Abstract
Exosomes are small extracellular vesicles released by cells for cell–cell communication. They play important roles in cancer development, metastasis, and drug resistance. Exosomal proteins have been demonstrated by many studies as promising biomarkers for cancer screening, diagnosis, and monitoring. Among many detection techniques, surface plasmon resonance (SPR) is a highly sensitive, label-free, and real-time optical detection method. Commercial prism-based wavelength/angular-modulated SPR sensors afford high sensitivity and resolution, but their large footprint and high cost limit their adaptability for clinical settings. Recently, a nanoplasmonic exosome (nPLEX) assay was developed to detect exosomal proteins for ovarian cancer diagnosis. However, comparing with conventional SPR biosensors, the broad applications of nanoplasmonic biosensors are limited by the difficult and expensive fabrication of nanostructures. We have developed an intensity-modulated, compact SPR biosensor (25 cm × 10 cm × 25 cm) which uses a conventional SPR sensing mechanism and does not require nanostructure fabrication. Calibration from glycerol showed that the compact SPR biosensor offered sensitivity of 9.258 × 103%/RIU and resolution of 8.311 × 10−6 RIU. We have demonstrated the feasibility of the compact SPR biosensor in lung cancer diagnosis using exosomal epidermal growth factor receptor (EGFR) and programmed death-ligand 1 (PD-L1) as biomarkers. It detected a higher level of exosomal EGFR from A549 nonsmall cell lung cancer (NSCLC) cells than BEAS-2B normal cells. With human serum samples, the compact SPR biosensor detected similar levels of exosomal EGFR in NSCLC patients and normal controls, and higher expression of exosomal PD-L1 in NSCLC patients than normal controls. The compact SPR biosensor showed higher detection sensitivity than ELISA and similar sensing accuracy as ELISA. It is a simple and user-friendly sensing platform, which may serve as an in vitro diagnostic test for cancer.
Keywords: surface plasmon resonance, biosensor, exosome, in vitro diagnostics, cancer
Graphical Abstract
Exosomes are nanosized vesicles (30–150 nm) released by all kinds of cells into the extracellular environment. They are stably present in all bodily fluids such as blood, urine, and saliva. Exosomes carry nucleic acids (DNAs, RNAs), proteins, and lipids from their parent cells, and are capable of transferring these materials to recipient cells for intercellular communication.1–3 Exosomes are actively involved in cancer development, metastasis, and drug resistance, which makes them promising biomarkers for cancer screening, diagnosis, and monitoring.4,5 Recently, exosomal proteins have shown very high sensitivity and specificity in diagnosing many cancers. For example, exosomal CD151, CD171, and tetraspanin 8 in plasma distinguished lung cancer patients from healthy control.6 Exosomal LRG1 was found to be a potential urinary biomarker for the detection of nonsmall cell lung cancer.7 Levels of exosomal glypican-1 informed pancreatic tumor burden, and distinguished early- and late-stage pancreatic cancer patients from healthy controls and patients with benign pancreatic diseases.8 Exosomal CA-125, EpCAM, CD24 combination biomarkers distinguished ovarian cancer patients from healthy controls with very high sensitivity and specificity.9
Many techniques have been applied to measure the levels of exosomal proteins, such as Western blot, enzyme-linked immunosorbent assay (ELISA), flow cytometry, surface plasmon resonance (SPR), and LC-MS.10–12 Among these techniques, SPR is a highly sensitive and real-time optical detection method. It is a label-free technology and does not require tedious sample handling steps, making it very attractive for exosomal protein analysis. For instance, Rupert et al. used the Biacore 2000 SPR instrument (GE healthcare) to measure the CD63 expression of exosomes derived from the human mast cell line, HMC-1.2.13 When the exosome concentrations were in the picomolar range, SPR showed high signal-to-noise ratio (SNR) of ~100, which was comparable with ELISA assay. Grasso et al. used the Biacore 3000 SPR instrument (GE healthcare) to detect six surface proteins (CD63, CD9, CD24, CD44, EpCAM, and HER2) of exosomes isolated from culture medium of breast cancer cells and from plasma samples of healthy controls, demonstrating that SPR can serve as a bioanalytical procedure for clinical applications.11 More recently, Sina et al. utilized a custom-built SPR platform and detected >10-fold higher HER2+ exosome subpopulation in serum samples from HER2+ breast cancer patients than HER2- patients and healthy controls.14
Commercial prism-based, wavelength/angular-modulated SPR sensors can provide a resolution of 10−7 refractive index unit (RIU) for single point detection, corresponding to a minimum detectable surface concentration of ~0.3 pg/mm2.15,16 However, their large detection spot and the need of a bulky coupling prism limit their effectiveness for compact and miniaturized biosensing and restrain their adaptability to clinical settings.17 In the past decade, miniaturized versions of prism-based SPR sensors have been reported to enable applications requiring integrated, low-cost, compact devices for rapid bioanalytical measurements.18,19 For example, Texas Instruments developed a portable SPR system, Spreeta 2000, in 1999–2000.20,21 Recently, several portable, prism-based SPR biosensors have been developed to detect antibodies,22,23 pathogens,24 and bacteria;25 however, their applications in the detection of exosomal proteins have not yet been explored.
In addition to the portable SPR biosensors developed based on conventional SPR sensing mechanism, nanoplasmonic biosensors that employ nanoscale topographies are a new category of SPR based biosensors. Im et al. developed a portable nanoplasmonic exosome (nPLEX) assay based on transmission SPR through periodic nanohole arrays.26,27 The nPLEX assay analyzed ascites samples from ovarian cancer patients and noncancerous cirrhosis patients (controls). With exosomal CD24 and EpCAM as the biomarkers, the nPLEX assay distinguished ovarian cancer patients from controls with 97% diagnostic accuracy, demonstrating that SPR biosensors are potent in vitro diagnostics for cancer. However, comparing with conventional SPR biosensors, the broad applications of nanoplasmonic biosensors are limited by the difficult and expensive fabrication of nanostructures.
In this work, we have developed an intensity-modulated, compact SPR biosensor that uses conventional SPR sensing mechanism (i.e., SPR on a simple gold-coated surface) and does not require the fabrication of any nanostructures. We have evaluated its sensing performance in detecting exosomal proteins as biomarkers for cancer diagnosis. Obviously, miniaturized SPR sensors cannot compete with high-end SPR systems in sensitivity, but because exosomes are ~100 nm in diameter and should be easier to resolve, the superior resolution provided by commercial high-end SPR systems are unnecessary in this particular sensing application. Our compact SPR biosensor has shown high enough sensitivity/resolution in detecting exosomal proteins. We have demonstrated better detection sensitivity of compact SPR biosensor than ELISA. Besides, the small footprint of the compact SPR biosensor (25 cm × 10 cm × 25 cm) makes it easy to be adapted to clinical settings.
