Instruments Group) | https://imaris.oxinst.com/ |
Highlights.
Tem cells rapidly infiltrate the liver during Plasmodium infection
Tem recruitment is antigen- and liver Trm-independent and unique to the liver
CD8 T cell-intrinsic LFA-1 and phagocytes orchestrate Tem recruitment to the liver
Tem cell recruitment facilitates clearance of liver-stage malaria infection
ACKNOWLEDGMENTS
We would like to thank the members of the Harty lab for helpful discussion; Ivan Badovinac, Barbara Badovinac, Carter Seely, Tavia Dreismeier, Megan Lindeman, and others for maintaining the insectary; and Sheridan Young for providing editorial direction. Pb sporozoites were sourced in part from the NYU Langone Health Insectary Core and Parasite Culture and the SporoCore at the University of Georgia, Athens, GA. Data herein were obtained from the Flow Cytometry Facility Core and Radiation and Free Radical Research (RFRR) Core (supported by funding from NIH P30 CA086862), which are Carver College of Medicine Core Research Facilities/Holden Comprehensive Cancer Center Core Laboratory at the University of Iowa. RNA sequencing raw data and count tables were obtained from the University of Minnesota Genomics Center (UMGC) and Informatics Institute (UMII), with special thanks to Juan E. Abrahante Lloréns. This work was supported by grants from the NIH (AI42767, AI985515, and AI100527 to J.T.H.; AI114543 to J.T.H. and V.P.B.; A125446 and A127481 to N.S.B.; GM134880 to V.P.B.; T32 HL007 to S.M.A.; T32 AI007511 to N.V.B-B. and I.J.J.; T32 AI007485 to I.J.J.; T32 AI007343 to S.L.U.; and T32 AI007485 to M.N.L.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109956.
DECLARATIONS OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agnandji ST, Lell B, Fernandes JF,Abossolo BP, Methogo BG, Kabwende AL, Adegnika AA, Mordmüller B, Issifou S, Kremsner PG, et al. ; RTS,S Clinical Trials Partnership (2012). A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N. Engl. J. Med 367, 2284–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, Jacobs H, Haanen JB, and Schumacher TN (2014). T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Messingham KA, Jabbari A, Haring JS, and Harty JT (2005). Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat. Med 11, 748–756. [DOI] [PubMed] [Google Scholar]
- Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. (2018). Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol 19, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges da Silva H, Wang H, Qian LJ, Hogquist KA, and Jameson SC (2019). ARTC2.2/P2RX7 Signaling during Cell Isolation Distorts Function and Quantification of Tissue-Resident CD8+ T Cell and Invariant NKT Subsets. J. Immunol 202, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brempelis KJ, and Crispe IN (2016). Infiltrating monocytes in liver injury and repair. Clin. Transl. Immunology 5, e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NS, Schmidt NW, and Harty JT (2010). Differential effector pathways regulate memory CD8 T cell immunity against Plasmodium berghei versus P. yoelii sporozoites. J. Immunol 184, 2528–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibrián D, and Sánchez-Madrid F (2017). CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol 47, 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IA, Amino R, Kelemen RK, Kuo SC, Tse SW, Radtke A, Mac-Daniel L, Ganusov VV, Zavala F, and Ménard R (2013). In vivo imaging of CD8+ T cell-mediated elimination of malaria liver stages. Proc. Natl. Acad. Sci. USA 110, 9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IA, Tse SW, and Zavala F (2014). CD8+ T cells eliminate liver-stage Plasmodium berghei parasites without detectable bystander effect. Infect. Immun 82, 1460–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousens LP, and Wing EJ (2000). Innate defenses in the liver during Listeria infection. Immunol. Rev 774, 150–159. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Healer J, Marapana D, and Marsh K (2016). Malaria: Biology and Disease. Cell 167, 610–624. [DOI] [PubMed] [Google Scholar]
