Abstract
The expression of CLC-K1 and CLC-K2, two kidney-specific CLC chloride channels, is transcriptionally regulated on a tissue-specific basis. Previous studies have shown that a GA element near their transcriptional start sites is important for basal and cell-specific activities of the CLC-K1 and CLC-K2 gene promoters. To identify the GA-binding proteins, the human kidney cDNA library was screened by a yeast one-hybrid system. A novel member of the Cys2-His2 zinc finger gene designated KKLF (for “kidney-enriched Krüppel-like factor”) and the previously isolated MAZ (for “myc-associated zinc finger protein”) were cloned. KKLF was found to be abundantly expressed in the liver, kidneys, heart, and skeletal muscle, and immunohistochemistry revealed the nuclear localization of KKLF protein in interstitial cells in heart and skeletal muscle, stellate cells, and fibroblasts in the liver. In the kidneys, KKLF protein was localized in interstitial cells, mesangial cells, and nephron segments, where CLC-K1 and CLC-K2 were not expressed. A gel mobility shift assay revealed sequence-specific binding of recombinant KKLF and MAZ proteins to the CLC-K1 GA element, and the fine-mutation assay clarified that the consensus sequence for the KKLF binding site was GGGGNGGNG. In a transient-transfection experiment, MAZ had a strong activating effect on transcription of the CLC-K1–luciferase reporter gene. On the other hand, KKLF coexpression with MAZ appeared to block the activating effect of MAZ. These results suggest that a novel set of zinc finger proteins may help regulate the strict tissue- and nephron segment-specific expression of the CLC-K1 and CLC-K2 channel genes through their GA cis element.
CLC-K1 and CLC-K2 are two kidney-specific members of the CLC chloride channel family (1, 35). Both are present in the plasma membranes of tubular cells in the kidney (36, 37), and it has been speculated that both serve as routes for transepithelial chloride transport. Mutations of CLCNKB (the human homologue of rat CLC-K2) were recently found in patients with Bartter's syndrome (30), and the CLC-K1 gene knockout in mice results in nephrogenic diabetes insipidus (21), confirming the important role of these channels in chloride transport in the kidneys. Although the two clones are highly homologous (80% amino acid identity in the rat sequence and 90% in the human sequence), their intrarenal localizations are completely different (39). Accordingly, the analysis of transcriptional regulation of these two genes is expected to elucidate mechanisms of kidney-specific and nephron segement-specific gene expression. To this end, we previously isolated the promoters of the rat CLC-K1 (34) and CLC-K2 (26) genes. Surprisingly, proximal 5′-flanking regions that include the transcriptional start sites are highly homologous and characterized by a GA element, GGGGAGGGGAGGGGGAGGG (26). Reporter gene assays and gel retardation assays (26, 34) revealed that this GA element is indispensable for the basal promoter activities of both genes, suggesting that one or more proteins binding to this element may be involved in the kidney-specific expression of the CLC-K1 and CLC-K2 genes.
In the present study, we isolated two cDNAs that bind to the GA element, i.e., MAZ, the previously isolated myc-associated zinc finger protein, and KKLF, a novel kidney-enriched Krüppel-like factor. MAZ and KKLF have opposite effects on the CLC-K1 promoter activity, suggesting that the kidney-specific expression of CLC-K genes may be regulated by a series of zinc finger proteins through the GA element. The spatial pattern of KKLF expression overlapped with negative expressions of CLC-K1 and CLC-K2 in the kidneys, supporting the idea that KKLF may contribute to the strict nephron segment-specific expression of the CLC-K genes in vivo. Furthermore, we also found that KKLF repressed the promoter activity of the α2(I) collagen gene. Given the localization of KKLF in interstitial fibroblasts in cardiac and skeletal muscle and in potentially fibrogenic cells such as the mesangial cells in the kidneys or stellate cells in the liver, it is reasonable to assume that KKLF may be involved in the fibrogenesis in these organs. In a unilateral ureteral obstruction (UUO) model of mouse kidney, a well-characterized model of progressive tubulointerstitial fibrosis, a rapid decrease of KKLF and subsequent increase of α2(I) collagen expression were observed, suggesting that KKLF is involved in type I collagen synthesis and tissue fibrosis.
MATERIALS AND METHODS
Yeast one-hybrid screening.
cDNA encoding proteins binding to the GA element of the rat CLC-K1 gene (34) was cloned using a yeast one-hybrid system (MATCHMAKER One-Hybrid System; Clontech, Palo Alto, Calif.). Briefly, sense and antisense strands of three tandem repeats of the GA element (AGCCGGGGAGGGGGAGGGGAGGGTGTTG) were synthesized, annealed, and cloned into the pHISi-1 vector (GA-pHISi-1). The yeast strain YM4271 transformed with GA-pHISi-1 was selected on synthetic dropout medium minus histidine (SD/−His) and used as a parent cell for library screening. Plasmid DNA (20 μg) in the pACT2 vector was prepared from a human kidney cDNA (106 colonies) library having the GAL4 activation domain (Clontech) and then introduced into GA-pHISi-1-transformed YM4271 cells and selected on an SD/−His/−Leu plate with 15 mM 3-aminotriazole. Plasmid DNA was rescued from selected yeast colonies, and the sequences of isolated cDNAs were compared with those in GenBank. To clone a rat homologue of isolated cDNAs, a rat kidney cDNA library (35) was screened by plaque hybridization as described previously (35) using each isolated human cDNA as a probe.
Recombinant KKLF and MAZ protein expression and electrophoretic mobility shift assay (EMSA).
Human and rat KKLF sequences (including amino acid residues 305 to 415) were subcloned into the pGEX6P-1 vector (Amersham Pharmacia Biotech), and five zinc finger domains of MAZ (including amino acid residues 298 to 497) were cloned into the pGEX4T vector. Recombinant glutathione S-transferase (GST), GST-KKLF, and GST-MAZ were expressed in Escherichia coli BL21(DE3). Briefly, transformed bacteria grown at 37°C overnight in Luria broth medium containing ampicillin (LB ampicillin) (50 μg/ml) were diluted 1:100 in fresh LB ampicillin and incubated for 2 h at 37°C. Recombinant protein expression was induced for an additional 3 h in the presence of 1 mM isopropyl-1-thio-β-galactoside. Extracts were prepared by sonication and purified by glutathione-Sepharose affinity binding. Gel mobility shift assays were performed as described previously (26, 34). Briefly, synthesized sense (AGCCGGGGAGGGGGAGGGGAGGGTGTTG) and antisense oligonucleotides were annealed and radiolabeled at the 5′ end with polynucleotide kinase and [γ-32P]ATP. After GST fusion proteins (1 to 50 ng) or tissue nuclear extracts (∼10 μg) were incubated with 0.5 ng of radiolabeled DNA at room temperature for 30 min in a buffer containing 25 mM HEPES-KOH (pH 7.9), 90 mM KCl, 0.5 mM EDTA-NaOH (pH 8.0), 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10% glycerol, 1 μg of poly(dI-dC), and, if indicated, competitors, they were electrophoresed through 6 or 4% nondenaturing polyacrylamide gels (19:1 acrylamide-to-bisacrylamide ratio) containing 10% glycerol in TGE buffer (50 mM Tris-HCl [pH 8.5], 380 mM glycine, 2 mM EDTA-NaOH [pH 8.0]) for 2 h at 4°C. Upon completion of electrophoresis, the gels were dried and the protein-DNA complexes were visualized by autoradiography. For the supershift assay using anti-KKLF antibody, 2 μl of affinity purified antibody was included in the binding-reaction mixture.
Plasmid, transient transfection, and reporter gene assay.
Isolated cDNA clones were subcloned into the pCDNA3 vector for the transactivation assay. Luciferase reporter plasmids (10 μg) in the pGL2 vector (Promega) containing the various 5′-flanking regions of the rat CLC-K1 gene (34), the pCDNA3 vector containing an isolated cDNA insert, and the pSV-β-gal vector (5 μg) (Promega) were introduced into cultured mammalian cells by electroporation. When cells plated on 150-mm plastic dishes reached 60 to 70% confluence, they were detached with 0.25% trypsin–EDTA, neutralized with complete medium, pelleted by brief centrifugation, resuspended in 500 μl of K-PBS buffer (30.8 mM NaCl, 120.7 mM KCl, 1.46 mM KH2PO4, 8.1 mM Na2HPO4, 10 mM MgCl2) containing plasmid DNAs, transferred to a cuvette (0.4 mm wide), and electroporated at settings of 370 V and 960 μF. At 10 min after electroporation, the cells were resuspended in prewarmed complete medium and seeded in a 60-mm-diameter dish. At 48 h after transfection, the cells were harvested for the luciferase and β-galactosidase assay (Promega). Transfection efficiency was corrected using β-galactosidase activity. The human α2(I) collagen promoter was isolated by PCR (11) and ligated to the pGL-2 basic vector.