Lung cancer was selected as the disease model because it is one of the most lethal diseases for both men and women worldwide. Current screening and diagnosis methods, such as low dose computed tomography (CT), bronchoscopy, and tissue biopsy, are compromised by high false positive rate, low sensitivity, invasive procedure, and high cost. Detection of exosomal proteins via compact SPR biosensor has great potential to become a complementary and companion diagnostics for lung cancer. Epidermal growth factor receptor (EGFR) and programmed death-ligand 1 (PD-L1) are two protein biomarkers overexpressed in many lung tumors. The levels of EGFR and PD-L1 are closely correlated with lung cancer stage and metastasis, and the diagnostic and prognostic value of EGFR and PD-L1 has been validated in large cohorts of lung cancer patients and lung cancer cell-derived exosomes.27–34 Therefore, we selected EGFR and PD-L1 as the biomarkers in this study. We have demonstrated the feasibility of using compact SPR biosensor to detect the exosomal EGFR and PD-L1 in cell culture medium and serum samples from lung cancer patients and normal controls. We have also demonstrated that the compact SPR biosensor showed better sensing performance than ELISA.
EXPERIMENTAL SECTION
Setup of the Compact SPR Biosensor System.
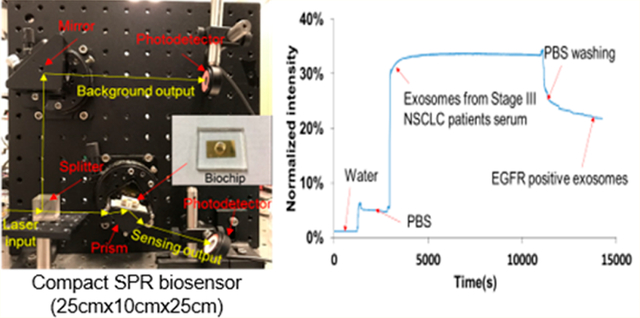
As shown in Figure 1a, the compact SPR biosensor consisted of a prism, a small rotation stage, a continuous wave solid-state laser at 785 nm, a splitter, and two photodetectors. The input laser beam was split into two beams by an optical splitter (50%/50%). One laser beam (50% of laser energy) went through the prism to launch SPR at the gold surface to interact with exosomes bound on the surface. The other laser beam (50% laser energy) was used as the reference beam. The reflection intensity and reference intensity were recorded by two photodetectors and used to quantify the expression of exosomal proteins.
Figure 1.
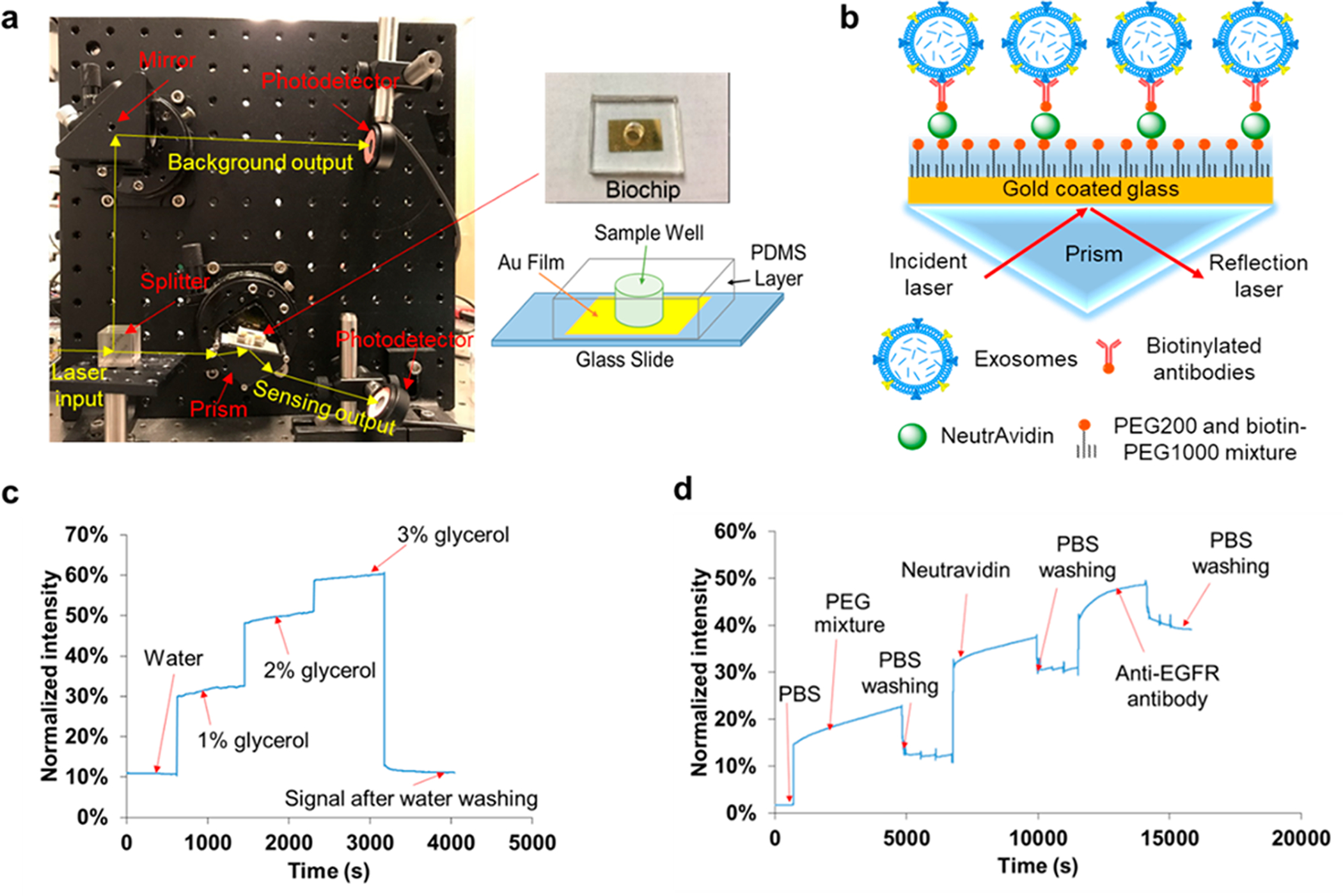
(a) Setup of compact SPR biosensor (left) and the photo and schematic diagram of the biochip (right). (b) Sensing mechanism of compact SPR biosensor. (c) Calibration of sensing performance of compact SPR biosensor with 1%, 2%, and 3% glycerol solutions. (d) Real-time SPR response upon the addition of PBS, PEG mixture, Neutravidin, and anti-EGFR antibodies during the biochip surface modification.
Fabrication of SPR Biochip.
Glass slides (Fisher Scientific, 12-550-A3) were cleaned subsequently in acetone, isopropyl alcohol, and deionized water (DI water) with 10 min sonication for each step. A 2 nm Ti adhesion layer followed by a 49 nm Au thin film were deposited on the sensing area of the glass slide by electron-beam evaporation (Indel system) at the deposition rate of 1 Å/s. A polydimethylsiloxane (PDMS) layer with a hole of 6 mm in the middle was bound on the glass surface to serve as the sample well.
Biochip Surface Modification with Antibodies.
The surface of the biochip was first coated with the mixture of methyl-polyethylene glycol-thiol (PEG200, MW = 200 g/mol, Thermo Fisher Scientific, Rockford, IL, 26132) and biotinylated-polyethylene glycol-thiol (biotin-PEG1000, MW = 1000 g/mol, Nanocs, Boston, MA, PG2-BNTH-1K) at molar ratio of 3:1 and the concentration of 10 mM in PBS by incubation at room temperature for 1 h. After washing off unbounded PEG by PBS, 0.05 mg/mL NeutrAvidin (Thermo Fisher Scientific, 31000) was added to react with biotin for 1 h at room temperature. The unreacted NeutrAvidin was washed off by PBS. Finally, biotinylated anti-EGFR antibodies (Abcam, Cambridge, MA, ab24293), biotinylated anti-PD-L1 antibodies (Thermo Fisher Scientific, 13-5983-82) and biotinylated anti-IgG antibodies (Thermo Fisher Scientific, 13-4714-85) were added at the concentration of 0.05 mg/mL and incubated at room temperature for 1 h to attach the antibodies on the biochip surface through biotin–avidin interaction. After the removal of unbounded antibodies by PBS washing, the biochip was ready for use.