- Crispe IN (2012). Migration of lymphocytes into hepatic sinusoids. J. Hepatol 57, 218–220. [DOI] [PubMed] [Google Scholar]
- Crompton PD, Moebius J, Portugal S, Waisberg M, Hart G, Garver LS, Miller LH, Barillas-Mury C, and Pierce SK (2014). Malaria immunity in man and mosquito: insights into unsolved mysteries of a deadly infectious disease. Annu. Rev. Immunol 32, 157–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahy DB, Anthony SM, Jensen IJ, Hartwig SM, Shan Q, Xue HH, Harty JT, Griffith TS, and Badovinac VP (2017). Polymicrobial sepsis impairs bystander recruitment of effector cells to infected skin despite optimal sensing and alarming function of skin resident memory CD8 T cells. PLoS Pathog. 13, e1006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll KL, and Harty JT (2014). Correlates of protective immunity following whole sporozoite vaccination against malaria. Immunol. Res 59, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll KL, Pewe LL, Kurup SP, and Harty JT (2016). Discriminating Protective from Nonprotective Plasmodium-Specific CD8+ T Cell Responses. J. Immunol 196, 4253–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V, Cozijnsen A, Collins N, et al. (2016). Liver-Resident Memory CD8+ T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity 45, 889–902. [DOI] [PubMed] [Google Scholar]
- Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, and von Andrian UH (2016). The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 45, 1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilas S, Valencia-Hernandez AM, Enders MH, Heath WR, and Fernandez-Ruiz D (2020). Resident Memory T Cells and Their Role within the Liver. Int. J. Mol. Sci 21, 8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gola A, Silman D, Walters AA, Sridhar S, Uderhardt S, Salman AM, Halbroth BR, Bellamy D, Bowyer G, Powlson J, et al. (2018). Prime and target immunization protects against liver-stage malaria in mice. Sci. Transl. Med 10, eaap9128. [DOI] [PubMed] [Google Scholar]
- Griffith JW, Sokol CL, and Luster AD (2014). Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol 32, 659–702. [DOI] [PubMed] [Google Scholar]
- Guidotti LG, Inverso D, Sironi L, Di Lucia P, Fioravanti J, Ganzer L, Fiocchi A, Vacca M, Aiolfi R, Sammicheli S, et al. (2015). Immunosurveillance of the liver by intravascular effector CD8(+) T cells. Cell 161, 486–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, and Vignali DA (2006). Generation of T-cell receptor retrogenic mice. Nat. Protoc 1, 406–417. [DOI] [PubMed] [Google Scholar]
- Holz LE, Prier JE, Freestone D, Steiner TM, English K, Johnson DN, Mollard V, Cozijnsen A, Davey GM, Godfrey DI, et al. (2018). CD8+ T Cell Activation Leads to Constitutive Formation of Liver Tissue-Resident Memory T Cells that Seed a Large and Flexible Niche in the Liver. Cell Rep. 25, 68–79.e4. [DOI] [PubMed] [Google Scholar]
- Hyde SR, Stith RD, and McCallum RE (1990). Mortality and bacteriology of sepsis following cecal ligation and puncture in aged mice. Infect. Immun 58, 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka AS, Lyke KE, DeZure A, Berry AA, Richie TL, Mendoza FH, Enama ME, Gordon IJ, Chang LJ, Sarwar UN, et al. (2016). Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat. Med 22,614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, and Masopust D (2018). Understanding Subset Diversity in T Cell Memory. Immunity 48, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen IJ, Winborn CS, Fosdick MG, Shao P, Tremblay MM, Shan Q, Tripathy SK, Snyder CM, Xue HH, Griffith TS, et al. (2018). Polymicrobial sepsis influences NK-cell-mediated immunity by diminishing NK-cell-intrinsic receptor-mediated effector responses to viral ligands or infections. PLoS Pathog. 14, e1007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen IJ, McGonagill PW, Lefebvre MN, Griffith TS, Harty JT, and Badovinac VP (2020). Worry and FRET: ROS Production Leads to Fluorochrome Tandem Degradation and impairs Interpretation of Flow Cytometric Results. Immunity 52, 419–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, and Crispe IN (2004). Passive and active mechanisms trap activated CD8+ T cells in the liver. J. Immunol 172, 5222–5229. [DOI] [PubMed] [Google Scholar]