Northern blot and dot blot analysis of KKLF.
The poly(A)+ RNA Northern blot membrane for human tissues and RNA Master Blot were purchased from Clontech and probed with the whole human KKLF.
Generation of anti-KKLF antiserum, Western blotting, and immunohistochemistry.
The carboxyl-terminal peptide of rat KKLF (14 residues, CHRFPRSSRAVRAIN-COOH) was synthesized, conjugated with keyhole limpet hemocyanin at an additional cysteine residue of the amino terminus, and used to raise a rabbit polyclonal antiserum which was affinity purified soon thereafter. In vitro-translated KKLF using TnT-coupled wheat germ extract (Promega) (10 μl) and tissue extracts (10 μg) was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and immunoblotted with rabbit anti-KKLF affinity-purified antiserum (1:200 dilution) or preimmune serum. Following incubation with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins (Dako), KKLF was visualized by enhanced chemiluminescence (Amersham Corp.). Tissue samples from the liver, heart, and kidneys of rats were prepared as follows. Samples (∼500 mg) of tissues homogenized in 1.5 ml of phosphate-buffered saline containing 1% Igepal CA-630 (Sigma), 0.5% sodium deoxycholate, 0.1% SDS, and 0.1 mg of phenylmethylsulfonyl fluoride were centrifuged at 15,000 × g, and the supernatant was recovered as total-cell lysate.
Immunohistochemistry was performed using a TSA-Indirect kit (NEN) as specified by the manufacturer. Anti-KKLF antiserum was used at a dilution of 1:100. KKLF was double stained with each marker protein by introducing its antiserum into the final reaction mixture of the TSA-Indirect system. These marker proteins were detected by a 1:200 dilution of Cy3-conjugated anti-rabbit or anti-mouse immunoglobulin antibody (Sigma) and visualized by confocal microscopy (LMS-510 instrument; Carl Zeiss). Although anti-KKLF antibody was also generated in rabbits, a preliminary experiment confirmed that the KKLF protein could be detected only by the TSA-Indirect system and not by the usual indirect-immunofluorescence method using Cy3-conjugated anti-rabbit immunoglobulin antibody as a secondary antibody. Anti-rat major histocompatibility complex class II (MHC-II) antibody and anti-CD 73 (ecto-5′-nucleotidase) were purchased from Antigen America Inc. and Alexis Biochemicals, respectively. Anti-desmine monoclonal antibody was from Oncogene Research Products, and anti-von Willebrand factor monoclonal antibody and anti-ED 2 monoclonal antibody were from Dako and BMA, respectively.
RT-PCR analysis of MAZ expression along rat nephron segments.
Microdissection of rat nephron segments was performed as described previously (32). Reverse-transcribed cDNA from dissected nephrons was divided for use with PCR templates of rat MAZ, rat KKLF, rat CLC-K1, rat CLC-K2, and β-actin. PCR was performed with the following profile: 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 30 cycles. CGACATAAGCTGTCGCATTCG and AAGCTGAGCTCAGCATCTTGC were used as PCR primers to detect rat MAZ, and TCTCCAGGACATCTTGGCAGG and CTGTTCTGACCCCAACGCTG were used to detect rat CLC-K1. PCR primers for rat CLC-K1 covered nucleotides 1894 to 2129 (35) and were individually designed in different exons (exons 17 and 19) to prevent amplification from genomic DNA. PCR primers for rat MAZ were designed based on the partial cDNA sequence of rat MA Z obtained by screening the rat kidney cDNA library with human MAZ as a probe. The amplified region of rat MAZ corresponded to nucleotides 908 to 1236 of human MAZ (3). We confirmed that there was no amplification from genomic DNA by using this primer set in a preliminary experiment. The primers for rat CLC-K2 were TGTTCGTGACGTCACGAGGC and CCAAGGGTCCGATGTGACAG, which together produced a 257-bp PCR product corresponding to nucleotides 2040 to 2297 of rat CLC-K2 cDNA (1). The rat KKLF primers for reverse transcription-PCR (RT-PCR) were TGCGAATTGCGCCTGTGCCCATTG and GTACTGCGCGGCTGCTTCGTG, which together covered nucleotides 1111 to 1512 of the cDNA. β-Actin primers for RT-PCR were purchased from Clontech. To confirm the specificity of the PCR products of rat MAZ and rat KKLF, Southern hybridization was performed using primers GGACGAGAAGCCCTACCAGTG for MAZ and CCTAGGGATCCTGAGCCAATA for KKLF.
UUO of mouse kidney.
To provide the UUO model, the left ureter was ligated as described previously (10) and total RNA was harvested from the ureter-obstructed kidney and contralateral kidney on days 1, 3, 5, 7, and 10. Total RNA was also obtained from kidneys of sham-operated mice.
Nucleotide sequence accession numbers.
The accession numbers of human and rat KKLF are AB029254 and AB020597, respectively.
RESULTS
KKLF and MAZ cloning.
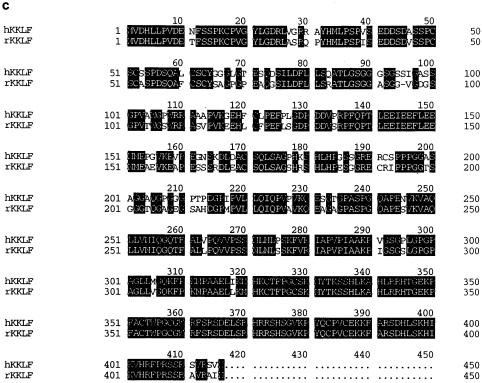
Screening of 106 clones from human kidney cDNA library resulted in the growth of several colonies on the SD/−His/−Leu plate with 15 mM 3-aminotriazole. Plasmids were rescued into DH5α competent cells, subcloned into the pcDNA3 vector, and sequenced. One clone contained a ∼2.5-kb insert that was shown by partial sequencing to be the same as the cDNAs previously isolated as human MAZ (3) or hamster Pur-1 (13). Sequencing of the total insert confirmed that it contained the whole open reading frame. Another isolated clone, which we later named KKLF, contained a ∼2.5-kb insert. The KKLF cDNA contained a single open reading frame of 1,245 bp which encoded a polypeptide of 415 amino acids with a predicted molecular mass of 44 kDa (Fig. 1a). The sequence around the ATG codon, ccagcAUGg, almost completely matched (8 of 9 nucleotides) the consensus sequence [cc(g/a)ccAUGg] proposed by Kozak (16, 17). The deduced KKLF amino acid sequence contained three zinc finger motifs (Cys2-His2) at the carboxyl terminus which were separated by a 7-amino-acid interfinger spacer similar to the H/C link consensus sequence, (T/S)GEKP(Y/F)X. Based on these features, KKLF can be classified as a member of the Krüppel family of proteins. A BLAST search of the GeneBank database also revealed that KKLF has zinc fingers similar to those of EKLF, LKLF, GKLF/EZF, BKLF, CPBP/Zf9, BTEB-2, and UKLF (2, 5, 8, 15, 20, 22, 27, 29, 31), as shown in Fig. 1b. Apart from the zinc finger domains, there was no known motif in the amino acid sequence of KKLF, but there were serine-rich and proline-rich stretches at amino acid residues 47 to 57 and 194 to 216, respectively. In addition to these clusters, KKLF was rich in proline and serine residues. In fact, proline and serine residues constitute 14 and 11% of the amino acid residues outside the zinc finger domains, respectively. There were also clusters of negatively charged amino acids at amino acid residues 41 to 45 and 142 to 150. To simplify the following immunohistochemical study to localize KKLF in vivo, the rat homologue of KKLF was obtained by screening a rat kidney cDNA library with human KKLF as a probe. This homologue also had a ∼2.5-kb insert and encoded a 415-amino-acid sequence that showed an 84% nucleotide and amino acid identity to human KKLF (Fig. 1c). Human chromosome mapping was also performed using human KKLF cDNA as a probe. Human KKLF was mapped at 3q21-q22 (data not shown).
FIG. 1.
Sequence of the KKLF cDNA and protein. (a) Sequence of the human KKLF cDNA and deduced amino acid sequence. Three zinc finger domains are underlined. H-C linkers are double underlined. The proline-rich domain (amino acid residues 194 to 216) is in boldface. The serine-rich domain (amino acid residues 47 to 57) is in boldface and underlined. Acidic domains are shown in italics. (b) Comparison of the amino acid sequence of human KKLF with amino acid sequences of related zinc finger proteins. The numbers to the left of the sequences indicate the identity to the amino acid sequence of KKLF. (c) Amino acid sequence of rat KKLF and its alignment with human KKLF. Identical amino acids are highlighted.
Gel mobility shift assay of recombinant MAZ and KKLF proteins.