Cell Culture.
A549 nonsmall cell lung cancer (NSCLC) cells and BEAS-2B normal human bronchial epithelial cells were obtained from the American Type Culture Collection (Manassas, VA). The cells were cultured in RPMI 1640 media (Thermo Fisher Scientific, 11875-093) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific, 26140-079) and 1× penicillin streptomycin (PS, Thermo Fisher Scientific, 15140-122). The cells were seeded at 2 × 105 cells/mL in P100 Petri dishes (Greiner Bio-one, Monroe, NC) and incubated in a CO2 incubator at 37 °C. The cells were subcultured every 2 days.
Exosome Isolation from Cell Culture Medium.
A549 cells and BEAS-2B cells were first cultured in regular cell culture medium, i.e., RPMI 1640 medium supplemented with 10% FBS and 1× PS. When the cell culture reached 90% confluence, the regular cell culture medium was removed. The cells were washed with PBS twice and then cultured in starving medium, i.e., RPMI 1640 basal medium without FBS and PS. Two days later, exosomes were isolated from starving medium using total exosome isolation kit (from cell culture medium; Thermo Fisher Scientific, 4478359) following manufacturer’s protocol with minor revision. Briefly, the starving medium was centrifuged at 4000 g for 30 min to remove cells and centrifuged at 10 000 g for 1 h to remove cell debris. Then one part of total exosome isolation kit was added into two parts of cell culture medium. The mixture was incubated at 4 °C overnight, and then centrifuged at 10 000 g for 1 h at 4 °C to pellet exosomes. The supernatant was removed and the exosome pellet was resuspended in PBS at the cell culture medium to PBS volume ratio of 50:1.
Human Serum Samples.
Human serum samples were collected from healthy donors and NSCLC patients prior to treatment. Deidentified human serum samples were obtained from Roswell Park Comprehensive Cancer Center (Buffalo, NY) after the approval from University at Buffalo Institutional Review Board (UB IRB).
Exosome Isolation from Human Serum Samples.
Total exosome isolation kit (from serum, Thermofisher Scientific, 4478360) was used to isolate exosome from serum samples following the manufacturer’s protocol with minor revision. Briefly, serum samples were centrifuged at 10 000 g for 30 min to remove debris. Then, one part of the total exosome isolation kit was added into five parts of serum samples. The mixture was incubated at 4 °C for 30 min and centrifuged at 10 000 g for 1 h at 4 °C to pellet exosomes. The supernatant was removed and the exosome pellet was resuspended in PBS at the serum to PBS volume ratio of 1:1.
Characterization of Exosomes.
Exosomes isolated from cell culture media were diluted 100-fold with PBS. Exosomes isolated from human serum samples were diluted 10 000-fold with PBS. Then the size, size distribution, and number concentration of exosomes were measured using the nanoparticle tracking analysis (NTA) system (NanoSight, LM10, Malvern Instruments Ltd., Westborough, MA). For all measurements, the detection threshold was set at 6, the screen gain was set at 8 and the camera level was set at 14. For exosome imaging, 7 μL of exosome solution was loaded on 400 mesh carbon-coated copper grids, followed by negative staining using 2% uranyl acetate. The morphology of exosomes was observed by JEOL 2010 transmission electron microscopy (TEM).
Exosomal Protein Characterization by Compact SPR Biosensor.
To measure the expression of exosomal proteins by compact SPR biosensor, DI water was first applied to the biochip to set the SPR angle and collect the baseline intensity signals (Iwater). Then, DI water was replaced with PBS, and the intensity signals of PBS (IPBS) were recorded. Exosomes were subsequently loaded after removing PBS, and incubated on the biochip at room temperature to allow the capture of exosomes by antibodies. Finally, the biochip was washed with PBS three times to remove unbounded exosomes, and the intensity signals of captured exosomes (Iexosome) were recorded.
The expression of exosomal protein (E) was calculated using the following equation:
| (1) |
Exosomal Protein Characterization by ELISA.
The expression of exosomal EGFR and exosomal PD-L1 was measured by ELISA (Abcam, ab100505; Thermo Fisher Scientific, BMS2212) following manufacturers’ protocols. Briefly, exosomes were isolated from cell culture media or serum samples as described above. Exosomes were diluted and added in ELISA plates. For EGFR ELISA assay, the plates were incubated at 4 °C overnight. For PD-L1 ELISA assay, the plates were incubated for 2 h at room temperature. After washing off unbounded exosomes, biotinylated detection antibodies was added and incubated with exosomes for 1 h at room temperature. After washing off excessive detection antibodies, horseradish peroxidase (HRP)–streptavidin was added. EGFR antibody coated plates and PD-L1 antibody coated plates were incubated for 45 and 30 min at room temperature, respectively. Then, 3,3′,5,5′-tetramethylbenzidine (TMB) one-step substrate reagent was added and incubated for 30 min in the dark. Finally, the stop solution was added and the absorbance at 450 nm was read by microplate reader immediately.
Scanning Electron Microscopy (SEM).
Field emission scanning electron microscope (FESEM) with Oxford energy-dispersive X-ray spectrometer (EDS - Hitachi SU70) was used to characterize the exosomes captured by the antibodies. Briefly, after unbounded exosomes were washed off by PBS, the biochip was air-dried at room temperature. The biochip was then coated with carbon and the exosomes were observed by SEM.
RESULTS AND DISCUSSION
Setup and Sensing Mechanism of Compact SPR Biosensor.
The setup of the compact SPR biosensor is shown in Figure 1a with all compact optical components assembled on an optical breadboard. A photo and the schematic diagram of the SPR biochip are also shown in Figure 1a. The key module of the compact SPR biosensor is the central rotation stage, the compact prism, and the biochip, which has footprint of 6.35 cm (L) × 6.35 cm (W) × 6.35 cm (H). The excitation source is a Coherent OBIS laser with the dimension of 7 cm (L) × 4 cm (W) × 3.8 cm (H). With the laser source included, the total footprint of our system is 25 cm (L) × 25 cm (W) × 10 cm (H), which was highly compact comparing to commercial units. The solid state laser may be integrated with the central module using the optical fiber interface to further reduce the footprint to as small as 10 cm (L) × 10 cm (W) × 10 cm (H) or even smaller.
In the compact SPR biosensor, the splitter separated the 785 nm laser beam into two beams, i.e., a sensing beam (50% laser energy) and a reference beam (50% laser energy). The sensing beam went through the prism and reached the biochip that sat on the small rotation stage. By adjusting the angle of the stage, the surface plasmon (SP) mode was launched at the gold surface, which interacted with biomolecules bound on the surface. The binding of exosomal proteins to antibodies changed local refractive index, affected the optical properties of the SP modes, changed the intensity of the reflected laser beam, and thus permitted optical detection of exosomal proteins (Figure 1b). The intensity of reference beam was used to normalize the reflection intensity signals. In this work, normalized intensity (i.e., reflection intensity/reference intensity ×100%) was reported as SPR signals.
