Figure 6. Global analysis of the HEK293T-PAD2 citrullinome.
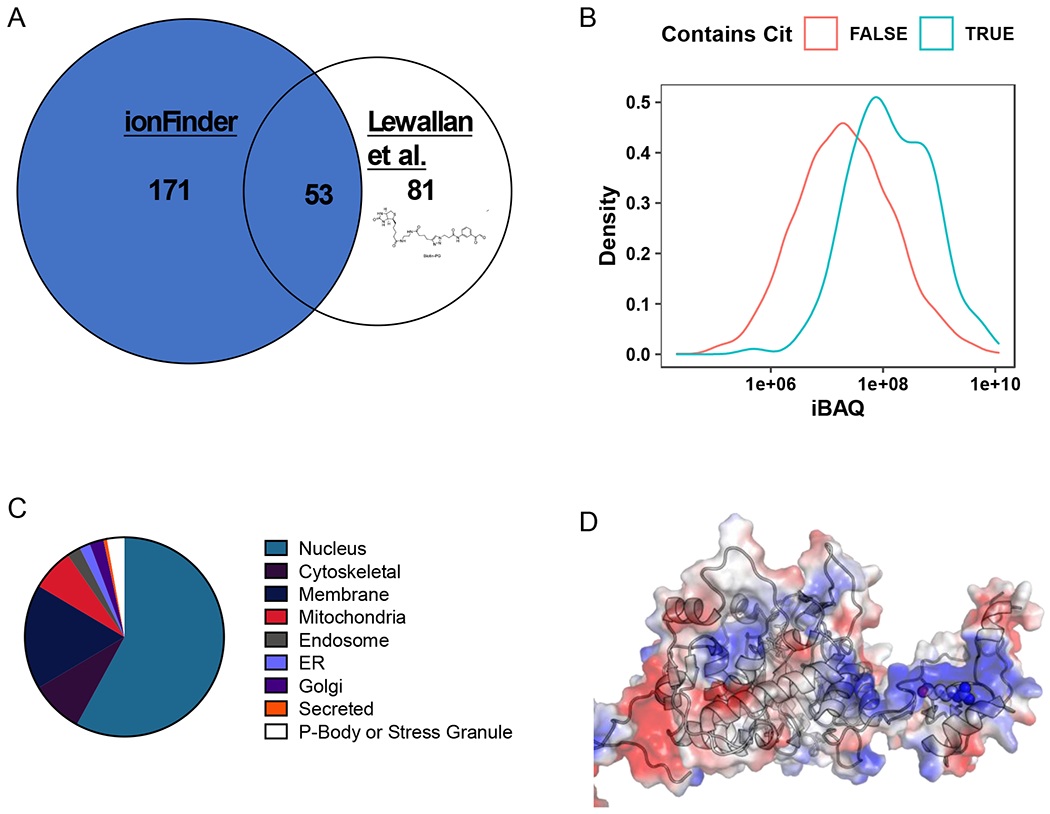
A) Comparison of the citrullinome identified by ionFinder and Lewallen et al.13 B) Kernel density estimate plot depicting the probability distribution of protein families, TRUE and FALSE, abundance based on cumulative iBAQ values from fractionated samples. The iBAQ at the maximum density in the plot was 7.45e+07 for Cit-containing proteins and 1.94e+07 for non-Cit-containing proteins. Thus, the maximum probability density of protein iBAQ values was 3.8 times greater for Cit-containing proteins than non-Cit containing proteins. C) Cellular localization of the citrullinated proteins identified by ionFinder, demonstrating that many of the proteins are, as expected, nuclear. D) Crystallographic depiction of IMPDH2 electrostatic distribution (PDB 1NF7) showing the location of Arg224 in the apo state of the enzyme.44 Arg 224 is in an exposed, and positively charged region, making it a target for citrullination by PAD2. When GTP is bound, Arg224 is no longer as accessible, as it lies in the nucleotide binding pocket near the dimer interface.