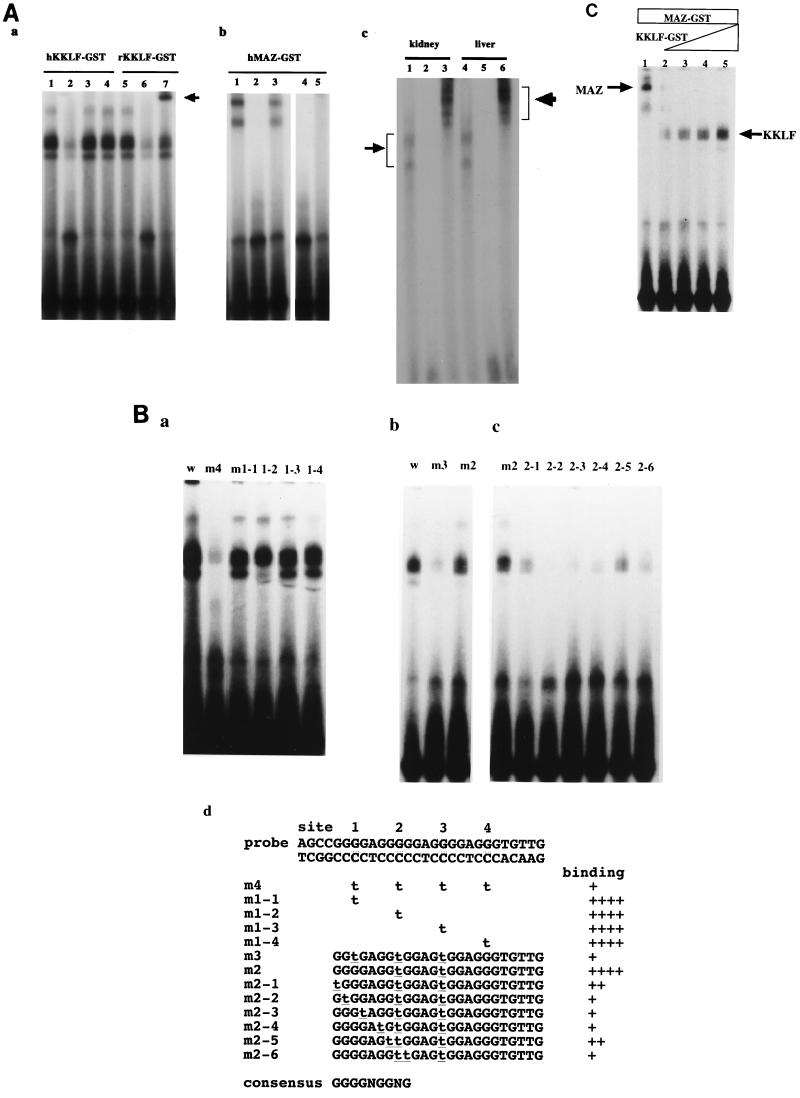
To confirm that MAZ and KKLF could bind to the GA element, three zinc fingers of human and rat KKLF and five zinc fingers of human MAZ were expressed as fusion proteins with GST, incubated with the 32P-labeled GA element, and electrophoresed in a nondenaturing polyacrylamide gel. As shown in Fig. 2A panel a, incubation of human and rat KKLF fusion proteins with the GA element resulted in the formation of retarded complexes (lane 1 and 5) that could be competed with a 100-fold molar excess of unlabeled GA element (lane 2 and 6). A 100-fold molar excess of cold SpI oligonucleotide (lane 3) did not compete with the GA element, suggesting that KKLF could not bind to the SpI site (GGCGGG). Inclusion of anti-rat KKLF antibody in the EMSA reaction mixture shifted the rat KKLF-GA element complex (lane 7, indicated by an arrow), confirming that the antibody described in this study could truly bind to rat KKLF and be used for the supershift assay. Since the full-length KKLF-GA complex migrated very slowly in the 6% gel, we used a 4% gel and ran it for 6 h in EMSA. Fig. 2A panel c shows that the kidney and liver nuclear extracts produced faint retarded bands with the GA element (lanes 1 and 4) that were competed with a 100-fold molar excess of the cold probe (lanes 2 and 5) and supershifted with anti-KKLF antibody (lanes 3 and 6). These results clearly indicated that a native KKLF from kidney and liver could bind the GA element of the CLC-K1 promoter.
FIG. 2.
(A) Gel mobility shift assay with the CLC-K1 GA element and recombinant extracts of MAZ-GST and KKLF-GST proteins. (Panel a) Purified human and rat KKLF-GST extracts (30 ng) were mixed with the 32P-labeled GA element (AGCCGGGGAGGGGGAGGGGAGGGTGTTG) of the CLC-K1 gene promoter (34) and electrophoresed in a 6% nondenaturing polyacrylamide gel. hKKLF-GST (lanes 1 to 4) and rKKLF-GST (lanes 5 to 7) have 30 ng of human and rat KKLF-GST in the reaction, respectively. Lanes 1 and 5, without the cold competitor; lanes 2 and 6, with a 100-fold molar excess of cold competitor; lane 3, with a 100-fold molar excess of SpI probe (Promega); lanes 4 and 6, with anti-rat KKLF antiserum added. (Panel b) Purified human MAZ-GST extracts (30 ng) were mixed with the 32P-labeled GA element (lane 1). The formation of DNA-MAZ complexes was abolished with a 100-molar excess of cold probe (lane 2), ME1a1 probe (GAAAAAGAAGGGAGGGGAGGGATC) (lane 4), or CD4 probe (CTGGGGGTGGGAGGGAGGGACTCCT) (lane 5). In lane 3, the m4 probe with four mutations (Fig. 2B panel d) in the wild GA-element was labeled with 32P and used for EMSA. The MAZ binding was reduced by the mutations. (Panel c) Rat kidney and liver nuclear extracts were incubated with the 32P-labeled GA element (lanes 1 and 4) and resolved in a 4% polyacrylamide gel. The faint retarded bands were observed. These bands were competed with the cold probe (lanes 2 and 5) and supershifted by the anti-rat KKLF antiserum (lanes 3 and 6). (B) Mutational analysis of the KKLF binding site in the GA element. Mutants m1 and m4 (panel a), m3 (panel b), and m2 (panel c) were used. The structure of each mutant probe is shown in panel d. All probes were labeled with 32P, and equal radioactivity was included in the binding reaction mixtures. (C) Competitive binding of KKLF and MAZ at the GA element. In all reactions, a constant amount of MAZ-GST (30 ng) was incubated with increasing amounts of human KKLF-GST (lane 1, 0 ng; lane 2, 1 ng; lane 3, 3 ng; lane 4, 30 ng; lane 5, 50 ng).
Previous studies revealed that MAZ binds to GGGGAGGGG sequences (3, 9, 24), i.e., sequences identical to the GA element of the CLC-K1 promoter. Accordingly, it stands to reason that the GA element forms the retarded band with MAZ-GST shown in Fig. 2A panel b (lane 1). These two bands (Fig. 2A-b) were completely blocked by the 100-fold molar excess of cold probe (lane 2). The ME1a1 probe (3) (lane 4) or the CD4 promoter probe (6) (lane 5), previously shown to bind to MAZ, also blocked the formation of the retarded bands. In lane 3 of Fig. 2A panel b, m4 probe containing four mutations (underlined) introduced into the wild-type GA element (AGCCGGtGAGGtGGAGtGGAGtGTGTTG) (Fig. 2B panel d) was labeled with 32P and used for EMSA. A significant decrease of binding was observed in EMSA using the m4 probe, which was consistent with the previously reported consensus sequence for the MAZ binding site (3, 9, 24).
To determine the KKLF binding-site consensus sequence, a series of mutations was introduced into the wild-type GA element (Fig. 2B panel d) and the binding ability of the mutations was determined by EMSA. Figure 2B panel a shows that the m4 probe with four mutations (sites 1, 2, 3, and 4, [Fig. 2B panel d]) had a significantly reduced binding ability just as for MAZ (Fig. 2A panel b, lane 3). However, probes that had a single mutation at any one of these four sites (m1-1 to m1-4) could bind to KKLF. Next, we restored a mutation of the m4 probe at site 4. This m3 probe, which had three mutations, at sites 1, 2, and 3, still showed a significantly reduced ability to bind to KKLF, just as for the m4 probe (Fig. 2B panel b). Next, we further restored the mutation of the m3 probe at site 1 and named the new probe m2 (Fig. 2B panel b, lane 3). Surprisingly, the binding ability of m2 was almost equal to that of the wild-type probe (lanes 1 and 3), suggesting that this G residue at site 1 was located at a crucial position in the m2 probe in terms of KKLF binding. According to the report by Klevit, KKLF is supposed to have a consensus sequence, NNGGNGNGG (14). To verify this, we introduced seven kinds of single mutations into the m2 probe around site 1. A single mutation at any site significantly reduced the binding of KKLF, as shown in Fig. 2B panel c. Based on these data, GGGGNGGNG was determined experimentally to be a consensus KKLF binding sequence (Fig. 2B panel d).