To calibrate the sensing performance of compact SPR biosensor, we introduced glycerol solutions with different concentrations (1%, 2%, and 3%) and refractive indices to measure the reflection intensity change. As shown in Figure 1c, continuous increase of reflection intensity was observed with increased concentrations of glycerol solutions. The signal of reflection intensity returned to the background level after the glycerol was washed off with DI water. By considering the SNR, the compact SPR biosensor obtained sensitivity of 9.258 × 103%/RIU and resolution of 8.311 × 10−6 RIU, which is better than Spreeta 2000 (resolution of 10−3–10−4 RIU)35 and comparable with a typical intensity-modulated SPR sensor (resolution of (2–5) × 10−6 RIU).36,37 Although the sensitivity and resolution of compact SPR biosensor are approximately 2 orders of magnitude lower than those of commercial high-end SPR systems, we have demonstrated below that this sensing capability is sufficient to resolve exosomes for lung cancer diagnosis.
Preparation and Surface Modification of the SPR Biochip.
To prepare the SPR biochip, a 2 nm Ti film was deposited on the glass slide followed by a 49 nm Au film. The Ti film was used to enhance the adhesion of the Au film and thus to optimize the reliability of the biochip. Then, a PDMS layer with a hole in the middle was attached to the glass slide to generate the sample wells (diameter of 6 mm). With this design, the users can simply use a pipet to add/remove samples into/from the sample wells. This design does not require extra training and equipment (such as syringe pumps), and is compatible with most standard clinic sample handling processes.
To detect exosomal proteins, the biochip surface was modified with the mixture of PEG200 and biotin-PEG1000, Neutravidin, and biotinylated antibodies. The surface modification can be monitored by our compact SPR biosensor. As shown in Figure 1d, when the mixture of PEG200 and biotin-PEG 1000 (molar ratio of 3:1, 10 mM in PBS) replaced PBS, the reflection intensity increased from ~2% to ~15%. When unbounded PEG mixture was removed by PBS washing, the reflection intensity reduced and stabilized at ~11%. The difference of reflection intensity, ~9%, before and after PBS washing demonstrated the successful coating of PEG mixture on the biochip surface. Similarly, we observed ~19% and ~9% net increases after the surface was modified with Neutravidin and biotinylated anti-EGFR antibodies, respectively, indicating the successful conjugation of antibodies on the biochip surface through biotin–avidin interaction. Since the biochip surface was modified with the mixture of PEG200 and biotin-PEG1000 at molar ratio of 3:1 and the antibodies were conjugated on the surface through biotin–avidin interaction, 25% of the biochip surface can be used to capture exosomes. Apparently, the molar ratio of PEG200 and biotin-PEG1000 determines the exosome capture efficiency. We selected the molar ratio of 3:1 because it was reported to provide the highest capture yield.27 The molar ratio of PEG200 and biotin-PEG1000 may need to be optimized in the future with our compact SPR biosensor to maximize the exosome capture efficiency.
Characterization of Cell-Derived Exosomes by Compact SPR Biosensor.
To demonstrate the feasibility of exosomal protein characterization, the compact SPR biosensor was used to measure the EGFR levels of exosomes isolated from A549 cell culture medium. A representative real-time SPR response curve was shown in Figure 2a. DI water and PBS were first applied on the biochip to obtain baseline signals for normalization. A549 exosomes were added at the concentration of 4 × 1010 exosomes/mL. An immediate increase of reflection intensity was observed because the refractive index of exosomes is significantly different from PBS. The intensity signal was stabilized at ~13.4% at ~10 000 s, suggesting that the steady-state was reached. After the unbounded exosomes were washed off by PBS, as expected, the reflection intensity was reduced to ~9.5% with the SNR of 274. This was SPR signal from EGFR positive exosomes. Based on the reflection intensities of water (~2.0%), PBS (~6.5%), and EGFR positive exosomes (~9.5%), the expression of exosomal EGFR was calculated to be ~0.7 using eq 1. Moreover, SEM was used to confirm the capture of EGFR positive exosomes by anti-EGFR antibodies (Figure 2b). These results demonstrated the successful capture and characterization of exosomes using the compact SPR biosensor.
Figure 2.
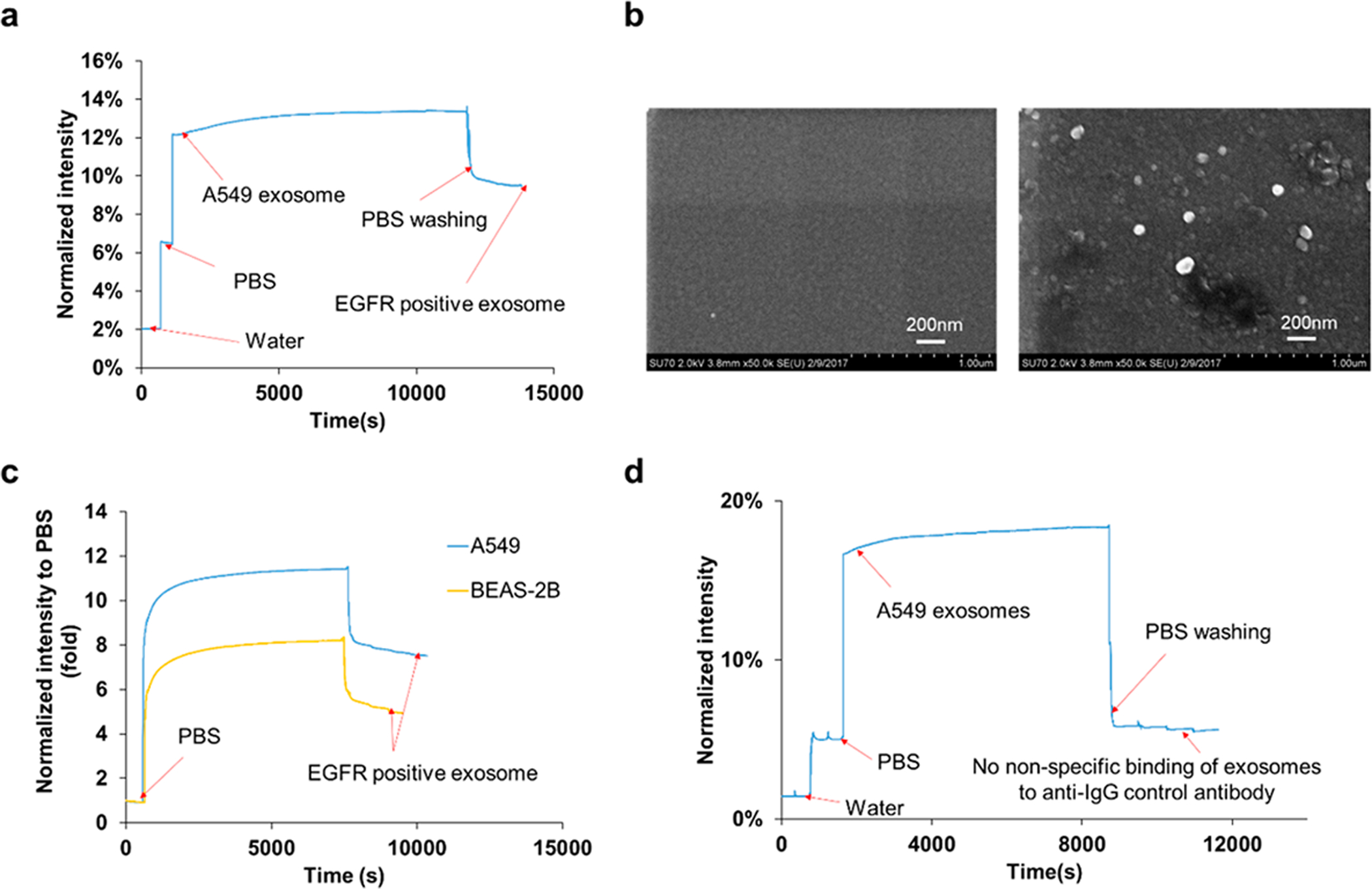
(a) Representative real-time response of compact SPR biosensor upon the addition of water, PBS, A549 exosomes at concentration of 4 × 1010 exosomes/mL, and PBS washing buffer. (b) SEM images of the biochip surface before (left) and after (right) the capture of EGFR positive exosomes. (c) Exosomes derived from A549 lung cancer cells showed higher exosomal EGFR expression than those from BEAS-2B normal cells. Both A549 exosomes and BEAS-2B exosomes were applied on the biochip at concentration of 4 × 1010 exosomes/mL. (d) With anti-IgG control antibodies modified biochip, no significant nonspecific binding of A549 exosomes was observed. A549 exosomes were applied on the biochip at much higher concentration, i.e., 2 × 1011 exosomes/mL.
