Abstract
Background
The highly transmissible Delta variant of SARS-CoV-2 (B.1.617.2), first identified in India, is currently replacing pre-existing variants in many parts of the world. To help guide public health policies it is important to monitor efficiently its spread. Genome sequencing is the gold standard for identification of Delta, but is time-consuming, expensive, and unavailable in many regions.
Objective
To develop and evaluate a rapid, simple and inexpensive alternative to sequencing for Delta identification.
Methods
A double-mismatch allele-specific RT-PCR (DMAS-RT-PCR) was developed. The technique exploits allele-specific primers, targeting two spike gene mutations, L452R and T478K, within the same amplicon. The discriminatory power of each primer was enhanced by an additional mismatch located at the fourth nucleotide from the 3′ end. Specificity was assessed by testing well characterised cell culture-derived viral isolates and clinical samples, most of which had previously been fully sequenced.
Results
In all cases the results of viral genotyping by DMAS-RT-PCR were entirely concordant with the results of sequencing, and the assay was shown to discriminate reliably between the Delta variant and other variants (Alpha and Beta), and ‘wild-type’ SARS-CoV-2. Influenza A and RSV were non-reactive in the assay. The sensitivity of DMAS-RT-PCR matched that of the diagnostic SARS-CoV-2 RT-qPCR screening assay. Several samples that could not be sequenced due to insufficient virus were successfully genotyped by DMAS-RT-PCR.
Conclusion
The method we describe would be simple to establish in any laboratory that can conduct PCR assays and should greatly facilitate monitoring of the spread of the Delta variant globally.
Keywords: SARS-CoV-2, Delta variant, allele-specific real time RT-PCR, Spike gene mutations, genotyping, molecular diagnostics
Abbreviations
- DMAS-RT-PCR
Double-mismatch allele-specific real time reverse transcription polymerase chain reaction
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
1. Introduction
The global outbreak of COVID-19 which started in China towards the end of 2019 was formally recognised as a pandemic by the World Health Organisation in March 2020. The causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been intensively studied since then and more than four million genome sequences have been accumulated and made freely available [1]. As SARS-CoV-2 has evolved, many variants have emerged in different parts of the world and four of these, Alpha, Beta, Gamma and Delta have been designated variants of concern (VOC), primarily on the basis of their increased transmissibility, increased virulence and/or ability to evade immunity. The first three VOC to emerge were the Alpha variant (B.1.1.7), originally identified in the UK, the Beta variant (B.1.351), first identified in South Africa and the Gamma variant (P.1), first identified in Brazil [2], [3], [4]. The Delta variant (B.1.617.2) was first detected in India in December 2020 and rapidly increased in prevalence, replacing pre-existing lineages and becoming the most common variant in that country by April 2021. Since then, it has also become the dominant variant in the UK and is out-competing pre-existing variants in many other countries [5,6].
The competitive advantage of the Delta variant is thought to be due to a combination of significantly increased transmissibility and a degree of resistance to both natural and vaccine-induced immunity. There is also evidence that infection with the Delta variant is associated with higher viral loads, more severe disease, and a greater risk of hospitalisation [7]. It is, therefore, important to monitor the spread of the Delta variant globally to inform public health policies and interventions [8]. The gold standard methodology for identifying Delta is full genome sequencing. Although this gives very detailed information and the opportunity to discover new variants, it has the disadvantage of being comparatively expensive, complex, and time-consuming (days or weeks), and is not available in many locations due to lack of expertise and resources. Alternative methods offering rapid, high-throughput, low-cost genotyping for efficient surveillance of the Delta variant are therefore required.
Detection of the S gene by the Applied Biosystems TaqPath Covid-19 PCR assay has been used in the UK as a surrogate marker for the Delta variant [9] to differentiate it from the pre-existing Alpha variant, which is characterised by ‘spike gene target failure’ (SGTF) due to the 69–70del S gene mutation. However, this proxy marker method is by no means specific for Delta because both Beta and Gamma variants would also be detected by this strategy [10]. In contrast, the double-mismatch allele-specific real time RT-PCR (DMAS-RT-PCR) method that we describe here does not exhibit such non-specificity because it targets two separate Delta spike gene mutations, L452R and T478K, within the same amplicon.