Since the consensus KKLF binding sequence was found to be similar to the MAZ binding sequence, we tested whether MAZ and KKLF competitively bind to the GA element. As shown in Fig. 2C, the incubation of an increasing amount of KKLF-GST with a constant amount of MAZ abolished MAZ binding to the GA element. Tenfold less KKLF than MAZ was enough to abolish the MAZ binding, indicating that KKLF had a higher affinity to the GA element than did MAZ.
Transactivation assay.
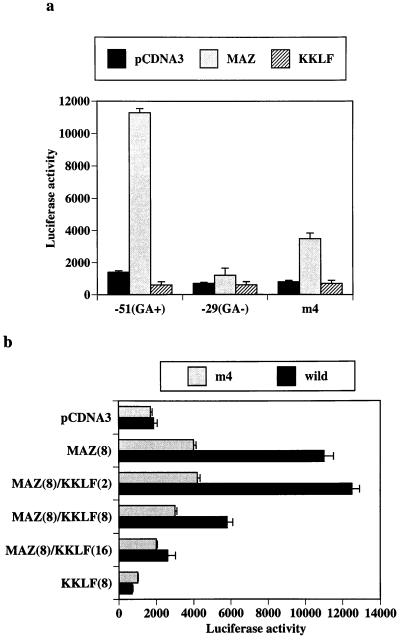
To assay the transactivating effect of cloned cDNAs, we selected NIH 3T3 as the host cell of transfection since the CLC-K1 promoter activities in NIH 3T3 cells were minimal in our previous study (34). As shown in Fig. 3a, cotransfection of MAZ resulted in a clear stimulation of the CLC-K1 promoter (positions −51 to +75). The construct which did not contain the GA element (positions −29 to +75) did not respond to the cotransfection of MAZ, suggesting that MAZ transactivated the CLC-K1 promoter through the GA element. MAZ activation of CLC-K1 promoter was also significantly reduced in the mutant CLC-K1 promoter (m4) that had four mutations in the GA element, which was consistent with the reduced binding in the EMSA study (Fig. 2A panel b). On the other hand, cotransfection of KKLF did not affect the baseline promoter activity in NIH 3T3 cells. As shown previously (34), the proximal CLC-K1 promoter (positions −51 to +75) showed basal promoter activity in some cell lines. In the preliminary experiment using MDCK cells, cotransfection of KKLF almost completely repressed the basal promoter activity of the CLC-K1 promoter (data not shown), and on this basis KKLF did appear to act as a repressor. In addition, EMSA confirmed the competitive binding of MAZ and KKLF to the GA element (Fig. 2C). Accordingly, we tested whether KKLF inhibited MAZ activity by transfecting expression plasmids for both into NIH 3T3 cells. As shown in Fig. 3b, when the two expression vectors were transfected in equivalent amounts, the MAZ-induced transactivation was inhibited to ∼40%. Increasing the amounts of KKLF further diminished and completely blocked the MAZ effect. This repressive effect of KKLF on MAZ activity was also observed in the m4 construct (Fig. 3b). Although EMSA showed a significant decrease of KKLF binding to the m4 probe, as with MAZ (Fig. 2B panel a), a faint retarded band was still observed, which may account for the repressive effect of KKLF in the m4 construct. These results indicated that KKLF acted as a functional competitor of MAZ through the competitive binding to the GA element.
FIG. 3.
Transfection of NIH 3T3 cells with MAZ and KKLF expression vectors and CLC-K1-luciferase reporter plasmids. (a) Human MAZ and KKLF in pCDNA3 (10 μg) were transfected with −51, −29, or mutant (m4) CLC-K1-luciferase reporter plasmids (34) (10 μg) and pSV-β-Gal (3 μg). Empty vector (pCDNA3) was cotransfected to maintain equivalent total plasmid amounts in each transfection. The −51 CLC-K1 luciferase reporter contains the GA element, but the −29 reporter does not. The m4 CLC-K1 luciferase reporter contains the four mutations in the −51 GA CLC-K1 luciferase plasmid. Luciferase and β-galactosidase reporter gene activities were measured 48 h after transfection. Data are relative light units divided by β-galactosidase activity from three different experiments (mean and standard error of the mean). (b) MAZ expression vector (8 μg) was cotransfected with different amounts (2 to 16 μg) of KKLF expression vector, −51 wild or m4CLC-K1 luciferase reporter plasmids (8 μg), and pSV-β-Gal (3 μg). Numbers in parentheses indicate the amounts of plasmids used for transfection.
Tissue distribution of human KKLF.
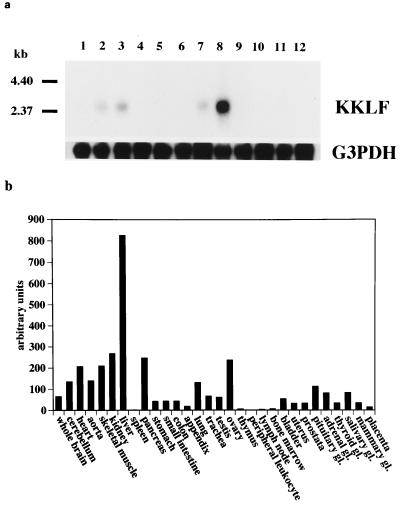
To determine the tissue distribution of KKLF, Northern blot analysis was performed using a full-length KKLF cDNA as a probe. As shown in Fig. 4a, a 2.5-kb mRNA was detected and its highest expression was observed in the liver. Moderate levels were also noted in the kidneys, heart, and skeletal muscle. Based on a quantitative dot blot analysis of the relative expression of KKLF using a human RNA Master Blot kit (Clontech), we obtained information on a more detailed pattern of KKLF expression among human tissues (Fig. 4b). As shown in the Northern analysis, the liver showed the highest expression among the tissues examined. In contrast to EKLF, KKLF expression was not detected in bone marrow and lymphoid tissues. Since the gene name LKLF is already used to designate lung enriched Krüppel-like factor, we called this liver-enriched Krüppel-like factor “Kidney-enriched Krüppel-like factor” (KKLF) in consideration of its isolation from the kidneys and high level of expression in that organ. Additional examination of the tissue distribution of rat KKLF by Northern analysis and RNase protection assay showed tissue distributions quite similar to those of human KKLF (data not shown).
FIG. 4.
Tissue distribution of human KKLF. (a) Northern analysis of human KKLF. Each lane contains polyA(+) RNAs (2 μg) from various human tissues. Lanes: 1, brain; 2, heart; 3, skeletal muscle; 4, colon; 5, thymus; 6, spleen; 7, kidney; 8, liver; 9, small intestine; 10, placenta; 11, lung; 12, leukocyte. (b) Relative KKLF expression. A dot blot analysis of 0.5 μg of poly(A)+ RNA was performed using the human RNA Master Blot (Clontech). Hybridization signals were quantified using an image analyzer (BAS-2500; Fuji Corp.)
Immunohistochemistry of KKLF.
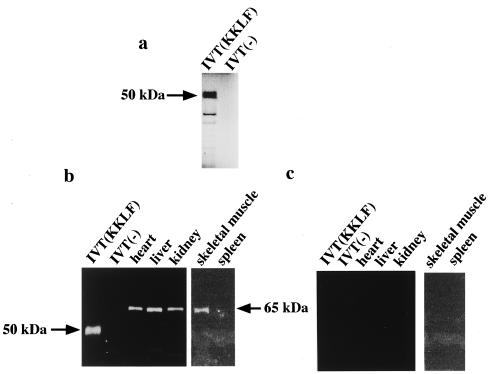
To better localize the cells expressing the KKLF gene, especially in the kidney cells already known to express CLC-K1 and CLC-K2, we generated anti-rat KKLF antiserum using a carboxyl-terminal peptide. Western blotting of in vitro-translated KKLF was performed with the antibody to confirm its specificity. In vitro-translated KKLF with [35S]methionine (Fig. 5a) showed an apparent molecular mass of ∼50 kDa, and this matched the calculated molecular mass of 44 kDa. The parallel samples with a cold complete amino acid mixture but without hot methionine were subjected to SDS-polyacrylamide gel electrophoresis Western blotted onto a nitrocellulose membrane, and incubated with anti-KKLF antibody (1:200). As shown in Fig. 5b, the anti-KKLF antibody recognized a band of ∼50 kDa only in one lane where in vitro-translated KKLF was present, thereby confirming the specificity of the antibody. This antibody also recognized a single protein of ∼65 kDa in the liver, heart, skeletal muscle, and kidneys but not in the spleen (Fig. 5b), which was consistent with the result of KKLF mRNA expression. Preimmune serum did not recognize in vitro-translated KKLF and a 65-kDa protein in rat tissues (Fig. 5c). Posttranslational modifications of KKLF in vivo may be possible based on the ∼15-kDa difference between the native protein in tissues and the in vitro-translated product. A similar phenomenon (14-kDa difference) was observed for another Krüppel-like factor, Zf9 (27). Although these modifications may include both phosphorylation and glycosylation, no glycosylation site was found in residues conserved in human and rat KKLF. Potential phophorylation sites for protein kinase C were found at amino acid residues 14 to 16 and 408 to 410.