To demonstrate the capability of distinguishing cancer-cell-derived exosomes and normal-cell-derived exosomes, exosomes derived from BEAS-2B normal bronchial epithelial cells were added on the biochip at the same concentration of 4 × 1010 exosomes/mL as A549 exosomes, and the expression of exosomal EGFR was measured using the compact SPR biosensor. As shown in Figure 2c, the EGFR expression of A549 exosomes was ~1.6-fold higher than that of BEAS-2B exosomes, indicating that exosomal EGFR successfully distinguished A549 cancer-cell-derived exosomes from BEAS-2B normal-cell-derived exosomes.
To exclude the potential nonspecific binding of exosomes, the biochip surface was modified with anti-IgG control antibodies. A549 exosomes were added to the biochip at much higher concentration, i.e., 2 × 1011 exosomes/mL, and incubated on the biochip for 2 h at room temperature. As expected, significant increase in reflective intensity was observed right after A549 exosomes were added on the biochip. No further intensity increase was observed during the 2 h incubation. After three PBS washes, the reflection intensity reduced back to the PBS level, suggesting that nonspecific binding of exosomes to anti-IgG control antibodies was negligible (Figure 2d).
Sensing Performance Comparison between Compact SPR Biosensor and ELISA.
To demonstrate the superior performance of compact SPR biosensor, we compared its sensing performance with that of conventional ELISA. A serial dilution of A549 exosomes was prepared with increasing concenrations from 0 to 4 × 1010 exosomes/mL. The expression of exosomal EGFR was measured by ELISA and the compact SPR biosensor. As shown in Figure 3a, ELISA detected exosomal EGFR at the A549 exosome concentration of 4 × 1010 exosomes/mL with the coefficient of variation (CV) of 54.3%. We also increased the A549 exosome concentrations up to 40 × 1010 exosomes/mL, the ODs at 450 nm were below the limit of detection of ELISA (see Figure S1 in Supporting Information); therefore, ELISA was not able to measure exosomal EGFR levels accurately. However, the compact SPR biosensor detected exosomal EGFR at the A549 concentration of 4 × 1010 exosomes/mL with the SNR of 274 and CV of 9.8% (Figure 3b). Besides, the compact SPR biosensor successfully detected exosomal EGFR in A549 exosomes at the concentration as low as 2 × 1010 exosomes/mL with SNR of 27 and CV of 16.6%. Since the SPR biochip and ELISA kit used same anti-EGFR antibodies (abcam, ab24293), the exosome capture efficiency in both assays should be similar. Therefore, our results demonstrated that the detection sensitivity of compact SPR biosensor is better than that of ELISA. The compact SPR biosensor showed higher detection senstivitity may be because the sensing target, i.e., exosomes, has diameter ~100 nm, which is relatively large and easier to detect by SPR mechanism than the colorimetric detection mechanism in ELISA.
Figure 3.
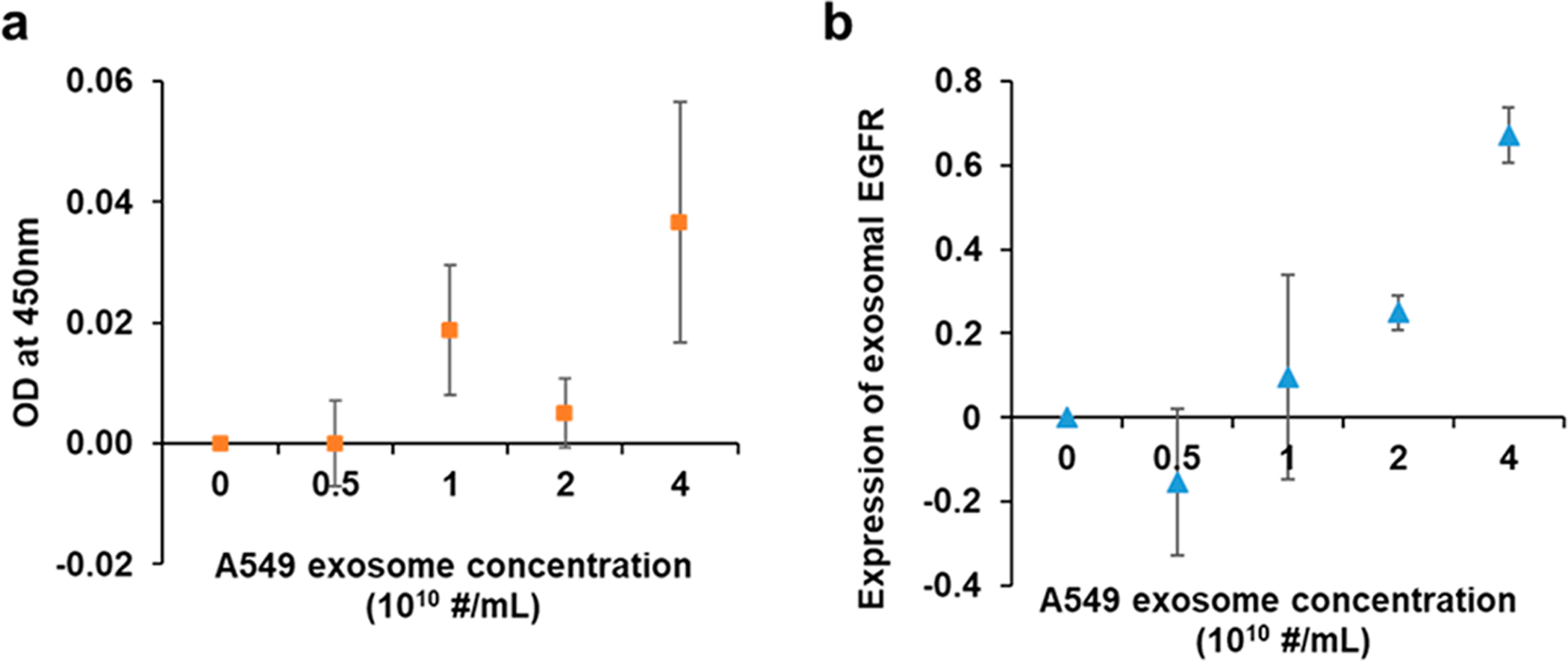
Exosomal EGFR expression measured by ELISA (a) and compact SPR biosensor (b) at increasing A549 exosome concentrations from 0 to 4 × 1010 exosomes/mL (n = 3).