- Jongo SA, Shekalaghe SA, Church LWP, Ruben AJ, Schindler T, Zenklusen I, Rutishauser T, Rothen J, Tumbo A, Mkindi C, et al. (2018). Safety, Immunogenicity, and Protective Efficacy against Controlled Human Malaria Infection of Plasmodium falciparum Sporozoite Vaccine in Tanzanian Adults. Am. J. Trop. Med. Hyg 99, 338–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, and Tacke F (2016). Hepatic macrophages in homeostasis and liver diseases: from pathogenesis to novel therapeutic strategies. Cell. Mol. Immunol 13, 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamran P, Sereti KI, Zhao P, Ali SR, Weissman IL, and Ardehali R (2013). Parabiosis in mice: a detailed protocol. J. Vis. Exp (80) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier JE, Miller SC, Smith J, Lu B, Gerard C, Cookenham T, Roberts AD, and Woodland DL (2008). The chemokine receptor CCR5 plays a key role in the early memory CD8+ T cell response to respiratory virus infections. Immunity 29, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraal G, van der Laan LJ, Elomaa O, and Tryggvason K (2000). The macrophage receptor MARCO. Microbes Infect. 2, 313–316. [DOI] [PubMed] [Google Scholar]
- Kumar KA, Sano G, Boscardin S, Nussenzweig RS, Nussenzweig MC, Zavala F, and Nussenzweig V (2006). The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature 444, 937–940. [DOI] [PubMed] [Google Scholar]
- Kurup SP, Anthony SM, Hancox LS, Vijay R, Pewe LL, Moioffer SJ, Sompallae R, Janse CJ, Khan SM, and Harty JT (2019a). Monocyte-Derived CD11c(+) Cells Acquire Plasmodium from Hepatocytes to Prime CD8 T Cell Immunity to Liver-Stage Malaria. Cell Host Microbe. 25, 565–577.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup SP, Butler NS, and Harty JT (2019b). T cell-mediated immunity to malaria. Nat. Rev. Immunol 19, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda JC, Pham OH, Depew CE, Fong KD, Lee BS, Rixon JA, and McSorley SJ (2021). Circulating immunity protects the female reproductive tract from Chlamydia infection. Proc. Natl. Acad. Sci. USA 118, e2104407118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurens MB (2020). RTS,S/AS01 vaccine (Mosquirix™): an overview. Hum. Vaccin. Immunother 16, 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, and Kubes P (2008). Leukocyte adhesion in the liver: distinct adhesion paradigm from other organs. J. Hepatol 48, 504–512. [DOI] [PubMed] [Google Scholar]
- Lee WY, Moriarty TJ, Wong CH, Zhou H, Strieter RM, van Rooijen N, Chaconas G, and Kubes P (2010). An intravascular immune response to Borrelia burgdorferi involves Kupffer cells and iNKT cells. Nat. Immunol 11, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre MN, and Harty JT (2020). You Shall Not Pass: Memory CD8 T Cells in Liver-Stage Malaria. Trends Parasitol. 36, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liehl P, Zuzarte-Luís V, Chan J, Zillinger T, Baptista F, Carapau D, Konert M, Hanson KK, Carret C, Lassnig C, et al. (2014). Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med 20, 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N, Gillessen S, Wilson SB, Sheehan C, Mihm M, and Dranoff G (2000). Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 60, 3239–3246. [PubMed] [Google Scholar]
- Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. (2016). Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463. [DOI] [PubMed] [Google Scholar]
- Malik A, Egan JE, Houghten RA, Sadoff JC, and Hoffman SL (1991). Human cytotoxic T lymphocytes against the Plasmodium falciparum circumsporozoite protein. Proc. Natl. Acad. Sci. USA 88, 3300–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MD, and Badovinac VP (2018). Defining Memory CD8 T Cell. Front. Immunol 9, 2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, and Soerens AG (2019). Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol 37, 521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, and O’Garra A (2015). Type I interferons in infectious disease. Nat. Rev. Immunol 15, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara HA, Cai Y, Wagle MV, Sontani Y, Roots CM, Miosge LA, O’Connor JH, Sutton HJ, Ganusov VV, Heath WR, et al. (2017). Upregulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci. Immunol. 2, eaaj1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehal WZ, Juedes AE, and Crispe IN (1999). Selective retention of activated CD8+ T cells by the normal liver. J. Immunol 163, 3202–3210. [PubMed] [Google Scholar]