FIG. 5.
In vitro translation of rat KKLF and Western blot of rat KKLF protein. (a) In vitro translation of rat KKLF. Rat KKLF protein was synthesized in vitro using TnT-coupled wheat germ extract in the presence of [35S]methionine. A 10-μl volume was analyzed by electrophoresis in an SDS-polyacrylamide gel, dried, and autoradiographed. IVT(KKLF) and IVT(−) indicate a rat KKLF (∼50 kDa) and negative control, respectively. (b and c) Western blots of in vitro-translated rat KKLF and various rat tissues. A 10-μl volume of the in vitro translation reaction mixture of rat KKLF cDNA in pCDNA3 and an empty pCDNA3 vector with cold methionine were electrophoresed together with 10 μg of tissue extracts from rat heart, liver, kidney, skeletal muscle, and spleen in an SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and reacted with anti-KKLF affinity-purified antiserum (1:200) (b) or preimmune serum (c). In the in vitro translation reaction of rat KKLF, the anti-KKLF antibody recognized a ∼50-kDa band which matched the apparent molecular mass of KKLF shown in panel A. In tissue samples, the antibody recognized a ∼65 kDa protein.
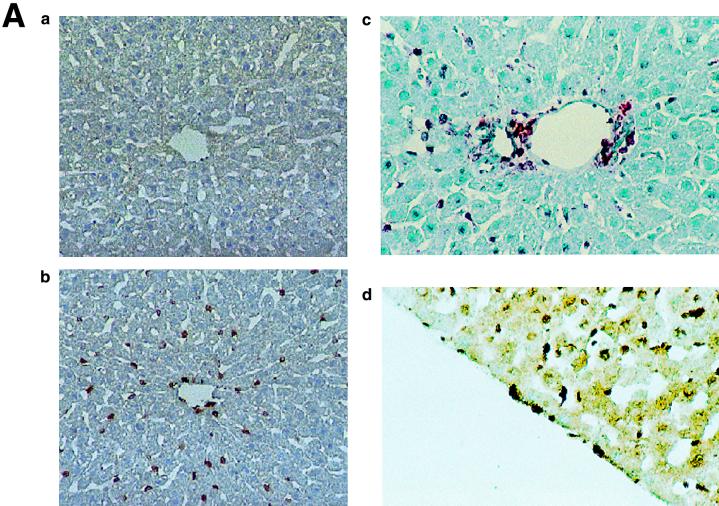
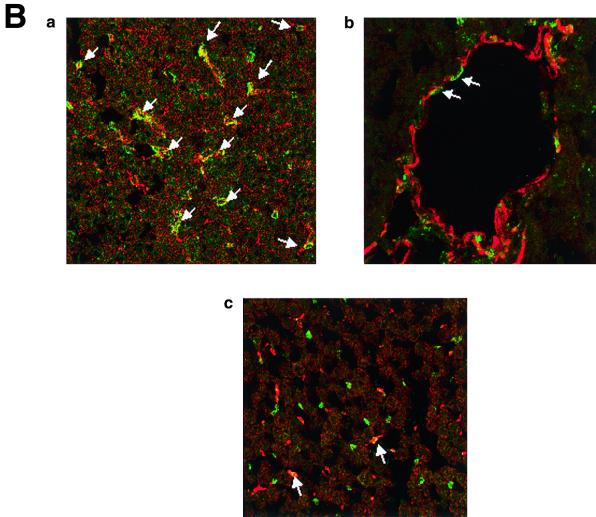
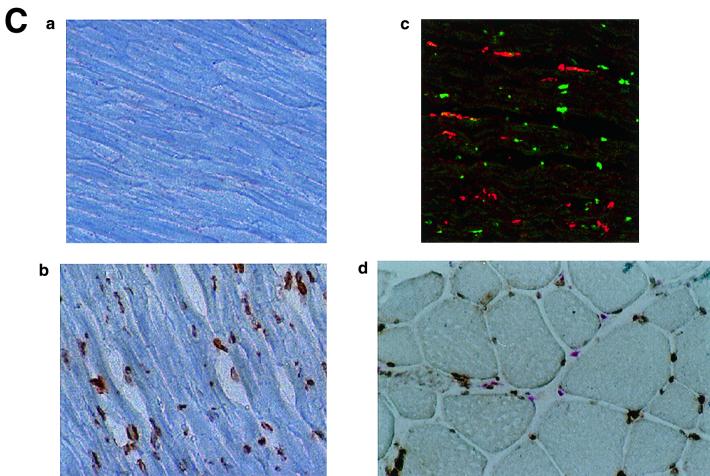
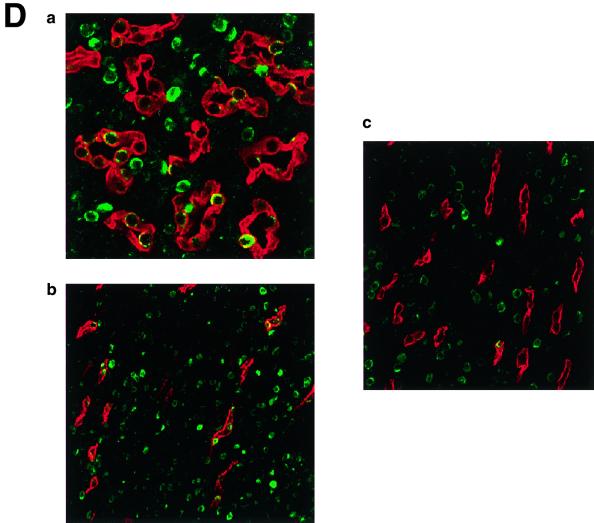
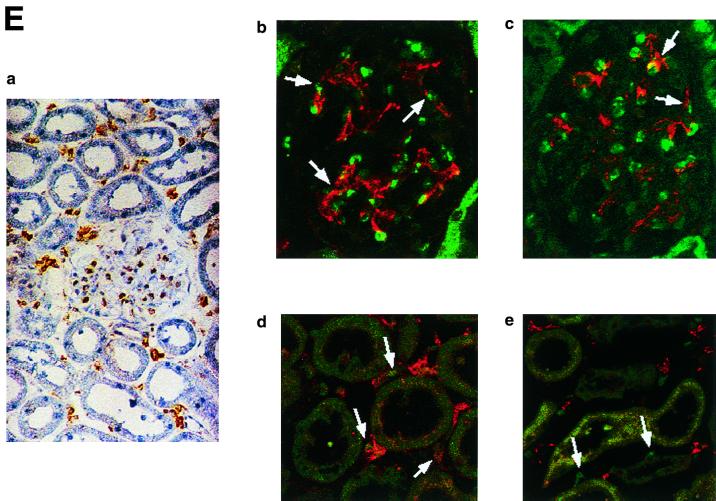
The immunohistochemistry of KKLF in the liver is shown in Fig. 6A and B. The nuclei of cells in the sinusoid were stained (Fig. 6A panel b), and double immunostaining of KKLF and desmin (Fig. 6B panel a) showed that most of the stained cells in the sinusoid were stellate cells. ED-2-positive Kupffer cells only occasionally colocalized with KKLF (Fig. 6B panel c). In addition to the stellate cells, KKLF staining was observed in subcapsular fibroblasts (Fig. 6A panel d), second-layer cells in the central vein (Fig. 6B panel b), and portal fibroblasts (Fig. 6A panel c). Most of the stained nuclei in the heart (Fig. 6C panels b and c) and skeletal muscle (panel d) were observed between the muscle cells and were not colocalized with ED-2 positivity, which suggested that the KKLF-positive interstitial cells in the heart and skeletal muscle were not resident macrophages but fibroblasts. In the kidneys, the KKLF staining was observed in the nuclei of cells in the inner medulla (Fig. 6D panels a through c), and there were also numerous stained cells in the glomeruli and interstitium in the cortex (Fig. 6E panel a). To examine whether KKLF and CLC-K1 colocalized in the inner medulla, double-immunofluorescence staining was performed. As shown in Fig. 6D panels a through c, colocalization of KKLF with AQP-2 (panel a) and AQP-1 (panel b) confirmed the presence of KKLF in the inner medullary collecting ducts and the thin descending limb of Henle's loop, but there was no colocalization with CLC-K1 (panel c). KKLF-positive cells that were negative for AQP-1, AQP-2, and CLC-K1 were also observed by simultaneous staining of all four proteins (data not shown), indicating that KKLF was also present in the interstitial cells in the inner medulla. KKLF staining in the glomeruli was mostly associated with desmin staining (Fig. 6E panel b, indicated by arrows) and occasionally associated with Von Willebrand facor staining (panel c, indicated by arrows), suggesting that KKLF was present predominantly in the mesangial cells in the glomeruli. In the interstitial cells of the kidney cortex, KKLF-positive cells were also positive for ecto-5′-nucleotidase (Fig. 6E panel d), a marker for fibroblasts in the renal cortex (12), but they were not colocalized with MHC-II antigen (panel e), a marker for dendritic cells, another predominant cell type in the interstitium of the renal cortex (12).