Characterization of Human Serum-Derived Exosomes by Compact SPR Biosensor.
To demonstrate the clinical utility of compact SPR biosensor, serum samples from lung cancer patients (n = 5) and normal controls (n = 5) were used. Table 1 summarized the patient characteristics. Exosomes were isolated from 50 μL serum samples and resuspended in 50 μL PBS. The exosome size and number concentrations were measured using the nanoparticle tracking analysis (NTA) system. Figure 4a showed a representative size distribution of exosomes isolated from the serum sample and the TEM image of the exosomes. The size of exosomes from all serum samples was around 100 nm (Table 1). No significant difference was observed in exosome number concentration between lung cancer patients and normal controls, suggesting that exosome number concentration might not be a sensitive biomarker to distinguish lung cancer patients from normal controls (Figure 4b). The expression levels of exosomal EGFR were measured by both compact SPR biosensor and ELISA. A representative real-time SPR response curve for the detection of exosomal EGFR was shown in Figure 4c. Significant increase in reflection intensity was observed, demonstrating the successful detection of exosomal EGFR. Both compact SPR biosensor and ELISA effectively detected exosomal EGFR from 50 μL serum samples (Figure 4d,e). However, neither compact SPR biosensor nor ELISA detected significant difference in the exosomal EGFR levels between lung cancer patients and normal controls, which may be due to the small sample size. Although exosomal EGFR has been shown to be a promising biomarker for lung cancer by a few studies, the field is still in its infancy and no conclusive results have been reached yet. For example, Yamashita et al. detected higher exosomal EGFR levels in 5 out of 9 lung cancer patient serum samples compared to normal controls, while Jakobsen et al. detected lower exosomal EGFR levels in plasma samples from lung cancer patients (n = 109) compared to normal controls (n = 110).30,31 Therefore, the diagnostic value of exosomal EGFR needs to be validated with a large cohort of patients. In order to compare the sensing accuracy between compact SPR biosensor and ELISA, the exosomal EGFR level of normal control #8 was used as the normalization factor to normalize the expression of exosomal EGFR measured by each assay. As shown in Figure 4f, no significant difference was observed in the normalized exosomal EGFR levels between the compact SPR biosensor and ELISA, suggesting that compact SPR biosensor afforded same sensing accuracy as ELISA.
Table 1.
Summary of Patient Characteristic
| ID | morphology | race | gender | age | stage | exosome size (nm) | exosome concentration (1012 exosome/mL) |
|---|---|---|---|---|---|---|---|
| 1 | Adenocarcinoma, NOS | White | M | 65–69 | III | 109 | 5.20 |
| 2 | Adenocarcinoma, NOS | White | F | 75–79 | IV | 91 | 5.26 |
| 3 | Adenocarcinoma, NOS | White | F | 60–64 | IIA | 115 | 3.35 |
| 4 | Adenocarcinoma, NOS | White | F | 55–59 | IIIA | 95 | 2.87 |
| 5 | Adenocarcinoma, NOS | White | F | 55–59 | IV | 128 | 2.73 |
| 6 | Normal | White | M | 70–74 | 78 | 3.99 | |
| 7 | Normal | White | M | 65–69 | 85 | 8.15 | |
| 8 | Normal | White | F | 70–74 | 118 | 4.54 | |
| 9 | Normal | White | M | 75–79 | 111 | 3.34 | |
| 10 | Normal | White | F | 50–54 | 99 | 5.62 |
Figure 4.
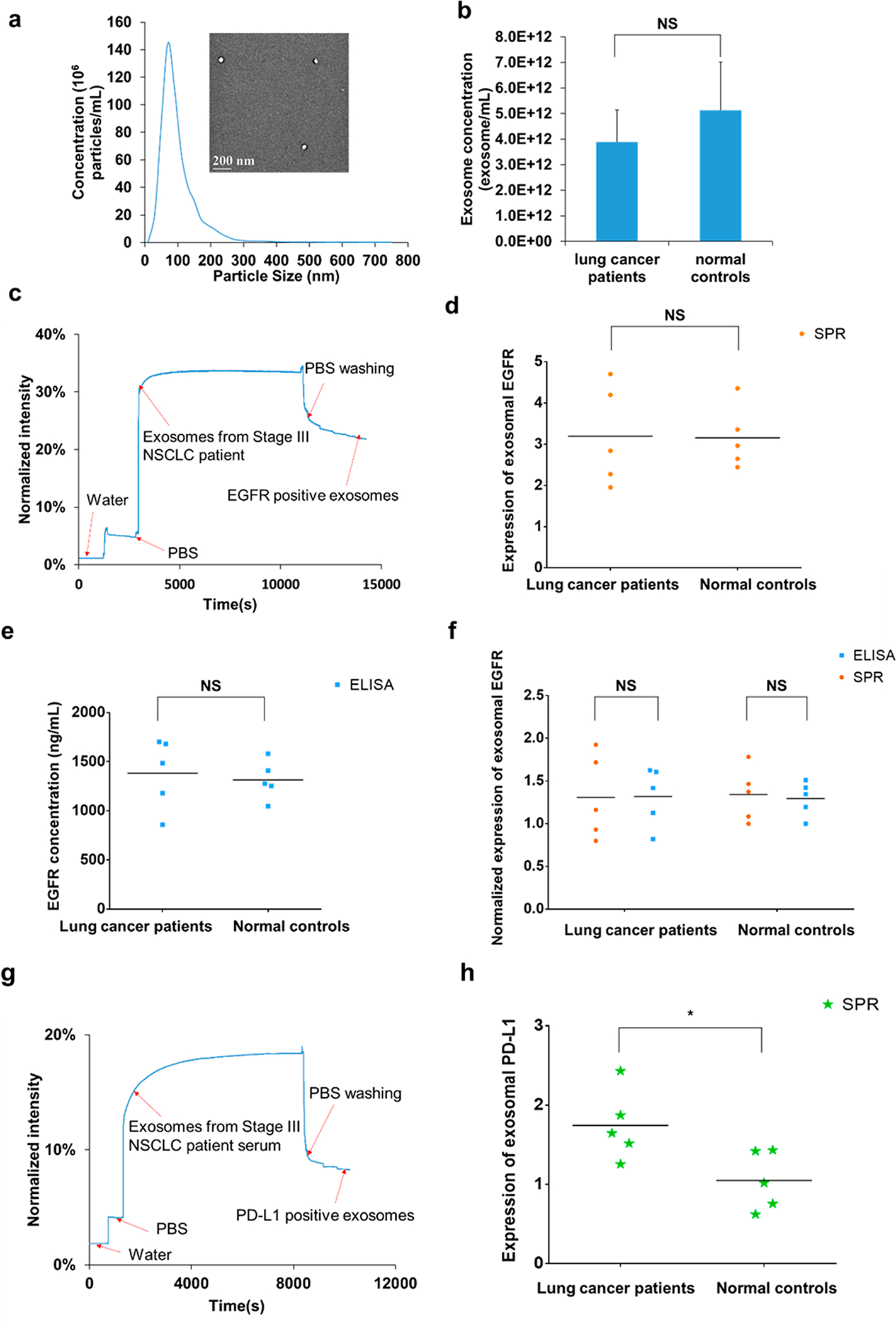
(a) Representative size distribution of exosomes isolated from a serum sample (normal control #10) measured by NTA and the morphology of exosomes imaged by TEM (inset). (b) No significant difference was observed in the number concentrations of exosomes between lung cancer patients and normal controls. (c) Representative real-time response curve of compact SPR biosensor detecting exosomal EGFR in serum sample from a Stage III lung cancer patient. Expression of exosomal EGFR in serum samples measured by (d) compact SPR biosensor and (e) ELISA. (f) Compact SPR biosensor showed the same detection accuracy as ELISA in measuring exosomal EGFR levels in serum samples. To compare compact SPR biosensor with ELISA, all data were normalized to normal control #8. (g) Representative real-time response curve of compact SPR biosensor detecting exosomal PD-L1 in serum sample from a Stage III lung cancer patient. (h) Expression of exosomal PD-L1 in serum samples measured by compact SPR biosensor. ELISA was not able to detect exosomal PD-L1 levels in serum samples. (50 μL serum sample was used in all measurements. n = 5. *: p < 0.05. NS: no significant difference).
