Figure 5. Specificity of autocitrullination.
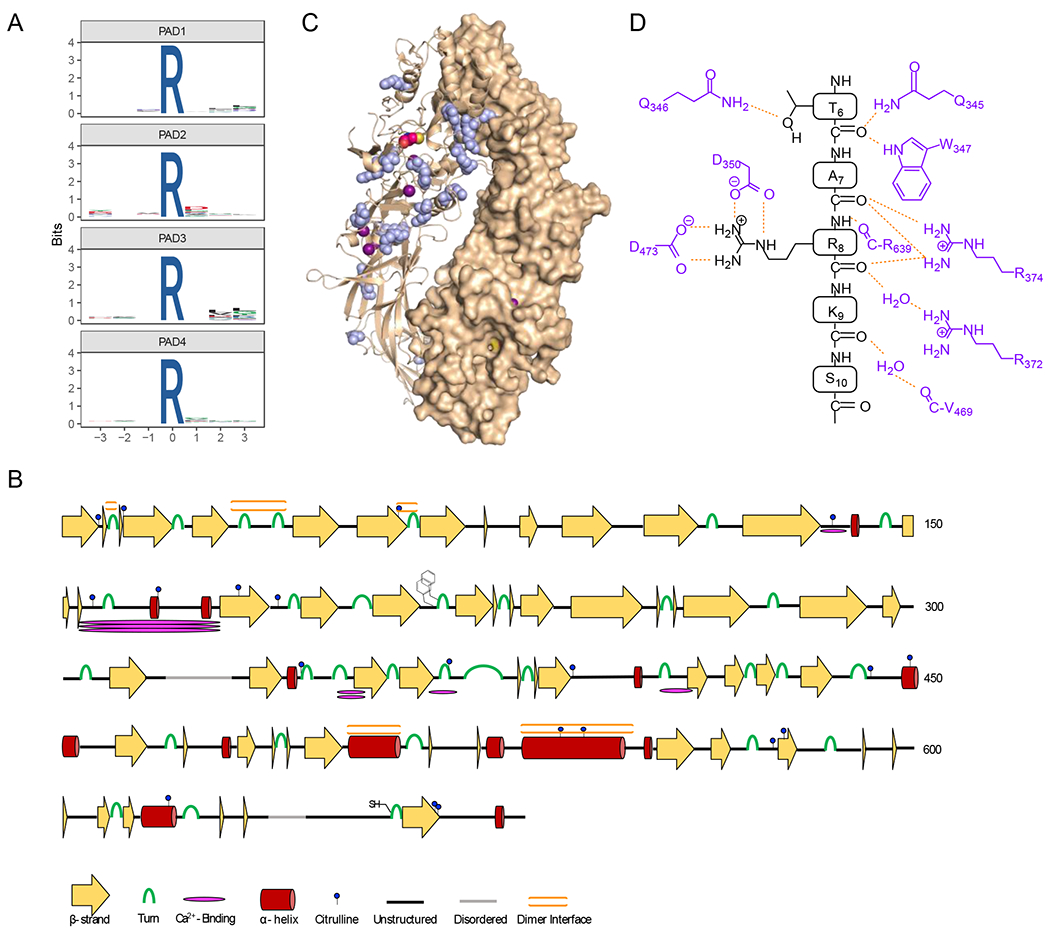
A) WebLogos depicting the substrate specificity determinants of autocitrullination by PAD1, PAD2, PAD3, and PAD4 (top to bottom). B) Secondary structural analysis of PAD2 autocitrullination shows that many of the sites of citrullination do not fall on canonical secondary structural regions. The cartoon highlights the sites of citrullination, different secondary structures, the FF-motif which plays a role in nuclear localization, and the location of the active site cysteine. C) Structure of the holo PAD2 dimer (PDBID: 4N2C) with citrullinated Arg residues shown in blue. D) Cartoon showing how histone H3 binds in the active site of PAD4 (PDB ID: 2DEW). The interactions are primarily between the enzyme and the substrate backbone, indicating that specific side chains do not make a major contribution to substrate specificity.