The DMAS-RT-PCR technique was first described in 2019 for the genotyping of cancer cell lines [11]. It was subsequently employed for genotyping cacao plant clones [12] but, to our knowledge, has not previously been exploited for genotyping RNA viruses. The very high discriminating power of the technique is achieved by the use of allele-specific primers which, in addition to the 3′ terminal mismatch, have a second artificial mismatch located at the fourth nucleotide from the 3′ end. In the present study, we have validated this approach by designing and evaluating a DMAS-RT-PCR assay that can reliably discriminate between the Delta variant and other SARS-CoV-2 variants. The assay was successfully tested on nose and throat swab samples, and on cell culture-derived SARS-CoV-2 isolates that had previously been characterised by full genome sequencing.
2. Materials and methods
2.1. Cell cultured viral isolates
Purified SARS-CoV-2 RNA was extracted from Vero cell supernatants, aliquoted and stored at −80 °C until use. The following samples were obtained: IC19 (hCoV-19/England/IC19/2020|EPI_ISL_475572|2020–03–17), a B.1 lineage virus with the D614G spike mutation but otherwise the same as the original “wild-type” Wuhan virus. Alpha #246 (hCoV-19/England/205080610/2020|EPI_ISL_723001), an example of the B.1.1.7 variant. Beta #65 (hCoV-19/England/205280030/2020|EPI_ISL_770441|2020–12–24) and Beta #78, examples of the B.1.351 variant. Delta #395 and Delta #02510, examples of the B.1.617.2 variant. The identities of all the above viruses had been confirmed by full genome sequencing. Influenza A virus PR8 (A/Puerto Rico/8/1934(H1N1) RNA and human respiratory syncytial virus (RSV) strains PP3L and PP3KL were also obtained.
2.2. Clinical samples
Anonymised, residual nose and throat swab samples in virus transport medium were obtained from staff and students at Imperial College London, and from household contacts of individuals with COVID-19 (REC reference 20/NW/0231, IRAS ID: 282820). Extracted RNA from the samples was stored at −80 °C for between 1 day and 8 months before being used in the present study. Initial screening of the samples for SARS-CoV-2 had been performed using a duplex RT-qPCR assay which targets both the E gene (Charité assay) and a human RNA transcript, RNase P (CDC assay) as an internal sample sufficiency control [13].
2.3. Sequencing
To establish their viral genotype, clinical samples in which SARS-CoV-2 had been detected were sequenced using an Illumina iSeq 100 next-generation sequencing system. In 6 of the samples, the viral concentration was too low for successful sequencing.
2.4. RNA extraction
Viral RNA was extracted from clinical samples with a CyBio Felix liquid handing robot (Analytik Jena) and InnuPREP Virus DNA/RNA Kit (Analytik Jena), used according to manufacturer's instructions. RNA was eluted in 50 µl of RNase Free Water and stored at −80 °C until use.
2.5. Primer and probe design
The Delta variant has multiple mutations distributed throughout its genome of which at least five are located within the S gene that encodes the spike protein [14]. Some of these individual mutations are shared by other variants but we noted that the combination of spike mutations L452R and T478K (nucleotide positions T22917G and C22995A, respectively) within the receptor binding domain can effectively differentiate Delta from other variants. Fortuitously, these point mutations are located close enough to each other to permit them to be targeted by a pair of allele-specific primers that generate an amplicon of a length (122 bp) suitable for efficient PCR amplification and detection by a fluorescently-labelled hydrolysis probe. Lefever [11] demonstrated that the discriminatory power of allele-specific primers could be significantly enhanced by adding a second mismatched nucleotide four nucleotides from the terminal mismatch at the 3′ end. We, therefore, designed a pair of double-mismatch allele-specific primers (DMAS) according to these principles. Alignments of SARS-CoV-2 VOC spike gene sequences downloaded from the GISAID database [1] were performed using MEGA version 7.0.21. The primers (Table 1 ) were checked by in silico PCR (https://genome.ucsc.edu/cgi-bin/hgPcr) to rule out unwanted reactivity with the human genome, and by NCBI BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cg) to exclude reactivity with other respiratory viruses including human coronaviruses 229E, OC43 and NL63. Melting temperature (Tm) estimations and checks to rule out primer dimers or significant secondary structure were also performed (IDT OligoAnalyzer, https://eu.idtdna.com/calc/analyser)
Table 1.