- Meiser A, Mueller A, Wise EL, McDonagh EM, Petit SJ, Saran N, Clark PC, Williams TJ, and Pease JE (2008). The chemokine receptor CXCR3 is degraded following internalization and is replenished at the cell surface by de novo synthesis of receptor. J. Immunol 180, 6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton J, Patterson AM, Gardner L, Schmutz C, and Ashton BA (2002). Leukocyte extravasation: chemokine transport and presentation by the endothelium. Blood 100, 3853–3860. [DOI] [PubMed] [Google Scholar]
- Nolz JC (2015). Molecular mechanisms of CD8(+) T cell trafficking and localization. Cell. Mol. Life Sci 72, 2461–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig RS, Vanderberg J, Most H, and Orton C (1967). Protective immunity produced by the injection of x-irradiated sporozoites of plasmodium berghei. Nature 216, 160–162. [DOI] [PubMed] [Google Scholar]
- Olsen TM, Stone BC, Chuenchob V, and Murphy SC (2018). Prime-and-Trap Malaria Vaccination To Generate Protective CD8+ Liver-Resident Memory T Cells. J. Immunol 201, 1984–1993. [DOI] [PubMed] [Google Scholar]
- Olson JA, McDonald-Hyman C, Jameson SC, and Hamilton SE (2013). Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38, 1250–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn JF, Hobbs SJ, Mooster JL, Khan TN, Kilgore AM, Harbour JC, and Nolz JC (2019). Central memory CD8+ T cells become CD69+ tissue-residents during viral skin infection independent of CD62L-mediated lymph node surveillance. PLoS Pathog. 15, e1007633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet MG, Cockburn IA, Chen YC, and Zavala F (2008). Protective CD8T cells against Plasmodium liver stages: immunobiology of an ‘unnatural’ immune response. Immunol. Rev 225, 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki LS, Gwadz RW, and Godson GN (1984). Simple centrifugation method for rapid separation of sporozoites from mosquitoes. J. Parasitol 70, 831–833. [PubMed] [Google Scholar]
- Panés J, Perry MA, Anderson DC, Manning A, Leone B, Cepinskas G, Rosenbloom CL, Miyasaka M, Kvietys PR, and Granger DN (1995). Regional differences in constitutive and induced ICAM-1 expression in vivo. Am. J. Physiol 269, H1955–H1964. [DOI] [PubMed] [Google Scholar]
- Pearen MA, Lim HK, Gratte FD, Fernandez-Rojo MA, Nawaratna SK, Gobert GN, Olynyk JK, Tirnitz-Parker JEE, and Ramm GA (2020). Murine Precision-Cut Liver Slices as an Ex Vivo Model of Liver Biology. J. Vis. Exp (157) [DOI] [PubMed] [Google Scholar]
- Peng H, Wisse E, and Tian Z (2016). Liver natural killer cells: subsets and roles in liver immunity. Cell. Mol. Immunol 13, 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal S, Carret C, Recker M, Armitage AE, Gonçalves LA, Epiphanio S, Sullivan D, Roy C, Newbold CI, Drakesmith H, and Mota MM (2011). Host-mediated regulation of superinfection in malaria. Nat. Med 17, 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai D, Sylvester K, Reddy A, Ganley JG, Wirth J, Cullen QE, Dave T, Kato N, Dave SS, and Derbyshire ER (2018). Plasmodium parasite exploits host aquaporin-3 during liver stage malaria infection. PLoS Pathog. 14, e1007057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protzer U, Maini MK, and Knolle PA (2012). Living in the liver: hepatic infections. Nat. Rev. Immunol 12, 201–213. [DOI] [PubMed] [Google Scholar]
- Rai D, Pham NL, Harty JT, and Badovinac VP (2009). Tracking the total CD8 T cell response to infection reveals substantial discordance in magnitude and kinetics between inbred and outbred hosts. J. Immunol 183, 7672–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, and Smith AD; American Association for the Study of Liver Diseases (2009). Liver biopsy. Hepatology 49, 1017–1044. [DOI] [PubMed] [Google Scholar]