FIG. 6.
(A) KKLF in the liver. Cryostat sections of rat liver were incubated with preimmune serum (a) or anti-KKLF antibody (1:100) (b to d) in the TSA-Indirect system, and KKLF was visualized with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (b and d) or 3-amino-9-ethyl carbazole (c). The sections were counterstained with hematoxylin. Cells in siunusoids (b), portal fibroblasts (c), and subcapsular fibroblasts (d) were stained. (B) Double immunostaining of KKLF with desmin and ED-2 in the liver. (a) KKLF staining in sinusoids (green) was associated with desmin staining (red) and became yellowish (arrows). (b) The nuclei of desmin-positive (red) second-layer cells were also positive for KKLF (arrows). (c) ED-2 stainings identifying Kupffer cells (red) were only occasionally associated with KKLF staining (green). (C) KKLF staining in the heart and skeletal muscle. (a and b) Cryostat sections of rat heart were incubated with preimmune serum (a) or anti-KKLF antibody (1:100) (b) in the TSA-Indirect system, and KKLF was visualized with DAB (b). KKLF staining was observed in the interstitial cells (brown). (c) Double staining of KKLF (green) and ED-2 (red) in the heart. Most of the KKLF-positive cells were ED-2 negative. (d) KKLF staining in the skeletal muscle. KKLF visualized by DAB staining (brown) was observed in the interstitial cells between the muscle fibers and was not associated with the ED-2 staining (pinkish staining by Fast red). (D) Immunohistochemistry of rat KKLF in the inner medulla of the rat kidney. Double immunofluorescence of KKLF (green) and other marker proteins (red) in the inner medulla: AQP-2 (a) AQP-1 (b), or CLC-K1 (c) KKLF is colocalized with AQP-1 and AQP-2 but not with CLC-K1. Magnifications, ×400 (a) and ×200 (b and c). (E) KKLF in the glomeruli and cortex. (a) KKLF visualized with DAB was observed in the interstitial cells and cells in glomeruli in the cortex Magnification, ×200. (b) KKLF staining (green [arrows]) in the nuclei of glomeruli was mostly surrounded by desmin staining (red). (c) Only a small portion of KKLF staining (arrows) was associated with Von Willebrand factor staining (red). (d) Most of the KKLF staining in the interstitium was surrounded by ecto-5′-nucleotidase staining (arrows). (e) Most of the MHC-II-positive dendritic cells (red) did not have KKLF staining (green).
RT-PCR of rat MAZ and KKLF along nephron segments.
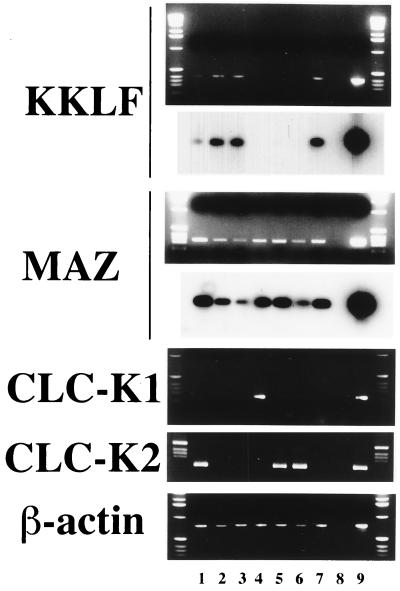
Human MAZ was reported to be expressed ubiquitously (3). RT-PCR analysis using manually dissected nephron segements was performed to confirm that MAZ is also expressed in the nephron segments together with CLC-K1 and CLC-K2. Dissected nephron segments were permeabilized with Triton X-100 and directly used as the templates of cDNA synthesis. The resultant cDNAs were divided for PCR of rat MAZ, rat CLC-K1, rat CLC-K2, rat KKLF, and β-actin. As shown in Fig. 7, CLC-K1 mRNA was detected only in the thin ascending limb of Henle's loop (35, 36) while CLC-K2 expression was detected in the glomeruli, thick ascending limb of Henle's loop, and distal tubules. This was consistent with the data from our in situ hybridization study (39). Just as for β-actin, the expression of rat MAZ was detected in all nephron segments tested (Fig. 7). This finding confirmed the ubiquitous nature of MAZ expression and suggested that MAZ could interact with the GA elements of the CLC-K1 and CLC-K2 gene promoters and activate the transcription of these genes in the nephron. Detection of the KKLF mRNA in the glomeruli, thin descending limb of Henle's loop, and inner medullary collecting ducts supported the results of the immunohistochemical study. The positive signal in the proximal tubules in RT-PCR analysis might have been due to contamination of the sample with the interstitial fibroblasts. KKLF expression along nephron segments appeared to be inversely correlated with CLC-K1 and CLC-K2 expression.
FIG. 7.
RT-PCR of rat MAZ and rat KKLF along nephron segments The upper panel of rKKLF shows ethidium bromide staining of RT-PCR products; 402-bp PCR products were detected. The lower panel of rKKLF shows a Southern blot of the gel in the upper panel probed with a 32P-labeled rKKLF-specific oligonucleotide probe and visualized by autoradiography. The upper panel of rMAZ shows ethidium bromide staining of RT-PCR; 330-bp PCR products were detected. The lower panel of rMAZ shows a Southern blot of the gel in the upper panel. Lanes: 1, glomeruli; 2, proximal tubules; 3, thin descending limb of Henle's loop; 4, thin ascending limb of Henle's loop; 5, thick ascending limb of Henle's loop; 6, cortical collecting ducts; 7, inner medullary collecting ducts; 8, glomeruli without RT; 9, cDNA as a positive control for PCR.
KKLF expression in the kidney of the UUO model and effect of KKLF on the promoter activity of human α2(I) collagen gene promoter.
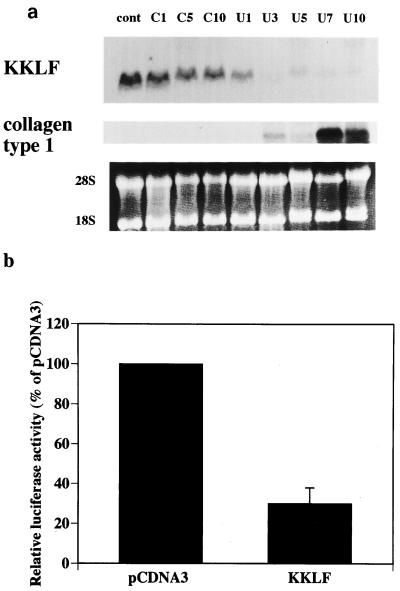
Immunohistochemistry revealed predominant expression of KKLF in the fibroblasts in several organs. To investigate the role of KKLF in the fibroblasts, we tested the expression of KKLF in UUO mouse kidneys, a well-characterized model of progressive interstitial fibrosis in the kidney (10). KKLF expression began to decrease 1 day after ureteral obstruction and could not be detected in Northern blots from 3 days after obstruction onward (Fig. 8a). This decrease of KKLF expression preceded the type I collagen expression, suggesting that KKLF expression in normal fibroblasts may be involved in the regulation of type I collagen expression. To address this issue, we tested the effect of KKLF expression on the human α2(I) collagen promoter (Fig. 8b). Cotransfection of the KKLF expression vector with the reporter gene driven by the human α2(I) collagen promoter inhibited the human α2(I) collagen promoter activity (Fig. 8b), clearly indicating that KKLF repressed the human α2(I) collagen promoter.
FIG. 8.