The levels of exosomal PD-L1 in serum samples were also measured by both compact SPR biosensor and ELISA. As shown in Figure 4g, the SPR biosensor successfully detected exosomal PD-L1 in 50 μL serum samples. Besides, higher exosomal PD-L1 levels were observed in serum samples from lung cancer patients than normal controls (Figure 4h), suggesting that exosomal PD-L1 might be a promising biomarker for lung cancer diagnosis. It should be noted that the utility of exosomal PD-L1 as a diagnostic biomarker needs to be further validated in large cohorts of patients, in patients with additional lung cancer histology (such as squamous NSCLC, small cell lung cancer), and in patients with earlier stages of disease (such as stage I). However, ELISA was not able to detect exosomal PD-L1 in 50 μL serum samples, indicating that compact SPR biosensor is more sensitive than ELISA, agreeing well with the results shown in Figure 3. The reason ELISA could not detect exosomal PD-L1 may be because the low abundance of exosomal PD-L1 in serum samples. From SPR results, the expression of exosomal PD-L1 was about 30% to 50% of the expression of exosomal EGFR.
In the future, the operating parameters of the compact SPR biosensor, such as sample volume and incubation time, need to be optimized for each specific exosomal protein biomarker to further enhance the biomarker’s detection sensitivity and specificity. The compact SPR biosensor will also be optimized to incorporate multiwell design and realize high-throughput detection. The possibility of on-chip exosome enrichment and purification will be investigated to eliminate the off-chip exosome isolation process, and therefore serum samples could be directly used in the biosensor. The sensing performance of compact SPR biosensor and the diagnostic/prognostic value of exosomal protein biomarkers need to be validated in large cohorts of patients with broad characteristics, such as different stage of diseases (Stage I, II, III, and IV) and lung cancer histology (adenocarcinoma NSCLC, squamous NSCLC, and small cell lung cancer).
CONCLUSIONS
We have developed a highly sensitive, compact SPR biosensor to capture exosomes and characterize exosomal proteins for cancer diagnosis. Glycerol solutions were used to calibrate the sensing performance of the compact SPR biosensor, which showed detection sensitivity of 9.258 × 103%/RIU and resolution of 8.311 × 10−6 RIU. With lung cancer as the disease model, compact SPR biosensor successfully detected the expression levels of EGFR in A549 exosomes at concentration as low as 2 × 1010 exosomes/mL with SNR of 27 and CV of 16.6%, which was much more sensitive than ELISA (4 × 1010 exosomes/mL with CV of 54.3%). The compact SPR biosensor also detected higher EGFR levels in exosomes derived from A549 cancer cells than those from BEAS-2B normal cells, suggesting that it was capable of distinguishing lung cancer from normal control. The clinical translational potential of compact SPR biosensor was evaluated using serum samples from lung cancer patients and normal controls. Although no significant difference was observed in the expression of exosomal EGFR between lung cancer patients and normal controls, the compact SPR biosensor showed comparable sensing accuracy as ELISA. The compact SPR biosensor successfully detected exosomal PD-L1 in 50 μL serum samples; however, ELISA was not able to detect exosomal PD-L1 in the same amount of serum samples, further demonstrating that compact SPR biosensor had higher detection sensitivity than ELISA. In addition, compact SPR biosensor detected higher exosomal PD-L1 levels in serum samples from lung cancer patients than normal controls, suggesting exosomal PD-L1 might be a promising biomarkers for lung cancer diagnosis.
The compact SPR biosensor is a simple and user-friendly sensing platform that offers sensitive, label-free, real-time, and cost-effective detection of exosomal protein biomarkers. It may serve as a liquid biopsy assay that complements more invasive and expensive diagnostic tests and assists in cancer screening, diagnosis, treatment response monitoring, and prognosis.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the funding support from National Cancer Institute of the National Institutes of Health under award number 5R33CA191245 to Y. W. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR001412 to University at Buffalo. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors acknowledge the funding support from National Science Foundation under award number IIP-1718177, ECCS-1507312, and CMMI-1562057. The authors thank the support from National Science Foundation under award number CBET-1337860 which funds the nanoparticle tracking analysis (NTA) system (NanoSight, LM10, Malvern Instruments Ltd). Authors thank the Data Bank and BioRepository Shared Resource (DBBR) at Roswell Park Comprehensive Cancer Center for providing the human serum samples.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.8b00230.