Details of primer and probe sequences used for the DMAS-RT-PCR assay.
| Oligonucleotide function | Oligonucleotide name | Sequence* | Length & Tm⁎⁎ |
|---|---|---|---|
| Forward primer for Reaction ‘A’. Reaction ‘A’ detects non-Delta SARS-CoV-2 |
Non-Delta_DMAS_F | 5′ GGTTGGTGGTAATTATAATTCCCT | 24 nt 59.7 °C |
| Reverse primer for Reaction ‘A’. Reaction ‘A’ detects non-Delta SARS-CoV-2 |
Non-Delta_DMAS_R | 5′ CCTTCAACACCATTACAACGTG | 22 nt 60.2 °C |
| Forward primer for Reaction ‘B’. Reaction ‘B’ detects the Delta variant of SARS-CoV-2 |
Delta_DMAS_F | 5′ GGTTGGTGGTAATTATAATTCCCG | 24 nt 60.5 °C |
| Reverse primer for Reaction ‘B’. Reaction ‘B’ detects the Delta variant of SARS-CoV-2 |
Delta_DMAS_R | 5′ CCTTCAACACCATTACAACGTT | 22 nt 59.9 °C |
| Common probe for both Reaction ‘A’ and Reaction ‘B’. Probe located on antisense strand |
Common_A& B_Prb |
5′ FAM-TCTCTCAAAAGGTTTGAGATTAGACTTCC-BHQ‡ | 29 nt 64.9 °C |
*Underlined nucleotides represent the second mismatch deliberately inserted to increase specificity. ‘C’ was chosen in each case as the least stable artificial mismatch on the basis of reported mismatch stability ranking [11]. The terminal 3′ nucleotide being targeted is shown highlighted in bold italic.
⁎⁎Tm calculated for the matched target using IDT OligoAnalyzer tool with qPCR parameter set and assuming primer concentration of 400 nM and probe concentration of 250 nM. Tm calculated with the deliberate internal mismatched base omitted.
‡FAM = 6-Carboxyfluorescein BHQ = Black Hole Quencher.
2.6. Double-mismatch allele-specific real time RT-PCR (DMAS-RT-PCR) assay
The method requires two parallel RT-PCR reactions carried out in separate wells. Reaction ‘A’ is designed to detect any SARS-CoV-2 sequence other than the Delta variant, i.e., non-Delta. Reaction ‘B’ is designed to detect only the Delta variant. The primers used for Reaction ‘A’ and Reaction ‘B’ are detailed in Table 1. The amplicons generated in both reactions are detected by a common fluorescently labelled hydrolysis probe (Table 1). Primers and probes were synthesized by Integrated DNA Technologies IDT, Belgium. Five μL RNA template was used in a 20 μL reaction containing 5 μL of 4x TaqMan Fast virus 1-step mastermix (Applied Biosystems), primers at 400 nM and probe at 250 nM. Thermal cycling was performed in a Bio-Rad CFX real-time PCR system with reverse transcription at 54 °C for 10 min, followed by 94 °C for 3 min, then 40 cycles of 94 °C for 15 s and 58 °C for 30 s. Data were processed using Bio-Rad CFX maestro 2.0 software with baseline subtracted curve fit, fluorescence drift correction, automatically calculated baseline cycles and manual threshold settings. Each run included IC19 RNA and Delta #395 RNA, both diluted a million-fold, as positive controls. Several no-template nuclease-free water negative controls were also included in each run.
2.7. Data interpretation
The assay is designed primarily for genotyping rather than for quantification and, therefore, does not require a calibration curve. Interpretation of the results is as follows: If in Reaction ‘A’ a sample generates a lower Ct value (i.e., threshold cycle) than in Reaction ‘B’ it indicates that the sample is not the Delta variant. Conversely, if in Reaction ‘B’ a sample generates a lower Ct value than in Reaction ‘A’ it indicates that the sample is the Delta variant. If neither reaction generates a Ct value of less than 40 cycles, this is interpreted as ‘SARS-CoV-2 not detected’.