(a) Northern blot of a UUO mouse kidney probed with rKKLF (top panel) and human type I collagen (middle panel) probes. Lanes: cont, total RNA from sham-operated mice kidney; C1, C5, and C10, RNAs from contralateral kidneys from UUO mice sacrificed 1, 3, and 5 days, respectively, after UUO; U1 to U10, RNAs from obstructed kidneys 1 to 10 days, respectively, after UUO. Each lane contains 10 μg of total RNA. The bottom panel shows ethidium bromide staining of the RNAs. (b) Effect of KKLF expression on the human α2(I) collagen gene promoter. The human α2(I) collagen gene promoter cloned in the pGL-2 basic vector (20 μg) was cotransfected with either pCDNA3.1+ empty vector or rat KKLF expression vector in pCDNA3.1+ (20 μg) in NIH 3T3 cells. At 48 h after transfection, luciferase activity was measured and corrected by β-galactosidase activity (n = 3, means and standard deviations).
DISCUSSION
In previous reports we characterized an essential element of the rat CLC-K1 and CLC-K2 gene promoters that contributes to the basal and cell-specific activities of these genes (26, 34). We also demonstrated in these reports that this GA element is recognized by one or more distinct proteins (26, 34). To identify possible transcription factors that bind to the GA element, we have screened a human kidney cDNA library by using a yeast one-hybrid system and isolated two cDNAs that encode polypeptides with zinc finger motifs. The first polypeptide was MAZ, a previously discovered protein (3) that transactivated the CLC-K1 gene promoter, and the second polypeptide was KKLF, a novel Krüppel-like factor that carried a three-zinc-finger (Cys2-His2) motif at its C-terminal domain and repressed the promoter activity. A human MAZ and its mouse homolog, mPur-1, were previously shown to bind to the GGGAGGG motif of the c-myc P2 promoter (3) and the GAGA box of the insulin promoter (13), respectively. Since the GA element of the CLC-K1 and CLC-K2 gene promoters is GGGGAGG(G)GGAGGGGAG, it stands to reason that MAZ can bind to the GA element. Cotransfection assay and EMSA clearly demonstrated that MAZ interacted with the GA element of the CLC-K1 promoter and transactivated the CLC-K1 promoter. Although Bossone et al. reported that MAZ is expressed at lower levels in the kidneys than in other tissues, we could confirm MAZ expression in the kidneys by Northern analysis (data not shown). We also detected its expression in all nephron segments, including the thin ascending limb of Henle's loop, where CLC-K1 is expressed, and the thick ascending limb of Henle's loop and the distal nephron segments, where CLC-K2 is expressed. Based on these results, we speculated that MAZ could be one of the transcription factors that activate the expression of the CLC-K1 and CLC-K2 genes in vivo. On the other hand, cotransfection of KKLF with MAZ appeared to block the strongly activating effect of MAZ. The consensus sequence of the KKLF binding site identified in this study, GGGGNGGNG, overlaps the consensus sequence of the MAZ binding site, GGGGAGGGG. Accordingly, we speculated that the MAZ binding competed with the KKLF binding, and we confirmed this by EMSA (Fig. 2C). This finding suggests that the interplay at the GA element between zinc finger proteins that repress or activate transcription may constitute kidney-specific expression of the CLC-K1 and CLC-K2 genes. Double immunostaining of KKLF and other marker proteins in rat inner medulla revealed that KKLF was present in the nuclei of inner medullary collecting ducts and the thin descending limb of Henle's loop, i.e., nephron segments where no expression of CLC-K1 and CLC-K2 is observed. Given that KKLF is never expressed in any of the cells or tissues where the expression of CLC-K1 and CLC-K2 is also absent, the combination of KKLF and MAZ alone may not account for all of the mechanisms of kidney-specific expression of CLC-K1 and CLC-K2. However, the spatial pattern of KKLF expression along the nephron overlaps the negative expression of the CLC-K1 and CLC-K2, as confirmed by RT-PCR analysis. Accordingly, KKLF might suppress CLC-K1 and CLC-K2 expression in inner medullary collecting ducts and the thin descending limb of Henle's loop in vivo, thereby contributing to the strict nephron segment-specific expression of CLC-K1 and CLC-K2. A recent pool of evidence suggets that strict cell- and tissue-specific gene expression is attained not by a single positive regulatory factor but by a combination of activators and repressors. For example, RINZF blocks the activating effect of SpI through the CACC element of the gastrin promoter (33), and β-enolase repressor factor 1, a Krüppel-like factor, can compete with other positively regulating factors on the BEE-1 element of the β-enolase gene (25). In agreement with these studies, the present study revealed that the GA element, although originally identified as a positive cis element, has both positive and negative effects on the CLC-K1 promoter.
Apart from the Cys2-His2 zinc finger domains, the KKLF coding sequence bears little resemblance to other reported zinc finger genes. For example, there were no Krüppel, leucine zipper, Ets, or basic helix-loop-helix domains. However, KKLF contains serine-rich or proline-rich sequences, which are involved in the transactivation functions of many transcription factors (23) and are considered typical features of repression motifs present in suppressor factors like Krüppel and WT1 (18, 19). Since this proline-rich sequence is shared by proteins closely related to KKLF, namely, EKLF (22), LKLF (2), GKLF (29), and BTEB2 (31), KKLF can be definitively classed a new Krüppel-like factor. In addition to these potential regulatory domains, there is a glutamic acid cluster at amino acid residues 142 to 150 (EEIEEFLEE) in KKLF. Several reports have indicated that a high charge is a common feature of repression motifs (4, 28, 38). On this basis, the glutamic acid cluster may be considered a repressive element of KKLF.
Although we named this new Krüppel-like factor KKLF for “kidney-enriched Krüppel like factor”), its expression is actually most abundant in the liver and more prevalent in heart and skeletal muscle than in the kidneys. Immunohistochemistry revealed that KKLF in the liver is present not in hepatocytes but in cells in the sinusoid. The various cell types in the sinusoid include Kupffer cells, stellate cells, and endothelial cells. Given that KKLF is not found in the lymphoid tissues such as the lymph nodes and spleen, it is less likely to be present in Kupffer cells. In fact, double staining of KKLF with ED-2, a marker for Kupffer cells, showed that only a few ED-2-positive cells were also KKLF positive. Most KKLF staining was associated with desmin staining, clearly suggesting that the KKLF-positive sinusoidal cells were stellate cells. KKLF was also present in the subcapsular and portal fibroblasts, suggesting that KKLF may be preferentially expressed in fibroblasts rather than in lymphoid cells. This may be true in the heart and skeletal muscles. In the heart and skeletal muscles, the stained nuclei appeared to be present between the muscle fibers rather than in the muscle cells themselves. The stained nuclei were not ED-2 positive, suggesting that the interstitial cells in the heart and skeletal muscles were also fibroblasts. Also, KKLF was apparently present in the interstitial cells in the kidney cortex. Most of the cells in the interstitium of the kidney cortex were fibroblasts and dendritic cells, cell types identified by ecto-5′-nucleotidase and MHC-II markers, respectively (12). Double immunostaining clearly revealed that KKLF-positive cells were positive for ecto-5′-nucleotidase, suggesting that KKLF-positive cells in the kidney cortex were also fibroblasts. The KKLF-positive cells in the glomeruli were speculated to be mesangial cells, since mesangial cells have properties similar to the stellate cells in the liver. As expected, KKLF-positive nuclei were mostly associated with desmin-positive mesangial matrix. These results suggest that KKLF may play an important role in fibroblasts and other potentially fibrogenic cells in the liver, heart, skeletal muscles, and kidneys. Zf9, a Krüppel-like factor present in stellate cells in the liver, transactivates a collagen α1(I) promoter (27) and is speculated to be involved in hepatic fibrosis. c-Krox, a Krüppel-like protein present in the skin, binds to the GGGAGGG sequence in type I collagen genes (7). Ihn et al. reported that G-rich elements in the human α2(I) collagen promoter had repressor activity (11). Accordingly, it would be interesting to determine whether KKLF is involved in the regulation of collagen gene promoters. As shown in Fig. 8b, cotransfection of KKLF significantly repressed the human type I collagen gene promoter. In addition, KKLF expression was dramatically decreased in the UUO kidneys, and after this decrease an abundant expression of type I collagen was noted. This evidence implies that KKLF could function as a repressor of type I collagen expression and an important regulator of tissue fibrosis. The regulation of KKLF expression in the other fibrosis models in other organs will help to confirm whether or not this is true.
In summary, MAZ and a novel Krüppel-like factor, KKLF, were cloned as GA element binding proteins. This study suggests that both positively and negatively regulating zinc finger proteins may interact at the GA element and may be involved in the strict transcriptional control of the CLC-K1 and CLC-K2 genes. KKLF may also be involved in the regulation of type I collagen expression in fibroblasts in vivo.