- Romero P, Maryanski JL, Corradin G, Nussenzweig RS, Nussenzweig V, and Zavala F (1989). Cloned cytotoxic T cells recognize an epitope in the circumsporozoite protein and protect against malaria. Nature 341, 323–326. [DOI] [PubMed] [Google Scholar]
- Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, Ego KM, Bruni CM, Deng Z, Schlachetzki JCM, et al. (2019). Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity 51, 655–670.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwein RW (2009). Clinical malaria vaccine development. Immunol. Lett 122, 115–117. [DOI] [PubMed] [Google Scholar]
- Schenkel JM, Fraser KA, Vezys V, and Masopust D (2013). Sensing and alarm function of resident memory CD8+ T cells. Nat. Immunol 14, 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NW, Podyminogin RL, Butler NS, Badovinac VP, Tucker BJ, Bahjat KS, Lauer P, Reyes-Sandoval A, Hutchings CL, Moore AC, et al. (2008). Memory CD8 T cell responses exceeding a large but definable threshold provide long-term immunity to malaria. Proc. Natl. Acad. Sci. USA 105, 14017–14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield L, Villaquiran J, Ferreira A, Schellekens H, Nussenzweig R, and Nussenzweig V (1987). Gamma interferon, CD8+ T cells and antibodies required for immunity to malaria sporozoites. Nature 330, 664–666. [DOI] [PubMed] [Google Scholar]
- Scholzen T, and Gerdes J (2000). The Ki-67 protein: from the known and the unknown. J. Cell. Physiol 182, 311–322. [DOI] [PubMed] [Google Scholar]
- Seder RA, Chang LJ, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, Holman LA, James ER, Billingsley PF, Gunasekera A, et al. ; VRC 312 Study Team (2013). Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science 341, 1359–1365. [DOI] [PubMed] [Google Scholar]
- Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, and Jameson SC (2013). Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol 14, 1285–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter B, Pewe LL, Kaech SM, and Harty JT (2013). Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity 39, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA, Dustin ML, Kishimoto TK, and Marlin SD (1987). Thelymphocyte-associated LFA-1, CD2, and LFA-3 molecules: cell adhesion receptors of the immune system. Annu. Rev. Immunol 5, 223–252. [DOI] [PubMed] [Google Scholar]
- Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, Southern PJ, and Masopust D (2015). Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Hernandez AM, Ng WY, Ghazanfari N, Ghilas S, de Menezes MN, Holz LE, Huang C, English K, Naung M, Tan PS, et al. (2020). A Natural Peptide Antigen within the Plasmodium Ribosomal Protein RPL6 Confers Liver TRM Cell-Mediated Immunity against Malaria in Mice. Cell Host Microbe 27, 950–962.e7. [DOI] [PubMed] [Google Scholar]
- Van Braeckel-Budimir N, Varga SM, Badovinac VP, and Harty JT (2018). Repeated Antigen Exposure Extends the Durability of Influenza-Specific Lung-Resident Memory CD8+ T Cells and Heterosubtypic Immunity. Cell Rep. 24, 3374–3382.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan LJ, Döpp EA, Haworth R, Pikkarainen T, Kangas M, Elomaa O, Dijkstra CD, Gordon S, Tryggvason K, and Kraal G (1999). Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol 162, 939–947. [PubMed] [Google Scholar]
- Vaughan AM, and Kappe SHI (2017). Malaria Parasite Liver Infection and Exoerythrocytic Biology. Cold Spring Harb. Perspect. Med 7, a025486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization; (2020). World Malaria Report 2020: 20 Years of Global Progress and Challenges (World Health Organization; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the RNA sequencing results (Figure 4) have the NCBI identifier GSE178821 and are publicly available as of the date of publication.
No unique code was generated in this study.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.