3. Results
3.1. Cell cultured viral isolates
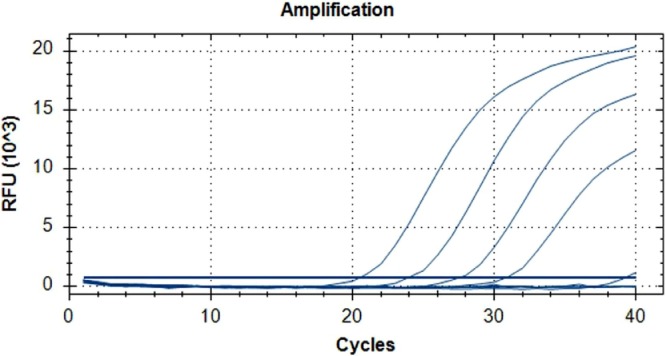
Viral RNA samples extracted from cell culture supernatants were serially diluted in ten-fold steps to cover the range 1:1000 to 1:1,000,000. Diluted samples were used as templates for Reaction ‘A’ and Reaction ‘B,’ as described under Methods. Fig 1 illustrates that, as predicted, Delta variant RNA (Delta #02510), even at the highest 1:1,000,000 dilution, was efficiently amplified and detected by Reaction ‘B’. In contrast, Reaction ‘A’ was only able to amplify the most concentrated 1:1000 dilution, but all of the higher dilutions remain undetectable. The 1:1000 dilution amplification curve crossed the threshold at cycle 39.3 in Reaction ‘A’, almost 19 cycles later than in Reaction ‘B’. The other Delta variant isolate, Delta #395, generated very similar results and was also readily detectable at a 1:1,000,000 dilution by Reaction ‘B’ but not detectable at all by Reaction ‘A’, even at the lowest 1:1000 dilution tested. Conversely, all of the non-Delta viral isolates tested (IC19, Alpha #246, Beta #65, Beta #78) were readily detected by Reaction ‘A’ at all dilutions including the highest 1:1,000,000 dilution, but not detected at any dilution by Reaction ‘B’. These findings are summarised in Table 2.
Fig 1.
Typical amplification curves generated by Reactions ‘A’ and ‘B’ with dilution series of Delta variant RNA. A ten-fold dilution series of viral isolate Delta #02510 RNA covering the range 1:1000 to 1:1,000,000 was amplified by Reaction ‘A’ and Reaction ‘B’. Reaction ‘B’ generated the four curves on the left with Ct values of 20.5, 24.0, 27.6 and 30.9. In contrast, Reaction ‘A’ only just detected the lowest 1:1000 dilution with a Ct value of 39.3 (the very small curve on the extreme right). The three higher dilutions were undetected by Reaction ‘A’..
Table 2.
DMAS-RT-PCR findings on cell cultured SARS-CoV-2 isolates.
| Viral Isolate | Genotype by sequencing | Reaction ‘A’ for non-Delta | Reaction ‘B’ for Delta | Genotype by DMAS-RT-PCR |
|---|---|---|---|---|
| IC19 | ‘wild type’* | Detected at all dilutions⁎⁎ | Not detected at any dilution | Not Delta variant |
| Alpha #246 | Alpha variant | Detected at all dilutions⁎⁎ | Not detected at any dilution | Not Delta variant |
| Beta #65 | Beta variant | Detected at all dilutions⁎⁎ | Not detected at any dilution | Not Delta variant |
| Beta #78 | Beta variant | Detected at all dilutions⁎⁎ | Not detected at any dilution | Not Delta variant |
| Delta #395 | Delta variant | Not detected at any dilution⁎⁎ | Detected at all dilutions | Delta variant |
| Delta #02510 | Delta variant | Detected at 1:1000 only; 18.8 cycles later than Reaction ‘B’ | Detected at all dilutions | Delta variant |
*IC19 is a B.1 lineage virus similar to the original ‘wild-type’ Wuhan virus but with the D614G spike mutation.
⁎⁎Ten-fold dilutions 1:1000, 1:10,000, 1:100,000 and 1:1000,000 were tested for each isolate.
3.2. Clinical samples
The results generated by testing clinical samples were consistent with those generated by testing cultured viral isolates, in that samples confirmed to be Delta by sequencing, or presumed to be Delta by virtue of the date that they were taken, were efficiently amplified and detected by Reaction ‘B’ but not by Reaction ‘A’. Similarly, samples confirmed non-Delta by sequencing were efficiently amplified and detected by Reaction ‘A’ but not by Reaction ‘B’. These results are summarised in Table 3 .