REFERENCES
- 1.Adachi S, Uchida S, Ito H, Hata M, Hiroe M, Marumo F, Sasaki S. Two isoforms of a chloride channel predominantly expressed in thick ascending limb of Henle's loop and collecting ducts of rat kidney. J Biol Chem. 1994;269:17677–17683. [PubMed] [Google Scholar]
- 2.Anderson K P, Kern C B, Crable S C, Lingrel J B. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossone S A, Asselin C, Patel A J, Marcu K B. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc Natl Acad Sci USA. 1992;89:7452–7456. doi: 10.1073/pnas.89.16.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowell I G, Hurst H C. Transcriptional repression by the human bZIP factor E4BP4: definition of a minimal repression domain. Nucleic Acids Res. 1994;22:59–65. doi: 10.1093/nar/22.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crossley M, Whitelaw E, Perkin A, Williams G, Fujiwara Y, Orkin S H. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a mojor CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan D D, Stupakoff A, Hedrick S M, Marcu K B, Siu G. A myc-associated zinc finger protein binding site is one of four important functional regions in the CD4 promoter. Mol Cell Biol. 1995;15:3179–3186. doi: 10.1128/mcb.15.6.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galera P, Park R-W, Ducy P, Mattei M-G, Karsenty G. c-Krox binds to several sites in the promoter of both mouse type I collagen genes. Structure/function study and developmental expression analysis. J Biol Chem. 1996;271:21331–21339. doi: 10.1074/jbc.271.35.21331. [DOI] [PubMed] [Google Scholar]
- 8.Garrett-Sinha L A, Eberspaecher H, Seldin M F, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 9.Her S, Ann Bell R, Bloom A K, Siddall B J, Wong D L. Phenylethanolamine N-methyltransferase gene expression. J Biol Chem. 1999;274:8698–8707. doi: 10.1074/jbc.274.13.8698. [DOI] [PubMed] [Google Scholar]
- 10.Hughes J, Johnson R J. Role of Fas (CD95) in tubulointerstitial disease induced by unilateral ureteric obstruction. Am J Physiol Ser F. 1999;277:F26–F32. doi: 10.1152/ajprenal.1999.277.1.F26. [DOI] [PubMed] [Google Scholar]
- 11.Ihn H, Ohnishi K, Tamaki T, Leroy E C, Trojanowska M. Transcriptional regulation of the human a2(I) collagen gene. J Biol Chem. 1996;271:26717–26723. doi: 10.1074/jbc.271.43.26717. [DOI] [PubMed] [Google Scholar]
- 12.Kaissling B, Le Her M. Characterization and distribution of interstitial cell types in the renal cortex of rats. Kidney Int. 1994;45:709–720. doi: 10.1038/ki.1994.95. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy G C, Rutter W J. Pur-1, a zinc-finger protein that binds to purine-rich sequences, transactivates an insulin promoter in heterologous cells. Proc Natl Acad Sci USA. 1992;89:11498–11502. doi: 10.1073/pnas.89.23.11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klevit R E. Recognition of DNA by Cys2,His2 zinc finger. Science. 1991;253:1367. doi: 10.1126/science.1896847. , 1393. [DOI] [PubMed] [Google Scholar]
- 15.Koritschoner N P, Bocco J L, Panzetta-Dutari G, Dumur C I, Flury A, Patrito L C. A novel human zinc finger protein that interacts with the core promoter element of a TATA-box-less gene. J Biol Chem. 1997;272:9573–9580. doi: 10.1074/jbc.272.14.9573. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licht J D, Grossel M J, Figge J, Hansen U M. Drosophila Kruppel protein is a transcriptional repressor. Nature. 1990;346:76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- 19.Madden S L, Cook D M, Rauscher F J. A structure-function analysis of transcriptional repression mediated by the WT1, Wilms' tumor suppressor protein. Oncogene. 1993;8:1713–1720. [PubMed] [Google Scholar]
- 20.Matsumoto N, Friedrich L, Aldabe R, Zhang W, Ramirez F, Yoshida T, Terada M. Cloning of the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J Biol Chem. 1998;273:28229–28237. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 21.Matsumura Y, Uchida S, Kondo Y, Miyazaki H, Ko S B H, Hayama A, Morimoto T, Liu W, Arisawa M, Sasaki S, Marumo F. Overt nephrogenic diabetes insipidus in mice lacking the CLC-K1 chloride channel. Nat Genet. 1999;21:95–98. doi: 10.1038/5036. [DOI] [PubMed] [Google Scholar]
- 22.Miller I J, Bieker J J. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Alder H, Gu Y, Prasad R, Canaani O, Kamada N, Gale R P, Lange B, Crist W M, Nowell P C, Croce C M, Canaani E. Genes on chromosomes 4, 9, and 19 involved in 11q23 abnormalities in acute leukemia share sequence homology and/or common motifs. Proc Natl Acad Sci USA. 1993;90:4631–4635. doi: 10.1073/pnas.90.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks C L, Shenk T. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and SpI. J Biol Chem. 1996;271:4417–4430. doi: 10.1074/jbc.271.8.4417. [DOI] [PubMed] [Google Scholar]
- 25.Passantino R, Antona V, Barbieri G, Rubino P, Melchionna R, Cossu G, Feo S, Giallongo A. Negative regulation of b enolase gene transcription in embryonic muscle is dependent upon a zinc finger factor that binds to the G-box within the muscle-specific enhancer. J Biol Chem. 1998;273:484–494. doi: 10.1074/jbc.273.1.484. [DOI] [PubMed] [Google Scholar]
- 26.Rai T, Uchida S, Sasaki S, Marumo F. Isolation and characterization of kidney-specific CLC-K2 chloride channel gene promoter. Biochem Biophys Res Commun. 1999;261:432–438. doi: 10.1006/bbrc.1999.1038. [DOI] [PubMed] [Google Scholar]
- 27.Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, Friedman S L. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci USA. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha S, Brickman J M, Lehming N, Ptashne M. New eukaryotic transcriptional repressors. Nature. 1993;363:642–652. doi: 10.1038/363648a0. [DOI] [PubMed] [Google Scholar]
- 29.Shields J M, Christy R J, Yang V W. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon D B, Bindra R S, Mansfield T A, Nelson-Williams C, Mendonca E, Stone R, Schurman S, Nayir A, Alpay H, Bakkaloglu A, Rodriguez-Soriano J, Morales J M, Sanjad S A, Taylor C M, Pilz D, Brem A, Trachtman H, Griswold W, Richard G A, John E, Lifton R P. Mutations in the chloride channel gene, CLCNKB, cause Bartter's syndrome type III. Nat Genet. 1997;17:171–178. doi: 10.1038/ng1097-171. [DOI] [PubMed] [Google Scholar]
- 31.Sogawa K, Imataka H, Yamasaki Y, Kusume H, Fujii-Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527–1532. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terada Y, Tomita K, Nonoguchi H, Marumo F. Polymerase chain reaction localization of constitutive nitric oxide synthase and soluble guanylate cyclase messenger RNAs in microdissected rat nephron segments. J Clin Investig. 1992;90:659–665. doi: 10.1172/JCI115908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tillotson L G. RIN ZF, a novel zinc finger gene, encodes proteins that bind to the CACC element of the gastrin promoter. J Biol Chem. 1999;274:8123–8128. doi: 10.1074/jbc.274.12.8123. [DOI] [PubMed] [Google Scholar]
- 34.Uchida S, Rai T, Yatsushige H, Matsumura Y, Kawasaki M, Sasaki S, Marumo F. Isolation and characterization of kidney-specific ClC-K1 chloride channel gene promoter. Am J Physiol. 1998;274:F602–F610. doi: 10.1152/ajprenal.1998.274.3.F602. [DOI] [PubMed] [Google Scholar]
- 35.Uchida S, Sasaki S, Furukawa T, Hiraoka M, Imai T, Hirata Y, Marumo F. Molecular cloning of a chloride channel that is regulated by dehydration and expressed predominantly in kidney medulla. J Biol Chem. 1993;268:3821–3824. [PubMed] [Google Scholar]
- 36.Uchida S, Sasaki S, Nitta K, Uchida K, Horita S, Nihei H, Marumo F. Localization and functional characterization of rat kidney-specific chloride channel, CLC-K1. J Clin Investig. 1995;95:104–113. doi: 10.1172/JCI117626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandewalle A, Cluzeaud F, Bens M, Kieferle S, Steinmeyer K, Jentsch T J. Localization and induction by dehydration of ClC-K chloride channels in the rat kidney. Am J Physiol Ser F. 1997;272:F678–F688. doi: 10.1152/ajprenal.1997.272.5.F678. [DOI] [PubMed] [Google Scholar]
- 38.Witzgall R, O'Leary E, Leaf A, Onaldi D, Bonventre J V. The Kruppe-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci USA. 1994;91:4514–4518. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa M, Uchida S, Yamauchi A, Miyai A, Tanaka Y, Sasaki S, Marumo F. Localization of rat CLC-K2 chloride channel mRNA in the kidney. Am J Physiol. 1999;276:F552–F558. doi: 10.1152/ajprenal.1999.276.4.F552. [DOI] [PubMed] [Google Scholar]