Standard curve of human EGFR generated by ELISA (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Kowal J; Tkach M; Thery C Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol 2014, 29, 116–25. [DOI] [PubMed] [Google Scholar]
- (2).Yanez-Mo M; Siljander PR; Andreu Z; Zavec AB; Borras FE; Buzas EI; Buzas K; Casal E; Cappello F; Carvalho J; Colas E; Cordeiro-da Silva A; Fais S; Falcon-Perez JM; Ghobrial IM; Giebel B; Gimona M; Graner M; Gursel I; Gursel M; Heegaard NH; Hendrix A; Kierulf P; Kokubun K; Kosanovic M; Kralj-Iglic V; Kramer-Albers EM; Laitinen S; Lasser C; Lener T; Ligeti E; Line A; Lipps G; Llorente A; Lotvall J; Mancek-Keber M; Marcilla A; Mittelbrunn M; Nazarenko I; Nolte-’t Hoen EN; Nyman TA; O’Driscoll L; Olivan M; Oliveira C; Pallinger E; Del Portillo HA; Reventos J; Rigau M; Rohde E; Sammar M; Sanchez-Madrid F; Santarem N; Schallmoser K; Ostenfeld MS; Stoorvogel W; Stukelj R; Van der Grein SG; Vasconcelos MH; Wauben MH; De Wever O Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Tkach M; Thery C Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164 (6), 1226–1232. [DOI] [PubMed] [Google Scholar]
- (4).Azmi AS; Bao B; Sarkar FH Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013, 32 (3–4), 623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Li W; Li C; Zhou T; Liu X; Liu X; Li X; Chen D Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16 (1), 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sandfeld-Paulsen B; Jakobsen KR; Baek R; Folkersen BH; Rasmussen TR; Meldgaard P; Varming K; Jorgensen MM; Sorensen BS Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J. Thorac. Oncol 2016, 11 (10), 1701–1710. [DOI] [PubMed] [Google Scholar]
- (7).Li Y; Zhang Y; Qiu F; Qiu Z Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 2011, 32 (15), 1976–1983. [DOI] [PubMed] [Google Scholar]
- (8).Melo SA; Luecke LB; Kahlert C; Fernandez AF; Gammon ST; Kaye J; LeBleu VS; Mittendorf EA; Weitz J; Rahbari N; Reissfelder C; Pilarsky C; Fraga MF; Piwnica-Worms D; Kalluri R Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523 (7559), 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhao Z; Yang Y; Zeng Y; He M A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16 (3), 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Nawaz M; Camussi G; Valadi H; Nazarenko I; Ekstrom K; Wang X; Principe S; Shah N; Ashraf NM; Fatima F; Neder L; Kislinger T Nat. Rev. Urol 2014, 11 (12), 688–701. [DOI] [PubMed] [Google Scholar]
- (11).Grasso L; Wyss R; Weidenauer L; Thampi A; Demurtas D; Prudent M; Lion N; Vogel H The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Anal. Bioanal. Chem 2015, 407 (18), 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Welton JL; Khanna S; Giles PJ; Brennan P; Brewis IA; Staffurth J; Mason MD; Clayton A Proteomics analysis of bladder cancer exosomes. Mol. Cell. Proteomics 2010, 9 (6), 1324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rupert DL; Lasser C; Eldh M; Block S; Zhdanov VP; Lotvall JO; Bally M; Hook F Determination of exosome concentration in solution using surface plasmon resonance spectroscopy. Anal. Chem 2014, 86 (12), 5929–36. [DOI] [PubMed] [Google Scholar]
- (14).Sina AA; Vaidyanathan R; Dey S; Carrascosa LG; Shiddiky MJ; Trau M Real time and label free profiling of clinically relevant exosomes. Sci. Rep 2016, 6, 30460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Homola J Springer Ser. Chem. Sens. Biosens 2006, 4, 3–44. [Google Scholar]
- (16).Homola J Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev 2008, 108 (2), 462–93. [DOI] [PubMed] [Google Scholar]
- (17).Raether H Surface plasmons on smooth and rough surfaces and on gratings; Springer: 1988. [Google Scholar]
- (18).Schasfoort RB Handbook of surface plasmon resonance; Royal Society of Chemistry: 2017. [Google Scholar]
- (19).Melendez J; Carr R; Bartholomew DU; Kukanskis K; Elkind J; Yee S; Furlong C; Woodbury R A commercial solution for surface plasmon sensing. Sens. Actuators, B 1996, 35 (1–3), 212–216. [Google Scholar]
- (20).Elkind J; Stimpson D; Strong AA; Bartholomew D; Melendez J Integrated analytical sensors: the use of the TISPR-1 as a biosensor. Sens. Actuators, B 1999, 54 (1), 182–190. [Google Scholar]
- (21).Soelberg SD; Chinowsky T; Geiss G; Spinelli CB; Stevens R; Near S; Kauffman P; Yee S; Furlong CE A portable surface plasmon resonance sensor system for real-time monitoring of small to large analytes. J. Ind. Microbiol. Biotechnol 2005, 32 (11–12), 669–74. [DOI] [PubMed] [Google Scholar]
- (22).Liu Y; Liu Q; Chen S; Cheng F; Wang H; Peng W Surface plasmon resonance biosensor based on smart phone platforms. Sci. Rep 2015, 5, 12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang XL; Liu Y; Fan T; Hu N; Yang Z; Chen X; Wang ZY; Yang J Design and Performance of a Portable and Multichannel SPR Device. Sensors 2017, 17 (6), 1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tokel O; Yildiz UH; Inci F; Durmus NG; Ekiz OO; Turker B; Cetin C; Rao S; Sridhar K; Natarajan N; Shafiee H; Dana A; Demirci U Portable microfluidic integrated plasmonic platform for pathogen detection. Sci. Rep 2015, 5, 9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wang S; Xie J; Jiang M; Chang K; Chen R; Ma L; Zhu J; Guo Q; Sun H; Hu J The Development of a Portable SPR Bioanalyzer for Sensitive Detection of Escherichia coli O157:H7. Sensors 2016, 16 (11), 1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Im H; Shao H; Park YI; Peterson VM; Castro CM; Weissleder R; Lee H Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol 2014, 32, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Im H; Shao H; Weissleder R; Castro CM; Lee H Nanoplasmonic exosome diagnostics. Expert Rev. Mol. Diagn 2015, 15, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Petrelli F; Maltese M; Tomasello G; Conti B; Borgonovo K; Cabiddu M; Ghilardi M; Ghidini M; Passalacqua R; Barni S; Brighenti M Clinical and Molecular Predictors of PD-L1 Expression in Non-Small-Cell Lung Cancer: Systematic Review and Meta-analysis. Clin. Lung Cancer 2018, 19 (4), 315–322. [DOI] [PubMed] [Google Scholar]
- (29).Zhang M; Li G; Wang Y; Wang Y; Zhao S; Haihong P; Zhao H; Wang Y PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis. Sci. Rep 2017, 7 (1), 10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yamashita T; Kamada H; Kanasaki S; Maeda Y; Nagano K; Abe Y; Inoue M; Yoshioka Y; Tsutsumi Y; Katayama S; Inoue M; Tsunoda S Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013, 68 (12), 969–973. [PubMed] [Google Scholar]
- (31).Jakobsen KR; Paulsen BS; Baek R; Varming K; Sorensen BS; Jorgensen MM Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J. Extracell. Vesicles 2015, 4, 26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sandfeld-Paulsen B; Aggerholm-Pedersen N; Baek R; Jakobsen KR; Meldgaard P; Folkersen BH; Rasmussen TR; Varming K; Jorgensen MM; Sorensen BS Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol 2016, 10 (10), 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Clark DJ; Fondrie WE; Yang A; Mao L Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J. Proteomics 2016, 133, 161–169. [DOI] [PubMed] [Google Scholar]
- (34).Pirker R; Pereira JR; von Pawel J; Krzakowski M; Ramlau R; Park K; de Marinis F; Eberhardt WE; Paz-Ares L; Störkel S; Schumacher KM; von Heydebreck A; Celik I; O’Byrne KJ EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012, 13 (1), 33–42. [DOI] [PubMed] [Google Scholar]
- (35).Chinowsky TM; Quinn JG; Bartholomew DU; Kaiser R; Elkind JL Performance of the Spreeta 2000 integrated surface plasmon resonance affinity sensor. Sens. Actuators, B 2003, 91 (1–3), 266–274. [Google Scholar]
- (36). http://www.horiba.com/scientific/products/surface-plasmon-resonance-imaging-spri/.
- (37).Spoto G; Minunni M Surface Plasmon Resonance Imaging: What Next? J. Phys. Chem. Lett 2012, 3, 2682–2691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