Table 3.
Concordance between SARS-CoV-2 genotyping by sequencing and by DMAS-RT-PCR assay.
| Sample No. | Genotype by sequencing | E gene PCR Ct | Reaction ‘A’ Ct (non-Delta) | Reaction ‘B’ Ct (Delta) | Genotype by DMAS-RT-PCR | Comments |
|---|---|---|---|---|---|---|
| 1 | Not done | 19.2 | 37.4 | 20.0 | Delta variant | Expected to be Delta* |
| 2 | Not done | 19.5 | 37.7 | 20.4 | Delta variant | Expected to be Delta* |
| 3 | Not done | 29.6 | Negative | 30.2 | Delta variant | Expected to be Delta* |
| 4 | Not done | 31.1 | Negative | 31.6 | Delta variant | Expected to be Delta* |
| 5 | Not done | 23.6 | Negative | 25.0 | Delta variant | Expected to be Delta* |
| 6 | Not done | 30.1 | Negative | 31.4 | Delta variant | Expected to be Delta* |
| 7 | Not done | 34.0 | Negative | 36.6 | Delta variant | Expected to be Delta* |
| 8 | Insufficient virus | 34.9 | Negative | 34.2 | Delta variant | Expected to be Delta* |
| 9 | Alpha variant | 19.2 | 19.9 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 10 | Alpha variant | 27.8 | 27.6 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 11 | Alpha variant | 28.3 | 28.6 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 12 | Alpha variant | 24.8 | 25.3 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 13 | Alpha variant | 26.0 | 25.3 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 14 | Alpha variant | 26.5 | 26.4 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 15 | Alpha variant | 25.2 | 27.1 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 16 | Alpha variant | 23.3 | 25.6 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 17 | Alpha variant | 23.8 | 23.7 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 18 | Alpha variant | 24.2 | 24.2 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 19 | Alpha variant | 35.6 | 33.3 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 20 | Alpha variant | 21.7 | 22.0 | 36.3 | Not Delta | Non-Delta SARS-CoV-2 |
| 21 | Alpha variant | 29.3 | 29.6 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 22 | Alpha variant | 28.2 | 28.3 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 23 | Alpha variant | 27.6 | 28.0 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 24 | Alpha variant | 30.6 | 31.1 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 25 | Alpha variant | 22.9 | 23.5 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 26 | Alpha variant | 24.0 | 25.2 | Negative | Not Delta | Non-Delta SARS-CoV-2 |
| 27 | Delta variant | 30.1 | Negative | 31.2 | Delta variant | Genotypes agree$ |
| 28 | Delta variant | 23.2 | Negative | 24.2 | Delta variant | Genotypes agree$ |
| 29 | Insufficient virus | 33.7 | Negative | 34.2 | Delta variant | Expected to be Delta* |
| 30 | Delta variant | 32.0 | Negative | 32.5 | Delta variant | Genotypes agree$ |
| 31 | Delta variant | 25.4 | Negative | 25.5 | Delta variant | Genotypes agree$ |
| 32 | Delta variant | 19.0 | Negative | 19.6 | Delta variant | Genotypes agree$ |
| 33 | Insufficient virus | 34.3 | Negative | 35.5 | Delta variant | Expected to be Delta* |
| 34 | Delta variant | 23.5 | Negative | 24.6 | Delta variant | Genotypes agree$ |
| 35 | Delta variant | 21.4 | Negative | 21.9 | Delta variant | Genotypes agree$ |
| 36 | Delta variant | 19.5 | Negative | 20.5 | Delta variant | Genotypes agree$ |
| 37 | Delta variant | 18.5 | 38.4 | 19.6 | Delta variant | Genotypes agree$ |
| 38 | Delta variant | 18.1 | Negative | 19.3 | Delta variant | Genotypes agree$ |
| 39 | Delta variant | 30.7 | Negative | 30.9 | Delta variant | Genotypes agree$ |
| 40 | Insufficient virus | 35.5 | Negative | 35.3 | Delta variant | Expected to be Delta* |
| 41 | Insufficient virus | 33.9 | Negative | 33.0 | Delta variant | Expected to be Delta* |
| 42 | Insufficient virus | 36.4 | Negative | 38.0 | Delta variant | Expected to be Delta* |
*Expected to be the Delta variant based on the date of the sample (approximately 99% of samples were Delta variant in UK during July 2021).
$Genotypes obtained by sequencing and by DMAS-RT-PCR are in agreement.
Seven of the 42 samples (Sample 1 to Sample 7) had not been submitted for sequencing but were expected to be the Delta variant because they were taken from patients during July 2021 when approximately 99% of SARS-CoV-2 infections in the UK were caused by Delta. DMAS-RT-PCR genotyping confirmed that, as expected, all were in fact the Delta variant. Of the remaining 35 samples where sequencing was intended, 6 had such low levels of virus that sequencing was not possible. Sample numbers 8, 29, 33, 40, 41 and 42 all had E gene PCR Ct values greater than 33 cycles. However, all these low virus level samples were successfully genotyped by DMAS-RT-PCR, and all proved to be the Delta variant as expected on the basis of the sample dates. In summary, the DMAS-RT-PCR assay was able to correctly identify all 24 clinical samples that were known or assumed to be Delta variant by sequencing or by sample date, and it also correctly identified all 18 samples that had been confirmed to be non-Delta by sequencing.
We observed that four samples (1, 2, 20 and 37) with very high viral concentrations, corresponding to Ct values ≤ 22 cycles, also produced a weak signal in the non-matching Reaction. For example, Sample 1, a Delta variant with a Reaction ‘B’ Ct value of 20 cycles, generated a very high Ct value of 37.4 in Reaction ‘A’. This is presumed to be due to a very minor degree of non-specific hybridisation of the non-matching primers despite the double mismatch design. This in no way compromises the ability of the DMAS-RT-PCR robustly to distinguish between Delta and non-Delta variants because there is always a very wide gap between the Ct value generated by the two Reactions ‘A’ and ‘B’. Typically, this Ct difference is around 17 cycles (range 14.3 to 18.8 cycles).
There was no evidence of false-positive results being generated by the DMAS-RT-PCR assay. No-template controls were consistently negative and 13 clinical samples that had been negative in the E gene PCR used for SARS-CoV-2 screening were also found to be negative by both Reaction ‘A’ and Reaction ‘B’, i.e., ‘SARS-CoV-2 not detected’.
3.3. Sensitivity
The sensitivity of the DMAS-RT-PCR matched that of the diagnostic SARS-CoV-2 RT-qPCR screening assay which targets the E gene (Charité assay) [13]. All 42 of the clinical samples detected by the E gene PCR screening assay were also detected by the DMAS-RT-PCR and the Ct values generated by the two assays were very similar (Table 3). The mean difference for Alpha variant samples was only 0.3 cycles and the mean difference for Delta variant samples was 0.7 cycles. Rowan et al. [13] demonstrated that the E gene PCR assay has a limit of detection of around 6 SARS-CoV-2 RNA copies per reaction and so we assume that the sensitivity of the DMAS-RT-PCR is approximately the same. This is equivalent to a limit of detection of approximately 150 viral genome copies per ml of viral transport medium.
3.4. Specificity
The high specificity of the DMAS-RT-PCR assay enabled it to discriminate reliably between Delta variant and non-Delta variant SARS-CoV-2 with both viral isolates and clinical samples. Influenza A and RSV were selected for testing as examples of other common respiratory viruses. The DMAS-RT-PCR did not generate any non-specific signal with either Influenza A virus PR8 or two RSV strains, PP3L and PP3KL.
4. Discussion
The DMAS-RT-PCR assay that we describe differs from previous applications of the technique in that it targets RNA rather than DNA, employs two DMAS primers rather than one, and uses a fluorescently-labelled hydrolysis probe for detection instead of a DNA-binding dye such as SYBR Green. The use of the hydrolysis probe adds an additional layer of specificity to the assay and reduces the chance of false positive signals resulting from primer dimers etc. However, in circumstances where resources are limited, the cost of the assay could be further reduced by replacing the hydrolysis probe with SYBR Green detection. Whether this would have a significant impact on the performance of the assay has yet to be established.
The results presented in this study clearly demonstrate that the Delta variant can be reliably detected and differentiated from other SARS-CoV-2 variants without having to resort to costly, complex, and time-consuming genome sequencing. Resource limitations mean that even in countries where genome sequencing is available it can only be used on a small proportion of positive samples. DMAS-RT-PCR could provide a comparatively inexpensive, simple, rapid, highly sensitive and specific non-commercial alternative to sequencing for epidemiological surveillance of Delta in many settings. An additional advantage of DMAS-RT-PCR is that it is capable of genotyping SARS-CoV-2 when the level of virus in clinical samples is too low for successful sequencing. Counter-intuitively, in countries such as the UK where the Delta variant is already dominant, one might consider using DMAS-RT-PCR for rapidly identifying the small minority of non-Delta cases and subjecting these to genome sequencing to increase the chance discovering novel, emergent non-Delta variants. In conclusion, the DMAS-RT-PCR assay that we describe would be simple to establish in any laboratory that can conduct PCR assays and should greatly facilitate monitoring of the spread of the SARS-CoV-2 Delta variant globally.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors
Author contributions
Jeremy A. Garson: Conceptualisation, Methodology, Supervision, Writing - original draft, Writing - review and editing, Final approval of submitted version. Samuel Badru: Investigation, Methodology, Writing - review and editing, Final approval of submitted version. Eleanor Parker: Investigation, Project administration, Supervision, Final approval of submitted version. Richard S. Tedder: Conceptualisation, Project administration, Writing - review and editing, Final approval of submitted version. Myra O. McClure: Project administration, Writing - review and editing, Final approval of submitted version.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to the NIHR BRC at Imperial College Healthcare Trust for its support of this study. We thank Professor Wendy Barclay and Dr John Tregoning for kindly providing viral isolates, and Professor Ajit Lalvani for support. We also acknowledge the assistance provided by Carolina Rosadas, Patricia Watber and Lucy Mosscrop in sample processing, inactivation and RNA extractions. Sequencing and sequence analysis was kindly performed by Simon Dustan, Anjna Badhan and Michael Crone, Department of Infectious Disease, Imperial College.
References
- 1.GISAID 2021. https://www.gisaid.org/.
- 2.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. 2021eabg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. 2021. [DOI] [PubMed] [Google Scholar]
- 4.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., et al. SARS-CoV-2 B1.617.2 Delta variant replication and immune evasion. Nature. 2021 doi: 10.1038/s41586-021-03944-y. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. 2021. [DOI] [PubMed] [Google Scholar]
- 7.Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00475-8. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brito A.F., Semenova E., Dudas G., Hassler G.W., Kalinich C.C., Kraemer M.U.G., et al. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv. 2021 doi: 10.1101/2021.08.21.21262393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Public Health England SARS-CoV-2 variants of concern and variants under investigation in England. Technical Briefing 17. 25th June. 2021 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1001354/Variants_of_Concern_VOC_Technical_Briefing_17.pdf. [Google Scholar]
- 10.Vogels C.B.F., Breban M.I., Alpert T., Petrone M.E., Watkins A.E., Ott I.M., et al. PCR assay to enhance global surveillance for SARS-CoV-2 variants of concern. medRxiv. 2021 doi: 10.1101/2021.01.28.21250486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefever S., Rihani A., Van der Meulen J., Pattyn F., Van Maerken T., Van Dorpe J., et al. Cost-effective and robust genotyping using double-mismatch allele-specific quantitative PCR. Sci. Rep. 2019;9:2150. doi: 10.1038/s41598-019-38581-z. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Wever J., Everaert H., Coppieters F., Rottiers H., Dewettinck K., Lefever S., et al. The development of a novel SNP genotyping assay to differentiate cacao clones. Sci. Rep. 2019;9:9512. doi: 10.1038/s41598-019-45884-8. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowan A.G., May P., Badhan A., Herrera C., Watber P., Penn R., et al. Optimized protocol for a quantitative SARS-CoV-2 duplex RT-qPCR assay with internal human sample sufficiency control. J. Virol. Methods 294 2021. 2021;294 doi: 10.1016/j.jviromet.2021.114174. DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suratekar R., Ghosh P., Niesen M.J.M., Donadio G., Anand P., Soundararajan V., et al. High diversity in Delta variant across countries revealed via genome-wide analysis of SARS-CoV-2 beyond the Spike protein. bioRxiv. 2021 doi: 10.1101/2021.09.01.458647. [DOI] [PMC free article] [PubMed] [Google Scholar]